Abstract
The retinoblastoma protein (Rb), which typically functions as a transcriptional repressor of E2F‐regulated genes, represents a major control hub of the cell cycle. Here, we show that loss of the Arabidopsis Rb homolog RETINOBLASTOMA‐RELATED 1 (RBR1) leads to cell death, especially upon exposure to genotoxic drugs such as the environmental toxin aluminum. While cell death can be suppressed by reduced cell‐proliferation rates, rbr1 mutant cells exhibit elevated levels of DNA lesions, indicating a direct role of RBR1 in the DNA‐damage response (DDR). Consistent with its role as a transcriptional repressor, we find that RBR1 directly binds to and represses key DDR genes such as RADIATION SENSITIVE 51 (RAD51), leaving it unclear why rbr1 mutants are hypersensitive to DNA damage. However, we find that RBR1 is also required for RAD51 localization to DNA lesions. We further show that RBR1 is itself targeted to DNA break sites in a CDKB1 activity‐dependent manner and partially co‐localizes with RAD51 at damage sites. Taken together, these results implicate RBR1 in the assembly of DNA‐bound repair complexes, in addition to its canonical function as a transcriptional regulator.
Keywords: Arabidopsis, DNA‐damage response, RAD51, retinoblastoma
Subject Categories: Cell Cycle; DNA Replication, Repair & Recombination; Plant Biology
Introduction
To ensure survival and reproductive fitness, all organisms have evolved mechanisms to withstand, at least temporarily, environmental stresses such as heat, heavy metals, or radiation. Many of these stresses affect genome integrity and hence not only threaten the vitality of the individual organisms but also the chances of survival of their offspring. The sessile nature of plants likely demanded the development of powerful DNA‐damage response (DDR) mechanisms to deal with DNA strand breaks and other changes in DNA structure. Indeed, plant cells appear to tolerate much higher concentrations of DNA‐damaging agents than animals (Yokota et al, 2005). Nonetheless, unfavorable environmental conditions are still responsible for major yield losses in crops as for instance seen in the case of aluminum toxicity in acidic soils (Von Uexküll & Mutert, 1995; Bulanova et al, 2001). Thus, unraveling and modulating DDR pathways have a great yet unexplored potential to contribute to food security in the world.
When challenged by DNA damage, cells in multicellular organisms often choose between three major responses: arrest of cell division followed by repair of the damage and resumption of the cell cycle, terminal differentiation and exit from the cell cycle, or programmed cell death (Nowsheen & Yang, 2012). Plant cells were found to be able to follow a fourth route by increasing the ploidy of the cell through endoreplication, that is, terminal differentiation that is coupled to cell‐cycle activity (Adachi et al, 2011). In all these cases, the cellular response to DNA damage appears to be tightly interwoven with cell‐cycle control.
The core components of the cell‐cycle machinery are generally conserved. In plants as well as other eukaryotes, the central regulators are cyclin‐dependent kinases (CDKs) that, in heterodimeric complexes with their cyclin partners, promote entry and progression through the cell cycle (Morgan, 1997; De Veylder et al, 2007; Harashima et al, 2013). In addition to a Cdk1/Cdk2 homolog, called CDKA;1, plants also contain a unique class of cell‐cycle CDKs, named B‐type CDKs, which are divided into a B1 class and B2 class in Arabidopsis. Especially B1‐type CDKs have been functionally analyzed revealing that they act as auxiliary kinases to A1‐type kinases during development and play an important role in homology‐dependent DNA repair (Xie et al, 2010; Cruz‐Ramirez et al, 2012; Nowack et al, 2012; Weimer et al, 2012, 2016a).
The Rb ortholog in the model plant Arabidopsis is called RETINOBLASTOMA‐RELATED 1 (RBR1), and many of the functions of the animal Rb protein appear to be conserved in plants (Sabelli & Larkins, 2009; Gutzat et al, 2012; Desvoyes et al, 2014; Kuwabara & Gruissem, 2014; Harashima & Sugimoto, 2016). The canonical function of Rb‐like proteins in plants and animals is the restriction of the entry into S‐phase by binding to and inhibiting the function of E2F transcription factors. Phosphorylation by CDKA;1 inhibits RBR1 resulting in the release of E2F that in turn activates S‐phase‐promoting genes that include F‐BOX‐LIKE 17 (FBL17), CELL DIVISION CONTROL 6 (CDC6), MINICHROMOSOME MAINTENANCE 5 (MCM5), and ORIGIN RECOGNITION COMPLEX 3 (ORC3) (Desvoyes et al, 2006; Nowack et al, 2012; Zhao et al, 2012; Sabelli et al, 2013). Besides this well‐understood role in S‐phase entry, Rb‐type proteins fulfill many other functions, for example, in cell differentiation, cell migration, and metabolism (Sabelli & Larkins, 2009; Gutzat et al, 2012; Dick & Rubin, 2013; Desvoyes et al, 2014; Kuwabara & Gruissem, 2014; Harashima & Sugimoto, 2016).
The Rb pathway has also been found to be involved in the DDR. For instance, liver cells of Rb‐deficient mice are hypersensitive against the genotoxic and carcinogenic compound diethylnitrosamine, manifesting in an early exit from quiescence and faulty reentry into the cell cycle combined with elevated levels of DNA strand breaks (Reed et al, 2010). Similarly, the Rb pathway has also been implicated in DDR in plants where loss of RBR1 in the quiescent center of the stem‐cell reservoir of the root tip led to hypersensitivity to the DNA double‐strand break (DSB) inducing compound zeocin resulting in severe growth reduction and early differentiation (Cruz‐Ramírez et al, 2013). However, the role of Rb‐type proteins in DDR outside the stem‐cell niche is currently not clear. A first hint for a general function of RBR1 in DNA repair came from the observation that loss of E2FA, a major target of RBR1 in Arabidopsis, results in hypersensitivity against DSB‐inducing agents (Roa et al, 2009; Lang et al, 2012).
To study the role of RBR1 in DDR at the organismic level in plants, we used a recently identified temperature‐sensitive mutant allele of RBR1 (Ebel et al, 2004; Chen et al, 2011; Nowack et al, 2012). We find that RBR1 controls three aspects of DDR. First, loss of RBR1 sensitizes cells to die after exposure to genotoxic drugs. Second, RBR1 represses the expression of several DDR genes. Hence, in dividing cells, when RBR1 is inactivated by phosphorylation of CDKs, not only proliferation genes but also DDR genes become upregulated. This may result in a poised repair state and prepare cells for DNA damage that can occur during the cell cycle. Finally, we show that RBR1 also has a direct role in DNA repair. Depending on CDKB1 activity, RBR1 accumulates at damaged DNA sites, where it partially co‐localizes with the RADIATION SENSITIVE 51 (RAD51) protein, a recombinase involved in homology‐dependent DNA repair. Importantly, RBR1 is required for DNA repair since in rbr1, the number of RAD51 foci is strongly reduced.
Results
Lack of functional RBR1 leads to hypersensitivity against DNA DSB inducing agents and aluminum
To analyze the role of RBR1 in DDR, we decided to use the previously described rbr1‐2 mutant (Ebel et al, 2004; Chen et al, 2011). This mutant was earlier found to behave in a temperature‐dependent manner; that is, growth at temperatures below 18°C gives rise to a stronger mutant phenotype (restrictive temperature) than growth at high temperatures, that is, 24°C or above (permissive temperature) (Nowack et al, 2012). Correspondingly, we found that RBR1 protein levels in rbr1‐2 are considerably reduced at the restrictive temperature (Fig EV1). In the following, we will refer to “rbr1 mutants” as to rbr1‐2 plants grown at the restrictive temperature if not indicated otherwise.
Figure EV1. rbr1‐2 is a temperature‐dependent mutant.
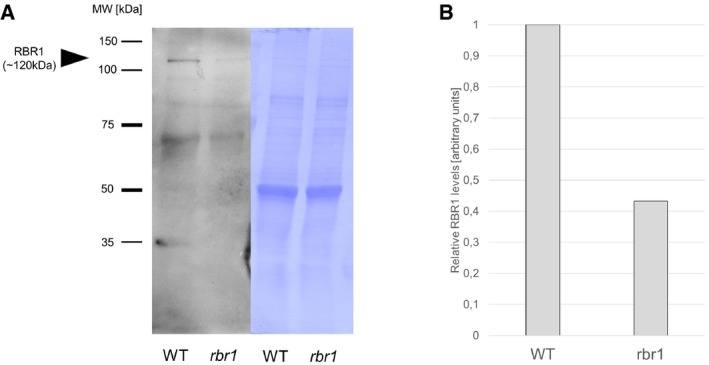
- Western blot of RBR1 protein in WT and rbr1 plants at the restrictive temperature of 17°C.
- Relative quantification of the signal intensity of the RBR1 bands (gray mean values measured by ImageJ) in (A).
Given that the main role of Rb proteins is the restriction of entry into DNA replication phase (S‐phase), we first tested whether rbr1 mutants are sensitive to hydroxyurea (HU, also called hydroxycarbamide). HU depletes the dNTP pools in a cell by inhibiting ribonucleotide reductase leading to an arrest of replication forks and subsequent DNA breakage (Yarbro, 1992). The response to replication stress is controlled by an intra‐S‐phase checkpoint that includes the checkpoint kinases ATR and WEE1 (Culligan et al, 2006; Cools et al, 2011). While loss‐of‐function mutants of ATR and WEE1 exhibited a strong hypersensitive phenotype with reduced cotyledon size and almost no root growth when grown on medium containing HU, consistent with previous data (Culligan et al, 2004; Cools et al, 2011), rbr1 showed no significant differences compared to the wild type (Fig 1A and B).
Figure 1. The rbr1 mutant is hypersensitive to bleomycin but not to hydroxyurea.
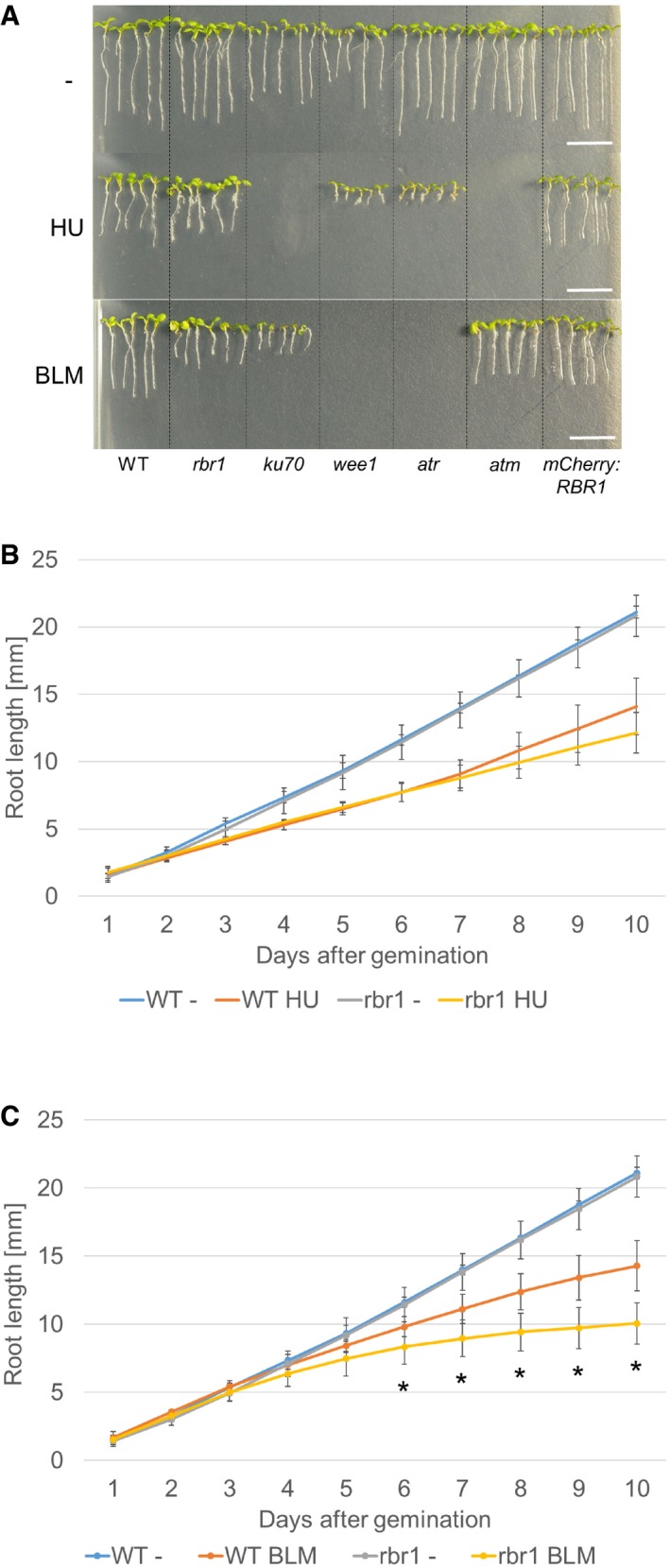
- Root length of plants germinated and grown on medium containing 1 mM HU, 6 μg/ml BLM, or no supplement (‐). Broken lines were added to visually separate the different plant lines. Scale bars: 10 mm.
- Comparison of root growth of the wild‐type and rbr1 mutant plants germinated and grown on medium containing 1 mM HU or no supplement (‐). Error bars signify the standard deviation in three independent experiments.
- Comparison of root growth of the wild‐type and rbr1 plants germinated and grown on medium containing 6 μg/ml BLM or no supplement (‐). Error bars signify the standard deviation in three independent experiments. The asterisks indicate a P‐value lower than 0.05 in Student's t‐test.
Next, we challenged plants with bleomycin (BLM), which induces DSBs that trigger an ATM‐dependent checkpoint and are repaired with the help of KU70/KU80 protein (Riha et al, 2002; Gallego et al, 2003; Yao et al, 2013; Furukawa et al, 2015). Consistent with previous reports, atm and ku70 mutants were hypersensitive to this compound. Importantly, we found that the growth of rbr1 was significantly reduced (Student's t‐test P < 0.05) in comparison with the wild type on BLM‐containing plates (Fig 1A and C).
As a third drug, we tested cisplatin (cisPt) in our root growth assays. CisPt is a DNA‐cross‐linking agent that eventually leads to the generation of DSBs. The damage inflicted by cisPt is typically repaired by homologous recombination repair that functions from S‐phase onward throughout G2 phase, when a sister chromatid is available for repair. As a positive control, we used atr mutants since inhibition of ATR was shown to lead to cisPt hypersensitivity in mammalian cells (Sangster‐Guity et al, 2011). Due to the relative instability of the compound, we germinated the seeds on medium without cisPt for 3 days and then transferred the seedlings to cisPt‐containing medium on which root growth was monitored. Similar to BLM‐containing media, root growth of rbr1 mutants was also severely reduced on media with cisPt, highlighting the importance of RBR1 for the cellular response to DSB‐causing drugs (Appendix Fig S1).
To test for a possible biological importance under natural environmental conditions, we finally tested the growth of rbr1 mutants on plates containing aluminum (Al), which has been previously found to also damage DNA (Bulanova et al, 2001; Murali Achary & Panda, 2010). Al is the third most abundant element in the Earth's crust and is the most common metal. Below a pH of 5, Al becomes soluble in form of Al(OH)2 +, AlOH2+, and Al3+ ions, which have found to be the primary growth‐limiting factors for plants on acid soils that can be found on more than 50% of the world's arable land. For instance in the Morava river area, concentrations in the range of 0.2–60 mg (0.007–2.2 mmol) mobile Al per 100 g soil have been measured (Foy, 1992; Krstic et al, 2012). As a positive control, we used mutants in the ALUMINUM‐SENSITIVE 3 (ALS3) gene, which encodes an ABC transporter‐like protein and was previously found to be required for tolerance of Arabidopsis plants to Al (Larsen et al, 2005). We found that growth of rbr1 mutants was also reduced on this real‐world genotoxin in comparison with the wild type (Fig 2A and B).
Figure 2. The rbr1 mutant is hypersensitive to aluminum.
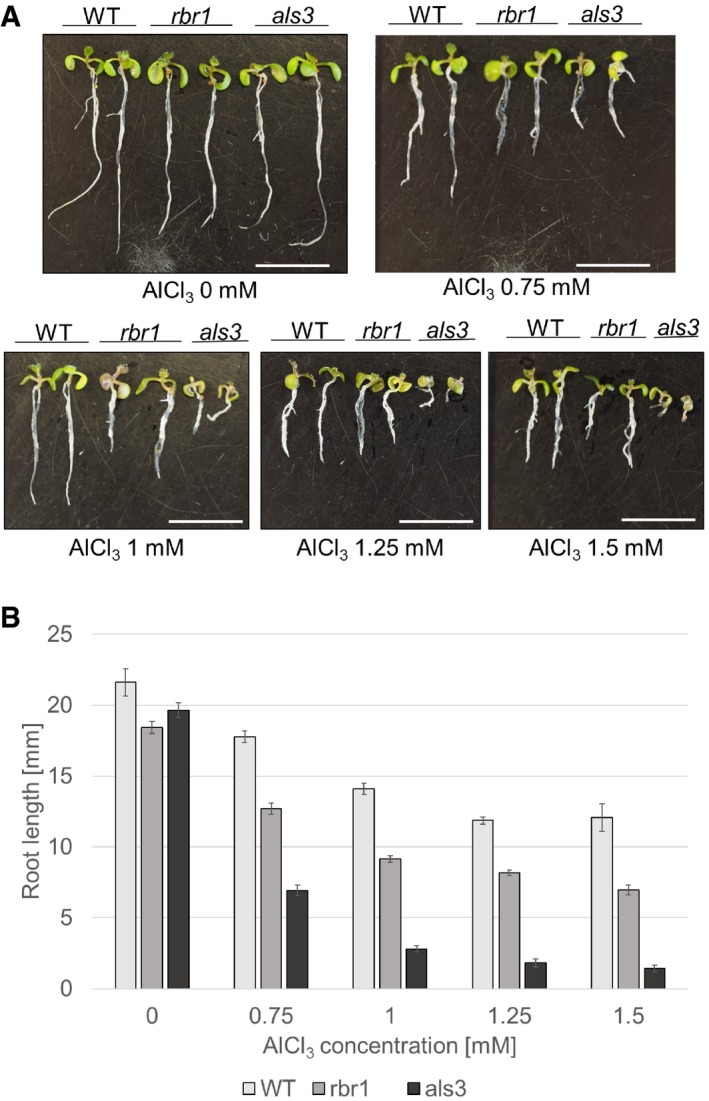
- Root length of 10‐day‐old plants germinated and grown on medium containing AlCl3 in four different concentrations (0.75, 1, 1.25, and 1.5 mM) or no supplement (‐) compared to the aluminum‐sensitive mutant als3. Scale bars: 10 mm.
- Comparison of root growth of 10‐day‐old wild‐type, rbr1, and als3 plants germinated and grown on medium containing 0.75, 1, 1.25, or 1.5 mM AlCl3 or no supplement (‐). Error bars signify the standard deviation in three independent experiments.
RBR1 is necessary for the maintenance of meristem integrity after DNA damage
To understand the cellular basis of the reduced root growth of rbr1 under DNA‐damaging conditions, we analyzed the root tips of rbr1 plants grown for 10 days on BLM‐containing plates. As control for the treatment, we used ku70 mutants. Similar to ku70, rbr1 mutants already showed slightly elevated cell death in comparison with the wild type in untreated roots. When grown on BLM‐containing medium, rbr1 root tips exhibited massive cell death resembling ku70 mutants and clearly distinct from wild‐type roots in which only occasionally dying cells were observed (Fig 3A). The appearance of differentiated root hairs close to the root tips suggested that root meristem function ceased in rbr1 (Furukawa et al, 2015).
Figure 3. Bleomycin treatment results in a distorted root tip architecture and elevated cell death in roots of rbr1 plants.
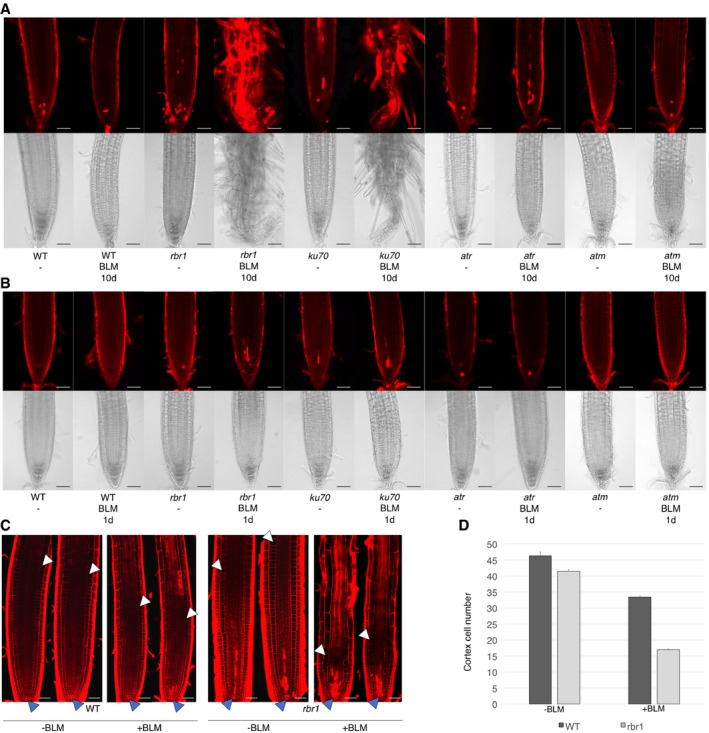
-
A, BCell death in root tips upon drug treatment. (A) Root tip phenotypes of 10‐day‐old seedlings germinated and grown on medium containing BLM (6 μg/ml) or no supplement (‐). (B) Root tip phenotypes of 8‐day‐old seedlings germinated and grown on medium containing no supplement and transferred to medium containing BLM (6 μg/ml) for 1 day. Root tip phenotypes after 2 days of incubation can be seen in Appendix Fig S1A. Upper rows show cell death visualized by propidium iodide staining; lower rows show brightfield microscopic images of root tips. Scale bars: 50 μm.
-
CRoot apical meristems of roots treated with 6 μg/ml BLM or mock for 24 h. Dark arrowheads mark the stem‐cell niche and white arrowheads the position of the first elongating cortex cell. Scale bars: 50 μm.
-
DNumber of cortex cells from the stem‐cell niche to the first elongating cortex cell of roots treated with 6 μg/ml BLM or mock for 24 h. Standard deviation from 15 plants and three replicates (total is 45 plants).
Since this severe phenotype of rbr1 mutants on BLM precluded a quantitative cellular analysis, we allowed plants to germinate and grow on medium without DNA stress and after 8 days of growth transferred rbr1 and the controls to medium supplemented with BLM. When analyzed after 1 and 2 days of treatment, no dying cells could be observed in wild‐type roots (Fig 3B and Appendix Fig S1A). While atr and atm mutant roots only had a few dead cells, rbr1 and ku70 both showed many dying cells. Next, we counted the number of unexpanded cortex cells in the root meristem as a measure of meristem activity and size. Consistent with the apparent loss of meristem activity under long‐term exposure to BLM and the high level of cell death, we found that the meristem size in rbr1 mutants was already reduced by half under short‐term BLM treatment compared to only small changes observed in the wild type (Fig 3C and D).
Similar phenotypes were obtained when rbr1 mutants were grown on cisPt‐containing plates (Appendix Fig S1B and C). Consistent with wild‐type‐like growth on HU, however, cell death and loss of meristem activity were not observed in rbr1 mutants grown on HU‐containing plates indicating the specificity of the response to DSB‐inducing drugs (Appendix Fig S2).
RBR1 is involved in DDR
The reduced growth on media containing DSB‐inducing drugs suggested that rbr1 mutants have increased levels of DNA damage. To test this, we analyzed the level of DNA fragmentation of rbr1 mutants by comet assays upon exposure to BLM (Fig 4A and B). First, we determined the level of fragmentation in plants grown on media without genotoxic drugs. This revealed that rbr1 mutants have already slightly elevated level of DNA damage in untreated conditions. When grown on media with BLM, rbr1 mutant plants had a significantly higher level of DNA fragmentation than wild‐type plants during BLM treatment and after a recovery phase (30 and 75% higher, respectively; Student's t‐test P < 0.01) (Fig 4A and B).
Figure 4. Plants with reduced levels of functional RBR1 show elevated levels of DNA damage following bleomycin (BLM) treatment.
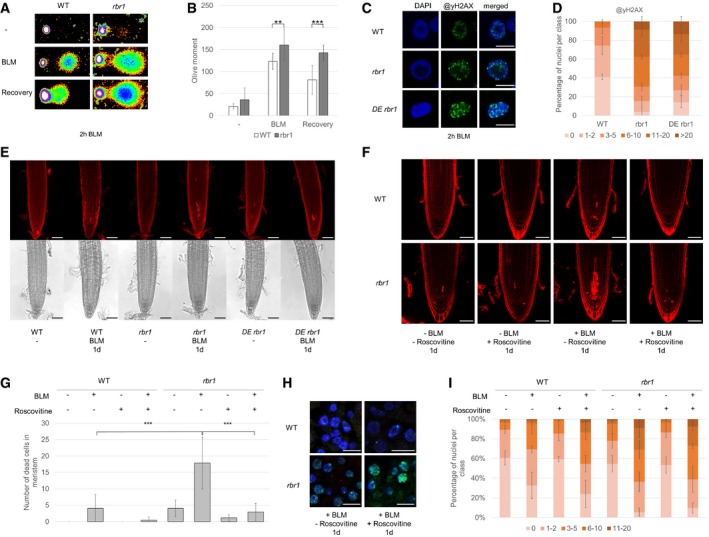
- Fragmentation of nuclear DNA following BLM treatment visualized by neutral comet assays. Shown are false color depictions of representative comets generated by the TriTek CometScore Software. Twenty‐one‐day‐old plantlets were incubated for 1 h in liquid medium with or without 30 μg/ml BLM. For recovery, BLM‐stressed plantlets were rinsed and incubated for 20 min in medium without BLM.
- Olive moment of BLM‐stressed plants and plants after the 20 min recovery period as calculated by the TriTek CometScore software. Error bars indicate the standard deviation in three independent experiments. Two asterisks indicate significance higher than 99% and three asterisks higher than 99.9% as calculated by Student's t‐test.
- Immunofluorescence analysis of γH2AX accumulation (green) in root tip spreads following 2 h of 30 μg/ml BLM treatment in the wild‐type, rbr1, and rbr1 DE plants with DAPI staining (DNA, blue). Scale bars: 5 μm.
- Quantification of γH2AX foci in WT, rbr1, and rbr1 DE plants after BLM treatment. One hundred nuclei per line per experiment were filed into six classes according to their counted number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10, 11–20, or more than 20 γH2AX foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
- Cell death in response to BLM treatment in root tips of rbr1 DE plants compared to rbr1 and wild‐type plants. Six‐day‐old plants were transferred for 1 day to medium containing 0.6 μg/ml BLM. Scale bars: 50 μm.
- PI staining of root tips of wild‐type and rbr1 plants treated for 24 h with BLM (0.6 μg/ml) and/or Roscovitine (1 μM). Scale bars: 50 μm.
- Quantification of dead cells in the plane of the quiescent center of root apical meristems of wild‐type and rbr1 plants treated for 24 h with BLM (0.6 μg/ml) and/or Roscovitine (1 μM). Error bars indicate the standard deviation. Three asterisks indicate significance higher than 99.9% in three replicates as calculated by Student's t‐test.
- Immunofluorescence analysis of γH2AX accumulation (green) in root tip spreads of wild‐type and rbr1 plants treated for 24 h with BLM (0.6 μg/ml) and/or Roscovitine (1 μM) with DAPI staining (DNA, blue). Scale bars: 5 μm.
- Quantification of γH2AX foci in root tip spreads of wild‐type and rbr1 plants treated for 24 h with BLM (0.6 μg/ml) and/or Roscovitine (1 μM). A table of significance levels can be found in Appendix Table S1. Error bars indicate the standard deviation. The experiment was done in three replicates and 50 nuclei per replicate were analyzed.
Phosphorylation of histone H2AX by the checkpoint kinases ATM and ATR is one of the first responses to DNA strand breaks and is often used as a quantitative measure of DNA damage. After immunofluorescence staining using an antibody against phosphorylated H2AX (γH2AX), we found that the nuclei of untreated rbr1 mutants had slightly more foci than the wild type corresponding to our comet assays (Fig EV2). When treated with BLM, over 70% of rbr1 nuclei showed more than five foci per nucleus and over 30% even more than ten foci per nucleus, compared to only 30% of wild‐type nuclei with one to two foci per nucleus (Fig 4C and D).
Figure EV2. Quantification of γH2AX foci in untreated WT and rbr1 plants.
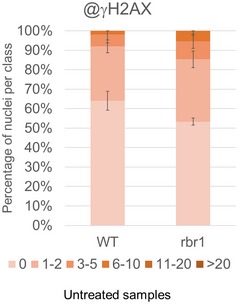
- Fifty nuclei per line per experiment were filed into six classes according to their counted number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10, 11–20, or more than 20 γH2AX foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
Since DNA fragmentation is also a consequence of cell death (Van Hautegem et al, 2015), we envisioned at least three different possibilities for the role of RBR1 under DNA‐damage conditions. First, loss of RBR1 might sensitize cells to die due to strongly increased cell‐cycle activity in the mutant and possibly compromised cell‐cycle checkpoints. In this scenario, RBR1 would not be involved in DNA repair itself and blocking the cell cycle would hence be expected to lead to the restoration of both cell viability and DNA integrity. Second, RBR1 could be involved in DNA repair and its loss would result in DNA lesions that would finally cause cell death, similar to what is seen in ku70 mutants grown on BLM‐containing media. Third, as a combination of hypotheses one and two, RBR1 might sensitize cells to die after inflicted DNA damage and could be at the same time involved in DNA repair, possibly even at different levels of DNA repair. However, a function in repair might be covered by the strong occurrence of cell death.
To narrow down the function of RBR1 in DNA damage, we made use of a previously described weak loss‐of‐function mutant of CDKA;1. In this mutant, a cdka;1 null mutant is rescued with CDKA;1 expression construct in which the two residues, Thr14 and Tyr15, in the regulatory loop of CDKA;1 are exchanged to Asp and Glu, designated PRO CDKA;1 :CDKA;1 T14D;Y15E cdka;1, which we will abbreviate here as DE (Dissmeyer et al, 2009). DE mutants have a much reduced cell‐cycle activity resulting in plants of reduced size that are comprised of fewer cells than the wild type (Dissmeyer et al, 2009). DE mutants themselves are not sensitive to BLM or cisplatin (Weimer et al, 2016a). Previously, it has been found that introgressing rbr1 into DE restored a failure to undergo asymmetric divisions in the root meristem of DE plants (Weimer et al, 2012). Analyzing the double mutant rbr1 DE revealed that it accumulated no more dying cells after growth on BLM‐containing media than the wild type (Fig 4E). In contrast, we found that the number of γH2AX foci in rbr1 DE was similar to that found in rbr1 and not restored to wild‐type levels (Fig 4C and D).
To further test and quantify the dependency of cell death onto cell proliferation in rbr1 mutants, we used the drug Roscovitine, which specifically inhibits Cdk1/2‐type kinases (Meijer et al, 1997; Planchais et al, 1997). The cell death was then quantified in wild‐type plants and rbr1 mutants treated with BLM for 1 day with or without concomitant application of Roscovitine and in comparison with plants treated with Roscovitine alone (Fig 4F and G). These experiments showed that the application of Roscovitine alone did not induce cell death in wild‐type plants (Fig 4F and G). In contrast, the cell death of rbr1 mutants, even in the absence of BLM, was slightly reduced when Roscovitine was applied, that is, from in average four to one dead cell per root meristem. Moreover, Roscovitine could reduce the cell death of wild‐type plants treated with BLM from four dead cells to no dying cell in the root meristem. Especially the cell death in rbr1 mutants grown on BLM (in average 18 dead cells per root meristem) could be suppressed to almost wild‐type levels (three dead cells per root meristem) when these plants were treated at the same time with Roscovitine (Fig 4G).
Next, we quantified the number of γH2AX foci in wild‐type plants and rbr1 mutants (Fig 4H and I). In the wild type, we observed a slight increase in γH2AX foci when roots were treated with BLM and Roscovitine than when treated with BLM alone; that is, more than 30% of the nuclei had more than two γH2AX foci when treated with BLM versus more than 45% of the nuclei that had more than two γH2AX foci when treated with both BLM and Roscovitine. This increase could be due a partial inhibition of B1‐type CDKs, which have been previously found to be sensitive to BLM (Weimer et al, 2016a). In rbr1, the number γH2AX foci in BLM‐treated versus BLM‐ and Roscovitine‐treated roots did not change; for example, more than 60% of the nuclei of rbr1 roots grown on medium with BLM had more than two γH2AX foci and almost the same percentage showed more than two γH2AX foci on medium containing both BLM and Roscovitine. This analysis demonstrates that Roscovitine, in contrast to its ability to suppress cell death, did not reduce the number of γH2AX foci, neither in the wild type nor in rbr1 mutants.
Taken together, the rbr1 cell‐death phenotype is largely dependent on CDK activity/cell‐cycle progression. Moreover, the elevated levels of γH2AX foci in rbr1 DE and in rbr1 mutants treated with Roscovitine in comparison with the wild type indicate that RBR1 has a cell‐cycle‐independent function in DNA repair.
Loss of RBR1 function leads to transcriptional upregulation of DDR genes
Given RBR1's main function is as a transcriptional regulator, a straightforward explanation for the increased DNA damage in rbr1 mutants could be due to failure to express DDR genes such as RAD51. This regulation could be through a de‐repression of a repressor of DDR genes. Alternatively, it has been recently found that Rb is constitutively bound to DP1 (the binding partner of E2F) throughout the cell cycle in the green algae Chlamydomonas and hence is presumably part of an active transcriptional complex similar to the situation in human cells in which Rb is required for the expression of apoptotic genes (Ianari et al, 2009; Olson et al, 2010).
To test for a possible transcriptional role of RBR1 in DDR, we selected 20 genes associated with major DDR pathways in Arabidopsis and followed their expression by quantitative RT‐PCR upon BLM treatment in the wild type and rbr1 mutants (Fig 5A). Two genes, MSH5 and PCNA1, were slightly (1.5‐ and twofold) yet significantly upregulated in rbr1 mutants, irrespective of whether the plants were grown on media containing BLM or not. However, both genes were not upregulated in the wild type after exposure to the DNA‐damaging drug. Five genes (AHP2, BRCA1, PARP2, RAD51, and TSO2) were upregulated by the BLM treatment in the wild type. The transcript levels of all these genes are already elevated in rbr1 compared to wild type under non‐DNA‐damaging conditions. Furthermore, all five genes could still be induced in rbr1 mutants, reaching expression levels comparable to the ones seen in wild‐type plants grown under DNA‐damaging conditions (RAD51 and TSO2) or became even slightly stronger expressed (AHP2, BRCA1, PARP2). To test for a direct regulation, we performed chromatin immunoprecipitation (ChIP) experiments demonstrating that a fragment around 100 bp upstream of the translational start site of RAD51 is bound by RBR1 (Fig 5B and C).
Figure 5. RBR1 is regulating the expression of DNA‐damage response genes.
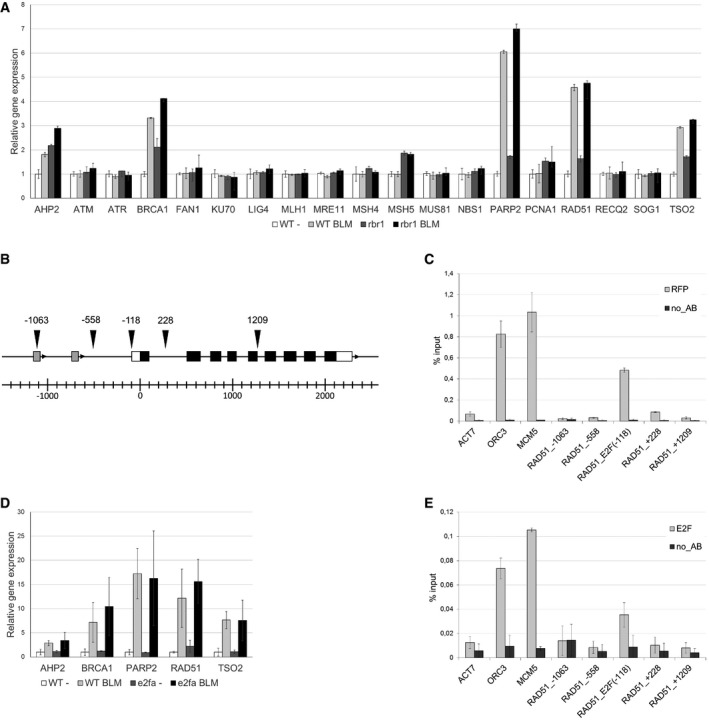
- Relative expression analysis of DNA‐damage response factors in 10‐day‐old BLM‐treated (2 h, 30 μg/ml) and untreated rbr1 seedlings compared to the wild type. Error bars indicate the standard deviation in three independent biological replicates.
- Genomic region of the RAD51 gene. Arrowheads indicate the positions of the amplified fragments in the ChIP assay. Numbers indicate distance to the translational start site. White boxes represent the UTRs and black boxes the coding exons of the RAD51 gene. Gray boxes indicate unrelated genes upstream of RAD51.
- RBR1 ChIP. Transgenic plants expressing PRO RBR1 mCherry:RBR1 were used in a ChIP assay with a DsRed antibody. ORC3 and MCM5 were used as positive controls, ACT7 as a negative control. The labeling of the RAD51 fragments corresponds to the positions indicated in (B). Error bars indicate the standard deviation in two independent biological replicates.
- Relative expression analysis of DNA‐damage response factors in 10‐day‐old BLM‐treated (2 h, 30 μg/ml) and untreated e2fa seedlings compared to the wild type. Error bars indicate the standard deviation in three biological replicates.
- E2F ChIP. Transgenic plants expressing PRO RBR1 mCherry:RBR1 were used in a ChIP assay with an E2FA antibody. ORC3 and MCM5 were used as positive controls, ACT7 as a negative control. The labeling of the RAD51 fragments corresponds to the positions indicated in (B). Error bars indicate the standard deviation in two independent biological replicates.
While we currently cannot exclude that these DNA‐damage genes are upregulated in rbr1 mutants due to the occurring cell death and elevated levels of DNA fragmentation, our ChIP data suggest that RBR1 functions as a conventional (negative) regulator of RAD51 and likely four additional DDR genes. Moreover, a de‐repression by RBR1 appears not to be the key step in transcriptional activation of these genes under DNA‐damaging conditions. To further explore the role of E2F‐RBR module in expression of DDR genes, we performed quantitative expression analyses in e2fa mutants, previously shown to be sensitive to BLM (Lang et al, 2012). While we could reproduce the reduced growth of e2fa mutants on plates with BLM, we did not find major differences in the expression of AHP2, BRCA1, PARP2, RAD51, and TSO2 upon growth on DNA‐damaging and control media between the e2fa mutant and the wild type (Fig 5D). At the same time, we could precipitate the same fragment of the RAD51 promoter in ChIP experiments with E2FA as with RBR1 (Fig 5E). Thus, although we can neither exclude at this moment that additional DDR genes might be downregulated in rbr1 mutants nor exclude that cell‐specific regulation of expression might mask a downregulation in root meristem cells, the most plausible explanation appears to be that the hypersensitivity of rbr1 plants to BLM, cisPt, and aluminum is not due to a failure to transcriptionally activate DDR genes.
RBR1 accumulates in nuclear foci after BLM treatment
In light of a possible posttranscriptional role, we decided to generate a new genomic reporter line for RBR1 in which we introduced mCherry immediately 5′ to the start codon of RBR1 while maintaining the remaining genomic context, that is, 5′ region, introns, and 3′ region. This construct was introduced into heterozygous rbr1 null mutants (rbr1‐3) as homozygous rbr1‐3 is not viable. In the progeny, homozygous rbr1‐3 mutants that carried the genomic reporter were recovered (designated as mCherry:RBR1 plants) and were indistinguishable from wild‐type plants in their growth (Fig 1A). Further analyses of the mCherry:RBR1 plants with rbr1 mutants grown on media with BLM revealed that our genomic RBR1 reporter largely rescued the rbr1 mutant phenotype (Fig 1A).
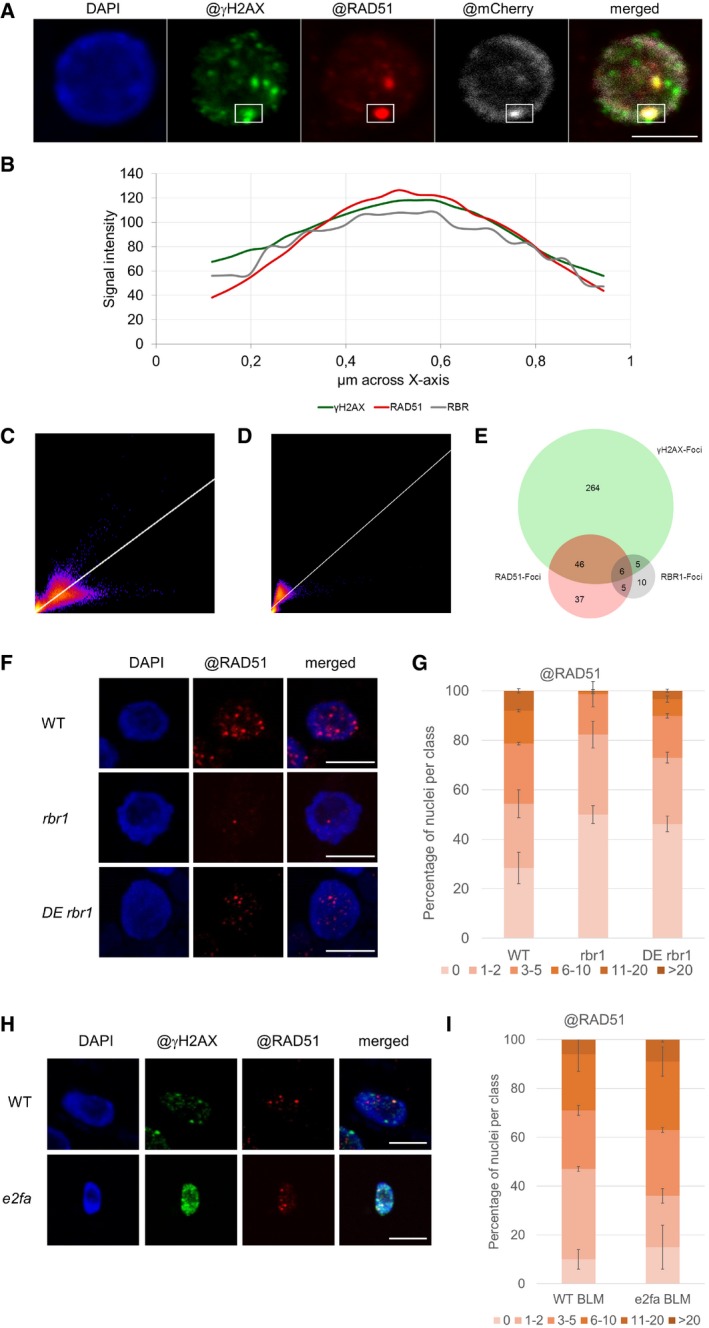
In root cells of mCherry:RBR1 plants, the mCherry:RBR1 fusion protein is predominantly nuclear‐localized where it is evenly distributed consistent with Arabidopsis protoplasts transfected with RBR1‐GFP, as reported before (Henriques et al, 2010). Interestingly, mCherry:RBR1 accumulated in foci upon growth on BLM‐containing media (Fig 6A). Although the individual RBR1 foci were typically large and only few per cell, this accumulation pattern was reminiscent to the formation of γH2AX foci after DNA damage (Fig 4C). Indeed, carrying out double immunofluorescence detection with an antibody against mCherry and γH2AX revealed co‐appearance of mCherry:RBR1 with γH2AX at a subset of γH2AX foci (Figs 6B and EV3, and Appendix Fig S3). Scanning through the overlapping region in one optical section revealed that the signal intensities for RBR1 and γH2AX very highly correlated (Fig 6C). Analyses via three different co‐localization algorithms (Pearson's coefficient 0.821, Manders coefficients M1 = 1.0 (fraction of γH2AX overlapping RBR1) and M2 = 0.995 (fraction of RBR1 overlapping γH2AX), and Costes randomization (200 rounds)‐based co‐localization r = 0.82) indicated a high level of overlap between the foci analyzed. Together, this points to a co‐localization of RBR1 and γH2AX and suggests a potential local role for RBR at DNA repair sites.
Figure 6. RBR1 accumulates in response to DNA stress at DNA breaks.
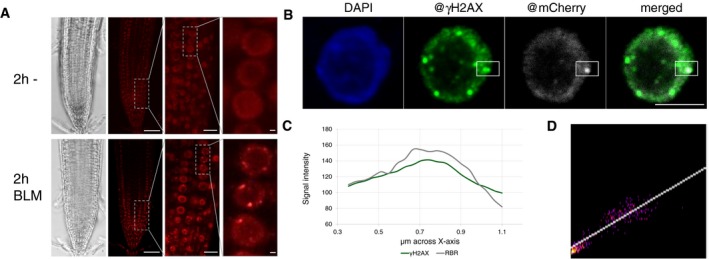
- In vivo accumulation of mCherry:RBR1 in root tips of BLM‐treated (2 h, 30 μg/ml) PRO RBR1 mCherry:RBR1 rbr1‐3 plants in comparison with untreated plants. First image from left depicts brightfield microscopic image, and second to fourth depicts the mCherry fluorescence signal. The third and fourth images are magnifications of the areas framed in the second and third image, respectively. Scale bars: 50, 10, and 1 μm, respectively, from left to right.
- Localization of RBR1 (gray) and γH2AX foci (green) in immunostained spreads of BLM‐treated (6 h, 6 μg/ml BLM) roots tips of PRO RBR1 mCherry:RBR1 rbr1‐3 plants, counterstained with DAPI (DNA, blue). The box marks the co‐localized foci. Scale bars: 5 μm.
- Signal intensity distribution of the total amount of pixels in the box of the stained nucleus in (B).
- 2D correlation histogram of the co‐localized γH2AX and RBR1 foci in (B). Pearson's coefficient: 0.821, Manders coefficients M1 = 1.0 (fraction of γH2AX overlapping RBR1) and M2 = 0.995 (fraction of RBR1 overlapping γH2AX), Costes randomization (200 rounds)‐based co‐lcalization: r = 0.82.
Figure EV3. Co‐localization of RBR1 and RAD51 to γH2AX foci.
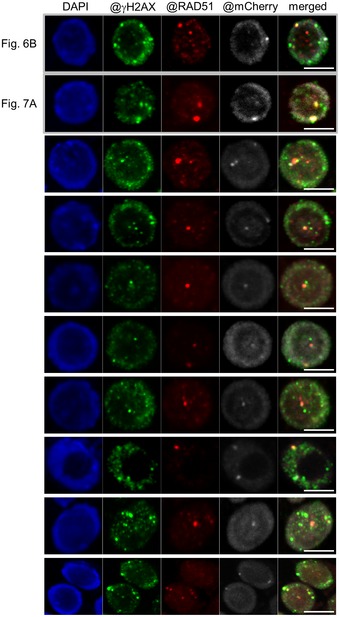
- Co‐localization of γH2AX (green) and RAD51 (red) to RBR1 foci (gray) in immunostained spreads of BLM‐treated root tips of PRO RBR1 mCherry:RBR1 rbr1‐3 plants, counterstained with DAPI (DNA, blue). Note that the examples shown in Figs 6 and 7 were also added to this figure (the first two rows) for better comparison. Scale bars: 5 μm.
RBR1 is necessary for RAD51 localization to DNA after BLM treatment
The RBR1 foci also resemble the localization pattern of RAD51 that is recruited to γH2AX sites after DNA damage (Kurzbauer et al, 2012). RAD51 belongs to the conserved family of RecA proteins and catalyzes homology‐dependent recombination repair in mitotic and meiotic cells (Lin et al, 2006). Loss of RAD51‐type proteins sensitizes plants toward DSB‐inducing and DNA‐cross‐linking drugs such as BLM and cisPt, respectively (Osakabe et al, 2002, 2005; Bleuyard & White, 2004; Abe et al, 2005; Bleuyard et al, 2005; Li et al, 2005; Charbonnel et al, 2010). Our results above indicated that RAD51 is upregulated in the wild type and rbr1 mutants upon growth on BLM‐containing media. Using antibodies against RAD51 and mCherry we found that in mCherry:RBR1 plants, RBR1 and RAD51 also partially co‐localized in foci after growth on BLM‐containing media (Fig 7A and B). Inclusion of the anti‐γH2AX antibody revealed that, indeed, all three proteins co‐localized at a subset of foci (Figs 7A and B and EV3, and Appendix Fig S3). The analyses of co‐localization coefficients for these overlaps underlined again strong correlation between RAD51 and γH2AX signals (Fig 7C; Pearson's coefficient: 0.899, Manders coefficients M1 = 0.923 (fraction of γH2AX overlapping RAD51) and M2 = 0.984 (fraction of RAD51 overlapping γH2AX), Costes randomization (200 rounds)‐based co‐localization: r = 0.9) as well as between RAD51 and RBR1 (Fig 7D; Pearson's coefficient: 0.87, Manders coefficients M1 = 0.9 (fraction of RAD51 overlapping RBR1) and M2 = 0.79 (fraction of RBR1 overlapping RAD51), Costes randomization (200 rounds)‐based co‐localization: r = 0.84). However, only six out of 26 RBR1 foci analyzed in more than 10 cells showed an overlap of all three marks (Fig 7E). Moreover, all three marks could also appear independently of any of the other marks. This, together with the finding that mCherry foci are less frequent than RAD51 foci, which occurred by themselves less often as γH2AX foci, indicates a dynamic formation of these foci.
Figure 7. RBR1 is needed for the accumulation of DNA‐damage response factor RAD51 at breakage sites.
- Co‐localization of γH2AX foci (green) and RAD51 (red) to RBR1 (gray) in immunostained spreads of BLM‐treated root tips (6 h, 6 μg/ml BLM) of PRO RBR1 mCherry:RBR1 rbr1‐3 plants, counterstained with DAPI (DNA, blue). See Appendix Fig S3 for more examples. Scale bars: 5 μm.
- Signal intensity distribution of the total amount of pixels in the box of the stained nucleus in (A).
- 2D correlation histogram of the co‐localized γH2AX and RAD51 foci in (A). Pearson's coefficient: 0.899, Manders coefficients M1 = 0.923 (fraction of γH2AX overlapping RAD51) and M2 = 0.984 (fraction of RAD51 overlapping γH2AX), Costes randomization (200 rounds)‐based co‐localization: r = 0.9.
- 2D correlation histogram of the co‐localized RAD51 and RBR11 foci in (A). Pearson's coefficient: 0.87, Manders coefficients M1 = 0.9 (fraction of RAD51 overlapping RBR1) and M2 = 0.79 (fraction of RBR1 overlapping RAD51), Costes randomization (200 rounds)‐based co‐localization: r = 0.84.
- Venn diagram showing the co‐localization of γH2AX, RAD51, and RBR1 foci in the nuclei shown in Fig EV3.
- Immunofluorescence localization of RAD51 (red) in rbr1 and rbr1 DE in spreads of BLM‐treated (2 h, 30 μg/ml) root tips compared to the wild type. DNA is counterstaining with DAPI (blue fluorescence). Scale bars: 5 μm.
- Quantification of RAD51 foci in the wild‐type, rbr1, and rbr1 DE plants after BLM treatment. One hundred nuclei per line per experiment were filed into six classes according to their counted number of RAD51 foci: nuclei containing no RAD51 foci, 1–2, 3–5, 6–10, 11–20, or more than 20 RAD51 foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
- Immunofluorescence analysis of γH2AX (green) and RAD51 (red) accumulation in root tip spreads following BLM treatment in the wild‐type and e2fa plants. DNA counterstaining with DAPI in blue. Scale bars: 5 μm.
- Quantification of RAD51 foci in WT and e2fa plants after BLM treatment. One hundred nuclei per line per experiment were filed into six classes according to their counted number of RAD51 foci: nuclei containing no RAD51 foci, 1–2, 3–5, 6–10, 11–20, or more than 20 RAD51 foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
To explore whether the presence of RBR1 affects locally acting DNA repair complexes, we studied the localization of RAD51 in rbr1. To quantify RAD51 foci after growth on BLM‐containing media, we considered six classes: nuclei without any foci, nuclei with one to two foci, nuclei with three to five foci, nuclei with six to ten foci, nuclei with eleven to 20 foci, and nuclei with more than 20 foci. In the wild‐type root cells, more than 40% of the nuclei exhibited more than three RAD51 foci. In contrast, only < 20% of the nuclei of rbr1 mutant roots had more than three foci and the class of nuclei with more than eleven foci, found in wild type in 5% of the nuclei, is below 1% in rbr1 mutant (Fig 7D and E). Consistent with our previous observation that reduction in proliferation activity did not restore the number of γH2AX foci, we also found that RAD51 foci were reduced in rbr1 DE plants to a similar level as in rbr1 itself with approximately 25% of nuclei with more than three RAD51 foci and < 5% with more than eleven foci (Fig 7D and E).
To further test the importance of RBR1 for RAD51 localization, we used a previously described RNAi knockdown line against RBR1, called amiGO (Cruz‐Ramírez et al, 2013). This line was also highly sensitive to DNA damage as seen by the strong accumulation of γH2AX foci in comparison with the wild type when treated with BLM (Fig EV4A). Similar to the analysis of rbr1‐2 plants, we found that the percent of nuclei with more than three RAD51 foci was severely reduced to < 25% in BLM‐treated amiGO plants in comparison with the wild type in which this class was larger than 60% (Fig EV4B). We did even not find any nuclei that had more than eleven foci (approximately 5% in the wild type) corroborating the results obtained with the rbr1‐2 allele.
Figure EV4. Distribution of γH2AX and RAD51 foci in amiGO RBR1 line.
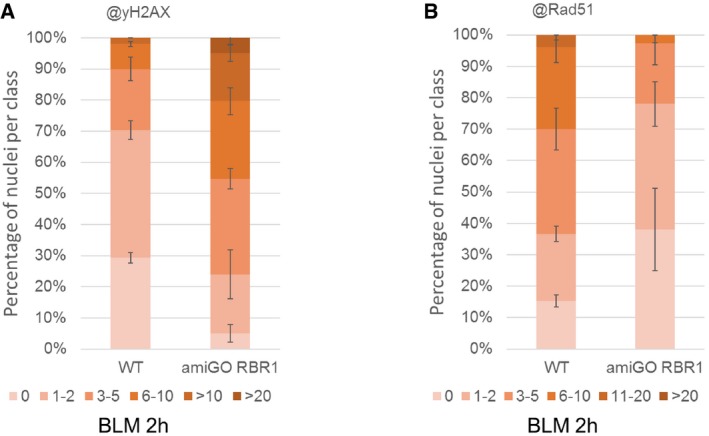
- Quantification of γH2AX foci in WT and amiGO RBR1 plants after BLM treatment. Fifty nuclei per line per experiment were filed into six classes according to their counted number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10, 11–20, or more than 20 γH2AX foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
- Quantification of RAD51 foci in WT and amiGO RBR1 plants after BLM treatment. Fifty nuclei per line per experiment were filed into six classes according to their counted number of RAD51 foci: nuclei containing no RAD51 foci, 1–2, 3–5, 6–10, 11–20, or more than 20 RAD51 foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
Given that e2fa is also sensitive to BLM, we asked whether E2FA is required for RAD51 localization as well. Therefore, we analyzed the pattern of γH2AX and RAD51 foci by immune staining in the e2fa‐2 mutant allele in which the transactivation domain is eliminated (from here on simply called e2fa). Consistent with its growth reduction, we could detect a similar increase in γH2AX foci in response to BLM in e2fa as in rbr1 (Figs 7H and EV5B). In contrast, we did not observe a decrease in the number of RAD51 signals compared to the wild type (Fig 7H and I). This shows that the correct RAD51 localization depends on RBR1 and is independent of E2FA.
Figure EV5. e2fa mutant is hypersensitive against BLM .
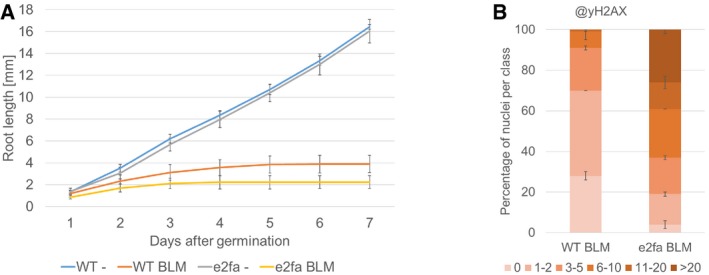
- Comparison of root growth of wild‐type and e2fa plants germinated and grown on medium containing 6 μg/ml BLM or no supplement (‐). Error bars signify the standard deviation in three independent experiments.
- Quantification of γH2AX foci in WT and e2fa plants after BLM treatment. One hundred nuclei per line per experiment were filed into six classes according to their counted number of γH2AX foci: nuclei containing no γH2AX foci, 1–2, 3–5, 6–10, 11–20, or more than 20 γH2AX foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
RBR1 localization after BLM treatment is dependent on CDKB1‐CYCB1 protein kinases
Recent work has shown that the plant‐specific CDKB1 kinases in conjunction with its cyclin partner CYCB1 are major regulators of DDR in plants and in particular control homologous recombination repair (Weimer et al, 2016a,b). Plants defective in CDKB1 or CYCB1 show similar phenotypes in response to DSB‐inducing agents as rbr1 mutants, especially a reduced occurrence of RAD51 foci after treatment with genotoxic agents. Hence, we wanted to know whether CDKB1 kinases and RBR1 act in the same pathways. To this end, we tested the epistasis of these factors by combining the rbr1‐2 allele with cdkb1;1 cdkb1;2 double mutants. The homozygous rbr1 cdkb1;1 cdkb1;2 triple mutants did not have obvious defects beyond a slightly reduced root growth, also apparent in the cdkb1 double mutant (Fig 8A). On BLM‐containing medium, the cdkb1 mutants exhibited shorter roots than rbr1 plants (Fig 8B). Root growth of the rbr1 cdkb1;1 cdkb1;2 triple mutants, however, was comparable to that of the cdkb1;1 cdkb1;2 double mutant and no additive effects were observed, suggesting that CDKB1 acts in the same genetic pathway as RBR1 during DDR.
Figure 8. RBR1 localization is dependent on CDKB1.
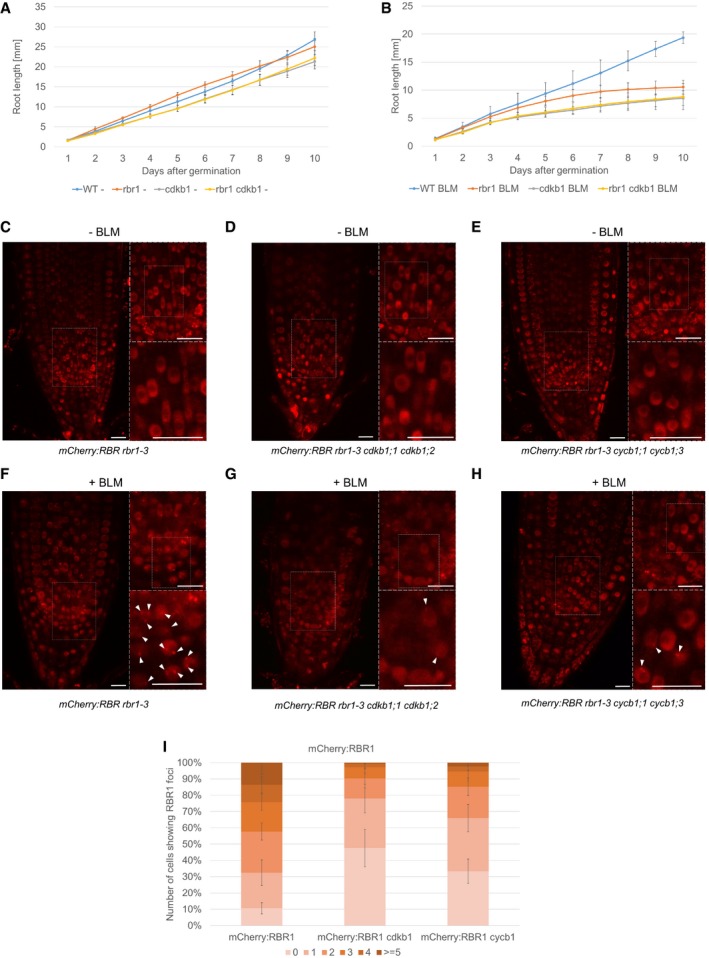
-
A, BComparison of root growth of the wild‐type, rbr1, cdkb1, and rbr1 cdkb1 triple‐mutant plants germinated and grown on medium containing no supplement (A) or 0.6 μg/ml BLM (B). Error bars signify the standard deviation in three independent experiments.
-
C–EIn vivo accumulation of mCherry:RBR1 in root tips of non‐treated PRO RBR1 mCherry:RBR1 rbr1‐3 (C), PRO RBR1 mCherry:RBR1 rbr1‐3 cdkb1;1 cdkb1;2 (D), and PRO RBR1 mCherry:RBR1 rbr1‐3 cycb1;1 cycb1;3 (E) plants. White boxes in the left part of each panel are shown in the upper right inlay, and boxed area in the upper inlay is shown in the lower inlay. Scale bars: 10 μm.
-
F–HIn vivo accumulation of mCherry:RBR1 in root tips of BLM‐treated (6 h, 6 μg/ml) in PRO RBR1 mCherry:RBR1 rbr1‐3 (F), PRO RBR1 mCherry:RBR1 rbr1‐3 cdkb1;1 cdkb1;2 (G), and PRO RBR1 mCherry:RBR1 rbr1‐3 cycb1;1 cycb1;3 (H) plants. White boxes in the left part of each panel are shown in the upper right inlay, and boxed area in the upper inlay is shown in the lower inlay. Arrowheads point to RBR1 foci. Scale bars: 10 μm.
-
IQuantification of RBR1 foci in PRO RBR1 mCherry:RBR1 rbr1‐3, PRO RBR1 mCherry:RBR1 rbr1‐3 cdkb1;1 cdkb1;2, and PRO RBR1 mCherry:RBR1 rbr1‐3 cycb1;1 cycb1;3 plants after BLM treatment (6 h, 6 μg/ml). Fifty nuclei per line per experiment were filed into six classes according to their counted number of RBR51 foci: nuclei containing no foci, 1–2, 3–5, 6–10, 11–20, or more than 20 foci, respectively. Three independent experiments were analyzed. Error bars indicate the standard deviation.
To explore a possible dependency of RBR1 localization onto the activity of CDKB1‐CYCB1 complexes, we introgressed the mCherry:RBR1 reporter line into cdkb1;1 cdkb1;2 and cycb1;1 cycb1;3 double mutants. The resulting plants with the genotypes mCherry:RBR1 rbr1‐3 cdkb1;1 cdkb1;2 and mCherry:RBR1 rbr1‐3 cycb1;1 cycb1;3 (designated as mCherry:RBR1 cdkb1 and mCherry:RBR1 cycb1, respectively) were analyzed for the formation of RBR1 foci in the root tip after DNA stress. While in mCherry:RBR1 plants most nuclei contained two or more foci (Fig 8I), the number of foci dropped in mCherry:RBR1 cdkb1 and mCherry:RBR1 cycb1 to one or no foci giving rise to the conclusion that CDKB1‐CYCB1 activity is needed for the efficient recruitment of RBR1 into foci (Fig 8G–I).
Discussion
Here, we have studied the role of the Arabidopsis Rb homolog RBR1 during DNA damage. The work on RBR1 is complicated since rbr1 null mutants cannot be easily generated due to an essential role of RBR1 in the female gametophyte (Ebel et al, 2004). Transient inactivation/downregulation of RBR1 in various Arabidopsis tissues and/or cell types, for instance by gene silencing or the expression of viral proteins that bind and inhibit Rb‐type proteins, revealed that RBR1 is required in many if not all cells (Gutzat et al, 2012; Desvoyes et al, 2014; Harashima & Sugimoto, 2016). Hence, an analysis of the complete loss of RBR1 function at the organism level is probably impossible. We therefore used here a temperature‐sensitive (cold‐sensitive) rbr1 mutant (rbr1‐2) grown at the restrictive temperature, that is, colder than 18°C (Ebel et al, 2004; Chen et al, 2011; Nowack et al, 2012). However, it is important to note that rbr1‐2 even when grown at the restrictive temperature is not a null but a hypomorphic mutant as indicted by the presence of residual levels of RBR1 protein in the mutant.
Screening different compounds that cause different types of DNA damage at different phases of the cell cycle revealed that rbr1 mutants are especially sensitive to the DSB‐inducing drug BLM and the DNA‐cross‐linking compound cisPt. Interestingly, rbr1 mutants were also hypersensitive to Al, which was previously suggested to act as a genotoxin (Bulanova et al, 2001; Murali Achary & Panda, 2010). The mechanism by which Al damages DNA is still unclear. The observation that rbr1 reacts similarly to the application of cisPt and BLM, which both cause DNA breaks, gives rise to the speculation that Al might have a similar yet likely milder impact on DNA, consistent with the observation of chromosome breaks in plants grown on aluminum (Bulanova et al, 2001). Given that Al is the primary growth‐limiting factor for many plants in acid soils (Foy, 1992), understanding and modulating RBR1 function in DDR also have possible implications for agricultural application.
Cell survival versus DNA repair
The most obvious phenotype of rbr1 mutants when grown on BLM‐, cisPt‐, or Al‐containing media is prevalent cell death in the root meristem. Even untreated rbr1 mutants showed a slightly increased number of dying cells in comparison with wild‐type roots (Fig 3), consistent with previous experiments in which tobacco leaf cells were found to die after the endogenous RBR was silenced (Jordan et al, 2007). Loss of meristem activity in BLM‐treated rbr1 mutants then causes several alterations in the root morphology, for example, the appearance of root hairs at the root tip. Loss of Rb function is also known to lead to cell death in animals as E2F1 directly activates cell‐death programs, a function that is also repressed by Rb binding (Blagosklonny, 1999; Abrams, 2002). In this context, it is interesting that E2FA was reported to be required for effector‐triggered programmed cell death in plant immunity (Wang et al, 2014). However, the cell‐death execution machinery between plants and animals does not appear to be very well conserved, and it remains to be seen how the cell death in rbr1 mutants is brought about (Van Hautegem et al, 2015).
The strong cell‐death phenotype also obscured the function of RBR1 in response to DNA damage. Using a hypomorphic cdka;1 mutant allele and the application of the CDK inhibitor Roscovitine allowed us to uncouple the cell‐death phenotype from a function of RBR1 in DDR since the double mutant rbr1 cdka;1 containing the transgene Pro CDKA;1 :CDKA;1 T14DY15E (short rbr DE) showed no signs of increased cell death but still had many DNA lesions as revealed by the detection γH2AX foci. Similarly, BLM‐induced cell death in rbr1 and notably in BLM‐treated wild‐type plants could be suppressed by the concomitant application of Roscovitine. Thus, execution of cell death depends on CDKA;1 activity and/or cell‐cycle progression. At the same time, this experiment revealed a more direct role of RBR1 in DDR.
The observation that reducing CDKA;1 activity rescues the cell‐death phenotype in rbr1 could be explained by slowing down an enhanced progression through the cell cycle in rbr1 that may cause DNA damage and/or does not allow sufficient time to repair damaged DNA, ultimately triggering a cell‐survival checkpoint (Fig 9). However, increasing cell‐proliferation rates by overexpression of CYCD3;1 was not found to be sufficient to trigger cell death (Horvath et al, 2017). A not mutually exclusive hypothesis is that CDKA;1 activity might be specifically needed for the execution of a cell‐death program and indeed activated Cdks have been implicated in triggering cell death (Shi et al, 1994; Yu et al, 1998; Konishi et al, 2002). Likely, loss of RBR1 would also enhance CDKA;1 activity further since RBR1 was found to repress the F‐box protein FBL17, which in turn promotes the degradation of CDK inhibitors of the INHIBITOR/INTERACTOR OF CDKS/KIP‐RELATED‐PROTEIN (ICK/KRP) class (Kim et al, 2008; Gusti et al, 2009; Zhao et al, 2012) (Fig 9). Since CDKA;1 was found to be the predominant target of this class of inhibitors, their loss results in an increased CDK activity (Nakai et al, 2006; Zhao et al, 2012). A dual function of RBR1 in suppressing cell death as well as in DDR also offers a possible explanation for the massive cell death seen upon growth on media with genotoxic compounds: an increased level of DNA damage due to the absence of RBR1 as DDR component might trigger a cell‐death program that may be usually suppressed by RBR1 action.
Figure 9. Hypothetical model of RBR1's multifacetted role in DNA‐damage response.
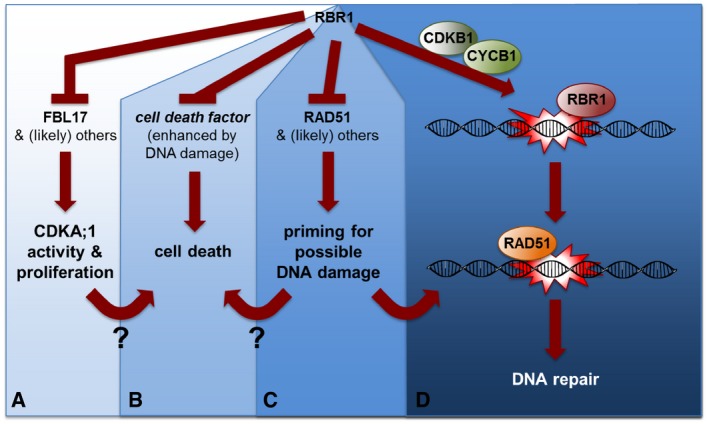
- RBR1 directly represses the expression of the F‐box‐like protein FBL17 as one the central most proliferation‐control genes (Zhao et al, 2012). FBL17, as a part of an SCF complex, mediates the degradation of CDK inhibitors of the KRP class (not shown) and by that promotes CDKA;1 activity (target of KRP action) (Kim et al, 2008; Gusti et al, 2009; Noir et al, 2015).
- RBR1 directly or indirectly promotes cell survival in an as yet unknown way, for example, by suppressing a cell‐death factor (direct scenario). The RBR1‐dependent cell death is proliferation/CDKA;1‐activity dependent since reduction in CDKA;1 activity restores viability but not the sensitivity toward genotoxic drugs. How proliferation/CDKA;1 activity could promote cell death is not yet understood. It is also not understood whether the execution of cell death upon RBR1 loss represents a safety belt in proliferating cells to promote their removal, preventing the propagation of mutations to daughter cells.
- RBR1 directly represses the expression of the DDR gene RAD51 (shown here). Hence, inactivation of RBR1 by CDKA;1 phosphorylation presumably not only promotes the expression of cell‐proliferation genes such as FBL17 but also DDR genes. This possibly primes proliferating cells for likely occurring damage during the cell cycle. The priming may also contribute to the execution of cell death in an unknown mechanism.
- RBR1 may bind to DNA lesions as revealed here by its partial co‐localization to γH2AX foci. There, RBR1 possibly acts to assemble local repair complexes. RBR1 is especially needed for the recruitment of RAD51 to damaged DNA. Notably, this function is independent of E2FA. For proper RBR1 localization in foci, the action of the CDKB1‐CYCB1 complexes, which play a major role DDR in plants, is needed.
Transcriptional versus structural role of RBR1 in DDR
An obvious possibility of RBR1 function during DNA damage is a transcriptional control of DDR genes. Support for such a possibility comes from the observation that Rb from mammals can be a component of active E2F1 complexes. Moreover, binding of Rb to E2F1 was found to be required for the full induction of proapoptotic gene expression in response to genotoxic stress (Ianari et al, 2009). Similarly, Rb in the green algae Chlamydomonas appears to be a component of transcriptionally active complexes (Olson et al, 2010). Here, we found that out of 20 genes known to participate in DDR, the expression of five (AHP2, BRCA1, PARP2, RAD51, and TSO2) was influenced by the presence of functional RBR1. However, these five genes were repressed and not activated by RBR1, consistent with the canonical role of RBR1 as a transcriptional repressor. Thus, somewhat similar to animals, proliferating cells in which RBR1 is inactivated would allow not only the expression of cell‐cycle‐promoting genes activated by E2F but also DDR genes. This is consistent with genomewide transcriptional studies of synchronized Arabidopsis cells that revealed that all five DDR genes (AHP2, BRCA1, RAD51, PARP2, and TSO2) have their expression maximum in S‐phase (Menges et al, 2003).
However, to untangle the effect of direct RBR1 repression versus increased cell‐cycle activity and elevated levels of DNA damage in rbr1 mutants even without the application of DSB‐inducing drugs is complicated since reduction in RBR1 activity in cdka;1 mutants did often not result in the restoration of expression levels of RBR1 targets such a PROLIFERATION NUCLEAR ANTIGEN 1 (PNCA1) (Nowack et al, 2012). This hints at a possible feedback regulation between RBR1 and CDKA;1 and/or other roles of CDKA;1 in controlling transcription as suggested by the recent finding that major transcriptional waves during the cell cycle of Chlamydomonas depend on CDKA;1 activity (Tulin & Cross, 2015).
In any case, our data suggest that the impact of E2F and RBR as transcriptional regulators during DNA damage is low since in e2fa and rbr1 mutants, the expression of DDR genes could be strongly induced upon DNA damage. This indicates that other transcription factors take over and an obvious candidate here would be SUPPRESSOR OF GAMMA‐IRRADIATION 1 (SOG1), which has been found to regulate many genes after DNA damage, including BRCA1 and RAD51 (Yoshiyama et al, 2009). Therefore, the E2F‐dependent activation of DDR genes via a reduction of RBR1 in proliferating cells might prime them for rapid induction if damage occurs (Fig 9). This would also be consistent with our observation that no transcriptional change was found in e2fa mutants in comparison with the wild type. In addition, E2FA function could be backed up by a redundant/compensatory action of other E2F genes in Arabidopsis, for example, E2FB, E2FC, and three DP‐E2F‐Like (DEL) genes. However, it still remains unclear why e2fa single mutants are already hypersensitive to DNA‐damaging drugs.
In addition to its transcriptional role, we revealed here that RBR1 re‐localizes to foci after DNA damage, which at least partially overlap with γH2AX and RAD51 foci. This hints at a structural role for RBR1 in DDR (Fig 9). In animals, a recent study reported that direct Rb binding is necessary for KU70 and KU80 to participate in a DNA repair pathway through non‐homologous end joining (NHEJ) of broken DNA ends in human cancer cells (Cook et al, 2015). Moreover, Rb was shown to physically interact with the BRCA1 protein (Aprelikova et al, 1999; Yarden & Brody, 1999; Fan et al, 2001). BRCA1 is part of DNA‐damage repair complexes that bind to DSBs. Furthermore, Rb was identified in a complex, together with BRCA1, that cleaves trapped topoisomerase II from lesions, leading to the hypothesis that Rb can provide a platform for repair factors to bind to damage sites (Xiao & Goodrich, 2005). Noteworthy, Arabidopsis BRCA1 partially overlaps with RBR1 foci, and RBR1 and BRCA1 can physically interact (Horvath et al, 2017).
The transcriptional function of RBR1 and its emerging local role at DNA might lead to an intriguing interplay in DDR; that is, RBR1 is phosphorylated by CDKs in proliferating cells resulting in the activation of E2F and the expression of not only cell‐proliferation genes needed for DNA replication but also DDR genes (Fig 9). At the same time, RBR1 becomes available as a potential repair protein giving rise to a poised repair state. The recruitment of RBR1 to foci was here found to depend on the activity of CDKB1‐CYCB1 complexes, which have been found to play an important role during homology‐dependent DNA repair and are activated in a SOG1‐dependent manner after DNA damage (Weimer et al, 2016a,b). Since RBR1 was previously shown to be a putative substrate of CDKB1‐CYCB1 complexes by in vitro kinase assays (Harashima & Schnittger, 2012), it is tempting to speculate that CDKB1‐dependent phosphorylation is instrumental for the activation and/or recruitment of RBR1 to DNA in response to BLM (Fig 9).
Here, we have revealed that the presence of RBR1 is crucial for RAD51 localization, shedding new light onto the assembly of repair complexes. It remains to be seen whether Rb also controls RAD51 localization in yeast and animals. Interestingly, reduction in RBR1 activity did not interfere with RAD51 localization in Arabidopsis meiosis during which RAD51 is involved in homologous recombination (Chen et al, 2011). Recent analysis of a RAD51 separation‐of‐function mutant by Da Ines et al showed that the DNA‐damage repair function of RAD51 in mitosis can be uncoupled from the recombination function in meiosis (Da Ines et al, 2013). In light of our data, it is tempting to speculate that RBR1 contributes to the differential activity of RAD51 in mitosis versus meiosis.
Taken together, RBR1 seems to function in the Arabidopsis DDR in at least three, likely interconnected pathways: as a cell‐cycle checkpoint‐dependent inhibitor of programmed cell death, as a transcriptional regulator possibly contributing to a priming effect, and finally as a mediator of RAD51 recruitment to lesions.
Materials and Methods
Plant material and growth conditions
Plants were grown on half concentrated MS medium with 0.5% sucrose and 0.8% agar under long day (16‐h light/8‐h dark) conditions at 17°C if not noted otherwise. DNA‐damaging supplements were added as needed. All mutants used in this study are in Col‐0 background and were described previously: atm: atm‐2 (Garcia et al, 2003), atr: atr‐2 (Culligan et al, 2004), cdka;1: CDKA;1 T14D;Y15E (Dissmeyer et al, 2009), ku70 (Riha et al, 2002), rad51 (Li et al, 2004), rbr1: rbr1‐2 (Ebel et al, 2004), rbr1‐3 (Ebel et al, 2004), wee1 (De Schutter et al, 2007). All genotypes were determined by PCR, and primers are indicated in Appendix Table S2.
Root growth assay
For the analysis of root growth on HU (1 mM final, Sigma‐Aldrich) or bleomycin (BLM, 6 μg/ml, Duchefa), plants were germinated and grown on vertical plates. Different batches of BLM showed different levels of potency. Appropriate controls were included in every experiment. Figures 1, 3A and B, 4A–E, 5A, 6, 7, 8 and EV1, and Appendix Figs S1 and S2 were obtained with the same batch of BLM. Figures 3C and D, 4F–I, 5D, EV3–EV5, and Appendix Fig S3 were done using different batches. The position of the root tip was marked daily for each plant. After 10 days, plates were photographed and root length was measured using the Simple Neurite Tracer plug‐in of the Fiji distribution package of the ImageJ software. The final values were calculated by determining the arithmetic mean of the root length values of three biological replicates, which were themselves the average of at least 20 plants. Root growth analysis on cisplatin (cisPt) medium was done similarly with the difference that plants were germinated on medium lacking cisPt and after 3 days transferred to medium with cisPt (15 μM). The measurements were done over the following 3 days. CisPt preparation was done fresh for each use, and the powder was dissolved in water by thoroughly pipetting up and down.
Propidium iodide staining
Staining of cell wall and dead cells for microscopy was done by submerging seedling for 1 min in a 10 μg/ml PI/water solution and rinsing shortly in water afterward.
Comet assays
The evaluation of DNA damage was done by a neutral/neutral comet assay. Seedlings were grown for 21 days and then transferred to ½ MS liquid medium (control) or ½ MS liquid medium containing 30 μg/ml BLM. After 1 h of incubation, a fraction of the treated plants were separated. The remaining plants were shortly dried on paper towels and immediately frozen in liquid nitrogen. The separated plants were washed three times with ½ MS and transferred to ½ MS liquid medium containing no BLM for recovery. After 30 min of incubation, these plants were also briefly dried and frozen. The preparation of the comet slides was performed according to Menke et al (2001) and stained with 3× GelRed Nucleic Acid Stain (Biotium, Hayward, CA, USA) diluted in 0.1M NaCl. The comets were observed, and pictures taken on an AXIO Imager Z1 fluorescence microscope with an AXIO Cam MRm (Carls Zeiss, Jena, Germany). The analysis of the pictures was done utilizing the TriTek Comet Score software, and 200 comets per sample were measured.
Immunofluorescence staining
Six‐ to ten‐day‐old plants were transferred to ½ MS liquid medium containing 30 μg/ml BLM, 50 μM cisPt, or no supplement. Incubation time was 2 h. Immunostaining of root tip spreads was subsequently performed as described earlier in Friesner et al (2005). A rabbit anti‐plant γH2AX antibody, provided by Dr. Charles White, was used in a 1:600 dilution. As secondary antibody, a goat Alexa Fluor® 488 anti‐rabbit (Life Technologies, Carlsbad, CA, USA) was used in a 1:300 dilution. For the observation of RAD51, we used a rat anti‐RAD51, provided by Dr. Peter Schlögelhofer, in a 1:500 dilution with Alexa Fluor® 588 anti‐rat (Life Technologies, Carlsbad, CA, USA) or Cy3 anti‐rat (Thermo Fisher Scientific; Cat.# A‐10522) at 1:300, and for the mCherry detection, we used an anti‐mCherry antibody (Abcam; ab205402) at 1:600 with goat anti‐chicken IgY (H+L) Alexa Fluor® 647 (Thermo Fisher Scientific; Cat.# A‐21449) at 1:300.
ChIP experiments
Two‐week‐old seedlings of PRO RBR1 :RBR1:mRFP‐expressing plants growing on ½ MS plates were used. Chromatin was sheared with a Bioruptor sonicator (Cosmo Bio) twice for 15 min with a 50% duty cycle and high power output to obtain 200‐ to 1,000‐bp DNA fragments. Immunoprecipitation was performed using the DsRed polyclonal antibody (Clontech) together with protein A‐magnetic beads (Millipore). The E2FA antibody was described previously (Heyman et al, 2011). Negative controls were performed without antibody. DNA was recovered using Magna ChIP spin filters according to the manufacturer's instructions (Millipore). Then, 0.5 or 1 μl of a one‐fifth dilution of ChIP DNA was analyzed by ChIP PCR or quantitative real‐time PCR using gene‐specific primers, respectively (see Appendix Table S2). Two biological and three technical replicates were performed for ChIP–quantitative PCR using MCM5 and ORC3 as positive controls and heterochromatic region primers as a negative control (see Appendix Table S2).
Expression analysis
Ten‐day‐old plants were used, either untreated or treated with 30 μg/ml BLM for 2 h, then immediately frozen in liquid nitrogen, and temporarily stored at −80°C. RNA was extracted using NucleoSpin RNA Plant Kit (MACHEREY‐NAGEL). A total of 10–20 seedlings were pooled for each sample. RNA concentration and purity were tested using nanodrop‐photometric quantification (Thermo Scientific). For cDNA synthesis, Superscript Vilo Mastermix (Invitrogen) was used. Quantitative real‐time PCR (qRT‐PCR) was done using the Roche LightCycler 480 system. Oligonucleotides were designed with QuantPrime (http://quantprime.mpimp-golm.mpg.de; Arvidsson et al, 2008) and used in final concentration of 0.5 μM each (primers are listed in Appendix Table S2). Three biological replicates with three technical replicates each were processed. Cq calling was done using the second derivative maximum method. Target‐specific efficiencies were calculated as the mean of all reaction‐specific efficiencies for a given target. Reaction‐specific efficiencies were deduced using LinRegPCR 2015.2 (http://LinRegPCR.nl; Ramakers et al, 2003; Ruijter et al, 2009). Data were quality‐controlled, normalized against at least two reference genes, and statistically evaluated using qbasePLUS 3.0 (http://www.biogazelle.com/products/qbaseplus; Hellemans et al, 2007). Suitable reference genes (At1g02410, At4g26410, AT4G30520, At5g36210) were identified using the genevestigator tool RefGenes (Hruz et al, 2008).
Image analyses
Imaging was done with a LSM700 confocal microscope (Carl Zeiss, Jena, Germany) and a Leica TCS SP8 confocal microscope (Carl Zeiss, Jena, Germany) at 40× magnification. The objective used was a HC PL APO CS2 40×/1.10 WATER with a numerical aperture of 1.1. The excitation light for the fluorophores was emitted by a diode 405 nm laser, an argon laser at 488 nm, a DPSS laser (561 nm), and a HeNe laser (594 nm). For the pixel intensity plot, the pixel brightness through a region of interest was measured using ImageJ and plotted against the X dimension. The fluorescence co‐localization was analyzed via the ImageJ modules Coloc2 (beta version) and JACoP. Venn diagrams were generated by the software BioVenn (Hulsen et al, 2008).
Author contributions
SB, HH, PC, MH, DB, KS, and AS conceived and designed the experiments. SB, HH, PC, DB, and KS performed the experiments. AS contributed material and reagents. SB, HH, PC, MH, DB, KS, and AS analyzed the data. SB and AS wrote the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Charles White (CNRS, Clermont‐Ferrand, France) for providing us with an anti‐plant γH2AX antibody, Peter Schlögelhofer (MFPL, Vienna, Austria) for an anti‐RAD51 antibody, Willhelm Gruissem (ETH, Zurich, Switzerland) for an anti‐RBR1 antibody, Lieven DeVeylder (VIB, Gent, Belgium) for an anti‐E2FA antibody, and Ben Scheres (University of Wageningen, the Netherlands) for making the RBR1 amiGO line available for us. We are grateful to Caroline Alice Sjogren (UC Riverside, Riverside, USA) for teaching us the preparation of aluminum media. We kindly acknowledge John Larkin (LSU, Baton Rouge, USA) for critical reading and helpful comments on the article. We thank Beatrix Horvath, Ben Scheres and Laszlo Bögre and their coworkers for sharing their unpublished data with us and for the open discussion on the role of RBR1 in DDR. We thank Prof. Dr. Catherine Schmidt (IBMP, Strasbourg, France) for hosting of S.B. This work was supported by a postdoctoral fellowship of the German Research Foundation (DFG) to S.B. and the ERA‐CAPS grant ALUCIDATE from the German Research Foundation (DFG) (SCHN 736/6‐1) to A.S.
The EMBO Journal (2017) 36: 1279–1297
See also: BM Horvath et al (May 2017)
References
- Abe K, Osakabe K, Nakayama S, Endo M, Tagiri A, Todoriki S, Ichikawa H, Toki S (2005) Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams JM (2002) Competition and compensation: coupled to death in development and cancer. Cell 110: 403–406 [DOI] [PubMed] [Google Scholar]
- Adachi S, Minamisawa K, Okushima Y, Inagaki S, Yoshiyama K, Kondou Y, Kaminuma E, Kawashima M, Toyoda T, Matsui M, Kurihara D, Matsunaga S, Umeda M (2011) Programmed induction of endoreduplication by DNA double‐strand breaks in Arabidopsis . Proc Natl Acad Sci USA 108: 10004–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprelikova ON, Fang BS, Meissner EG, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen RA, Liu ET (1999) BRCA1‐associated growth arrest is RB‐dependent. Proc Natl Acad Sci USA 96: 11866–11871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño‐Pachón DM, Mueller‐Roeber B (2008) QuantPrime–a flexible tool for reliable high‐throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV (1999) A node between proliferation, apoptosis, and growth arrest. Bioessays 21: 704–709 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J 23: 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI (2005) Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Bulanova NV, Synzynys BI, Koz'min GV (2001) Aluminum induces chromosome aberrations in wheat root meristem cells. Genetika 37: 1725–1728 [PubMed] [Google Scholar]
- Charbonnel C, Gallego ME, White CI (2010) Xrcc1‐dependent and Ku‐dependent DNA double‐strand break repair kinetics in Arabidopsis plants. Plant J 64: 280–290 [DOI] [PubMed] [Google Scholar]
- Chen Z, Higgins JD, Hui JT, Li J, Franklin FC, Berger F (2011) Retinoblastoma protein is essential for early meiotic events in Arabidopsis . EMBO J 30: 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook R, Zoumpoulidou G, Luczynski MT, Rieger S, Moquet J, Spanswick VJ, Hartley JA, Rothkamm K, Huang PH, Mittnacht S (2015) Direct involvement of retinoblastoma family proteins in DNA repair by non‐homologous end‐joining. Cell Rep 10: 2006–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, Van den Daele H, Van Isterdael G, Schnittger A, De Veylder L (2011) The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz‐Ramirez A, Diaz‐Trivino S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, Caballero‐Perez J, Horvath B, Long Y, Mahonen AP, Zhang H, Xu J, Murray JA, Benfey PN, Bako L, Maree AF et al (2012) A bistable circuit involving SCARECROW‐RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz‐Ramírez A, Díaz‐Triviño S, Wachsman G, Du Y, Arteága‐Vázquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V, Scheres B (2013) A SCARECROW‐RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol 11: e1001724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K, Tissier A, Britt A (2004) ATR regulates a G2‐phase cell‐cycle checkpoint in Arabidopsis thaliana . Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB (2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Da Ines O, Degroote F, Goubely C, Amiard S, Gallego ME, White CI (2013) Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet 9: e1003787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Joubes J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inze D, De Veylder L (2007) Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inze D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez‐Parra E, Xie Q, Chua NH, Gutierrez C (2006) Cell type‐specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol 140: 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, de Mendoza A, Ruiz‐Trillo I, Gutierrez C (2014) Novel roles of plant RETINOBLASTOMA‐RELATED (RBR) protein in cell proliferation and asymmetric cell division. J Exp Bot 65: 2657–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Rubin SM (2013) Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol 14: 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, Pusch S, De Schutter K, Kamei CL, Nowack M, Novak B, Duan GL, Zhu YG, De Veylder L, Schnittger A (2009) Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21: 3641–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W (2004) Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780 [DOI] [PubMed] [Google Scholar]
- Fan S, Yuan R, Ma YX, Meng Q, Goldberg ID, Rosen EM (2001) Mutant BRCA1 genes antagonize phenotype of wild‐type BRCA1. Oncogene 20: 8215–8235 [DOI] [PubMed] [Google Scholar]
- Foy CD (1992) Soil chemical factors limiting plant root growth In Limitations to plant root growth, Hatfield JL, Stewart BA. (eds), pp 97–149. Advances in soil sciences 19. New York: Springer; [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB (2005) Ionizing radiation‐dependent gamma‐H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3‐related. Mol Biol Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Angelis KJ, Britt AB (2015) Arabidopsis DNA polymerase lambda mutant is mildly sensitive to DNA double strand breaks but defective in integration of a transgene. Front Plant Sci 6: 357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego ME, Bleuyard JY, Daoudal‐Cotterell S, Jallut N, White CI (2003) Ku80 plays a role in non‐homologous recombination but is not required for T‐DNA integration in Arabidopsis . Plant J 35: 557–565 [DOI] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P (2009) The Arabidopsis thaliana F‐box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4: e4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Gruissem W (2012) Emerging roles of RETINOBLASTOMA‐RELATED proteins in evolution and plant development. Trends Plant Sci 17: 139–148 [DOI] [PubMed] [Google Scholar]
- Harashima H, Schnittger A (2012) Robust reconstitution of active cell‐cycle control complexes from co‐expressed proteins in bacteria. Plant Methods 8: 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23: 345–356 [DOI] [PubMed] [Google Scholar]
- Harashima H, Sugimoto K (2016) Integration of developmental and environmental signals into cell proliferation and differentiation through RETINOBLASTOMA‐RELATED 1. Curr Opin Plant Biol 29: 95–103 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real‐time quantitative PCR data. Genome Biol 8: R19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1‐E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Van den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Alvim Kamei CL, De Jaeger G, Koncz C, De Veylder L (2011) Arabidopsis ULTRAVIOLET‐B‐INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase‐promoting complex/cyclosome. Plant Cell 23: 4394–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath BM, Kourova H, Nagy S, Nemeth E, Magyar Z, Papdi C, Ahmad Z, Sanchez‐Perez GF, Perilli S, Blilou I, Pettkó‐Szandtner A, Darula Z, Meszaros T, Binarova P, Bogre L, Scheres B (2017) Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J 36: 1261–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta‐analysis of transcriptomes. Adv Bioinformatics 2008: 420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W (2008) BioVenn – a web application for the comparison and visualization of biological lists using area‐proportional Venn diagrams. BMC Genom 9: 488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, Haigis K, Gulino A, Lees JA (2009) Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell 15: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CV, Shen W, Hanley‐Bowdoin LK, Robertson DN (2007) Geminivirus‐induced gene silencing of the tobacco retinoblastoma‐related gene results in cell death and altered development. Plant Mol Biol 65: 163–175 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG (2008) Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Konishi Y, Lehtinen M, Donovan N, Bonni A (2002) Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell 9: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Krstic D, Djalovic I, Nikezic D, Bjelic D (2012) Aluminium in acid soils: chemistry, toxicity and impact on maize plants In Food production – approaches, challenges and tasks, Aladjadjiyan A. (ed.), pp 231–242. Rijeka, Croatia: Intech; [Google Scholar]
- Kurzbauer MT, Uanschou C, Chen D, Schlögelhofer P (2012) The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis . Plant Cell 24: 2058–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara A, Gruissem W (2014) Arabidopsis RETINOBLASTOMA‐RELATED and Polycomb group proteins: cooperation during plant cell differentiation and development. J Exp Bot 65: 2667–2676 [DOI] [PubMed] [Google Scholar]
- Lang J, Smetana O, Sanchez‐Calderon L, Lincker F, Genestier J, Schmit AC, Houlné G, Chabouté ME (2012) Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol 194: 353–363 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJ, Jones CA, Williams KM, Cancel JD (2005) ALS3 encodes a phloem‐localized ABC transporter‐like protein that is required for aluminum tolerance in Arabidopsis . Plant J 41: 353–363 [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann‐Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double‐stranded break repair in Arabidopsis . Plant Physiol 138: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H (2006) Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA 103: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG, Moulinoux JP (1997) Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin‐dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem 243: 527–536 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome‐wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Menke M, Chen I, Angelis KJ, Schubert I (2001) DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat Res 493: 87–93 [DOI] [PubMed] [Google Scholar]
- Morgan DO (1997) Cyclin‐dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Murali Achary VM, Panda BB (2010) Aluminium‐induced DNA damage and adaptive response to genotoxic stress in plant cells are mediated through reactive oxygen intermediates. Mutagenesis 25: 201–209 [DOI] [PubMed] [Google Scholar]
- Nakai T, Kato K, Shinmyo A, Sekine M (2006) Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2‐associated kinases, including plant‐specific B‐type cyclin‐dependent kinase. FEBS Lett 580: 336–340 [DOI] [PubMed] [Google Scholar]
- Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P (2015) The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F‐box protein FBL17. Plant Cell 27: 1461–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A (2012) Genetic framework of cyclin‐dependent kinase function in Arabidopsis . Dev Cell 22: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Nowsheen S, Yang ES (2012) The intersection between DNA damage response and cell death pathways. Exp Oncol 34: 243–254 [PMC free article] [PubMed] [Google Scholar]
- Olson BJ, Oberholzer M, Li Y, Zones JM, Kohli HS, Bisova K, Fang SC, Meisenhelder J, Hunter T, Umen JG (2010) Regulation of the Chlamydomonas cell cycle by a stable, chromatin‐associated retinoblastoma tumor suppressor complex. Plant Cell 22: 3331–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Yoshioka T, Ichikawa H, Toki S (2002) Molecular cloning and characterization of RAD51‐like genes from Arabidopsis thaliana . Plant Mol Biol 50: 71–81 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, Ito Y, Kato T, Tabata S, Kurei S, Yoshioka Y, Machida Y, Seki M, Kobayashi M, Shinozaki K, Ichikawa H, Toki S (2005) Arabidopsis Rad51B is important for double‐strand DNA breaks repair in somatic cells. Plant Mol Biol 57: 819–833 [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Tréhin C, Perennes C, Bureau JM, Meijer L, Bergounioux C (1997) Roscovitine, a novel cyclin‐dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY‐2 cell suspension. Plant J 12: 191–202 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption‐free analysis of quantitative real‐time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Reed CA, Mayhew CN, McClendon AK, Knudsen ES (2010) Unique impact of RB loss on hepatic proliferation: tumorigenic stresses uncover distinct pathways of cell cycle control. J Biol Chem 285: 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE (2002) Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J 21: 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montané MH, Houlné G, Chabouté ME (2009) Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis . Plant Physiol 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2009) Regulation and function of retinoblastoma‐related plant genes. Plant Sci 177: 540–548 [Google Scholar]
- Sabelli PA, Liu Y, Dante RA, Lizarraga LE, Nguyen HN, Brown SW, Klingler JP, Yu J, LaBrant E, Layton TM, Feldman M, Larkins BA (2013) Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma‐related pathway in maize endosperm. Proc Natl Acad Sci USA 110: E1827–E1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster‐Guity N, Conrad BH, Papadopoulos N, Bunz F (2011) ATR mediates cisplatin resistance in a p53 genotype‐specific manner. Oncogene 30: 2526–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Nishioka WK, Th'ng J, Bradbury EM, Litchfield DW, Greenberg AH (1994) Premature p34cdc2 activation required for apoptosis. Science 263: 1143–1145 [DOI] [PubMed] [Google Scholar]
- Tulin F, Cross FR (2015) Cyclin‐dependent kinase regulation of diurnal transcription in Chlamydomonas. Plant Cell 27: 2727–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hautegem T, Waters AJ, Goodrich J, Nowack MK (2015) Only in dying, life: programmed cell death during plant development. Trends Plant Sci 20: 102–113 [DOI] [PubMed] [Google Scholar]
- Von Uexküll HR, Mutert E (1995) Global extent, development and economic impact of acid soils. Plant Soil 171: 1–15 [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X (2014) A noncanonical role for the CKI‐RB‐E2F cell‐cycle signaling pathway in plant effector‐triggered immunity. Cell Host Microbe 16: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Nowack MK, Bouyer D, Zhao X, Harashima H, Naseer S, De Winter F, Dissmeyer N, Geldner N, Schnittger A (2012) RETINOBLASTOMA RELATED1 regulates asymmetric cell divisions in Arabidopsis . Plant Cell 24: 4083–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D, Sugimoto K, Koncz C, Doerner P, Umeda M, Schnittger A (2016a) The plant‐specific CDKB1‐CYCB1 complex mediates homologous recombination repair in Arabidopsis . EMBO J 35: 2068–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Schnittger A (2016b) Specialization of CDK regulation under DNA damage. Cell Cycle 16: 143–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Goodrich DW (2005) The retinoblastoma tumor suppressor protein is required for efficient processing and repair of trapped topoisomerase II‐DNA‐cleavable complexes. Oncogene 24: 8105–8113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, Sack FD, Grotewold E (2010) Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22: 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Bilichak A, Titov V, Golubov A, Kovalchuk I (2013) Genome stability of Arabidopsis atm, ku80 and rad51b mutants: somatic and transgenerational responses to stress. Plant Cell Physiol 54: 982–989 [DOI] [PubMed] [Google Scholar]
- Yarbro JW (1992) Mechanism of action of hydroxyurea. Semin Oncol 19: 1–10 [PubMed] [Google Scholar]
- Yarden RI, Brody LC (1999) BRCA1 interacts with components of the histone deacetylase complex. Proc Natl Acad Sci USA 96: 4983–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Shikazono N, Tanaka A, Hase Y, Funayama T, Wada S, Inoue M (2005) Comparative radiation tolerance based on the induction of DNA double‐strand breaks in tobacco BY‐2 cells and CHO‐K1 cells irradiated with gamma rays. Radiat Res 163: 520–525 [DOI] [PubMed] [Google Scholar]
- Yoshiyama K, Conklin PA, Huefner ND, Britt AB (2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Jing T, Liu B, Yao J, Tan M, McDonnell TJ, Hung MC (1998) Overexpression of ErbB2 blocks Taxol‐induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinase. Mol Cell 2: 581–591 [DOI] [PubMed] [Google Scholar]
- Zhao X, Harashima H, Dissmeyer N, Pusch S, Weimer AK, Bramsiepe J, Bouyer D, Rademacher S, Nowack MK, Novak B, Sprunck S, Schnittger A (2012) A general G1/S‐phase cell‐cycle control module in the flowering plant Arabidopsis thaliana . PLoS Genet 8: e1002847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


