Abstract
Many studies have indicated that Macrophage migration inhibitory factor (MIF)-173G/C gene polymorphisms are associated with susceptibility to pulmonary tuberculosis (PTB). Additionally, some studies have suggested that there are higher levels of serum MIF in patients with PTB than the controls. However, the results of these studies were underpowered. The current study aimed to precisely evaluate the association between the MIF-173G/C polymorphism and serum MIF concentrations with PTB. Therefore, a systematic literature search was preformed to identify studies involving the indicated association. Eleven articles (1316 cases and 1272 controls) were included in the study. The results indicated that the MIF-173G/C polymorphism was significantly associated with PTB susceptibility, especially in Asians. Interestingly, the results further detected that circulating MIF levels were significantly higher in patients with PTB than in healthy controls, but this was only the case among Asians. Moreover, the statistical significance was also similar to that of the high quality group. The present study indicated that the MIF-173G/C polymorphism may contribute to the development of PTB. Furthermore, significantly higher serum MIF levels were observed in PTB patients than in controls, which further indicated that the MIF may play an important role in PTB progression, particularly in Asians.
Introduction
Tuberculosis (TB) is a serious global health problem that is mainly caused by the bacillus Mycobacterium tuberculosis (Mtb). According to a recent World Health Organization (WHO) report, there were an estimated 9.6 million new TB cases and 1.5 million TB deaths globally in 20141. Currently, South-East Asia and the Western Pacific Regions account for 58% of the world’s TB cases1. India, China and Indonesia account for 23%, 10% and 10% of the total global cases, respectively1. Although approximately one-third of the world’s population is infected with Mtb, only approximately 10–15% of those infected have a risk of developing active disease at some later stage in life2. It is well known that a series of factors contribute to the risk of progression to infection and disease, which mainly include malnutrition, smoking, diabetes, alcohol use, human immunodeficiency virus (HIV) infection, socioeconomic status and environmental pollution, among others3, 4. Additionally, many studies have indicated that host genetic factors are also important determinants5.
Macrophage migration inhibitory factor (MIF), a 12.5-kDa protein, has been widely studied. Fifty years ago, MIF was first identified as a soluble factor that is produced by activated T lymphocytes that inhibit the random migration of macrophages in vitro, contributing to delayed hypersensitivity reactions6. During subsequent decades, many studies have confirmed MIF expression in a variety of cells and tissues (such as the pituitary, macrophages, dendritic cells, and neutrophils), except activated T cells7. In addition, MIF has been considered to have a variety of biologic functions, including macrophage activation, tumoricidal activities, glucocorticoid negativeregulation, pro-inflammatory activity and catalytic activity8–10. More recently, studies have found the MIF may played a vital role in the pathogenesis of infectious diseases, such as sepsis and Gram-negative bacterial infection11. Furthermore, significantly higher MIF concentrations were observed in patients with pulmonary tuberculosis (PTB) than in the healthy population12. In vitro, MIF has also been found to play an important role in restraining virulent Mtb growth13.
The human MIF gene is located on chromosome 22q11.2. One polymorphism (−173G/C, rs755622) in the MIF gene promoter with potential functional relevance has been identified14. A previous study indicated that individual subjects carrying a MIF-173C allele had significantly higher MIF production in the blood15. Several studies have demonstrated that the MIF-173G/C genetic variation may be associated with autoimmune diseases and cancer susceptibility16, 17. Moreover, the MIF-173G/C gene polymorphism was found to increase the risk of PTB18, 19. However, small sample sizes of some studies possibly lacked sufficient power to assess the true value. Therefore, to the best of our knowledge, a question of whether the MIF-173G/C gene polymorphism and serum MIF levels are associated with PTB risk has not been systematically explored.
A meta-analysis is the statistical analysis of a large collection of results from multiple original studies to synthesize their findings. It has the advantage of encompassing large subject numbers, increasing the ability to detect small but important effects. In the current study, we performed a systematic review and meta-analysis to accurately investigate the impact of the MIF-173G/C gene polymorphism and serum MIF levels on PTB susceptibility.
Results
Study characteristics
Depending on the search strategy, we identified 113 articles when we initially searched the PubMed, Embase, CNKI, and Wanfang databases as well commercial Internet search engines. As shown in Fig. 1, sixteen studies were excluded because they were duplicate studies. Sixty-two articles were excluded based on their titles and abstracts. After full-view screening, fourteen articles were excluded because they were not relevant to PTB risk in relation to the MIF-173G/C gene polymorphism and/or serum MIF concentrations. Two articles were excluded because they were reviews. Five articles were excluded because they were potential repeat studies. Two studies were excluded because they were animal experiments. Another single article was not included in the meta-analysis because it was not designed as a case-control study. Finally, 11 eligible articles12, 18–27 were included in the current meta-analysis. Nine of the 11 included articles were in English12, 18–24, 27 and two were in Chinese25, 26. Among the studies, seven articles12, 19, 23–27 were conducted in Asian populations, three studies18, 20, 21 were conducted in Caucasians and one study22 was conducted in Africans. Kibiki et al. recruited PTB-HIV patients as participants to participate in this clinical study22. The included subjects in the four studies were free of HIV12, 18, 20, 26, whereas the comorbidity (HIV) was not addressed in another six studies19, 21, 23–25, 27. Additionally, three articles were of moderate quality (score = 6)24, 26, 27, and the other included studies were of high quality according to the Newcastle-Ottawa Scale quality score evaluation (see Methods)12, 18–23, 25. The characteristics of the collected studies are listed in Tables 1 and 2.
Figure 1.
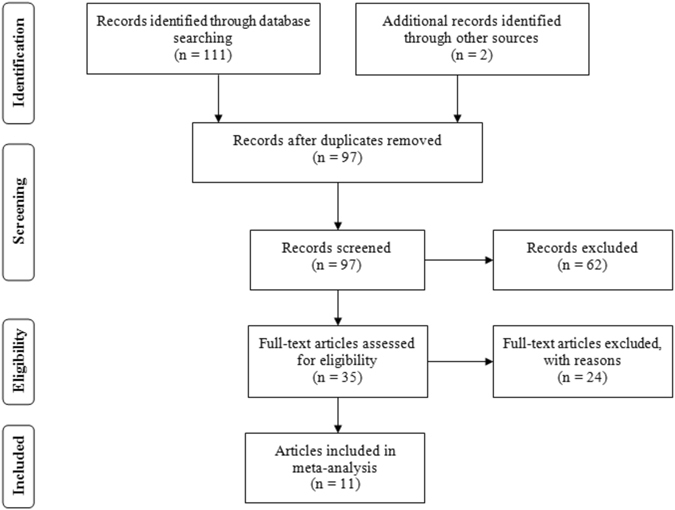
The flow diagram of the included and excluded studies.
Table 1.
Characteristics of the studies involving an association between the MIF-173G/C polymorphism and PTB risk included in the meta-analysis.
| Author | Year | Coutry | Ethnicity | P1/C2 | Age | HIV | Case | Control | Method | Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | CC | CG | GG | |||||||||
| Gomez LM20 | 2007 | Colombia | Caucasian | 230/235 | 40 ± 16/43 ± 16 | No | 15 | 95 | 120 | 16 | 86 | 133 | PCR-Sequencing | 7 |
| Hashemi M21 | 2015 | Iran | Caucasian | 161/142 | 50.6 ± 20.5/47.3 ± 15.4 | NR | 9 | 53 | 99 | 5 | 32 | 105 | PCR-RFLP4 | 8 |
| Kuai SG23 | 2015 | China | Asian | 47/50 | NR3 | NR | 25 | 22 | 16 | 34 | PCR-RFLP | 7 | ||
| Li Y19 | 2012 | China | Asian | 215/245 | 43.8 ± 5.76/40.2 ± 4.90 | NR | 32 | 74 | 109 | 16 | 61 | 168 | PCR-RFLP | 7 |
| Liu AH27 | 2015 | China | Asian | 200/100 | 40.56 ± 17.39/38.65 ± 9.15 | NR | 4 | 116 | 80 | 6 | 37 | 57 | PCR-RFLP | 6 |
| Sadki K18 | 2010 | Morocco | Caucasian | 123/154 | 33.30 ± 13.09/32.89 ± 11.20 | No | 21 | 45 | 57 | 9 | 63 | 82 | RT-PCR5 | 7 |
1patient; 2control; 3not report; 4polymerase chain reaction-restriction fragment length polymorphism; 5real time-polymerase chain reaction.
Table 2.
Characteristics of the studies involving the association between the serum MIF levels and PTB included in the meta-analysis.
| Author | Year | Country | Ethnicity | P1/C2 | Age | HIV | Case | Control | Unit | Method | Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD4 | N | Mean | SD | N | ||||||||||
| Dai XL25 | 2011 | China | Asian | 41/41 | 21-42/19-44 | NR | 43.24 | 29.29 | 41 | 20.86 | 11.99 | 41 | ng/ml | ELISA4 | 7 |
| Kibiki GS22 | 2007 | Tanzania | African | 27/46 | 39.2 ± 8.9/39.3 ± 10.6 | Yes | 130.50 | 71.11 | 27 | 136.4 | 104.3 | 46 | ng/ml | ELISA | 7 |
| Kibiki GS22 | 2007 | Tanzania | African | 25/25 | 33.2 ± 13.8/38.7 ± 10.9 | No | 47.60 | 87.48 | 25 | 39.30 | 23.93 | 25 | ng/ml | ELISA | 7 |
| Kuai SG23 | 2015 | China | Asian | 47/50 | NR3 | NR | 17.19 | 6.64 | 47 | 9.36 | 5.29 | 50 | ng/ml | ELISA | 7 |
| Li Y24 | 2012 | China | Asian | 151/149 | NR | NR | 705.2 | 67.98 | 151 | 355.3 | 57.29 | 149 | pg/ml | ELISA | 6 |
| Liu AH27 | 2014 | China | Asian | 200/100 | 40.3 ± 17.9/28.7 ± 9.2 | NR | 9.67 | 7.24 | 200 | 4.57 | 1.89 | 100 | ng/ml | ELISA | 6 |
| Yamada G12 | 2002 | Japan | Asian | 34/30 | 54 ± 20/47 ± 10 | No | 19.84 | 11.27 | 34 | 4.38 | 1.34 | 30 | ng/ml | ELISA | 7 |
| Zhao GX26 | 2012 | China | Asian | 62/55 | 37.5 ± 12.3/35.4 ± 10.5 | No | 465.46 | 51.15 | 65 | 273.82 | 37.48 | 55 | pg/ml | ELISA | 6 |
1patient; 2control; 3not reported; and 4enzyme-linked immunosorbent assay.
Association between PTB and the MIF-173G/C gene polymorphism
In total, six studies (976 cases and 926 controls) reported an association between the MIF-173G/C gene polymorphism and PTB susceptibility. In the overall meta-analysis, the fixed-effect model was used in the dominant (CC + CG vs. GG), co-dominant (CG vs. GG), and allele (C vs. G) genetic models, while the random-effect model was applied in the co-dominant (CC vs. GG) and recessive (CC vs. CG + GG) genetic models. As summarized in Table 3, the results indicated that there was evidence for significant associations between PTB and the MIF-173G/C gene polymorphism in the dominant (CC + CG vs. GG, OR = 1.65, 95%CI = 1.37–1.99, P < 0.001), co-dominant (CG vs. GG, OR = 1.54, 95%CI = 1.26–1.88, P < 0.001), and allele (C vs. G, OR = 1.49, 95%CI = 1.28–1.74, P < 0.001) genetic models. In a stratified analysis by specific ethnicity, there were significant associations between the MIF-173G/C gene polymorphism and PTB risk for the dominant, co-dominant (CG vs. GG), and allele genetic models in Asians, but that was only the case for the dominant and allele genetic models (Fig. 2 and Table 3) in Caucasians. When a subgroup analysis was performed by the study quality specific effect, there were significant associations between the MIF-173G/C gene polymorphism and PTB risk in the high quality group (CC + CG vs. GG, OR = 1.60 95%CI = 1.31–1.95, P < 0.001) (Table 3). No publication bias was evaluated by either the Begg’s (P = 0.452) or Egger’s test (P = 0.419, Fig. 3A).
Table 3.
Summary of the overall and subgroup analysis results from different comparative genetic models.
| Genetic model | Overall | Asian | Caucasian | High quality | ||||
|---|---|---|---|---|---|---|---|---|
| OR1 (95%CI2) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | |
| CC + CG vs. GG | 1.65 (1.37–1.99) | <0.001 | 2.11 (1.59–2.79) | <0.001 | 1.36 (1.06–1.75) | 0.015 | 1.60 (1.31–1.95) | <0.001 |
| CC vs. CG + GG | 1.46 (0.73–2.93) | 0.286 | 0.97 (0.13–7.23) | 0.975 | 1.70 (0.77–3.77) | 0.188 | 1.91 (1.09–3.36) | 0.024 |
| CC vs. GG | 1.73 (0.90–3.32) | 0.098 | 1.32 (0.21–8.21) | 0.763 | 1.83 (0.87–3.85) | 0.110 | 2.18 (1.48–3.22) | <0.001 |
| CG vs. GG | 1.54 (1.26–1.88) | <0.001 | 2.01 (1.46–2.77) | <0.001 | 1.28 (0.99–1.67) | 0.061 | 1.43 (1.14–1.78) | 0.002 |
| C vs. G | 1.49 (1.28–1.74) | <0.001 | 1.70 (1.18–2.46) | 0.004 | 1.34 (1.10–1.64) | 0.004 | 1.53 (1.16–2.01) | 0.002 |
1odds ratio and 2confidence interval.
Figure 2.
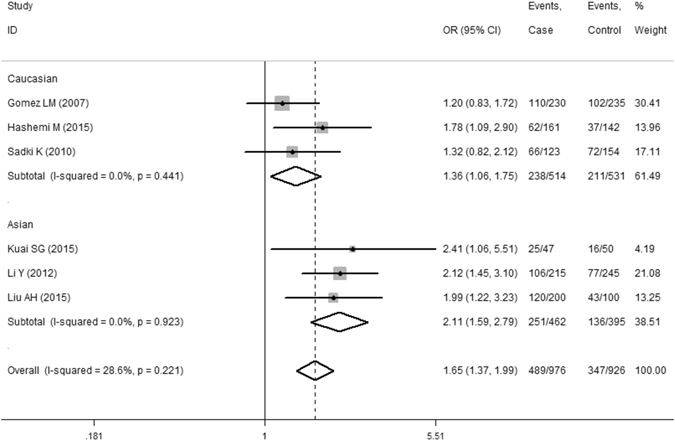
Meta-analysis results of the association between the PTB risk and MIF-173G/C gene polymorphism (CC + CG vs. GG).
Figure 3.
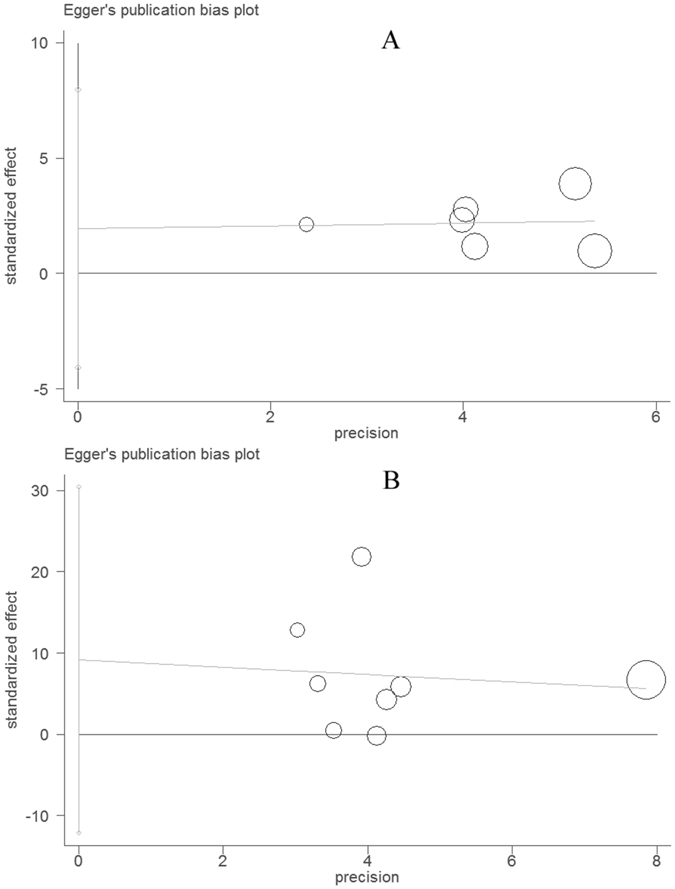
Funnel plot for evaluating publication bias on the association between the PTB risk and MIF-173G/C gene polymorphism (A) as well as on the association between the serum MIF levels and PTB (B). Each circle represents a separate study for the indicated association.
Association between PTB and the serum MIF levels
All 8 eligible case-control studies (587 cases and 496 controls) from seven articles showed an association between PTB and the serum MIF concentrations. Overall, the meta-analysis results indicated that the serum MIF levels in patients with PTB were significantly higher than those in healthy controls (SMD = 1.85, 95%CI = 0.62–3.09, P = 0.003) (Table 4 and Fig. 4). However, a non-ignorable heterogeneity among studies was observed (I2 = 98.3%). Although we conducted the overall meta-analysis by the random-effect model, the results are required for further analysis. As a result, we performed subgroup analyses by ethnicity-specific effects. The serum MIF levels in Asian patients with PTB were significantly higher than those in healthy controls (SMD = 2.46, 95%CI = 0.96–3.96, P = 0.001), but not in Africans (SMD = 0.02, 95%CI = −0.34–0.38, P = 0.921). The statistical significance was similar to that of the high quality group when we performed subgroup analysis according to the study quality (SMD = 0.84, 95%CI = 0.17–1.52, P = 0.014). Moreover, we further executed a sensitivity analysis by sequentially excluding studies from the meta-analysis to investigate the influence of each study on the pooled results. The results of the sensitivity analysis revealed that the pooled OR was not materially altered (Fig. 5). There was no any publication bias as calculated by the Begg’s (P = 0.266) and Egger’s tests (P = 0.332, Fig. 3B).
Table 4.
The pooled results of the serum MIF levels in PTB patients compared with healthy controls.
| Samples (patients/controls) | Number of studies | SMD1 | 95%CI2 | P | I2 | Model | |
|---|---|---|---|---|---|---|---|
| Overall | 587/496 | 8 | 1.85 | 0.62–3.09 | 0.003 | 98.3 | Random |
| Asian | 535/425 | 6 | 2.46 | 0.96–3.96 | 0.001 | 98.5 | Random |
| African | 52/71 | 2 | 0.02 | −0.34–0.38 | 0.921 | 0 | Fixed |
| High quality | 174/192 | 5 | 0.84 | 0.17–1.52 | 0.014 | 89.1 | Random |
1standardized mean difference and 2confidence interval.
Figure 4.
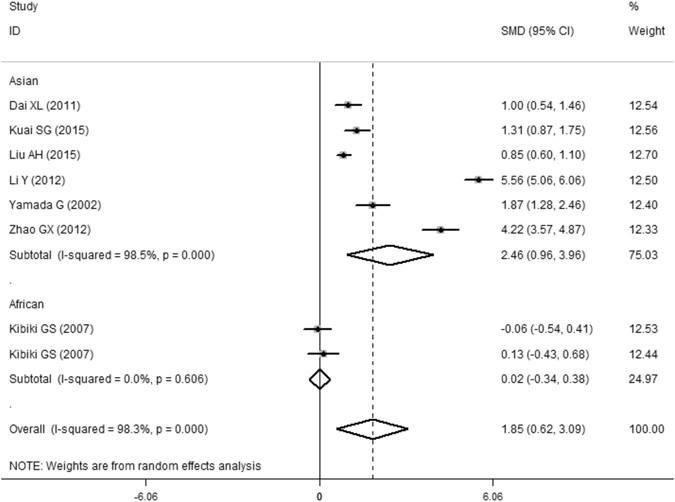
Meta-analysis results of the association between the serum MIF levels and PTB.
Figure 5.
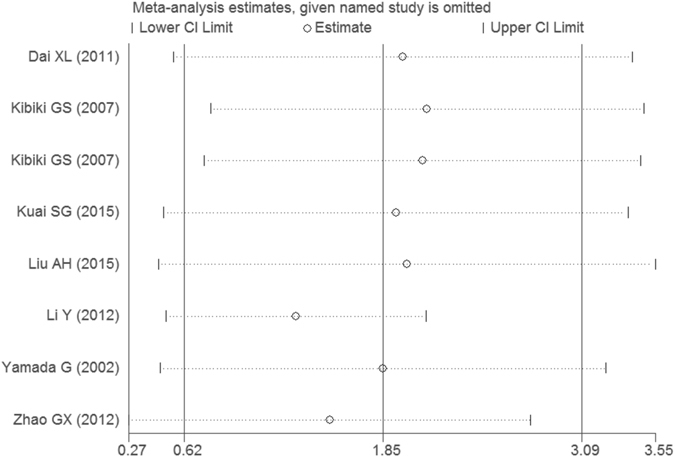
Sensitivity analysis result of the association between the serum MIF levels and PTB.
Discussion
In the current meta-analysis, six case-control studies involving the relationship between the MIF-173G/C gene polymorphism and PTB risk were included. The overall results indicated that the individuals with the variant C allele in the MIF gene had increased PTB susceptibility. In addition, subgroup analyses found that the MIF-173G/C gene polymorphism was associated with PTB susceptibility in Asians, and the study quality did not significantly impact the results.
Interestingly, there were eight primary case-control studies that compared the blood MIF concentrations in patients with PTB and in healthy controls. The overall results of this meta-analysis indicated that patients with PTB had higher serum MIF levels than controls. However, we found a very significant heterogeneity (I2 = 98.3%) between the studies in the meta-analysis, which may be attributed to many factors, as follows: (1) different demographic and genetic characteristics of the Caucasian, Asian and African populations; (2) different quality of the included studies; (3) different co-morbidity in each study; and (4) different diagnosis and inclusion criteria for each primary study. Although the heterogeneity can be taken into account with the random-effect model, it would increase the probability of Type I error. To further identify the reasons for heterogeneity, we first carried out a sensitivity analysis by sequentially excluding each study. Statistically similar results were obtained, suggesting the stability of the meta-analysis. Second, we conducted subgroup analysis by ethnicity and study quality.
On the topic of the association between PTB and the serum MIF levels, the included studies were mainly conducted in Asians (six case-control studies). Subgroup analysis by ethnicity found that Asian patients with PTB had higher serum MIF levels than the healthy population. However, the blood MIF concentrations did not increase in Africans with PTB compared to the control subjects. We do not need to note that the result may have insufficient power to reveal a reliable value because only one article in an African population was included. Additionally, we also observed that patients with PTB still had higher serum MIF concentrations than in the healthy population when we combined the high quality studies. The heterogeneity was still detected after stratified analysis. Although the heterogeneity was still detected after stratified analyses by ethnicity and study quality, subgroup analyses may provide more precise results than the overall analysis.
MIF is produced by a variety of cells and tissues, and it is rapidly released after exposure to microorganisms (toxins and cell wall components) and pro-inflammatory mediators as well as in response to stress7, 9, 28. Additionally, MIF had been shown to play a crucial role in the trafficking and regulation of innate and adaptive immunity9, 29, 30. In previous studies, we found that the MIF gene variant may be associated with susceptibility to cancer and renal disease for which cytokines and immuno-regulation play an important role in pathogenesis17, 31.
Furthermore, many earlier studies found that macrophages and activated T lymphocytes release MIF, which promotes a range of pro-inflammatory cytokines, such as TNF-α after Mtb infection32, 33. More recently, a study suggested that the MIF knock-out mice succumbed more quickly with a higher Mtb burden, decreased innate immunity cytokine expression (such as TNF-α and interleukin-12), and impaired Mtb killing34. Another study demonstrated that MIF produced by infected human macrophages inhibited the growth of virulent Mtb 13. In the clinical study conducted by Yamada and co-workers, the authors found that the mean levels of blood MIF were significantly higher in those with PTB than in healthy participants. Additionally, they suggested that the circulating MIF values significantly correlated with serum interferon-γ, which is one of the most important cytokines contributing to protective immunity against Mtb 12. Similar results were found by Li and co-colleagues in a Chinese population24. Fortunately, our meta-analysis results were consistent with the previous consequences, suggesting that MIF may played a crucial role in immune responses to individual infection with Mtb. Additionally, Donn et al. reported that the serum levels of MIF were significantly higher in individual carrying a MIF-173C allele15. Interestingly, we also found that the MIF-173G/C gene polymorphism may be a risk factor contributing to PTB susceptibility in the current meta-analysis. Although the mechanism by which serum MIF and the MIF-173G/C gene polymorphism are involved in PTB development in humans remains completely unclear, the results of the present and previous studies may help us to identify new molecular markers for TB diagnosis and targets for treatment.
There are several limitations to this meta-analysis. First, even if no publication bias was observed by Egger’s test, only published studies were identified in a few databases. As a result, there may be other biases in the present study. Second, we are unable to further perform subgroup analysis to investigate the other factors (gene-environment interaction, gender and age, etc.) that may affect our results because individual level data are not available. Third, because most studies have not reported the comorbidity (HIV), we did not perform a subgroup analysis on the comorbidity-specific effect. Despite these limitations, we minimized the likelihood of bias through the entire process by establishing a detailed protocol and performing study identification, data selection, and statistical analysis as well as controlling for publication bias. In any case, the reliability of the results is assured.
In conclusion, the current study suggested that the MIF-173G/C gene polymorphism may be a risk factor contributing to PTB susceptibility. Moreover, significantly higher serum MIF levels were detected in PTB patients than in controls, which further indicates MIF possibly played an important role in developing PTB, particularly in Asians. We strongly recommend that researchers design more rigorously and uniformly case-control or cohort studies to confirm the results in the future.
Methods
Literature search
We performed a systematic literature search in the PubMed, Embase, and Wanfang databases and China National Knowledge Internet (CNKI) to identify studies involving the association between the MIF-173G/C gene polymorphism and/or blood MIF concentrations with PTB susceptibility up to October 12, 2016. The key terms in the search were as follows: (“tuberculosis” OR “pulmonary tuberculosis” or “TB” or “PTB”) and (“macrophage migration inhibitory factor” or “MIF”). The language was restricted to English and Chinese. Additionally, we conducted a Web-based search of all types of commercial Internet search engines (such as Google and Baidu) using the same technique. Furthermore, the reference lists of the obtained articles were also reviewed.
Study selection
The inclusion criteria were as follows: (1) the study should be designed as a case–control study; (2) the study should evaluate the association between the MIF-173G/C gene polymorphism and/or serum MIF levels and PTB risk; (3) the available data for calculating the odds ratio (OR) and standardized mean difference (SMD) with a 95% confidence interval (CI) should be provided in the primary study; and (4) the study subjects should be human. The exclusion criteria were as follows: (1) lack of a control cohort; (2) review and overlapping study; and (3) the study does not show the available data and other essential information.
Quality score evaluation
The qualities of the included studies were assessed by the Newcastle-Ottawa Scale (case-control study), a scale that is used to estimate quality based on three aspects, including selection, comparability and exposure in the primary study. The total score ranged from 0 to 9 (0–3, 4–6, and 7–9 were considered low, moderate, and high quality, respectively).
Date extraction
Two independent authors (Xiang Tong and Zhipeng Yan) collected the detailed information and data from each study by a pre-designed data extraction Excel form. If there was a disagreement, the third author (Qilong Zhou) settled the disagreement. The information and data were extracted as follows: first author, publication year, country, ethnicity, sample size, age, genotype distribution, serum MIF levels, comorbidity (HIV) and test method.
Statistical methods
In the present study, the OR and 95% CI were used to investigate the effect strength of the association between the MIF-173G/C gene polymorphism and PTB susceptibility, while the SMD was applied to compare the serum MIF levels in the patients with PTB and healthy controls. We calculated the heterogeneity using the χ2 based Q-test and I-squared (I2) statistics test. The pooled effect size (OR and SMD) was assessed by the random-effect model if heterogeneity was considered statistically significant (I2 value more than 50% and P value less than 0.10). If not, the fixed-effect model was used. To evaluate the ethnicity and study quality specific effects, we also performed subgroup analysis by different specific effects.
In addition, publication bias was evaluated by several methods. The Begg’s and Egger’s tests were used to assess publication bias35, 36. Visual inspection of asymmetry in funnel plots was also performed to further investigate the publication bias. All data analyses were performed by STATA 12.0 software37.
Acknowledgements
This work was supported by the Bureau of Human Resources and Social Security of Sichuan Province grant ([2015]100-13).
Author Contributions
Hong Fan designed the study, coordinated the study and directed its implementation. Xiang Tong, Zhipeng Yan and Qilong Zhou searched the publications, extracted the data and wrote the manuscript. Xiang Tong, Zhipeng Yan, Sitong Liu, Jing Han, and Yao Ma were responsible for data synthesis and figure creation. Sitong Liu and Xue Yang wrote the abstract and made the tables. All of the authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xiang Tong, Zhipeng Yan and Qilong Zhou contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Global tuberculosis report 2015. http://www.who.int/tb/publications/global_report/en/ (2015).
- 2.Maher, D. The natural history of Mycobacterium tuberculosis infection in adults. Tuberculosis, 129–132 (2009).
- 3.Narasimhan P, Wood J, Macintyre CR, Mathai D. Risk factors for tuberculosis. Pulm Med. 2013;2013:828939. doi: 10.1155/2013/828939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonucci G, et al. Risk Factors for Tuberculosis in HIV-lnfected Persons: A Prospective Cohort Study. JAMA. 1995;274:143–148. doi: 10.1001/jama.1995.03530020061033. [DOI] [PubMed] [Google Scholar]
- 5.Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun. 2003;4:4–11. doi: 10.1038/sj.gene.6363915. [DOI] [PubMed] [Google Scholar]
- 6.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 7.Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calandra T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377:68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 9.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linge HM, Ochani K, Lin K, Lee JY, Miller EJ. Age-dependent alterations in the inflammatory response to pulmonary challenge. Immunol Res. 2015;63:209–215. doi: 10.1007/s12026-015-8684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang TY, et al. High levels of serum macrophage migration inhibitory factor and interleukin 10 are associated with a rapidly fatal outcome in patients with severe sepsis. Int J Infect Dis. 2014;20:13–17. doi: 10.1016/j.ijid.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Yamada G, et al. Elevated levels of serum macrophage migration inhibitory factor in patients with pulmonary tuberculosis. Clin Immunol. 2002;104:123–127. doi: 10.1006/clim.2002.5255. [DOI] [PubMed] [Google Scholar]
- 13.Oddo M, Calandra T, Bucala R, Meylan PR. Macrophage migration inhibitory factor reduces the growth of virulent Mycobacterium tuberculosis in human macrophages. Infect Immun. 2005;73:3783–3786. doi: 10.1128/IAI.73.6.3783-3786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renner P, Roger T, Calandra T. Macrophage migration inhibitory factor: gene polymorphisms and susceptibility to inflammatory diseases. Clin Infect Dis. 2005;41(Suppl 7):S513–519. doi: 10.1086/432009. [DOI] [PubMed] [Google Scholar]
- 15.Donn R, et al. Mutation screening of the macrophage migration inhibitory factor gene: positive association of a functional polymorphism of macrophage migration inhibitory factor with juvenile idiopathic arthritis. Arthritis Rheum. 2002;46:2402–2409. doi: 10.1002/art.10492. [DOI] [PubMed] [Google Scholar]
- 16.Arisawa T, et al. Functional promoter polymorphisms of the macrophage migration inhibitory factor gene in gastric carcinogenesis. Oncol Rep. 2008;19:223–228. [PubMed] [Google Scholar]
- 17.Tong X, et al. The MIF-173G/C gene polymorphism increase gastrointestinal cancer and hematological malignancy risk: evidence from a meta-analysis and FPRP test. Int J Clin Exp Med. 2015;8:15949–15957. [PMC free article] [PubMed] [Google Scholar]
- 18.Sadki K, et al. Analysis of MIF, FCGR2A and FCGR3A gene polymorphisms with susceptibility to pulmonary tuberculosis in Moroccan population. J Genet Genomics. 2010;37:257–264. doi: 10.1016/S1673-8527(09)60044-8. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, et al. Association of tuberculosis and polymorphisms in the promoter region of macrophage migration inhibitory factor (MIF) in a Southwestern China Han population. Cytokine. 2012;60:64–67. doi: 10.1016/j.cyto.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Gomez LM, et al. Macrophage migration inhibitory factor gene influences the risk of developing tuberculosis in northwestern Colombian population. Tissue Antigens. 2007;70:28–33. doi: 10.1111/j.1399-0039.2007.00843.x. [DOI] [PubMed] [Google Scholar]
- 21.Hashemi, M. et al. Macrophage migration inhibitory factor-173 G/C polymorphism is associated with an increased risk of pulmonary tuberculosis in Zahedan. Southeast Iran (2015). [DOI] [PMC free article] [PubMed]
- 22.Kibiki GS, et al. Serum and BAL macrophage migration inhibitory factor levels in HIV infected Tanzanians with pulmonary tuberculosis or other lung diseases. Clin Immunol. 2007;123:60–65. doi: 10.1016/j.clim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Kuai, S. G. et al. Functional p olymorphisms in the gene encoding macrophage migration inhibitory factor (MIF) are associated with active pulmonary tuberculosis. Infect Dis (Lond), 1–7, 10.3109/23744235.2015.1107188 (2015). [DOI] [PubMed]
- 24.Li Y, Zeng Z, Deng S. Study of the relationship between human MIF level, MIF-794CATT5-8 microsatellite polymorphism, and susceptibility of tuberculosis in Southwest China. Braz J Infect Dis. 2012;16:383–386. doi: 10.1016/j.bjid.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Dai XL, et al. Relationship between human serum MIF level and TB susceptibility (Chinese) China Tropical Medicine. 2011;11:783–784. [Google Scholar]
- 26.Zhao G, Wang XD. Clinical Study of Macrophage Migration Inhibitory Factor and Granulocyte-Macrophage Colony Stimulating Factor in Active Pulmonary Tuberculosis (Chinese) Chinese Journal of General Practice. 2012;10(230):247. [Google Scholar]
- 27.Liu A, et al. Single nucleotide polymorphisms in cytokine MIF gene promoter region are closely associated with human susceptibility to tuberculosis in a southwestern province of China. Infect Genet Evol. 2016;39:219–224. doi: 10.1016/j.meegid.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Bernhagen J, et al. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365:756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- 29.Baugh JA, Bucala R. Macrophage migration inhibitory factor. Crit Care Med. 2002;30:S27–35. doi: 10.1097/00003246-200201001-00004. [DOI] [PubMed] [Google Scholar]
- 30.Martiney JA, et al. Macrophage migration inhibitory factor release by macrophages after ingestion of Plasmodium chabaudi-infected erythrocytes: possible role in the pathogenesis of malarial anemia. Infect Immun. 2000;68:2259–2267. doi: 10.1128/IAI.68.4.2259-2267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong X, et al. Macrophage migration inhibitory factor-173G/C gene polymorphism increases the risk of renal disease: a meta-analysis. Nephrology (Carlton) 2015;20:68–76. doi: 10.1111/nep.12353. [DOI] [PubMed] [Google Scholar]
- 32.Bacher M, et al. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc Natl Acad Sci USA. 1996;93:7849–7854. doi: 10.1073/pnas.93.15.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baugh JA, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3:170–176. doi: 10.1038/sj.gene.6363867. [DOI] [PubMed] [Google Scholar]
- 34.Das R, et al. Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110:E2997–3006. doi: 10.1073/pnas.1301128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutton, A. J. et al. Methods for meta-analysis in medical research. Vol. 348 (Wiley Chichester, 2000).
- 36.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaimani A, Mavridis D, Salanti G. A hands-on practical tutorial on performing meta-analysis with Stata. Evid Based Ment Health. 2014;17:111–116. doi: 10.1136/eb-2014-101967. [DOI] [PubMed] [Google Scholar]


