Abstract
Efficient neuronal function depends on the continued modulation of the local neuronal proteome. Local protein synthesis plays a central role in tuning the neuronal proteome at specific neuronal regions. Various aspects of translation such as the localization of translational machinery, spatial spread of the newly translated proteins, and their site of action are carried out in specialized neuronal subcompartments to result in a localized functional outcome. In this review, we focus on the various aspects of these local translation compartments such as size, biochemical and organelle composition, structural boundaries, and temporal dynamics. We also discuss the apparent absence of definitive components of translation in these local compartments and the emerging state‐of‐the‐art tools that could help dissecting these conundrums in greater detail in the future.
Keywords: compartments, local translation, nascent protein, plasticity, spatial spread
Subject Categories: Neuroscience, Protein Biosynthesis & Quality Control
Glossary
- 3′UTR
3′ untranslated region
- Acot7
acyl‐coA thioesterase 7
- AHA
azidohomoalanine
- AMPAR
α‐amino‐3‐hydroxy‐5‐methyl‐4‐isoxazolepropionic acid receptor
- APV
amino‐5‐phosphonovaleric acid
- Arc
activity‐regulated cytoskeleton‐associated protein
- Arp
actin‐related protein
- ATF4
activating transcription factor 4
- ATP
adenosine triphosphate
- Aβ1‐42
amyloid β peptide 1‐42
- BDNF
brain‐derived neurotrophic factor
- CaMK2a
calcium/calmodulin‐dependent protein kinase 2 alpha
- CB1
cannabinoid receptor type 1
- CHX
cycloheximide
- DCC
deleted in colorectal cancer
- DHPG
dihydroxyphenylglycine
- DIV
days in vitro
- DRG
dorsal root ganglion
- E5
embryonic day 5
- EphA2
ephrin type‐A receptor 2
- ER
endoplasmic reticulum
- ERGIC
endoplasmic reticulum–Golgi intermediate compartment
- FISH
fluorescence in situ hybridization
- FMRP
fragile X mental retardation 1
- FRAP
fluorescence recovery after photobleaching
- FRET
fluorescence resonance energy transfer
- FUNCAT
fluorescent non‐canonical amino acid tagging
- GFP
green fluorescent protein
- GluA1
glutamate receptor subunit A1
- KO
knock out
- Kv3.1a
voltage‐dependent potassium channel 3.1a
- LTP
long‐term potentiation
- Lys
lysine
- MAG
myelin‐associated glycoprotein
- MAP2
microtubule‐associated protein 2
- mGluR
metabotropic glutamate receptors
- miRNA
micro ribonucleic acid
- mRNA
messenger ribonucleic acid
- NA
not applicable
- NGF
nerve growth factor
- NLS
nuclear localization signal
- NMDAR
N‐methyl‐d‐aspartate receptor
- NT3
neurotrophin‐3
- PAGFP
photoactivatable green fluorescent protein
- PALM
photoactivated localization microscopy
- PRPs
plasticity‐related proteins
- PSD‐95
postsynaptic density protein 95
- Puro‐PLA
puromycylation‐proximity ligation assay
- RanBP1
Ran‐specific guanosine triphosphatase activating protein
- RNA
ribonucleic acid
- RNA‐seq
ribonucleic acid sequencing
- Rpl35
ribosomal protein l35
- Sema3A
semaphorin 3A
- Thy1
thymus cell surface antigen 1
- tRNA
transfer ribonucleic acid
- WIN
(R)‐(+)‐[2,3‐dihydro‐5‐methyl‐3‐(4‐morpholinylmethyl)pyrrolo[1,2,3‐de]‐1,4‐benzoxazin‐6‐yl]‐1‐naphthalenylmethanone mesylate
Introduction
Protein synthesis is essential for the maintenance and regulation of the cellular proteome. The discovery of the mechanism of protein synthesis 60 years ago led to the understanding that the decoding of information from mRNA to protein is carried out by “adaptor” RNAs and catalyzed by enzymes in cell extracts—later identified as tRNAs and ribosomes, respectively 1, 2. In the past 20–30 years, it has become clear that the translation of mRNA to protein is not only regulated temporally in a cell type‐dependent manner, but also has a strong subcellular component. Once thought to occur exclusively in the somatic space close to the nucleus, protein translation has been demonstrated far from the central perinuclear region in decentralized local domains—a process referred to as local translation 3. One cell type that has been studied extensively in the context of local translation is the neuron. Neurons are highly polarized cells with specialized morphologies. Efficient neuronal function is mediated by the collection and integration of signals received by dendrites, processing, and “decision‐making” in the soma, and then transmission of information to the axons (Fig 1). Axons communicate to adjacent neurons at synapses where chemical transmitters released from the presynaptic terminal bind to receptors at the postsynaptic terminal of a dendrite (Fig 1). A single neuron can receive signals at several thousand independent synapses, and the strength of the signal transmission can be regulated at the level of single inputs. The highly polarized morphology and function of neurons and the continuous demand to adapt to external stimuli make local translation a key process in the regulation of neuronal physiology 3.
Figure 1. Neuron and its structural compartments.
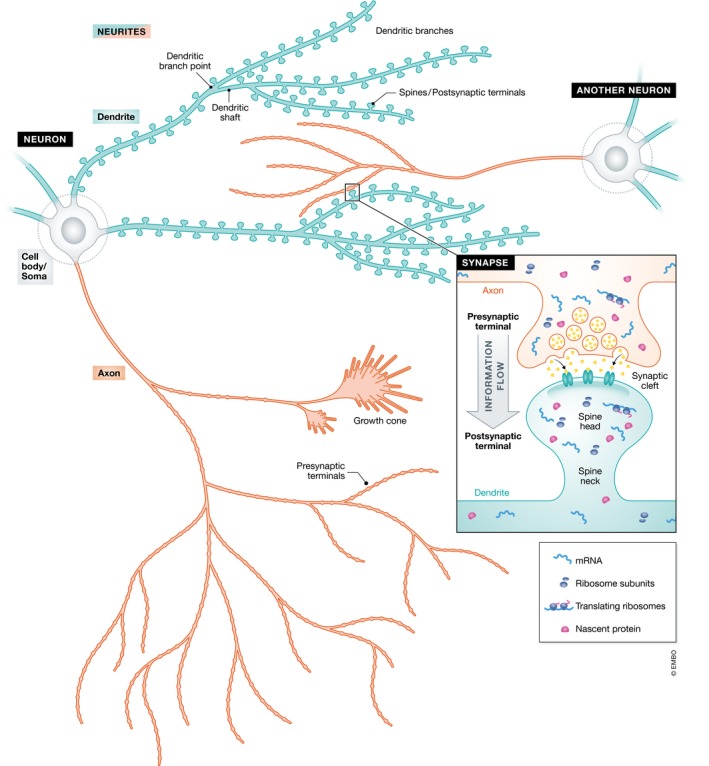
Morphology of the neuron showing its cell body (gray) and neurites–composed of dendrites (blue) and axons (red). The inset shows a synapse formed between the presynaptic terminal of one neuron (red) and the postsynaptic terminal of another neuron (blue).
Local protein synthesis provides a means to locally establish, maintain, and modify the synaptic proteome. Classical studies set the foundation for this idea by showing that protein synthesis constituents and machinery are present in or around synapses. Among the first players detected in dendrites were the mRNAs for microtubule‐associated protein 2 (MAP2) 4, calcium/calmodulin‐dependent protein kinase 2 alpha (CaMK2a) 5, activity‐regulated cytoskeleton‐associated protein (Arc) 6, and polyribosomes 7, 8, 9. The demonstration of protein synthesis in severed neurites (axons and/or dendrites) and soma‐free biochemical preparations further supported this concept 10, 11, 12, 13. Furthermore, the functional significance of these observations came with the demonstration that local protein synthesis is involved in some forms of synaptic plasticity and learning 14, 15, 16, 17. This led to the idea that local translation could drive the synthesis of a specific set of “plasticity‐related proteins” (PRPs) and that their identity could be unraveled by the characterization of localized mRNAs—the local transcriptome.
Both high‐throughput and single‐molecule candidate approaches have been developed to characterize the local transcriptome and the newly synthesized proteome 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29. The characterization of local transcriptomes by RNA‐seq and microarrays by various groups 18, 30, 31, 32, 33, 34, 35, 36, 37 revealed that the comprehensive set of localized mRNAs is as large as 2,550 mRNAs—in the neuropil alone 18. Thus, it is important to also characterize the local translatome—the fraction of mRNAs that get actively translated to carry out neuronal function. To address this question, high‐throughput methods exploiting the association of ribosomes with mRNAs have been implemented 38, 39, 40, 41, 42, 43, 44, 45, 46. In addition, the use of mRNA tracking along with nascent protein visualization has enabled single‐molecule, real‐time visualization of translation and its kinetics 22, 23, 26, 27, 28, 29.
All of the above techniques have set the stage to probe different facets of local translation and address a new generation of questions. For example, where exactly is a protein translated, what is its functional fate and what is its spatial range of action within subneuronal regions—in other words, what are the relevant compartments?
Space redefined: What are the relevant compartments?
We define the term compartments here as spatially restricted domains within which cell biological machines carry out a function. In the context of protein synthesis, we consider that if new proteins are synthesized locally in subneuronal regions, they should be spatially restricted to sustain their functional activity in a localized fashion. How does one define a relevant compartment for local translation (translation compartment)? And how might these cell biological compartments map onto functional compartments for information processing?
First, it is important to discriminate between the site of synthesis (source compartment) and the site of action of the nascent protein (effector compartment) (Fig 2A–E). The source and the effector compartments could be within a few microns of each other (e.g., within a dendritic branch) or hundreds of microns apart (Fig 2A–E). We also consider specific features of compartments: Do they possess defined structural boundaries? What roles do cellular organelles such as ribosomes, mitochondria, and secretory pathway machinery play? Are these compartments dynamic—do they form, adapt, and/or disassemble in response to local cues? All these characteristics operate in unison to constitute a functional outcome.
Figure 2. Local translation compartments.
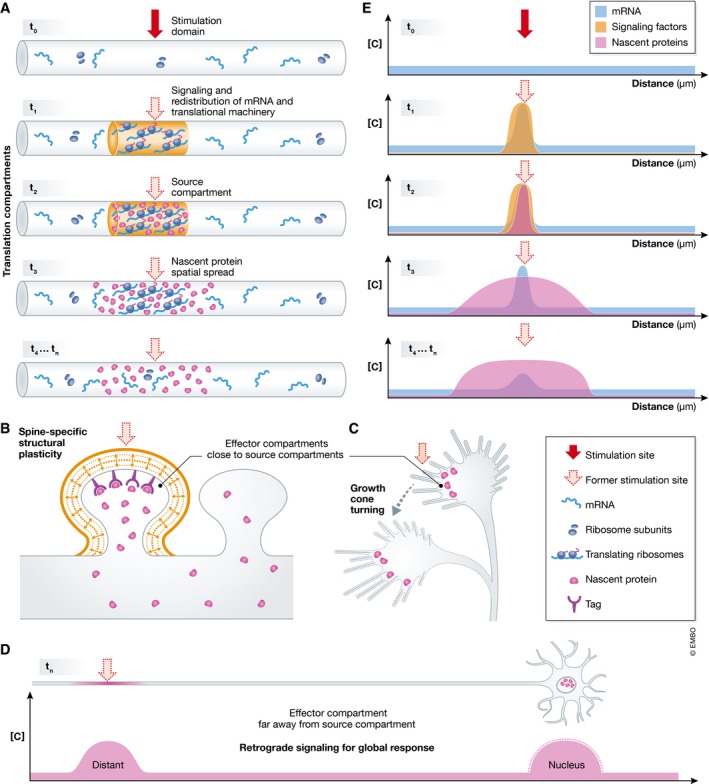
(A) On receiving a focal stimulus (red filled arrow)—stimulation domain (t 0)—mRNA and translational machinery are redistributed to the stimulation site (t 1). This local redistribution of mRNA and translational machinery, on overlap with signaling events (orange) form the site of synthesis for nascent proteins (magenta)—source compartment (t 2)—near the former stimulation site (red dotted arrow). The nascent proteins quickly spread over time. This nascent protein spatial spread (t 3) gradually increases and might reach a compartment of stable size defined by unknown factors (t 4, t n). The site of action of the nascent proteins—effector compartment—is restricted within a smaller region of the nascent protein spatial spread and can be either close to the source compartment as in a spine (B) and the growth cone (C) or hundreds of microns apart as in the nucleus (D). All these translation compartments operate in unison to elicit a functional outcome, for example, spine‐specific structural plasticity (B), growth cone turning (C), and retrograde signaling for global response (D). (E) Graphical representation showing the concentration ([C]) of mRNA (blue), signaling factors (orange), and nascent proteins (magenta) plotted at various time points t 0, t 1, t 2, t 3, t 4, t n.
This review's focus is on the emerging picture of the spatio‐temporal organization of such compartments relevant for local translation. In addition, we summarize some unsolved issues—the apparent absence of expected translational components in some compartments and the unexplored prerequisites such as local energy reserves for translation.
Translation compartments in dendrites
Local translation was initially studied by comparing the translational capacity of structurally defined classical compartments, namely the cell body and neurites. The protein synthesis observed in neurites was mainly attributed to dendrites, owing to the low levels of mRNAs and conflicting data on the presence of local translational machinery in axons 47, 48. Furthermore, the capacity of dendrites, isolated from the cell body, to independently synthesize new proteins upon stimulation strongly supported this view 10.
In order to understand how translation compartments are formed, it is important to describe the two general prerequisites that have to be fulfilled to allow localized translation: (i) the mRNA of interest and the translational machinery have to be present at the site of action, and (ii) the stimulus to induce translation has to be sensed and transferred to the translational machinery. The overlap of these prerequisites defines the site of translation. In some cases, while mRNA is widely distributed in the neurites by efficient mRNA transport, the presence of overlapping local signals leads to minor local redistribution of mRNA and its subsequent translation. In other cases, the mRNA localization itself is confined and targeted to specific subneuronal regions; in this case, even global stimulation would lead to only restricted sites of translation, despite the widespread availability of receptors to sense the stimulus. One example is the spatial coding for the delivery of BDNF mRNAs to different parts of somata and dendrites. Under basal conditions, BDNF mRNA localization is mainly somatic 49, 50, and the translated BDNF protein distribution is regulated by the secretory pathway. Upon activity, BDNF mRNA levels can be upregulated, and their selective distribution to proximal or distal dendrites is achieved by a code in their 5′ non‐coding regions. This selective distribution of BDNF mRNA results in restricted sites of putative BDNF translation in dendrites even upon global stimulation 49.
Dendrites comprise several recognizable structural compartments such as the dendritic spine, spine neck, dendritic shaft, branch points, and dendritic branches (Fig 1). Measured translation compartments, however, are often not limited to these structural boundaries. They seem to exist as a continuum of spatial domains either restricted within part of these structures or spanning across them. It is only beginning to be understood where specific proteins are synthesized, and what the limits of their spatial spread and subsequent function are in the context of these structural boundaries.
Spines
Spines are nodes where dendrites receive information from adjacent neurons (Fig 1). Spine heads, the sites of most excitatory synapses, are diffusionally and electrically restricted from their respective dendritic shafts by thin spine necks 51, 52. The spine neck acts as a diffusion barrier to proteins and small molecules, and the ease of diffusion is modulated by activity 53, 54, 55, 56. This compartmentalization is likely important for spine‐specific synaptic modulation, as local stimulation of spine heads shows spine‐specific structural plasticity 52, 57, 58. It is not clear, however, if local translation of proteins is confined to spines (Fig 3A). During tetanic stimulation, the enrichment of polyribosomes in spines compared to dendritic shafts supports this view 59. Moreover, upon a global increase in basal translation, translation hot spots were observed in some spines in addition to hot spots in dendritic shafts 60 (Table 1). In order to achieve local stimulation, a clever approach was recently developed to stimulate multiple adjacent spines and visualize β‐actin mRNA and its translation simultaneously. This experiment revealed the recruitment of β‐actin mRNAs and newly synthesized β‐actin protein near the stimulated region. However, these translation hot spots were not spine‐specific 29 (Table 1) and direct evidence for spine‐specific translation is still lacking (Fig 3A). This is partly because translation has not yet been successfully monitored in the presence of localized single‐spine‐restricted stimulation. However, it is possible that the translation compartment measured upon single‐spine stimulation is not restricted to a spine, but the functional outcome of the translation event is spine‐specific. For instance, a stimulated spine could selectively trap a newly synthesized protein even if the respective mRNA and translation machinery are localized outside the spine 61, 62, 63 (Fig 3B). If that were the case, what would be the spatial spread of the nascent protein? And what influences the spatial limit of its action—the effector compartment?
Figure 3. Subcompartments in dendrites.
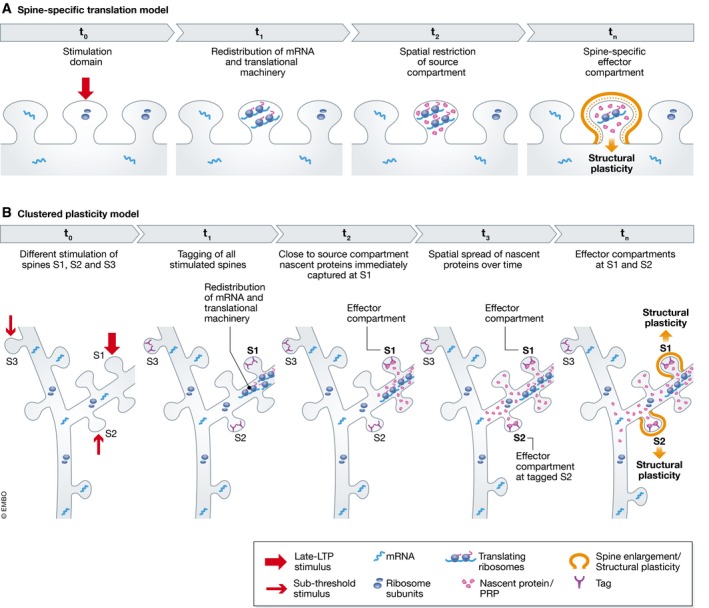
(A) Concept of spine‐specific translation: A spine‐specific stimulation (t 0) could result in redistribution of mRNA and translational machinery to the stimulated spine (t 1). On overlapping with spine‐specific signaling events, this would result in the synthesis of nascent proteins—source compartment—whose spatial spread is restricted within the spine, due to diffusional restriction by the spine neck (t 2). This would lead to a spine‐specific effector compartment and subsequent functional outcome—structural plasticity (t n). (B) Clustered plasticity model: Spine S1 receives a late‐LTP stimulus, and spines S2 and S3 receive a subthreshold stimulus (t 0). All three spines get tagged but mRNA and translational machinery redistribute only close to spine S1 that received the late‐LTP stimulus (t 1). The newly translated proteins—source compartment—are instantly captured at spine S1 (t 2) and with time, the spatial spread of the nascent proteins increases allowing for its additional capture at adjacent tagged spine S2 (t 3). Both the tagged spines S1 and S2 that capture nascent proteins—effector compartment—undergo spine‐specific structural plasticity—functional outcome (t n). However, only tagged spines clustered within a nascent protein spatial spread of ~50 μm show this functional outcome—tagged spine (S3) present beyond this spatial spread does not.
Table 1.
Translation compartment sizes estimated from literature data
| Article | Condition | Stimulating factor | Readout (measure)a | Additional comments | Compartment size | Translation/stimulation domain ratiob |
|---|---|---|---|---|---|---|
| I. Dendrite | ||||||
| 63 | Local glutamate uncaging 30 pulses, 0.5 Hz plus bath application with forskolin | Late‐LTP induction | Spine volume change/structural plasticity (GFP fluorescence) (R) | Organotypic slice cultures from Thy1‐GFP mice, 8–16 DIV; protein synthesis inhibitor sensitive | 50 μm | 50 |
| 19 | Spot perfusion 60 μm for 30 min | BDNF—modulator of neuronal activity | Nascent proteins (fluorescent AHA) (E) | Rat hippocampal neurons 17 DIV | 200 μm; whole dendritic stretch imaged showed an increase | 3 |
| 29 | Local glutamate uncaging 10 pulses, 0.5 Hz, 6 μm | Glutamate—AMPAR activation | Nascent β‐actin (halotag of β‐actin detected by dye) (R) | Mouse hippocampal neurons 14–21 DIV; uncaging 100 μm away from cell body | 18 μm | 2 |
| β‐actin mRNA (GFP tagged to β‐actin mRNA binding protein) (R) | 6 μm | 1 | ||||
| 64 | Spot perfusion 3–10 μm for 15 min | Dihydrexidine—dopamine receptor agonist | Nascent proteins (fluorescent puromycin signal) (E) | Rat hippocampal neurons 14–21 DIV; perfusion 100 μm away from cell body | 3–10 μm | 1 |
| 130 | Spot perfusion 25–59 μm for 90–105 min | APV—NMDAR mini blockade | Surface GluR1 (immunofluorescence) (E) | Rat hippocampal neurons > 14 DIV | 25–59 μm | 1 |
| 66 | Local dendritic perfusion < 50 μm for 10 min inmicrofluidic chambers | DHPG—mGluR activation | Enrichment of newly transcribed Arc mRNA (FISH) (E) | Rat hippocampal neurons 15–20 DIV | < 5 μm | < 0.1 |
| 10 | Bath application on optically isolated dendrites after photobleaching soma | BDNF—modulator of neuronal activity | Nascent myristoylated GFP with CaMK2a 3′UTR (GFP fluorescence) (R) | Rat hippocampal neurons 14–21 DIV | Whole dendritic stretch imaged ~150 μm | NA |
| 60 | Bath application for 3 min | DHPG—mGluR activation | Nascent PSD‐95 (Venus‐PSD95 fluorescence super resolved by PALM) (R) | Mouse hippocampal neurons 12–16 DIV; newly synthesized PSD‐95 did not colocalize with preexisting translational site | PSD‐95 translation “hot spots” enriched in spines and dendritic shaft | NA |
| FMRP‐KO | Basal neuronal activity | Increase in basal translation | ||||
| 67 | None (visualized PSD95 redistribution following photoactivation for 60 min) | Basal neuronal activity in anesthetized mice | Photoactivated PSD95‐not nascent protein (PAGFP fluorescence) (R) | Mouse pyramidal neurons of somatosensory cortex E16 | ~10–15 μm | NA |
| 68 | Local glutamate uncaging 20 pulses, 1 Hz, 2 μm2 | Dicer activation | miRNA maturation (FRET sensor ratio) (R) | Rat hippocampal neurons 14–21 DIV | 1–2 μm | 1–2 |
| Nascent CaMK2a repression (Puro‐PLA signal) (E, R) | ~20 μm | 14 | ||||
| II. Axon | ||||||
| 89 | Focal bead challenge 4.5 μm overnight | NGF, NT3, MAG, Sema3A—growth factors/guidance cues | β‐actin, peripherin, Kv3.1a mRNAs (FISH) (E) | Rat dorsal root ganglion cells injury conditioned in vivo for 7 days and cultured overnight | 5–20 μm | 1–4 |
| 106 | Focal bead challenge 5 μm in axonal compartment of microfluidic chambers for 24 h | Poly‐D‐Lys—for induction of presynaptic terminals | β‐catenin mRNA (FISH) (E) | Rat hippocampal neuron axons > 10 DIV | 5 μm | 1 |
| For 15 min–3 h | β‐catenin protein (immunofluorescence) (E) | Protein synthesis inhibitor sensitive β‐catenin increase | 5 μm | 1 | ||
| 113 | Nerve crush lesion in vivo ~1 mm | Injury | Newly synthesized NLS‐binding protein (fluorescent NLS‐peptide binding) (E) | Sciatic nerve crush generated nascent protein is retrogradely transported as a domain | ~1 mm | ~1 |
| 108 | Local perfusion 150 μm, 5 × 5 min pulses, soma‐free neurons | Serotonin—for long‐term facilitation | Nascent dendra with sensorin 5′3′UTR (photoswitchable dendra fluorescence) (R) | Aplysia sensory neuron‐motor neuron cocultures ~3 DIV; translational hot spots observed only in perfusion area | 10–20 μm hot spots | ~0.07–0.13 |
| 114 | None (visualized recovery of RanBP1 following photobleaching in isolated axons) | Basal neuronal activity | Nascent myristoylated GFP with RanBP1 3′UTRs (GFP FRAP) (R) | Transfected DRG neurons; reporter recovery after FRAP is sensitive to the UTR variant used and to protein synthesis inhibitors | 5–10 μm | NA |
| 110 | During axon branching, bath application | NGF—growth factor | Nascent myristoylated GFP with β‐actin, cortactin, Arp 3′UTRs (GFP FRAP) (R) | Chicken dorsal root ganglion neurons 7 DIV; majority of GFP reporter hot spots colocalize with mitochondria but not all mitochondria localize with hot spots | ~5 μm | NA |
| β‐actin mRNA (FISH) (E) | β‐actin mRNA accumulation along some mitochondria | ~5 μm | NA | |||
| 36 | Axonal compartment bath application in microfluidic chambers for 24 h and 48 h | Aβ1‐42—for induction of Alzheimer's pathogenicity | Nascent proteins (fluorescent AHA) (E) | Rat hippocampal neurons 9–10 DIV; measured hot spots sensitive to protein synthesis inhibitor | ~5–10 μm hot spots | NA |
| 109 | Bath application for 25 min | WIN–cannabinoid receptor CB1 agonist | Newly synthesized protein (FUNCAT) (E) | Hippocampal neuron culture; synthesis is increased in the whole neuron with hot spots visible around CB1‐positive terminals | 2–5 μm | NA |
| 107 | Bath application for 24 h in soma‐free neurons | Netrin 1—chemotropic factor | Sensorin protein (immunofluorescence) (E) | Aplysia sensory neuron‐motor neuron cocultures 3 DIV | 10 μm hot spots | NA |
| III. Growth cone | ||||||
| 104 | In tissue commissural axon midline crossing | Midline crossing–guidance cue | GFP with EphA2 3′UTR (GFP fluorescence) (R) | Commissural neurons; E5 chicken spinal cord; sharp 20 μm domain upon experiencing the cue, translation domain widens as growth cone crosses | 20 μm; 80–100 μm | NA |
| 97 | Bath application for 5 min | Netrin‐1–chemotropic factor | Nascent β‐actin (immunofluorescence) (E) | Xenopus stage 24 retinal ganglion cell growth cone; β‐actin hot spots correspond to growth cone filopodia; CHX‐sensitive | 5–10 μm | NA |
| Bath application for 10 min | Nascent kaede with β‐actin 3′UTR (photoswitchable kaede fluorescence) (R) | Kaede hot spots in growth cone and filopodia | 5–15 μm | NA | ||
| Asymmetric gradient, 90° to the direction of axon shaft for 5 min | β‐actin protein accumulation (immunofluorescence) (E) | Protein synthesis inhibitor sensitive β‐actin fluorescence; signal is higher near the gradient source | < 20 μm domains | NA | ||
| 99 | Bath application for 5 min | Slit conditioned medium—guidance cue | Cofilin protein accumulation (immunofluorescence) (E) | Xenopus retinal explants culture stage 35/36; protein synthesis inhibitor‐sensitive cofilin increase in filopodia | 5–10 μm | NA |
| 41 | None (visualized recovery of Acot7 following photobleaching) | Basal neuronal activity | Nascent myristoylated GFP with Acot7 axon‐exon (GFP FRAP) (R) | Xenopus retinal ganglion cell growth cone | < 10 μm | NA |
Translation compartments were measured either by monitoring endogenous nascent proteins (E) or reporter constructs (R).
Ratio of the translation compartment size (protein or mRNA) to the stimulation domain (perfusion, local uncaging, focal stimulation, etc.). The calculated ratios have been rounded off to the closest whole number, where applicable. For Govindarajan et al 63, the stimulation domain size for uncaging was assumed as 1 × 1 μm2. Literature listed in each segment—dendrite, axon, growth cone—is in decreasing order of the calculated ratio, followed by chronology.
Dendritic compartments
In order to probe the spatial spread of nascent proteins, several experiments have been conducted to monitor the translation compartment size in response to local stimulation of a dendritic stretch (Table 1). By translation compartment size, we mean the measured sizes (in microns) of different aspects of translation—particularly the spatial spread of nascent proteins and in some cases the redistribution of mRNA or translation markers (Fig 2A and Table 1). The measured sizes range from 3 to 60 μm and are dependent on the size of the locally perfused region, diffusivity of the perfused stimulating factor and duration of the perfusion 19, 64, 65, 66 (Table 1). In order to understand the spatial relationship between the stimulation domain size and the resulting translation compartment, we examined the size ratio of the translation compartment to the stimulation domain (translation to stimulation domain ratio, Table 1). The translation to stimulation domain ratio of nascent proteins ranges between 1 and 3 across different experiments, implying a close spatial relationship between the stimulation and its concomitant localized translation compartment. The reported translation compartments were measured using a wide range of methodologies, with some monitoring endogenous nascent proteins and the others relying on reporter constructs (Table 1). Interestingly, a recent study showed that for the same stimulation, the size of the nascent protein spatial spread (~18 μm) is slightly larger than the mRNA redistribution domain (~6 μm) 29 (Table 1, Fig 2A and E). This could be due to the fast spread of the nascent protein within a larger predestined spatial domain, compared to a narrower mRNA localization domain marking the point of translation. A similar observation was made following single‐spine PSD‐95 photoactivation, where the photoactivated proteins redistribute and stay “captured” in spines within a defined spatial range of 10–15 μm of a dendritic shaft 67 (Table 1). Moreover, measurements of the regulation of local translation by miRNAs during single‐spine stimulation showed a ~20 μm larger domain of protein repression compared to a 1–2 μm narrower domain of miRNA maturation 68 (Table 1). These experiments argue that the site of action of nascent proteins (effector compartment) is defined but not limited to the site of synthesis (source compartment) or structural boundaries. It is likely that the intensity of the stimulation, the amount of nascent protein made, the nature of the nascent protein (transmembrane, cytoskeletal, or cytosolic), its diffusional property, and the number of competing slots for trapping the nascent protein influence its spatial spread and functional outcome.
The functional consequence of the spatial spread of nascent proteins was first demonstrated in dendrites by experiments probing the “clustered plasticity” model 63, 69. According to this model, a spine receiving a stimulus that leads to long‐term structural plasticity (late‐LTP stimulus) (S1, Fig 3B) gets “tagged” in a protein synthesis‐independent manner and also leads to the synthesis of PRPs. These newly synthesized PRPs are subsequently “captured” at the tagged spine leading to spine‐specific structural changes (S1, Fig 3B). However, it was not clear if these PRPs were synthesized locally in the dendrites 63, 69 or in the soma 61, 62. In order to address this, another spine was “tagged” by a subthreshold stimulus—insufficient to induce PRP synthesis by itself (S2, S3, Fig 3B). It was observed that the subthreshold‐tagged spine (S2) was able to show spine‐specific structural plasticity by capturing PRPs, only when it was clustered within a distance of ~50 μm from the late‐LTP‐induced spine (S1) 63. Spines distributed beyond this spatial distance (S3) did not show acquired plasticity, suggesting that PRP synthesis cannot be somatic but has to be local. This effective distance of the cluster is dependent on the number of neighboring tagged synapses that compete for the limited pool of PRPs. The more neighboring tagged synapses, the more promptly the PRPs are captured, thereby shortening the spatial spread of the nascent proteins and the size of the effector compartment. The observed time dependence of this clustering also suggests the transient nature of these compartments. The exact location where PRPs are synthesized in dendrites is still not known. Recent tools available to simultaneously visualize mRNA and translation 22, 26, 27, 28, 29, in combination with a functional readout for synaptic clustering, should enable careful investigation of the dynamic spatial spread and functional outcome of these translation compartments.
Mapping functional compartments for information processing
Functional evidence for clustered plasticity is not only limited to observations in cultured neurons, but also has been observed in various animal model systems 70, 71, 72. While it is not yet clear if the observed clustered plasticity is protein synthesis‐dependent, the observation of synaptic clustering during learning paradigms supports this view 73, 74. Synaptic clustering in a dendritic branch is thought to increase local spike initiation by non‐linear summation resulting in enhanced dendritic excitability and effective information processing 51, 75, 76, 77, 78, 79, 80. On the other hand, studies have also demonstrated that spines that respond to similar sensory inputs are widely distributed throughout the dendritic arbor and are therefore non‐clustered 81, 82, 83, 84, 85. While this is still a subject of debate 86, the investigation of local translation during learning paradigms in animal model systems should facilitate the understanding of the molecular mechanisms underlying the functional mapping of synaptic inputs and the subsequent information processing.
Translation compartments in axons
Significant developments in the ability to separate axons from dendrites 34, 35, 36, 37, 93, 94, 95, 96 led to the demonstration of local translation in axons, in addition to dendrites. In fact, among the earliest evidences for neuronal local translation was the incorporation of radioactive amino acids into proteins in the isolated squid giant axon 11 and the detection of protein synthesis machinery in the squid axoplasm 90, 91, 92. Also, the dependence of synaptic plasticity in Aplysia neurons on presynaptic local translation suggested that the axon is capable of protein synthesis 16. Despite this evidence, there remained skepticism about generality of axonal translation with some arguing that these model systems represented special cases. The apparent low basal content of mRNA and protein synthesis machinery in axons also contributed to this skepticism. The current emerging theme is that axons make use of local translation primarily when they act as signal receivers in response to local cues 3, 93, 94, 95, 96. For example, axonal translation is observed during development in response to guidance or maturation cues, under injury conditions and presumably during synaptic activity. Where in the axon is a signal sensed and how does this relate to the site of local translation? What is the site of action of the newly synthesized proteins? Broadly speaking two opposing scenarios have been observed in axons for locally synthesized proteins in response to a local signal: (i) spatially restricted use for local response and (ii) distant use for global response.
Before discussing these local and global effects of local translation, it is important to look at the factors that influence the size of the axonal source compartment. In general, the dynamics of mRNA distribution depend not only on the transcript/protein of interest but also on the strength and type of stimulation 89, 97. When axonal segments were focally stimulated using ligand‐coated beads, mRNAs were redistributed in both transcript‐ and cue‐dependent manner. In one study, a growth factor NT3‐dependent increase in β‐actin mRNA was confined to a narrow domain of 5 μm, corresponding to the bead size, while for other cues, it spread over wider regions (~20 μm) of the axon around the stimulation site 89. Interestingly, focal cues that stimulated the downregulation of the transcript resulted in a wider domain size (> 20 μm) than a cue that resulted in upregulation of the transcript (~5 μm). This study also showed that the subsequent translation responses correlated with the regions of mRNA distribution. However, it was not addressed if axons use these nascent proteins for local function.
Local use of translated proteins: the case of growth cone guidance
Locally restricted use of axonally translated proteins was first observed in growth cones of the developing axon 94, 97, 98, 99, 100, 101. Growth cones are axon‐end structures that pioneer axonal growth in response to attractive and repulsive chemical guidance cues. They fulfill this role even when axons are severed from cell bodies and the response to several cues is protein synthesis‐dependent 94, 97, 98, 99, 100, 101. The growth cone is easy to identify morphologically, and excellent assays have been developed to measure growth cone responses. This has allowed researchers to address the spatial relationship between the compartment that senses the cue, the induced signaling compartment, the resulting source compartment (e.g., redistribution of mRNA and translation markers), spatial spread of nascent proteins, and the functional outcome (e.g., growth cone turning and collapse). Even with the bath application of guidance cues, the compartment sizes involved in cue sensing, local translation, and the functional outcome are often restricted to the growth cone itself (~40 μm) or to parts of the growth cone like filopodia (5–10 μm) 81, 97, 99 (Table 1). Application of cues as point source gradients on one side leads to a directional response of the growth cone 94, 97, 98, 99. For example, the introduction of Netrin‐1 as point source resulted in the asymmetric distribution of translational markers for β‐actin synthesis. Gradients within the growth cone concentrated the translational markers in a < 20 μm zone toward the cue and correlated with the direction of the growth cone response 97. The overlap of signaling compartments and β‐actin mRNA redistribution to filopodia resulted in asymmetric local translation of β‐actin. However, the turning response was absent when β‐actin mRNA was knocked down. Thus, these data suggest that the overlap of different compartments contributes to the direction‐specific growth cone response.
A similar pattern has been observed for a number of other cues in which the growth cone responses involve the overlap of several different signals or processes 94, 100, 102. Local translation, which is likely restricted to the growth cone, is also necessary for restoring ongoing growth cone responsiveness 98, axon elongation, and membrane remodeling during development 96, 103. Moreover, during axonal pathfinding, sharp upregulation of translation (within ~20 μm, Table 1) was observed upon contact of growth cones with an intermediate target 104. Taken together, these data show that many local remodeling responses are mediated by local translation and its local use within compartments smaller than 20 μm, resulting in a fast, autonomous functional outcome in the growth cone.
Local use of translated proteins: branch maturation, synapse maturation, and synaptic plasticity
During growth, axons branch out as they navigate toward their postsynaptic targets and make contacts that eventually mature into presynaptic boutons. The axonally localized mRNA population is regulated during development and presynapse formation 34, 35, 37, 41, 105, 106. Focal bead‐triggered presynapse formation was associated with enriched β‐catenin mRNA within the newly formed terminal with a compartment size of ~5 μm, corresponding to the size of the artificial trigger 106. A translation‐dependent increase in β‐catenin protein was also confined to the newly formed presynaptic terminals and was essential for regulation of vesicle recycling. Evidence that presynaptic local translation is involved in synaptic plasticity was first shown in Aplysia neurons during long‐term facilitation 16, 107, 108. In soma‐free neurons, translation hot spots of ~10–20 μm were observed in response to local perfusion or bath application of the stimulus, in spite of a neuron‐wide distribution of mRNA (Table 1). Here, translation hot spots likely corresponded to presynaptic varicosities. In addition, in the adult hippocampus, the involvement of presynaptic local translation in synaptic plasticity was shown during cannabinoid signaling 109. Translation markers were found within 1–2 μm of presynaptic terminals; however, the spatial spread of the nascent proteins was not determined. In cultured neurons, in addition to a global increase in protein synthesis after cannabinoid receptor stimulation, strikingly, an apparent enrichment of nascent proteins at cannabinoid receptor‐positive terminals was observed in axons (~2–5 μm) 109 (Table 1).
Axon branching also requires local translation. Interestingly, mitochondria were recruited near the sites of branch point formation; the size (~5 μm, Table 1) and location of detected local translation compartments correlated with that of the mitochondria. This mitochondrial recruitment was found to be a prerequisite for local translation and subsequent branch stabilization 110, 111.
Local source and global effector compartments: a case of retrograde signaling
As discussed so far, local translation provides a source for proteins that are used as close as 1–20 μm from their site of synthesis to elicit an effect. However, in other cases, the effector compartment can be more than hundreds of microns to millimeters away from the source compartment. Retrograde signaling to the nucleus involving locally synthesized proteins is used in different situations by axons to elicit global responses via transcriptional changes. The response to axonal injury is one well‐studied example where the main retrograde signaling response relies on local translation near the injury site 112. The initial size of the source compartment and the subsequent spatial spread of nascent proteins in response to injury are difficult to measure in tissue. In the peripheral nervous system, a smart binding assay reporting protein synthesis and subsequent retrograde transport labeled a compartment of 1 mm at the site of lesion in the sciatic nerve. The size of the compartment was unchanged after 6 h but was shifted several millimeters toward the cell body, indicating precise localization, timing, and transport of the newly synthesized protein 113. In cell culture, monitoring translation of one of the injury‐related mRNAs in isolated axons revealed a translation compartment as narrow as 5–10 μm 114.
In addition to lesion events, local translation with retrograde signaling is elicited in response to neurotrophic factors by growth cones and in neuronal subtype specification 115, 116. Evidence that axon‐site‐synthesis‐to‐nucleus signaling plays a role in mature central nervous system and Alzheimer's pathology came recently with the demonstration that injection of Aβ1‐42 into the dentate gyrus resulted in increased levels of ATF4 mRNA and protein in forebrain neuronal axons that project to the dentate gyrus 36. Local synthesis of the transcription factor ATF4, followed by retrograde transport and a nuclear transcriptional change, led to neurodegeneration. Consistent with other studies, hot spots of local translation with a compartment size of ~5–10 μm were apparent (Table 1).
Local translation of transcription factors and their retrograde transport to the nucleus partially explains how a global response in the neuron can be evoked 36, 115, 117, 118. Often non‐transcription factors are locally synthesized, triggering the assembly of defined transport complexes and their modification by coincident local signaling events 113, 117, 119, 120. This has led to the understanding that these locally synthesized non‐transcription factors could carry a signature about the synthesis site to the nucleus. It should be noted that dendrites also employ synapse‐to‐nucleus signaling of transcription factors to elicit global responses 121, 122, 123. There is evidence that local translation also plays a role in dendritic synapse‐to‐nucleus signaling 122.
In all of the above cases, the translation event (source compartment) and the elicited function (effector compartment) are uncoupled in space and time. This uncoupling might therefore facilitate the visualization of the various translation compartments, including the gradual changes in the spatial spread of nascent proteins, and their dynamics.
Unanswered questions
In the field of local translation, there remain some unresolved issues. For example, in many cases, components of the downstream processing machinery required for canonical protein translation are apparently missing in subneuronal compartments. It is important to address these issues, as they might serve as important factors in defining the site and size of local translation compartments. Besides, as described below for membrane protein processing, resolving such issues also sheds light on the potential function of local translation.
The membrane protein conundrum
There is ample evidence for the dendritic and axonal localization of mRNAs that code for membrane and secreted proteins 18. Moreover, there is experimental data on their local translation, their potential to reach the plasma membrane, and the capacity of isolated dendrites to glycosylate them 25, 64, 104, 124, 125, 126, 127, 128, 129, 130. Membrane and secreted proteins require insertion into the endoplasmic reticulum (ER) for proper protein folding, followed by passage via the ER–Golgi intermediate compartment (ERGIC) and to the Golgi apparatus for extensive glycosylation. ER, ER exit sites, and ERGIC carriers have been found throughout dendrites and axons 131, 132, 133. The ER network is highly non‐uniform in dendrites with interspersed regions of high complexity and diffusion‐restricted zones close to large spines and at branch points, indicating hot spots of local processing 134. In addition, highly mobile carriers of the dendritic ERGIC system are restricted in their mobility by neuronal activity 132.
The Golgi apparatus, however, is localized primarily in the soma raising the question: How are membrane and secretory proteins processed locally in response to local stimuli? A specialized dendritic Golgi compartment termed “Golgi outposts” 132, 135, 136, consisting of discrete, static elements with a Golgi ministack morphology are occasionally observed at primary dendritic branch points. However, this compartment is scarce and found in < 20% of mature hippocampal neurons 136. Consistent with immunoelectron microscopic studies describing undefined, heterogeneous membrane compartments near postsynaptic sites in distal dendrites 137, 138, a tubulo‐vesicular carrier system called the “Golgi satellite” was recently characterized in dendrites 139. The movement of Golgi satellites is controlled by synaptic activity. However, the lack of an entire Golgi enzyme repertoire in Golgi satellites was postulated to lead to altered glycosylation patterns and less efficient membrane delivery of membrane proteins. Indeed, a recent study demonstrated the large‐scale presence of immature N‐glycans at the neuronal plasma membrane 126. The differential immature‐to‐mature glycosylation patterning in subsets of neuronal membrane proteins was found to be important for receptor function and protein turnover, indicating that locally synthesized membrane proteins are likely endowed with distinct features and a signature for the site of synthesis 126.
Ribosomes and their puzzling aspects
Ribosome localization in neuronal compartments is essential for local translation, but their biogenesis occurs in the nucleolus and nucleoplasm 140, 141, 142. It has therefore been puzzling to observe a consistently large fraction of localized mRNAs coding for ribosomal proteins by various high‐throughput approaches 18, 31, 34, 41. The functional significance and whether these mRNA populations are translated into proteins remain unknown. The synthesis of single ribosomal proteins could serve as replacement of specific ribosomal protein subunits for local repair or maintenance. On the other hand, the local translation of ribosomal proteins could result in distinct ribosomal species with different translational properties compared with the somatically synthesized and assembled ones. Evidence for ribosomal heterogeneity has been shown in yeast where mass spectrometric analysis revealed ribosomal heterogeneity that influences protein translation rate 143. Ribosomal heterogeneity is also important for translational control of specific transcripts—Rpl35 mutant mouse embryos showed a defect in the translation of a select group of transcripts important for skeletal patterning during development, while global protein expression remained unaffected 144. Various other ribosomal proteins have also been shown to have tissue‐specific functions 145. Thinking further, this opens up the intriguing possibility of a spatially controlled regulatory step in local translation, achieved by the tethering of specific ribosomal species at select neuronal subcompartments. This is supported by evidence that ligand–receptor interactions (Netrin and its receptor DCC) can regulate the translational state of ribosomes in a localized fashion 146. Tethering of ribosomes and select mRNAs to subcellular sites and organelles has been observed in non‐neuronal cells 42, 43, 147, 148. While it is accepted that secretory and membrane protein mRNAs are translated by ribosomes associated with the ER, whether cytosolic protein mRNAs might also be translated at the ER is still a matter of debate 42, 148, 149, 150. It remains to be determined whether the contradictory findings of cytosolic protein translation at the ER could perhaps reflect an additional layer of translational regulation. Evidence also exists for cytosolic protein mRNAs tethered to the ER that do not undergo translation 151. It is possible that the local heterogeneity of ribosomes and RNA‐binding proteins at the ER could contribute to mRNA tethering and subsequent translation regulation. Similarly, other organelles like mitochondria and endosomes tether specific subsets of mRNAs and serve as translation platforms (discussed below), but the role of ribosome composition in this context has not yet been determined. The ability of all these organelles to attach to the cellular transport machinery 147, 152, 153, 154 could in addition contribute to the dynamic nature and size of source compartments. Careful characterization of unique ribosomal species from subcellular compartments and organelles will shed light on the structural consequence of their specific biochemical composition, their transcript preference, and their physiological significance in local translation.
In contrast to the observation of polyribosome redistribution in dendritic spines following tetanic stimulation 59, the paucity of polyribosomes in axons has long been a major detriment to the idea of axonal translation. Several findings have shed light on this enigma suggesting the need to reconsider long‐standing concepts. In the sciatic nerve, axons contain low numbers of ribosomes. Upon sciatic nerve injury, Schwann cells (myelinating glial cells in the peripheral nervous system) deliver ribosomes including polyribosomes via membrane protrusions and multilamellar vesicles to the sciatic nerve axons, after which axonal ribosome numbers increase by orders of magnitude 155. Consistent with this, an electron microscopy study suggested that spinal cord axons receive ribosomes from central nervous system glia 156. The modes of delivery are not fully understood but broadly assumed to be from direct connections or exosome transfer 157. The origin of local translation material from non‐autonomous sources could also explain the observation of clustered, submembranous distribution of putative source compartments named periaxoplasmic ribosomal plaques, ~20 μm in size, in some axonal preparations 158, 159, 160, 161.
In addition, recent studies have opened up the possibility of monosomes being more translationally active than previously assumed. This was revealed by monosome isolation and translatome profiling in yeast, where most monosomes were translationally active with a bias toward the translation of low abundance regulatory proteins 162. Real‐time imaging of mRNA translation also showed that the fraction of mRNA translated by monosomes is highly transcript‐dependent 22. This new approach now provides the means to address whether axonal translation could be largely carried out by monosomes that might be present at higher copy numbers in axons.
The intercellular route—solving the delivery problem?
The demonstration of the intercellular delivery of ribosomes from Schwann cells to axons 155 as discussed above was one of the studies that indicated that other cells can serve as a source for translation machinery components, mRNAs, regulatory/processing molecules, or even newly synthesized proteins in axons or at synapses 157, 163, 164, 165, 166, 167, 168, 169, 170. The intercellular exchange of material between cells by multivesicular body‐derived exosomes and other extracellular vesicles is now regarded as an integral part of an organism's physiological and pathophysiological repertoire. Proteins, mRNAs, miRNAs, and multiprotein complexes like ribosomes represent extracellular vesicle cargoes. As such, it is not surprising that extracellular vesicles have been shown to complement the intercellular communication system in neurons 171, 172. At the Drosophila neuromuscular junction, multivesicular bodies fuse with the plasma membrane of presynaptic boutons at extrasynaptic sites and the expelled exosomes travel in the extracellular space to interact with receptors in the muscle's subsynaptic reticulum 166, 167. Schwann cells and oligodendrocytes (myelinating cells in close contact with axons) also communicate with axons via exosomes. Dedifferentiated Schwann cells and oligodendrocytes release exosomes for enhancing axonal regeneration and neurotransmitter‐triggered neuronal uptake, respectively 164, 165, 168, 169. Notably, neurons also release exosomes in an activity‐dependent manner 173, 174, 175, and these exosomes bind to presynaptic terminals 176, potentially providing a source of material for local translation and source compartment formation. Intriguingly, active endocannabinoids, involved in regulating synaptic plasticity by targeting presynaptic cannabinoid receptors 109, 177, 178, were found associated with extracellular vesicles 179. In fungi, Septin mRNAs were translated on the endosomal surface during transport 147, 152 potentially serving as a means to sort newly translated proteins into exosomes.
In addition, tunneling nanotubes (direct connection between cells) have also been implicated as routes of material delivery between cells 180, 181. Suggestive evidence for trans‐endocytosis of spinules (small structures extending mostly from postsynaptic spines into presynaptic terminals) provided by an electron microscopy study also offers a route for material transfer ideally suited for synaptic plasticity 182. Spinules, likely containing ribosomes, showed activity‐related changes in their numbers 181, 182. Future experiments combining genetic manipulation, highly sensitive detection methods, and sophisticated sample preparation will determine how such intercellular material transfer relates to local translation.
The energy question
Similar to ribosomal protein mRNAs, transcripts for nuclear‐encoded mitochondrial proteins were also found in large numbers in local transcriptomes 18, 183. Although it is not clear if the full set of mitochondrial transcripts is translated locally, it has been demonstrated that the local translation of some of them is crucial for mitochondrial and neuronal function 183, 184, 185. A large fraction of nuclear‐encoded mitochondrial mRNAs and ribosomes associated with the mitochondrial outer membrane in yeast 43, 186, 187, 188, 189, 190, and flies 191 indicate that the machinery required to carry out local changes in the mitochondrial proteome is present. Furthermore, proteomic analyses of neurons show that the mitochondrial proteome, especially the subset important for energy metabolism, is regulated during synaptic plasticity 192, 193. Moreover, a recent study of the mitochondrial translatome in yeast reveals that the translation of proteins relevant for energy metabolism is dynamically regulated based on the nutrient source 194. If the local mitochondrial proteome is indeed dynamically regulated according to local energy demands, distinct mitochondrial species unique to specific subneuronal compartments must exist. This is supported by the observation of a specially tuned mitochondrial proteome in synaptic mitochondria compared with non‐synaptic mitochondria 195.
Given the availability of the translational machinery associated with mitochondria, including miRNAs for translational regulation 184, 196, 197, is it possible that translation of proteins on the mitochondrial surface is not just limited to mitochondrial proteins? In other words, could mitochondria serve as local platforms for the translation of other proteins? As discussed earlier, the localization of β‐actin mRNA and its corresponding translation hot spots along the length of mitochondria during axon branching is consistent with this view 110 (Table 1).
Another important facet of mitochondria that is largely unexplored in the context of local translation is its potential to supply energy. Proteins are turned over at high rates at synapses. At the glutamatergic postsynaptic density and synaptic vesicle pool alone, about 2,670 and 13,800 proteins are turned over per neuron per minute, respectively 198. Considering such high protein turnover rates, the energy required to synthesize these proteins at a rate of 4 ATP molecules per peptide bond 199 represents a high local energy demand indicating the need for compartmentalized energy sources. Considerable evidence suggests that mitochondria function in spatially defined compartments to power local energy demands. For example, mitochondrial motility is regulated by neuronal activity in dendrites 200 and axons 154 and they redistribute into active spines during spine morphogenesis and plasticity 201. Most importantly, stalling of mitochondria near the base of axonal branches is essential for local translation 110. If mitochondria power local translation, it is not clear if they function as compartmentalized entities serving specific translation compartments. The stabilization of mitochondrial localization at presynaptic sites 202, 203 and dendritic branch points 204 during development and plasticity supports this view. Careful examination of such stable mitochondrial compartments that might power local translation is warranted in the future. On the other hand, the importance of local translation for mitochondrial function 183, 185 reveals a striking mutual dependence between local translation and mitochondrial function.
Although mitochondria produce ATP at a higher yield than glycolysis 205, during high energy demands glycolysis seems to play a complementary role 206, 207, 208, 209. Given the cytosolic nature of glycolytic enzymes, local energy provision by glycolysis requires their spatial compartmentalization in local subcellular regions—called the “glycolytic metabolon”. These metabolons observed in synaptic vesicles 207, 210, 211, postsynaptic densities 212, and nerve endings 213 are thought to power local biological processes. More importantly, high energy demanding states such as neuronal stimulation, hypoxia, and inhibition of mitochondrial respiration drive the transient compartmentalization of these glycolytic metabolons within ~1–2 μm compartments of presynaptic terminals for fueling synaptic function 208. In the context of local translation, it would therefore be useful to investigate whether these glycolytic metabolons exist in dendrites and whether they serve as energy supply.
Summary
Given their complex morphology, neurons have evolved mechanisms to independently remodel their local proteome for efficient neuronal function. Local translation and proteome remodeling is a complex phenomenon carried out in several phases. Each phase (mRNA redistribution, signaling events, translation of proteins) occupies its own spatial compartment ultimately operating in unison to result in a functional outcome. In some cases, translational hot spots of 5–20 μm size have been observed in dendrites and axons following global neuronal stimulation. Similar sizes of translation compartments have also been observed following local stimulation. The repeated observation of this apparent translation compartment size argues that it could be characteristic of local translation processes although what defines this size remains to be determined. In particular, it should be clarified if the measured size of the translation compartment is driven by the biology or the methodology used. It is certain that structural boundaries are not the sole defining parameter, as in most cases the translation compartments were spread over or restricted within a substructure. Current emerging tools should enable clarifying this question in the near future.
Conflict of interest
The authors declare that they have no conflict of interest.
Box 1:In need of answers.
- For a given stimulus:
- What are the specific sets of mRNAs recruited and the subsequent proteins newly made?
- How different are the spatial spreads of these recruited mRNAs and their respective nascent proteins?
- What are the temporal details of the following processes: redistribution of translational machinery, protein translation, the ensuing spatial spread of the nascent proteins, and their functional outcome?
How does stimulus strength influence the size of the different translation compartments—source compartment, spatial spread of the nascent protein, effector compartment, and functional outcome?
Is the site of local translation static, in other words, what is the contribution of dynamic structures such as mitochondria, endosomes, and polysomes in local translation?
Do the proteins synthesized in local translation compartments carry special characteristics and what significance does it have on local biological processes? For instance, given that locally translated membrane proteins (especially receptors) are immaturely glycosylated 126, could this facilitate local modulation of their physiological properties when compared with their somatically synthesized counterparts?
Does local translation of ribosomal mRNA result in the synthesis of special subsets of ribosomal subunits? If yes, do translational compartments possess specialized ribosomes made of special subsets of ribosomal subunits?
What is the contribution of translational machinery delivered from adjacent cellular sources 155 in local translation?
How are the energy demands of various translation compartments met? Do compartmentalized energy sources exist? If yes, what is the spatial compartment they serve and what is their significance in local translation?
EMBO Reports (2017) 18: 693–711
See the Glossary for abbreviations used in this article.
References
- 1. Hoagland MB, Keller EB, Zamecnik PC (1956) Enzymatic carboxyl activation of amino acids. J Biol Chem 218: 345–358 [PubMed] [Google Scholar]
- 2. Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC (1958) A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231: 241–257 [PubMed] [Google Scholar]
- 3. Holt CE, Schuman EM (2013) The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80: 648–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garner CC, Matus A (1988) Different forms of microtubule‐associated protein 2 are encoded by separate mRNA transcripts. J Cell Biol 106: 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT (1990) In situ hybridization histochemistry of Ca2+/calmodulin‐dependent protein kinase in developing rat brain. J Neurosci 10: 1788–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steward O, Wallace CS, Lyford GL, Worley PF (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21: 741–751 [DOI] [PubMed] [Google Scholar]
- 7. Bodian D (1965) A suggestive relationship of nerve cell RNA with specific synaptic sites. Proc Natl Acad Sci USA 53: 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steward O, Fass B (1983) Polyribosomes associated with dendritic spines in the denervated dentate gyrus: evidence for local regulation of protein synthesis during reinnervation. Prog Brain Res 58: 131–136 [DOI] [PubMed] [Google Scholar]
- 9. Steward O, Levy WB (1982) Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci 2: 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30: 489–502 [DOI] [PubMed] [Google Scholar]
- 11. Giuditta A, Dettbarn WD, Brzin M (1968) Protein synthesis in the isolated giant axon of the squid. Proc Natl Acad Sci USA 59: 1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rao A, Steward O (1991) Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: analysis of proteins synthesized within synaptosomes. J Neurosci 11: 2881–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Torre ER, Steward O (1992) Demonstration of local protein synthesis within dendrites using a new cell culture system that permits the isolation of living axons and dendrites from their cell bodies. J Neurosci 12: 762–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber KM, Kayser MS, Bear MF (2000) Role for rapid dendritic protein synthesis in hippocampal mGluR‐dependent long‐term depression. Science 288: 1254–1257 [DOI] [PubMed] [Google Scholar]
- 15. Kang H, Schuman EM (1996) A requirement for local protein synthesis in neurotrophin‐induced hippocampal synaptic plasticity. Science 273: 1402–1406 [DOI] [PubMed] [Google Scholar]
- 16. Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER (1997) Synapse‐specific, long‐term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell 91: 927–938 [DOI] [PubMed] [Google Scholar]
- 17. Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M (2002) Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36: 507–519 [DOI] [PubMed] [Google Scholar]
- 18. Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM (2012) The local transcriptome in the synaptic neuropil revealed by deep sequencing and high‐resolution imaging. Neuron 74: 453–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci USA 103: 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hodas JJ, Nehring A, Hoche N, Sweredoski MJ, Pielot R, Hess S, Tirrell DA, Dieterich DC, Schuman EM (2012) Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT). Proteomics 12: 2464–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL et al (2016) Real‐time quantification of single RNA translation dynamics in living cells. Science 352: 1425–1429 [DOI] [PubMed] [Google Scholar]
- 23. Pichon X, Bastide A, Safieddine A, Chouaib R, Samacoits A, Basyuk E, Peter M, Mueller F, Bertrand E (2016) Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J Cell Biol 214: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vlatkovic IS, Schuman EM (2016) Local translation in dendrites In Dendrites, Stuart G, Spruston N, Häusser M. (eds), pp 129–157. Oxford: Oxford University Press; [Google Scholar]
- 25. tom Dieck S, Kochen L, Hanus C, Heumuller M, Bartnik I, Nassim‐Assir B, Merk K, Mosler T, Garg S, Bunse S et al (2015) Direct visualization of newly synthesized target proteins in situ . Nat Methods 12: 411–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang C, Han B, Zhou R, Zhuang X (2016) Real‐time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu B, Eliscovich C, Yoon YJ, Singer RH (2016) Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan X, Hoek TA, Vale RD, Tanenbaum ME (2016) Dynamics of translation of single mRNA molecules in vivo . Cell 165: 976–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoon YJ, Wu B, Buxbaum AR, Das S, Tsai A, English BP, Grimm JB, Lavis LD, Singer RH (2016) Glutamate‐induced RNA localization and translation in neurons. Proc Natl Acad Sci USA 113: E6877–E6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyashiro K, Dichter M, Eberwine J (1994) On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci USA 91: 10800–10804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moccia R, Chen D, Lyles V, Kapuya E, Yaping E, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M et al (2003) An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci 23: 9409–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC (2006) Identification of process‐localized mRNAs from cultured rodent hippocampal neurons. J Neurosci 26: 13390–13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhong J, Zhang T, Bloch LM (2006) Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci 7: 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE (2010) Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci 30: 15464–15478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW (2009) Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci 29: 4697–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U (2014) Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell 158: 1159–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW (2011) Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA 17: 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez‐Farinas M, Schwarz C, Stephan DA, Surmeier DJ et al (2008) A translational profiling approach for the molecular characterization of CNS cell types. Cell 135: 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS (2009) Cell‐type‐specific isolation of ribosome‐associated mRNA from complex tissues. Proc Natl Acad Sci USA 106: 13939–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shigeoka T, Jung H, Jung J, Turner‐Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE (2016) Dynamic axonal translation in developing and mature visual circuits. Cell 166: 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jan CH, Williams CC, Weissman JS (2014) Principles of ER cotranslational translocation revealed by proximity‐specific ribosome profiling. Science 346: 1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams CC, Jan CH, Weissman JS (2014) Targeting and plasticity of mitochondrial proteins revealed by proximity‐specific ribosome profiling. Science 346: 748–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML et al (2008) Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kratz A, Beguin P, Kaneko M, Chimura T, Suzuki AM, Matsunaga A, Kato S, Bertin N, Lassmann T, Vigot R et al (2014) Digital expression profiling of the compartmentalized translatome of Purkinje neurons. Genome Res 24: 1396–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bagni C, Mannucci L, Dotti CG, Amaldi F (2000) Chemical stimulation of synaptosomes modulates alpha ‐Ca2+/calmodulin‐dependent protein kinase II mRNA association to polysomes. J Neurosci 20: RC76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alvarez J, Giuditta A, Koenig E (2000) Protein synthesis in axons and terminals: significance for maintenance, plasticity and regulation of phenotype. With a critique of slow transport theory. Prog Neurobiol 62: 1–62 [DOI] [PubMed] [Google Scholar]
- 48. Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS (1998) Sorting of beta‐actin mRNA and protein to neurites and growth cones in culture. J Neurosci 18: 251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baj G, Leone E, Chao MV, Tongiorgi E (2011) Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc Natl Acad Sci USA 108: 16813–16818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Will TJ, Tushev G, Kochen L, Nassim‐Assir B, Cajigas IJ, Tom Dieck S, Schuman EM (2013) Deep sequencing and high‐resolution imaging reveal compartment‐specific localization of BDNF mRNA in hippocampal neurons. Sci Signal 6: rs16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harnett MT, Makara JK, Spruston N, Kath WL, Magee JC (2012) Synaptic amplification by dendritic spines enhances input cooperativity. Nature 491: 599–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tonnesen J, Katona G, Rozsa B, Nagerl UV (2014) Spine neck plasticity regulates compartmentalization of synapses. Nat Neurosci 17: 678–685 [DOI] [PubMed] [Google Scholar]
- 53. Araya R, Vogels TP, Yuste R (2014) Activity‐dependent dendritic spine neck changes are correlated with synaptic strength. Proc Natl Acad Sci USA 111: E2895–E2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bloodgood BL, Sabatini BL (2005) Neuronal activity regulates diffusion across the neck of dendritic spines. Science 310: 866–869 [DOI] [PubMed] [Google Scholar]
- 55. Grunditz A, Holbro N, Tian L, Zuo Y, Oertner TG (2008) Spine neck plasticity controls postsynaptic calcium signals through electrical compartmentalization. J Neurosci 28: 13457–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ngo‐Anh TJ, Bloodgood BL, Lin M, Sabatini BL, Maylie J, Adelman JP (2005) SK channels and NMDA receptors form a Ca2+‐mediated feedback loop in dendritic spines. Nat Neurosci 8: 642–649 [DOI] [PubMed] [Google Scholar]
- 57. Kwon HB, Sabatini BL (2011) Glutamate induces de novo growth of functional spines in developing cortex. Nature 474: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matsuzaki M, Honkura N, Ellis‐Davies GC, Kasai H (2004) Structural basis of long‐term potentiation in single dendritic spines. Nature 429: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ostroff LE, Fiala JC, Allwardt B, Harris KM (2002) Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron 35: 535–545 [DOI] [PubMed] [Google Scholar]
- 60. Ifrim MF, Williams KR, Bassell GJ (2015) Single‐molecule imaging of PSD‐95 mRNA translation in dendrites and its dysregulation in a mouse model of fragile X syndrome. J Neurosci 35: 7116–7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frey U, Morris RG (1997) Synaptic tagging and long‐term potentiation. Nature 385: 533–536 [DOI] [PubMed] [Google Scholar]
- 62. Frey U, Morris RG (1998) Weak before strong: dissociating synaptic tagging and plasticity‐factor accounts of late‐LTP. Neuropharmacology 37: 545–552 [DOI] [PubMed] [Google Scholar]
- 63. Govindarajan A, Israely I, Huang SY, Tonegawa S (2011) The dendritic branch is the preferred integrative unit for protein synthesis‐dependent LTP. Neuron 69: 132–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith WB, Starck SR, Roberts RW, Schuman EM (2005) Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron 45: 765–779 [DOI] [PubMed] [Google Scholar]
- 65. Sutton MA, Schuman EM (2005) Local translational control in dendrites and its role in long‐term synaptic plasticity. J Neurobiol 64: 116–131 [DOI] [PubMed] [Google Scholar]
- 66. Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM (2010) Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron 66: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gray NW, Weimer RM, Bureau I, Svoboda K (2006) Rapid redistribution of synaptic PSD‐95 in the neocortex in vivo . PLoS Biol 4: e370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sambandan S, Akbalik G, Kochen L, Rinne J, Kahlstatt J, Glock C, Tushev G, Alvarez‐Castelao B, Heckel A, Schuman EM (2017) Activity‐dependent spatially localized miRNA maturation in neuronal dendrites. Science 355: 634–637 [DOI] [PubMed] [Google Scholar]
- 69. Govindarajan A, Kelleher RJ, Tonegawa S (2006) A clustered plasticity model of long‐term memory engrams. Nat Rev Neurosci 7: 575–583 [DOI] [PubMed] [Google Scholar]
- 70. Kleindienst T, Winnubst J, Roth‐Alpermann C, Bonhoeffer T, Lohmann C (2011) Activity‐dependent clustering of functional synaptic inputs on developing hippocampal dendrites. Neuron 72: 1012–1024 [DOI] [PubMed] [Google Scholar]
- 71. Makino H, Malinow R (2011) Compartmentalized versus global synaptic plasticity on dendrites controlled by experience. Neuron 72: 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Takahashi N, Kitamura K, Matsuo N, Mayford M, Kano M, Matsuki N, Ikegaya Y (2012) Locally synchronized synaptic inputs. Science 335: 353–356 [DOI] [PubMed] [Google Scholar]
- 73. Fu M, Yu X, Lu J, Zuo Y (2012) Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo . Nature 483: 92–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McBride TJ, Rodriguez‐Contreras A, Trinh A, Bailey R, Debello WM (2008) Learning drives differential clustering of axodendritic contacts in the barn owl auditory system. J Neurosci 28: 6960–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Harnett MT, Magee JC, Williams SR (2015) Distribution and function of HCN channels in the apical dendritic tuft of neocortical pyramidal neurons. J Neurosci 35: 1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Larkum ME, Nevian T (2008) Synaptic clustering by dendritic signalling mechanisms. Curr Opin Neurobiol 18: 321–331 [DOI] [PubMed] [Google Scholar]
- 77. Lavzin M, Rapoport S, Polsky A, Garion L, Schiller J (2012) Nonlinear dendritic processing determines angular tuning of barrel cortex neurons in vivo . Nature 490: 397–401 [DOI] [PubMed] [Google Scholar]
- 78. Magee JC (2000) Dendritic integration of excitatory synaptic input. Nat Rev Neurosci 1: 181–190 [DOI] [PubMed] [Google Scholar]
- 79. Magee JC (2011) Observations on clustered synaptic plasticity and highly structured input patterns. Neuron 72: 887–888 [DOI] [PubMed] [Google Scholar]
- 80. Smith SL, Smith IT, Branco T, Hausser M (2013) Dendritic spikes enhance stimulus selectivity in cortical neurons in vivo . Nature 503: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen X, Leischner U, Rochefort NL, Nelken I, Konnerth A (2011) Functional mapping of single spines in cortical neurons in vivo . Nature 475: 501–505 [DOI] [PubMed] [Google Scholar]
- 82. Hill DN, Varga Z, Jia H, Sakmann B, Konnerth A (2013) Multibranch activity in basal and tuft dendrites during firing of layer 5 cortical neurons in vivo . Proc Natl Acad Sci USA 110: 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jia H, Rochefort NL, Chen X, Konnerth A (2010) Dendritic organization of sensory input to cortical neurons in vivo . Nature 464: 1307–1312 [DOI] [PubMed] [Google Scholar]
- 84. Jia H, Varga Z, Sakmann B, Konnerth A (2014) Linear integration of spine Ca2+ signals in layer 4 cortical neurons in vivo . Proc Natl Acad Sci USA 111: 9277–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Varga Z, Jia H, Sakmann B, Konnerth A (2011) Dendritic coding of multiple sensory inputs in single cortical neurons in vivo . Proc Natl Acad Sci USA 108: 15420–15425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. DeBello WM, McBride TJ, Nichols GS, Pannoni KE, Sanculi D, Totten DJ (2014) Input clustering and the microscale structure of local circuits. Front Neural Circuits 8: 112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, Riccio A (2010) An NGF‐responsive element targets myo‐inositol monophosphatase‐1 mRNA to sympathetic neuron axons. Nat Neurosci 13: 291–301 [DOI] [PubMed] [Google Scholar]
- 88. Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha‐Chowdhury R, Leung KM, Holt CE, Fawcett JW (2009) Axonal mRNAs: characterisation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci 42: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL (2007) Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol 178: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Giuditta A, Cupello A, Lazzarini G (1980) Ribosomal RNA in the axoplasm of the squid giant axon. J Neurochem 34: 1757–1760 [DOI] [PubMed] [Google Scholar]
- 91. Giuditta A, Menichini E, Perrone Capano C, Langella M, Martin R, Castigli E, Kaplan BB (1991) Active polysomes in the axoplasm of the squid giant axon. J Neurosci Res 28: 18–28 [DOI] [PubMed] [Google Scholar]
- 92. Giuditta A, Metafora S, Felsani A, Del Rio A (1977) Factors for protein synthesis in the axoplasm of squid giant axons. J Neurochem 28: 1393–1395 [DOI] [PubMed] [Google Scholar]
- 93. Batista AF, Hengst U (2016) Intra‐axonal protein synthesis in development and beyond. Int J Dev Neurosci 55: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Campbell DS, Holt CE (2001) Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32: 1013–1026 [DOI] [PubMed] [Google Scholar]
- 95. Eng H, Lund K, Campenot RB (1999) Synthesis of beta‐tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci 19: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR (2009) Axonal elongation triggered by stimulus‐induced local translation of a polarity complex protein. Nat Cell Biol 11: 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE (2006) Asymmetrical beta‐actin mRNA translation in growth cones mediates attractive turning to netrin‐1. Nat Neurosci 9: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ming GL, Wong ST, Henley J, Yuan XB, Song HJ, Spitzer NC, Poo MM (2002) Adaptation in the chemotactic guidance of nerve growth cones. Nature 417: 411–418 [DOI] [PubMed] [Google Scholar]
- 99. Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C (2006) Signaling mechanisms underlying Slit2‐induced collapse of Xenopus retinal growth cones. Neuron 49: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR (2005) Local translation of RhoA regulates growth cone collapse. Nature 436: 1020–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ (2006) An essential role for beta‐actin mRNA localization and translation in Ca2+‐dependent growth cone guidance. Nat Neurosci 9: 1265–1273 [DOI] [PubMed] [Google Scholar]
- 102. Deglincerti A, Liu Y, Colak D, Hengst U, Xu G, Jaffrey SR (2015) Coupled local translation and degradation regulate growth cone collapse. Nat Commun 6: 6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gracias NG, Shirkey‐Son NJ, Hengst U (2014) Local translation of TC10 is required for membrane expansion during axon outgrowth. Nat Commun 5: 3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Brittis PA, Lu Q, Flanagan JG (2002) Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell 110: 223–235 [DOI] [PubMed] [Google Scholar]
- 105. Dubacq C, Jamet S, Trembleau A (2009) Evidence for developmentally regulated local translation of odorant receptor mRNAs in the axons of olfactory sensory neurons. J Neurosci 29: 10184–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Taylor AM, Wu J, Tai HC, Schuman EM (2013) Axonal translation of beta‐catenin regulates synaptic vesicle dynamics. J Neurosci 33: 5584–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kim S, Martin KC (2015) Neuron‐wide RNA transport combines with netrin‐mediated local translation to spatially regulate the synaptic proteome. Elife 4: e04158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC (2009) Synapse‐ and stimulus‐specific local translation during long‐term neuronal plasticity. Science 324: 1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Younts TJ, Monday HR, Dudok B, Klein ME, Jordan BA, Katona I, Castillo PE (2016) Presynaptic protein synthesis is required for long‐term plasticity of GABA release. Neuron 92: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Spillane M, Ketschek A, Merianda TT, Twiss JL, Gallo G (2013) Mitochondria coordinate sites of axon branching through localized intra‐axonal protein synthesis. Cell Rep 5: 1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Spillane M, Ketschek A, Donnelly CJ, Pacheco A, Twiss JL, Gallo G (2012) Nerve growth factor‐induced formation of axonal filopodia and collateral branches involves the intra‐axonal synthesis of regulators of the actin‐nucleating Arp2/3 complex. J Neurosci 32: 17671–17689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rishal I, Fainzilber M (2014) Axon‐soma communication in neuronal injury. Nat Rev Neurosci 15: 32–42 [DOI] [PubMed] [Google Scholar]
- 113. Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van‐Minnen J, Twiss JL et al (2003) Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40: 1095–1104 [DOI] [PubMed] [Google Scholar]
- 114. Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal‐Ruder Y, Ben‐Yaakov K, Hieda M et al (2008) Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR (2008) Intra‐axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol 10: 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ji SJ, Jaffrey SR (2012) Intra‐axonal translation of SMAD1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron 74: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ben‐Yaakov K, Dagan SY, Segal‐Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M et al (2012) Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J 31: 1350–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yan D, Wu Z, Chisholm AD, Jin Y (2009) The DLK‐1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell 138: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Perlson E, Hanz S, Ben‐Yaakov K, Segal‐Ruder Y, Seger R, Fainzilber M (2005) Vimentin‐dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron 45: 715–726 [DOI] [PubMed] [Google Scholar]
- 120. Perry RB, Doron‐Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, Coppola G, McDonald MK, Gomes C, Geschwind DH et al (2012) Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron 75: 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ch'ng TH, Uzgil B, Lin P, Avliyakulov NK, O'Dell TJ, Martin KC (2012) Activity‐dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell 150: 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Crino P, Khodakhah K, Becker K, Ginsberg S, Hemby S, Eberwine J (1998) Presence and phosphorylation of transcription factors in developing dendrites. Proc Natl Acad Sci USA 95: 2313–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, Konig I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P et al (2008) Caldendrin‐Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol 6: e34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gonzalez SA, Affranchino JL (2016) Processing, fusogenicity, virion incorporation and CXCR4‐binding activity of a feline immunodeficiency virus envelope glycoprotein lacking the two conserved N‐glycosylation sites at the C‐terminus of the V3 domain. Arch Virol 161: 1761–1768 [DOI] [PubMed] [Google Scholar]
- 125. Grigston JC, VanDongen HM, McNamara JO III, VanDongen AM (2005) Translation of an integral membrane protein in distal dendrites of hippocampal neurons. Eur J Neurosci 21: 1457–1468 [DOI] [PubMed] [Google Scholar]
- 126. Hanus C, Geptin H, Tushev G, Garg S, Alvarez‐Castelao B, Sambandan S, Kochen L, Hafner AS, Langer JD, Schuman EM (2016) Unconventional secretory processing diversifies neuronal ion channel properties. Elife 5: e20609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC (2004) Activity‐dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci 7: 244–253 [DOI] [PubMed] [Google Scholar]
- 128. Kacharmina JE, Job C, Crino P, Eberwine J (2000) Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc Natl Acad Sci USA 97: 11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Torre ER, Steward O (1996) Protein synthesis within dendrites: glycosylation of newly synthesized proteins in dendrites of hippocampal neurons in culture. J Neurosci 16: 5967–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sutton MA, Ito HT, Cressy P, Kempf C, Woo JC, Schuman EM (2006) Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell 125: 785–799 [DOI] [PubMed] [Google Scholar]
- 131. Hanus C, Ehlers MD (2016) Specialization of biosynthetic membrane trafficking for neuronal form and function. Curr Opin Neurobiol 39: 8–16 [DOI] [PubMed] [Google Scholar]
- 132. Hanus C, Kochen L, Tom Dieck S, Racine V, Sibarita JB, Schuman EM, Ehlers MD (2014) Synaptic control of secretory trafficking in dendrites. Cell Rep 7: 1771–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Krijnse‐Locker J, Parton RG, Fuller SD, Griffiths G, Dotti CG (1995) The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol Biol Cell 6: 1315–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cui‐Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD (2012) Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell 148: 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Horton AC, Ehlers MD (2003) Dual modes of endoplasmic reticulum‐to‐Golgi transport in dendrites revealed by live‐cell imaging. J Neurosci 23: 6188–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Horton AC, Racz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD (2005) Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron 48: 757–771 [DOI] [PubMed] [Google Scholar]
- 137. Gardiol A, Racca C, Triller A (1999) Dendritic and postsynaptic protein synthetic machinery. J Neurosci 19: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Pierce JP, Mayer T, McCarthy JB (2001) Evidence for a satellite secretory pathway in neuronal dendritic spines. Curr Biol 11: 351–355 [DOI] [PubMed] [Google Scholar]
- 139. Mikhaylova M, Bera S, Kobler O, Frischknecht R, Kreutz MR (2016) A dendritic Golgi satellite between ERGIC and retromer. Cell Rep 14: 189–199 [DOI] [PubMed] [Google Scholar]
- 140. Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H (2001) Maturation and intranuclear transport of pre‐ribosomes requires Noc proteins. Cell 105: 499–509 [DOI] [PubMed] [Google Scholar]
- 141. Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E (2002) 60S pre‐ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Warner JR (2001) Nascent ribosomes. Cell 107: 133–136 [DOI] [PubMed] [Google Scholar]
- 143. Slavov N, Semrau S, Airoldi E, Budnik B, van Oudenaarden A (2015) Differential stoichiometry among core ribosomal proteins. Cell Rep 13: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M (2011) Ribosome‐mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Xue S, Barna M (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG (2010) Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell 141: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Baumann S, Konig J, Koepke J, Feldbrugge M (2014) Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep 15: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Jagannathan S, Reid DW, Cox AH, Nicchitta CV (2014) De novo translation initiation on membrane‐bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA 20: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Jan CH, Williams CC, Weissman JS (2015) LOCAL TRANSLATION. Response to Comment on “Principles of ER cotranslational translocation revealed by proximity‐specific ribosome profiling”. Science 348: 1217 [DOI] [PubMed] [Google Scholar]
- 150. Reid DW, Nicchitta CV (2015) Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol 16: 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Kraut‐Cohen J, Gerst JE (2010) Addressing mRNAs to the ER: cis sequences act up!. Trends Biochem Sci 35: 459–469 [DOI] [PubMed] [Google Scholar]
- 152. Haag C, Steuten B, Feldbrugge M (2015) Membrane‐coupled mRNA trafficking in fungi. Annu Rev Microbiol 69: 265–281 [DOI] [PubMed] [Google Scholar]
- 153. Wagner W, Brenowitz SD, Hammer JA III (2011) Myosin‐Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nat Cell Biol 13: 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Wang XN, Schwarz TL (2009) The mechanism of Ca2+‐dependent regulation of kinesin‐mediated mitochondrial motility. Cell 136: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Court FA, Hendriks WT, MacGillavry HD, Alvarez J, van Minnen J (2008) Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J Neurosci 28: 11024–11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li YC, Cheng CX, Li YN, Shimada O, Atsumi S (2005) Beyond the initial axon segment of the spinal motor axon: fasciculated microtubules and polyribosomal clusters. J Anat 206: 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Twiss JL, Fainzilber M (2009) Ribosomes in axons–scrounging from the neighbors? Trends Cell Biol 19: 236–243 [DOI] [PubMed] [Google Scholar]
- 158. Koenig E, Martin R (1996) Cortical plaque‐like structures identify ribosome‐containing domains in the Mauthner cell axon. J Neurosci 16: 1400–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Koenig E, Martin R, Titmus M, Sotelo‐Silveira JR (2000) Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J Neurosci 20: 8390–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kun A, Otero L, Sotelo‐Silveira JR, Sotelo JR (2007) Ribosomal distributions in axons of mammalian myelinated fibers. J Neurosci Res 85: 2087–2098 [DOI] [PubMed] [Google Scholar]
- 161. Sotelo‐Silveira J, Crispino M, Puppo A, Sotelo JR, Koenig E (2008) Myelinated axons contain beta‐actin mRNA and ZBP‐1 in periaxoplasmic ribosomal plaques and depend on cyclic AMP and F‐actin integrity for in vitro translation. J Neurochem 104: 545–557 [DOI] [PubMed] [Google Scholar]
- 162. Heyer EE, Moore MJ (2016) Redefining the translational status of 80S monosomes. Cell 164: 757–769 [DOI] [PubMed] [Google Scholar]
- 163. Crispino M, Chun JT, Cefaliello C, Perrone Capano C, Giuditta A (2014) Local gene expression in nerve endings. Dev Neurobiol 74: 279–291 [DOI] [PubMed] [Google Scholar]
- 164. Frohlich D, Kuo WP, Fruhbeis C, Sun JJ, Zehendner CM, Luhmann HJ, Pinto S, Toedling J, Trotter J, Kramer‐Albers EM (2014) Multifaceted effects of oligodendroglial exosomes on neurons: impact on neuronal firing rate, signal transduction and gene regulation. Philos Trans R Soc Lond B Biol Sci 369: 20130510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Fruhbeis C, Frohlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Mobius W, Goebbels S, Nave KA et al (2013) Neurotransmitter‐triggered transfer of exosomes mediates oligodendrocyte‐neuron communication. PLoS Biol 11: e1001604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Koles K, Nunnari J, Korkut C, Barria R, Brewer C, Li Y, Leszyk J, Zhang B, Budnik V (2012) Mechanism of evenness interrupted (Evi)‐exosome release at synaptic boutons. J Biol Chem 287: 16820–16834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, Gherbesi N, Budnik V (2009) Trans‐synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Kramer‐Albers EM, Bretz N, Tenzer S, Winterstein C, Mobius W, Berger H, Nave KA, Schild H, Trotter J (2007) Oligodendrocytes secrete exosomes containing major myelin and stress‐protective proteins: trophic support for axons? Proteomics Clin Appl 1: 1446–1461 [DOI] [PubMed] [Google Scholar]
- 169. Lopez‐Verrilli MA, Picou F, Court FA (2013) Schwann cell‐derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 61: 1795–1806 [DOI] [PubMed] [Google Scholar]
- 170. Giuditta A, Chun JT, Eyman M, Cefaliello C, Bruno AP, Crispino M (2007) Axonal and presynaptic RNAs are locally transcribed in glial cells. Riv Biol 100: 203–219 [PubMed] [Google Scholar]
- 171. Kramer‐Albers EM, Hill AF (2016) Extracellular vesicles: interneural shuttles of complex messages. Curr Opin Neurobiol 39: 101–107 [DOI] [PubMed] [Google Scholar]
- 172. Budnik V, Ruiz‐Canada C, Wendler F (2016) Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci 17: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Faure J, Lachenal G, Court M, Hirrlinger J, Chatellard‐Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V et al (2006) Exosomes are released by cultured cortical neurones. Mol Cell Neurosci 31: 642–648 [DOI] [PubMed] [Google Scholar]
- 174. Goldie BJ, Dun MD, Lin M, Smith ND, Verrills NM, Dayas CV, Cairns MJ (2014) Activity‐associated miRNA are packaged in Map1b‐enriched exosomes released from depolarized neurons. Nucleic Acids Res 42: 9195–9208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Lachenal G, Pernet‐Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, Blot B, Haase G, Goldberg Y, Sadoul R (2011) Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci 46: 409–418 [DOI] [PubMed] [Google Scholar]
- 176. Chivet M, Javalet C, Laulagnier K, Blot B, Hemming FJ, Sadoul R (2014) Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J Extracell Vesicles 3: 24722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Castillo PE, Younts TJ, Chavez AE, Hashimotodani Y (2012) Endocannabinoid signaling and synaptic function. Neuron 76: 70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Younts TJ, Chevaleyre V, Castillo PE (2013) CA1 pyramidal cell theta‐burst firing triggers endocannabinoid‐mediated long‐term depression at both somatic and dendritic inhibitory synapses. J Neurosci 33: 13743–13757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Gabrielli M, Battista N, Riganti L, Prada I, Antonucci F, Cantone L, Matteoli M, Maccarrone M, Verderio C (2015) Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep 16: 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Agnati LF, Fuxe K (2014) Extracellular‐vesicle type of volume transmission and tunnelling‐nanotube type of wiring transmission add a new dimension to brain neuro‐glial networks. Philos Trans R Soc Lond B Biol Sci 369: 20130505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Petralia RS, Wang YX, Mattson MP, Yao PJ (2015) Structure, distribution, and function of neuronal/synaptic spinules and related invaginating projections. Neuromolecular Med 17: 211–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Spacek J, Harris KM (2004) Trans‐endocytosis via spinules in adult rat hippocampus. J Neurosci 24: 4233–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Hillefors M, Gioio AE, Mameza MG, Kaplan BB (2007) Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol 27: 701–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Aschrafi A, Kar AN, Natera‐Naranjo O, MacGibeny MA, Gioio AE, Kaplan BB (2012) MicroRNA‐338 regulates the axonal expression of multiple nuclear‐encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci 69: 4017–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Yoon BC, Jung H, Dwivedy A, O'Hare CM, Zivraj KH, Holt CE (2012) Local translation of extranuclear lamin B promotes axon maintenance. Cell 148: 752–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Corral‐Debrinski M, Blugeon C, Jacq C (2000) In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol 20: 7881–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kellems RE, Allison VF, Butow RA (1975) Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol 65: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Marc P, Margeot A, Devaux F, Blugeon C, Corral‐Debrinski M, Jacq C (2002) Genome‐wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 3: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Gadir N, Haim‐Vilmovsky L, Kraut‐Cohen J, Gerst JE (2011) Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae . RNA 17: 1551–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Saint‐Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C (2008) Yeast mitochondrial biogenesis: a role for the PUF RNA‐binding protein Puf3p in mRNA localization. PLoS One 3: e2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, Lu B (2015) PINK1 and parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab 21: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Dahlhaus M, Li KW, van der Schors RC, Saiepour MH, van Nierop P, Heimel JA, Hermans JM, Loos M, Smit AB, Levelt CN (2011) The synaptic proteome during development and plasticity of the mouse visual cortex. Mol Cell Proteomics 10: M110.005413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Piccoli G, Verpelli C, Tonna N, Romorini S, Alessio M, Nairn AC, Bachi A, Sala C (2007) Proteomic analysis of activity‐dependent synaptic plasticity in hippocampal neurons. J Proteome Res 6: 3203–3215 [DOI] [PubMed] [Google Scholar]
- 194. Couvillion MT, Soto IC, Shipkovenska G, Churchman LS (2016) Synchronized mitochondrial and cytosolic translation programs. Nature 533: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Volgyi K, Gulyassy P, Haden K, Kis V, Badics K, Kekesi KA, Simor A, Gyorffy B, Toth EA, Lubec G et al (2015) Synaptic mitochondria: a brain mitochondria cluster with a specific proteome. J Proteomics 120: 142–157 [DOI] [PubMed] [Google Scholar]
- 196. Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y et al (2014) MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Natera‐Naranjo O, Aschrafi A, Gioio AE, Kaplan BB (2010) Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 16: 1516–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Cohen LD, Zuchman R, Sorokina O, Muller A, Dieterich DC, Armstrong JD, Ziv T, Ziv NE (2013) Metabolic turnover of synaptic proteins: kinetics, interdependencies and implications for synaptic maintenance. PLoS One 8: e63191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Waterlow JC (1984) Protein turnover with special reference to man. Q J Exp Physiol 69: 409–438 [DOI] [PubMed] [Google Scholar]
- 200. MacAskill AF, Rinholm JE, Twelvetrees AE, Arancibia‐Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT (2009) Miro1 is a calcium sensor for glutamate receptor‐dependent localization of mitochondria at synapses. Neuron 61: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Li Z, Okamoto K, Hayashi Y, Sheng M (2004) The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119: 873–887 [DOI] [PubMed] [Google Scholar]
- 202. Lewis TL Jr, Turi GF, Kwon SK, Losonczy A, Polleux F (2016) Progressive decrease of mitochondrial motility during maturation of cortical axons in vitro and in vivo . Curr Biol 26: 2602–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Smit‐Rigter L, Rajendran R, Silva CA, Spierenburg L, Groeneweg F, Ruimschotel EM, van Versendaal D, van der Togt C, Eysel UT, Heimel JA et al (2016) Mitochondrial dynamics in visual cortex are limited in vivo and not affected by axonal structural plasticity. Curr Biol 26: 2609–2616 [DOI] [PubMed] [Google Scholar]
- 204. Faits MC, Zhang CM, Soto F, Kerschensteiner D (2016) Dendritic mitochondria reach stable positions during circuit development. Elife 5: e11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Pfeiffer T, Schuster S, Bonhoeffer S (2001) Cooperation and competition in the evolution of ATP‐producing pathways. Science 292: 504–507 [DOI] [PubMed] [Google Scholar]
- 206. Rangaraju V, Calloway N, Ryan TA (2014) Activity‐driven local ATP synthesis is required for synaptic function. Cell 156: 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Hinckelmann MV, Virlogeux A, Niehage C, Poujol C, Choquet D, Hoflack B, Zala D, Saudou F (2016) Self‐propelling vesicles define glycolysis as the minimal energy machinery for neuronal transport. Nat Commun 7: 13233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Jang S, Nelson JC, Bend EG, Rodriguez‐Laureano L, Tueros FG, Cartagenova L, Underwood K, Jorgensen EM, Colon‐Ramos DA (2016) Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron 90: 278–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Pathak D, Shields LY, Mendelsohn BA, Haddad D, Lin W, Gerencser AA, Kim H, Brand MD, Edwards RH, Nakamura K (2015) The role of mitochondrially derived ATP in synaptic vesicle recycling. J Biol Chem 290: 22325–22336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelieres FP, Marco S, Saudou F (2013) Vesicular glycolysis provides on‐board energy for fast axonal transport. Cell 152: 479–491 [DOI] [PubMed] [Google Scholar]
- 211. Ikemoto A, Bole DG, Ueda T (2003) Glycolysis and glutamate accumulation into synaptic vesicles. Role of glyceraldehyde phosphate dehydrogenase and 3‐phosphoglycerate kinase. J Biol Chem 278: 5929–5940 [DOI] [PubMed] [Google Scholar]
- 212. Wu K, Aoki C, Elste A, Rogalski‐Wilk AA, Siekevitz P (1997) The synthesis of ATP by glycolytic enzymes in the postsynaptic density and the effect of endogenously generated nitric oxide. Proc Natl Acad Sci USA 94: 13273–13278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Knull HR, Bronstein WW, DesJardins P, Niehaus WG Jr (1980) Interaction of selected brain glycolytic enzymes with an F‐actin‐tropomyosin complex. J Neurochem 34: 222–225 [DOI] [PubMed] [Google Scholar]


