Summary
The parasitic weeds Striga asiatica and Striga hermonthica cause devastating yield losses to upland rice in Africa. Little is known about genetic variation in host resistance and tolerance across rice genotypes, in relation to virulence differences across Striga species and ecotypes.
Diverse rice genotypes were phenotyped for the above traits in S. asiatica‐ (Tanzania) and S. hermonthica‐infested fields (Kenya and Uganda) and under controlled conditions.
New rice genotypes with either ecotype‐specific or broad‐spectrum resistance were identified. Resistance identified in the field was confirmed under controlled conditions, providing evidence that resistance was largely genetically determined. Striga‐resistant genotypes contributed to yield security under Striga‐infested conditions, although grain yield was also determined by the genotype‐specific yield potential and tolerance. Tolerance, the physiological mechanism mitigating Striga effects on host growth and physiology, was unrelated to resistance, implying that any combination of high, medium or low levels of these traits can be found across rice genotypes.
Striga virulence varies across species and ecotypes. The extent of Striga‐induced host damage results from the interaction between parasite virulence and genetically determined levels of host–plant resistance and tolerance. These novel findings support the need for predictive breeding strategies based on knowledge of host resistance and parasite virulence.
Keywords: grain yield, Oryza glaberrima, Oryza sativa, photosynthesis, post‐attachment resistance, predictive breeding, witchweed
Introduction
Species of the Striga genus (Orobanchaceae family) are obligate hemi‐parasitic plants that parasitize roots of host plants via a specialized organ called the haustorium (Musselman, 1980). Striga spp. (henceforward referred to as Striga) are most prevalent in tropical Africa, where they pose serious threats as weeds in rain‐fed cereal production systems (Parker, 2012). The most important species in rice are Striga asiatica and Striga hermonthica (Rodenburg et al., 2010). Together with the related but less widespread Striga aspera (Willd.) Benth., they constrain rain‐fed rice production in 38 African countries, with an estimated incidence rate of 12% (Rodenburg et al., 2016). Average Striga‐inflicted yield losses of rice in farmers’ fields range between 21% and 80% (Elliot et al., 1993; N'Cho, 2014). The extent of these losses is a function of many factors, including the Striga infestation level, environmental conditions and the genetic interaction between host‐plant genotype and parasite ecotype (host–parasite specificity), which determines the level of Striga resistance and tolerance.
Host‐plant resistance to Striga is defined as the ability to reduce or prevent infection (Shew & Shew, 1994), while tolerance refers to the extent to which effects of infection on the host plant are mitigated (Caldwell et al., 1958). Mechanisms that prevent or reduce Striga seed germination rates are categorized as pre‐attachment resistance, while those that prevent or reduce the success of root penetration or establishment of the vascular connection between host and parasite are called post‐attachment resistance (Yoder & Scholes, 2010). As a consequence of the large genetic variation within and between Striga ecotypes (populations), complete host‐plant resistance (immunity) against this parasite is rare. As host damage can be inflicted by a few parasitic infections, varieties with partial Striga resistance should also have good levels of tolerance to avoid yield losses in the field (Rodenburg & Bastiaans, 2011).
A number of studies have shown the existence of genetic variation in resistance to different ecotypes of Striga across a range of rice genotypes. For example, Harahap et al. (1993) showed that four genotypes of Oryza sativa were partially resistant to S. hermonthica in western Kenya. Riches et al. (1996) and Johnson et al. (1997), identified five genotypes of the African rice species Oryza glaberrima and two O. sativa genotypes with partial resistance against an ecotype of S. aspera and S. hermonthica in northern Côte d'Ivoire. Under controlled environments, Jamil et al. (2011) identified a number of interspecific New Rice for Africa (NERICA) cultivars with pre‐attachment resistance against S. hermonthica, while Cissoko et al. (2011) identified cultivars with post‐attachment resistance (within the same germplasm pool) against an ecotype of S. hermonthica and S. asiatica.
These studies assessed either resistance among a relatively diverse group of rice genotypes against one Striga species or ecotype in the field (Harahap et al., 1993; Jamil et al., 2012), or resistence among a genetically related group of rice genotypes (i.e. the NERICAs) against different Striga species or ecotypes (Cissoko et al., 2011; Jamil et al., 2011; Rodenburg et al., 2015) under controlled environment conditions. Thus, these and other studies have resulted in a pool of known genotypes resistant to a specific ecotype of Striga, but limited information on how broad‐spectrum resistance is against genetically different species and ecotypes, and how expression of resistance is affected by environmental variability. Moreover, none of the previous studies has conclusively established genetic variation in Striga tolerance in rice and the potential mechanistic background of tolerance in infected hosts. Identification of genetic sources of broad‐spectrum resistance and effective tolerance in rice germplasm is critical for marker‐assisted and conventional breeding programmes to develop useful cultivars for affected rice farmers.
The objectives of this study were therefore to determine whether Striga resistance among a diverse set of rice genotypes (some with previously identified resistance to Striga) is specific to a particular Striga species or ecotype, or broad‐spectrum; whether resistance against Striga is sufficient to maintain high rice grain yields under Striga‐infested conditions in different environments; and whether genetic variation in Striga tolerance exists in rice and through which host‐plant morphological or physiological traits this can be assessed. Achievement of the last objective would also shed light on the mechanisms underlying tolerance to this parasite.
Materials and Methods
Field screening trials
Twenty rice genotypes of different species and origins (Table 1), 19 of which had putative Striga resistance, were grown in Striga‐infested plots at Kyela, Tanzania (Striga asiatica (L.) Kuntze), and at Namutumba, Uganda, and Mbita, Kenya (both Striga hermonthica Benth.). The interspecific rice cultivar NERICA‐2 was included as a Striga‐resistant and high‐yielding check against which the performances of all other genotypes were compared across sites, Striga ecotypes and years. The cultivar IAC165 (Oryza sativa ssp. japonica), originally from Brazil, was included as a Striga‐susceptible check. Seeds of all rice genotypes were obtained from the Africa Rice Center (AfricaRice), Cotonou, Benin. Seeds of S. asiatica and S. hermonthica were collected in the previous season from plants parasitizing rice at Kyela, Tanzania (Sa‐Kyela) and Namutumba, Uganda (Sh‐Namutumba) and maize (Zea mays L.) at Mbita, Kenya (Sh‐Mbita) in farmers’ fields surrounding the experimental field sites. These seeds were used to supplement the existing soil seed bank in the field trials as well as for the controlled environment studies.
Table 1.
Upland rice genotypes used in the study; their full names, species, origins, presumed reaction types to Striga and literature sources
| Genotypes | Species and origin | Reaction type | Source |
|---|---|---|---|
| ACC102196 | Oryza glaberrima (Liberia) | R | 1 , 2 |
| Agee | O. glaberrima (Ghana) | R | 3 |
| Anakila | O. glaberrima (Mali) | R | 3 |
| CG14 | O. glaberrima (Senegal) | R | 4 , 5 , 6 , 7 , 8 |
| Makassa | O. glaberrima (Sierra Leone) | R | 1 , 2 |
| MG12 | O. glaberrima (Mali) | R | 2 |
| Ble Chai | Oryza sativa ssp. indica (Thailand) | R | 9 |
| IAC165 | O. sativa ssp. indica (Brazil) | S | 3 , 4 , 5 , 6 |
| IR49255‐B‐B‐5‐2 | O. sativa ssp. indica (Philippines) | R | 2 , 6 , 9 |
| IR38547‐B‐B‐7‐2‐2 | O. sativa ssp. indica (Philippines) | R | 9 |
| UPR‐103‐80‐1‐2 | O. sativa (origin unknown) | R | 9 |
| WAB56‐50 | O. sativa ssp. japonica (Côte d'Ivoire) | I | 4 , 6 , 8 |
| WAB56‐104 | O. sativa ssp. japonica (Côte d'Ivoire) | I | 4 , 5 , 6 , 8 |
| WAB928‐22‐2‐A‐A‐B | O. sativa, ssp. japonica × indica (Côte d'Ivoire) | R | 10 |
| SCRID090‐60‐1‐1‐2‐4 | O. sativa ssp. japonica, cv FOFIFA161 × interspecific cv NERICA‐3 (Madagascar) | R | 11 |
| WAB935‐5‐A‐2‐A‐A‐B | Interspecific, cv IR47 × cv CG20 (Côte d'Ivoire) | R | 10 |
| WAB880‐1‐32‐1‐1‐P2‐HB‐1‐1‐2‐2 | Interspecific, cv WAB56‐50 × cv CG14 (Côte d'Ivoire) | R | 11 |
| NERICA‐2* | Interspecific, cv WAB56‐104 × cv CG14 (Côte d'Ivoire) | R | 4 , 5 , 6 , 8 |
| NERICA‐4 | Interspecific, cv WAB56‐104 × cv CG14 (Côte d'Ivoire) | R | 4 , 5 , 6 , 8 |
| NERICA‐10 | Interspecific, cv WAB56‐104 × cv CG14 (Côte d'Ivoire) | R | 4 , 5 , 6 , 8 |
Text in bold indicates how genotype names are abbreviated.
1Riches et al. (1996); 2Johnson et al. (1997); 3Jamil et al. (2012); 4Cissoko et al. (2011); 5Jamil et al. (2011); 6Rodenburg et al. (2015); 7Kaewchumnong & Price (2008); 8Samejima et al. (2016); 9Harahap et al. (1993); 10C. Riches, NRI (pers. comm.); 11L. M. Raboin, Cirad (pers. comm.); R, Resistant; I, Intermediate; S, Susceptible; all interspecifics are offspring of crosses between Oryza glaberrima and O. sativa ssp. japonica. *NERICA‐2 is the Striga‐resistant and high‐yielding check.
The S. asiatica field screening trials were conducted during the rainy seasons (February/March–July) of 2014 and 2015 in Mbako (9°35ʹS, 33°48ʹE; 525 m above sea level, asl), in Kyela District, Mbeya Region in southern Tanzania (Supporting Information Table S1). Kyela District is a S. asiatica‐endemic upland rice‐growing area. This screening trial was executed in an already infested farmer's field. Rainfall data were obtained from a rain gauge installed in the middle of the field (Table S1; Fig. 1).
Figure 1.
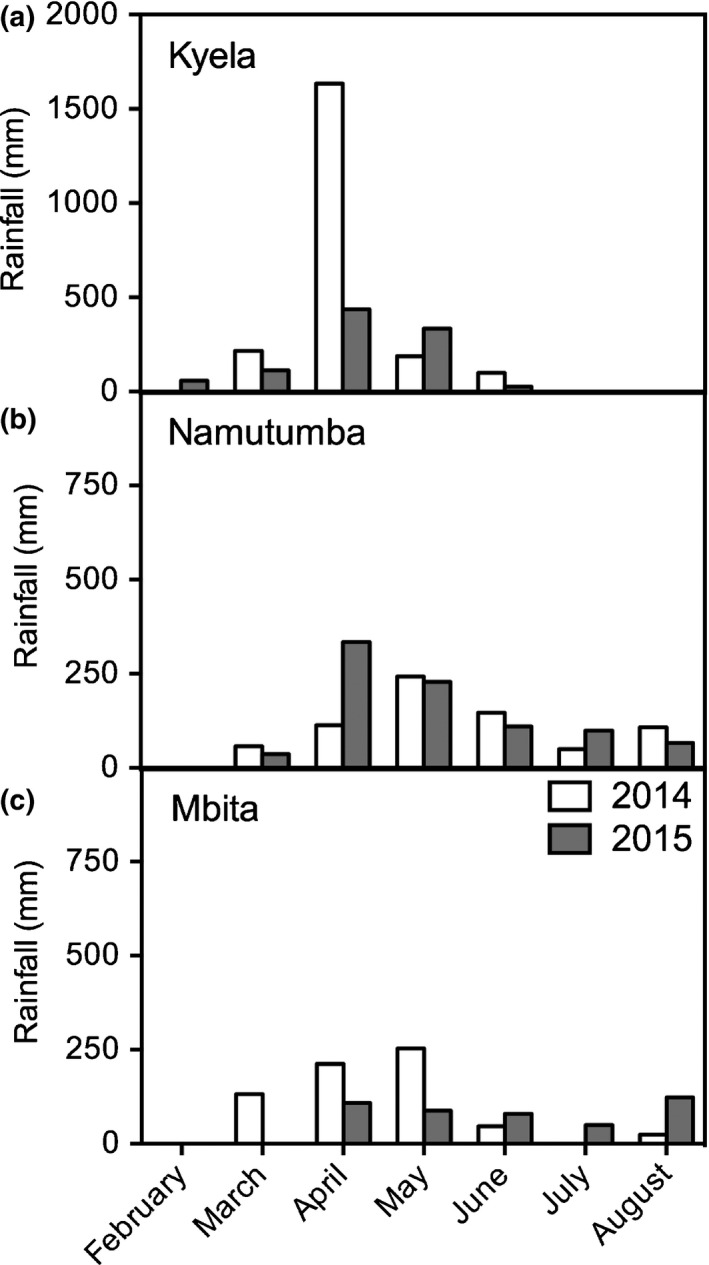
Rainfall data for field sites in (a) Kyela (Tanzania), (b) Namutumba (Uganda) and (c) Mbita (Kenya) in 2014 and 2015.
The S. hermonthica field screening trials were conducted during the long rainy seasons of 2014 and 2015 (March–August/September) at two locations: in a farmer's field in Nsinze, Namutumba District, Uganda (00°51ʹN, 33°41ʹE; 1125 m asl); and at the farm of the International Centre of Insect Physiology and Ecology (ICIPE) at Mbita (0°43ʹS, 34°20ʹE; 1141 m asl), in Suba District, western Kenya (Table S1). Both trials were laid out on heavily Striga‐infested fields. Rainfall data in Namutumba were obtained from a nearby meteorological station, and in Mbita from ICIPE's meteorological station at the experimental farm (Table S1; Fig. 1).
All field trials were laid out in a randomized block design with six replicates. At Kyela each plot, representing an individual genotype, measured 1.25 × 3.75 m (4.69 m2) and contained five rows of 15 hills with an inter‐hill distance of 0.25 × 0.25 m (Table S1). At Mbita and Namutumba, each plot measured 1.25 × 2.75 m (3.44 m2) with five rows of 11 hills with the same hill and row distances as in Kyela. Plots were separated by one open row of 0.25 m to avoid neighbour effects. Each replicate was separated by a 1.25‐m alley.
Each plot received supplementary Striga seeds that were mixed with 200 g of white sand and incorporated in the upper 5–10 cm of soil. An amount of 0.21 g of S. asiatica seed m−2 (germination rate: 70%) was provided at Kyela, in both years. At Namutumba, the S. hermonthica seed infestation rate was 0.29 g m−2 in 2014 and 0.26 g m−2 in 2015 (germination rate 90%) and at Mbita this was 0.29 g m−2 (germination rate 90%) in both years.
For crop establishment and weed control, we followed procedures described in Rodenburg et al. (2015). At all sites, fertilizer was applied at 35 d after sowing (DAS). In Kyela, nitrogen−phosphorus−potassium (N‐P‐K; 20 : 10 : 10) was applied at an equivalent rate of 100 kg ha−1, while at Namutumba and Mbita, N‐P‐K (17 : 17 : 17) was applied at a rate of 50 kg ha−1 (Table S1).
The number of Striga plants that emerged within the central area of each plot (comprising 27 rice hills) was recorded regularly. At Kyela, counting was carried out at 71 and 88 DAS and at harvest in 2014, and at 43, 54, 71, 82 and 105 DAS and at harvest in 2015. In Namutumba and Mbita, Striga plants were counted bi‐weekly, at 43, 57, 71, 85, 99/100 DAS and at harvest in both years. These data enabled the assessment of the maximum number of emerged Striga plants (NS max), a measure of Striga resistance in the field (Rodenburg et al., 2005). At harvest, emerged Striga plants within each observation area of 27 hills in each plot were collected, oven‐dried at 70°C for 48 h, and weighed on digital weighing scales, for the assessment of Striga biomass dry weight.
At harvest, the height of the rice plants growing in the central nine hills was measured from ground level to the tip of the tallest panicle. Rice panicles were harvested from the same central 27 hills of each plot and air‐dried for 2 wk, after which rice grains were separated from the panicles and weighed. Grain moisture content was assessed, using a digital grain moisture meter (Model SS‐7; Satake Eng. Co., Tokyo, Japan), to correct rice grain dry weights to 14% moisture content. Rice straw biomass dry weights were assessed for plants from the central nine hills within each 27‐hill harvest area, and included all aboveground rice biomass, except panicles. The straw was oven‐dried at 70°C for 48 h before weighing.
Resistance ranking of rice genotypes under controlled environmental conditions
To determine the impact of the field environment on the resistance ranking of the genotypes, a subset of 11 genotypes were phenotyped for post‐attachment resistance under semi‐controlled environment conditions at a screen house at AfricaRice in Dar es Salaam, Tanzania (with the Kyela ecotype of S. asiatica – Sa‐Kyela – and the Mbita ecotype of S. hermonthica – Sh‐Mbita) and fully controlled environment conditions in a screening facility at the University of Sheffield (with the Namutumba ecotype of S. hermonthica – Sh‐Namutumba) with parasite seeds collected from the field sites. Striga‐infected rice plants of each genotype were grown in a rhizotron system as described by Cissoko et al. (2011). Four replicates were evaluated for each genotype × Striga spp. combination. The genotypes tested were NERICA‐2, NERICA‐4, WAB928, WAB880, WAB56‐50, WAB56‐104, IR38547, Ble Chai, SCRID090, CG14 and the susceptible check IAC165. Ble Chai was missing in the rhizotron screen against S. asiatica because of germination failure. Quantification of post‐attachment resistance levels was based on total number of attachments and mean parasite biomass dry weight (assessed after oven‐drying at 50°C for 48 h) per host root system for each genotype at 21 d after inoculation (DAI).
Determining the tolerance levels of the rice genotypes
A pot experiment was carried out in the screen house of AfricaRice, from October 2015 until February 2016, using natural incoming light (70% of light intensity outside the screen house). Plastic 10‐l pots (height: 27.5 cm; diameter: 25 cm) were filled with a sand : soil mixture at a ratio of 2 : 1. The soil was collected from the experimental farm of Sokoine University of Agriculture, in Morogoro, and the sand was collected from the shores of the Ruvu River, adjacent to the Ruvu irrigated rice scheme. This mixture contained 0.17% N, 6.7 ppm P and 228 ppm K, and had a pH (H2O) of 6.8 (Crop Nutrition Laboratory Services Ltd, Nairobi, Kenya). The pot experiment comprised two Striga levels (Striga‐infested and Striga‐free) and nine rice genotypes, following a randomized complete block design with four replications. It included the O. glaberrima genotypes ACC102196, CG14 and Makassa, the O. sativa genotypes IR38547, WAB56‐104, WAB928 and IAC165, and the interspecific genotypes WAB935 and NERICA‐10.
Thirty‐six pots (half of the experiment) were infested with S. asiatica seeds, and the other half contained Striga‐free soil (control treatments). For the Striga‐infestation treatment, the upper 10 cm of soil was mixed with 0.050 g of viable S. asiatica seeds. During the 10 d after Striga infestation, the soil in each pot was kept between field capacity and saturation to allow Striga seed preconditioning. Fertilizer was applied at a rate equivalent to 100 kg of N‐P‐K (17 : 17 : 17) ha−1 (c. 1.2 g per pot), and mixed with the upper 10 cm of soil during Striga infestation. Rice was sown at a rate of six seeds per pot (10 d after Striga infestation) and thinned to three plants per pot at 14 DAS. Throughout the experiment, in all pots soil moisture levels were maintained between field capacity and saturation.
Rice plant height from ground level to the tip of the tallest leaf (at 43 and 57 DAS) or panicle (at maturity) was measured to assess maximum height. At maturity, rice grains obtained from the three plants in each pot were threshed, air‐dried for 10 d and weighed. The grain moisture content of each sample was assessed to standardize grain weights to 14% moisture content. At harvest, rice straw (leaf, stem and rachis) was collected from each pot, oven‐dried and weighed to establish total aboveground straw biomass dry weight. Emerged Striga plants were counted every 3 d starting after the first Striga emergence in each pot, to assess maximum aboveground Striga numbers (NS max).
Photosynthesis was measured with the Li‐Cor 6400XT from Li‐Cor Bioscience (Lincoln, NE, USA). Light‐saturated leaf CO2 assimilation rates (A max) of rice were measured at 1200 μmol m−2 s−1 (photosynthetically active radiation (PAR); over the waveband 400–700 nm) at c. 30, 45 and 60 DAS (± 2 d). On each occasion, measurements were conducted on four consecutive days, with one full replicate per day, between 11:00 and 15:00 h. The same plants were used for repeated measurements. Measurements were always made halfway along the length of the youngest fully expanded leaf. During the measurements, leaf temperature ranged between 29.1 and 39.6°C. Relative humidity in the leaf chamber was controlled to stay within the range of 35–50%. The inlet CO2 concentration was set at 400 ppm and depletion never exceeded 20 ppm.
Statistical analyses
Before analyses, data were checked for homoscedasticity and normality following Sokal & Rohlf (1995). Following these tests, field data on rice grain and Striga dry weights were analysed using a linear mixed model. We tested whether there was a significant location × year × genotype interaction effect, and, where this was the case, we fitted a model for each location (Kyela, Mbita and Namutumba) separately, and tested whether there was a significant year × genotype interaction effect within each location. We first performed a log‐likelihood ratio test for the homogeneity of variance and, when the variance was not constant, we took into account the heterogeneity of the variances. When the year × genotype interaction effect was significant (P < 0.05), we fitted a model for each year separately, where genotype was considered a fixed effect and block, nested in replicate, and replicate were considered random effects. For parameters for which there was a significant cultivar effect, Dunnett's method (Dunnett, 1955) was used to compare each genotype with NERICA‐2, which was used as a control. For analyses of the maximum number of emerged Striga plants (NS max), a generalized linear mixed model (McCullagh & Nelder, 1989) was used under the assumption of a Poisson distribution. Least‐squares means (LS‐Means) and associated SE derived from the linear mixed model were computed. Spearman rank correlations for parameters measured in the field were calculated between LS‐Means of NS max and Striga dry weight (DWStriga), between NS max and rice grain yield, and between rice grain yield and rice plant height.
The rhizotron and pot data were analysed following checks for homoscedasticity and normality. ANOVAs were followed by a comparison of means using Tukey's honest significant difference test. Striga‐inflicted losses in plant height (Height) and light‐saturated photosynthesis (A max) were calculated relative to the Striga‐free control for each genotype as
(X c, the mean Striga‐free control value of parameter X, calculated over four replicates; X s,i, the value of parameter X of a Striga‐infected plant of replicate i.) The Striga‐inflicted losses data were analysed using a generalized linear mixed model with a binomial distribution. All data were analysed using Sas/stat software, Version 9.2 of the Sas System for Windows (SAS Institute, 2011).
Results
Resistance levels among diverse rice genotypes exposed to different Striga species and ecotypes
Year by rice genotype interaction effects on NS max were highly significant (P < 0.001) at all sites, requiring analysis per year (Table S2). At all sites and in all years, rice genotype had a highly significant (P < 0.001) effect on NS max (Table S2). The genotype ranking showed that the resistant check NERICA‐2 was always among the most resistant genotypes (Fig. 2). Highly significant (P < 0.001) correlations were found between NS max and Striga biomass in all field trials (Table S3).
Figure 2.
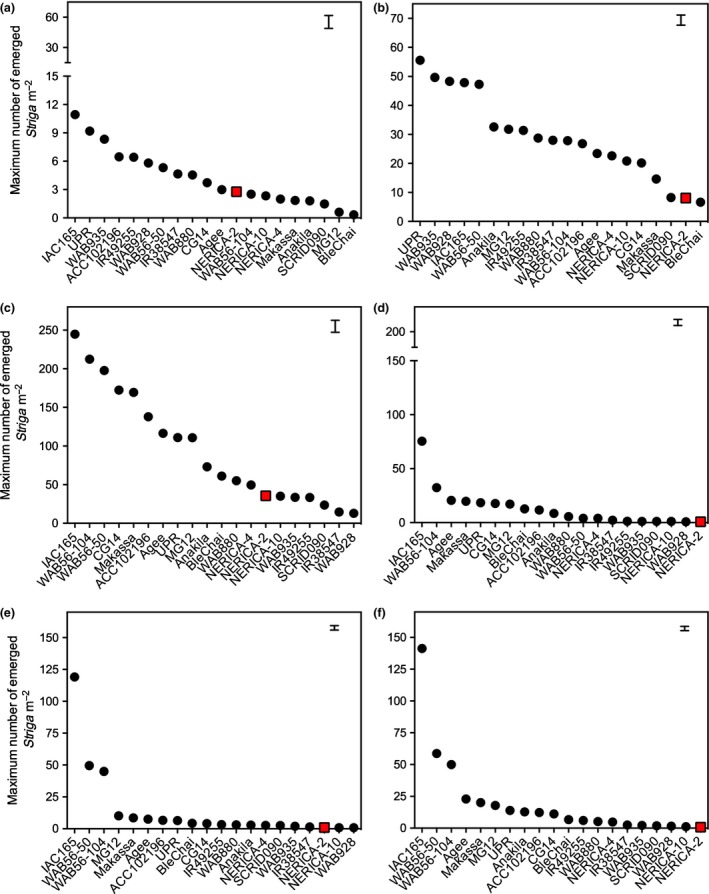
Maximum number of emerged Striga plants m−2 per rice cultivar for field trials at Kyela, Tanzania under Striga asiatica infestation in Kyela in (a) 2014 and (b) 2015, Striga hermonthica infestation in Mbita, Kenya in (c) 2014 and (d) 2015 and S. hermonthica infestation in Namutumba, Uganda in (e) 2014 and (f) 2015. Bars represent ± SE of the least squares (LS) means. Red boxes indicate the position of resistant check genotype NERICA‐2.
In Kyela in 2015, a year with generally high S. asiatica infection levels, only SCRID090 and Ble Chai showed similar levels of resistance to NERICA‐2. All other genotypes had significantly (P < 0.01) higher infection levels. Twelve genotypes proved moderately resistant, with Makassa, CG14, NERICA‐10, NERICA‐4 and Agee being the most resistant. Five genotypes were clearly susceptible: UPR, WAB935, WAB928, IAC165 and WAB56‐50 (Fig. 2b). In 2014, a year with generally lower infection levels in Kyela, the genotype ranking was similar, although 11 genotypes showed resistance levels equivalent to that of NERICA‐2, and two genotypes, MG12 and Ble Chai, were significantly (P < 0.01) more resistant. Six genotypes (UPR, WAB935, WAB928, IAC165, IR49255 and ACC102196) were significantly (P < 0.05) more susceptible (Fig. 2a).
In Mbita in 2014, a year with generally high infection levels, three genotypes, NERICA‐10, WAB935 and IR49255, had similar resistance levels to S. hermonthica to NERICA‐2, and three genotypes, SCRID090, IR38547 and WAB928, were significantly (P < 0.01) more resistant. Thirteen genotypes had significantly (P < 0.01) higher infection levels than NERICA‐2. Four of them (NERICA‐4, WAB880, Ble Chai and Anakila) proved moderately resistant, while five (IAC165, WAB56‐104, WAB56‐50, CG14 and Makassa) were very susceptible (Fig. 2c). The ranking in 2015, with generally lower S. hermonthica infection levels, was similar but showed less differentiation between genotypes. Five genotypes (NERICA‐10, WAB935, IR49255, SCRID090 and WAB928) had similar infection levels to NERICA‐2, and the remaining genotypes were all significantly (P < 0.05) less resistant (Fig. 2d). In Namutumba, the years were more similar in terms of S. hermonthica infection level (Fig. 2e,f). In 2014 only five genotypes had similar resistance levels to NERICA‐2. In 2015, four of them, WAB935, SCRID090, WAB928 and NERICA‐10, were as resistant as NERICA‐2. All other genotypes were more susceptible (Fig. 2e).
In rhizotron screens (controlled environments), genotype rankings on Striga numbers and Striga biomass dry weight were similar to those observed in the field (Figs 2, 3). In both field and rhizotron screens, WAB928 showed susceptibility to S. asiatica (Kyela), but high resistance against both S. hermonthica ecotypes (Mbita and Namutumba) (Figs 3, 4). Similarly, although less pronounced, IR38547 was generally susceptible to S. asiatica (Kyela) in both rhizotron and field screening but resistant to both ecotypes of S. hermonthica (Figs 3, 4).
Figure 3.
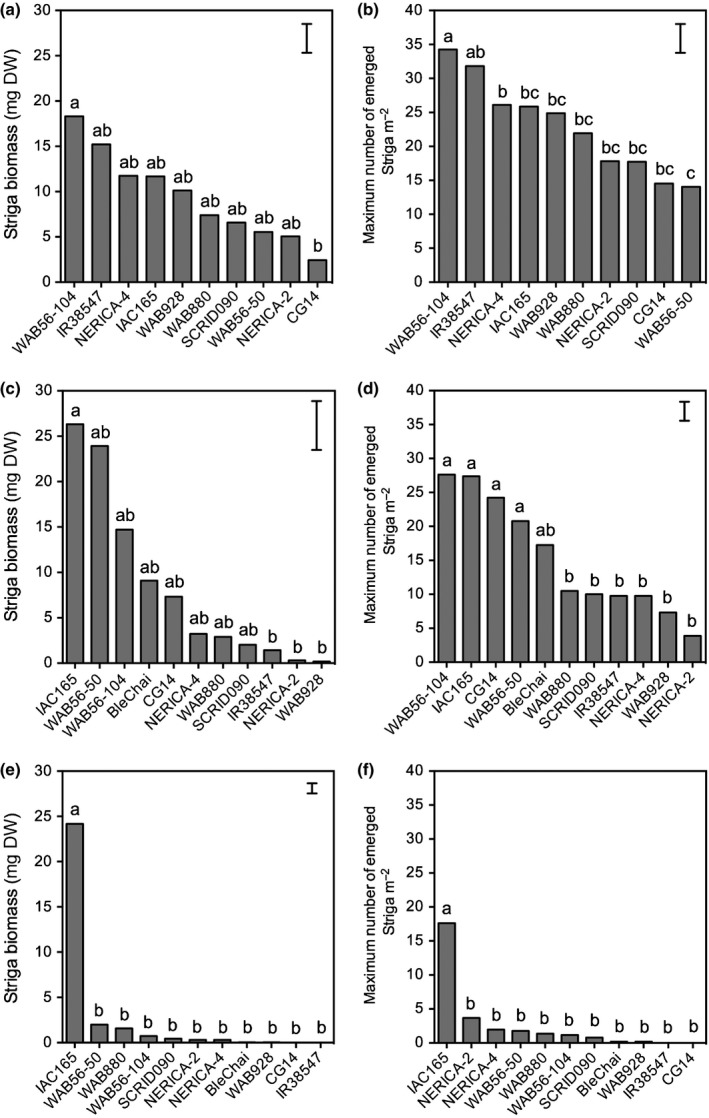
Striga numbers and biomass on a subset of rice genotypes observed in the rhizotron system infected with either (a, b) Striga asiatica from Kyela (Sa‐Ky), (c, d) Striga hermonthica from Mbita (Sh‐Mb) or (e, f) Striga hermonthica from Namutumba (Sh‐Na). Bars represent ± SE of the least squares (LS) means. Bars with different lowercase letters are significantly different (P < 0.05).
Figure 4.

Resistance phenotypes of WAB935, WAB928, IR38547 and IAC165, screened in the rhizotron systems infected with either Striga asiatica from Kyela (Sa‐Ky), Striga hermonthica from Mbita (Sh‐Mb) or Striga hermonthica from Namutumba (Sh‐Na). Bars: main images, 0.5 cm; inset image, 500 μm.
Genotype‐specific crop yields across environments, Striga species and ecotypes
At each site, significant year by genotype interaction effects on rice grain yields under Striga‐infested conditions were observed (Table S4), requiring analyses per year. In each year and at each site, highly significant (P < 0.0001) genotype effects on rice grain yields under Striga‐infested conditions were observed. Similarly, significant correlations between rice yield and rice plant height were observed in Kyela and Namutumba (Table S3).
Under conditions of generally low S. asiatica infection levels, at Kyela in 2014, the resistant check genotype NERICA‐2 had the highest rice grain yields (Fig. 5a) but yields were generally low. In 2015, the rice grain yields were much higher overall, despite the higher overall Striga infection levels (Fig. 5b). ACC102196, Agee, Anakila, Makassa and CG14, all O. glaberrima, had significantly (P < 0.05) higher grain yields, while WAB935, IR38547 and WAB928 had significantly (P < 0.01) lower yields than check genotype NERICA‐2.
Figure 5.
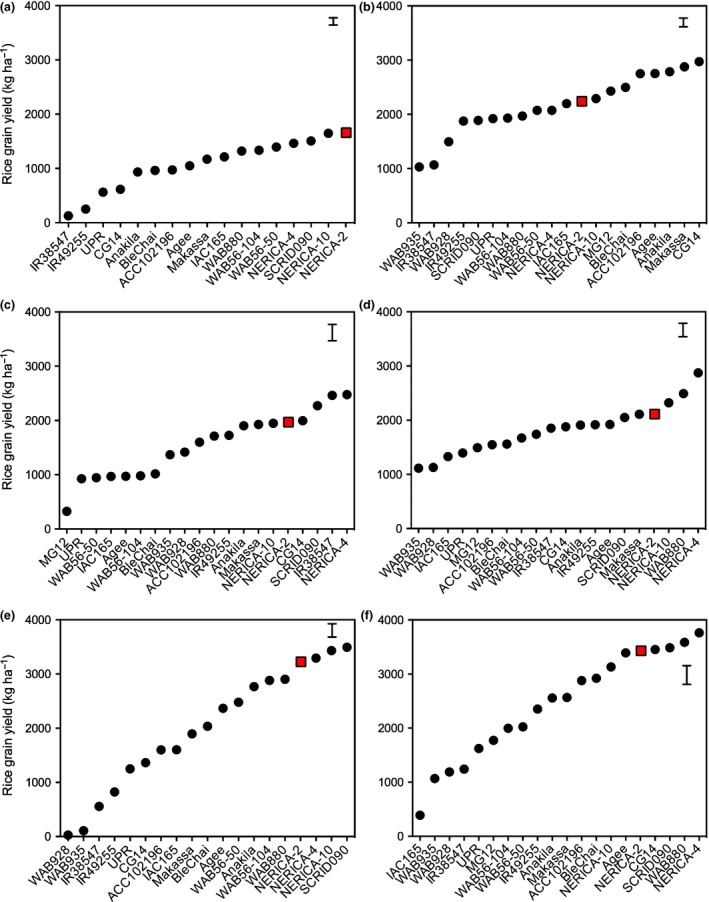
Rice grain weights under Striga asiatica infestation in Kyela in (a) 2014 and (b) 2015, Striga hermonthica infestation in Mbita, Kenya in (c) 2014 and (d) 2015 and S. hermonthica infestation in Namutumba, Uganda in (e) 2014 and (f) 2015. Bars represent ± SE of the least squares (LS) means. Red boxes indicate the position of resistant check genotype NERICA‐2.
In Mbita, rice grain yields under S. hermonthica‐infested conditions were comparable across years (Fig. 5c,d). A large number of genotypes (12 in 2014 and 13 in 2015) were statistically as high yielding as NERICA‐2 and only one genotype (NERICA‐4 in 2015) had a significantly (P < 0.05) higher yield than NERICA‐2. The remaining seven (in 2014) and five (in 2015) had significantly (P < 0.05) lower yields than NERICA‐2.
In Namutumba, grain yields as high as that of NERICA‐2 were obtained from six genotypes in 2014 (WAB56‐104, Anakila, NERICA‐10, SCRID090, WAB880 and NERICA‐4) and eight genotypes in 2015 (ACC102196, Ble Chai, NERICA‐10, Agee, CG14, SCRID090, WAB880 and NERICA‐4) (Fig. 5d,e). All other genotypes had significantly lower grain yields. NERICA‐2 and SCRID090 were the most stable in yield across years, while a number of genotypes showed high yield variation between years. Across sites and years, NERICA‐2, ‐4 and ‐10, and to a lesser extent SCRID090 showed stable high yields when grown under Striga‐infested conditions.
In situations with generally high Striga infection levels (Kyela 2015 and Mbita 2014), genotype means of rice grain yields showed significant (P < 0.05) negative correlations with means of maximum aboveground Striga numbers (NS max) (Table S3). No significant correlations were observed in other experiments. Comparing NS max to rice grain yields, per genotype, with reference lines (horizontal and vertical) through NERICA‐2 enables the identification of genotypes in four quadrants, relative to the check genotype (Fig. 6): quadrant I comprises genotypes that are more susceptible but also higher yielding than NERICA‐2 under Striga‐infested conditions; quadrant II contains genotypes that are more susceptible and lower yielding than NERICA‐2 under Striga infestation; genotypes in quadrant III are more resistant but lower yielding than NERICA‐2 under Striga infestation and genotypes in quadrant IV are more resistant and higher yielding than NERICA‐2 under Striga infestation.
Figure 6.
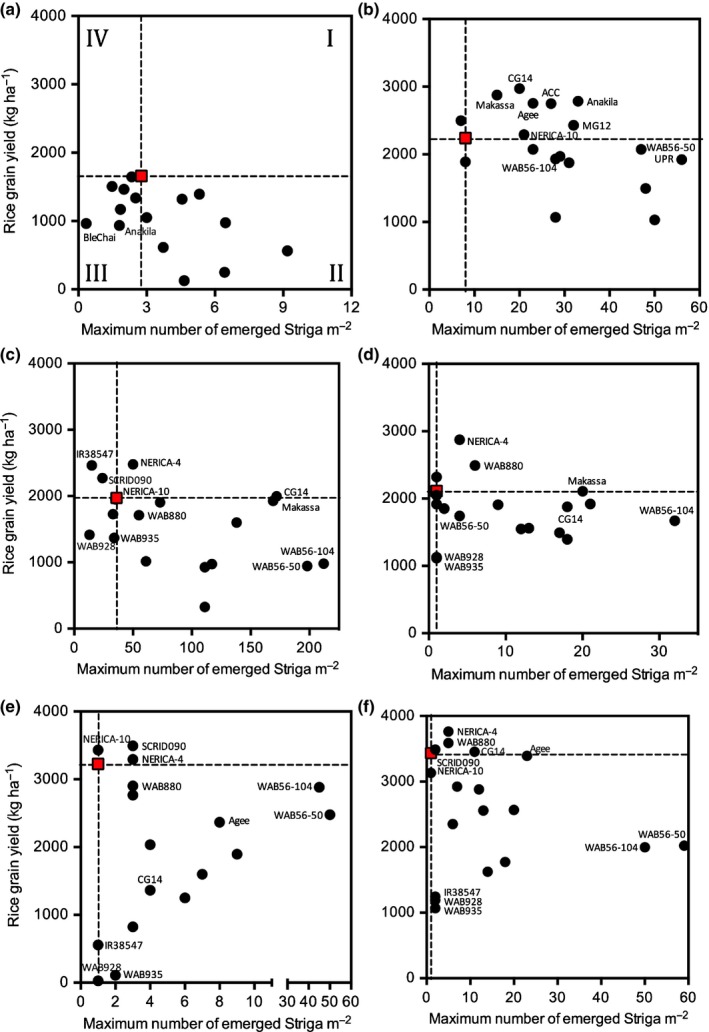
Rice grain yields plotted against maximum number of emerged Striga plants under Striga asiatica infestation in Kyela in (a) 2014 and (b) 2015, Striga hermonthica infestation in Mbita, Kenya in (c) 2014 and (d) 2015 and S. hermonthica infestation in Namutumba, Uganda in (e) 2014 and (f) 2015. Red boxes indicate the position of resistant check genotype NERICA‐2. Roman numerals in (a) refer to different quadrants with groups of genotypes relative to NERICA‐2.
Very few genotypes were found in quadrant IV in any of the years and sites. In Kyela in 2014, there was no genotype with a better performance than NERICA‐2 and in 2015, only Ble Chai had a similar resistance level and as high a yield as NERICA‐2. In Mbita, in 2014, only IR38547 and SCRID090 performed similarly to NERICA‐2. The performance of NERICA‐10 was similar to that of NERICA‐2 in terms of S. hermonthica resistance and yield in both years in Mbita and in Namutumba but none of the genotypes performed better than NERICA‐2.
Seven genotypes were identified in quadrant I in Kyela in 2015. Six of them were O. glaberrima genotypes (Makassa, CG14, Agee, ACC102196, Anakila and MG12) and the other one was an interspecific (NERICA‐10). At the S. hermonthica‐infested field sites, NERICA‐4 was consistently as high or higher yielding despite higher Striga infection levels than NERICA‐2. Other genotypes that featured in this quadrant under S. hermonthica infestation were WAB880, CG14, SCRID090, Makassa and Agee. WAB56‐50 and ‐56‐104, the O. sativa parents of the NERICA genotypes, were almost always in quadrant II, with higher Striga infection levels and lower yields than NERICA‐2. Some genotypes were as resistant as or more resistant than NERICA‐2, but had much lower yields (quadrant III), notably Ble Chai and Anakila in Kyela in 2014, WAB928 and WAB935 in Mbita and WAB928, WAB935 and IR38547 in Namutumba.
Genetic variation in Striga tolerance and host‐plant morphological and physiological traits depicting tolerance
To shed light on the role of tolerance in host‐plant performance, a pot experiment was conducted with Striga‐free compared with Striga‐infested plants of a subset of genotypes, with CG14, ACC102196, Makassa and NERICA‐10 as potential tolerant lines and with S. asiatica from Kyela as the parasite ecotype. Given the significant correlations between rice grain yield and rice plant height in the field (Table S3), we used plant height as a proxy for crop performance in the pot experiment.
Highly significant (P < 0.01) negative effects of Striga on plant height were observed on 43‐d‐old rice plants (Table S5). There was a significant infection by genotype interaction effect on rice plant height at 43 (P = 0.024) and 57 (P = 0.0004) DAS as well as on the maximum plant height (P = 0.0038). Significant genotypic differences were observed in Striga‐inflicted (maximum) height (P < 0.0001; F = 17.6) losses relative to uninfected control plants. Height losses across genotypes ranged from 32% (ACC102196) to 63% (IAC165).
Significant (P < 0.05) negative effects of Striga infection on leaf photosynthesis were observed on 30‐d‐old rice plants and even more pronounced effects were observed 15 d later (Table S5). Infection by genotype effects on leaf photosynthesis were only significant at 45 DAS. When compared within genotypes, four genotypes showed a significantly (P < 0.05) lower rate of photosynthesis in leaves of Striga‐infected plants compared with the Striga‐free controls at 30 DAS (Fig. 7b). Photosynthesis of genotypes ACC102196, Makassa, CG14, WAB928 and WAB935 was not significantly affected at 30 DAS. At 45 DAS, all but one genotype (ACC102196) showed highly significantly (P < 0.001) reduced photosynthesis levels in Striga‐infected compared with Striga‐free plants (Fig. 7c). In ACC102196, the reduction of photosynthesis was also significant (P < 0.05) but less pronounced compared with other genotypes.
Figure 7.
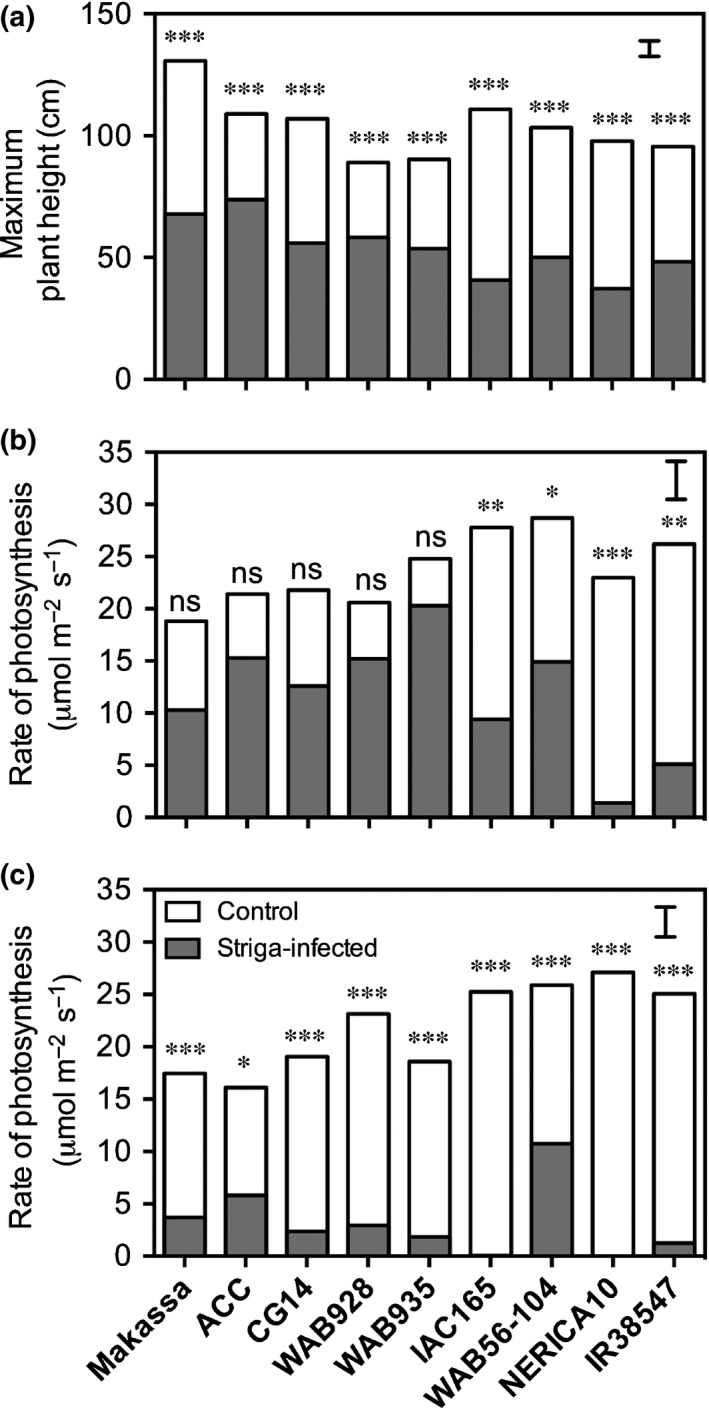
(a) Maximum rice plant height and (b, c) light‐saturated photosynthesis at (b) 30 and (c) 45 d after sowing (DAS) for a subset of genotypes grown in Striga asiatica‐infested (grey bars) and Striga‐free control (white bars) pots. Significant within‐genotype differences between infected and uninfected plants are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Error bars indicate ± SE.
For an accurate assessment of tolerance, the genotype‐specific differences in Striga infection levels should be considered. In the pot experiment, CG14 (NS max = 10) was significantly (F = 15.7; P < 0.0001) more resistant than six other genotypes. Only NERICA‐10 (NS max = 15) and IR38547 (NS max = 16) were equally resistant. With an NS max of 43 Striga plants, WAB928 was significantly more susceptible than any other genotype except ACC102196 (NS max = 31). The latter was as susceptible as IAC165, WAB935, WAB56‐104 and Makassa (NS max = 22–27).
The extent of height losses was relatively independent of infection level (Fig. 8a). A number of genotypes with small to moderate height losses compared with controls (ACC102196, WAB928 and WAB935) had high Striga infection levels, while some genotypes with the greatest height losses (e.g. NERICA‐10) were among the least infected. Comparison of relative height losses between genotypes with similar infection levels indicated genotype differentiation in Striga effects. For NERICA‐10, Striga infection had a greater effect on plant height than equally resistant CG14. Given their high infection levels, ACC102196 and to a lesser extent WAB928 were less affected by Striga in terms of plant height reduction.
Figure 8.
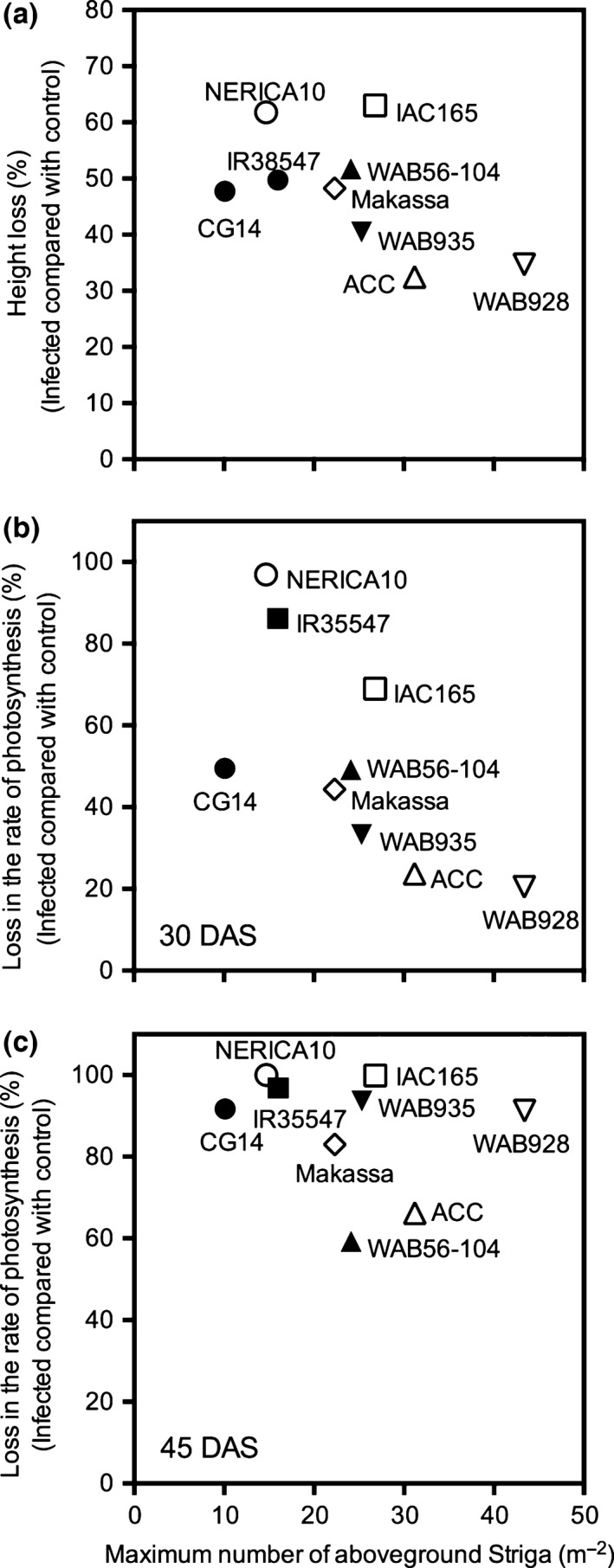
Striga asiatica‐inflicted losses (%) in (a) maximum rice plant height and (b, c) light‐saturated photosynthesis at (b) 30 and (c) 45 d after sowing (DAS), relative to the Striga‐free control plants, plotted against the maximum number of emerged Striga plants, for a subset of genotypes grown in the pot experiment. Genotypes are indicated by different symbols.
Losses in photosynthesis were also relatively independent of infection level (Fig. 8b,c). Again, the more resistant genotypes seemed to incur higher losses than the more susceptible ones, with more pronounced differences at 30 DAS (Fig. 8b) compared with 45 DAS (Fig. 8c). At 30 DAS, NERICA‐10 and IR38547 showed the greatest negative effects of Striga. Relatively low (≤ 50%) Striga‐inflicted losses in photosynthesis at 30 DAS were observed with CG14, Makassa, WAB56‐104, WAB935, ACC1021196 and WAB928. Most notable were ACC1021196 and WAB928, as they showed the smallest effects despite the highest Striga infection levels (Fig. 8b). Fifteen days later, at 45 DAS, Striga effects on photosynthesis were more severe, leading to near‐total losses in the majority of genotypes, irrespective of Striga infection level. Only WAB56‐104 and ACC102196 maintained these losses well below 80% (Fig. 8c).
Discussion
Does Striga resistance occur among genotypes and is it Striga species‐ or ecotype‐specific or broad‐spectrum?
At each site, a relatively large number of resistant rice germplasms were confirmed or newly identified. However, the level of resistance of rice genotypes in the field varied with Striga species and ecotype, as well as between sites and years. Climate variations affected overall Striga infection levels across genotypes, as shown before by Johnson et al. (1997). In the current study this is illustrated by the differences in overall infection levels between 2014 and 2015 at Kyela and Mbita which were associated with clear differences in rainfall between the years. This was further supported by the observation that in Namutumba, where rainfall was comparable in the two years, Striga infection levels were similar. However, environmental effects did not alter the expression of resistance; at low infection levels, it just became more difficult to distinguish between the resistance levels of the genotypes. In years of high infection, quantitative differences in resistance were more obvious and the resistance rankings corresponded well with those obtained in the controlled environment experiments.
The genotype rankings in the S. hermonthica‐infested sites at Mbita and Namutumba were similar. Consistent broad‐spectrum resistance against S. hermonthica (hence against both ecotypes) was observed among a large number of genotypes including NERICA‐2, ‐4 and ‐10, WAB928, ‐935 and ‐880, IR38547 and ‐49255, SCRID090, Ble Chai and Anakila. In Kyela, many genotypes resistant to S. asiatica were also found (i.e. Ble Chai, NERICA‐2, ‐4, and ‐10, SCRID090, CG14, Makassa and Agee). The resistance in the NERICA cultivars confirms previous findings (Cissoko et al., 2011; Jamil et al., 2011; Rodenburg et al., 2015; Samejima et al., 2016).
While resistance rankings at Namutumba and Mbita (S. hermonthica) were very similar, differences were observed in overall infection levels between the ecotypes at both sites. The field infection levels in Namutumba were similar and rather low in both years, while in Mbita the infection levels were similar to the levels in Namutumba in 2015 but much higher in 2014. The experiments carried out under controlled environment conditions showed similar results; higher infection levels overall were observed with S. hermonthica from Mbita than with S. hermonthica from Namutumba, suggesting that the Mbita ecotype was more virulent than the Namutumba ecotype for these rice genotypes. Between Striga species there were even more notable differences. First, against S. asiatica very few, if any, genotypes showed effective post‐attachment resistance in the rhizotron experiment, to the extent shown against the two S. hermonthica ecotypes where parasitic biomass approached zero on a number of genotypes. Second, while there was some overlap in the genotypes of rice with good resistance against S. asiatica and S. hermonthica, there were also some striking differences in species‐specific reaction types. For example, WAB928 and WAB935 were very resistant to both ecotypes of S. hermonthica but were among the most susceptible to S. asiatica in the field. Similarly, IR38547‐B‐B‐7‐2‐2 and IR49255‐B‐B‐5‐2 were resistant to S. hermonthica ecotypes but moderately susceptible to the S. asiatica ecotype. This observation on contrasting reaction types between Striga species was confirmed with WAB928 and IR38547 under controlled environment conditions.
Thus, our data show that some genotypes of rice exhibited ecotype‐specific resistance, others exhibited resistance against the two ecotypes of S. hermonthica (but not the ecotype of S. asiatica) and two genotypes (NERICA‐2 and SCRID090) showed very strong and reliable broad‐spectrum resistance across both parasite species and ecotypes. Three others (NERICA‐4 and ‐10 and Ble Chai) were also consistently among the more resistant to both Striga species and ecotypes. To determine whether the observed differences in the resistance of specific rice genotypes against the two S. hermonthica ecotypes and the S. asiatica ecotype reflect a Striga species difference requires follow‐up studies with a wider range of ecotypes screened under controlled environment conditions.
Is resistance against Striga enough to maintain high rice grain yields under Striga‐infested conditions in different environments?
Encouragingly, the rice genotypes that exhibited good broad‐spectrum resistance were among the high‐yielding and farmer‐preferred varieties and thus could be introduced and promoted more widely in Striga‐prone areas. Moreover, they provide valuable additional sources for resistance breeding. Effective breeding, using marker‐assisted selection (MAS), would, however, require the identification of the genes or quantitative trait loci (QTLs) underlying the Striga resistance, as demonstrated by Swarbrick et al. (2009).
Some rice genotypes, that is, WAB935 and ‐928 in Kyela and IAC165, WAB56‐50 and WAB56‐104 in Mbita, were very susceptible and had low grain yields. Conversely, a number of rather susceptible genotypes, for example O. glaberrima genotypes Makassa, CG14, Agee and ACC102196, still had good grain yields despite relatively high infection levels. The yields obtained by some of the other less resistant genotypes, including Anakila, Agee and CG14 (all O. glaberrima), appeared more variable across years. Yield stability under Striga‐infested conditions seems therefore to be one of the merits of resistance, but more data are required to support such a conclusion. Yield performance of the NERICA cultivars, for instance, could also be the result of their general environmental adaptation and high yield potential (Saito et al., 2012; Sekiya et al., 2013).
Correlations between rice grain yields and Striga numbers (both S. asiatica and S. hermonthica) were only significant under situations of high parasite pressure. This confirms previous studies, both with rice (Rodenburg et al., 2015) and with sorghum (Sorghum bicolor (L.) Moench) (Rodenburg et al., 2005). The inconsistency of this correlation indicates that resistance, responsible for reduced Striga infection levels, is not the sole determinant of high yields under Striga‐infested field conditions. In years with lower Striga infection levels, tolerance and yield potential, rather than resistance, seem to be important, confirming previous studies by Rodenburg et al. (2005, 2015).
Does genetic variation in tolerance to Striga exist in rice germplasm and which host‐plant morphological or physiological traits can be used to predict tolerance?
In studies to identify tolerance in maize (Pierce et al., 2003) and sorghum (Bebawi & Farah, 1981; Showemimo, 2003), the extent of stunting of the host plant is often used as an indicator for tolerance. In our study, the usefulness of the reduction in height of the main stem of Striga‐infected rice (as a percentage of the uninfected plant) could be assessed as this parameter ranged between c. 30% and 65% as a function of the rice genotype. Based on this measure, ACC102196 and WAB928 proved to be more tolerant than the other cultivars. This is in agreement with the yield data from the field trials for ACC101196, but not for WAB928 (which was very low yielding). Possibly the yield potential or environmental adaptation of WAB928 is suboptimal, causing a low baseline yield level, but this requires additional investigation.
In this study, measurement of the rate of photosynthesis at 30 DAS was a better discriminator of tolerance (lower levels of Striga damage) between the rice genotypes than the reduction in height of infected plants. The ability of maize and sorghum varieties to maintain high rates of photosynthesis under Striga infection has also proved a good indicator for physiological tolerance (Gurney et al., 2002; Rodenburg et al., 2008). The rice cultivars began to exhibit the damaging effects of Striga very quickly after attachment, confirming previous findings (e.g. Cechin & Press, 1994; Watling & Press, 2001). Measurements of the rate of photosynthesis also became less discriminating and predictive of tolerance with time after infection, illustrating the need to make these measurements early during the host–parasite interaction.
The O. glaberrima genotypes Makassa, ACC102196 and CG14 showed good tolerance in comparison to many of the O. sativa genotypes used in this study. They showed no significant Striga‐induced reductions in leaf photosynthesis at early stages of the host–parasite interaction when CO2 assimilation rates of other genotypes were already severely reduced. CG14 and Makassa were also relatively high yielding in Mbita, even in a year when infection levels were generally high such as 2014. All the O. glaberrima genotypes screened at Kyela showed higher yields at higher S. asiatica infection levels than NERICA‐2. As the species O. glaberrima is generally not high yielding (Dingkuhn et al., 1998), these relatively high yields, despite high infection levels, are probably indeed the outcome of effective physiological tolerance. This observation also agrees with that of Johnson et al. (1997), who found lower levels of Striga damage on Striga‐infected O. glaberrima cultivars compared with O. sativa cultivars. Based on these observations, O. glaberrima germplasm may be a good source of ‘tolerance’ genes that could be exploited for breeding this trait into Striga‐resistant cultivars.
Resistance and tolerance are not often found together in the same genotype. Some susceptible cultivars show high levels of tolerance to Striga damage, for example ACC102196 and WAB928 in the current study, while some cultivars with good resistance are highly sensitive to one or two parasite attachments, for example NERICA‐10. A similar combination of high resistance but high sensitivity was observed in the sorghum genotype N13 (Rodenburg et al., 2005). Thus, in order to control Striga and maintain high yields, both tolerance and resistance are required in cultivars recommended to farmers. The high genetic variability of the parasite seed bank means that even strongly resistant cultivars may be infected by a few Striga individuals, leading to yield losses if the genotypes do not possess some degree of tolerance. Conversely, tolerant genotypes will allow the build‐up of the Striga seed bank if they do not possess some degree of resistance. Thus, varieties with both resistance and tolerance, grown in combination with other control measures, will provide a feasible and durable solution to farmers and delay the evolution of virulence in parasite populations.
Conclusions
This is the first study to compare the resistance levels of the same suite of rice genotypes in three regions of Africa infested by different genetic ecotypes of S. hermonthica and S. asiatica. First, we have shown that the resistance ranking of rice genotypes in the field was very similar to that under controlled environment conditions, thus demonstrating that resistance was genetically determined. Second, some rice genotypes exhibited broad‐spectrum resistance to all the ecotypes of Striga, while others exhibited ecotype‐specific resistance. Finally, the resistance rankings of rice genotypes at Mbita and Namutumba – both areas infested with S. hermonthica ecotypes – were similar, suggesting that the parasite virulence genes in these populations were similar. This contrasted with the virulence profile of the S. asiatica ecotype, as some of the rice genotypes exhibited different resistance rankings. We have shown that tolerance was also genetically determined, and the level of tolerance varied across genotypes, as evident from the extent to which Striga‐inflicted losses in plant height and photosynthesis were differentially mitigated across genotypes, but it was independent of the level of resistance in these genotypes. Thus, the grain yield of a given rice genotype obtained in a Striga‐infested field is the result of the inherent yield potential of that genotype and the level of host resistance and tolerance against the field‐specific parasite species and ecotype. These novel findings provide invaluable information for molecular and conventional rice breeders, and strongly support the need for predictive breeding strategies to be employed for affected staple crops such as rice. For such a predictive breeding approach, knowledge of the molecular genetic background of host resistance and tolerance can be coupled to that of the prevailing parasite ecotype in a specific region in order to breed cultivars with effective defence.
Author contributions
J.R., M.C., J.B., C.A.O.M., and J.D.S. planned and designed the research; J.R., M.C., N.K., R.I. and I.M. performed experiments and collected data; I.D. analysed data; J.R. M.C., C.A.O.M., I.D. and J.D.S. wrote the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Table S1 Overview of experimental conditions of the field trials conducted at Kyela, Tanzania, Namutumba, Uganda and Mbita, Kenya (2014 and 2015)
Table S2 ANOVA output on maximum emerged Striga numbers observed in the field at three sites (Kyela, Mbita and Namutumba) in the years 2014 and 2015 with 20 rice genotypes
Table S3 Spearman rank correlations between LS‐Means of maximum aboveground Striga numbers and aboveground Striga biomass dry weights at harvest, between NS max and rice grain yields, and between rice grain yields and rice plant height for the field data in Kyela, Mbita and Namutumba in both seasons (2014 and 2015)
Table S4 ANOVA output on rice grain yields and rice straw dry weights observed in the field at three sites (Kyela, Mbita and Namutumba) in the years 2014 and 2015 with 20 rice genotypes
Table S5 ANOVA output on maximum rice plant height, and plant height at 43 and 57 d after sowing (DAS), and photosynthesis at 30 and 45 DAS, with Striga infection and rice genotype as sources of variation
Acknowledgements
This work was funded by the Biotechnology and Biological Sciences Research Council, the Department for International Development and (through a grant to BBSRC) the Bill & Melinda Gates Foundation, under the Sustainable Crop Production Research for International Development programme, a joint initiative with the Department of Biotechnology of the Government of India's Ministry of Science and Technology (grant no. BB/J011703/1).
Contributor Information
Jonne Rodenburg, Email: j.rodenburg@cgiar.org.
Julie D. Scholes, Email: j.scholes@sheffield.ac.uk
References
- Bebawi FF, Farah AF. 1981. Effects of parasitic and non‐parasitic weeds on sorghum. Experimental Agriculture 17: 415–418. [Google Scholar]
- Caldwell RM, Schafer JF, Compton LE, Patterson FL. 1958. Tolerance to cereal leaf rust. Science 128: 714–715. [DOI] [PubMed] [Google Scholar]
- Cechin I, Press MC. 1994. Influence of nitrogen on growth and photosynthesis of a C3 cereal, Oryza sativa, infected with the root hemiparasite Striga hermonthica . Journal of Experimental Botany 45: 925–930. [Google Scholar]
- Cissoko M, Boisnard A, Rodenburg J, Press MC, Scholes JD. 2011. New Rice for Africa (NERICA) cultivars exhibit different levels of post‐attachment resistance against the parasitic weeds Striga hermonthica and Striga asiatica . New Phytologist 192: 952–963. [DOI] [PubMed] [Google Scholar]
- Dingkuhn M, Jones MP, Johnson DE, Sow A. 1998. Growth and yield potential of Oryza sativa and O. glaberrima upland rice cultivars and their interspecific progenies. Field Crops Research 57: 57–69. [Google Scholar]
- Dunnett CW. 1955. A multiple comparison procedure fro comparing several treatments with a control. Journal of the American Statistical Association 50: 1096–1121. [Google Scholar]
- Elliot PC, Clarisse RN, Beby R, Josue HR. 1993. Weeds in rice in Madagascar. International Rice Research Notes 18: 53–54. [Google Scholar]
- Gurney AL, Taylor A, Mbwaga A, Scholes JD, Press MC. 2002. Do maize cultivars demonstrate tolerance to the parasitic weed Striga asiatica? Weed Research 42: 299–306. [Google Scholar]
- Harahap Z, Ampong Nyarko K, Olela JC. 1993. Striga hermonthica resistance in upland rice. Crop Protection 12: 229–231. [Google Scholar]
- Jamil M, Charnikhova T, Houshyani B, van Ast A, Bouwmeester HJ. 2012. Genetic variation in strigolactone production and tillering in rice and its effect on Striga hermonthica infection. Planta 235: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil M, Rodenburg J, Charnikhova T, Bouwmeester HJ. 2011. Pre‐attachment Striga hermonthica resistance of New Rice for Africa (NERICA) cultivars based on low strigolactone production. New Phytologist 192: 964–975. [DOI] [PubMed] [Google Scholar]
- Johnson DE, Riches CR, Diallo R, Jones MJ. 1997. Striga on rice in West Africa; crop host range and the potential of host resistance. Crop Protection 16: 153–157. [Google Scholar]
- Kaewchumnong K, Price AH. 2008. A study on the susceptibility of rice cultivars to Striga hermonthica and mapping of Striga tolerance quantitative trait loci in rice. New Phytologist 180: 206–216. [DOI] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. 1989. Generalized linear models. London, UK: Chapman & Hall. [Google Scholar]
- Musselman LJ. 1980. The biology of Striga, Orobanche and other root‐parasitic weeds. Annual Review of Phytopathology 18: 463–489. [Google Scholar]
- N'Cho SA. 2014. Socio‐economic impacts and determinants of parasitic weed infestation in rainfed rice systems of sub‐Saharan Africa. PhD thesis, Wageningen University Wageningen, the Netherlands.
- Parker C. 2012. Parasitic weeds: a world challenge. Weed Science 60: 269–276. [Google Scholar]
- Pierce S, Mbwaga AM, Press MC, Scholes JD. 2003. Xenognosin production and tolerance to Striga asiatica infection of high‐yielding maize cultivars. Weed Research 43: 139–145. [Google Scholar]
- Riches CR, Johnson DE, Jones MP 1996. The selection of resistance to Striga species in upland rice In: Moreno MT, Cubero JI, Berner D, Joel D, Musselman LJ, Parker C, eds. Advances in parasitic research. Proceedings of the Sixth International Parasitic Weed Symposium. Cordoba, Spain: Dirección general de investigación agraria, Servicio de publicaciones y divulgación, 673–680. [Google Scholar]
- Rodenburg J, Bastiaans L. 2011. Host‐plant defence against Striga spp.: reconsidering the role of tolerance. Weed Research 51: 438–441. [Google Scholar]
- Rodenburg J, Bastiaans L, Schapendonk AHCM, van der Putten PEL, van Ast A, Dingemanse NJ, Haussmann BIG. 2008. CO2‐assimilation and chlorophyll fluorescence as indirect selection criteria for host tolerance against Striga. Euphytica 160: 75–87. [Google Scholar]
- Rodenburg J, Bastiaans L, Weltzien E, Hess DE. 2005. How can field selection for Striga resistance and tolerance in sorghum be improved? Field Crops Research 93: 34–50. [Google Scholar]
- Rodenburg J, Cissoko M, Kayeke J, Dieng I, Khan ZR, Midega CAO, Onyuka EA, Scholes JD. 2015. Do NERICA rice cultivars express resistance to Striga hermonthica (Del.) Benth. and Striga asiatica (L.) Kuntze under field conditions? Field Crops Research 170: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenburg J, Demont M, Zwart SJ, Bastiaans L. 2016. Parasitic weed incidence and related economic losses in rice in Africa. Agriculture Ecosystems and Environment 235: 306–317. [Google Scholar]
- Rodenburg J, Riches CR, Kayeke JM. 2010. Addressing current and future problems of parasitic weeds in rice. Crop Protection 29: 210–221. [Google Scholar]
- Saito K, Sokei Y, Wopereis MCS. 2012. Enhancing rice productivity in West Africa through genetic improvement. Crop Science 52: 484–493. [Google Scholar]
- Samejima H, Babiker AG, Mustafa A, Sugimoto Y. 2016. Identification of Striga hermonthica‐resistant upland rice varieties in Sudan and their resistance phenotypes. Frontiers in Plant Science 7: 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute . 2011. The SAS system for Windows; version 9.2. Cary, NC, USA: SAS Institute Inc. [Google Scholar]
- Sekiya N, Khatib KJ, Makame SM, Tomitaka M, Oizumi N, Araki H. 2013. Performance of a number of NERICA cultivars in Zanzibar, Tanzania: yield, yield components and grain quality. Plant Production Science 16: 141–153. [Google Scholar]
- Shew HD, Shew BB. 1994. Host resistance In: Campbell CL, Benson DM, eds. Epidemiology and management of root diseases. Berlin, Germany: Springer‐Verlag, 244–275. [Google Scholar]
- Showemimo FA. 2003. Selection criteria for combining high yield and Striga resistance in Sorghum. Tropicultura 21: 157–159. [Google Scholar]
- Sokal RR, Rohlf FJ. 1995. Biometry. New York, NY, USA: W. H. Freeman & Co. [Google Scholar]
- Swarbrick PJ, Scholes JD, Press MC, Slate J. 2009. A major QTL for resistance of rice to the parasitic plant Striga hermonthica is not dependent on genetic background. Pest Management Science 65: 528–532. [DOI] [PubMed] [Google Scholar]
- Watling JR, Press MC. 2001. Impacts of infection by parasitic angiosperms on host photosynthesis. Plant Biology 3: 244–250. [Google Scholar]
- Yoder JI, Scholes JD. 2010. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Current Opinion in Plant Biology 13: 478–484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Table S1 Overview of experimental conditions of the field trials conducted at Kyela, Tanzania, Namutumba, Uganda and Mbita, Kenya (2014 and 2015)
Table S2 ANOVA output on maximum emerged Striga numbers observed in the field at three sites (Kyela, Mbita and Namutumba) in the years 2014 and 2015 with 20 rice genotypes
Table S3 Spearman rank correlations between LS‐Means of maximum aboveground Striga numbers and aboveground Striga biomass dry weights at harvest, between NS max and rice grain yields, and between rice grain yields and rice plant height for the field data in Kyela, Mbita and Namutumba in both seasons (2014 and 2015)
Table S4 ANOVA output on rice grain yields and rice straw dry weights observed in the field at three sites (Kyela, Mbita and Namutumba) in the years 2014 and 2015 with 20 rice genotypes
Table S5 ANOVA output on maximum rice plant height, and plant height at 43 and 57 d after sowing (DAS), and photosynthesis at 30 and 45 DAS, with Striga infection and rice genotype as sources of variation


