Abstract
The CRISPR‐Cas system is an adaptive and heritable immune response that destroys invading foreign nucleic acids. The effector complex of the Type III CRISPR‐Cas system targets RNA and DNA in a transcription‐coupled manner, but the exact mechanism of DNA targeting by this complex remains elusive. In this study, an effector Csm holocomplex derived from Thermococcus onnurineus is reconstituted with a minimalistic combination of Csm1121334151, and shows RNA targeting and RNA‐activated single‐stranded DNA (ssDNA) targeting activities. Unexpectedly, in the absence of an RNA transcript, it cleaves ssDNA containing a sequence complementary to the bound crRNA guide region in a manner dependent on the HD domain of the Csm1 subunit. This nuclease activity is blocked by a repeat tag found in the host CRISPR loci. The specific cleavage of ssDNA without a target RNA suggests a novel ssDNA targeting mechanism of the Type III system, which could facilitate the efficient and complete degradation of foreign nucleic acids.
Keywords: CRISPR, Csm complex, DNase, RNase, Thermococcus onnurineus
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; RNA Biology
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR‐associated (Cas) systems eliminate invading phages and plasmids in prokaryotes 1, 2, 3, 4. CRISPR loci are composed of variable spacer sequences derived from genetic invaders and a series of short repeat sequences 4, 5. Transcripts from CRISPR loci are processed to generate short RNAs, termed crRNAs, that assemble with Cas proteins into a large nuclease effector complex 6. Based on the cas gene content and the mechanism of action, CRISPR‐Cas systems are classified into six major types (types I–VI) and at least 16 subtypes 7. The multi‐protein Cascade complex of the Type I system detects foreign nucleic acids and then recruits the Cas3 protein to degrade DNA using its HD nuclease domain 8, 9. Type II/V systems also target DNA, but recognition and cleavage are mediated by a ribonucleoprotein effector containing a single protein, Cas9 or Cpf1 10, 11, 12. The Cascade complex of Type I and the Csm and/or Cmr effector complexes of Type III are phylogenetically related multi‐subunit ribonucleoprotein complexes, and in the holocomplex assembly, both form a common twisted helical structure with homologous components 13, 14, 15, 16. In contrast, the Type I and Type II systems target exclusively DNA, and the Type III system targets RNA or/and DNA 3, 17, 18, 19, 20, 21, 22
The Type III system can be further classified into subtypes (subtypes III‐A–III‐D) or mainly into Type III‐A and Type III‐B 14, 23. The Csm complexes of Type III‐A from Sulfolobus solfataricus, Thermus thermophilus, Streptococcus thermophilus, Staphylococcus aureus and Staphylococcus epidermis have been characterized in vitro and in vivo 3, 19, 21, 22, 23, 24. The Csm complex consists of five subunits (Csm1–5) and a crRNA, while the Cmr complex of Type III‐B consists of six subunits (Cmr1–6) and a crRNA 22, 25, 26. The crRNA is composed of eight nucleotides (nt) at its 5′ end, termed the 5′ handle, and 30–45 nt, termed the guide sequence, derived from a spacer sequence of CRISPR. The common tag at the 5′ end of the crRNA derived from the repeat sequence of the CRISPR is processed by Cas6 and recognized by the Csm4 or Cmr3 subunit in the complexes 27, 28. The Csm/Cmr complexes possess an RNase activity that cleaves target RNAs at the complementary guide region of crRNA at 6‐nt intervals by means of multiple copies of Csm3 in the Csm complex 21, 22 or Cmr4 in the Cmr complex 29
The DNA targeting mechanism of the Type III system is a recent topic of ongoing investigation 30, 31, 32. The target cleavage by the Type III‐A system requires directional transcription that accompanies the displacement of the target double‐stranded DNA (dsDNA) and the production of RNA transcripts 33. The Csm complex from S. epidermis degrades the RNA transcripts and the non‐template DNA strand through the transcription‐coupled targeting mechanism in vitro and in vivo by the nuclease activity of the Csm1 subunit 19. The Csm complex from S. thermophilus exhibits a similar non‐specific degradation of DNA in an RNA transcript‐dependent manner with the catalytic site in the HD domain of Csm1 30. The Cmr complexes from P. furiosus and T. maritima cleave DNA and protect cells from plasmid invasion in a transcription‐coupled manner by the HD domain of the Cmr2 subunit 22, 32. For the Type I and Type II systems, the protospacer adjacent motif (PAM) sequence plays a key role in discriminating between self and non‐self DNA to recognize foreign nucleotides and facilitates the unwinding of the target DNA. Type III systems are unique in that they lack a PAM motif 13, 34, 35, 36. Instead of recognizing a distinct PAM sequence, the Csm complexes in Type III systems might check for complementarity between the repeat‐derived region of the crRNA and the target DNA strand 37. However, the exact mechanisms of the DNA targeting in Type III systems remain elusive, particularly those mechanisms underlying the positioning of the effector complex with respect to the separated ssDNA strands during transcription and the conformational change for the activation of nuclease activity.
In this study, we reconstituted the Csm effector holocomplex of Thermococcus onnurineus NA1 and found that this effector complex has a novel single‐stranded DNA (ssDNA)‐specific nuclease activity in addition to the previously known RNA targeting and RNA‐activated DNA targeting activities. We report the characterization of this nuclease activity, designated as the RNA transcript‐independent ssDNA targeting, which could ensure the direct recognition and degradation of the non‐template strand of foreign DNA without affecting self DNA.
Results
In vitro assembly of the ToCsm complex
The hyperthermophilic archaeon, T. onnurineus NA1, has six CRISPR loci and two Cas systems, Type III‐A and putative Type IV, according to a CRISPR database (Fig EV1A and B) 38, 39. The Type III‐A‐specific csm gene cluster (Ton_0892–Ton_0898) is located between CRISPR locus 3 and locus 4 (Fig 1A). We sought to reconstitute the effector holocomplex (ToCsm complex) composed of five different subunits (ToCsm1–ToCsm5) and a crRNA. We found that the ToCsm1–Csm4 and ToCsm2–Csm5 subcomplexes could be produced in a soluble form in E. coli by coexpression of the respective subunits, while each individual subunit was insoluble. In contrast, ToCsm3 alone exhibited reasonable solubility. Incubation of these recombinant subcomplexes and the protein did not result in the formation of the ToCsm holocomplex. Subsequently, we synthesized crRNAs that contain a common 8‐nt repeat sequence 27, 37 at CRISPR locus 3, 5′‐GUGGAAAG‐3′, and a spacer sequence with varying length ranging from 30 to 40 nt. Each of these crRNAs was incubated with purified ToCsm1–Csm4, ToCsm2–Csm5 and ToCsm3 at 60°C for 20 min. The mixtures showed a major single peak corresponding to an approximately 280 kDa mass in size‐exclusion chromatography (SEC), indicating holocomplex assembly in the presence of a crRNA (Figs 1B and EV1C). Among the crRNAs, a 38‐nt crRNA yielded the most significant peak in the SEC analysis and was thus selected for further functional studies. This peak fraction contained all five subunits of the ToCsm complex as shown by denaturing SDS polyacrylamide gel electrophoresis (PAGE) and displayed a single band on a native polyacrylamide gel. This fraction also contained the crRNA as identified by denaturing urea–PAGE (Fig 1C). A multi‐angle light scattering (MALS) analysis indicated that this fraction corresponds to a molecular mass of 273.4 (± 1.32%) kDa, which is consistent with the SEC analysis (Fig 1D). These data demonstrated that the ToCsm complex could be reconstituted in vitro in a stable, highly pure form with the recombinant components and the 38‐nt crRNA.
Figure EV1. Identification of the CRISPR‐Cas systems in T. onnurineus and in vitro assembly of the ToCsm complex.
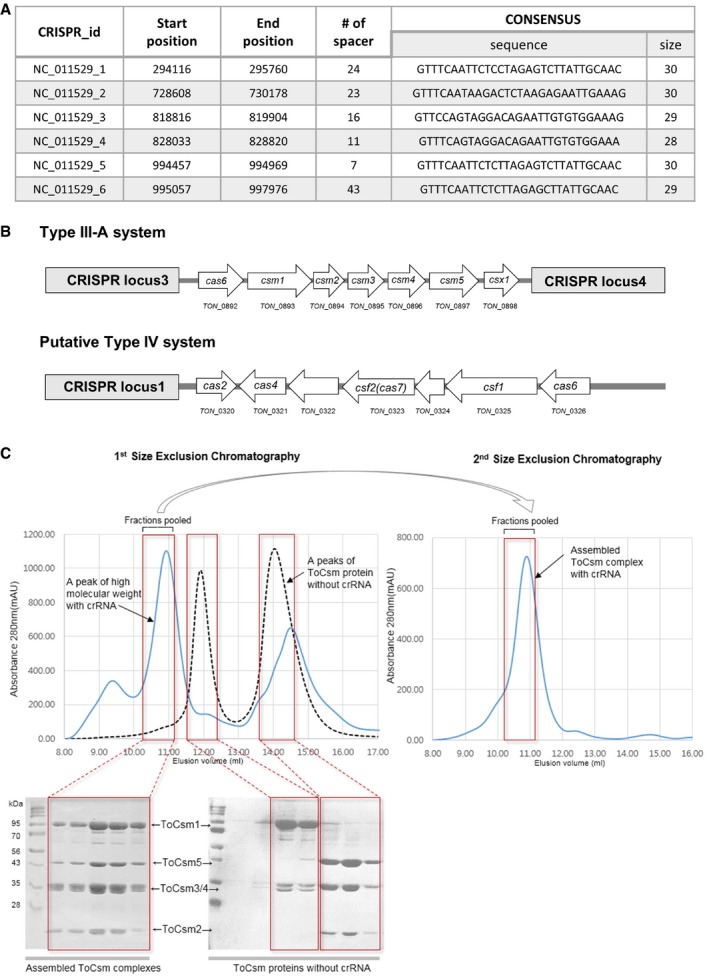
- Six CRISPR loci identified in the genome of T. onnurineus NA1 from the CRISPR database (http://crispr.u-psud.fr/).
- The organization of the cas locus in the Type III and the putative Type IV systems. CRISPR loci 1, 3 and 4 are labelled (grey box). White arrows indicate the ORFs.
- The dotted lines and the blue lines correspond to the elution profile of the ToCsm complex in the absence or presence of the crRNA, respectively. SDS–PAGE analyses of the SEC fractions are shown in the red boxes. The pooled fractions are indicated.
Figure 1. In vitro assembly of the ToCsm complex.
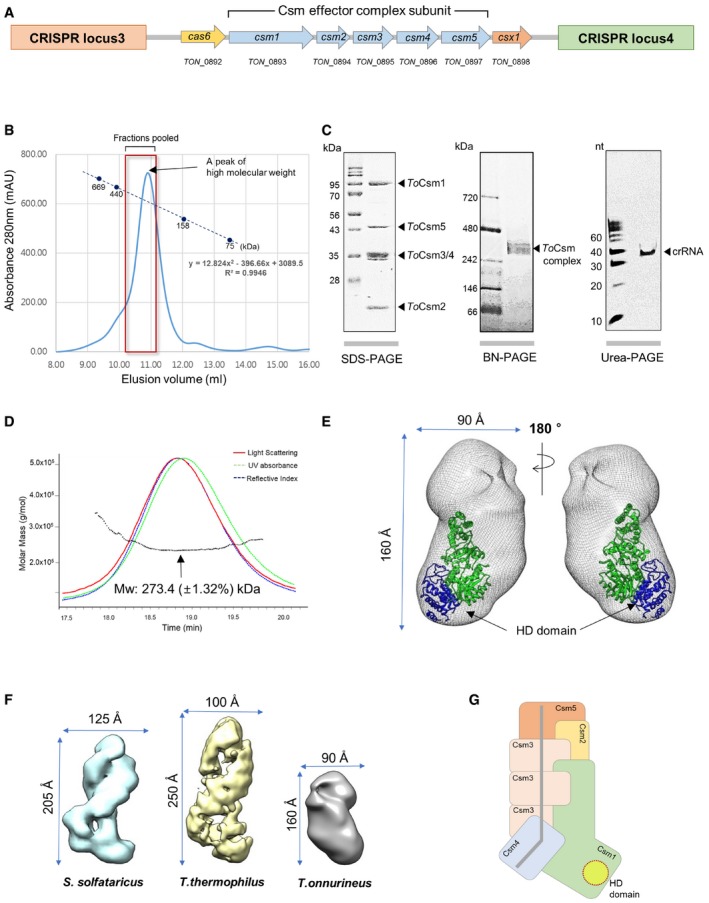
- Organization of the cas locus of the Type III‐A system in T. onnurineus. The ORFs between CRISPR loci 3 and 4 are shown. Blue arrows represent the csm genes encoding the ToCsm complex.
- Size‐exclusion chromatography. The elution profile of the ToCsm complex is presented with the deduced molecular weight (280 kDa). The size markers are thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa) and conalbumin (75 kDa).
- SDS–PAGE, blue native (BN)–PAGE and urea–PAGE analyses.
- Multi‐angle light scattering (MALS) analysis of the ToCsm complex. SEC combined with MALS allowed the calculation of the molecular weight (Mw) distributions (dotted black line).
- Docking of the ToCsm1 structure (PDB ID: 4UW2) into the EM map of the ToCsm complex (EMD‐3454). The HD domain of the ToCsm1 monomer is coloured blue (see Fig EV2D for the docking of the Cmr complex).
- Comparison of the EM structures of the Csm complexes. Lengths and widths of the Csm complexes are shown.
- Schematic representation of the ToCsm complex bound to crRNA. The active sites of the HD domain of the ToCsm1 (dotted circle) and the crRNA (grey line) are highlighted.
To determine the overall structure of the complex, we performed single‐particle electron microscopy (EM) for three‐dimensional reconstruction. We recorded 24 images and extracted 7,644 individual particles (Fig EV2A). Reference‐free 2D image classification allowed an initial appreciation of an elongated‐helical shape of the complex with the longest dimension of 160 Å (Fig EV2B). The projections of the initial model map agreed well with the corresponding 2D averages. The three‐dimensional reconstruction of the ToCsm complex revealed elongated and twisted blobs with dimensions of 160 × 90 × 90 Å. The final resolution of the maps was ~25 Å, calculated at 0.5 Fourier shell correlation (three sigma) (Fig EV2C). The crystal structure of the Cmr complex lacking the Cmr1 subunit (PDB ID: 3X1L) agrees well with the EM density of the ToCsm complex (Fig EV2D) 28. Based on the position of Cmr2, which corresponds to Csm1, the crystal structure of ToCsm1 was fitted into the EM map, which positioned its HD domain at the bottom of the protruding lobe (Fig 1E). The complex was significantly smaller than that of S. solfataricus or T. thermophilus 16, 22 (Fig 1F). The stoichiometry of the ToCsm complex was estimated as 1121334151 based on the EM density fitting and the molecular mass of each component. This is the smallest Csm complex reported to date (Fig 1G). In this complex, three copies of ToCsm3 are likely to form the backbone to shape the twisted helical geometry, as similarly observed in the previously determined structures of the Csm and Cmr complexes 22, 28.
Figure EV2. EM reconstruction of the ToCsm complex.
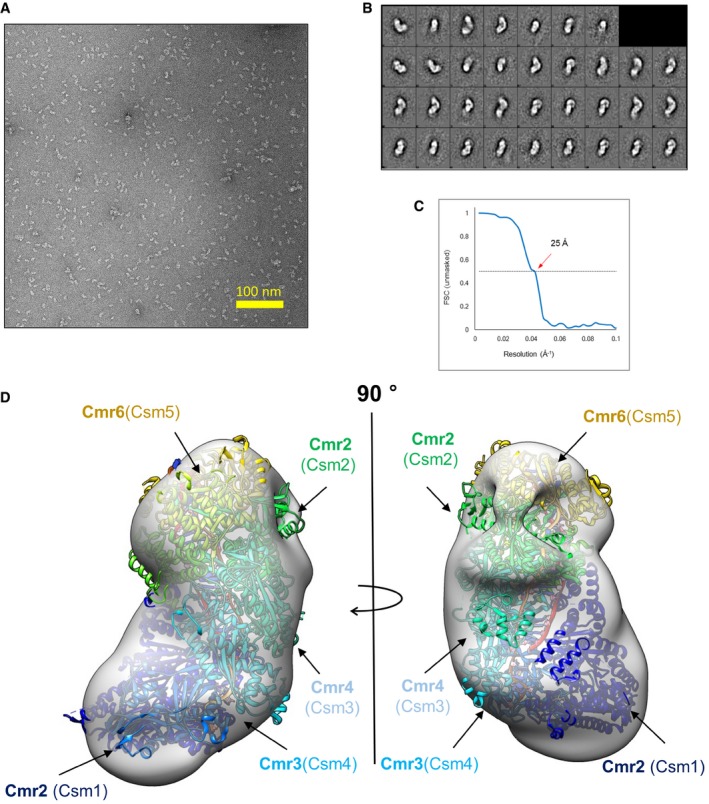
- Raw micrograph.
- Free 2D image classification.
- Fourier shell correlation (FSC) plot. The resolution was estimated at an FSC = 0.5 cut‐off.
- Fit of the Cmr complex structure (PDB ID: 3X1L) into the EM map of the ToCsm complex (EMD‐3454). The Cmr subunits are shown in different colours. The corresponding Csm subunits are indicated in brackets.
RNase activity of the ToCsm complex
Csm complexes bind and cleave RNA transcripts that contain a sequence complementary to the guide region of the bound crRNA (termed the target sequence) 21, 22. To examine the RNA targeting activity of the ToCsm complex, we first synthesized a 40‐nt RNA substrate containing a middle 30‐nt target sequence and a 5‐nt non‐complementary sequence at both ends with a radio‐label at the 5′ end. This ssRNA substrate, termed target RNA, was incubated with the effector complex, and the reaction mixture was analysed in a time course on a denaturing gel. Three major cleaved products were observed that differ from each other by 6 or 12 nt: 30‐, 24‐ and 18‐nt RNA fragments (Figs 2A and EV3A). The cleavage pattern suggested that the target 40‐nt RNA is cleaved predominantly at site 2 or 3. In accordance with previous studies, the ToCsm complex could not cleave an RNA substrate containing a non‐complementary 30‐nt sequence, termed non‐target RNA, under the same reaction conditions, indicating that the sequence complementarity between the target RNA and the crRNA is essential for target RNA cleavage (Fig EV3B). This is consistent with the properties of the Csm complexes from S. epidermidis, S. thermophilus and T. thermophilus 19, 21, 22. Previously, Csm3 was identified as the catalytic subunit responsible for the endoribonuclease activity of the Type III‐A system in S. thermophilus 21. We generated an active‐site mutant of ToCsm3 containing a D36A substitution (ToCsm3D36A) and examined its effect on the RNase activity. The Csm3D36A mutant did not affect the assembly of ToCsm but abolished the RNA cleavage activity of the effector complex. (Fig EV3C). These data show that the ToCsm complex shared the same backbone‐mediated RNA cleavage activity as observed with other Type III effector complexes. Next, to examine whether the RNase activity might be affected by mutations in the ToCsm1 subunit, which are known to abolish the RNA‐coupled DNase activity, we produced ToCsm complexes reconstituted with a ToCsm1 mutant, containing H14A/D15N double substitutions in its HD domain (ToCsm1H14A/D15N, HDm), D587A/D588A double substitutions in its GGDD motif (ToCsm1D587A/D588A, DDm) or H14A/D15N/D587A/D588A quadruple substitutions (ToCsm1H14A/D15N/D587A/D588A, HD/DDm). All three ToCsm1 mutant‐containing ToCsm complexes cleaved the target RNA without a notable difference compared with the wild‐type ToCsm complex, demonstrating that these mutations affecting the DNA targeting activity of ToCsm1 do not affect the RNA cleavage activity of the effector complex (Fig EV3D).
Figure 2. RNase activity and target RNA‐activated ssDNase activity.
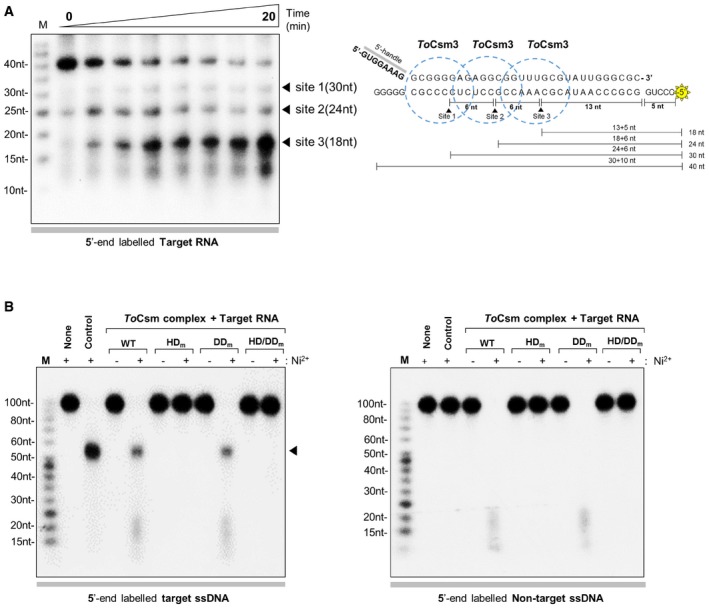
- Cleavage of 40‐nt target RNA by the ToCsm complex in a time course (0, 0.5, 1, 3, 5, 10, 15, 20 min). Triangles indicate the cleaved fragments on a urea polyacrylamide gel (left). Cleavage sites and generated fragments are mapped on the target RNA sequence (right). Dotted circles represent the region covered by the ToCsm3 subunit according to previous reports 19, 21, 22. The RNA substrate was 5′ end‐labelled with 32P (yellow asterisk).
- Target RNA‐activated DNA cleavage by the wild‐type (WT) and mutant ToCsm complex. The mutant ToCsm complex containing a ToCsm1 mutant (HD m, ToCsm1H14A/D15N; DD m, ToCsm1D587A/D588A; or HD/DD m, ToCsm1H14A/D15N/D587A/D588A). Target ssDNA substrate (left) and non‐target ssDNA substrate (right). “Control” indicates the reaction with wild‐type ToCsm complex in the absence of target RNA. The arrow on the left panel indicates the largest cleavage product.
Source data are available online for this figure.
Figure EV3. RNase activity and target RNA‐activated ssDNase activity.
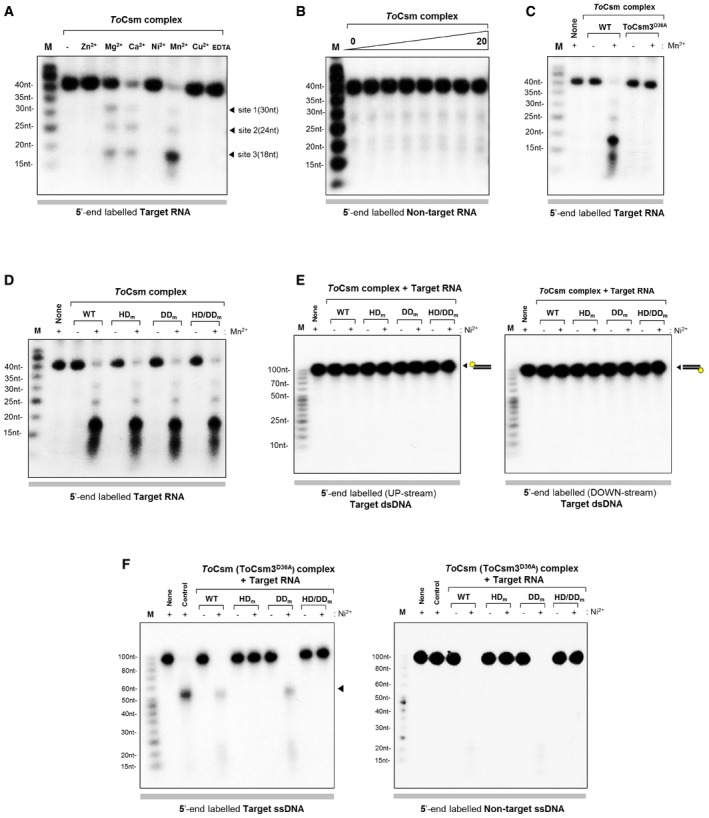
- Metal ion‐dependent RNase activity. The RNase activity was measured in the presence of 5 mM ZnSO 4, MgCl2, CaCl2, NiSO 4, MnCl2, CuSO 4 or EDTA.
- Cleavage assay. Non‐target RNA was reacted with the ToCsm complex in the presence of 5 mM Mn2+.
- Cleavage of target RNA by wild‐type (WT) and mutant ToCsm complex (ToCsm3D36A). The metal ion cofactor Mn2+ was included in all of the reactions.
- Cleavage of target RNA by the wild‐type ToCsm complex and the mutant ToCsm complex containing a ToCsm1 mutant (HD m, ToCsm1H14A/D15N; DD m, ToCsm1D587A/D588A; or HD/DD m, ToCsm1H14A/D15N/D587A/D588A).
- Cleavage assay. Target dsDNA was reacted with the wild‐type or mutant ToCsm complex containing the ToCsm1 mutant (HD m, ToCsm1H14A/D15N; DD m, ToCsm1D587A/D588A; or HD/DD m, ToCsm1H14A/D15N/D587A/D588A) in the presence of the target RNA. Two dsDNA substrates were labelled at one of the two 5′ ends, respectively.
- Cleavage assay. Target ssDNA (left) and non‐target ssDNA (right) were incubated with the ToCsm complex containing ToCsm3D36A and wild‐type or mutant ToCsm1 (HD m, DD m or HD/DD m) in the presence of the target RNA. “Control” represents the reaction with the wild‐type ToCsm complex in the absence of the target RNA. The arrow in the left panel indicates the largest cleavage fragment.
Source data are available online for this figure.
Target RNA‐activated DNase activity of the ToCsm complex
We previously reported that the ToCsm1 subunit alone degraded single‐stranded DNA (ssDNA) non‐specifically 40. However, when the ToCsm1 subunit was assembled in the ToCsm complex, its nuclease function was inhibited, and the complex could not degrade non‐specific ssDNA substrate. Recently, the Csm complex from S. thermophilus and the Cmr complex from T. maritima or P. furiosus were reported to degrade ssDNA only in the presence of a complementary RNA transcript 30, 31, 32. To test whether the target RNA reactivates the DNase activity of the ToCsm complex, we prepared 5′ radio‐labelled linear 100‐nt ssDNA substrates containing a sequence either complementary or non‐complimentary to the bound crRNA, termed target ssDNA and non‐target ssDNA, respectively. First, the ToCsm complex was pre‐incubated with the synthesized 40‐nt RNA, the same substrate used for the RNA cleavage assay, in the absence of any divalent metal ions. Then, the ssDNA substrate was added to the pre‐incubated ToCsm complex in the presence of Ni2+ ion, a known metal cofactor for the nuclease activity of ToCsm1 40. The ToCsm complex degraded the ssDNA substrates containing either the complementary sequence or the non‐complementary sequence. The ToCsm complex containing ToCsm1 (HDm) or ToCsm1 (HDm/DDm) did not show any cleavage activity towards the ssDNA substrate, whereas the ToCsm complex containing ToCsm1 (DDm) degraded the substrate in the presence of the 40‐nt target RNA (Fig 2B). In comparison, the ToCsm complex did not degrade dsDNA substrates at all, regardless of the presence of a complementary sequence (Fig EV3E). To examine whether this DNase activity could be affected by the active site mutation (D36A) on the ToCsm3 subunit that abolishes its RNA cleavage activity, we reconstituted the ToCsm complex containing ToCsm3D36A and measured the DNase activity. The resulting mutant ToCsm complex cleaved the ssDNA substrate without a notable difference from the wild‐type complex, indicating that the RNase activity of the ToCsm3 subunit did not affect the DNA cleavage activity of the ToCsm complex (Fig EV3F). Together, these results demonstrate that the active site located at the HD domain of the ToCsm1 subunit exhibits target RNA‐activated nuclease activity towards ssDNA but not towards dsDNA. This non‐sequence‐specific DNase activity in the presence of the target RNA would completely degrade the ssDNA substrate.
Target RNA‐independent binding of ssDNA via crRNA
In the target RNA‐activated DNase assay, we noted that the ToCsm complex limitedly degraded linear ssDNA substrate containing a target sequence, producing a large uncleaved fragment (Fig 2B). In contrast, the complex degraded ssDNA substrate without a target sequence into much smaller fragments. An observation of the different cleavages of the two substrates led us to examine whether, in the absence of a target RNA, the ToCsm complex may directly target ssDNA substrate containing the complementary sequence. We synthesized 5′ radio‐labelled linear ssDNA, RNA and dsDNA, each containing a 30‐nt complementary sequence inserted in the centre, and analysed their binding to the ToCsm complex by an electrophoretic mobility shift assay (EMSA). To avoid degradation by metal‐dependent DNase or RNase activity, a metal chelator was added to the reaction buffer. The ToCsm complex significantly shifted the target ssDNA, indicating a direct binding unaided by a target RNA. This is in a sharp contrast with ToCsm1‐Csm4, ToCsm2‐Csm5 subcomplexes and ToCsm3 protein, which did not result in a mobility shift of these nucleic acids (Fig EV4A and B). The dsDNA does not appear to interact with the ToCsm complex, as no shift was observed regardless of the presence or absence of the complementary sequence (Fig EV4C). The ToCsm complex bound to the target ssDNA and RNA with a dissociation constant (K D) of approximately 1.5 nM (ssDNA) and 1.3 nM (RNA), respectively (Fig 3A–C). An ssDNA version containing a non‐complementary sequence exhibited only a marginal shift at high concentration of the ToCsm complex (Fig 3D and E). The ToCsm complexes containing the ToCsm1 (HDm), ToCsm1 (DDm) or ToCsm1 (HD/DDm) mutant interacted with the target ssDNA without a notable difference compared with the wild‐type ToCsm complex, demonstrating that these mutations in ToCsm1 do not affect the interaction of the effector complex with the target nucleotide (Fig EV4D). A significantly diminished mobility shift of the target ssDNA was observed by pre‐incubation of the ToCsm complex with the target RNA. In contrast, pre‐incubation of the complex with a non‐target RNA did not affect the mobility shift (Fig 3F and G). EMSAs performed in the presence of competing nucleotides indicated that the ToCsm complex binds the target ssDNA and RNA, indistinguishably (Fig 3H and I). Together, these data indicate that the interaction of the ToCsm complex with the target ssDNA is mediated by the bound crRNA via complementary base paring between its guide sequence and the target sequence in the ssDNA.
Figure EV4. EMSA analysis of the binding properties of the ToCsm complex.
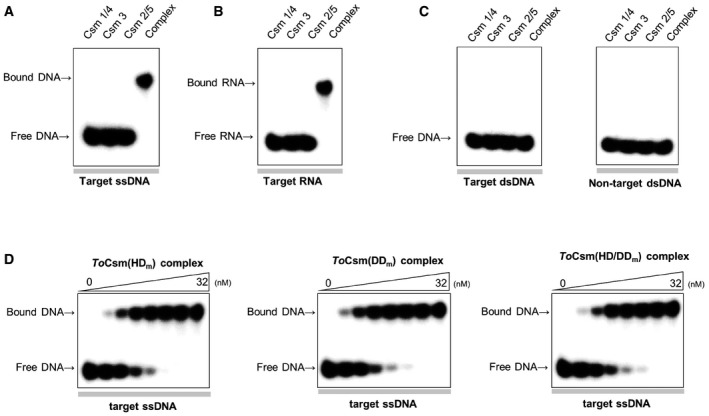
-
A–CAnalysis of the interaction between the ToCsm protein and target ssDNA (A), target RNA (B) and dsDNA (C).
-
DAnalysis of the interaction between the mutant ToCsm complex and target ssDNA. Mutant ToCsm complex containing the ToCsm1 mutant (HD m, DD m and HD/DD m).
Source data are available online for this figure.
Figure 3. Binding properties of ToCsm complex to target ssDNA and RNA .
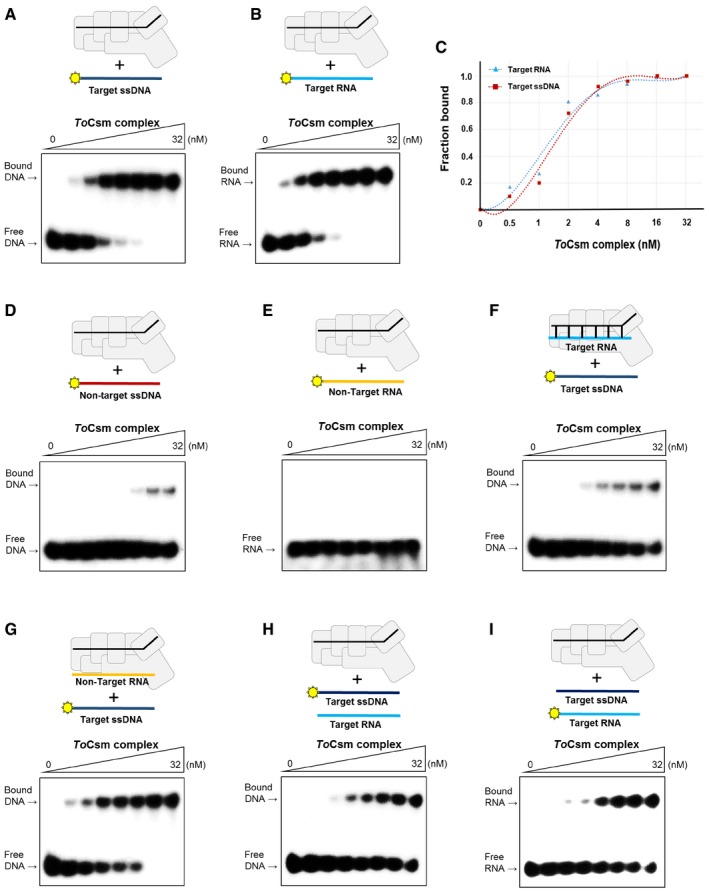
-
A, BEMSA analysis of the interaction between the ToCsm complex and the target ssDNA (A) or the target RNA (B).
-
CBinding affinity of ToCsm complex for target ssDNA and target RNA. The fraction of target bound to the titrating ToCsm complex in (A) and (B). The KD value obtained from the curve fitting is indicated. ToCsm complex to target ssDNA KD ˜1.5 ± 0.37 nM; ToCsm complex to target RNA KD ˜1.3 ± 0.32 nM.
-
D, EEMSA analysis of the interaction between the ToCsm complex and the non‐target ssDNA (D) or the non‐target RNA (E).
-
F, GInteraction between the labelled target ssDNA and the ToCsm complex that was pre‐incubated with unlabelled target RNA (F) or unlabelled non‐target RNA (G) at a molar ratio of 1:1.
-
H, ICompetition binding of the labelled target ssDNA and the unlabelled target RNA (H) or the unlabelled target ssDNA and the labelled target RNA (I) to the increasing amount of the ToCsm complex.
Source data are available online for this figure.
Target RNA‐independent cleavage of ssDNA
To examine whether, in the absence of a target RNA, the ToCsm complex can cleave ssDNA, which contains a target sequence, we prepared two different crRNAs: M13 crRNA containing a sequence complementary to the circular M13mp18 ssDNA and 3.3 crRNA containing a non‐complementary sequence. The ToCsm complex assembled with M13 crRNA or 3.3 crRNA was incubated with M13mp18. The ToCsm complex loaded with M13 crRNA completely degraded M13mp18, whereas the effector complex loaded with 3.3 crRNA did not. Under the same reaction conditions, the M13 crRNA‐loaded ToCsm complex did not degrade ΦX174, a circular ssDNA but with a different sequence (Fig 4A). These data clearly indicate that the ToCsm complex possesses a crRNA‐guided nuclease activity in the absence of a target RNA. To locate the active site of the ToCsm complex responsible for the nuclease activity, we performed cleavage assay with the ToCsm complex containing the ToCsm1 (HDm), ToCsm1 (DDm) or ToCsm1 (HD/DDm) mutant. The ToCsm1 (HDm)‐ or ToCsm1(HD/DDm)‐containing ToCsm complex could not degrade M13mp18, while the effector complex containing ToCsm1 (DDm) completely degraded the substrate (Fig 4A). Mn2+ or Ni2+ supported the nuclease activity, while Mg2+ or Ca2+ (required for the RNase activity of the ToCsm3 subunit) did not (Fig 4B). In addition, the effector complex containing ToCsm3D36A cleaved the ssDNA substrate without a notable difference from the wild‐type complex (Fig 4C). These results demonstrate that the HD domain of the ToCsm1 subunit is the active site for the target RNA‐independent ssDNA cleavage.
Figure 4. Target RNA‐independent cleavage of target ssDNA .
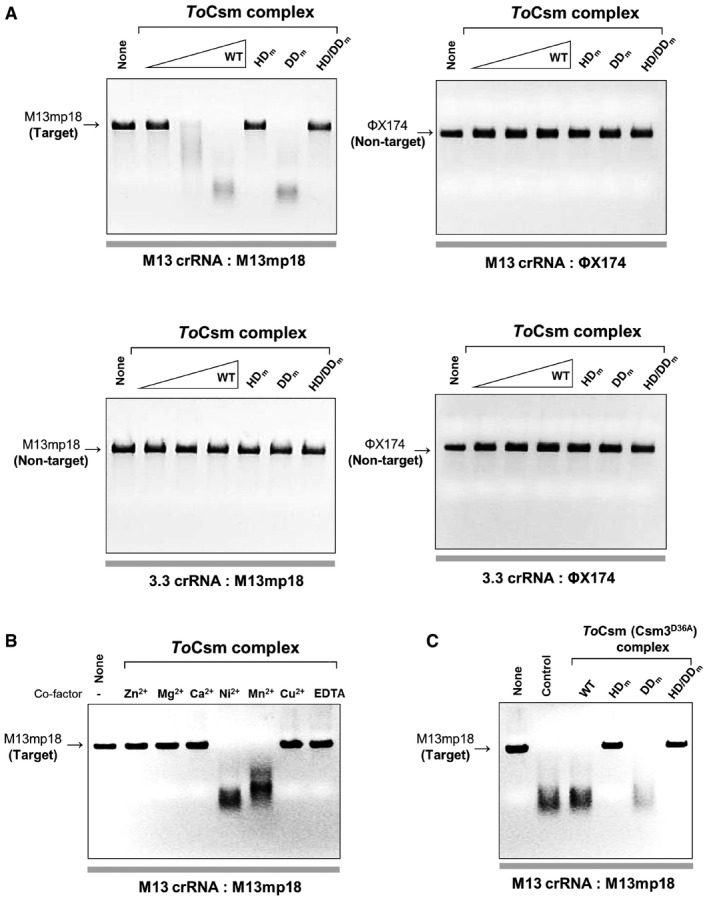
- Cleavage of circular ssDNA by the ToCsm complex loaded with the M13 crRNA or 3.3 crRNA. The ToCsm complexes containing a ToCsm1 mutant are indicated as in Fig 2B. M13 crRNA contains a sequence complementary to M13mp18. The crRNA‐DNA substrate pairs are indicated at the bottom of the polyacrylamide gel. The ToCsm complexes were incubated with the ssDNA substrate in the presence of 5 mM Ni2+.
- Metal ion‐dependency test. The ToCsm complex loaded with M13 crRNA was reacted with M13mp18 ssDNA in the presence of the indicated metal ions.
- Cleavage of circular ssDNA by the ToCsm complexes containing ToCsm3D36A and wild‐type (WT) or mutant ToCsm1 (HD m, DD m or HD/DD m). “Control” represents the reaction with the wild‐type ToCsm complex.
Source data are available online for this figure.
Identification of the target RNA‐independent ssDNA cleavage sites
To gain further insight into the mechanism of the target ssDNA cleavage in the absence of a target RNA, we synthesized a linear radio‐labelled 100‐nt ssDNA containing a target sequence in the middle, a 10‐nt non‐complementary sequence at the 5′ end and a 60‐nt non‐complementary sequence at the 3′ end of the DNA. Upon reaction with the ToCsm complex assembled with the M13 crRNA, this ssDNA substrate, termed target ssDNA, was cleaved to yield a 52–54‐nt fragment (Figs 5A and EV5A). Cleavage occurred at the 3′ flanking side of the complementary segment, approximately 12–14 nt downstream from position 1 of the guide region of the crRNA (Fig 5B). The ToCsm complex containing ToCsm1 (HDm) or ToCsm1 (HDm/DDm) did not show any cleavage activity towards the 5′ radio‐labelled target ssDNA, whereas the ToCsm complex containing ToCsm1 (DDm) degraded this substrate (Fig 5C). In contrast, ssDNA substrate containing a non‐complementary sequence or dsDNA was not cleaved (Fig EV5B and C). We noted that the same 100‐nt ssDNA, but with a 3′ radio‐labelled, exhibited a near complete degradation, producing 10–25‐nt fragments (Fig 5D). The ToCsm complex containing ToCsm3D36A exhibited the same DNA cleavage pattern as the wild‐type effector complex (Fig 5E). These data suggest that the ToCsm complex cleaves the target ssDNA substrate at the 3′ side of the complementary sequence and degrades the 3′ end fragment.
Figure 5. ssDNA targeting mechanism by the ToCsm complex.
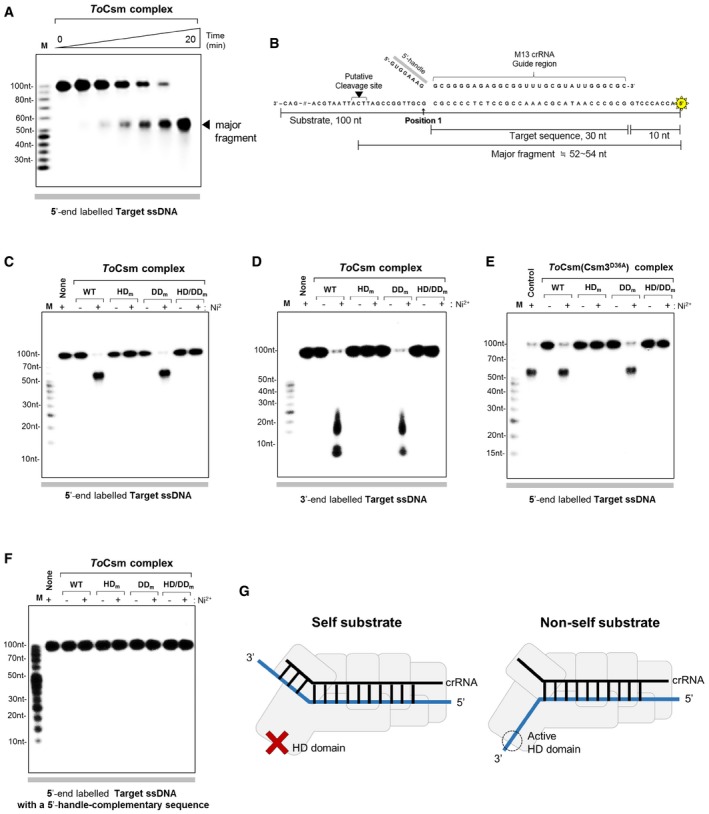
- Cleavage of linear target ssDNA by the ToCsm complex in a time course (0, 0.5, 1, 3, 5, 10, 20 min). The radio‐labelled substrate is indicated at the bottom of the polyacrylamide gel. The triangle indicates the major cleaved fragment.
- The major fragment is mapped on the sequence of the linear ssDNA substrate. Position 1 denotes the 3′ end of the target sequence. 5′ radio‐labelling (asterisk) and the approximate length of the major fragment are indicated.
- Cleavage of 5′ radio‐labelled linear target ssDNA by the wild‐type (WT) and mutant ToCsm complex. The ToCsm complexes containing a ToCsm1 mutant are indicated as in Fig 2B.
- Cleavage of 3′ radio‐labelled linear target ssDNA by the wild‐type (WT) and mutant ToCsm complex.
- Cleavage of 5′ radio‐labelled linear target ssDNA by the ToCsm complexes containing ToCsm3D36A and wild‐type or mutant ToCsm1. “Control” represents the reaction with the wild‐type ToCsm complex.
- Cleavage of 5′ radio‐labelled linear target ssDNA containing the 8‐nt sequence complementary to the 5′ handle of the crRNA.
- A model for the discrimination of self and non‐self ssDNA by the ToCsm complex. The non‐self substrate ssDNA does not contain a sequence complementary to the 5′ handle of the crRNA (right). The putative closed and open states of the active site of the HD domain are indicated by a cross and a dotted circle, respectively.
Source data are available online for this figure.
Figure EV5. DNA cleavage by the ToCsm complex.
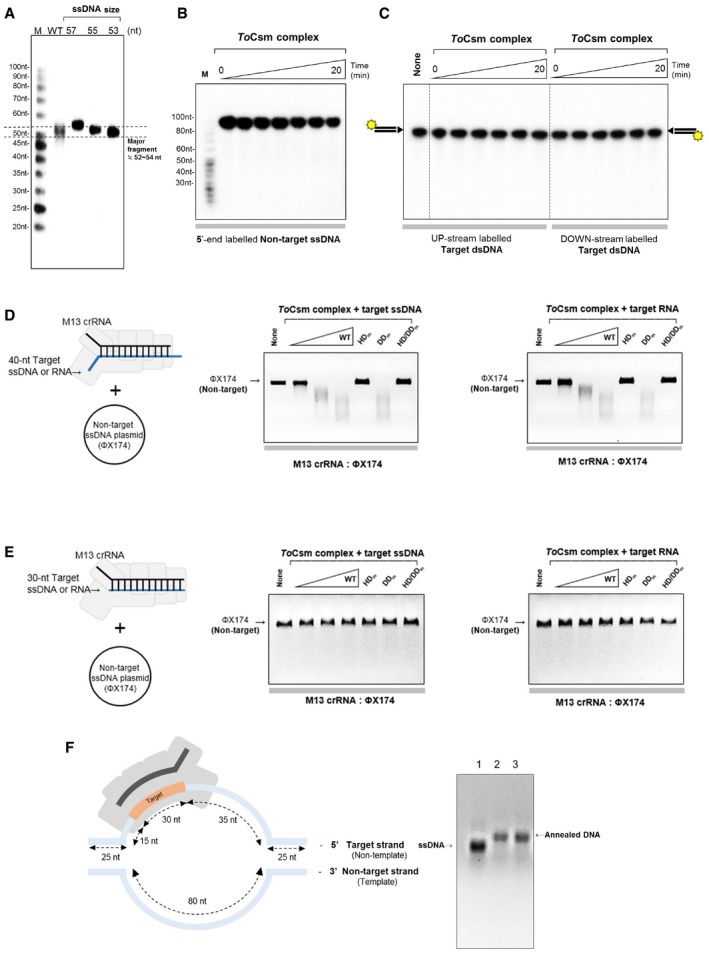
-
ASize analysis of the major fragment generated from the target ssDNA. WT represents the fragment produced by the wild‐type ToCsm complex. “ssDNA size” indicates synthetic oligonucleotides (57, 55 and 53 nt) analysed by 15% denaturing urea–PAGE. The dotted line indicates the size of the major product.
-
BCleavage assay. 5′ radio‐labelled non‐target ssDNA was reacted with the ToCsm complex in a time course.
-
CCleavage assay. Two target dsDNA substrates (with identical sequence), each radio‐labelled at one of the two 5′ ends, were reacted with the ToCsm complex.
-
D, ETrans‐cleavage of non‐target ssDNA by the ToCsm‐target ssDNA (RNA) complex. ToCsm complex was incubated with target ssDNA or RNA (mixed molar ratio; 1:1) and then complex with increasing concentration react circular non‐target ssDNA. The ToCsm complexes containing a ToCsm1 mutant are indicated as in Fig 2B. The designed 40‐nt target ssDNA (RNA) contains a 30‐nt target sequence in the centre (D) and designed 30‐nt target ssDNA (RNA) only contains a 30‐nt target sequence (E).
-
FSchematic drawing of an artificial DNA duplex bound to the ToCsm complex. The designed duplex contains 25‐nt complementary sequence at both ends. The target strand contains a 30‐nt target sequence in the centre, and the non‐target strand contains an 80‐nt sequence not complementary to the target strand. The two DNA strands were annealed and analysed on a 2% agarose gel. Lane 1: 130‐nt target ssDNA, lane 2: 130‐bp annealed DNA and lane 3: 130‐bp annealed DNA containing a 5′‐handle‐complementary sequence. The target strand and the non‐target strand correspond to the non‐template strand and the template strand of a transcription bubble, respectively.
Source data are available online for this figure.
Suppression of the target RNA‐independent ssDNA cleavage by a repeat‐derived sequence
The repeat‐derived 5′ handle of the bound crRNA was reported to play a key role in self versus non‐self discrimination in the Type III system‐mediated immunity 37. In a trans‐cleavage assay where ssDNA substrates were employed as the substrates for the StCsm effector complex loaded with a target RNA, it was shown that the presence of a 3′ target sequence‐flanking region complementary to the 5′ handle of the bound crRNA prevented cleavage of the ssDNA substrates 30. To investigate whether the 3′ flanking region of the target sequence affects the target RNA‐independent DNA cleavage activity of the ToCsm complex, we employed an ssDNA substrate that contained a 3′ flanking sequence complementary to the 5′ handle of the bound crRNA. Notably, this ssDNA substrate was not degraded at all by the wild‐type ToCsm complex as well as the ToCsm complexes containing a defective ToCsm1 mutant (Fig 5F), suggesting that the presence of a 5′‐handle‐complementary sequence in ssDNA substrates blocks this nuclease activity of the ToCsm complex (Fig 5G). We next performed a trans‐cleavage assay where the ToCsm complex was preloaded with a short 40‐nt ssDNA or RNA containing a target sequence that is complementary to the guide sequence of the bound crRNA. These nucleotides shared the same sequence and contained a 5‐nt 3′ flanking region which is not complementary to the 5′ handle of the bound crRNA. Both the ToCsm complexes degraded the non‐target ΦX174 plasmid DNA (Fig EV5D). It is noted that the ToCsm complex containing ToCsm1 (HDm) or ToCsm1 (HD/DDm) could not degrade ΦX174, while the effector complex containing ToCsm1 (DDm) degraded this substrate. Intriguingly, ToCsm complex loaded with a target ssDNA or RNA lacking the 3′ flanking sequence could not degrade ΦX174 plasmid DNA (Fig EV5E). These data demonstrate that the nuclease activity of the HD domain is activated by nucleotide binding to the ToCsm complex, but the bound nucleotide should have a 3′ target sequence‐flanking segment that does not base pair with the 5′ handle of the bound crRNA.
Selective cleavage of partially unwound DNA duplex
Next, we examined whether the ToCsm complex could cleave partly unwound DNA duplex containing a target sequence. We prepared an artificial bubble DNA substrate containing 80‐nt unwound strands in the centre flanked by 25‐nt double‐stranded segments at both ends. In this DNA duplex, one strand contained a 30‐nt target sequence in the middle of the single‐stranded region (target strand), and the other strand (non‐target strand) contained an 80‐nt sequence not complementary to the target strand. This DNA duplex mimics a “transcription bubble” (Fig EV5F). In order to trace reaction products, two versions of the DNA duplex were prepared that were radio‐labelled at the 5′ end of either the target strand or the non‐target strand. Upon reaction with the ToCsm complex, the target strand was cleaved to yield approximately 85‐nt long fragments (Fig 6A), while the non‐target strand was not cleaved at all (Fig 6B). We also prepared a DNA duplex in which the target strand contained a sequence complementary to both the 5′ handle and the guide region of the crRNA and found that the ToCsm complex did not cleave the target strand at all (Fig 6C). Conclusively, the ToCsm complex is able to selectively cleave the target strand in partially unwound DNA that exposes the target sequence on this strand in the absence of a target RNA. This nuclease activity, however, was completely blocked by the presence of the 8‐nt 5′‐handle‐complementary sequence next to the target sequence. Likely, this inhibitory mechanism prevents the ToCsm complex from cleaving self DNA when the CRISPR loci are transcribed 19, 37.
Figure 6. Cleavage of target ssDNA in DNA duplex.
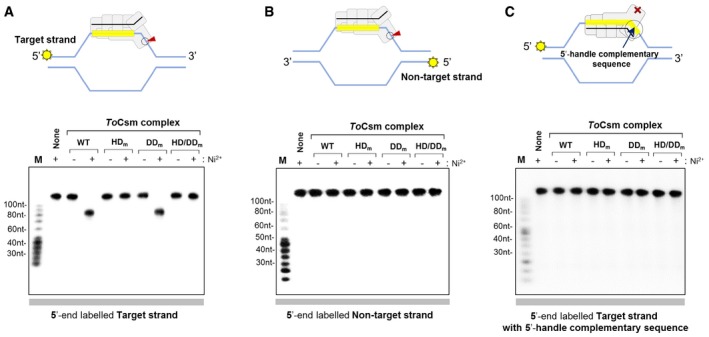
-
A–CCleavage of the target strand (A), the non‐target strand (B) and the target strand with the 5′‐handle‐complementary sequence (C) in DNA duplex. The bubble‐form DNA duplex (shown in cartoons) was generated by a central non‐complementary region in the two DNA strands (see text for details). The indicated ToCsm complexes were incubated with the 5′ end‐labelled duplex DNA, and the reaction products were analysed by autoradiography. The cartoons depict the expected cleavage or non‐cleavage of the target strand, depending on the presence or absence of the 5′‐handle‐complementary sequence. The red arrow indicates the cleavage at the active site of the HD domain of the ToCsm1 subunit.
Source data are available online for this figure.
Discussion
We reconstituted a ToCsm complex, which is conceivably the same as the endogenous functional form, as it possesses the RNase activity and the target RNA‐activated non‐sequence‐specific DNase activity observed for other Csm complexes 19, 23, 30. This effector complex was found to recognize a target sequence in ssDNA via the bound crRNA and cleave the substrate via the HD domain of the ToCsm1 subunit in a target RNA‐independent manner. Although the activated HD in the ToCsm complex upon binding to the 3′ flanking sequence of target ssDNA is able to cleave ssDNA substrate in a trans‐acting manner (Fig EV5D), the degradation of the target strand in the bubble‐shaped DNA duplex indicates a direct cis‐acting activity of this effector (Fig 6A and B). Given the observation that the Type III‐A system of S. epidermis tolerates lysogenization by temperate phages, but prevents their lytic phase 33, Csm complexes seem to function during transcription. Further investigation is required to know whether the ToCsm complex may target ssDNA in the transcription bubble and whether the cis‐acting activity that we observed in in vitro may be functionally relevant in cells.
The novel ssDNA targeting activity of the ToCsm complex, which needs further investigation in cells, provides a more complete picture of how transcription is coupled to the destruction of non‐self DNA. One effector complex activated by the RNA transcript produced during the transcription process non‐specifically cleaves any exposed DNA strands of the transcription bubble, and another effector complex activated by binding to the target sequence on the non‐template strand may cleave this strand directly (Fig 7A) 41. The destruction of the foreign DNA by target RNA‐activated ssDNase mechanism may not be sufficient alone; during the transcription of foreign DNA, the Type III effector complex, which cleaves ssRNA faster than it cleaves ssDNA 30, may dissociate from the tethering RNA transcript without cleaving the single‐stranded region of the transcription bubble. Notably, the delineated target RNA‐independent ssDNA cleavage could explain previously reported observations. First, this activity, which exclusively targets the non‐template DNA strand of DNA duplex, in contrast with the non‐discriminative target RNA‐activated ssDNase activity, explains why crRNA complementary to the non‐template DNA provides efficient immunity and why a spacer sequence on the non‐template strand is preferentially selected for cleavage during transcription 19, 33. Second, the location of the HD domain relative to the target sequence binding site on the ToCsm complex explains the previously identified cleavage sites on the non‐template DNA strand located at the 3′ side of the guide region of the bound crRNA (Fig 7B) 19. By the RNA transcript‐dependent ssDNAase activity, the non‐template strand, in principle, could be cleaved at random positions with respect the target sequence on it. However, it was observed that the non‐template strand cleavage mediated by the Type III‐A system of S. epidermis occurs only at the 3′ flanking side of the target sequence 19, which is consistent with the pattern of cleavage by the target RNA‐independent ssDNase activity observed in this study. Finally, the blocking of this nuclease activity by the presence of the 5′‐handle‐complementary sequence next to the target sequence provides a rationale for the previously observed protection of self DNA from cleavage by the Type III system 19, 37. The RNA transcript‐independent cleavage of target ssDNA by the ToCsm complex may be pivotal, or at least it could augment the previously known target RNA‐activated DNase activity for the efficient destruction of foreign DNA molecules in cells. Alternatively, the ToCsm complex could play a specific role against ssDNA viruses, rather than acting on genomic dsDNA. Recently, some Cas9s were reported to bind to ssDNA with higher affinity than to dsDNA, demonstrating a significant divergence of the CRISPR‐Cas systems 42.
Figure 7. Model for ssDNA targeting by the ToCsm complex.
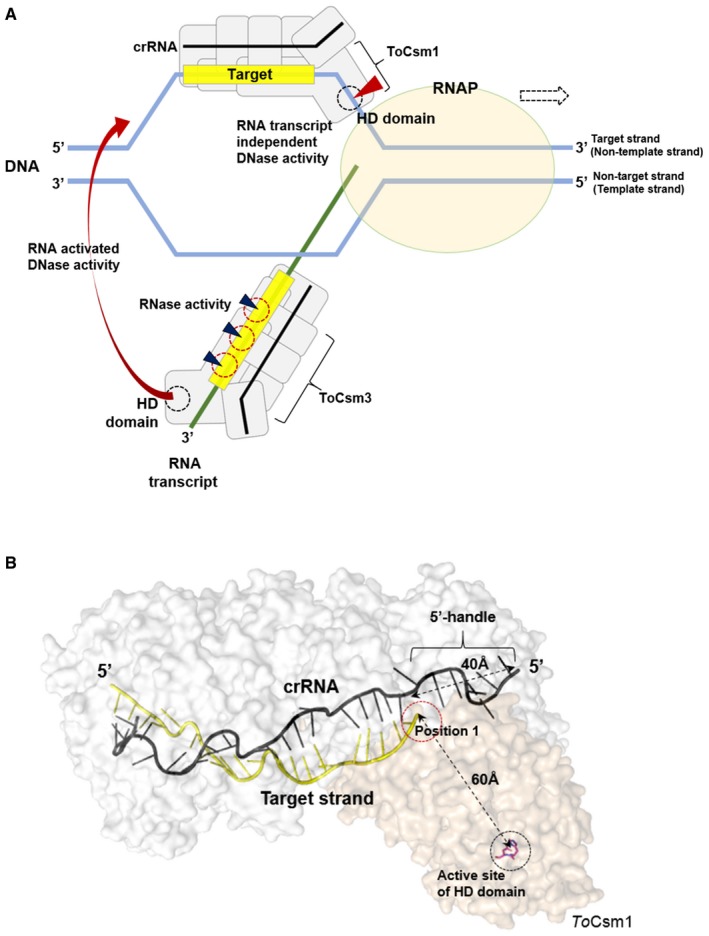
- Transcription‐coupled cleavages of foreign dsDNA and RNA by the ToCsm complex. RNAP stands for RNA polymerase. The blue triangle indicates the cleavage of the RNA transcript (green line) by the ToCsm3 subunit. Binding of the RNA transcript to the ToCsm complex activates the DNase activity of the HD domain, which cleaves the template or non‐template strands. Another ToCsm complex directly recognizes the exposed target sequence on the non‐template strand mediated by the bound crRNA (black line). Without a need for an RNA transcript, the complex cleaves the non‐template strand mediated by the ToCsm1 subunit (red triangle).
- The structural model of the ToCsm Complex with target ssDNA. The distance between position 1 of the target region and the active site of the HD domain is comparable to the length of nucleotides from the sequence of position 1 to the cleavage site. The ToCsm1 structure (PDB ID: 4UW2, wheat) was modelled to ToCsm complex EM map (EMD‐3454) and the Cmr complex structure (PDB ID: 3X1L, white) based on the position of Cmr2 to highlight the active site of the HD domain.
In summary, we reconstituted a highly pure and functionally active Type III effector complex, the smallest Csm complex observed so far with a minimum combination of subunits. Type I, II and III systems may share a common mechanism of crRNA‐mediated target DNA sequence recognition, although the mechanisms of target cleavage are different from each other. The RNA activation‐independent ssDNA targeting activity highlights the versatility of the Type III effector complex in T. onnurineus for the efficient and complete silencing of foreign nucleic acids.
Materials and Methods
Cloning and protein purification
The genes encoding ToCsm1–ToCsm5 (Ton_0893–0897) were amplified by PCR from T. onnurineus NA1 genomic DNA. The ToCsm1 and ToCsm4 genes were cloned into the BamHI/HindIII and the NdeI/KpnI sites of the pRSFDuet‐1 vector (Novagen), respectively. The ToCsm3 gene was inserted into the NcoI/SalI sites of pETDuet‐1 (Novagen), and the ToCsm2‐ToCsm5 genes were inserted into the NcoI/SalI and the NdeI/XhoI sites of pACYCDuet‐1 (Novagen). These Csm proteins were produced in the E. coli strain BL21‐CodonPlus (DE3) at 37°C by induction with 1 mM IPTG. E. coli cells were harvested and lysed. Following centrifugation, the supernatant was incubated in a hot water bath at 75°C for 10 min to remove E. coli proteins. After clarification, the supernatant was applied to a His‐Trap HP column (GE Healthcare) and a Superdex 200 increase 10/300 column (GE Healthcare) with a final elution buffer composed of 50 mM Tris–HCl (pH 8.0), 500 mM NaCl, 5 mM β‐mercaptoethanol and 5% glycerol.
In vitro assembly of the ToCsm complex and SEC‐MALS
Synthetic crRNAs were purchased from Integrated DNA Technologies. After initial optimization, the purified ToCsm1‐Csm4, ToCsm3, ToCsm2‐Csm5 and crRNA were mixed in a molar ratio of 1:3:1:1 and then incubated at 60°C for 20 min. After centrifugation at 10,000 g for 5 min, the protein sample was applied to a Superdex 200 increase 10/300 column (GE Healthcare) and equilibrated with 50 mM Tris–HCl (pH 8.0), 250 mM NaCl and 5% glycerol. Fractions containing the ToCsm complex were pooled and applied again to the Superdex 200 increase 10/300 column. The assembled samples and the size marker (GenDEPOT, Novex and Affymetrix) were analysed on 15% SDS gel, 4–12% BN–PAGE gel and 12% polyacrylamide denaturing 8 M urea gels. SEC–MALS was performed using a WTC‐050S5 SEC column with an in‐line DAWN HELIOS II system and an Optilab T‐rEX differential refractometer (Wyatt). The ToCsm complex (100 μl of 1 mg/ml) was dissolved in a buffer solution composed of 50 mM Tris–HCl (pH 8.0), 250 mM NaCl, 3 mM β‐mercaptoethanol. Data were collected and analysed using ASTRA 6 (Wyatt). The ToCsm complexes containing a mutant Csm1 subunit were reconstituted and purified in the same manner as the wild‐type ToCsm complex. The purified complex was frozen in liquid nitrogen and stored at −80°C.
Site‐directed mutagenesis of ToCsm1 and ToCsm3
The ToCsm1 mutants (HDm, ToCsm1H14A/D15N; DDm, ToCsm1D587A/D588A; and HD/DDm, ToCsm1H14A/D15N/D587A/D588A) and the ToCsm3 mutant (ToCsm3D36A) were generated with the site‐directed mutagenesis kit (Enzynomics). The mutations were verified by sequencing the respective genes (Solgent).
Electron microscopy
The ToCsm complex was negatively stained with 2% (w/v) uranyl acetate for 1 min on 400‐mesh carbon grids. Images were collected at a magnification of 50,000× at a final sampling of 2.07 Å/pixel with a defocus value of 0.5–1.5 μm on a 4 × 4 K CCD camera (Tietz Vieo and imaging Processing System) attached to Jeol JEM2100F field emission gun transmission electron microscope operated at 200 kV. Data processing was performed using the EMAN2 program 43. In total, 7,644 particles were selected and used to generate reference‐free 2D class averages. The initial model was built from 34 selected classes using the e2initialmodel.py program. Then, the model was further iteratively refined by the e2refine_easy.py program with a low‐pass filter (cut‐off = 0.04). The resolution was estimated as 25 Å from the last iteration of the unmasked 0.5 FSC curve. The 3D map of ToCsm has been deposited in the PDBe; EMD‐3454. The crystal structure of Cmr complex (PDB ID: 3X1L) into the refined 3D map of ToCsm (EMD‐3454) was docked by the “Fit in map” function of Chimera software 44.
Preparation of substrates
The RNA and DNA substrates were purchased from Integrated DNA Technologies and Bioneer, respectively (Table EV1). The DNA and the RNA substrates were 5′‐labelled with γP32‐ATP (Perkin Elmer) and T4 PNK enzyme (Enzynomics) at 37°C for 20 min, respectively. DNA substrates were 3′‐labelled with αP32‐dATP (Perkin Elmer) and TDT enzyme (Enzynomics) for 40 min at 37°C. Circular ssDNA of M13mp18 and ΦX174 Virion was purchased (New England BioLabs). To generate the bubble shape DNA, each oligonucleotide was mixed at a 1:1 molar ratio in reaction buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl2), heated to 95°C for 5 min and slowly cooled to room temperature for annealing.
Electrophoretic mobility shift assay
DNA/RNA binding assays were performed by incubating varying amounts (0, 0.5, 1, 2, 4, 8, 16, 32 nM) of the ToCsm complexes at 55°C for 20 min with 1 nM γP32‐ATP and 5′‐labelled 40‐nt DNA and/or 40‐nt RNA in binding buffer composed of 30 mM Tris–HCl (pH 8.0), 5% glycerol, 0.1 mg/ml BSA and 0.5 mM EDTA. The samples were loaded directly onto native 8% (w/v) polyacrylamide gel. Electrophoresis was carried out at room temperature at 100 V for 80 min using running buffer composed of 0.5× TBE and 0.1 mM EDTA. Gels were dried and visualized using an FLA‐5100 phosphorimager (Fujifilm). The K D of the ssDNA and RNA binding of the ToCsm complex was evaluated assuming the complex concentration at which half of the substrate is bound as a rough estimate of K D value.
RNA cleavage assay
The ToCsm complex (600 nM) and the radio‐labelled RNA substrate (2 nM) were incubated at 55°C for 5 min in reaction buffer A composed of 30 mM Tris–HCl (pH 8.0), 100 mM KCl, 100 mM NaCl and 3% glycerol. The reactions were initiated by the addition of 5 mM MnCl2 and incubated at 55°C for 20 min. The reactions were stopped by the addition of formamide loading buffer B composed of 95% formamide, 0.025% SDS, 0.01% bromophenol blue, 0.01% xylene cyanol and 1 mM EDTA, followed by heating at 95°C for 10 min. The samples and the size marker (Affymetrix) were analysed on 12.5% polyacrylamide denaturing 8 M urea gels and visualized by phosphorimaging (Fujifilm).
Target RNA‐dependent DNA cleavage assay
The ToCsm complex (600 nM) and the unlabelled 40‐nt target RNA (600 nM) were incubated at 55°C for 15 min in reaction buffer A with 5 mM NiSO4. The reaction was initiated by the addition of radio‐labelled 100‐nt DNA (2 nM) and incubated at 55°C for 20 min. The reaction was stopped by the addition of sample loading buffer B followed by heating at 95°C for 10 min. The samples and the size marker (Affymetrix) were analysed on 10% polyacrylamide denaturing 8 M urea gels and visualized by phosphorimaging (Fujifilm).
Target RNA‐independent DNA cleavage assay
Reactions with the ToCsm complex (30 nM) and the circular single‐stranded DNA substrates (5 nM) M13mp18 or ΦX174 were carried out in buffer A with 5 mM NiSO4. After incubation at 55°C for 20 min, the reactions were quenched by the addition of 6× DNA loading dye (Fermentas) and analysed on 1% agarose gels. For the radio‐labelled linear DNA substrates (2 nM), reactions were carried out with the ToCsm complex (600 nM) and stopped by the addition of formamide loading buffer B and heating at 95°C for 10 min. The samples and the size marker (Affymetrix) were analysed by 10% polyacrylamide denaturing 8 M urea gels and visualized by phosphorimaging (Fujifilm).
Trans‐ssDNA cleavage assay
The ToCsm complex (30 nM) and the unlabelled target ssDNA (30 nM) or target RNA (30 nM) were incubated at 55°C for 15 min in reaction buffer A with 5 mM NiSO4. The reaction was initiated by the addition of circular single‐stranded DNA substrates ΦX174 (5 nM), incubated at 55°C for 20 min and quenched by the addition of 6× DNA loading dye (Fermentas) before analysis on 1% agarose gels.
Author contributions
K‐HP, B‐HO and E‐JW conceived the idea. J‐HK provided scientific suggestions. K‐HP and YA performed protein purification and In vitro complex assembly. K‐HP, I‐YB, W‐CA, HN and YA performed experiments. EM work was carried out and analysed by T‐YJ, HH and J‐JS The manuscript was written by K‐HP, B‐HO and E‐JW.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Dr. Kang Sung‐Gyun in KIOST for providing the genomic DNA of T. onnurineus NA1. This work was partly supported by the Marine and Extreme Genome Research Center programme from the Ministry of Land, Transport and Maritime Affairs and partly by the National Research Foundation of Korea (Grant Number: NRF‐2014K1A3A1A49069194/NRF‐2015R1A2A2A03006970) and KRIBB Research Initiative Programme. J.‐J.S. and H.H. were supported by a STINT‐NRF special exchange grant (2014K2A3A1000137). T.‐Y.J. is a recipient of a postdoc fellowship from KAIST Institute.
EMBO Reports (2017) 18: 826–840
Contributor Information
Byung‐Ha Oh, Email: bhoh@kaist.ac.kr.
Eui‐Jeon Woo, Email: ejwoo@kribb.re.kr.
References
- 1. van der Oost J, Westra ER, Jackson RN, Wiedenheft B (2014) Unravelling the structural and mechanistic basis of CRISPR‐Cas systems. Nat Rev Microbiol 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- 3. Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bolotin A, Quinquis B, Sorokin A, Ehrlich SD (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 [DOI] [PubMed] [Google Scholar]
- 5. Terns MP, Terns RM (2011) CRISPR‐based adaptive immune systems. Curr Opin Microbiol 14: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J (2016) Diverse evolutionary roots and mechanistic variations of the CRISPR‐Cas systems. Science 353: aad5147 [DOI] [PubMed] [Google Scholar]
- 8. Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) Cas3 is a single‐stranded DNA nuclease and ATP‐dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K et al (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9‐crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71 [DOI] [PubMed] [Google Scholar]
- 12. Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E (2016) The CRISPR‐associated DNA‐cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532: 517–521 [DOI] [PubMed] [Google Scholar]
- 13. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF et al (2011) Evolution and classification of the CRISPR‐Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH et al (2015) An updated evolutionary classification of CRISPR‐Cas systems. Nat Rev Microbiol 13: 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R et al (2011) Structural basis for CRISPR RNA‐guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536 [DOI] [PubMed] [Google Scholar]
- 16. Rouillon C, Zhou M, Zhang J, Politis A, Beilsten‐Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF (2013) Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell 52: 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatoum‐Aslan A, Maniv I, Samai P, Marraffini LA (2014) Genetic characterization of antiplasmid immunity through a type III‐A CRISPR‐Cas system. J Bacteriol 196: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng W, Feng M, Feng X, Liang YX, She Q (2015) An archaeal CRISPR type III‐B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res 43: 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum‐Aslan A, Marraffini LA (2015) Co‐transcriptional DNA and RNA Cleavage during Type III CRISPR‐Cas Immunity. Cell 161: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (2009) RNA‐guided RNA cleavage by a CRISPR RNA‐Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V (2014) Programmable RNA shredding by the type III‐A CRISPR‐Cas system of Streptococcus thermophilus. Mol Cell 56: 506–517 [DOI] [PubMed] [Google Scholar]
- 22. Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K et al (2014) RNA targeting by the type III‐A CRISPR‐Cas Csm complex of Thermus thermophilus . Mol Cell 56: 518–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Graham S, Tello A, Liu H, White MF (2016) Multiple nucleic acid cleavage modes in divergent type III CRISPR systems. Nucleic Acids Res 44: 1789–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao L, Gao CH, Zhu J, Zhao L, Wu Q, Li M, Sun B (2016) Identification and functional study of type III‐A CRISPR‐Cas systems in clinical isolates of Staphylococcus aureus . Int J Med Microbiol 306: 686–696 [DOI] [PubMed] [Google Scholar]
- 25. Hale CR, Cocozaki A, Li H, Terns RM, Terns MP (2014) Target RNA capture and cleavage by the Cmr type III‐B CRISPR‐Cas effector complex. Genes Dev 28: 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV et al (2012) Structure and mechanism of the CMR complex for CRISPR‐mediated antiviral immunity. Mol Cell 45: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carte J, Wang R, Li H, Terns RM, Terns MP (2008) Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osawa T, Inanaga H, Sato C, Numata T (2015) Crystal structure of the CRISPR‐Cas RNA silencing Cmr complex bound to a target analog. Mol Cell 58: 418–430 [DOI] [PubMed] [Google Scholar]
- 29. Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgartner M, Bonneau F, Mann M, Conti E (2014) Structural model of a CRISPR RNA‐silencing complex reveals the RNA‐target cleavage activity in Cmr4. Mol Cell 56: 43–54 [DOI] [PubMed] [Google Scholar]
- 30. Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas C, Siksnys V (2016) Spatiotemporal control of type III‐A CRISPR‐Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell 62: 295–306 [DOI] [PubMed] [Google Scholar]
- 31. Estrella MA, Kuo FT, Bailey S (2016) RNA‐activated DNA cleavage by the Type III‐B CRISPR‐Cas effector complex. Genes Dev 30: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP (2016) Bipartite recognition of target RNAs activates DNA cleavage by the Type III‐B CRISPR‐Cas system. Genes Dev 30: 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldberg GW, Jiang W, Bikard D, Marraffini LA (2014) Conditional tolerance of temperate phages via transcription‐dependent CRISPR‐Cas targeting. Nature 514: 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hayes RP, Xiao Y, Ding F, van Erp PB, Rajashankar K, Bailey S, Wiedenheft B, Ke A (2016) Structural basis for promiscuous PAM recognition in type I‐E Cascade from E. coli . Nature 530: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA (2015) Cas9 specifies functional viral targets during CRISPR‐Cas adaptation. Nature 519: 199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Connell MR, Oakes BL, Sternberg SH, East‐Seletsky A, Kaplan M, Doudna JA (2014) Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516: 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marraffini LA, Sontheimer EJ (2010) Self versus non‐self discrimination during CRISPR RNA‐directed immunity. Nature 463: 568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jung TY, Park KH, An Y, Schulga A, Deyev S, Jung JH, Woo EJ (2016) Structural features of Cas2 from Thermococcus onnurineus in CRISPR‐cas system type IV. Protein Sci 25: 1890–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grissa I, Vergnaud G, Pourcel C (2007) The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jung TY, An Y, Park KH, Lee MH, Oh BH, Woo E (2015) Crystal structure of the Csm1 subunit of the Csm complex and its single‐stranded DNA‐specific nuclease activity. Structure 23: 782–790 [DOI] [PubMed] [Google Scholar]
- 41. Barnes CO, Calero M, Malik I, Graham BW, Spahr H, Lin G, Cohen AE, Brown IS, Zhang Q, Pullara F et al (2015) Crystal structure of a transcribing RNA polymerase II complex reveals a complete transcription bubble. Mol Cell 59: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma E, Harrington LB, O'Connell MR, Zhou K, Doudna JA (2015) Single‐stranded DNA cleavage by divergent CRISPR‐Cas9 enzymes. Mol Cell 60: 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46 [DOI] [PubMed] [Google Scholar]
- 44. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Source Data for Expanded View
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6


