This review by Morimoto and colleagues examines mechanisms by which protein homeostasis (proteostasis) is achieved in multicellular organisms and discusses the implications for health and disease.
Abstract
The proteostasis network (PN) regulates protein synthesis, folding, transport, and degradation to maintain proteome integrity and limit the accumulation of protein aggregates, a hallmark of aging and degenerative diseases. In multicellular organisms, the PN is regulated at the cellular, tissue, and systemic level to ensure organismal health and longevity. Here we review these three layers of PN regulation and examine how they collectively maintain cellular homeostasis, achieve cell type-specific proteomes, and coordinate proteostasis across tissues. A precise understanding of these layers of control has important implications for organismal health and could offer new therapeutic approaches for neurodegenerative diseases and other chronic disorders related to PN dysfunction.
Introduction
Proteome integrity is maintained by the proteostasis network (PN), which consists of interconnected systems that regulate protein synthesis, folding, transport, and degradation in every cell. The functionality of this network declines during aging, thus compounding the risk for diseases related to proteostasis dysfunction, such as neurodegenerative diseases, cardiomyopathies, and metabolic disorders (Balch et al., 2008). Studies in yeast and tissue culture have provided fundamental insights into the molecular mechanisms of PN function and regulation within single cells (Balchin et al., 2016). Multicellular organisms, however, consist of different cell types that are structurally and functionally diverse, reflecting distinct proteomes (Uhlén et al., 2015). Thus, the composition and functionality of the PN must be tailored to meet the specific needs of each cell and tissue type throughout development and adulthood. Differentiation, specialization, and spatial organization of cells in complex organisms also influence the ability of individual cells to sense and respond to stressful stimuli. Therefore, transcellular mechanisms are in place to orchestrate PN functionality across organs and tissues (van Oosten-Hawle and Morimoto, 2014). Here, we review the differential scales of proteostasis regulation from the cellular to the organismal level and discuss implications for human health.
The cellular proteostasis network
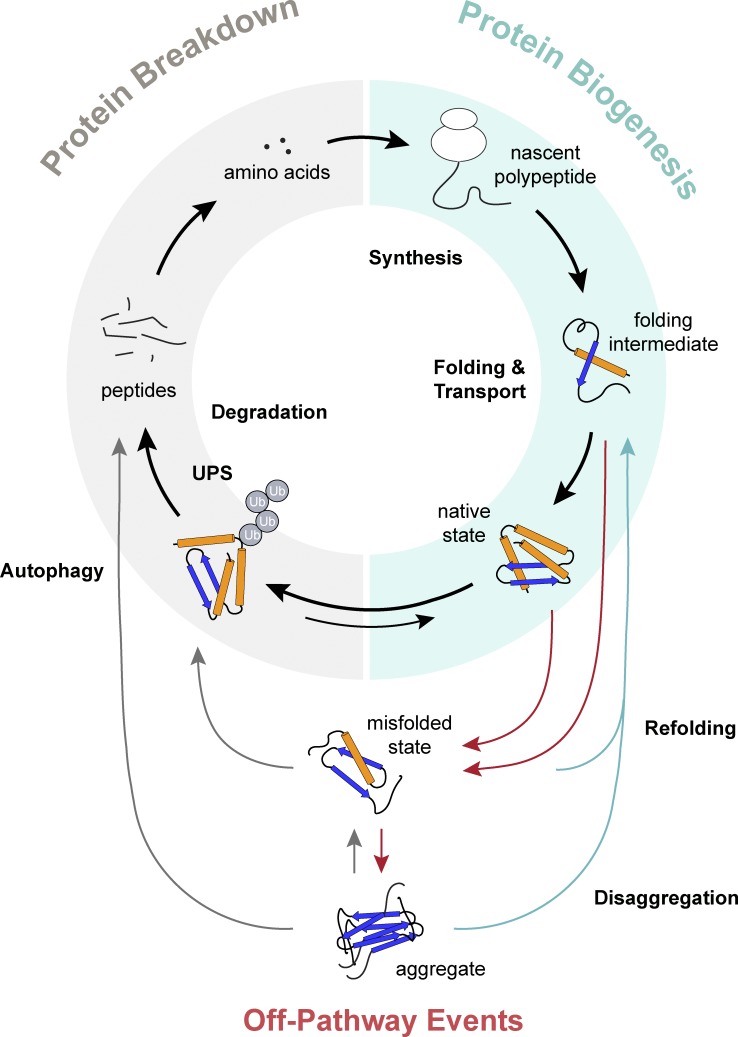
At the single-cell level, the PN comprises the molecular machineries and systems that are essential for all stages of protein biogenesis and breakdown (Balch et al., 2008). The generic view of the eukaryotic PN (Fig. 1) encompasses the following processes (Labbadia and Morimoto, 2015a): (a) translation, controlled by the ribosome and associated factors that regulate the synthesis of the nascent polypeptide chain; (b) protein folding, assisted cotranslationally and posttranslationally by molecular chaperones and cochaperones through cycles of substrate binding and release; (c) protein trafficking in the cytosol, across biological membranes, and within subcellular compartments; and (d) protein degradation by the ubiquitin-proteasome system (UPS), the autophagy/lysosomal pathways, and cellular proteases. The PN extends to all subcellular compartments, such as the ER, mitochondria, and nucleus, which possess generic as well as dedicated machineries that are specific to the respective microenvironment (Kaushik and Cuervo, 2015). These subcellular networks are highly interconnected and communicate with each other to promote proteostasis across the cell (Wolff et al., 2014).
Figure 1.
Overview of cellular proteostasis. Proteostasis encompasses the cellular processes that guide the synthesis, folding, transport, and degradation of all proteins. It is regulated by the PN, which consists of the translation machinery, molecular chaperones, UPS, and autophagy to maintain the overall flux of proteostasis (black arrows). Nonnative conformations produced by off-pathway events (red arrows) are recognized by quality control mechanisms to prevent the accumulation of abnormal proteins in the cell. Misfolded and aggregated proteins are either redirected to the folding pathway through disaggregation and refolding (blue arrows) or targeted to degradation systems (gray arrows).
Causes of protein misfolding
As illustrated in Fig. 1, imbalance in the overall flux of proteostasis, however transient, promotes off-pathway events that lead to the formation of damaged, misfolded, or aggregated protein species, which can be toxic to the cell (Balchin et al., 2016). Protein misfolding occurs continuously because of the inherently error-prone nature of biological systems. For example, errors in transcription, splicing, and translation can result in unstable or aberrant protein variants. The highly crowded cellular environment favors nonnative interactions during protein synthesis, refolding, and conformational changes (Ellis and Minton, 2006). The presence of intrinsically disordered regions within native proteins, as well as conformational changes associated with protein function or posttranslational modifications, and the formation of multimeric complexes, also put proteins at risk for adopting alternate nonnative structures (Uversky et al., 2008; Prabakaran et al., 2012). In addition to intracellular causes, proteotoxic stress induced by environmental fluctuations and physiological stimuli can rapidly affect the composition or integrity of the proteome. Off-pathway events are counteracted by chaperone machineries, which refold nonnative species and resolve protein aggregates, and degradation pathways, which destroy misfolded and aggregated proteins (Balchin et al., 2016).
Chaperone networks
Among PN components, the plethora of functions performed by molecular chaperones is central to protein fate and cellular proteostasis. Chaperones can function as ATP-independent holdases that interact with nonnative polypeptides to prevent aggregation, foldases that actively promote protein folding, or disaggregases that extract polypeptides from aggregates in an ATP-dependent manner. Chaperone activity is driven by specific associations with cochaperones and other partners that determine substrate specificity and the function of chaperone complexes (Kim et al., 2013). Members of the HSP40/J-protein cochaperone family, which regulate substrate binding to HSP70, are more abundant and exhibit higher sequence divergence than HSP70 family members (41 HSP40 vs. 11 HSP70 genes in the human genome), thereby amplifying the range of HSP70 functions (Kampinga and Craig, 2010). One example of this is the ability of DNAJC2 (HSP40) to recruit HSP70 to the ribosome, thus coupling translation to protein folding (Hundley et al., 2005; Otto et al., 2005). This ribosome-associated chaperone complex coordinates protein synthesis rate with cellular folding capacity under stress conditions (Koplin et al., 2010). Molecular chaperones are also important in determining the balance between protein folding and degradation. Cofactors with tetratricopeptide repeat (TPR) domains, such as CHIP (C terminus of Hsp70 interacting protein, also known as STUB1), and BAG domain–containing nucleotide exchange factors interact with the HSP70/HSP90 machinery and direct substrates for degradation by the UPS or lysosomes (Agarraberes and Dice, 2001; Demand et al., 2001; Gamerdinger et al., 2009). Furthermore, in metazoans, different combinations of HSP40 family members can associate with HSP70 and the nucleotide exchange factor HSP110 to form distinct disaggregase complexes capable of resolving various types of protein aggregates (Nillegoda et al., 2015). Thus, chaperones act as a hub that connects multiple branches of the PN to promote cellular proteostasis.
Protein degradation networks
In addition to their role in the regulated turnover of cellular proteins, degradation systems are essential for protein quality control and to limit the accumulation of abnormal proteins during stress conditions. While the UPS promotes the clearance of misfolded and damaged proteins, autophagy targets aggregated species for lysosomal degradation (Kaushik and Cuervo, 2015). Protein turnover by the UPS involves an enzymatic cascade that catalyzes the covalent attachment of ubiquitin moieties to substrates, followed by degradation of the polyubiquitinated substrates by the proteasome (Hershko and Ciechanover, 1998; Finley, 2009). Substrate specificity is conferred by the large family of ubiquitin ligases, which comprises nearly 600 genes in the human genome (Li et al., 2008). The ubiquitination machinery has important roles in the regulation of a myriad of cellular processes and, importantly, in linking protein degradation to different PN activities. For example, the ubiquitin ligase listerin associates with the ribosome and ubiquitinates nascent chains upon stalled translation to prevent the buildup of aberrant polypeptides (Bengtson and Joazeiro, 2010; Brandman et al., 2012). Ubiquitination is also key to the triage decision of the HSP70/HSP90 complex between substrate folding and proteasome-mediated degradation that is governed by the ubiquitin ligase activity of the cochaperone CHIP (Connell et al., 2001). In addition, the UPS is central to the quality control of ER proteins, which are polyubiquitinated, retrotranslocated, and cleared by cytosolic proteasomes in a process termed ER-associated degradation (Preston and Brodsky, 2017). Ubiquitination also aids the selective targeting of proteins to lysosomes by macroautophagy and in endosomal sorting mechanisms, indicating significant cross talk between these pathways (Kraft et al., 2010; Komander and Rape, 2012).
PN regulation by cell stress response pathways
Despite the remarkable ability of the PN to buffer off-pathway events, its activity can become limiting under sustained proteotoxic stress. To counteract the accumulation of nonnative species in the cell, the PN is highly dynamic, and the level of individual components can be adjusted upon changes in proteostasis load. The functionality of different branches of the PN is continuously monitored by multiple pathways, including the heat shock response (HSR), the unfolded protein response (UPR), and the oxidative stress response (OxR; Labbadia and Morimoto, 2015a). Each of these stress responses is controlled by distinct transcription factors that regulate gene expression in response to specific stresses. In addition, the transcriptional responses are generally coupled with a decrease in global protein synthesis through reduced RNA splicing and translation, thereby reducing the influx of newly synthesized proteins and allowing preferential translation of stress-responsive mRNAs until balance is restored (Harding et al., 2000; Biamonti and Caceres, 2009; Shalgi et al., 2013).
The HSR
The HSR is controlled by heat shock factor 1 (HSF1), which increases the expression of specific chaperones to enhance folding capacity in response to protein misfolding in the cytosol (Akerfelt et al., 2010). In its inactive state, monomeric HSF1 localizes to the cytosol or nucleus in association with the HSP70/HSP90 machinery. Nonnative proteins that form during stress conditions compete with HSF1 for chaperone binding. Free HSF1 forms homotrimers that bind with high affinity to heat shock elements and induce the transcription of its gene targets, including HSP70 and HSP90 (Baler et al., 1993; Shi et al., 1998; Zou et al., 1998). Attenuation of the HSR involves acetylation of HSF1, proteasomal turnover, and reassociation with chaperones (Westerheide et al., 2009; Raychaudhuri et al., 2014).
The UPRER
The UPR of the endoplasmic reticulum (UPRER) involves the transcription factors XBP1, ATF6, and ATF4 as separate response elements to implement three ER stress–responsive arms (Hetz et al., 2015). XBP1 is activated by the transmembrane endoribonuclease IRE1, which senses the folding environment inside the ER. Activation of ATF6, an ER-resident transmembrane protein, involves its relocation from the ER to the Golgi apparatus and subsequent proteolytic cleavage in response to ER stress. Both XBP1 and ATF6 induce prosurvival pathways that up-regulate genes involved in protein folding, ER-associated protein degradation, and lipid metabolism (Walter and Ron, 2011). Finally, activation of the kinase PERK inhibits protein translation via phosphorylation of the translation initiation factor eIF2α and leads to the activation of ATF4, which induces the expression of chaperones, autophagy components, and detoxifying enzymes. During prolonged ER stress, hyperactivation of IRE1, as well as induction of the proapoptotic transcription factor CHOP by ATF4, eventually triggers cell death, likely to remove damaged cells from the population (Tabas and Ron, 2011).
The UPRmt
Analogous to the UPRER, the mitochondrial UPR (UPRmt) is activated upon folding stress in the mitochondria via a conserved transcription factor, known as ATF5 in mammals and ATFS1 in Caenorhabditis elegans (Schulz and Haynes, 2015; Fiorese et al., 2016). In the absence of mitochondrial stress, ATFS1, which contains both a mitochondrial targeting sequence and a nuclear localization signal, is imported into mitochondria and degraded. Stress-induced impairment of mitochondrial import leads to nuclear translocation of ATFS1, which, together with the ubiquitin-like protein UBL5 and the transcription factor DVE1, activates the expression of genes involved in mitochondrial repair mechanisms, including protein folding and detoxification (Haynes et al., 2007; Nargund et al., 2012).
The OxR
Oxidative and xenobiotic stress activate the OxR, which controls the expression of redox-regulatory proteins and components involved in protein degradation. The OxR has two branches, which are mediated by the stress-responsive transcription factors NRF1/NFE2L1 and NRF2/NFE2L2 in mammals, whereas in C. elegans these functions are performed by SKN1 (Itoh et al., 1997; An and Blackwell, 2003; Radhakrishnan et al., 2010). The ER-resident transcription factor NRF1 undergoes proteolytic cleavage upon activation and controls the expression of proteasome subunits and other UPS components (Radhakrishnan et al., 2014; Sha and Goldberg, 2014). NRF2 is negatively regulated by the redox-sensitive ubiquitin ligase KEAP1 in the cytosol. Inactivation of KEAP1 by oxidative and electrophilic stress leads to the stabilization and nuclear translocation of NRF2, which induces the expression of antioxidant proteins and detoxification enzymes (Kensler et al., 2007).
Cross talk between cell stress responses
Although the HSR, UPR, and OxR pathways differ in their input and output, increasing evidence indicates that substantial cross talk exists between their signaling components. The HSR can be activated by ER stress, and it was further shown that HSF1 overexpression in IRE1-deficient cells relieves defects in ER proteostasis, suggesting an interplay between the HSR and the UPRER (Liu and Chang, 2008). SKN1 is also activated by the UPRER and has a central role in the transcriptional response to ER stress. Conversely, key UPRER signaling factors are involved in the activation of SKN1 during oxidative stress. Notably, these two SKN1-mediated responses as part of the UPRER and OxR have distinct but overlapping targets (Glover-Cutter et al., 2013). Recently, a mitochondrial-to-cytosolic stress response was identified in C. elegans that links mitochondrial proteostasis to the cytosolic folding environment (Kim et al., 2016). Therefore, multiple pathways cooperate during stress to mount an appropriate response to preserve the proteome across the cell.
Evidence for a tissue-specific PN
The essential role of the PN in shaping protein structure and function suggests a tight relationship between proteome and PN composition in a given cell. PN components have undergone substantial expansion during evolution, paralleling the increasing complexity and diversity of the proteome. This is exemplified by the near-linear relationship between the total number of genes and the number of chaperone genes belonging to HSP families in genomes (Powers and Balch, 2013). Hence, the increase of proteome size during evolution was accompanied by a diversification of chaperone machineries rather than simply an increase in chaperone levels, suggesting that chaperones may have coevolved with the proteome. The specific proteomes that support the organization and function of different cell types in multicellular organisms represent an additional layer of complexity to proteostasis regulation. This implies that chaperones and other PN components must be tailored to accommodate specialized proteomes in different cell and tissue types (Powers et al., 2009). Here we focus on muscle and secretory cells as two well-characterized examples of cell types that rely on specific PN composition.
Proteostasis in muscle cells
Muscle function depends on complex, highly ordered protein structures, mainly composed of actin and myosin filaments that provide contractile force. The proper folding of actin and myosin, their assembly into filaments, and maintenance of these dynamic structures, require both specialized and ubiquitous PN components. In C. elegans, the assembly of myosin filaments requires the muscle-specific chaperone UNC45, which binds to the unstructured motor domain to promote folding and recruits the ubiquitous HSP90 chaperone to assist myosin assembly (Barral et al., 1998, 2002; Kim et al., 2008). Homologs of UNC45 are also essential for myogenesis in zebrafish, mouse, and human cells, suggesting that the requirement for myosin-specific chaperones in muscle is conserved (Price et al., 2002; Wohlgemuth et al., 2007). The folding of monomeric actin is assisted by the ubiquitous chaperone prefoldin (also known as GimC), which prevents aggregation of nascent actin polypeptides, and by the ubiquitous TRiC chaperonin (also known as CCT), which encapsulates partially folded actin to complete its folding (Vainberg et al., 1998). Knockdown of different TRiC subunits in C. elegans showed specific activation of an HSR reporter in body wall muscle, indicating a high requirement for the activity of this ubiquitous chaperonin in muscle cells (Guisbert et al., 2013). The small heat-shock protein (sHSP) αB-crystallin promotes the folding of various filamentous proteins, including actin and the intermediate filament protein desmin, and is linked to several myopathies affecting skeletal and cardiac muscles (Bennardini et al., 1992; Singh et al., 2007). A recent study in C. elegans showed that the myogenic transcription factor HLH-1 (MyoD) drives the expression of muscle chaperones, thereby establishing muscle-specific proteostasis as part of the differentiation program (Bar-Lavan et al., 2016). Muscle maintenance also relies on specialized machineries for controlled protein degradation. For example, a chaperone complex consisting of the constitutively expressed HSP70 (HSC70) together with BAG3, HSPB8, and CHIP, targets damaged structural components to the autophagic system for degradation (Arndt et al., 2010). The muscle-specific ubiquitin ligases MAFbx/atrogin-1 and MuRF family members are induced and mediate breakdown of various muscle proteins during atrophic conditions, further demonstrating the need for specific PN activities in this tissue (Bodine et al., 2001; Gomes et al., 2001). Collectively, these findings indicate that a cell-type specific proteostasis environment is crucial for the establishment and maintenance of muscle cell function.
PN regulation in secretory cells
Secretory cells, such as insulin-secreting β cells and antibody-secreting plasma cells, produce high levels of specific proteins that need to be translated, processed through the ER and Golgi apparatus, and exported. High secretory capacity requires expansion of the ER and up-regulation of PN components to ensure proper synthesis, folding, and transport of the proteins to be secreted (Brewer and Hendershot, 2005). Differentiation of B cells into plasma cells, and associated expansion of the secretory apparatus, is dependent on the UPRER mediator XBP1 (Reimold et al., 2001; Shaffer et al., 2004). Interestingly, XBP1 activation during differentiation does not appear to arise from ER stress caused by increased immunoglobulin synthesis, but rather represents a programmed event integral to the differentiation process (Hu et al., 2009; Todd et al., 2009). XBP1-deficient mice exhibit defects in the development of multiple secretory organs, including liver, pancreas, and salivary glands, suggesting that the role of XBP1 in differentiation extends to other secretory tissues (Reimold et al., 2000; Lee et al., 2005). In pancreatic β cells, acute activation of the UPRER also adjusts the rates of protein synthesis, folding, and clearance in response to metabolic signals and changes in protein load (Harding et al., 2001; Scheuner et al., 2005). Thus, activation of the UPRER allows specific cell types to modulate the composition of the PN to support both cellular differentiation and function.
PN composition in different tissues
In support of the notion of tissue-specific proteomes to direct specialized cellular functions, a recent analysis of gene expression as part of the Human Protein Atlas project revealed that only 44% of protein-coding genes are expressed in all tissues (Uhlén et al., 2015). As shown in Fig. 2, the expression of genes corresponding to soluble, membrane-associated, secreted, and mitochondrially encoded proteins varies greatly across tissues. Some striking examples of tissue-specific enrichment are in the pancreas and muscle, which have elevated expression of secreted and mitochondrially encoded proteins, respectively, compared with the mean of all tissues (Fig. 2, compare A with C). Analysis of this dataset focusing on the chaperome, a previously defined set of genes encoding chaperones and cochaperones (Brehme et al., 2014), shows that the expression levels of individual classes also vary across tissues (Fig. 2 C). Certain classes are highly enriched in specific tissues, such as ER-specific chaperones, which constitute the major class of the expressed chaperome in the secretory tissues of the pancreas, small intestine, and liver (Fig. 2 C). The sHSPs are overrepresented in skeletal and cardiac muscle, consistent with their role in the folding of filament components. By comparison, the proportion of HSP70, HSP40, and HSP90 classes is relatively constant in all tissues (Fig. 2 C), in accordance with the central role of these chaperone machineries in proteome maintenance in all cells. However, members of these families can be enriched in specific tissues to support specialized functions. The expression of individual chaperones or PN components across human tissues showed highly variable profiles in previous studies, including core chaperones such as HSC70 and HSP90 (Hageman and Kampinga, 2009; Powers et al., 2009). Several HSP40 cochaperones also have tissue-specific expression patterns, such as HSJ1 (DNAJB2), which is preferentially expressed in neurons, whereas other family members showed testis-specific expression (Cheetham et al., 1992; Hageman and Kampinga, 2009). Notably, our analysis revealed that only 10% of chaperome genes show highly restrictive tissue expression (31 of 324 chaperome genes). Most of these are confined to reproductive organs, which follows the general trend observed for the entire proteome (Uhlén et al., 2015). The TPR and HSP40 cochaperone groups represent the majority of the tissue-enriched genes (11 and 7, respectively). As expected, expression of the functional homolog of UNC45 in humans, UNC45b, was restricted to skeletal and heart muscle, whereas tissue-enriched genes belonging to the ER-specific class were selectively expressed in secretory organs. Therefore, tissue specificities of the chaperome are determined by both differential expression of ubiquitous components and specific factors. These results indicate that tissue-specific proteomes are supported by profound differences in the overall composition of the chaperome, and we expect that this will apply also to other branches of the PN.
Figure 2.
Tissue expression profile of the human proteome and chaperome genes. Analysis of mRNA expression data from the Human Protein Atlas (Uhlén et al., 2015). (A) Combined expression data of 20,358 human protein-coding genes in 32 tissues are represented as the fraction of total transcripts encoding soluble, membrane-associated, secreted, mitochondrial-encoded and genes with isoforms belonging to more than one category. For each category, the number of genes included in the analysis is indicated in brackets. (B) Combined expression data of genes corresponding to molecular chaperones and cochaperones of the human chaperome (Brehme et al., 2014) in 32 tissues. The chaperome consists of the following groups of chaperones and cochaperones found in all compartments: HSP40, HSP70, HSP90, HSP60, prefoldin (PFD), sHSPs, and TPR domain-containing proteins, as well as organelle-specific chaperones of the ER (ER-specific) and mitochondria (MITO-specific, all nuclear encoded). It should be noted that the groups defined as HSP70 and HSP90 include both HSP chaperones and associated factors. Of the 332 chaperome genes defined by Brehme et al. (2014), 324 were present in the Human Protein Atlas dataset. (C) Tissue-specific expression of the proteome and the chaperome in selected tissues corresponding to bone marrow, liver, pancreas, skin, brain, small intestine, skeletal muscle, heart muscle, and adipose tissue. Expression data for the proteome and chaperome are represented as in A and B, respectively.
PN regulation in development
The aforementioned findings suggest that differential PN composition is essential for cell identity and is likely established early during differentiation. The PN undergoes major remodeling during development to support rapid growth and morphogenesis. HSF1 is essential for development in C. elegans, Drosophila melanogaster, and mice (Jedlicka et al., 1997; Xiao et al., 1999; Walker et al., 2003) and was recently shown to control a transcriptional program that is distinct from the HSR, which includes chaperone genes and other factors that promote protein biogenesis and anabolism (Li et al., 2016). This is reminiscent of the role of HSF1 during malignant transformation of cancer cells, in which it also drives a specific program that supports deregulated growth and proliferation (Mendillo et al., 2012). HSF1 is also required in mouse oocytes where it supports meiosis through the specific induction of the cytoplasmic HSP90α (Metchat et al., 2009). Similarly, the expression and activity of the sHSP SIP1 is restricted to oocytes and embryos in C. elegans (Fleckenstein et al., 2015). These observations further reinforce the notion that spatial and temporal regulation of PN components is critical for cell fate determination.
Coordination of proteostasis at the organismal level
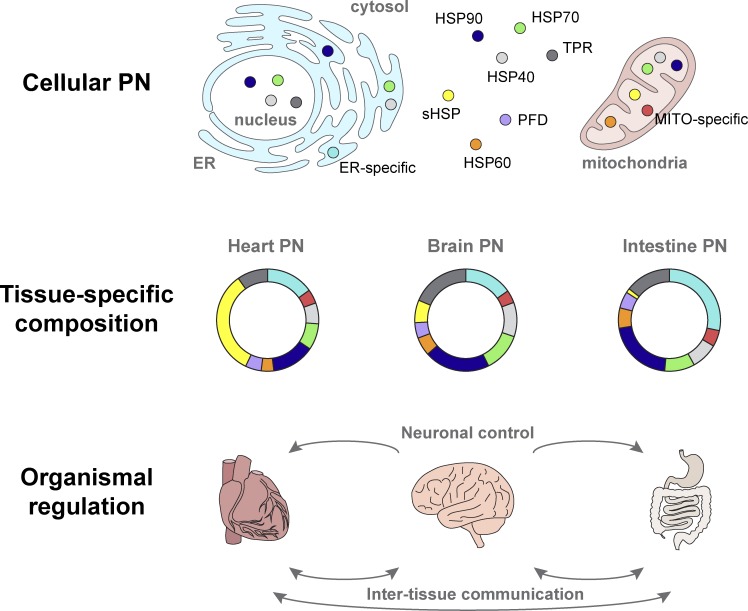
Living systems are continuously confronted with a broad range of environmental insults and physiological changes that can result in cellular stress and macromolecular damage. The complex organization of cells and tissues in multicellular organisms poses challenges to stress-induced PN remodeling, as cells greatly differ in their exposure and sensitivity to external and internal cues. Therefore, proteotoxic events are communicated between cells and tissues to activate repair mechanisms and prevent further damage to the organism. Cell-nonautonomous regulation of the PN represents a third layer, in addition to cellular and tissue-specific regulation, that allows systemic control of proteostasis (Fig. 3).
Figure 3.
Differential scales of proteostasis regulation in multicellular organisms. The PN is regulated at multiple scales from the cellular to the organismal level, which is illustrated here for the human chaperome (refer to Fig. 2 for details). At the cellular level, the PN consists of the molecular machineries required in all compartments to maintain proteostasis (top). Tissue-specific regulation of PN components tailors PN activity to tissue-specific functions (middle). Recent discoveries in invertebrate and vertebrate models suggest that the PN is also controlled across tissues and organs by neuronal activity and intertissue communication to regulate proteostasis at the organismal level (bottom).
Cell-nonautonomous regulation of the HSR
Mechanisms of cell-nonautonomous regulation of proteostasis are best characterized in the case of the HSR, which underlies systemic control by thermosensory neurons through serotonergic signaling in C. elegans (Prahlad et al., 2008; Tatum et al., 2015). Optogenetic activation of either thermosensory neurons or serotonergic neurons is in fact sufficient to activate the HSR in the absence of heat stress (Tatum et al., 2015). This neuronal circuitry constitutes an additional level of control upstream of the cellular HSR because it prevented cell-autonomous induction of HSF1 in response to misfolded proteins, possibly to protect cells from deleterious effects of chronic activation of the stress response (Prahlad and Morimoto, 2011). These findings suggest that efforts to achieve optimal proteostasis at the single-cell level are not necessarily beneficial at the level of the organism, which could explain the requirement for master regulators upstream of cellular stress responses. In addition to neuronal control of the HSR, chaperone expression is regulated through transcellular signaling, whereby local perturbation caused either by the tissue-specific expression of a misfolded protein or altered expression of a chaperone triggers compensatory responses in other tissues (van Oosten-Hawle et al., 2013). This process relies on the transcription factor PHA4/FOXA, which acts in both the signaling and the receiving tissues to regulate chaperone expression.
Cell-nonautonomous control of the UPR
Evidence from multiple organisms indicates that the UPRER and the UPRmt are also under cell-nonautonomous control. Perturbation of the mitochondrial electron transfer chain increases lifespan in both invertebrates and rodents through the activation of the UPRmt (Liu et al., 2005; Copeland et al., 2009; Durieux et al., 2011). Neuron-targeted disruption of mitochondrial function can lead to cell-nonautonomous activation of the UPRmt in nonneuronal tissues in C. elegans (Durieux et al., 2011). Mild perturbation of the electron transfer chain in Drosophila muscle also leads to a systemic response, which involves repression of insulin signaling and has beneficial effects on organismal health and lifespan (Owusu-Ansah et al., 2013). Similarly, the UPRER is induced in peripheral tissues when active XBP1 is overexpressed in neurons, which likely depends on neuronal activity (Taylor and Dillin, 2013). Induction of the UPRER in nonneuronal tissues during infection is mediated by sensory neurons in C. elegans, suggesting an organismal stress response to pathogens (Sun et al., 2011). Cell-nonautonomous regulation of cellular stress responses has also been observed in mice, where overexpression of active XBP1 in pro-opiomelanocortin neurons leads to activation of the UPRER in the liver (Williams et al., 2014).
Cell-nonautonomous regulation in longevity pathways
Several longevity pathways that increase stress resistance and proteostasis have been shown to be regulated cell nonautonomously in invertebrates. Dietary restriction increases lifespan and proteostasis in C. elegans, Drosophila, and mice (Mair and Dillin, 2008). The effects of dietary restriction on organismal health and longevity have been linked to an isoform of the oxidative stress transcription factor SKN1, which is specifically expressed in a subset of sensory neurons in C. elegans (Bishop and Guarente, 2007). Intestinal expression of DAF16/FOXO, the effector of the longevity insulin/IGF1 signaling pathway, also acts distantly on muscle tissue to enhance proteostasis (Zhang et al., 2013). Similarly, overexpression of dFOXO in Drosophila muscle influences proteostasis in retina, brain, and adipose tissues (Demontis and Perrimon, 2010).
Organismal health and longevity is also controlled by the reproductive system in C. elegans (Hsin and Kenyon, 1999). Signaling from the germ stem cells was shown to repress somatic cell stress responses and proteostasis capacity during early adulthood, at the onset of reproduction (Shemesh et al., 2013; Labbadia and Morimoto, 2015b). Germ stem cell–mediated regulation of the HSR in somatic tissues is associated with the accumulation of repressive chromatin marks at heat shock genes, which restricts HSF1-mediated induction of stress-responsive genes (Labbadia and Morimoto, 2015b). Enhanced proteostasis and extended lifespan in animals devoid of a germline rely on multiple transcription factors, including DAF16, PHA4, and SKN1, in addition to HSF1 (Lin et al., 2001; Lapierre et al., 2011; Steinbaugh et al., 2015). Removal of the germline also increases lifespan in Drosophila through modulation of insulin signaling, indicating that regulation of health and longevity by the reproductive system is conserved (Flatt et al., 2008).
Most of our understanding of cell-nonautonomous control of proteostasis comes from studies in invertebrate model organisms. However, evidence is starting to emerge that a similar process exists in mammals, where circulating factors have long been known to distantly regulate multiple aspects of physiology (Williams et al., 2014). Further investigations are required to identify these factors and the pathways that they regulate, and to determine whether they are conserved in humans.
Implications for neurodegenerative diseases
Although the dynamic nature of the PN allows organisms to buffer acute and chronic stresses, proteostasis capacity is limited and declines during aging. Many age-related pathologies, and in particular neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis (ALS), are characterized by the accumulation of abnormal proteins, which is associated with proteostasis dysfunction (Meijering et al., 2015; Mukherjee et al., 2015). Experimental evidence for the limited capacity of the PN came from studies in C. elegans, where expression of aggregation-prone polyglutamine proteins exposed the phenotypes of diverse temperature-sensitive mutations at the permissive temperature in different tissues (Gidalevitz et al., 2006). Misfolding of metastable proteins also occurs during normal aging, which coincides with the decline of cellular stress response in young adults (Ben-Zvi et al., 2009; Shemesh et al., 2013; Labbadia and Morimoto, 2015b). Studies in rodents revealed that the levels of molecular chaperones and stress responses are decreased in older animals, suggesting that reduced PN capacity is a hallmark of aging (Blake et al., 1991; Carnemolla et al., 2014). Changes in chaperone expression have also been observed in the human brain during aging (Brehme et al., 2014). Although levels of ATP-dependent chaperones (HSP60, HSP40, HSP70, and HSP90 families) decreased, a subset of ATP-independent chaperones (small HSPs and TPR-containing proteins) were induced, which suggests a major remodeling of the PN during aging. These changes were exacerbated in individuals with Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease, indicating that PN remodeling during aging may contribute to the onset and progression of these diseases (Brehme et al., 2014).
A distinctive feature of protein conformational diseases is the vulnerability of certain cell types, which may be explained by cell type–specific differences in PN function and regulation. The causative genes in many neurodegenerative diseases, such as Huntington’s disease and familial forms of Parkinson’s disease, Alzheimer’s disease, and ALS, are ubiquitously expressed, yet the toxicity of the mutant protein is apparently selective to certain subpopulations of neurons. It has been proposed that differences in inducibility of the HSR (Marcuccilli et al., 1996; Batulan et al., 2003), chaperone activity (Kim et al., 2002; Hay et al., 2004), and protein turnover rates (Tsvetkov et al., 2013) could contribute to this phenomenon. Cell type–specific vulnerability could also arise from different rates of proteostasis decline among tissues during aging (Labbadia and Morimoto, 2015a). An improved understanding of the functional basis for cell type–specific PN functionality will help to define disease-relevant changes in human pathologies. In addition, the recent identification of cell-nonautonomous regulation of proteostasis suggests that these mechanisms could impact disease susceptibility in human populations, provided they are conserved.
Strategies to remodel the PN in disease
Remodeling of the PN to compensate for the age-related decline in proteostasis offers a therapeutic strategy that could profoundly influence vulnerability to disease in humans. In support, studies in animal models have revealed that modulation of longevity pathways influences proteotoxicity. Genetic manipulation of IGF1/DAF2 and HSF1 pathways prolongs lifespan and protects against aggregation and toxicity of disease model proteins in C. elegans, Drosophila, and mice (Kaspar et al., 2003; Cohen et al., 2009; Kenyon, 2010; Neef et al., 2011). Genome-wide screens for modifiers of polyglutamine aggregation and toxicity have identified genes that are highly enriched in PN components, with molecular chaperones being an important class (Nollen et al., 2004; Bilen and Bonini, 2007; Silva et al., 2011). Accordingly, overexpression of chaperones and cochaperones from the HSP70 and HSP40 families ameliorates neurodegeneration in mouse models (Cummings et al., 2001; Adachi et al., 2003; Labbadia et al., 2012). Consistent with genetic studies, chemical compounds that modulate the activity of different branches of the PN promote proteostasis and are protective in various models of protein conformational diseases (Brandvold and Morimoto, 2015; Labbadia and Morimoto, 2015a). Small molecules that induce the HSR, UPRER, or OxR or act as allosteric modulators of HSP70 or pharmacological chaperones were shown to reduce aggregation and toxicity of multiple disease-linked proteins in animal models (Calamini et al., 2011; Wang et al., 2013; Makley et al., 2015; Bott et al., 2016; Plate et al., 2016).
Recent discoveries in invertebrate models have raised the possibility that cell-nonautonomous signaling also regulates protein aggregation and toxicity in misfolding diseases. Neuronal control of proteostasis regulates the aggregation of disease-linked polyglutamine and mutant SOD1 proteins in nonneuronal tissues (Garcia et al., 2007; Prahlad and Morimoto, 2011). Overexpression of DAF16 in the intestine also triggers a systemic response that ameliorates toxicity of Aβ peptide in muscle (Zhang et al., 2013). It is well established that nonneuronal cells such as glia control neuronal function and contribute to toxicity in several neurodegenerative diseases, including ALS and Huntington’s disease (Ilieva et al., 2009). Interestingly, it was recently shown that activation of the innate immune response in intestine suppresses rotenone-induced neurotoxicity in C. elegans, suggesting that neurodegeneration could be influenced cell nonautonomously by distal tissues in the periphery (Chikka et al., 2016). These findings suggest that protein folding in a damaged tissue could be restored by targeting other tissues or by acting on the systemic signals that distantly regulate proteostasis.
Concluding remarks
Multicellular organisms have evolved complex pathways to regulate the activity and composition of the PN at the cellular, tissue, and systemic level. Although generally viewed as a housekeeping network invariably present in all cells, the composition of the PN can differ greatly between tissues, with important implications for organismal health and disease. Invertebrate models have been instrumental for the discovery of tissue-specific PN components and pathways that regulate proteostasis throughout the organism. Evolutionary conservation of many key components of these pathways reinforces the use of model organisms as important tools in the study of PN regulation. Future efforts should address how cell type–specific PNs are established and how they respond to different forms of stress. Furthermore, unraveling the details of the tissue circuitry of systemic PN regulation and the directionality of transcellular signals, as well as their identity, could offer new therapeutic strategies in diseases related to PN dysfunction, including the possibility of treating one tissue by targeting another. Such an approach would overcome the inaccessibility of certain tissues to therapeutics and could therefore be of particular interest for the treatment of neurodegenerative diseases.
Acknowledgments
We thank the members of the Morimoto laboratory for valuable input during the preparation of this work, particularly Thomas Stoeger for helpful discussions and advice on the data analysis. We also thank Patricija van Oosten-Hawle, Johnathan Labbadia, and Jian Li for critical reading of the manuscript.
This study was supported by grants from the National Institutes of Health (National Institute on Aging [grants R37 AG026647 and AG049665] and National Institute of Mental Health), the Ellison Medical Foundation, the Glenn Family Foundation, the Chicago Biomedical Consortium, and the Daniel F. and Ada L. Rice Foundation to R.I. Morimoto and a postdoctoral fellowship from the National Ataxia Foundation to L.C. Bott.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ALS
- amyotrophic lateral sclerosis
- HSR
- heat shock response
- OxR
- oxidative stress response
- PN
- proteostasis network
- sHSP
- small heat shock protein
- TPR
- tetratricopeptide repeat
- UPR
- unfolded protein response
- UPS
- ubiquitin-proteasome system
References
- Adachi H., Katsuno M., Minamiyama M., Sang C., Pagoulatos G., Angelidis C., Kusakabe M., Yoshiki A., Kobayashi Y., Doyu M., and Sobue G.. 2003. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J. Neurosci. 23:2203–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarraberes F.A., and Dice J.F.. 2001. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114:2491–2499. [DOI] [PubMed] [Google Scholar]
- Akerfelt M., Morimoto R.I., and Sistonen L.. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11:545–555. 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J.H., and Blackwell T.K.. 2003. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17:1882–1893. 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Fürst D.O., Saftig P., Saint R., Fleischmann B.K., et al. . 2010. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20:143–148. 10.1016/j.cub.2009.11.022 [DOI] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., and Kelly J.W.. 2008. Adapting proteostasis for disease intervention. Science. 319:916–919. 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Balchin D., Hayer-Hartl M., and Hartl F.U.. 2016. In vivo aspects of protein folding and quality control. Science. 353:aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- Baler R., Dahl G., and Voellmy R.. 1993. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol. Cell. Biol. 13:2486–2496. 10.1128/MCB.13.4.2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Lavan Y., Shemesh N., Dror S., Ofir R., Yeger-Lotem E., and Ben-Zvi A.. 2016. A differentiation transcription factor establishes muscle-specific proteostasis in Caenorhabditis elegans. PLoS Genet. 12:e1006531 10.1371/journal.pgen.1006531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral J.M., Bauer C.C., Ortiz I., and Epstein H.F.. 1998. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J. Cell Biol. 143:1215–1225. 10.1083/jcb.143.5.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral J.M., Hutagalung A.H., Brinker A., Hartl F.U., and Epstein H.F.. 2002. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 295:669–671. 10.1126/science.1066648 [DOI] [PubMed] [Google Scholar]
- Batulan Z., Shinder G.A., Minotti S., He B.P., Doroudchi M.M., Nalbantoglu J., Strong M.J., and Durham H.D.. 2003. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J. Neurosci. 23:5789–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson M.H., and Joazeiro C.A.. 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 467:470–473. 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardini F., Wrzosek A., and Chiesi M.. 1992. Alpha B-crystallin in cardiac tissue. Association with actin and desmin filaments. Circ. Res. 71:288–294. 10.1161/01.RES.71.2.288 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A., Miller E.A., and Morimoto R.I.. 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA. 106:14914–14919. 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G., and Caceres J.F.. 2009. Cellular stress and RNA splicing. Trends Biochem. Sci. 34:146–153. 10.1016/j.tibs.2008.11.004 [DOI] [PubMed] [Google Scholar]
- Bilen J., and Bonini N.M.. 2007. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 3:1950–1964. 10.1371/journal.pgen.0030177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N.A., and Guarente L.. 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 447:545–549. 10.1038/nature05904 [DOI] [PubMed] [Google Scholar]
- Blake M.J., Fargnoli J., Gershon D., and Holbrook N.J.. 1991. Concomitant decline in heat-induced hyperthermia and HSP70 mRNA expression in aged rats. Am. J. Physiol. 260:R663–R667. [DOI] [PubMed] [Google Scholar]
- Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K., et al. . 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 294:1704–1708. 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- Bott L.C., Badders N.M., Chen K.L., Harmison G.G., Bautista E., Shih C.C., Katsuno M., Sobue G., Taylor J.P., Dantuma N.P., et al. . 2016. A small-molecule Nrf1 and Nrf2 activator mitigates polyglutamine toxicity in spinal and bulbar muscular atrophy. Hum. Mol. Genet. 25:1979–1989. 10.1093/hmg/ddw073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.W., Zhou S., King D., Shen P.S., Weibezahn J., et al. . 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151:1042–1054. 10.1016/j.cell.2012.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvold K.R., and Morimoto R.I.. 2015. The chemical biology of molecular chaperones—Implications for modulation of proteostasis. J. Mol. Biol. 427:2931–2947. 10.1016/j.jmb.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehme M., Voisine C., Rolland T., Wachi S., Soper J.H., Zhu Y., Orton K., Villella A., Garza D., Vidal M., et al. . 2014. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Reports. 9:1135–1150. 10.1016/j.celrep.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer J.W., and Hendershot L.M.. 2005. Building an antibody factory: A job for the unfolded protein response. Nat. Immunol. 6:23–29. 10.1038/ni1149 [DOI] [PubMed] [Google Scholar]
- Calamini B., Silva M.C., Madoux F., Hutt D.M., Khanna S., Chalfant M.A., Saldanha S.A., Hodder P., Tait B.D., Garza D., et al. . 2011. Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8:185–196. 10.1038/nchembio.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnemolla A., Labbadia J.P., Lazell H., Neueder A., Moussaoui S., and Bates G.P.. 2014. Contesting the dogma of an age-related heat shock response impairment: implications for cardiac-specific age-related disorders. Hum. Mol. Genet. 23:3641–3656. 10.1093/hmg/ddu073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham M.E., Brion J.P., and Anderton B.H.. 1992. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem. J. 284:469–476. 10.1042/bj2840469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikka M.R., Anbalagan C., Dvorak K., Dombeck K., and Prahlad V.. 2016. The mitochondria-regulated immune pathway activated in the C. elegans intestine is neuroprotective. Cell Reports. 16:2399–2414. 10.1016/j.celrep.2016.07.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E., Paulsson J.F., Blinder P., Burstyn-Cohen T., Du D., Estepa G., Adame A., Pham H.M., Holzenberger M., Kelly J.W., et al. . 2009. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 139:1157–1169. 10.1016/j.cell.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Höhfeld J., and Patterson C.. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93–96. 10.1038/35050618 [DOI] [PubMed] [Google Scholar]
- Copeland J.M., Cho J., Lo T. Jr., Hur J.H., Bahadorani S., Arabyan T., Rabie J., Soh J., and Walker D.W.. 2009. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr. Biol. 19:1591–1598. 10.1016/j.cub.2009.08.016 [DOI] [PubMed] [Google Scholar]
- Cummings C.J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H.T., Dillmann W.H., and Zoghbi H.Y.. 2001. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10:1511–1518. 10.1093/hmg/10.14.1511 [DOI] [PubMed] [Google Scholar]
- Demand J., Alberti S., Patterson C., and Höhfeld J.. 2001. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 11:1569–1577. 10.1016/S0960-9822(01)00487-0 [DOI] [PubMed] [Google Scholar]
- Demontis F., and Perrimon N.. 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 143:813–825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J., Wolff S., and Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R.J., and Minton A.P.. 2006. Protein aggregation in crowded environments. Biol. Chem. 387:485–497. 10.1515/BC.2006.064 [DOI] [PubMed] [Google Scholar]
- Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513. 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese C.J., Schulz A.M., Lin Y.F., Rosin N., Pellegrino M.W., and Haynes C.M.. 2016. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26:2037–2043. 10.1016/j.cub.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T., Min K.J., D’Alterio C., Villa-Cuesta E., Cumbers J., Lehmann R., Jones D.L., and Tatar M.. 2008. Drosophila germ-line modulation of insulin signaling and lifespan. Proc. Natl. Acad. Sci. USA. 105:6368–6373. 10.1073/pnas.0709128105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein T., Kastenmüller A., Stein M.L., Peters C., Daake M., Krause M., Weinfurtner D., Haslbeck M., Weinkauf S., Groll M., and Buchner J.. 2015. The chaperone activity of the developmental small heat shock protein Sip1 is regulated by pH-dependent conformational changes. Mol. Cell. 58:1067–1078. 10.1016/j.molcel.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Gamerdinger M., Hajieva P., Kaya A.M., Wolfrum U., Hartl F.U., and Behl C.. 2009. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28:889–901. 10.1038/emboj.2009.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S.M., Casanueva M.O., Silva M.C., Amaral M.D., and Morimoto R.I.. 2007. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes Dev. 21:3006–3016. 10.1101/gad.1575307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., and Morimoto R.I.. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 311:1471–1474. 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K.M., Lin S., and Blackwell T.K.. 2013. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 9:e1003701 10.1371/journal.pgen.1003701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., and Goldberg A.L.. 2001. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA. 98:14440–14445. 10.1073/pnas.251541198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert E., Czyz D.M., Richter K., McMullen P.D., and Morimoto R.I.. 2013. Identification of a tissue-selective heat shock response regulatory network. PLoS Genet. 9:e1003466 10.1371/journal.pgen.1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J., and Kampinga H.H.. 2009. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones. 14:1–21. 10.1007/s12192-008-0060-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., and Ron D.. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D.D., and Ron D.. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell. 7:1153–1163. 10.1016/S1097-2765(01)00264-7 [DOI] [PubMed] [Google Scholar]
- Hay D.G., Sathasivam K., Tobaben S., Stahl B., Marber M., Mestril R., Mahal A., Smith D.L., Woodman B., and Bates G.P.. 2004. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 13:1389–1405. 10.1093/hmg/ddh144 [DOI] [PubMed] [Google Scholar]
- Haynes C.M., Petrova K., Benedetti C., Yang Y., and Ron D.. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 13:467–480. 10.1016/j.devcel.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Hershko A., and Ciechanover A.. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hetz C., Chevet E., and Oakes S.A.. 2015. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 17:829–838. 10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H., and Kenyon C.. 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 399:362–366. 10.1038/20694 [DOI] [PubMed] [Google Scholar]
- Hu C.C., Dougan S.K., McGehee A.M., Love J.C., and Ploegh H.L.. 2009. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 28:1624–1636. 10.1038/emboj.2009.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley H.A., Walter W., Bairstow S., and Craig E.A.. 2005. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 308:1032–1034. 10.1126/science.1109247 [DOI] [PubMed] [Google Scholar]
- Ilieva H., Polymenidou M., and Cleveland D.W.. 2009. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 187:761–772. 10.1083/jcb.200908164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. . 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313–322. 10.1006/bbrc.1997.6943 [DOI] [PubMed] [Google Scholar]
- Jedlicka P., Mortin M.A., and Wu C.. 1997. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 16:2452–2462. 10.1093/emboj/16.9.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., and Craig E.A.. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar B.K., Lladó J., Sherkat N., Rothstein J.D., and Gage F.H.. 2003. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 301:839–842. 10.1126/science.1086137 [DOI] [PubMed] [Google Scholar]
- Kaushik S., and Cuervo A.M.. 2015. Proteostasis and aging. Nat. Med. 21:1406–1415. 10.1038/nm.4001 [DOI] [PubMed] [Google Scholar]
- Kensler T.W., Wakabayashi N., and Biswal S.. 2007. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47:89–116. 10.1146/annurev.pharmtox.46.120604.141046 [DOI] [PubMed] [Google Scholar]
- Kenyon C.J. 2010. The genetics of ageing. Nature. 464:504–512. 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Kim H.E., Grant A.R., Simic M.S., Kohnz R.A., Nomura D.K., Durieux J., Riera C.E., Sanchez M., Kapernick E., Wolff S., and Dillin A.. 2016. Lipid biosynthesis coordinates a mitochondrial-to-cytosolic stress response. Cell. 166:1539–1552.e16. 10.1016/j.cell.2016.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Löwe T., and Hoppe T.. 2008. Protein quality control gets muscle into shape. Trends Cell Biol. 18:264–272. 10.1016/j.tcb.2008.03.007 [DOI] [PubMed] [Google Scholar]
- Kim S., Nollen E.A., Kitagawa K., Bindokas V.P., and Morimoto R.I.. 2002. Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 4:826–831. 10.1038/ncb863 [DOI] [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., and Hartl F.U.. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82:323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Komander D., and Rape M.. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., and Deuerling E.. 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189:57–68. 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Peter M., and Hofmann K.. 2010. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12:836–841. 10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- Labbadia J., and Morimoto R.I.. 2015a The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84:435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., and Morimoto R.I.. 2015b Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell. 59:639–650. 10.1016/j.molcel.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., Novoselov S.S., Bett J.S., Weiss A., Paganetti P., Bates G.P., and Cheetham M.E.. 2012. Suppression of protein aggregation by chaperone modification of high molecular weight complexes. Brain. 135:1180–1196. 10.1093/brain/aws022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L.R., Gelino S., Meléndez A., and Hansen M.. 2011. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 21:1507–1514. 10.1016/j.cub.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Chu G.C., Iwakoshi N.N., and Glimcher L.H.. 2005. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24:4368–4380. 10.1038/sj.emboj.7600903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chauve L., Phelps G., Brielmann R.M., and Morimoto R.I.. 2016. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 30:2062–2075. 10.1101/gad.283317.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., and Joazeiro C.A.. 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 3:e1487 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Hsin H., Libina N., and Kenyon C.. 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28:139–145. 10.1038/88850 [DOI] [PubMed] [Google Scholar]
- Liu Y., and Chang A.. 2008. Heat shock response relieves ER stress. EMBO J. 27:1049–1059. 10.1038/emboj.2008.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jiang N., Hughes B., Bigras E., Shoubridge E., and Hekimi S.. 2005. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 19:2424–2434. 10.1101/gad.1352905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W., and Dillin A.. 2008. Aging and survival: The genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77:727–754. 10.1146/annurev.biochem.77.061206.171059 [DOI] [PubMed] [Google Scholar]
- Makley L.N., McMenimen K.A., DeVree B.T., Goldman J.W., McGlasson B.N., Rajagopal P., Dunyak B.M., McQuade T.J., Thompson A.D., Sunahara R., et al. . 2015. Pharmacological chaperone for α-crystallin partially restores transparency in cataract models. Science. 350:674–677. 10.1126/science.aac9145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcuccilli C.J., Mathur S.K., Morimoto R.I., and Miller R.J.. 1996. Regulatory differences in the stress response of hippocampal neurons and glial cells after heat shock. J. Neurosci. 16:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering R.A., Henning R.H., and Brundel B.J.. 2015. Reviving the protein quality control system: therapeutic target for cardiac disease in the elderly. Trends Cardiovasc. Med. 25:243–247. 10.1016/j.tcm.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Mendillo M.L., Santagata S., Koeva M., Bell G.W., Hu R., Tamimi R.M., Fraenkel E., Ince T.A., Whitesell L., and Lindquist S.. 2012. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 150:549–562. 10.1016/j.cell.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchat A., Akerfelt M., Bierkamp C., Delsinne V., Sistonen L., Alexandre H., and Christians E.S.. 2009. Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90α expression. J. Biol. Chem. 284:9521–9528. 10.1074/jbc.M808819200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Morales-Scheihing D., Butler P.C., and Soto C.. 2015. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med. 21:439–449. 10.1016/j.molmed.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund A.M., Pellegrino M.W., Fiorese C.J., Baker B.M., and Haynes C.M.. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 337:587–590. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef D.W., Jaeger A.M., and Thiele D.J.. 2011. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov. 10:930–944. 10.1038/nrd3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda N.B., Kirstein J., Szlachcic A., Berynskyy M., Stank A., Stengel F., Arnsburg K., Gao X., Scior A., Aebersold R., et al. . 2015. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 524:247–251. 10.1038/nature14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen E.A., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., and Plasterk R.H.. 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA. 101:6403–6408. 10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Conz C., Maier P., Wölfle T., Suzuki C.K., Jenö P., Rücknagel P., Stahl J., and Rospert S.. 2005. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. USA. 102:10064–10069. 10.1073/pnas.0504400102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W., and Perrimon N.. 2013. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 155:699–712. 10.1016/j.cell.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate L., Cooley C.B., Chen J.J., Paxman R.J., Gallagher C.M., Madoux F., Genereux J.C., Dobbs W., Garza D., Spicer T.P., et al. . 2016. Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. eLife. 5:5 10.7554/eLife.15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., and Balch W.E.. 2013. Diversity in the origins of proteostasis networks—A driver for protein function in evolution. Nat. Rev. Mol. Cell Biol. 14:237–248. 10.1038/nrm3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., and Balch W.E.. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78:959–991. 10.1146/annurev.biochem.052308.114844 [DOI] [PubMed] [Google Scholar]
- Prabakaran S., Lippens G., Steen H., and Gunawardena J.. 2012. Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 4:565–583. 10.1002/wsbm.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V., and Morimoto R.I.. 2011. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc. Natl. Acad. Sci. USA. 108:14204–14209. 10.1073/pnas.1106557108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V., Cornelius T., and Morimoto R.I.. 2008. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 320:811–814. 10.1126/science.1156093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston G.M., and Brodsky J.L.. 2017. The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem. J. 474:445–469. 10.1042/BCJ20160582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.G., Landsverk M.L., Barral J.M., and Epstein H.F.. 2002. Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J. Cell Sci. 115:4013–4023. 10.1242/jcs.00108 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S.K., Lee C.S., Young P., Beskow A., Chan J.Y., and Deshaies R.J.. 2010. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 38:17–28. 10.1016/j.molcel.2010.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan S.K., den Besten W., and Deshaies R.J.. 2014. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife. 3:e01856 10.7554/eLife.01856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Loew C., Körner R., Pinkert S., Theis M., Hayer-Hartl M., Buchholz F., and Hartl F.U.. 2014. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell. 156:975–985. 10.1016/j.cell.2014.01.055 [DOI] [PubMed] [Google Scholar]
- Reimold A.M., Etkin A., Clauss I., Perkins A., Friend D.S., Zhang J., Horton H.F., Scott A., Orkin S.H., Byrne M.C., et al. . 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., and Glimcher L.H.. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 412:300–307. 10.1038/35085509 [DOI] [PubMed] [Google Scholar]
- Scheuner D., Vander Mierde D., Song B., Flamez D., Creemers J.W., Tsukamoto K., Ribick M., Schuit F.C., and Kaufman R.J.. 2005. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 11:757–764. 10.1038/nm1259 [DOI] [PubMed] [Google Scholar]
- Schulz A.M., and Haynes C.M.. 2015. UPR(mt)-mediated cytoprotection and organismal aging. Biochim. Biophys. Acta. 1847:1448–1456. 10.1016/j.bbabio.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Z., and Goldberg A.L.. 2014. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 24:1573–1583. 10.1016/j.cub.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., et al. . 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Shalgi R., Hurt J.A., Krykbaeva I., Taipale M., Lindquist S., and Burge C.B.. 2013. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell. 49:439–452. 10.1016/j.molcel.2012.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh N., Shai N., and Ben-Zvi A.. 2013. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell. 12:814–822. 10.1111/acel.12110 [DOI] [PubMed] [Google Scholar]
- Shi Y., Mosser D.D., and Morimoto R.I.. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12:654–666. 10.1101/gad.12.5.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M.C., Fox S., Beam M., Thakkar H., Amaral M.D., and Morimoto R.I.. 2011. A genetic screening strategy identifies novel regulators of the proteostasis network. PLoS Genet. 7:e1002438 10.1371/journal.pgen.1002438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.N., Rao K.S., Ramakrishna T., Rangaraj N., and Rao ChM.. 2007. Association of alphaB-crystallin, a small heat shock protein, with actin: role in modulating actin filament dynamics in vivo. J. Mol. Biol. 366:756–767. 10.1016/j.jmb.2006.12.012 [DOI] [PubMed] [Google Scholar]
- Steinbaugh M.J., Narasimhan S.D., Robida-Stubbs S., Moronetti Mazzeo L.E., Dreyfuss J.M., Hourihan J.M., Raghavan P., Operaña T.N., Esmaillie R., and Blackwell T.K.. 2015. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. eLife. 4:4 10.7554/eLife.07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R., and Aballay A.. 2011. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science. 332:729–732. 10.1126/science.1203411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I., and Ron D.. 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13:184–190. 10.1038/ncb0311-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum M.C., Ooi F.K., Chikka M.R., Chauve L., Martinez-Velazquez L.A., Steinbusch H.W., Morimoto R.I., and Prahlad V.. 2015. Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr. Biol. 25:163–174. 10.1016/j.cub.2014.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.C., and Dillin A.. 2013. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 153:1435–1447. 10.1016/j.cell.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D.J., McHeyzer-Williams L.J., Kowal C., Lee A.H., Volpe B.T., Diamond B., McHeyzer-Williams M.G., and Glimcher L.H.. 2009. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J. Exp. Med. 206:2151–2159. 10.1084/jem.20090738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov A.S., Arrasate M., Barmada S., Ando D.M., Sharma P., Shaby B.A., and Finkbeiner S.. 2013. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat. Chem. Biol. 9:586–592. 10.1038/nchembio.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. . 2015. Proteomics. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Oldfield C.J., and Dunker A.K.. 2008. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 37:215–246. 10.1146/annurev.biophys.37.032807.125924 [DOI] [PubMed] [Google Scholar]
- Vainberg I.E., Lewis S.A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H.L., and Cowan N.J.. 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 93:863–873. 10.1016/S0092-8674(00)81446-4 [DOI] [PubMed] [Google Scholar]
- van Oosten-Hawle P., and Morimoto R.I.. 2014. Organismal proteostasis: role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 28:1533–1543. 10.1101/gad.241125.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P., Porter R.S., and Morimoto R.I.. 2013. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell. 153:1366–1378. 10.1016/j.cell.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G.A., Thompson F.J., Brawley A., Scanlon T., and Devaney E.. 2003. Heat shock factor functions at the convergence of the stress response and developmental pathways in Caenorhabditis elegans. FASEB J. 17:1960–1962. [DOI] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang A.M., Miyata Y., Klinedinst S., Peng H.M., Chua J.P., Komiyama T., Li X., Morishima Y., Merry D.E., Pratt W.B., et al. . 2013. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 9:112–118. 10.1038/nchembio.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide S.D., Anckar J., Stevens S.M. Jr., Sistonen L., and Morimoto R.I.. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 323:1063–1066. 10.1126/science.1165946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.W., Liu T., Kong X., Fukuda M., Deng Y., Berglund E.D., Deng Z., Gao Y., Liu T., Sohn J.W., et al. . 2014. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 20:471–482. 10.1016/j.cmet.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S.L., Crawford B.D., and Pilgrim D.B.. 2007. The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev. Biol. 303:483–492. 10.1016/j.ydbio.2006.11.027 [DOI] [PubMed] [Google Scholar]
- Wolff S., Weissman J.S., and Dillin A.. 2014. Differential scales of protein quality control. Cell. 157:52–64. 10.1016/j.cell.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Xiao X., Zuo X., Davis A.A., McMillan D.R., Curry B.B., Richardson J.A., and Benjamin I.J.. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18:5943–5952. 10.1093/emboj/18.21.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Judy M., Lee S.J., and Kenyon C.. 2013. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: Roles for GATA factors and lipid gene regulators. Cell Metab. 17:85–100. 10.1016/j.cmet.2012.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D.F., and Voellmy R.. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 94:471–480. 10.1016/S0092-8674(00)81588-3 [DOI] [PubMed] [Google Scholar]