The mechanisms of stem cell niche formation are largely unknown. Lai et al. show that proper formation of the Drosophila melanogaster adult ovarian germline stem cell niche requires a Hedgehog gradient, signaling through a Ci–Traffic Jam–E-cadherin regulatory axis, to direct segregation of intermingled cells by differential cell affinity.
Abstract
Stem cells require different types of supporting cells, or niches, to control stem cell maintenance and differentiation. However, little is known about how those niches are formed. We report that in the development of the Drosophila melanogaster ovary, the Hedgehog (Hh) gradient sets differential cell affinity for somatic gonadal precursors to specify stromal intermingled cells, which contributes to both germline stem cell maintenance and differentiation niches in the adult. We also report that Traffic Jam (an orthologue of a large Maf transcription factor in mammals) is a novel transcriptional target of Hh signaling to control cell–cell adhesion by negative regulation of E-cadherin expression. Our results demonstrate the role of Hh signaling in niche establishment by segregating somatic cell lineages for differentiation.
Introduction
Stem cells reside in microenvironments, called niches, formed to recruit stem cells during development and regulate stem cell identify and behavior in adults (Jones and Wagers, 2008). However, despite intense research into stem cell regulation by niches, little is known about how they are specified.
The Drosophila melanogaster ovary is an excellent model to address this issue, because the development of the adult ovary from the embryonic gonad involves only a small number of specific cell types, occurs progressively (Fig. 1 P; Gilboa, 2015), and contains well-characterized germline stem cells (GSCs) and niches (Wong et al., 2005). Through coalescence of primordial germ cells (PGCs; each with a unique membranous organelle, or fusome) and somatic gonadal precursors (SGPs) derived from the mesoderm (Williamson and Lehmann, 1996), the Drosophila ovary forms a sphere at the end of embryogenesis. During larval stages, with increased numbers of PGCs and SGPs and induced morphogenetic movements along the anterior–posterior and medial–lateral axis, the ovary forms a two-dimensional array of 16–20 stacks of somatic cells called terminal filaments (TFs; Sahut-Barnola et al., 1995). During pupariation, apical somatic cells migrate basally between TFs and through intermingled cells (ICs; which locate at the middle region of the gonad and interact with PGCs) and basal cells (which locate at the bottom of the gonad) to form 16–20 ovarioles (Cohen et al., 2002), functional units that produce eggs (Spradling, 1993). Basal cells form basal stalks that connect ovarioles to the oviduct (King et al., 1968). The anterior-most structure of the ovariole, the germarium (Fig. 2 C), houses two to three GSCs; each of their fusomes faces cap cells (a major GSC maintenance niche component), which are adjacent to basal TFs (Kirilly and Xie, 2007). GSC progeny are wrapped by escort cells (the differentiation niche) with long cellular processes and move toward the posterior of the germarium (Chen et al., 2011), where they are surrounded by a layer of follicle cells (Kirilly and Xie, 2007). The entire structure buds off from the germarium to form a new egg chamber, which develops into a mature egg. The loss of cap cells results in GSC loss (Song et al., 2007; Hsu and Drummond-Barbosa, 2009), and dysfunction of escort cells causes accumulation of undifferentiated GSCs (Jin et al., 2013; Wang et al., 2015), signifying the importance of these niches. However, how niche cap and escort cells are specified is unclear.
Figure 1.
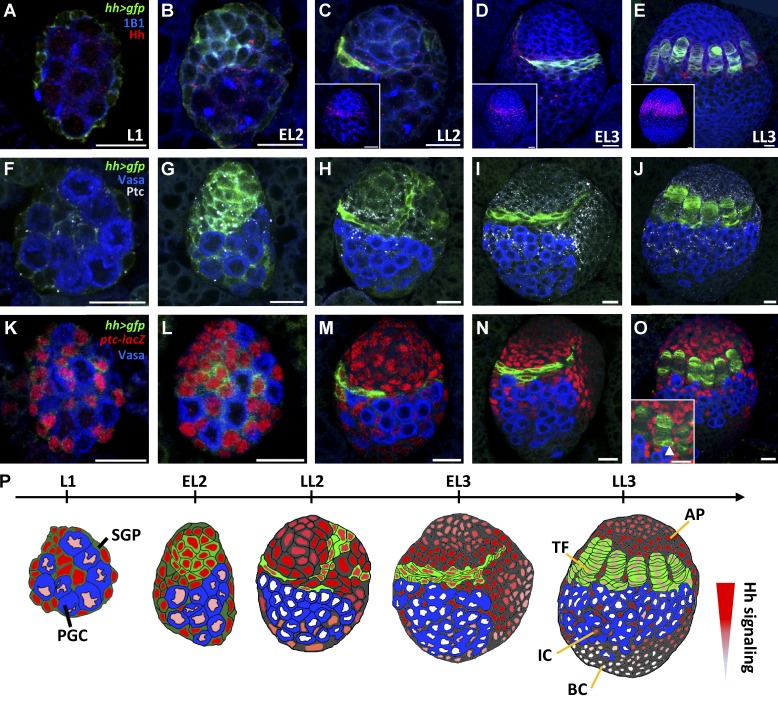
Expression patterns of Hh, Ptc, and ptc-lacZ in larval ovaries. Female gonads of L1 (A, F, and K), early L2 (EL2; B, G, and L), late-L2 (LL2; C, H, and M), early L3 (EL3; D, I, and N), and late-L3 (LL3) larvae (E, J, and O) with hh>gfp (green, A–O), 1B1 (blue, fusomes and somatic cell membranes, A–E), Hh (red, A–E), Vasa (blue, PGCs, F–O), Ptc (gray, F–J), and ptc-lacZ (red, an Hh signaling reporter, K–O). Insets in C–E show Hh distribution in gonads; inset in O shows a cap cell expressing hh-GAL4 and ptc-lacZ (arrowhead). Bars, 10 µm. (P) Schematic of Hh-producing and -receiving cells of larval ovaries. The red gradient indicates strength of Hh signaling. AP, apical cell; BC, basal cell.
Figure 2.
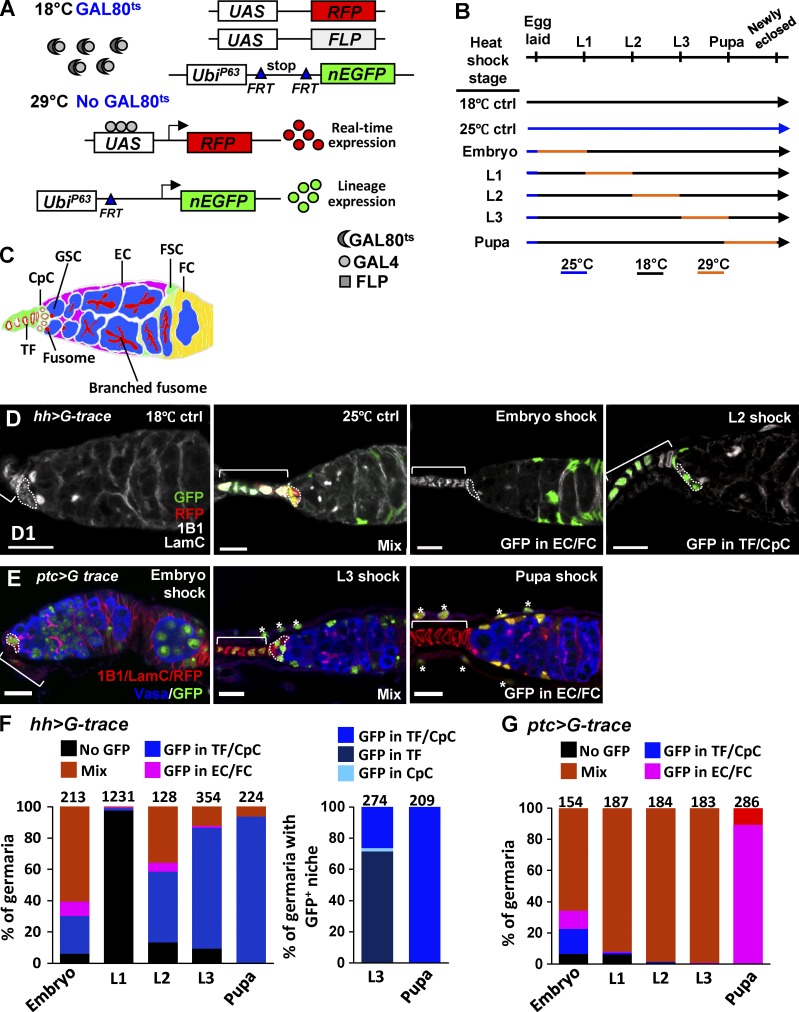
Gonadal hh-GAL4- and ptc-GAL4-expressing cells contribute to adult GSC niches. (A) Schematic of the G-TRACE system controlled by GAL80ts. At 18°C, GAL80ts suppresses the activity of GAL4, driven by a specific promoter. At 29°C, GAL80ts is degraded, and GAL4 activates expressions of RFP (representing real-time expression of GAL4) and Flipase (FLP). The expression of FLP in cells excises the Flipase recognition target (FRT)–flanked stop cassette separating the Ubi-p63E promoter and the nuclear EGFP (nEGFP) coding region to initiate GFP expression, which is maintained in all daughter cells. (B) Schematic of the strategy for tracing hh and ptc-GAL4-expressing cells during development. Embryos were collected at 25°C (blue lines) and maintained at 18°C (black lines), except at the stage of G-TRACE activation at 29°C (orange lines), and dissected 1 day after eclosion. (C) Drosophila germarium with germline stem cells (GSCs), terminal filaments (TFs), cap cells (CpCs), escort cells (ECs), follicle stem cells (FSCs), and follicle cells (FCs). (D) One-day (D1)–old germaria carrying hh>G-trace without activation (left) or activated throughout development (left middle), at the embryo (right middle) and L2 (right) stages with 1B1 (gray, fusomes) and LamC (gray, TF and cap cell nuclear envelopes). (E) D1 germaria carrying ptc>G-trace activated at the embryo (left), L3 (middle), and pupal (right) stages with 1B1 (red), LamC (red), and Vasa (blue). The germaria are grouped by those expressing GFP only in TFs and/or CpCs (GFP in TF/CpC, D, right), only in ECs and/or FCs (GFP in EC/FC, D, right middle, and E, right), or in both TFs and CpCs in addition to ECs and/or FCs, showing a mixed (Mix) pattern (D, left middle, and E, middle). Asterisks in E, middle and right, sheath cells. Bars, 10 µm. (F and G) Percentage of hh>G-trace (F) and ptc>G-trace germaria (G) activated at the indicated stage with no GFP, GFP in TFs/CpCs, GFP in EC/FCs, or a mixed (Mix) pattern. (F, right) Percentage of hh>G-trace germaria activated at the L3 or pupal stage with GFP only in TFs, in CpCs, or in both TFs and CpCs. Statistical differences in F and G were analyzed by χ2. Numbers of analyzed germaria are shown above each bar.
Hedgehog (Hh) signaling is highly conserved and controls several developmental processes (Huangfu and Anderson, 2006; Ruiz-Gómez et al., 2007; Ingham et al., 2011). Hh proteins are secreted ligands that bind to their receptor Patched (Ptc). In the absence of Hh, Ptc suppresses cell-surface localization of Smoothened (Smo), allowing Cubitus interruptus (Ci; an orthologue of Gli in mammals) to be proteolytically processed into a transcriptional repressor. On binding of Hh to Ptc, Ptc inhibition of Smo is relieved and Smo accumulates at the plasma membrane; thus, full-length Ci is released and acts as a transcriptional activator. Drosophila has one homologue for each component of the Hh signaling pathway (Ingham et al., 2011). hh transcripts and Hh proteins are expressed in TFs of late-L3 female larval gonads (Forbes et al., 1996; Gancz et al., 2011), but whether Hh signaling is involved in niche establishment is unexplored.
We show that Hh signaling specifies stromal ICs, which contribute to adult GSC niches. SGPs located posteriorly to TFs receive Hh signals to suppress E-cadherin expression, thus becoming ICs to intermingle with PGCs. Hh signaling–defective ICs exhibit a high level of E-cadherin expression, displaying epithelial basal cell characteristics, and do not intermingle with PGCs; thus, in such gonads, PGCs aggregate and form a large cluster. The loss of ICs further results in the reduction of adult niche cap and escort cells and GSCs. Conversely, hyperactivation of Hh signaling in SGPs forms ectopic ICs accompanied by increased PGCs and the absence of basal cells, resulting in the expansion of escort cells. We also report that Hh signaling maintains IC cell fate by Traffic Jam (Tj)–mediated suppression of E-cadherin expression. Clonal analysis reveals that smo-defective ICs exhibit increased E-cadherin expression but decreased Tj expression; in contrast, overexpression of tj in ICs suppresses E-cadherin expression. Decreased E-cadherin expression or enhanced tj expression in smo-defective ICs partially rescues the IC pool and soma-germline interaction defect. Furthermore, tj- or shotgun (shg; encoding E-cadherin)–knockdown cells are excluded from the IC region. These results suggest that Hh signaling controls the segregation of ICs and basal cells via control of cell-cell adhesion, an effect that is mediated by Tj–E-cadherin regulation. Finally, we report that Tj is a novel target of Ci in response to Hh signals. Collectively, our results show that Hh signaling controls the formation of GSC niche precursors through its developmental role in promoting IC fate by which Ci directly targets Tj. These studies add insight into how an organ uses differential cell affinity to generate niches.
Results
Hh proteins produced from TFs signal to apical somatic cells and ICs
To examine if Hh signaling is involved in niche formation, we first characterized Hh-producing and Hh-receiving cells in larval gonads. We used UAS-gfp driven by hh-GAL4 (Sarikaya and Extavour, 2015) to mark hh-expressing cells. hh-GAL4 was expressed in SGPs by the early L2 stage (Fig. 1, A, B, F, G, K, and L), and its expression was restricted to disc-shaped cells, forming a crown-like structure in the anterior portion of late-L2 and early-L3 larval gonads (Fig. 1, C, D, H, I, M, and N). In the late-L3 gonad, 7 to 10 disc-shaped cells formed a TF where hh-GAL4 was continuously expressed (Fig. 1, E, J, and O). Unlike hh-GAL4 expression, Hh expression showed punctate patterns in both SGPs and PGCs by the early-L2 stage (Fig. 1, A and B), whereas it was strongly expressed in TFs and gradually reduced in cells adjacent to TFs after the late-L2 stage (Fig. 1, C–E). We identified Hh signal–receiving cells by examining expression of Ptc, and ptc-lacZ, an Hh signaling reporter (Chen and Struhl, 1996). Ptc (expressed as punctate granules) and ptc-lacZ were expressed primarily in SGPs but also in PGCs of L1 and L2 gonads (Fig. 1, F, G, K, and L). In L3 gonads, Ptc and ptc-lacZ were excluded from TFs but strongly present in cells adjacent to TFs, including ICs (Fig. 1, H–J and M–O). Our results suggest that Hh acts as an autocrine signal in SGPs of early larval gonads, whereas in late larval gonads, Hh is mainly produced by TFs and signals to apical somatic cells and ICs (Fig. 1 P).
Gonadal Hh signaling–producing cells contribute to adult GSC maintenance niches
To identify cell types that are derived from gonadal Hh-producing and Hh-receiving cells, we used the GAL4 technique for real-time and clonal expression (G-TRACE) system to trace gonadal cells that expressed hh-GAL4 and ptc-GAL4, which mimic the expression of Hh (see Fig. 1) and Ptc (Fig. S1), respectively. G-TRACE combined GAL4/UAS, Flipase (FLP)/FLP recognition target (FRT), and fluorescent reporters to report real-time expression of GAL4 (UAS-RFP) and permanent labeling (ubi-GFP) of cells that were expressing GAL4 (Fig. 2 A; Evans et al., 2009). We also used a temperature-sensitive mutant of GAL80 (GAL80ts) to control activation of the G-TRACE cassette at the indicated stage (Fig. 2 B). hh>G-trace flies cultured at a permissive temperature, 18°C, throughout development showed no red fluorescent protein (RFP) or GFP expression in 1-d-old germaria (Fig. 2, C and D). At a nonpermissive temperature (25°C), RFP was expressed in TF and cap cells (maintenance niches), and GFP was present in TF, cap, escort (differentiation niche), and follicle cells, displaying a mixed pattern (Fig. 2, D, left middle). In addition, after activation at each stage, RFP expression mimicked hh-GAL4 expression (see Fig. 1), and GFP was well expressed in hh>G-trace gonads (Fig. S1, A–D). These results demonstrated the availability of the G-TRACE cassette, which was precisely controlled by GAL4 together with GAL80ts. G-TRACE driven by hh-GAL4 during embryogenesis resulted in 61% of germaria (n = 213) showing as GFP positive in a mixed pattern, but without RFP expression (because larvae were kept at 18°C after embryogenesis; Fig. 2, D [right middle] and F), suggesting the multipotency of embryonic SGPs. Of the germaria, 97% (n = 1,231) had no GFP expression when G-TRACE was activated at the L1 stage, reflecting the low expression of hh-GAL4 (see Figs. 1 F and S1 B). Notably, the proportion of germaria that expressed GFP in maintenance niches, induced at the L2 and pupal stages, increased from 45% (n = 128) to 94% (n = 224; Fig. 2, D [right middle] and F). A similar observation was found in late-L3 hh>G-trace gonads, in which GFP-positive cells marked after the L1 stage were more restricted in TFs (Fig. S1, E–H). We found that 72% of germaria (n = 274) with GFP expression in maintenance niches induced at the L3 stage (Fig. 2 F, right) carried GFP only in TFs, whereas 27% carried GFP in both TF and cap cells. However, all germaria (n = 209) expressed GFP in both TF and cap cells when GFP was induced at the pupal stage (Fig. 2 F, right). These results all indicate that Hh-producing cells contribute to adult niche cap cells.
Hh signaling–receiving cells contribute to both GSC maintenance and differentiation niches
G-TRACE activated by ptc-GAL4 during embryogenesis induced GFP expression in germ cells in 27% of germaria (n = 154; Fig. 2 E) and in a mixed pattern in 66% of germaria (n = 154; Fig. 2 G). After the embryonic stage, GFP expression induced at L1 and L2 stages exhibited a mixed pattern in most late-L3 gonads (Fig. S1, J and K) and 1-d-old germaria (Fig. 2 G). Cells marked GFP at the L3 stage were mostly excluded from TFs in the late-L3 gonad (Fig. S1 L) but exhibited a mixed pattern in D1 germaria (Fig. 2, middle). Further, 89% of germaria (n = 286) expressed GFP only in escort and follicle cells when G-TRACE was activated during pupation (Fig. 2, E [right] and G). Apical cells and basal cells are not incorporated into germaria (King et al., 1968; Cohen et al., 2002), suggesting that ptc-GAL4–expressing cells (ICs) are the source for niche cap and escort cells. We confirmed this by activating G-trace using tj-GAL4, which was expressed in ICs and basal cells at the late-L3 stage (Fig. S1, M–R). Notably, both hh-GAL4– and ptc-GAL4–expressing cells were able to contribute to niche cap cells, an observation supported by the results that forming niche cap cells expressed both Hh and ptc-lacZ (see Fig. 1 O). From these results, we conclude that ICs receive Hh signals and contribute to niche cap and escort cells.
Somatic Hh signaling maintains the IC pool for niche formation
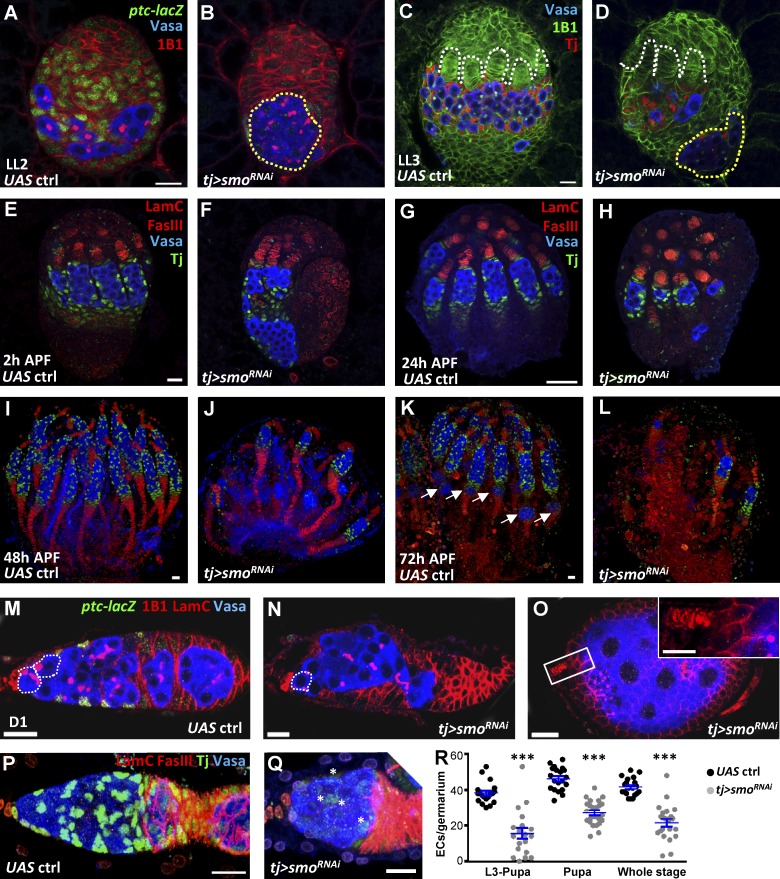
To investigate the role of Hh signaling in ICs, we used tj-GAL4 to drive a smoRNAi line (Bloomington) in ovaries at different developmental stages. In the late-L2 control gonad, ptc-lacZ was expressed in apical cells and ICs that were well intermingled with PGCs (Fig. 3 A), whereas ptc-lacZ expression was nearly abolished and PGCs clustered together in the tj>smoRNAi gonad (Fig. 3 B), showing the efficiency of the RNAi line. In the late-L3 control gonad, TFs were present and PGCs intermingled with ICs that expressed Tj (Fig. 3 C), whereas a large PGC cluster was located at the bottom of tj>smoRNAi gonads and a few PGCs interacted with ICs closer to TFs (Fig. 3 D and Fig. S2, A and B). Similar phenomena were observed when other SGP drivers, ptc-GAL4 or bab1-GAL4 (Bolívar et al., 2006), drove the same smoRNAi line or when tj-GAL4 drove another UAS-smoRNAi (NIG; Fig. S2, C–H). Significantly, tj>smoRNAi gonads carried fewer ICs (485 ± 194, n = 9) than did controls (716 ± 114, n = 7). MA33, another IC marker (Gilboa and Lehmann, 2006), was also dramatically reduced in ICs; instead, cells in the IC region expressed a level of F-actin similar to that of basal cells (Fig. S2, I–L). These results suggest that Hh signaling maintains the IC pool.
Figure 3.
Somatic Hh signaling controls soma–germline interaction and GSC niche formation. (A–D) Control (ctrl; A and C) and tj>smoRNAi gonads (B and D) of late-L2 (A and B) and late-L3 (C and D) larvae with Vasa (blue, PGCs), 1B1 (red in A and B, green in C and D, fusomes and somatic cell membranes), ptc-lacZ in A and B (green, an Hh signaling reporter), and Tj in C and D (red, ICs). White dashed lines, TFs; yellow dashed circles, PGC clusters. (E–L) Control (E, G, I, and K) and tj>smoRNAi pupal ovaries (F, H, J, and L) with Vasa (blue), LamC (red, TF and cap cell nuclear envelopes), FasIII (red, stalk cell membranes), and Tj (green) at 2 (E and F), 24 (G and H), 48 (I and J), and 72 h (K and L) after puparium formation (APF). Arrows in K, newly formed egg chambers. (M–Q) One-day (D1) –old control (M and P) and tj>smoRNAi ovaries (N, O, and Q) with ptc-lacZ in M–O (green), Vasa (blue, germ cells), 1B1 in M–O (red, fusomes and follicle cell membranes), LamC (red), FasIII in P and Q (red, follicle cell membranes), and Tj expression in P and Q (green, cap, escort and follicle cell nuclei). The inset in O shows an empty germarium; asterisks in Q show escort cells with weak Tj expression. Dashed circles, GSCs. Bars, 10 µm. (R) Escort cell (EC) number per germarium of D1 control and flies with smo-knockdown driven by tj-GAL4 from L3 to pupal stages, during the pupal stage, or throughout development (whole stage). Statistical differences were analyzed by two-tailed t-test. Error bars represent SEM. ***, P < 0.001.
At 2 h after puparium formation (APF), germaria were formed and ICs incorporated into each germarium; these ICs were identified as escort cells (Fig. 3, E and G), and at 48 h APF, germaria were completely formed with basal stalks, derived from basal cells, marked by Fasciclin III (FasIII; Fig. 3 I). At 72 h APF, a newly formed egg chamber was located posterior to the germarium (Fig. 3 K, arrows), indicating the onset of oogenesis. However, in the tj>smoRNAi ovary at 2 h APF, a large PGC cluster was still present, and only a few ICs intermingled with PGCs located closer to TFs (Fig. 3 F). Germaria carried fewer PGCs and escort cells, and each of them connected to a basal stalk in smo-knockdown ovaries (Fig. 3, H, J, and L). The PGC cluster was absent in the tj>smoRNAi ovary at 24 h APF. Germaria without PGCs likely undergo degeneration, thus forming fewer ovarioles in the adult. Indeed, 1-d-old tj>smoRNAi ovaries (n = 24) carried only 6.9 ± 2.4 ovarioles in comparison with 17.7 ± 1.7 ovarioles in control ovaries (tj>mCD8-gfp, n = 22, P < 0.001).
As a consequence, 1-d-old tj>smoRNAi germaria with the absence of ptc-lacZ had a severe reduction in GSCs and niche cap cells (Fig. 3, M–O; and Fig. S3 A), and some were empty with a large egg chamber (Fig. 3 O). In contrast to control germaria (42 ± 5, n = 20) germaria with smo knockdown throughout development carried fewer escort cells (26 ± 10, n = 21, P < 0.001; Fig. 3, P–R). A similar degree of escort cell reduction was observed in gonads with smo knockdown from L3 to pupal stages, whereas suppression of smo expression only during pupation caused less reduction of escort cells (Fig. 3 R), suggesting that Hh signaling is important at this period for there to be escort cells in the adult. The anterior-most PGCs receive bone morphogenetic protein signals from maintenance niches to adapt GSC fate (Song et al., 2004). Bone morphogenetic protein signaling in GSCs, as revealed by p-Mad expression and the number of p-Mad–positive GSCs, was reduced in late-L3 tj>smoRNAi gonads, as compared with controls (Fig. S3, B–E), indicating impairment of GSC recruitment. Our results clearly show that Hh signaling maintains ICs for niche formation, as well as GSC recruitment.
Forcing Hh signaling expands the IC population
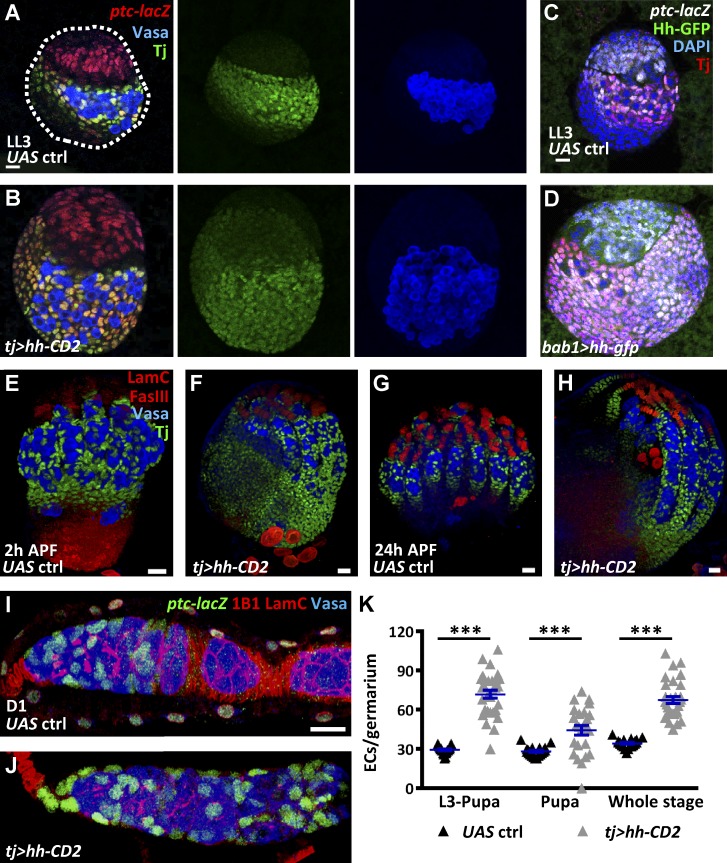
Conversely, hyperactivation of Hh signaling in SGPs caused enlarged gonads accompanied by expanded ICs and PGCs at the late-L3 stage (Fig. 4, A–D) when tj-GAL4 and bab1-GAL4 were used to drive UAS-hh-CD2 (membrane-bound Hh) and UAS-hh-gfp (diffusible Hh; Tanimoto et al., 2000; Torroja et al., 2004), respectively. In control gonads, ICs (Tj-positive cells) intermingled with PGCs and occupied the middle portion of the gonad, whereas basal cells did not express Tj and remained separate from PGCs. In contrast, in hh-overexpressing gonads, nearly every somatic cell, except apical and TF cells, expressed both ptc-lacZ and Tj and were well intermingled with PGCs. In addition, we did not detect ptc-lacZ expression in PGCs of tj>hh-CD2 or tj>hh-gfp gonads, suggesting that the expansion of PGCs resulted from hyperactivation of Hh signaling in ICs. These results indicate that Hh signaling has the capacity to direct SGPs to become ICs and suppresses the formation of basal cells.
Figure 4.
Overexpression of Hh promotes IC formation leading to expansion of escort cells. (A and B) Late-L3 (LL3) control (ctrl; A) and tj>hh-CD2 gonads (B) with Vasa (blue, PGCs), ptc-lacZ (red, an Hh signaling reporter), and Tj (green, ICs). Dashed line in A outlines the gonad. (C and D) LL3 control (ctrl; C) and bab1>hh-gfp gonads (D) with GFP (green), ptc-lacZ (gray), Tj (red), and DAPI (blue, DNA). (E–H) Control (E and G) and tj>hh-CD2 pupal ovaries (F and H) with Vasa (blue), LamC (red, TF and cap cell nuclear envelopes), FasIII (red, stalk cell membranes), and Tj (green) at 2 (E and F) and 24 h (G and H) after puparium formation (APF). (I and J) 1-d-old (D1) control (I) and tj>hh-CD2 ovaries (J) with ptc-lacZ (green, escort cells), Vasa (blue, germ cells), 1B1 (red, fusomes and follicle cell membranes), and LamC (red). Bars, 10 µm. (K) Escort cell (EC) number per germarium of D1 control and flies with hh-CD2 overexpression driven by tj-GAL4 from L3 to pupal stages, during the pupal stage, or throughout development (whole stage). Statistical differences were analyzed by two-tailed t-test. Error bars represent SEM. ***, P < 0.001.
As expected, the expanded ICs in hh-overexpressing ovaries resulted in ectopic escort cells in germaria (Fig. 4, E–K). Forcing hh expression in gonads from L3 to pupal stages, or throughout development, showed a comparable increase in escort cell numbers (Fig. 4 K), suggesting that somatic cells posterior to TFs are sensitive to Hh signaling for cell fate choice at the L3 to pupal stage. Similar results were obtained using another escort cell marker, PZ1444 (Margolis and Spradling, 1995; Kirilly et al., 2011; Fig. S3, F–H). Surprisingly, we did not observe an increased number of niche cap cells or GSCs in hh-overexpressing germaria (Fig. S3 I). Instead, escort cells were often found in the space where niche cap cells were originally located (Fig. 4 J). Combining the data showing ICs of tj>smoRNAi with decreased IC markers but increased basal cell markers (Fig. S2, I–L), we hypothesized that Hh signaling promotes ICs, which contribute GSC niches, but suppresses basal cells, which form basal stalks.
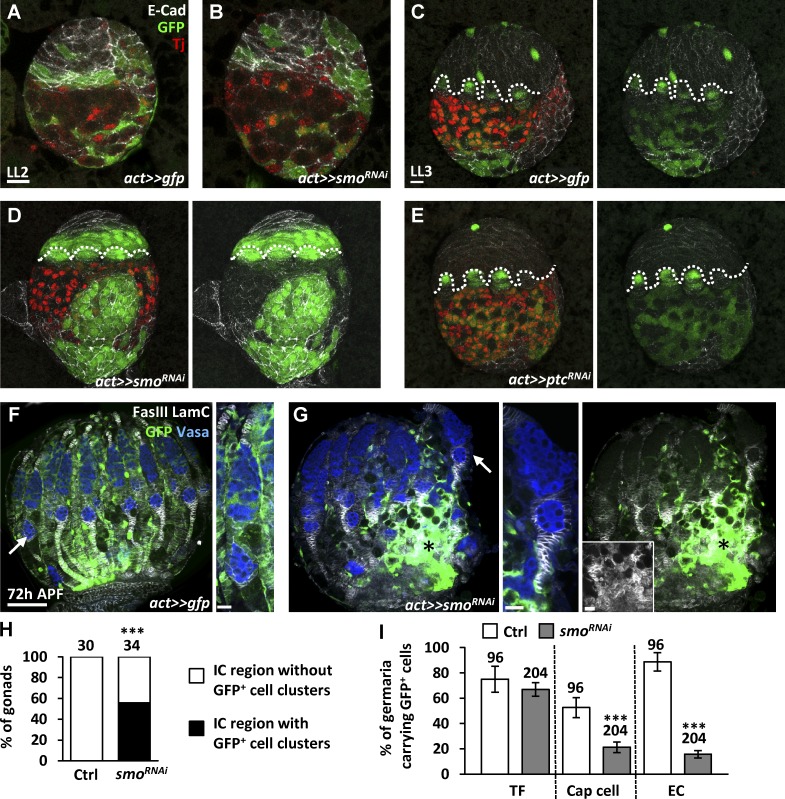
Hh signaling controls the cell fate decision between stromal ICs and epithelial basal cells
To test this hypothesis, we examined the cell property of smo-knockdown cells by examining the expression of Tj, an IC marker, which suppresses E-cadherin expression in follicle cells (Li et al., 2003). We generated smo-knockdown SGPs in early-L1 larvae by using a 10-min heat shock to activate flip-out GAL4 (actin promoter-FRT-CD2-FRT-GAL4) and traced them at different developmental stages (Fig. 5). smoRNAi was expressed when the stop codon flanked by two FRT sites was excised by FLP (smo-knockdown cells were characterized by the presence of GFP). We did not observe dramatic differences between control and smo-knockdown cells in late-L2 gonads (Fig. 5, A and B); at this stage, basal cells were not formed. However, late-L3 smo-knockdown cells were gathered in the IC region of 56% of mosaic gonads (n = 34) in comparison with control gonads (n = 30, P < 0.001; Fig. 5, C, D, and H). These smo-knockdown cells did not express Tj but expressed levels of E-cadherin comparable to that in basal cells (Fig. 5, C and D), suggesting that smo-knockdown cells exhibit basal cell characteristics. In contrast, ptc-knockdown cells (forced Hh signaling) generated at the early L1 stage were not only in the IC region but also in the basal region of the mosaic gonads (Fig. 5 E); these cells expressed Tj and E-cadherin at a level comparable to that in ICs and were well intermingled with PGCs (Fig. 5 E), which represents IC properties.
Figure 5.
Somatic Hh signaling controls cell fate decision between stromal ICs and epithelial basal cells. (A–E) Control (act>>gfp; A and C), smo- (B and D), and ptc-knockdown (E) mosaic gonads with GFP (green, flip-out clones), E-cad (gray), and Tj expression (red, ICs) of late-L2 (A and B) and late-L3 larvae (C–E). Dashed lines mark forming TFs. (F and G) Mock control (F) and smo-knockdown mosaic pupal ovaries (G) with GFP (green, flip-out clones), Vasa (blue, germ cells), LamC (gray, TF and cap cell nuclear envelopes), and FasIII (gray, stalk cell membranes) 72 h after puparium formation (APF). F, right, and G, middle, are an enlarged view of the germarium in (F) and (G, arrows). The inset in G, right, shows only the irregular arrangement of GFP+ stalk cells from the region indicated by asterisks in G, left, and G, right. Bars: (A–E; F, right; G, middle; and G, right, inset) 10 µm; (F, left, and G, left) 50 µm. (H) Percentage of gonads with or without GFP+ cell clusters in the IC region of late-L3 mock control and smo-knockdown mosaic gonads. (I) Percentage of germaria carrying GFP+ TFs, cap cells, and escort cells (ECs) in control and smo-knockdown mosaic pupal ovaries at 72 h APF. Gonad or germaria numbers analyzed are shown above each bar. ***, P < 0.001. Statistical differences analyzed by χ2 in H and two-tailed t-test in in I. Error bars represent SD in I.
If smo-knockdown cells in the IC region tend to adopt basal cell fate, those cells should mostly contribute to stalk cells (derived from basal cells), but not cap or escort cells. Indeed, smo-knockdown cells generated at the early-L1 stage were not favored to contribute to cap or escort cells, examined at 72 h APF (Fig. 5, F, G, and I). Many smo-knockdown cells expressed FasIII, a stalk cell marker, and accumulated at the bottom of the ovary (asterisk in Fig. 5 G). Furthermore, in tj>smoRNAi gonads, we did not detect enhanced cell death or decreased epidermal growth factor receptor (Egfr) signaling (Fig. S4, A–D), which is activated by Spitz produced from PGCs to maintain IC survival (Gilboa and Lehmann, 2006). Perhaps ICs closer to PGCs in tj>smoRNAi gonads were supported by Egfr signaling. These results further support our hypothesis that Hh signaling determines IC fate.
Hh signaling cell-autonomously controls expression of E-cadherin and Tj in ICs
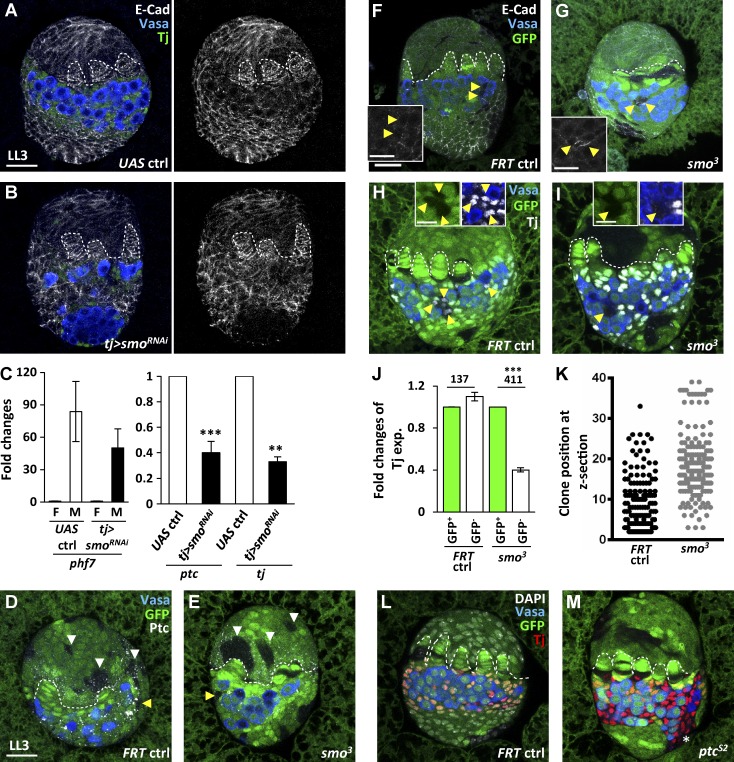
We next asked whether Hh signaling controls Tj expression in ICs. Consistently, tj>smoRNAi gonads carried fewer ICs that weakly expressed Tj and were associated with PGCs, whereas other somatic cells in the IC region did not express Tj but expressed high levels of E-cadherin characteristic of basal cells (Fig. 6, A and B). In addition, quantitative real-time RT-PCR results showed that late-L3 tj>smoRNAi female gonads expressed very low levels of the male marker phf7 (Yang et al., 2012) and decreased ptc (a direct target of Hh signaling) and tj transcripts compared with controls (Fig. 6 C). These results suggest that Hh signaling controls tj expression at a transcriptional level. Furthermore, smo3 mutant somatic cells (identified by the absence of GFP, generated by FRT-mediated mitotic recombination) exhibited decreased Ptc expression (Fig. 6, D and E), indicating impairment of Hh signaling. Expression of Tj in smo3 mutant ICs was reduced to 40% of that in neighboring normal GFP-positive ICs, whereas E-cadherin expression was increased (Fig. 6, F–J), indicating that Hh signaling directly controls Tj and E-cadherin expression levels in ICs.
Figure 6.
Somatic Hh signaling directly controls Tj and E-cadherin expression in ICs. (A and B) Late-L3 (LL3) control (ctrl; A) and tj>smoRNAi gonads (B) with E-cad (gray), Vasa (blue, germ cells), and Tj (green, ICs). (C, left) The mean fold changes of phf7 transcripts in male (M) gonads relative to those in female (F) gonads of control and tj>smoRNAi flies. (C, right) The mean fold changes of ptc and tj transcripts in tj>smoRNAi gonads relative to those in control gonads. (D–I) LL3 mock control (D, F, and H) and smo3 mutant mosaic gonads (E, G, and I) with GFP (green, wild-type cells), Vasa (blue), Ptc in D and E (gray), E-cad in F and G (gray), and Tj in H and I (gray). (J) The mean fold changes of Tj protein expression (exp.) in GFP− ICs relative to those of neighboring GFP+ wild-type ICs of LL3 mock and smo3 mutant mosaic gonads. Numbers of ICs analyzed are shown above each bar. (K) Clonal position in z-sections of control and smo3 mutant mosaic gonads from the site facing the midline toward the site facing the fat body (0–40 sections). (L and M) LL3 mock control (L) and ptcS2 mutant mosaic gonads (M) with GFP (green, wild-type cells), Vasa (blue), Tj (red), and DAPI (gray, DNA). The asterisk in M indicates the ptcS2 mutant clones located at the bottom region but expressing Tj. Dashed lines mark forming TFs. White and yellow arrowheads indicate clones present in the apical and IC regions, respectively. Bars: (insets) 10 µm; (A, D, and F) 20 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical differences in C and J were analyzed by two-tailed t-test. Error bars represent SD in C and SEM in J.
smo3 mutant ICs, compared with ICs in controls, were located closer to the lateral aspect of gonads, facing the fat body (high number in the z-section; Fig. 6 K), where TFs are formed later than in the medial side of the gonad facing the midline of the larva (Sahut-Barnola et al., 1995). This result suggests that smo3 mutant ICs may be pushed away from the forming germaria of the mosaic gonad, probably because the affinity of such cells is closer to that of cells receiving fewer Hh signals. In addition, mitotic recombination-induced ptcS2 mutant ICs expanded to the basal region and expressed a high level of Tj (Fig. 6, L and M), consistent with results from the flip-out clonal assay. Tj has also been reported to promote expression of Piwi, which interacts with Piwi-interacting RNA to induce gene silencing, including FasIII (Saito et al., 2009); however, FasIII was not detectable in the control ovary until the pupal stage (Figs. 3 and 4), and Piwi was not reduced in smo-knockdown or mutant cells (Fig. S4, E–K), probably because the remaining amount of Tj is enough to maintain Piwi expression. Our results indicate that Hh signaling cell-autonomously controls Tj expression in ICs, and this regulation is critical to maintain proper cell affinity of ICs.
Tj controls SGP adhesion to adopt IC fate
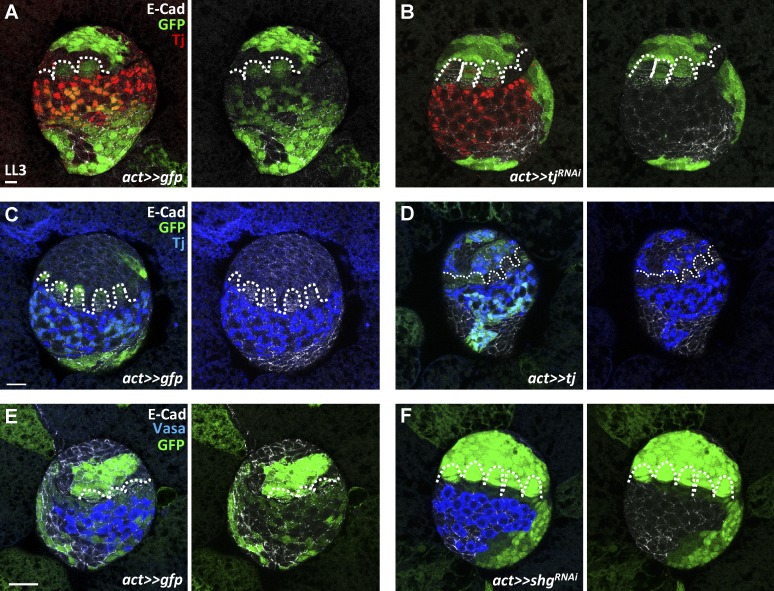
During embryogenesis or organogenesis, cells are segregated into distinct domains and then further differentiate into specific cell lineages (Krens and Heisenberg, 2011). We thus proposed that SGPs posterior to TFs might be segregated into IC and basal regions according to their cell affinity, at least in part through the regulation of Tj-mediated suppression of E-cadherin expression. To test this possibility, we generated tj-knockdown or tj-overexpressing somatic cells (GFP positive) in early-L1 larvae by flip-out GAL4 and traced them at the late-L3 stage (Fig. 7, A–D). In control gonads (n = 12; Fig. 7 A), GFP-positive control cells randomly distributed in apical, basal, and IC regions. However, most of the tj-knockdown somatic cells did not interact with PGCs and were localized to apical and basal regions of mosaic gonads (Fig. 7 B), where they expressed high levels of E-cadherin. Only 8% and 12% of act>> tjRNAi (II) (n = 32) and act>> tjRNAi (III) gonads (n = 37), respectively, carried tj-knockdown somatic cells in the IC region. tj-knockdown mosaic gonads did not exhibit reduced gonad size or carry condensed DNA accumulation (unpublished data), suggesting that tj-knockdown cells were unable to stay in the IC region, rather than suggesting cell death. In contrast, tj-overexpressing flip-out cells of late-L3 gonads formed a cluster with decreased E-cadherin expression (Fig. 7 C, and D), consistent with our previous finding that Tj negatively controls E-cadherin expression in ICs. Decreased E-cadherin expression in somatic cells in L1 larvae with flip-out GAL4 to drive RNAi against shg also caused cells to adhere together outside the IC region in the late-L3 stage (n = 18; Fig. 7, E and F). These results suggest that a proper level of E-cadherin expression mediated by Tj is required for SGPs to become ICs.
Figure 7.
Tj modulates E-cadherin expression for somatic cells to intermingle with PGCs and adopt IC cell fate. (A–F) Late-L3 mock control (ctrl; A, C, and E), tj-knockdown (B), tj-overexpressing (D), and shg-knockdown (F) mosaic gonads with GFP (green, flip-out clones), E-cad (gray), Tj in A–D (red in A and B, blue in C and D, ICs), and Vasa in E and F (blue, PGCs). tjRNAi and shgRNAi flip-out cells do not intermingle with PGCs and only locate in apical and basal regions of mosaic gonads. tj-overexpressing flip-out clones exhibit low expression of E-cadherin. Dashed lines mark forming TFs. Bars: (A and C) 10 µm; (E) 20 µm.
Decreasing shg or overexpressing tj in smo-knockdown somatic cells rescues soma–germline interaction and the IC population
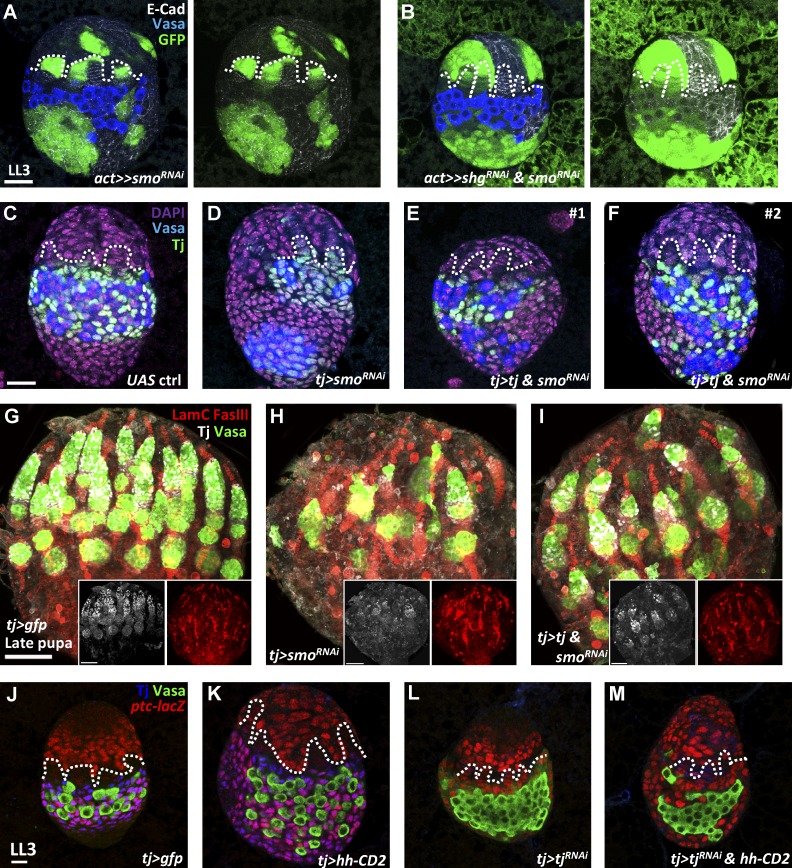
We next asked whether Hh signaling controls cell affinity for lineage specification via Tj–E-cadherin regulation. We first decreased E-cadherin expression in smo-knockdown somatic cells by using flip-out GAL4 to drive shgRNAi and smoRNAi and examined gonad morphogenesis at the late-L3 stage. smo-knockdown cells with increased E-cadherin expression adhered together (Fig. 8 A); strikingly, coknockdown shg and smo in somatic cells rescued the soma–germline interaction in 67% of gonads (n = 15; Fig. 8 B). In addition, compared with tj>smoRNAi gonads, overexpression of tj in smo-knockdown gonads by tj-GAL4 partially restored soma–germline interaction accompanied by small PGC clusters (Fig. 8, C–F), suggesting that ICs are rescued. Consequently, overexpression of tj dramatically increased Tj-positive cells in smo-knockdown ovaries at the late pupal stage (Fig. 8, G–I). Furthermore, in addition to GSCs and niche cap cells, the ovariole number in 1-d-old smo-knockdown ovaries was partially rescued by supplying exogenous Tj (12.6 ± 3.8, n = 24) in comparison to control (16.6 ± 1.7, n = 22 ovaries, P < 0.001) and tj>smoRNAi ovaries (6.9 ± 2.4, n = 24 ovaries, P < 0.001; Fig. 8, G–I). Lastly, elimination of tj expression in tj>hh gonads suppressed ectopic formation of ICs and PGCs, which was observed in tj>hh gonads, and formed PGC clusters (Fig. 8, J–M). These results indicate that Hh signaling controls IC fate via E-cadherin mediated by Tj.
Figure 8.
Somatic Hh signaling controls IC formation to establish GSC niches and ovary morphogenesis. (A and B) Late-L3 (LL3) smo-knockdown (A) and smo- and shg-knockdown (B) mosaic gonads with GFP (green, flip-out clones), E-cad (gray), and Vasa (blue, PGCs). (C–F) LL3 control (ctrl; C), tj>smoRNAi (D), and tj>tj&smoRNAi (E and F) gonads with Tj (green, ICs), Vasa (blue), and DAPI (magenta, DNA). Dashed lines mark forming TFs. (G–I) tj>gfp (G), tj>smoRNAi (H), and tj>tj&smoRNAi pupal ovaries (I) 72 h after puparium formation with LamC (red, TF and cap cell nuclear envelopes), FasIII (red, follicle and stalk cells), Tj (gray), and Vasa (green, germ cells). Insets in G–I with gray and red channels show that overexpression of tj in smo-knockdown ovaries partially restores GSC niche cells. (J–M) LL3 tj>gfp (J), tj>hh-CD2 (K), tj>tjRNAi (L), and tj> tjRNAi&hh-CD2 (M) gonads with ptc-lacZ (red, an Hh signaling reporter), Tj (blue), and Vasa (green). Knockdown tj expression in hh-overexpressing ovaries suppresses the ectopic formation of ICs and forms PGC clusters. Bars: (A and C) 20 µm; (G and insets) 50 µm; (J) 10 µm.
Canonical Hh signaling directly activates tj transcription via Ci
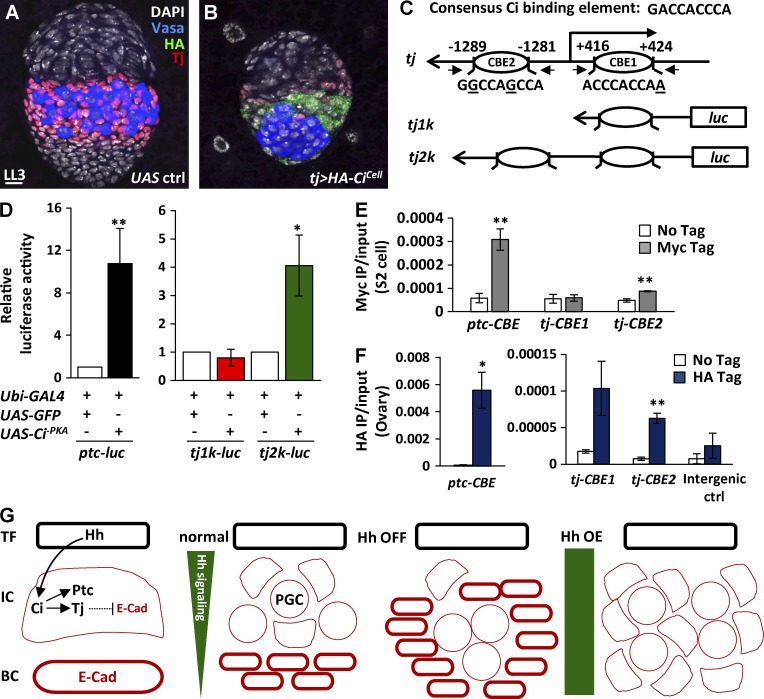
Hh signaling regulates transcription through Ci, a sequence-specific DNA-binding protein (Von Ohlen et al., 1997), suggesting that Hh signaling controls tj transcription via Ci. To test this, we overexpressed a repressor form of Ci tagged with hemagglutinin, HA-CiCell (Méthot and Basler, 1999), by tj-GAL4 (Fig. 9, A and B). Similar to tj>smoRNAi gonads, tj>HA-CiCell gonads exhibited a large PGC cluster and decreased ICs (Tj positive) at the late-L3 stage. We also found two putative Ci-binding elements (CBEs) of the tj gene; CBE1 (AACCACCCA) was +424 to +416 bp in the 5′ untranslated region, and CBE2 (GGCCAGCCA) was −1,289 to −1,281 bp upstream of the transcriptional start site (Fig. 9 C). These CBEs were similar to the CBE (GACCACCCA) of the ptc promoter, which shows the highest Ci-binding affinity (Ramos and Barolo, 2013). We first examined the effects of Ci binding on tj transcription by using a promoter activity assay (Fig. 9 D). We generated luciferase reporter genes driven by 1.044 and 2 kb of the tj promoter containing CBE1 (tj1k) and CBE1 and CBE2 (tj2k), respectively. These reporters were transfected into S2 cells with gfp or Ci-PKA, a constitutive active form of Ci (Han et al., 2015), and were activated by ubiquitin (ubi)-GAL4 (Fig. 9 D). The ptc promoter (−758 to +130 bp) containing a CBE was used as a positive control and exhibited an 11-fold increase of luciferase expression in cells with ubi>Ci-PKA compared with cells with ubi>gfp. The addition of Ci-PKA increased luciferase expression by fourfold in cells transfected with tj2k reporter, but not tj1k reporter. We further used chromatin immunoprecipitation (ChIP) to examine whether Ci-PKA (tagged with Myc) binds to the CBEs of the tj promoter in transfected or untransfected S2 cells (Fig. 9 E). We used qPCR to determine Ci-PKA occupancy at the CBE of the ptc promoter (as a positive control). Antibodies against Myc efficiently immunoprecipitated the CBE of the ptc promoter at an amount sevenfold higher in S2 cells expressing Myc-tagged Ci-PKA than in cells without transfection (negative control). The occupancy of Ci-PKA in CBE2, but not CBE1, of the tj promoter showed a twofold enrichment compared with the negative control. ChIP assays with 1-d-old ovaries carrying tj>HA-Ci-PKA showed similar results (Fig. 9 E). This result was in agreement with a previous study showing that the low affinity of the Ci-binding site is critical for the transcription of several Hh target genes (Ramos and Barolo, 2013). Our results indicate that Ci activates tj transcription directly, predominately via interaction with CBE2.
Figure 9.
Hh signaling activates tj transcription via Ci. (A and B) Late-L3 control (ctrl; A) and tj>HA-CiCell gonads (B) with Tj (red, ICs), Vasa (blue, PGCs), DAPI (gray, DNA), and HA (green). Bar, 10 µm. (C) Luciferase expression driven by the 1 kb (tj1k) and 2 kb tj promoters (tj2k) carrying Ci-binding element 1 (CBE1), and both CBE1 and CBE2, respectively. (D) Luciferase reporter assay. S2 cells were transiently transfected with ptc, tj1k, or tj2k luciferase reporter plus ubi-GAL4 together with UAS-GFP or UAS-Ci-PKA. The luciferase activity of ubi-GAL4 & UAS-GFP was set at 1. (E and F) ChIP analysis of Ci binding in S2 cells (E) and in 1-d-old ovaries (F). The chromatin from S2 cells with or without expressing Myc-Ci-PKA and ovaries with or without expressing HA-Ci-PKA was precipitated with antibodies against Myc and HA, respectively. Coprecipitated DNA was analyzed by qPCR using primers against positions containing CBE of the ptc gene, CBE1, and CBE2 of the tj gene. The amplicon for the intergenic region was used as a negative control. Statistical differences in D–F were analyzed by two-tailed t-test. Error bars represent SD in D and SEM in E and F. *, P < 0.05; **, P < 0.001. (G) Hh signaling controls Tj via Ci to suppress E-cadherin (E-Cad) for IC fate. BC, basal cell; OE, overexpression.
Discussion
The establishment of niches for stem cell recruitment, maintenance, and differentiation is a critical process during organ formation, but the mechanisms involved are unclear. We report that the fly ovary uses cell–cell adhesion via Hh signaling to select niche precursors (ICs), which express low levels of E-cadherin mediated by Tj and intermingle with PGCs (Fig. 9 G). Hh ligands are produced by TFs and are received by ICs, where Ptc is expressed and activated by Hh signaling, but not by basal cells, forming an Hh signaling gradient. Hh signaling activates Tj expression via Ci to suppress E-cadherin expression; in contrast, basal cells do not express Tj but have a high level of E-cadherin expression. When Hh signaling is disrupted, ICs become basal cells with reduced Tj and increased E-cadherin expression and do not intermingle with PGCs, leading to PGC clustering. Conversely, hyperactivation of Hh signaling activates Tj to suppress E-cadherin expression and converts all somatic cells posterior to TFs to ICs, which promotes PGC proliferation via unknown mechanisms. These findings add to our understanding of how organs use morphogen gradients to segregate cells for niche formation.
Hh signaling in ovary development
Although Hh signaling participates in many aspects of organ development, few studies report the role of Hh signaling in ovary development. In Drosophila, Hh is expressed in the gonadal mesoderm, acting as a diffusible chemoattractant to guide PGC migration to the somatic gonad during embryogenesis (Deshpande et al., 2001; Araújo, 2015); however, some research results contradict this hypothesis (Renault et al., 2009). Sato et al. have proposed that TFs and PGCs in L3 ovaries produce Hh, which is received by PGCs to regulate PGC proliferation (Sato et al., 2010). In addition, Fused, a Hh signal–transducing serine/threonine kinase, is expressed in PGCs to mediate Hh signals produced by TFs for the control of soma–germline interaction (Besse et al., 2005). However, mutations of smo in germ cells do not show a similar phenotype (Besse et al., 2005). Here, we report the role of Hh signaling in determining ICs, which contribute to adult niche cap and escort cells. Hh signaling may also coordinate with Notch signaling to specify cap cells derived from the anterior of ICs adjacent to TFs. First, overexpression of Delta or Notch during development induces the transformation of escort cells to cap cells (Song et al., 2007), indicating that Delta produced by apical cells and TFs drives ICs to become cap cells. Second, Hh signaling occurs in forming cap cells (Fig. 1 O, inset), whereas Hh signaling revealed by ptc-lacZ is absent from adult cap cells (Liu et al., 2015b), suggesting that Hh signaling needs to be turned off in cap cells after they are specified. Third, overexpression of Hh signaling in gonadal somatic cells expands the IC population and induces ectopic escort cells, but not cap cells, at the adult stage; instead, escort cells occupy the space where cap cells are located, suggesting that constitutive activation of Hh signaling disrupts the specification of cap cells.
In the mouse ovary, folliculogenesis occurs when an individual oocyte is surrounded by epithelial granulosa cells that recruit precursors of stromal theca cells for steroid production (Hirshfield, 1991). A recent paper also reported that specification of theca cells requires expression of Gli1 (Ci orthologue in mammals) in response to Hh from granulosa cells (Liu et al., 2015a). In addition, Tj is most similar to mammalian c-Maf and Maf B based on their high similarity in amino acid sequence and protein structure (Li et al., 2003). Large Maf proteins act as major regulators for cell differentiation in many tissues (Tsuchiya et al., 2015). Although the function of Maf proteins in mammal ovary is unexplored, c-Maf and MafB are expressed in stromal cells of mouse embryonic ovaries (Maatouk et al., 2012). These results suggest a conserved role of Hh signaling to large Maf transcription factors in the lineage determination of ovarian somatic cells. Furthermore, ovarian cell hyperproliferation induced by Hh (Fig. 4) or Tj overexpression (not depicted) is reminiscent of the mechanism by which aberrant activation of Hh signaling causes ovarian cancer (Russell et al., 2007).
Differential cell affinity in organ development
During tissue or organ development, the cell fate decision is followed by segregation of cells into distinct domains, where different cell lineages are further specified (Krens and Heisenberg, 2011). A general hypothesis is that tissue separation occurs through differential adhesion of cells (Fagotto, 2014). The most accepted experimental results to support this hypothesis involve the reformation of well-segregated cell populations from a mixed aggregate initially formed by cells that are dissociated from different embryonic tissues (Moscona and Moscona, 1952; Steinberg and Gilbert, 2004). These results also suggest that cells have the ability to recognize the identity of their neighbors and gather with cells of the same type. Several classic examples in both vertebrate and invertebrate systems have been used as paradigms for the analysis of tissue separation (Fagotto, 2014), such as embryonic parasegment boundaries and anterior–posterior and dorsoventral compartments of larval wing discs in Drosophila, as well as the formation of germ layers, somites, and rhombomeres in frog and fish. However, many other segregations in developing tissues have not been investigated, such as that of the ovary. Here, we report that the development of the Drosophila ovary requires such separation to specify stromal ICs and epithelial basal cells and that this process is controlled by an Hh signaling–Tj–E-cadherin axis. Interestingly, in the Drosophila wing disc, the regulation of cadherin Cad 99 by of Hh signaling is involved in setting the anterior–posterior compartment (Schlichting et al., 2005). In addition, Sonic Hh signaling negatively regulates E-cadherin expression in mouse gastric epithelial cells in the culture system (Xiao et al., 2010). These results suggest a common role for Hh signaling in the control of cell adhesion and its general participation in tissue separation.
Materials and methods
Drosophila strains and culture
Drosophila stocks were maintained at 22–25°C on standard medium, unless otherwise indicated. w1118 was used as a wild-type control. The following fly strains were used in this study: cold-sensitive mutant smo3 and hypomorphic allele ptcS2 have been described previously (Chen and Struhl, 1996). ptc-lacZ was used to monitor Hh signaling (Chen and Struhl, 1996). PZ1444 (a gift from T. Xie, Stowers Institute for Medical Research, Kansas City, MO) and MA33 (a gift from T. Schüpbac, Princeton University, Princeton, NJ) are enhancer trap lines in which lacZ is expressed in niche cap and escort cells (Margolis and Spradling, 1995; Kirilly et al., 2011) and in ICs (Gilboa and Lehmann, 2006), respectively. UAS-RNAi lines against smo (BL43134 and NIG-Fly 11561R-1), ptc (NIG-Fly 2411R-1), tj (Vienna Drosophila RNAi Center [VDRC] 30526, second chromosome [II], and 108255, third chromosome [III]), shg (VDRC 27081), and Ci (BL31320, BL28984, NIG-Fly 2125R-1, and VDRC 105620) were obtained from the VDRC, the National Institute of Genetics-Fly Stocks, or the Bloomington Drosophila Stock Center (BL). Sequences of the RNAi lines used in this study are available from the website of each stock center, and their efficiencies were described previously or tested here. In this study, we used the smoRNAi line from Bloomington, unless otherwise indicated. UAS-hh-gfp, UAS-hh-CD2, UAS-tj (a gift from D. Godt, University of Toronto, Toronto, Canada), UAS-HA-piwi (a gift from R. Lehmann, New York University School of Medicine, New York, NY), UAS-HA-CiCell (a gift from C.-T. Chien, Institute of Molecular and Cell Biology, Academia Sinica, Taipei, Taiwan), UAS-HA-Ci-PKA (a gift from J. Jiang, UT Southwestern Medical Center, Dallas, TX), tj-GAL4, bab1-GAL4, ptc-GAL4, and hh-GAL4 lines (a gift from M. Buszczak, UT Southwestern Medical Center, Dallas, TX) were described previously (Burke et al., 1999; Méthot and Basler, 1999; Tanimoto et al., 2000; Torroja et al., 2004; Bolívar et al., 2006; Hsu and Drummond-Barbosa, 2009; Gunawan et al., 2013; Tseng et al., 2014). Other genetic elements are described in FlyBase (http://flybase.bio.indiana.edu).
Developmental stage of larvae and pupae
Morphological definitions of the developmental stages of Drosophila followed those of Ashburner (2005). Flies were transferred into a fresh vial to lay eggs at 25°C for 3 h and were then removed. The vials were left at 25°C. After hatching (∼20 h after egg laying [AEL]), larvae were collected as first-instar larvae (L1). At this stage, SGPs intermingled with PGCs and did not form specific groups. We defined larvae with gonads exhibiting apical gonadal somatic cells as early second-instar larvae (early L2; ∼50 h AEL), larval gonads starting to form TFs as late-L2 (∼70 h AEL), and larvae that climbed up and down from the food as mid-third instar larvae (mid-L3; ∼100 h AEL). We defined the larvae that moved out of food but in which pupation had not started as late-L3 wandering larvae (∼125-140 h AEL). At this stage, most TFs were still forming and cap cells were starting to form. To obtain pupae that were at a synchronized developmental stage, we removed wandering larvae but kept white prepupae (referred to as 0 h prepupae) in the vial.
In addition to synchronization at hatched L1 and white prepupal stages, we also used gonadal somatic cell development as an indicator to stage the larval and pupal gonads of flies at 18°C and 29°C (Fig. 1).
Gal4-based cell lineage analysis
G-TRACE was used to trace lineages of GAL4-expressing cells (Evans et al., 2009). The embryos that were produced by crosses carrying the G-TRACE cassette (UAS-DsRed, UAS-flp; ubi>Stop>gfp/CyO) (a gift from H. Sun, Institute of Molecular and Cell Biology, Academia Sinica, Taipei, Taiwan) and flies carrying tub-GAL80ts with hh-GAL4, ptc-GAL4, or tj-GAL4 were collected within 3 h at 25°C. The embryos were cultured at 18°C until dissection (1 day after eclosion), except for the stage-specific heat shock at 29°C to induce FLP. For embryo heat shock, collected embryos were cultured at 29°C until hatching (∼16 h). To perform larval-stage shock, we cultured embryos at 18°C until hatching (∼37 h AEL) and collected newly hatched larvae (L1) within 30 min for stage synchronization. For L1 and L2 shocks, newly hatched L1 larvae (∼85 h AEL) were switched to 29°C for 24 h. For L3 shock, mild-L3 larvae that remained in the food but were climbing up and down from the food (∼135 h AEL) were switched to 29°C for 17 h. For pupa stage shock, white pupae (∼217 h AEL) were switched to 29°C until eclosion.
Genetic mosaic analysis
Genetic mosaics were generated by FLP/FRT-mediated mitotic recombination (Xu and Rubin, 1993). For conventional mosaic analysis, L1 larvae of the genotypes hs-flp122/+; smo3 FRT40A/ubi-gfp FRT40A, hs-flp122/+; ubi-gfp FRT40A/neoFRT40A, hs-flp122/+; FRT42D ptcS2/FRT42D ubi-gfp, and hs-flp122/+; FRT42D arm-lacZ/FRT 42D ubi-gfp were generated from standard crosses and subjected to heat shock for 1 h at 37°C, twice a day for 3 d. After heat shock, the larvae were cultured at 25°C until late L3 for dissection (smo3 mosaic mutant larvae were cultured at 18°C). For flip-out clone analysis, L1 larvae of genotypes hs-flp122/+; ptc-lacZ/+; UAS-smoRNAi/actin5c>Stop>GAL4 UAS-gfp, hs-flp122/+; UAS-ptcRNAi/actin5c>Stop>GAL4 UAS-gfp, hs-flp122/+; UAS-tjRNAi/actin5c>Stop>GAL4 UAS-gfp, hs-flp122/+; UAS-tj/+; actin5c>Stop>GAL4 UAS-gfp/+, hs-flp122/+; UAS-shgRNAi/+; UAS-smoRNAi/actin5c>Stop>GAL4 UAS-gfp and hs-flp122/+; actin5c>Stop>GAL4 UAS-gfp/+ were generated from standard crosses and subjected to heat shock for 10 min at 37°C. After heat shock, the larvae were cultured at 25°C until dissection. Homozygous mutant cells were identified by the absence of GFP in conventional mosaic analyses, and RNAi-expressing cells were recognized by the presence of GFP in flip-out clones.
Quantitative real-time PCR
Total RNA of male or female gonads of late-L3 larvae (∼25) was extracted with TRIzol reagent (Invitrogen). 1 µg total RNA was reverse transcribed with the Transcriptor First Strand cDNA Synthesis kit (Roche). Steady-state mRNA levels were determined by using the LightCycler 480 Probes Master combined with a Universal Probe library (Roche).
Each gene was analyzed using the primer pairs and probes listed below: tj: probe#52, 5′-CTGAACAAGCGGCTCCAT-3′ and 5′-CGTCGCTTCTGCTTCAGAC-3′; piwi: probe#54, 5′-CTGCCCGAGAGATACGACTT-3′ and 5′-CGACACTGTACCCTGACGAA-3′; fasIII: probe #101, 5′-TTGACACAAAACACATCCTCTACA-3′ and 5′-TTGATTTAATGTGTGGGCTGA-3′; ptc: probe#73, 5′-AGCCTAAGCCGTAACCCTATTT-3′ and 5′-CCATGAGAATCCATGAGAACG-3′; phf7: probe#15, 5′-CTGTTGGGCTCCATCTCG-3′ and 5′-AGTGCGGCGACGTAAGAC-3′; RpL19: probe #128, 5′-GAGCGTATTGCCACCAGGA-3′ and 5′-CGATCTCGTCCTCCTTAGCA-3′; RpL32: probe #117, 5′-CGGATCGATATGCTAAGCTGT-3′ and 5′-CGACGCACTCTGTTGTCG-3′.
Generation of Drosophila tj promoter constructs
We amplified 1.044 kb (tj1K, position −555 to +489) and 2 kb (tj2K, position −1,511 to +489) of the Drosophila tj promoter before the ATG translation start site containing one and two putative CBEs (−1,289 to −1,281 and +416 to +424), respectively, from a BAC clone containing the tj gene (RP98-28G24, BACPAC Resources Center) by PCR using the following primers: tj2K 5′-GGGGGG(KpnI site)ATACGAGCCAAAACAAATCG-3′ and 5′-GGGGGG(XhoI site)TGGATCGACCAGGGAC-3′; tj1K 5′-GGGGGG(KpnI site)GCAGATCAAAATATAATCGC-3′ and 5′-GGGGGG(XhoI site)TGGATCGACCAGGGAC-3′. The KpnI and XhoI sites were used to insert tj promoter fragments upstream of the firefly luciferase reporter gene in pGL4.15 (Promega).
Luciferase reporter assay
Drosophila S2 cells were cultured with Schneider’s Drosophila medium, containing 10% FBS and 10% streptomycin/penicillin. tj1K and tj2K of the tj promoter region were cloned into the pGL 4.15 vector. ptc-luciferase and RL-PolIII Renilla luciferase reporters were used as positive and internal controls, respectively. A total of 2.7 × 106 S2 cells were transfected with 1.1 µg reporter constructs (1 µg Luciferase and 0.1 µg Renilla) and 1 µg expression constructs (0.5 µg ubi-GAL4 and 0.5 µg UAS-GFP or UAS-Myc-Ci-PKA; a gift from J. Jiang) using Cellfectin II reagent (Invitrogen). After being transfected for 5 h, the cells were split into a 24-well plate and cultured for 72 h at 25°C. Luciferase activity was subsequently measured with a Dual-Glo luciferase assay kit (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. Data represent the mean ± SD of at least three independent experiments.
ChIP
ChIP assays were performed as previously described (Huang et al., 2014), with minor modification. S2 cells (2.5 × 106) were seeded per well in six-well plates and transfected with 1 µg expression constructs (0.5 µg ubi-GAL4 and 0.5 µg UAS-Myc-Ci-PKA) 1 day after seeding using Effectene transfection reagent (QIAGEN). Cells that were not transfected were used as the background control. At 48 h after transfection, the cells were harvested and fixed in Schneider’s insect medium containing 10% FBS and 1% formaldehyde for 10 min at room temperature; cross-linking was stopped by adding glycine to a final concentration of 125 mM. After two washes with PBS, the fixed cells were sonicated in 300 µl buffer A2 (15 mM Hepes, pH 7.5, 140 mM sodium chloride, 1 mM EDTA, 0.5 mM EGTA, 1% TritonX-100, 0.1% sodium deoxycholate, 0.1% SDS, 0.5% N-lauroyl sarcosine, 1 mM PMSF, 5 mM sodium fluoride, 5 mM sodium butyrate, and protease inhibitor cocktail; Roche) using a Bioruptor (Diagenode) for 15 min (30 s on/30 s off) on the high-power setting. After centrifugation at 4°C, 7.5 µl soluble chromatin was subjected to Western blot to check transfection, and 240 µl soluble chromatin was incubated with 1 µl mouse antimyc (9B11; Cell Signaling Technology) and 5 µl mouse antimyc (9E10; Santa Cruz Biotechnology, Inc.) antibodies overnight at 4°C. Antibody-bound chromatin was pulled down by magnetic protein A/G beads (EMD Millipore) for 2 h at 4°C, and the beads were washed five times with RIPA buffer (50 mM Hepes, pH 7.5, 1 mM EDTA, 0.7% sodium deoxycholate, 1% NP-40, 0.5 M LiCl, and 1 mM PMSF) and once with TE buffer containing 50 mM NaCl. The chromatin was eluted twice in TE buffer containing 1% SDS and 250 mM NaCl for 20 min at 65°C. After RNase A and proteinase K treatment, cross-linking was reversed overnight at 65°C. DNA was purified by phenol-chloroform extraction and ethanol precipitation. The quantitative PCR data obtained from the input and immunoprecipitated DNA samples were analyzed with the standard curve method.
ChIP of ovaries was performed with 180 pairs of ovaries from 1-d-old flies carrying UAS-HA-Ci-PKA (no tag control) alone or tj-GAL4 and UAS-HA-Ci-PKA. Ovaries were dissected in Shield insect medium and fixed in 950 µl PBS containing 1.8% formaldehyde for 10 min at room temperature. Crossing-linking was stopped by adding 50 µl 2.5 M glycine and incubating for 5 min. After two washes with PBS, fixed cells were sonicated in 400 µl buffer A2 at the same sonicator setting. After centrifugation, 280 µl soluble chromatin was incubated with 5 µl rat anti-HA (3F10; Roche) overnight at 4°C and processed for ChIP analysis.
The following primers were used to amplify fragments of the tj or ptc gene: tj-CBE1 (327–569 bp): 5′-TCTGTACTCCGTTTCCGCTG-3′ and 5′-AATTCCCGCATAGCCCCATT-3′; tj-CBE2 (−1,459 to −1,240 bp): 5′-AAGCGCCAAAGTGCGGATA-3′ and 5′-CCATTTCCGCCCACCTCC-3′; ptc CBE (−778 to −693 bp): 5′-AGCTGAACGTTTGGGTAGGG-3′ and 5′-CAAATAGCTCCGCCACGAGA-3′; intergenic control: 5′-GAGCAGACAACGCTCCAAGACCCAA-3′ and 5′-AAATTTTCCACCTACCTGCCGCACG-3′. The amplicon for the intergenic region is located at chr2L (18,415,855–18,415,978). Error bars represent SEM (*, P < 0.05; **, P < 0.001).
Immunostaining and fluorescence microscopy
Adult ovaries were dissected, fixed, and immunostained as described previously (Hsu et al., 2008). In brief, ovaries were dissected in Grace’s insect medium (Lonza) and fixed with 5.3% paraformaldehyde/Grace’s insect medium for 13 min with gentle agitation at room temperature. Ovaries were washed in PBST (0.1% Triton X-100 in PBS) 20 min for three times, and teased apart in PBST then incubate with blocking solution (5% bovine serum albumin and 0.05% normal goat serum in PBST) for 3 h at room temperature or 4°C overnight. Ovaries were incubated with primary antibodies (diluted in blocking solution) for 3 h at room temperature or 4°C overnight, and follow by PBST 30 min wash for three to four times. Then ovaries were incubated with secondary antibodies (diluted in blocking solution) for 3 h at room temperature or 4°C overnight, and follow by PBST 30 min wash for three to four times. For gonad dissection, larvae were transferred into a clear glass well, and their gonads and associated fat body were dissected in Grace’s insect medium (Lonza). The gonads and associated fat body were transferred into a 24-well cell culture dish with mesh inserts and fixed with 5.3% paraformaldehyde at room temperature for 13 min with gentle agitation. After washing, gonads were immunostained following a standard procedure, as described above. The following primary antibodies were used: rabbit anti-Hh (1:1,000; a gift from T. Tabata, Institute of Molecular and Cellular Biosciences, Tokyo, Japan), guinea pig anti-Tj (1:5,000; a gift from D. Godt), mouse anti-Piwi (1:500; a gift from M. Siomi, University of Tokyo, Tokyo, Japan), mouse anti–Hu-li tai shao (Drosophila adducing-related protein; 1:10; 1B1, Developmental Studies Hybridoma Bank [DSHB]), rat anti-Ci (1:10; 2A1; DSHB), mouse anti–Lamin C (1:25; LC28.26; DSHB), mouse anti-Ptc (1:100; DSHB), mouse anti-FasIII (1:50; 7G10; DSHB), rat anti–DE-cadherin (1:3; DCAD2; DSHB), rabbit anti-Vasa (1:500; Santa Cruz Biotechnology, Inc.), mouse anti–β-gal (1:1,000; Promega), rabbit anti-GFP (1:2,000; Torrey Pines), mouse anti-Myc (1:50; 9E10; Santa Cruz Biotechnology, Inc.), rat anti-HA (1:200; 3F10; Roche), rabbit anti–phospho (p)-ErK (1:200; 4370; Cell Signaling Technology), and rabbit anti-pMad (phospho S423+S425; 1:100; 52903; Abcam). Note that although Ci transcription was detected in every cell of the late-L3 gonad examined by ci-lacZ (Sato et al., 2010), we did not detect Ci expression in the larval gonad by using anti–Ci antibody (unpublished data), whose specificity has been demonstrated (Chen et al., 2000; Oh et al., 2015), probably because of its low expression. Alexa Fluor 488–, Alexa Fluor 568–, or Alexa Fluor 633–conjugated goat anti–mouse, anti–rabbit, anti–rat, and anti–guinea pig secondary antibodies (1:400; Molecular Probes or Abcam) were used. F-actin was stained by phalloidin (1:80, R415; Thermo Fisher Scientific). Samples were stained with 0.5 µg/ml DAPI (Sigma-Aldrich), mounted in 80% glycerol containing 20.0 µg/ml N-propyl gallate (Sigma-Aldrich), and analyzed using LSM 700 confocal microscopes (ZEISS).
Microscopy
Ovary or gonad samples were stored in mounting solution (80% glycerol containing 20.0 µg/ml N-propyl gallate) at 4°C before mounting. The immunostaining signals were detected using an upright confocal system (LSM700) with Axio imager 2 microscope (ZEISS) at 21–22°C. Confocal images were captured by the upright confocal system with a Plan-APOCHROMAT 63×/1.4 NA or 40×/1.3 NA oil objective lens and the acquisition software ZEN Image Brower (ZEISS). The images were explored and then processed in Photoshop 7.0.1 and Illustrator CS6 (Adobe Systems).
Statistical and quantification analyses
For GSC and cap cell analyses, GSCs were identified by the anterior position of their fusome (recognized by 1B1 labeling), which is juxtaposed to cap cells whose nuclear envelopes are ovoid and recognized by LamC labeling. For escort cell counting, we counted germarial somatic cells that were positive for ptc-lacZ or PZ1444 lines but negative for LamC (cap cells) and FasIII (follicle cell lineage). All statistical data were recorded in Excel (Microsoft) and graphed in either Excel or Prism 6.0 (GraphPad Software). p-values were calculated using two-sided unpaired t tests or χ2 in Excel or Prism. P < 0.05 was considered a statistically significant difference. Error bars represent the SD or SEM as described in the figure legends. For analyzing pMad expression in PGCs and Tj and Piwi expression in GFP-negative ICs of smo3 mutant mosaic gonads, ImageJ was used to measure the mean fluorescence intensity (arbitrary units) in confocal z-sections at the largest IC cell nuclear diameter. Tj and Piwi expression in GFP-negative ICs neighboring GFP-positive ICs were also measured as references. For IC and PGC counting, z-sections covering a whole gonad were reconstructed into a 3D image, and the numbers of Tj-positive cells or PGCs were analyzed by Imaris ×64 8.4.0 (Bitplane). For experiments analyzed with t tests, data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows G-trace activated by hh-GAL4, ptc-GAL4, or tj-GAL4 in ovaries at different developmental stages. Fig. S2 shows that knockdown somatic Hh signaling decreases ICs accompanied by PGC clusters. Fig. S3 shows that somatic Hh signaling controls niche formation, GSC recruitment, and maintenance. Fig. S4 shows that suppression of somatic Hh signaling does not induce cell death, decrease Egfr signaling, or decrease Piwi expression. Fig. S5 shows that overexpression of tj in smo-knockdown gonads partially rescues ovary morphogenesis, niche cap cells, and GSC numbers.
Supplementary Material
Acknowledgments
We thank T. Xie, J. Jiang, C. T. Chien, H. Sun, T. Schupbach, T. Tabata, D. Godt, M. Siomi, R. Lehma, and M. Buszczak; the Bloomington, National Institute of Genetics-Fly Stocks, and VDRC Stock Center; and the DSHB for Drosophila stocks and antibodies. We also thank S.-C. Hsu (Institute of Cellular and Organismic Biology Image Core, Academia Sinica) for assistance with image analysis and Y.-N. Lee, C.-A. Chen, C.-T. Chien, and H. Sun for comments on this article.
This work was supported by an Academia Sinica thematic grant and the Ministry of Science and Technology, Taiwan (grant 104-2311-B-001-029-MY3).
The authors declare no competing financial interests.
Author contributions: C.-M. Lai and H.-J. Hsu designed and interpreted the experiments and wrote the paper. F. Huang, K.-Y. Lin, and S.-H. Kao helped with the ChIP assay, cell culture, Western blot analysis, and molecular cloning. Y.-N. Chen analyzed the tj promoter.
Footnotes
Abbreviations used:
- AEL
- after egg laying
- APF
- after puparium formation
- CBE
- Ci-binding element
- ChIP
- chromatin immunoprecipitation
- Ci
- Cubitus interruptus
- Egfr
- epidermal growth factor receptor
- FLP
- Flipase
- FRT
- Flipase recognition target
- G-TRACE
- GAL4 technique for real-time and clonal expression
- GSC
- germline stem cell
- Hh
- Hedgehog
- IC
- intermingled cell
- PGC
- primordial germ cell
- Ptc
- Patched
- RFP
- red fluorescent protein
- SGP
- somatic gonadal precursor
- Smo
- Smoothened
- TF
- terminal filament
- Tj
- Traffic Jam
References
- Araújo S.J. 2015. The Hedgehog signalling pathway in cell migration and guidance: What we have learned from Drosophila melanogaster. Cancers (Basel). 7:2012–2022. 10.3390/cancers7040873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. 2005. Drosophila: A laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 1409 pp. [Google Scholar]
- Besse F., Busson D., and Pret A.-M.. 2005. Hedgehog signaling controls Soma-Germen interactions during Drosophila ovarian morphogenesis. Dev. Dyn. 234:422–431. 10.1002/dvdy.20537 [DOI] [PubMed] [Google Scholar]
- Bolívar J., Pearson J., López-Onieva L., and González-Reyes A.. 2006. Genetic dissection of a stem cell niche: The case of the Drosophila ovary. Dev. Dyn. 235:2969–2979. 10.1002/dvdy.20967 [DOI] [PubMed] [Google Scholar]
- Burke R., Nellen D., Bellotto M., Hafen E., Senti K.-A., Dickson B.J., and Basler K.. 1999. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 99:803–815. 10.1016/S0092-8674(00)81677-3 [DOI] [PubMed] [Google Scholar]
- Chen Y., and Struhl G.. 1996. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 87:553–563. 10.1016/S0092-8674(00)81374-4 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang S., and Xie T.. 2011. Restricting self-renewal signals within the stem cell niche: Multiple levels of control. Curr. Opin. Genet. Dev. 21:684–689. 10.1016/j.gde.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Chen Y., Goodman R.H., and Smolik S.M.. 2000. Cubitus interruptus requires Drosophila CREB-binding protein to activate wingless expression in the Drosophila embryo. Mol. Cell. Biol. 20:1616–1625. 10.1128/MCB.20.5.1616-1625.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E.D., Mariol M.-C., Wallace R.M.H., Weyers J., Kamberov Y.G., Pradel J., and Wilder E.L.. 2002. DWnt4 regulates cell movement and focal adhesion kinase during Drosophila ovarian morphogenesis. Dev. Cell. 2:437–448. 10.1016/S1534-5807(02)00142-9 [DOI] [PubMed] [Google Scholar]
- Deshpande G., Swanhart L., Chiang P., and Schedl P.. 2001. Hedgehog signaling in germ cell migration. Cell. 106:759–769. 10.1016/S0092-8674(01)00488-3 [DOI] [PubMed] [Google Scholar]
- Evans C.J., Olson J.M., Ngo K.T., Kim E., Lee N.E., Kuoy E., Patananan A.N., Sitz D., Tran P., Do M.-T., et al. . 2009. G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods. 6:603–605. 10.1038/nmeth.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F. 2014. The cellular basis of tissue separation. Development. 141:3303–3318. 10.1242/dev.090332 [DOI] [PubMed] [Google Scholar]
- Forbes A.J., Lin H., Ingham P.W., and Spradling A.C.. 1996. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 122:1125–1135. [DOI] [PubMed] [Google Scholar]
- Gancz D., Lengil T., and Gilboa L.. 2011. Coordinated regulation of niche and stem cell precursors by hormonal signaling. PLoS Biol. 9:e1001202 10.1371/journal.pbio.1001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L. 2015. Organizing stem cell units in the Drosophila ovary. Curr. Opin. Genet. Dev. 32:31–36. 10.1016/j.gde.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Gilboa L., and Lehmann R.. 2006. Soma-germline interactions coordinate homeostasis and growth in the Drosophila gonad. Nature. 443:97–100. 10.1038/nature05068 [DOI] [PubMed] [Google Scholar]
- Gunawan F., Arandjelovic M., and Godt D.. 2013. The Maf factor Traffic jam both enables and inhibits collective cell migration in Drosophila oogenesis. Development. 140:2808–2817. 10.1242/dev.089896 [DOI] [PubMed] [Google Scholar]
- Han Y., Shi Q., and Jiang J.. 2015. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc. Natl. Acad. Sci. USA. 112:6383–6388. 10.1073/pnas.1421628112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield A.N. 1991. Development of follicles in the mammalian ovary. Int. Rev. Cytol. 124:43–101. 10.1016/S0074-7696(08)61524-7 [DOI] [PubMed] [Google Scholar]
- Hsu H.-J., and Drummond-Barbosa D.. 2009. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc. Natl. Acad. Sci. USA. 106:1117–1121. 10.1073/pnas.0809144106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-J., LaFever L., and Drummond-Barbosa D.. 2008. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 313:700–712. 10.1016/j.ydbio.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Paulson A., Dutta A., Venkatesh S., Smolle M., Abmayr S.M., and Workman J.L.. 2014. Histone acetyltransferase Enok regulates oocyte polarization by promoting expression of the actin nucleation factor spire. Genes Dev. 28:2750–2763. 10.1101/gad.249730.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., and Anderson K.V.. 2006. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 133:3–14. 10.1242/dev.02169 [DOI] [PubMed] [Google Scholar]
- Ingham P.W., Nakano Y., and Seger C.. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12:393–406. 10.1038/nrg2984 [DOI] [PubMed] [Google Scholar]
- Jin Z., Flynt A.S., and Lai E.C.. 2013. Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated Dpp signaling. Curr. Biol. 23:1442–1448. 10.1016/j.cub.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., and Wagers A.J.. 2008. No place like home: Anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 9:11–21. 10.1038/nrm2319 [DOI] [PubMed] [Google Scholar]
- King R.C., Aggarwal S.K., and Aggarwal U.. 1968. The development of the female Drosophila reproductive system. J. Morphol. 124:143–166. 10.1002/jmor.1051240203 [DOI] [PubMed] [Google Scholar]
- Kirilly D., and Xie T.. 2007. The Drosophila ovary: An active stem cell community. Cell Res. 17:15–25. 10.1038/sj.cr.7310123 [DOI] [PubMed] [Google Scholar]
- Kirilly D., Wang S., and Xie T.. 2011. Self-maintained escort cells form a germline stem cell differentiation niche. Development. 138:5087–5097. 10.1242/dev.067850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krens S.F.G., and Heisenberg C.-P.. 2011. Cell sorting in development. In Current Topics in Developmental Biology. Force and Tension in Development. Vol. 95 1 Labouesse M, editor. Academic Press, San Diego: 189–213. [DOI] [PubMed] [Google Scholar]
- Li M.A., Alls J.D., Avancini R.M., Koo K., and Godt D.. 2003. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat. Cell Biol. 5:994–1000. 10.1038/ncb1058 [DOI] [PubMed] [Google Scholar]
- Liu C., Peng J., Matzuk M.M., and Yao H.H.C.. 2015a Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat. Commun. 6:6934 10.1038/ncomms7934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhong G., Chai P.C., Luo L., Liu S., Yang Y., Baeg G.H., and Cai Y.. 2015b Coordinated niche-associated signals promote germline homeostasis in the Drosophila ovary. J. Cell Biol. 211:469–484. 10.1083/jcb.201503033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk D.M., Mork L., Hinson A., Kobayashi A., McMahon A.P., and Capel B.. 2012. Germ cells are not required to establish the female pathway in mouse fetal gonads. PLoS One. 7:e47238 10.1371/journal.pone.0047238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J., and Spradling A.. 1995. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 121:3797–3807. [DOI] [PubMed] [Google Scholar]
- Méthot N., and Basler K.. 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 96:819–831. 10.1016/S0092-8674(00)80592-9 [DOI] [PubMed] [Google Scholar]
- Moscona A., and Moscona H.. 1952. The dissociation and aggregation of cells from organ rudiments of the early chick embryo. J. Anat. 86:287–301. [PMC free article] [PubMed] [Google Scholar]
- Oh S., Kato M., Zhang C., Guo Y., and Beachy P.A.. 2015. A comparison of Ci/Gli activity as regulated by Sufu in Drosophila and mammalian Hedgehog response. PLoS One. 10:e0135804 10.1371/journal.pone.0135804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A.I., and Barolo S.. 2013. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130018 10.1098/rstb.2013.0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault A.D., Ricardo S., Kunwar P.S., Santos A., Starz-Gaiano M., Stein J.A., and Lehmann R.. 2009. Hedgehog does not guide migrating Drosophila germ cells. Dev. Biol. 328:355–362. 10.1016/j.ydbio.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gómez A., Molnar C., Holguín H., Mayor F. Jr., and de Celis J.F.. 2007. The cell biology of Smo signalling and its relationships with GPCRs. Biochim. Biophys. Acta. 1768:901–912. 10.1016/j.bbamem.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Russell M.C., Cowan R.G., Harman R.M., Walker A.L., and Quirk S.M.. 2007. The hedgehog signaling pathway in the mouse ovary. Biol. Reprod. 77:226–236. 10.1095/biolreprod.106.053629 [DOI] [PubMed] [Google Scholar]
- Sahut-Barnola I., Godt D., Laski F.A., and Couderc J.-L.. 1995. Drosophila ovary morphogenesis: Analysis of terminal filament formation and identification of a gene required for this process. Dev. Biol. 170:127–135. 10.1006/dbio.1995.1201 [DOI] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., and Siomi M.C.. 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 461:1296–1299. 10.1038/nature08501 [DOI] [PubMed] [Google Scholar]
- Sarikaya D.P., and Extavour C.G.. 2015. The Hippo pathway regulates homeostatic growth of stem cell niche precursors in the Drosophila ovary. PLoS Genet. 11:e1004962 10.1371/journal.pgen.1004962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Ogata J., and Niki Y.. 2010. BMP and Hh signaling affects primordial germ cell division in Drosophila. Zool. Sci. 27:804–810. 10.2108/zsj.27.804 [DOI] [PubMed] [Google Scholar]
- Schlichting K., Demontis F., and Dahmann C.. 2005. Cadherin Cad99C is regulated by Hedgehog signaling in Drosophila. Dev. Biol. 279:142–154. 10.1016/j.ydbio.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Song X., Wong M.D., Kawase E., Xi R., Ding B.C., McCarthy J.J., and Xie T.. 2004. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 131:1353–1364. 10.1242/dev.01026 [DOI] [PubMed] [Google Scholar]
- Song X., Call G.B., Kirilly D., and Xie T.. 2007. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development. 134:1071–1080. 10.1242/dev.003392 [DOI] [PubMed] [Google Scholar]
- Spradling A.C. 1993. Developmental Genetics of Oogenesis in Drosophila. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. 70 pp. [Google Scholar]
- Steinberg M.S., and Gilbert S.F.. 2004. Townes and Holtfreter (1955): Directed movements and selective adhesion of embryonic amphibian cells. J. Exp. Zoolog. A Comp. Exp. Biol. 301:701–706. 10.1002/jez.a.114 [DOI] [PubMed] [Google Scholar]
- Tanimoto H., Itoh S., ten Dijke P., and Tabata T.. 2000. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol. Cell. 5:59–71. 10.1016/S1097-2765(00)80403-7 [DOI] [PubMed] [Google Scholar]
- Torroja C., Gorfinkiel N., and Guerrero I.. 2004. Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development. 131:2395–2408. 10.1242/dev.01102 [DOI] [PubMed] [Google Scholar]
- Tseng C.Y., Kao S.H., Wan C.L., Cho Y., Tung S.Y., and Hsu H.J.. 2014. Notch signaling mediates the age-associated decrease in adhesion of germline stem cells to the niche. PLoS Genet. 10:e1004888 (published erratum appears in PLoS Genet. 2015. 11:e1005766). 10.1371/journal.pgen.1004888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M., Misaka R., Nitta K., and Tsuchiya K.. 2015. Transcriptional factors, Mafs and their biological roles. World J. Diabetes. 6:175–183. 10.4239/wjd.v6.i1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Ohlen T., Lessing D., Nusse R., and Hooper J.E.. 1997. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc. Natl. Acad. Sci. USA. 94:2404–2409. 10.1073/pnas.94.6.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Gao Y., Song X., Ma X., Zhu X., Mao Y., Yang Z., Ni J., Li H., Malanowski K.E., et al. . 2015. Wnt signaling-mediated redox regulation maintains the germ line stem cell differentiation niche. eLife. 4:e08174 10.7554/eLife.08174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A., and Lehmann R.. 1996. Germ cell development in Drosophila. Annu. Rev. Cell Dev. Biol. 12:365–391. 10.1146/annurev.cellbio.12.1.365 [DOI] [PubMed] [Google Scholar]
- Wong M.D., Jin Z., and Xie T.. 2005. Molecular mechanisms of germline stem cell regulation. Annu. Rev. Genet. 39:173–195. 10.1146/annurev.genet.39.073003.105855 [DOI] [PubMed] [Google Scholar]
- Xiao C., Ogle S.A., Schumacher M.A., Schilling N., Tokhunts R.A., Orr-Asman M.A., Miller M.L., Robbins D.J., Hollande F., and Zavros Y.. 2010. Hedgehog signaling regulates E-cadherin expression for the maintenance of the actin cytoskeleton and tight junctions. Am. J. Physiol. Gastrointest. Liver Physiol. 299:G1252–G1265. 10.1152/ajpgi.00512.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., and Rubin G.M.. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237. [DOI] [PubMed] [Google Scholar]
- Yang S.Y., Baxter E.M., and Van Doren M.. 2012. Phf7 controls male sex determination in the Drosophila germline. Dev. Cell. 22:1041–1051. 10.1016/j.devcel.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.