Abstract
Wnt/β‐catenin signaling plays a key role in embryonic development, stem cell biology, and neurogenesis. However, the mechanisms of Wnt signal transmission, notably how the receptors are regulated, remain incompletely understood. Here we describe that the Parkinson's disease‐associated receptor GPR37 functions in the maturation of the N‐terminal bulky β‐propellers of the Wnt co‐receptor LRP6. GPR37 is required for Wnt/β‐catenin signaling and protects LRP6 from ER‐associated degradation via CHIP (carboxyl terminus of Hsc70‐interacting protein) and the ATPase VCP. GPR37 is highly expressed in neural progenitor cells (NPCs) where it is required for Wnt‐dependent neurogenesis. We conclude that GPR37 is crucial for cellular protein quality control during Wnt signaling.
Keywords: ER‐associated degradation, GPR37, LRP6, PAEL‐R, Wnt signaling
Subject Categories: Neuroscience, Protein Biosynthesis & Quality Control, Signal Transduction
Introduction
Low‐density lipoprotein receptor (LDLR)‐related protein 6 (LRP6) is a single transmembrane protein, which acts as a co‐receptor for Wnt ligands in cooperation with core receptors of the Frizzled (FZD) family 1, 2. LRP6 and its close ortholog LRP5 are essential for directing Wnt signaling toward the canonical or Wnt/β‐catenin pathway, which plays paramount roles in development and disease 3, 4, 5, 6, 7. Wnt proteins induce the formation of a ternary FZD‐LRP6 complex, followed by receptor clustering and phosphorylation in signalosomes. LRP6 activation triggers a series of events culminating in GSK3 inhibition, one consequence of which is the stabilization of β‐catenin, which translocates to the nucleus to regulate expression of Wnt target genes 1, 8.
The folding of cell surface receptors, such as LRP5/6, is monitored in the endoplasmic reticulum (ER) before they enter the secretory pathway. In mammalian cells, quality control mechanisms, including a variety of chaperones 9, monitor and promote correct protein folding, such that only the properly folded receptors leave the ER, while misfolded proteins either accumulate in the ER or are removed by ER‐associated degradation (ERAD) 10, 11. This quality control prevents defective receptors from reaching the cell surface, where they could disturb normal signaling.
LRP5/6 are particularly prone to ER misfolding due to their large extracellular domains containing more than 40 cysteine residues, which have to form correct disulfite bridges 12, 13. They belong to the LDLR protein family which all contain three types of repeated sequences: the complement‐type repeats (also known as type A repeats), epidermal growth factor (EGF) repeats, and YWTD repeats that fold into six‐bladed β‐propellers 14, 15. LRP6 has four β‐propeller/EGF repeats (E1‐4), which form two rigid blocks separated by a short hinge between domains E2 and E3 (see model in Synopsis) 16, 17. Wnt1, 2, 6, 7a/b, 9a/b, and 10a/b bind preferentially to domains E1‐E2, whereas Wnt3a binds to domain E3 17, 18, 19, 20. The LRP6 antagonist Dkk1 has several binding sites, one of them overlapping with the Wnt3a binding site on domain E3 and therefore competing with Wnt3a for binding 17, 18, 21. Mutations in the extracellular domains of LRP5/6, which result in impaired ligand binding or alterations in receptor cell surface expression, are linked to multiple human diseases such as bone density disorders 22, 23, 24, 25, cardiovascular diseases 26, aberrant eye development 22, and neurodegenerative diseases such as Alzheimer's disease 27, highlighting the importance of the Wnt co‐receptors 2.
Despite the pivotal roles of LRP5/6, how their folding and maturation are regulated remains incompletely understood. One important regulator of LRP5/6 folding is the ER‐resident chaperone mesoderm development (Mesd) 12, 13, 28. Mesd binds to the β‐propellers of newly synthesized LRP6, as well as other LDLRs, until they reach a more stable configuration upon translation of the succeeding EGF‐like repeats 12. In the absence of Mesd, LRP6 is retained in the ER, and Wnt/β‐catenin is inhibited 13.
Here we identify G‐protein‐coupled receptor 37 (GPR37) as a novel maturation factor of LRP6. GPR37 is a seven‐pass transmembrane protein, which is highly expressed in mouse brain and testis 29, 30, 31. GPR37 is a substrate of the E3 ligase Parkin, and hence is also known as Parkin‐associated endothelin‐like receptor (PAEL‐R) 32. Missense mutations of Parkin that are associated with a juvenile form of Parkinson's disease result in the accumulation of GPR37 in the brain of Parkinson's disease patients 32. Furthermore, overexpression of GPR37 leads to its accumulation in aggregates, ER stress, and neuronal cell death 32, 33. On the other hand, it has been suggested that native GPR37 has a neuroprotective role by binding of prosaposin and prosaptide 34. In addition, it has recently been proposed that GPR37 promotes oligodendrocyte differentiation and myelination via ERK signaling 35.
We show that GPR37 assists the maturation of LRP6 domains E1 and E2 in the ER. GPR37 protects LRP6 from ERAD via CHIP (carboxyl terminus of Hsc70‐interacting protein; also known as STUB1) and the ATPase VCP, thereby promoting Wnt/β‐catenin signaling. In neural progenitor cells (NPCs), both LRP6 and GPR37 are required to promote neuronal fate.
Results and Discussion
GPR37 is a novel positive regulator of Wnt/β‐catenin signaling
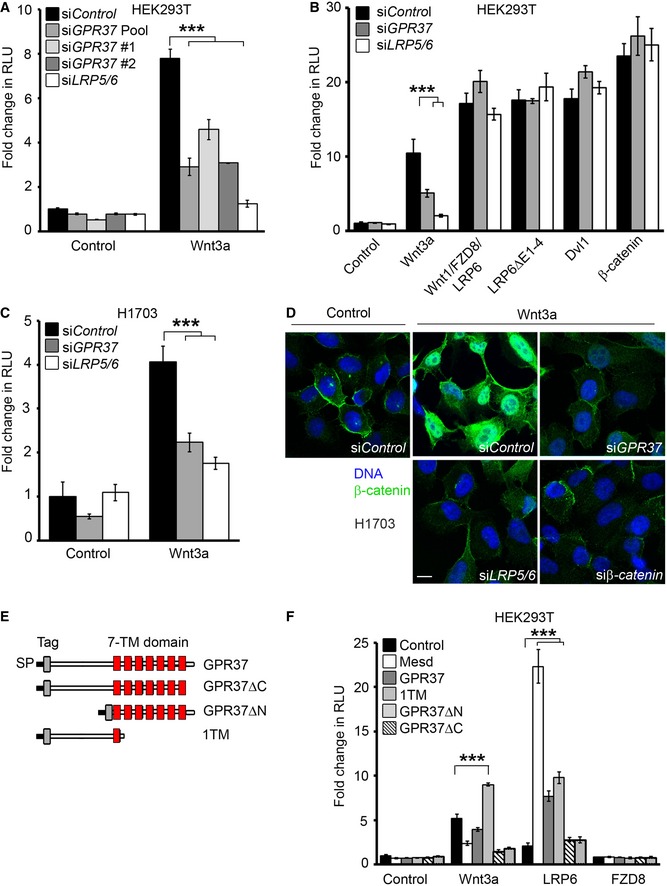
To uncover novel regulators of the Wnt/β‐catenin signaling pathway, we performed a genome‐wide siRNA screen 36, and identified GPR37 as a putative Wnt activator. Knockdown of GPR37 in HEK293T cells by two independent siRNAs inhibited Wnt3a‐induced signaling in Topflash reporter assays (Figs 1A and EV1A), without affecting BMP, TGFβ, or FGF signaling reporter activities (Fig EV1C–F). Depletion of GPR37 did not inhibit Topflash activity induced by the ligand–receptor complex (Wnt1/LRP6/FZD8), the constitutively active Wnt co‐receptor LRP6ΔE1‐4, or cytoplasmic effectors such as Dvl1 or β‐catenin (Fig 1B). Treatment with the proposed GPR37 ligand prosaptide 34 did not affect Wnt3a‐induced Topflash activity (Fig EV1G). A requirement for GPR37 in Wnt3a signaling was also observed in the lung cancer cell line H1703, where siGPR37 decreased Topflash reporter activity and prevented accumulation of β‐catenin to a similar extent as the depletion of the Wnt co‐receptors LRP5/6 (Figs 1C and D, and EV1B). These findings suggest that GPR37 is a positive regulator of Wnt/β‐catenin signaling, acting at the Wnt receptor level, and independent of its proposed peptide ligand.
Figure 1. GPR37 is a novel positive regulator of Wnt signaling.
-
A, BTopflash reporter assay in HEK293T cells upon knockdown of GPR37 using siRNA pool or single siRNAs, or siLRP5/6. Cells were stimulated with Wnt3a‐conditioned media or by transfection with the indicated constructs (mean values ± SD, n = 3; ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
-
CTopflash reporter assay in H1703 cells upon knockdown of GPR37 or LRP5/6. Cells were treated as indicated (mean values ± SD, n = 3; ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
-
DImmunofluorescence microscopy showing β‐catenin accumulation (green) in H1703 cells. Cells were transfected with the indicated siRNAs and stimulated with Wnt3a‐conditioned media for 3 h. Scale bar: 10 μm.
-
EGPR37 deletion constructs used in this study. All deletion constructs contain the Kremen signal peptide (SP) followed by an N‐terminal tag. The transmembrane region (TM) is shown in red.
-
FTopflash reporter assay in HEK293T cells upon overexpression of GPR37 deletion constructs shown in (E) in combination with the indicated constructs, and stimulated as indicated (mean values ± SD, n = 3; ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test). Note that the lack of activity in GPR37ΔC may be possibly due to misfolding of the seven transmembrane domains.
Figure EV1. GPR37 is required for Wnt/β‐catenin signaling.

-
A, BqPCR representing the knockdown efficiencies of the indicated single siRNAs or siRNA pool (siGPR37) used in Fig 1.
-
C–FReporter assays for Wnt (C), TGFβ (D), BMP4 (E), or FGF (F) signaling in HEK293T cells upon knockdown of GPR37 or LRP5/6. Cells were transfected as described in the Materials and Methods section, and stimulated with conditioned media (C) or the indicated recombinant proteins (D–F) (mean values ± SD, n = 3).
-
GTopflash reporter assay in HEK293T cells transfected with either control vector or GPR37. Cells were treated for 24 h with recombinant prosaptide (0–3 μM) in the presence or absence of Wnt3a‐conditioned media (mean values ± SD, n = 3).
In lack of adequate antibodies for endogenous human GPR37 protein, and to identify the GPR37 domains that function in Wnt signaling, we generated various GPR37 deletion constructs (Fig 1E). Overexpression of the N‐terminal region of GPR37 containing the first transmembrane region (GPR37‐1TM), but not the other constructs, enhanced Wnt3a‐induced reporter activity (Fig 1F). Furthermore, full‐length GPR37 and GPR37‐1TM, but not GPR37 lacking the N‐terminus (GPR37ΔN) or the intracellular C‐terminus (GPR37ΔC), promoted LRP6‐induced reporter activity similarly as Mesd, suggesting that GPR37 functions through LRP6. In contrast, none of the GPR37 constructs cooperated with Fzd8 in reporter assays. In addition, overexpression of GPR37‐1TM rescued the siGPR37‐mediated inhibition of Wnt signaling, attesting to the specificity of the siRNA and corroborating that expression of this deletion construct mediates a GPR37 gain of function (Figs 1F and EV2A). Notably, GPR37‐1TM was expressed at higher levels than GPR37 (Fig EV2B), possibly accounting for its stronger effect on Wnt3a‐induced reporter activity. Taken together, these data indicate that the most N‐terminal region of GPR37 is required for its function in Wnt signaling.
Figure EV2. GPR37‐1TM activates Wnt reporter assay.

- Topflash reporter assay in HEK293T cells upon siRNA knockdown of GPR37. Cells were transfected with control vector or GPR37‐1TM to rescue the siGPR37 effect, and treated as indicated (mean values ± SD, n = 3).
- Immunoblot of transfected V5‐GPR37 and V5‐GPR37‐1TM in HEK293T cells. Shown is a representative experiment performed five times.
GPR37 regulates Wnt signaling through LRP6
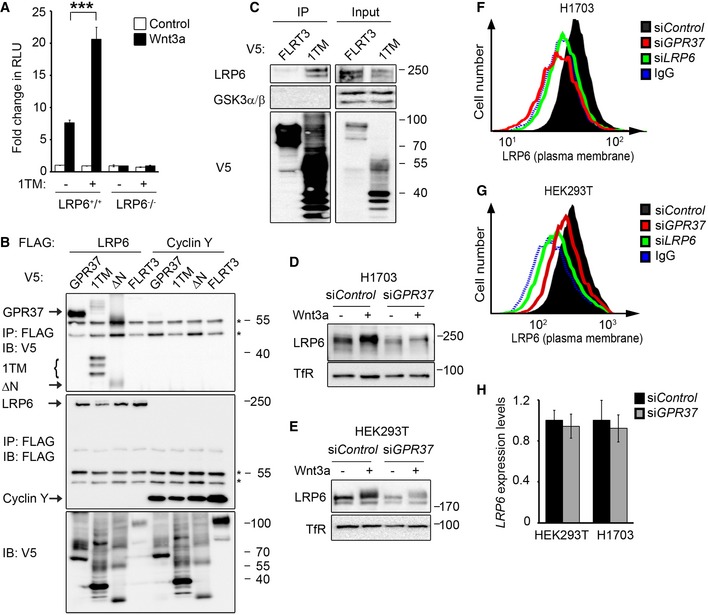
To further study the specificity of GPR37 to function through LRP6, we made use of HEK293T cells mutant for this Wnt co‐receptor. In LRP6−/− cells, overexpression of GPR37‐1TM and GPR37 did not activate Wnt signaling, unless LRP6 was reintroduced (Figs 2A and EV3A and B). Additionally, dose‐dependent knockdown of LRP6 decreased the effect of overexpressed GPR37‐1TM in Topflash reporter assays (Fig EV3C), supporting that GPR37 requires LRP6 to function in Wnt signaling.
Figure 2. GPR37 regulates LRP6 protein levels.
-
ATopflash reporter assay in LRP6 +/+ and LRP6 −/− HEK293T cells. Cells were transfected with GPR37‐1TM (1TM) or control vector and treated as indicated (mean values ± SD, n = 3; ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
-
BCo‐immunoprecipitation of overexpressed LRP6 and the indicated GPR37 constructs. Immunoblots of immunoprecipitates from HEK293T cells transfected with the indicated FLAG‐ and V5‐tagged constructs. IgG bands are indicated with an asterisk. Note that LRP6 binds preferentially the lower molecular weight band of GPR37‐1TM, which represents the ER form of GPR37‐1TM. Shown is a representative experiment carried out three times.
-
CCo‐immunoprecipitation of endogenous LRP6. Immunoblots of immunoprecipitates from HEK293T cells transfected with the indicated V5‐tagged constructs. GSK3 serves as a negative control. Shown is a representative experiment carried out five times.
-
D, EImmunoblots of H1703 (D) or HEK293T (E) cell lysates upon knockdown of GPR37. Transferrin receptor serves as a loading control. Shown are representative experiments carried out eight and seven times, respectively.
-
F, GFACS analyses of cell surface LRP6 protein levels upon knockdown of GPR37 or LRP6 in H1703 (F) or HEK293T (G) cells. Blue dotted line shows background signal upon staining with control IgGs. Shown are representative experiments carried out three times.
-
HqPCR analysis of LRP6 expression levels in HEK293T or H1703 cells upon knockdown of GPR37 (mean values ± SD, n = 3).
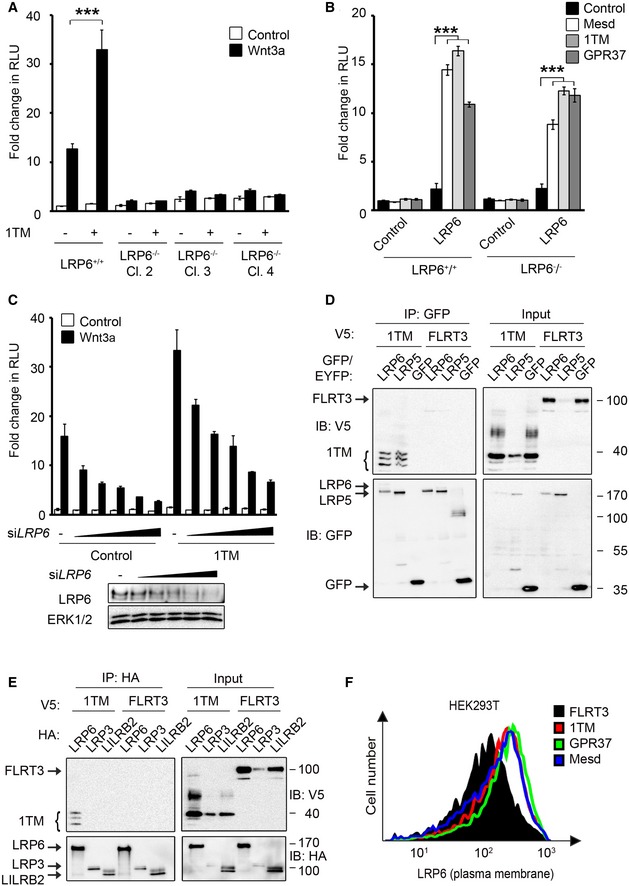
Figure EV3. GPR37 requires the Wnt co‐receptor LRP6 to function in Wnt signaling.
-
A, BTopflash reporter assays in LRP6+/+ HEK293T cells and three individual LRP6−/− HEK293T clones. Cells were transfected with the indicated constructs and treated as indicated (mean values ± SD, n = 3; ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
-
CUpper panel, Topflash reporter assays in HEK293T cells upon dose‐dependent siRNA knockdown of LRP6 (0–100 nM siRNA) and transfection with GPR37‐1TM. Cells were treated as indicated (SD, n = 3). Lower panel, immunoblots show the knockdown efficiencies of LRP6 in the Topflash assay.
-
D, ECo‐immunoprecipitation of GPR37‐1TM with LRP5 (D) and LRP3 (E). FLRT3 and LILRB2 transmembrane proteins serve as controls. Note that in (E) LRP6 but not LRP3 co‐immunoprecipitates GPR37‐1TM. Shown is a representative experiment carried out three times.
-
FFACS analysis of LRP6 cell surface levels upon transfection with the indicated constructs. Shown is a representative experiment carried out three times.
To examine whether GPR37 interacts with LRP6, we performed co‐immunoprecipitation (co‐IP) experiments. Overexpressed LRP6 co‐precipitated full‐length GPR37 and GPR37‐1TM, whereas only background binding for GPR37ΔN was observed (Fig 2B). Moreover, GPR37‐1TM also co‐precipitated with overexpressed LRP5 (Fig EV3D), but not with overexpressed LRP3 (Fig EV3E), suggesting that GPR37‐1TM preferentially binds to the Wnt co‐receptors. Furthermore, endogenous LRP6, but not the downstream component GSK3, co‐precipitated with GPR37‐1TM (Fig 2C). These data indicate that GPR37 interacts with LRP5/6 via its N‐terminal domain.
In line with the siGPR37 effect observed in Topflash, GPR37 depletion strongly reduced total LRP6 protein levels in H1703 and HEK293T cells (Fig 2D and E). Moreover, siGPR37 decreased cell surface LRP6 protein levels in both cell lines (Fig 2F and G), while LRP6 mRNA levels remained unaffected (Fig 2H). Conversely, overexpression of both full‐length GPR37 and GPR37‐1TM increased LRP6 cell surface levels, similarly as observed for Mesd (Fig EV3F). The results suggest that GPR37 functions in Wnt signaling by upregulating LRP6 protein levels post‐translationally.
GPR37 is required for LRP6 maturation
To delineate in which cellular compartment GPR37 and LRP6 functionally interact, we co‐transfected HEK293T cells with GPR37 and LRP6 in combination with plasmids encoding specific organelle markers. We observed co‐localization of LRP6 and GPR37 with the ER marker calreticulin, the Golgi marker TGNP and the trans‐Golgi marker SiT, and to lesser extent with the lysosomal marker cathepsin (Fig 3A). Similar results were obtained with GPR37‐1TM (Fig EV4A). This pattern of co‐localization suggests that GPR37 may mediate LRP6 maturation or transport from the ER. Consistently, GPR37‐1TM co‐precipitated preferentially the ER‐retained, immature form of LRP6 (Fig EV4B).
Figure 3. GPR37 promotes the maturation of LRP6 domains E1 and E2.
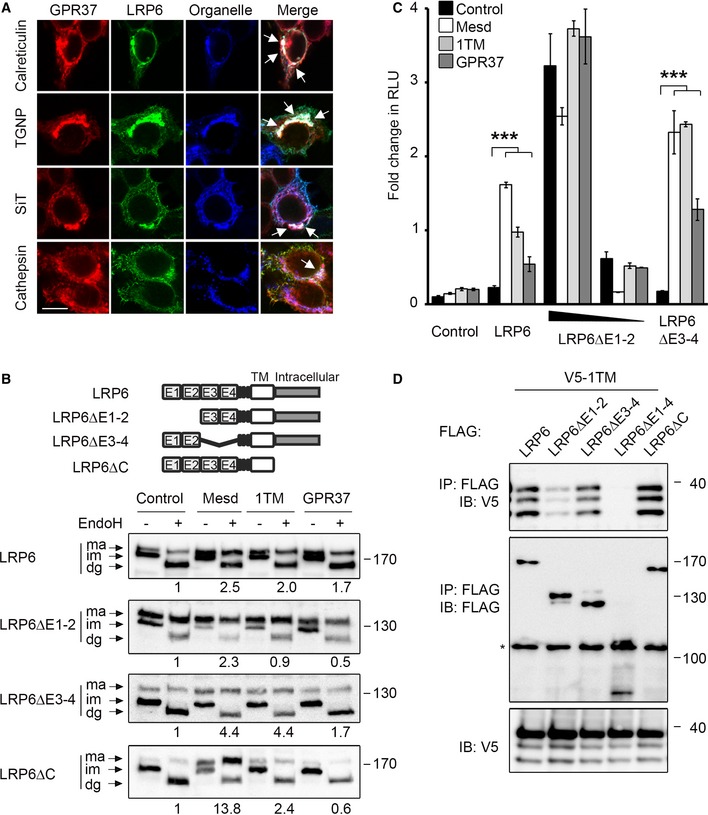
- Immunofluorescence microscopy of HEK293T cells transfected with V5‐GPR37 and HA‐LRP6 together with the organelle markers mCherry‐calreticulin (ER), mCherry‐TGNP (Golgi), mCherry‐SiT (trans‐Golgi), or mCherry‐cathepsin (lysosome), which are shown in blue. White signal in merge shows co‐localization of all three labels (arrows). Scale bar: 10 μm.
- Representative immunoblots of HEK293T cells transfected with the indicated LRP6 deletion constructs and co‐transfected with control vector, Mesd, GPR37‐1TM, or GPR37. Cell lysates were subjected to EndoH treatment as indicated. Ma, mature LRP6; im, immature; dg, deglycosylated. Numbers under lanes indicate the ratio of mature (ma) to immature (im) LRP6 band from densitometric analysis. Shown are representative experiments that were carried out three times.
- Topflash reporter assay in HEK293T cells transfected with the indicated LRP6 constructs and co‐transfected with control vector, Mesd, GPR37‐1TM, or GPR37. Cells were treated as indicated (mean values ± SD, n = 3; ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
- Co‐immunoprecipitation of GPR37‐1TM with LRP6 deletion constructs. Immunoblots of immunoprecipitates from HEK293T cells transfected with the indicated constructs. The asterisk indicates an unspecific band. Shown are representative experiments that were performed six times.
Figure EV4. GPR37‐1TM interacts with immature LRP6.
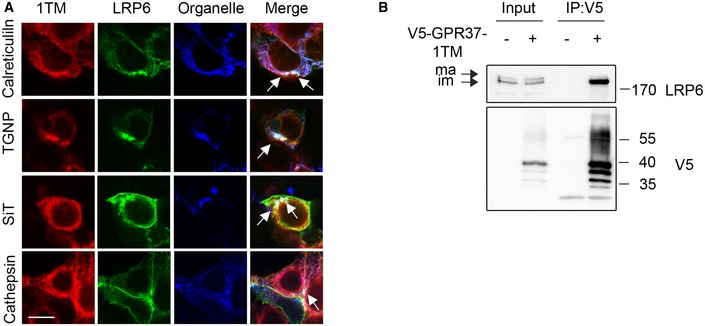
- Immunofluorescence microscopy of transfected LRP6 and V5‐GPR37‐1TM with the organelle markers calreticulin (ER), TGNP (Golgi), SiT (trans‐Golgi), or cathepsin (lysosomes), all shown in blue. White signal in merge shows co‐localization of all three constructs (arrows). Scale bar: 10 μm.
- Immunoblots of immunoprecipitated GPR37‐1TM with LRP6. Note that GPR37 only binds the immature (im, ER) form of LRP6, but not to the mature (ma) form. Shown is a representative experiment carried out three times.
LRP6 is a highly glycosylated protein, whose maturation and folding is facilitated by the ER‐resident chaperone Mesd 12, 13, 28, 37, 38. To examine whether GPR37 may also promote LRP6 maturation, we performed an endoglycosidase H (EndoH) assay. EndoH deglycosylates high‐mannose glycoproteins, which have not yet passed the Golgi apparatus, where they are further modified and become EndoH resistant. Overexpression of either GPR37‐1TM or Mesd strongly increased the level of mature, EndoH‐resistant LRP6 (Fig 3B). LRP6 contains four extracellular β‐propellers (E1–E4), and using deletion mutants, we found that GPR37‐1TM increased the levels of mature LRP6ΔE3‐4 and LRP6ΔC, but not of LRP6ΔE1‐2 (Fig 3B). Of note, E1‐2 folding/maturation may be particularly challenging since their deletion increased the ratio of mature to immature protein (Fig 3B). In addition, GPR37‐1TM, and to a lesser extent GPR37, cooperated with limiting amounts of LRP6ΔE3‐4, but not with LRP6ΔE1‐2, in Wnt reporter assays (Fig 3C). Furthermore, GPR37‐1TM co‐precipitated with LRP6, LRP6ΔE3‐4, and LRP6ΔC, but not with LRP6ΔE1‐4 or LRP6ΔE1‐2 (Fig 3D). Taken together, these data suggest that GPR37 promotes maturation of the E1‐E2 domains of LRP6.
GPR37 protects LRP6 from ERAD
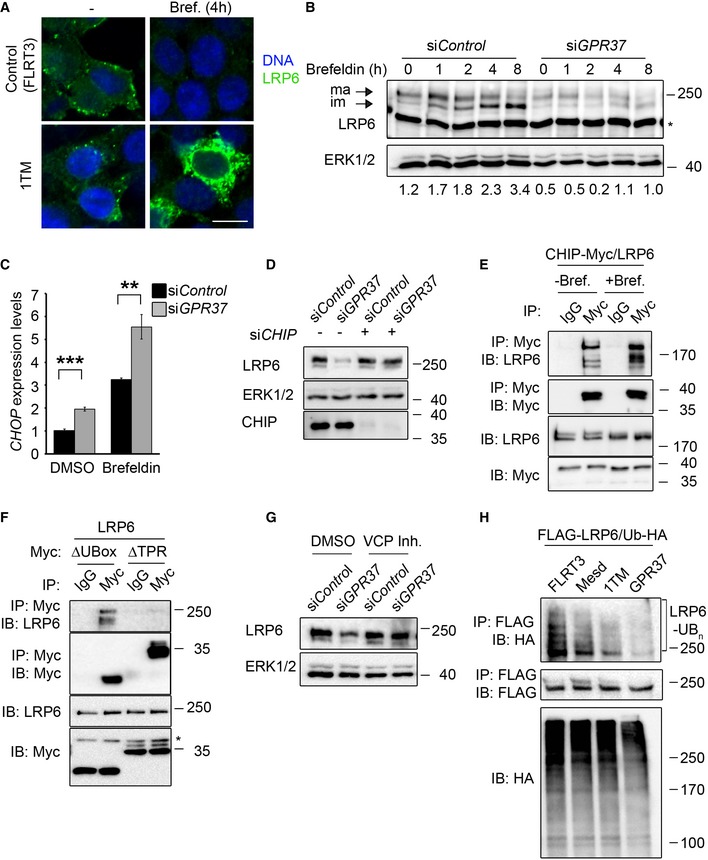
When protein misfolding occurs in the ER and cannot be resolved, proteins are exported from the ER to undergo ERAD 10, 11, 39. To address whether GPR37 protects LRP6 from ERAD, we used brefeldin A, which inhibits the transport of proteins from the ER to the Golgi, leading to fusion of both organelles and ER stress 40. Upon brefeldin A treatment, overexpressed LRP6 decreased in control cells, but strongly accumulated in the ER upon co‐transfection with GPR37‐1TM (Fig 4A), suggesting that GPR37‐1TM stabilized LRP6. In endogenous LRP6, the immature, lower molecular weight band accumulated under brefeldin A treatment (Fig 4B), which was impaired by siGPR37. Furthermore, expression of the ER stress marker CHOP 41 was increased upon siGPR37 under basal conditions and upon brefeldin A treatment (Fig 4C), consistent with GPR37 counteracting protein misfolding. These results support that GPR37 is required to stabilize LRP6 in the ER and to protect it from ERAD, notably under stress conditions.
Figure 4. GPR37 protects LRP6 from ER‐associated degradation (ERAD) via CHIP and VCP .
-
AImmunofluorescence microscopy of HEK293T cells transfected with HA‐LRP6 (green) upon co‐transfection with V5‐GPR37‐1TM or V5‐FLRT3 (Control), and treated with brefeldin A (Bref.) or vehicle (−) for 4 h as indicated. Scale bar: 10 μm.
-
BRepresentative immunoblots of endogenous LRP6 from H1703 cells upon knockdown of GPR37 and treatment with brefeldin A as indicated. Values under the immunoblots show the quantitation of immature, low molecular weight LRP6, normalized to the loading control ERK1/2. Asterisk indicates an unspecific band. Shown are representative experiments that were carried out three times.
-
CqPCR analysis of the ER stress marker CHOP upon knockdown of GPR37 and treatment with the ER stress‐inducing agent brefeldin A for 6 h (mean values ± SD, n = 3; **P < 0.01, ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
-
DImmunoblots of H1703 cells transfected with the indicated siRNAs. ERK1/2 serves as a loading control. Shown are representative experiments that were performed three times.
-
E, FImmunoblots of immunoprecipitates from HEK293T cells transfected with the indicated constructs. Cells were treated with brefeldin A (E and F) or vehicle for 4 h (E). Shown are representative experiments that were performed five times.
-
GImmunoblots of H1703 cells upon knockdown of GPR37 and treatment with the VCP inhibitor NMS‐873, or the vehicle DMSO, for 5 h. ERK1/2 serves as a loading control. Shown are representative experiments that were performed three times.
-
HLRP6 ubiquitination analysis. Representative immunoblots of immunoprecipitated FLAG‐LRP6 in HEK293T cells transfected as indicated and treated with the proteasomal inhibitor MG‐132 and the lysosomal inhibitor chloroquine for 6 h to induce accumulation of ubiquitinated LRP6 (LRP6‐Ubn). Ub‐HA, ubiquitin‐HA. Shown are representative experiments that were performed five times.
Misfolded proteins are recognized by chaperones that either protect them from stress, such as Mesd, or target them for degradation. CHIP (also known as STUB1) binds chaperones of the Hsc70 family and converts them from protein‐folding machines into factors that target misfolded proteins for degradation 42. For instance, CHIP interacts with Parkin and promotes degradation of misfolded GPR37 32, 43. Interestingly, CHIP depletion rescued LRP6 protein levels upon knockdown of GPR37 (Fig 4D). This suggests that CHIP also recognizes misfolded LRP6 and targets it for degradation. CHIP harbors two distinct motifs: A U‐Box domain with E3 ligase activity, and a tetratricopeptide repeat (TPR) domain. The latter is required for the interaction with chaperones that assist misfolded proteins to undergo proteasomal degradation 44. Co‐IP experiments showed binding of LRP6 to full‐length CHIP, which was increased upon ER stress (Fig 4E). LRP6 also bound to CHIP lacking the U‐Box (CHIPΔUBox), but not to CHIPΔTPR (Fig 4F). These findings support that CHIP interacts with LRP6 and targets it for degradation via the TPR domain.
Misfolded proteins in the ER are retro‐translocated to the cytoplasm to be poly‐ubiquitinated and thereby targeted for proteasomal degradation. Different routes for retro‐translocation exist, but all of them involve the ER exporter valosin‐containing protein (VCP/p97) 39. Treatment with the selective VCP inhibitor NMS‐873 fully rescued decreased LRP6 levels upon GPR37 depletion (Fig 4G), indicating that in the absence of GPR37, LRP6 is exported in a VCP‐dependent manner from the ER to the cytoplasm for degradation. Consistent with this model, overexpressed LRP6 showed substantial poly‐ubiquitination, which was strongly reduced by co‐expression of GPR37‐1TM and GPR37, and to a lower extent by Mesd (Fig 4H). We conclude that GPR37 protects LRP6 from CHIP‐ and VCP‐dependent ERAD.
GPR37 regulates neural fate in NPCs
In adult mice, Gpr37 is highly expressed in the brain and testis (31 and Fig 5A). Consistently, mice mutant for Gpr37 exhibit dopaminergic neuron and Sertoli cell defects 29, 45. We analyzed mouse E14 embryos and found that Gpr37 is also highly expressed in the developing brain, as well as in isolated NPCs (Fig 5B). NPCs are self‐renewing, multipotent cells that can differentiate into neurons, astrocytes, and oligodendrocytes 46. Wnt signaling promotes NPC proliferation, leads to neuronal differentiation, and prevents oligodendrocyte differentiation 47, 48, 49, 50.
Figure 5. GPR37 modulates neural fate in neural progenitor cells.

-
A, BqPCR expression analysis of Gpr37 in the indicated mouse tissues or isolated NPCs. Gpr37 expression was normalized against Gapdh (mean values ± SD, n = 3; one‐way ANOVA followed by Holm–Sidak test).
-
CImmunofluorescence microscopy of endogenous GPR37 (red) with LRP6, calreticulin (ER), TGN38 (Golgi), EEA1 (early endosome), clathrin (endocytic vesicles), or pan‐cadherin (cell junctions), shown in green, in NPCs from disassociated neurospheres. Yellow signal shows co‐localization (arrows). Scale bar: 10 μm.
-
DImmunofluorescence microscopy of endogenous LRP6 (green) and GPR37 (red) in neurospheres upon siGpr37 treatment. DNA (Hoechst) is shown in blue. Scale bar: 100 μm.
-
EImmunoblot analysis of LRP6 protein levels in NPC cells upon siRNA knockdown of GPR37 or LRP5/6. Note that siGpr37 reduces LRP6 protein levels, without affecting its mRNA expression (see Fig EV5D). Shown are representative experiments that were carried out three times.
-
F, GqPCR expression analysis of the Wnt target gene Sp5 (F) and the indicated neural fate markers (G) in NPCs upon siRNA knockdown of GPR37 or LRP5/6. Neurospheres were treated for 5 h with control or Wnt3a‐conditioned media. Expression was normalized against Hprt (mean values ± SE, n = 5; *P < 0.05, ***P < 0.001, one‐way ANOVA followed by Holm–Sidak test).
-
HImmunofluorescence microscopy of MAP2 (green) and NG2 (red) in differentiating NPCs. Neurospheres were treated with the indicated siRNA and cultured in neuronal differentiation medium (see Materials and Methods). Bottom right, percentage of MAP2‐positive cells per colony (mean values ± SE, n = 5; **P < 0.01, one‐way ANOVA followed by Holm–Sidak test). Scale bar: 100 μm.
To further examine the role of endogenous GPR37, we studied its function in NPCs from E14 mouse embryos, where the protein co‐localized with endogenous LRP6, as well as with calreticulin (ER) and TGN38 (Golgi) (Fig 5C), whereas only limited co‐localization was observed with EEA1 (early endosomes). This confirms that GPR37 co‐localizes with LRP6 in the ER and Golgi apparatus. Moreover, depletion of GPR37 reduced LRP6 protein levels in NPCs as well (Figs 5D and E, and EV5A–C), but not Lrp6 mRNA levels (Fig EV5D). Expression of the Wnt target gene Sp5 was decreased upon both siGpr37 and siLrp5/6 treatment (Fig 5F), consistent with GPR37 functioning as a positive regulator of Wnt/β‐catenin signaling in NPCs.
Figure EV5. GPR37 regulates neural fate in NPCs through Wnt signaling.
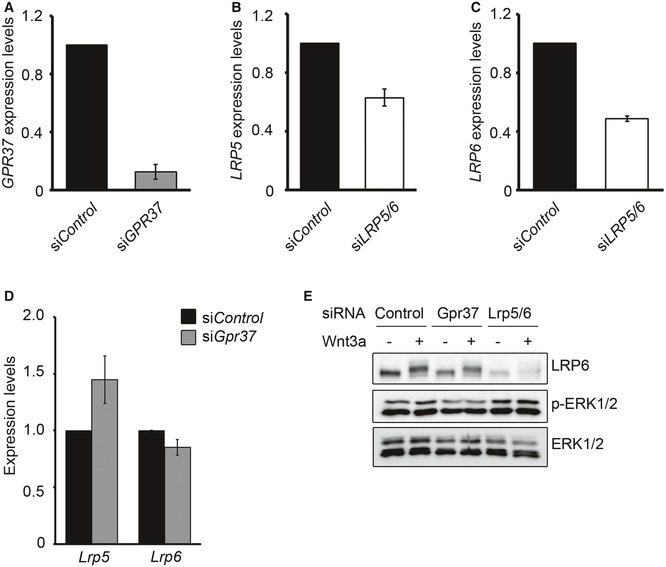
-
A–DqPCR showing the knockdown efficiencies of GPR37 (A), LRP5 (B), LRP6 (C), or LRP5 and LRP6 mRNA levels in NPCs upon knockdown of GPR37 (D) (mean values ± SE, n = 5).
-
EImmunoblots showing phospho‐ERK1/2 levels in NPCs upon siRNA knockdown of GPR37 or LRP5/6. Cells were treated with control or Wnt3a‐conditioned media for 5 h. Shown is a representative experiment performed three times.
To analyze the role of GPR37 in neurogenesis of NPCs, we examined neural marker gene expression upon siGPR37. Depletion of GPR37 decreased the expression of the neuronal fate marker Dcx and increased expression of the glial fate marker Cspg4, similar to siLrp5/6 (Fig 5G). To corroborate that GPR37 is required to promote Wnt‐driven neurogenesis of NPCs, we induced NPC differentiation by retinoic acid and monitored the terminal differentiation protein MAP2 (Fig 5H). Knockdown of either GPR37 or LRP5/6 reduced the number of MAP2‐positive cells. It has been proposed recently that GPR37 promotes oligodendrocyte differentiation and myelination via ERK signaling 35. However, phospho‐ERK1/2 levels were unaffected by depletion of GPR37 in NPCs (Fig EV5E). Taken together, these results indicate that GPR37 is required in Wnt signaling to promote neuronal fate in NPCs.
Wnt signaling plays critical roles in neurogenesis during brain development 47, 48, 49. Furthermore, Wnt signaling in NPCs of old mice induces neurogenesis in the hippocampus and counteracts cognitive decline 51. Our results indicate that GPR37 is required in NPCs to maintain normal LRP6 protein levels and Wnt signaling (Fig 5D and E), thereby promoting commitment to neuronal specification, while inhibiting glial fate (Fig 5F–H). Consistently, GPR37 is a negative regulator of oligodendrocyte differentiation and Gpr37‐deficient mice display brain defects, including a decrease in striatal dopamine levels, defects in motor behavior, anxiety, and learning deficiencies 35, 52, 53, 54, 55, which are even more severe in aged animals 53.
Gpr37‐deficient mice are viable, unlike Lrp5/6 mutants 56, suggesting that GPR37 acts as a cell‐specific LRP5/6 maturation factor. In other tissues, different chaperones, including Mesd or the GPR37‐homolog GPR37L1, may aid LRP5/6 folding. Like GPR37, GPR37L1 is highly expressed in the brain 45 and is able to protect primary astrocytes from cell death 34, suggesting that GPR37L1 and Mesd may partially compensate for GPR37 loss of function.
GPR37 in LRP6 folding and ER stress
In this study, we found that the Parkinson's disease‐associated receptor GPR37 functions in Wnt/β‐catenin signaling to promote maturation of LRP6. Mechanistically, the N‐terminal domain of GPR37, which is only found in placental mammals, binds and promotes the maturation of domains E1 and E2 of LRP6 in the ER. Depletion of GPR37 leads to LRP6 misfolding and ERAD via the ER exporter VCP, assisted by CHIP. Consequently, levels of mature LRP6 are reduced and Wnt/β‐catenin signaling is inhibited (see model in Synopsis). Interestingly, the N‐terminal domain of GPR37 undergoes metalloprotease‐dependent cleavage 57, which mediates its localization 34. However, this occurs during GPR37 transit through the Golgi, after it functions in the ER to promote LRP6 maturation.
GPR37 may function by directly stabilizing the E1‐2 domains of LRP6 during their folding. Alternatively, GPR37 may recruit additional maturation factors to LRP6, similarly to the ER chaperones calnexin/calreticulin, which recruit thiol‐oxidoreductases to their substrates 58. An intriguing question is why the domains E1‐2 of LRP6 would require a dedicated maturation factor. For instance, Mesd assists the folding of the domains E1‐4 of LRP6, as well as the β‐propellers of other LDLRs 12. The difference between GPR37 and Mesd activity may not be so much due to differences in structure of domains E1‐2 vs. E3‐4, as these are highly homologous 17. Instead, domains E1‐2 may be more susceptible to misfolding because they are the first to be translated and fold together as a single unit 59, which may be stabilized by inter‐domain interactions during folding of nascent LRP6 59, 60. Thus, GPR37 may act co‐translationally to channel the folding pathway of the nascent LRP5/6 polypeptides.
Mutations in the N‐terminal domains of LRP5/6 genes are implicated in multiple age‐related diseases, such as Alzheimer's disease, macular degeneration, or low/high bone mass 24, 25, 27, 61, 62. Thus, the requirement of GPR37 in Wnt signaling in vivo may be more relevant in old individuals or in folding‐associated diseases (proteopathies), which also increase with age due to defects in the protein quality control machinery 63, consistent with the severed phenotype of aged Gpr37‐deficient mice 53.
Parkinson's disease is a proteopathy in which GPR37 is implicated 32, 33. In light of our results, GPR37 may have a physiological neuroprotective role as chaperone, consistent with previous studies showing that GPR37 can protect primary astrocytes from cell death 34. Given the critical roles of Wnt signaling in promoting neural plasticity, neurogenesis, and cognition 51, 64, 65, it appears fruitful to investigate whether loss of GPR37 contributes to Parkinson's, as well as to other neuronal diseases, by reducing Wnt‐LRP5/6 signaling.
Materials and Methods
Cell culture
HEK293T cells (ATCC) were cultured in DMEM containing 10% fetal bovine serum (FBS), and supplemented with glutamine and penicillin/streptomycin. H1703 cells (ATCC) were maintained in RPMI containing 10% FBS and supplemented with glutamine, penicillin/streptomycin, and sodium pyruvate. Control and Wnt3a‐conditioned media were prepared as described 66.
Neural progenitor cells were obtained from E14 C57BL/6N embryos. Pregnant females were sacrificed by cervical dislocation and embryos were dissected. NPCs were isolated from disassociated telencephalons in Hank's buffer supplemented with 10 mM Hepes, 1% glutamine, 1% penicillin/streptomycin, 0.1% glucose, and 1% sodium pyruvate. Tissue lysates were treated with papain, trypsin, and DNase I to obtain single‐cell suspension. Neurosphere cultures were obtained by maintaining NPCs in ultra‐low attachment plates (Corning), and using NPC media (Neurobasal A, 2% B27 supplement without vitamin A (Gibco), 1% glutamine, 1% penicillin/streptomycin, 1% non‐essential amino acids, and 20 ng/ml EGF). For subsequent cell passages, cells were treated with trypsin to obtain single‐cell suspensions. All mouse experiments were performed according to federal and institutional guidelines.
For NPC differentiation, neurospheres were grown first in NPC media for 40 h, and then transferred to poly‐D‐lysine‐coated glass coverslips in 12‐well plates and cultured for 24 h in NPC differentiation media (Neurobasal A, 2% B27 supplement with vitamin A (retinoid acid; Gibco), 10% FBS, 1% glutamine, 1% penicillin/streptomycin).
Where indicated, cells were treated with 5 μg/ml brefeldin A (AppliChem) for 0–9 h, 10 μM chloroquine (Sigma‐Aldrich) for 6 h, 10 μM MG‐132 (Sigma‐Aldrich) for 6 h, 10 μM NMS‐873 (CalBiochem) for 5 h.
Small interfering RNA (siRNA)
Scrambled (siControl) and on‐target plus siRNA SmartPools against mouse and human GPR37, CHIP/STUB1, LRP5, and LRP6 were obtained from Thermo Scientific. Where indicated, individual targeted siRNAs against GPR37 were used. For knockdown experiments in HEK293T and H1703 cells, 50 nM of siRNA was delivered into cells using Dharmafect 1 transfection reagent and following the supplier's protocol. For transfection of NPCs with siRNA, 65 nM of on‐target plus siRNA was transfected using Lipofectamine 2000 and following the supplier's manual.
Expression constructs
Mesd, LRP6, and LRP6 deletion constructs (LRP6Δ1‐2, LRP6Δ13‐4, LRP6Δ1‐4, and LRP6ΔC) were as described previously 21. Human GPR37 was obtained from the DKFZ clone collection (Clone ID 131834988). Tagged constructs were generated by inserting GPR37 27‐613 (GPR37), GPR37Δ27‐264 (GPR37ΔN), GPR37Δ555‐613 (GPR37ΔC), or GPR37Δ290‐613 (GPR37‐1TM) into a pCS‐based vector containing an N‐terminal V5 or FLAG tag, followed by the Kremen signal peptide. The organelle markers mCherry‐TGNP‐N‐10, mCherry‐SiT‐N‐15, mCherry‐cathepsin‐B‐6, and mCherry‐calreticulin‐N‐16 were a gift from M. Davidson (Addgene plasmids #55145; #55133, #55007, #55006). WT‐CHIP‐Myc, ΔUBox‐CHIP‐Myc, and ΔTPR‐CHIP‐Myc plasmids were a generous gift from J.C. Schisler 67. Other constructs including reporter constructs used for Topflash assays or TGFβ, FGF, or BMP signaling were as described previously 36.
Antibodies
Rabbit polyclonal anti‐LRP6 antibody was as described previously 66. Other antibodies used were as follows: anti‐ERK1/2, anti‐transferrin receptor, anti‐FLAG M2, anti‐Map2 (M4403) (Sigma); anti‐β‐catenin, anti‐calnexin, anti‐TGN38, anti‐EEA1, anti‐clathrin (BD Transduction Laboratories); anti‐GFP (Antibodies online); anti‐LRP6 (MAB1505 and FAB1505A) used for FACS, anti‐pan‐cadherin (R&D Systems); anti‐HA (Roche); anti‐V5 (Invitrogen); anti‐Myc (9E10, DSHB); anti‐GPR37 (Proteintech); NG2 (H‐300) (Santa Cruz); anti‐phospho‐ERK1/2 (9101S, Cell Signaling); and anti‐GSK3α/β (Abcam). For Western blot analyses, antibodies were diluted in TBST containing 5% bovine serum albumin, 5 mM EDTA, and 1 mM EGTA.
Western blots
For isolation of total lysates, cells were harvested in PBS and lysed in lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 5 mM NaF, 1× proteinase inhibitor tablet (Roche), 1% Nonidet P‐40, 0.1% SDS, and 1 mM β‐mercaptoethanol). Cleared lysates were mixed with Laemmli buffer and analyzed by SDS–PAGE and Western blotting.
In Fig 2D, cell membrane fractions were prepared by lysing cells in TBS buffer supplemented with 0.025% saponin, 1× protease inhibitor mixture tablet (Roche), 2 mM EDTA, 10 mM β‐mercaptoethanol, and 10 mM NaF. Lysates were centrifuged at 14,000 g, and the membrane pellets were resuspended in lysis buffer. Cleared lysates were analyzed by SDS–PAGE and Western blotting. Western blots were imaged in a LAS‐3000 system (Fujifilm), processed with the manufacturer's software (Multi Gauge v3.2), and assembled using Photoshop (CS5, CS6).
For co‐immunoprecipitation experiments, transfected HEK293T cells were lysed in IP buffer (TBS, 1% Triton X‐100, 2 mM β‐mercaptoethanol, 1 mM MgCl2, 10 mM sodium pyrophosphate, 1× protease inhibitor mixture tablet (Roche), supplemented with 5 mM EDTA and 1 mM EGTA after clearing the lysates). Proteins were immunoprecipitated in IP buffer using conjugated anti‐FLAG beads (Sigma), anti‐HA beads (Roche), or anti‐V5‐beads (Sigma), or by the corresponding antibodies and protein A or G agarose (Santa Cruz). Immunopurified proteins were analyzed by SDS–PAGE and Western blot. In Fig 2B, HEK293T cells grown in 6‐well plates were transfected with cyclin Y (50 ng) or LRP6 (400 ng) and co‐transfected with V5‐GPR37 (200 ng), V5‐1TM (200 ng), V5‐FLRT3 (50 ng), or V5‐GPR37ΔN (200 ng). In Fig 2D, HEK293T cells grown in 15‐cm dishes were transfected with 5 μg V5‐FLRT3 or 0.8 μg V5‐1TM. In Fig EV3D, HEK293T cells grown in 6‐well plates were transfected with 300 ng EGFP‐LRP6 and GFP‐LRP5 or 50 ng GFP and co‐transfected with 200 ng V5‐FLRT3 or V5‐1TM. In Fig EV3E, HEK293T cells grown in 6‐well plates were transfected with 300 ng HA‐LRP6, LRP3‐HA, or LILRB2‐HA and co‐transfected with 200 ng V5‐1TM or V5‐FLRT3. For co‐IP experiments in Fig 4D, HEK293T cells in 6‐well plates were transfected with 200 ng V5‐1TM and co‐transfected with Flag‐tagged LRP6 (400 ng), LRP6Δ1‐2 (150 ng), LRP6Δ3‐4 (150 ng), LRP6Δ1‐4 (150 ng), or LRP6ΔC (400 ng), and the detergent used in the IP buffer was 1% CHAPS. For co‐IP experiments in Fig 4E and F, HEK293T cells were transfected in 6‐well plates with 250 ng LRP6 and 150 ng of either WT‐CHIP‐Myc, ΔUBox‐CHIP‐Myc, or ΔTPR‐CHIP‐Myc. For co‐IP experiments in Fig EV4B, HEK293T cells in 6‐well plates were transfected with 100 ng of V5‐1TM or pCS2 together with 300 ng LRP6.
To examine ubiquitination status of LRP6 (Fig 4H), HEK293T cells grown in 6‐well plates were transfected with 100 ng V5‐FLRT3, Mesd, V5‐1TM, or V5‐GPR37 and 350 ng FLAG‐LRP6. The next day, cells were co‐transfected with 300 ng ubiquitin‐HA. After 24 h, cells were treated with MG‐132 (10 μM) and chloroquine (10 μM) for 6 h to prevent degradation of proteins and harvested in ice‐cold PBS. Pellets were lysed in IP buffer supplemented with 5 mM N‐ethylmaleimide, and LRP6 was immunoprecipitated.
For LRP6 deglycosylation assays (Fig 3B), HEK293T cells were grown in 6‐well plates and transfected with 150 ng pCS2, 150 ng Mesd, 150 ng V5‐1TM, or 150 ng V5‐GPR37 together with 350 ng LRP6, 350 ng FLAG‐LRP6Δ1‐2, 350 ng FLAG‐LRP6Δ3‐4, or 350 ng FLAG‐LRP6Δ1‐4. After 24 h, cells were harvested in PBS and lysed in lysis buffer without NaF and supplemented with 1 mM EDTA. Cleared lysates were subjected to EndoH (Roche) treatment (0.25 U/ml) in 100 mM sodium acetate pH 5.5 for 30 min at 37°C. Samples were analyzed by SDS–PAGE and Western blotting.
Reporter assays
For Topflash assay, HEK293T or H1703 cells were grown in 96‐well plates and transfected with 5 ng Topflash and 1 ng Renilla and filled up to 50 ng DNA with empty vector (pCS2+) using X‐tremeGene 9 transfection reagent (Roche) following the supplier's protocol. Where indicated, siRNA‐mediated knockdown of the indicated genes was performed 24 h prior to Topflash/Renilla transfection. For epistasis experiments in Fig 1B, HEK293T cells were co‐transfected with Wnt1/LRP6/FZD8 (5 ng/3 ng/1 ng), Xenopus β‐catenin (1 ng), Dishevelled 1 (20 ng), or LRP6Δ1‐4 (10 ng). Where indicated, cells were stimulated with control or Wnt3a conditioned for 24 h. In Fig EV1G, HEK293T cells were co‐transfected with GPR37, and stimulated with 0, 0.5, 1, or 3 μM prosaptide (AnaSpec) in the presence of absence of Wnt3a media for 24 h. In Figs 2A and EV3A, LRP6−/− and WT HEK293T cells were co‐transfected with 5 ng of GRP37‐1TM. For cooperation studies in Figs 1F and EV3B, 24 ng LRP6 and 2 ng Frizzled 8 (Fig 1F) were co‐transfected with 2 ng Mesd or 5 ng GPR37 constructs, and cells were analyzed 48 h after transfection. Topflash assays in Fig 3C were performed with co‐expression of 24 ng LRP6, 1 ng or 10 ng LRP6Δ1‐2, and 24 ng LRP6Δ3‐4 together with 2 ng Mesd, 5 ng V5‐GPR37‐1TM, or 5 ng V5‐GPR37.
For TGFβ reporter assays (ARE‐Luc reporter assays) in Fig EV1D, HEK293T cells were transfected with ARE‐Luc (10 ng), Fast 1 (1 ng), and Renilla (8 ng). For BMP4 reporter assays in Fig EV1E, HEK293T cells were transfected with BRE‐luc (10 ng) and Renilla (10 ng). For FGF reporter assays (Gal‐Luc assays) in Fig EV1F, HEK293T cells were transfected with Ga‐luc reporter (10 ng), Gal‐ELK (2 ng), and Renilla (5 ng). Transfected cells were stimulated with recombinant TGFβ (2 ng/ml), BMP4 (10 ng/ml), or FGF8b (10 ng/ml) for 24 h, and luciferase activity was analyzed.
Immunofluorescence
For immunofluorescence (IF) experiments performed, cells were grown on poly‐D‐lysine‐coated glass coverslips in 12‐well plates. For endogenous IF in Fig 5C, NPCs were first dissociated using trypsin and plated for 4 h on poly‐D‐lysine‐coated glass coverslips in NPC media. Cells were fixed in 2–4% paraformaldehyde (PFA) in PBS, and permeabilized with 1% Triton in PBS. After blocking for 2 h in blocking buffer (PBS with 0.1% Triton (PBST) supplemented with 2% horse or goat serum), cells were incubated with the indicated antibodies in blocking buffer for 3 h in a humidity chamber at room temperature (RT). The coverslips were washed three times in PBST, and incubated with Alexa 488‐ or Alexa 546‐tagged secondary antibodies for 1 h at RT. Cells were washed in PBST and PBS, rinsed in water, and mounted prior to analysis.
For co‐localization studies in Figs 3A and EV4A, HEK293T cells were grown in 12‐well plates and transfected with 100 ng mCherry‐tagged calreticulin, SiT, TGNP, or cathepsin together with 150 ng HA‐LRP6 and 70 ng V5‐GPR37 or V5‐GPR37‐1TM, respectively. Twenty‐four hours after transfection, cells were fixed and stained as described above. To examine the effect of GPR37 on LRP6 in Fig 4A, HEK293T cells were cultured in 12‐well plates and transfected with 100 ng of V5‐1TM or control (V5‐FLRT3) and co‐transfected with 100 ng of LRP6.
Cells were imaged in a Nikon EZ‐C1, or Leica TCS SP5 II (Figs 3A and EV4A). The images were processed and assembled using Image J (1.43r) and Photoshop (CS5, CS6).
FACS analysis
To determine cell surface LRP6 levels, FACS analysis was performed as described previously 68. Briefly, cells were collected in Hank's buffer supplemented with 1 mM EDTA and pellets were resuspended in blocking solution (PBS containing 1% bovine serum albumin and 0.1% sodium azide). After blocking for 30 min at 4°C, cells were incubated with LRP6 antibody (R&D Systems) for 4 h at 4°C on a rotation wheel. Samples were washed in block solution and incubated with secondary antibody (goat anti‐mouse Alexa 488) supplemented with 0.1 mg/ml propidium iodide (PI), for 1 h at 4°C under rotation. Samples were washed in blocking solution and analyzed using FACSCalibur cytometer. In Fig EV3F, cells were co‐transfected with GFP and the indicated constructs, and stained with LRP6‐APC antibody (R&D Systems). GFP‐positive cells (transfected) were gated for analysis.
Quantitative real‐time PCR
HEK293T, H1703, or isolated NPCs (neurospheres) and the indicated tissues were directly collected in RNA lysis buffer supplemented with β‐mercaptoethanol. Cells were harvested for RNA isolation following the Nucleo Spin RNA (Macherey Nagel) manual. Reverse transcription was performed using Superscript II (Invitrogen), and the obtained cDNA was analyzed using a LightCycler 480 (Roche) and the indicated primers (Appendix Table S1).
Generation of LRP6−/− HEK293T cells
LRP6−/− HEK293T cells were generated by CRISPR/Cas9‐mediated gene editing 69 with the following gRNA: ATTATTGTCCCCCGATGGGC. Clonal colonies were obtained by limiting dilution, and deletions were confirmed by sequence analysis using CRISPR‐ID 70.
Statistics
Data are represented as mean values ± standard deviation (SD) or standard error (SE) of biological replicates (n = 3, or n = 5 for Fig 5). Sigma Plot software was used to determine statistical significance applying one‐way analysis of variance (ANOVA). Data were considered to be significant for following P‐values: *P < 0.05, **P < 0.01, ***P < 0.001, or no significant difference (n.s.).
Author contributions
BSB and SPA conceived, performed, and analyzed experiments. JH supervised animal husbandry and performed experiments. SK generated the LRP6−/− cell lines. CN and SPA conceptualized and supervised all aspects of the project. BSB, SPA, and CN wrote the manuscript with input from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank C.M. Cruciat, D. Ingelfinger, and M. Boutros for sharing genome‐wide siRNA screen data, the DKFZ for microscopy, and genomics core facilities for technical help, and A. Glinka, M. Davidson, and J.C. Schisler for reagents. This work was supported by the DFG.
EMBO Reports (2017) 18: 712–725
Contributor Information
Sergio P Acebron, Email: s.perezacebron@DKFZ-Heidelberg.de.
Christof Niehrs, Email: niehrs@DKFZ-Heidelberg.de.
References
- 1. MacDonald BT, He X (2012) Frizzled and LRP5/6 receptors for Wnt/β‐catenin signaling. Cold Spring Harb Perspect Biol 4: a007880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joiner DM, Ke J, Zhong Z, Xu HE, Williams BO (2013) LRP5 and LRP6 in development and disease. Trends Endocrinol Metab 24: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klaus A, Birchmeier W (2008) Wnt signalling and its impact on development and cancer. Nat Rev Cancer 8: 387–398 [DOI] [PubMed] [Google Scholar]
- 4. Clevers H, Nusse R (2012) Wnt/beta‐catenin signaling and disease. Cell 149: 1192–1205 [DOI] [PubMed] [Google Scholar]
- 5. Anastas JN, Moon RT (2013) WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 13: 11–26 [DOI] [PubMed] [Google Scholar]
- 6. Hikasa H, Sokol SY (2013) Wnt signaling in vertebrate axis specification. Cold Spring Harb Perspect Biol 5: a007955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Robertis EM (2010) Wnt signaling in axial patterning and regeneration: lessons from planaria. Sci Signal 3: pe21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niehrs C (2012) The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 13: 767–779 [DOI] [PubMed] [Google Scholar]
- 9. Braakman I, Hebert DN (2013) Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 5: a013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meusser B, Hirsch C, Jarosch E, Sommer T (2005) ERAD: the long road to destruction. Nat Cell Biol 7: 766–772 [DOI] [PubMed] [Google Scholar]
- 11. Ruggiano A, Foresti O, Carvalho P (2014) Quality control: ER‐associated degradation: protein quality control and beyond. J Cell Biol 204: 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Culi J, Springer TA, Mann RS (2004) Boca‐dependent maturation of beta‐propeller/EGF modules in low‐density lipoprotein receptor proteins. EMBO J 23: 1372–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines ME, Rosenquist T, Holdener BC (2003) Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell 112: 355–367 [DOI] [PubMed] [Google Scholar]
- 14. He X, Semenov M, Tamai K, Zeng X (2004) LDL receptor‐related proteins 5 and 6 in Wnt/beta‐catenin signaling: arrows point the way. Development 131: 1663–1677 [DOI] [PubMed] [Google Scholar]
- 15. Hussain MM, Strickland DK, Bakillah A (1999) The mammalian low‐density lipoprotein receptor family. Annu Rev Nutr 19: 141–172 [DOI] [PubMed] [Google Scholar]
- 16. Ahn VE, Chu ML, Choi HJ, Tran D, Abo A, Weis WI (2011) Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev Cell 21: 862–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng Z, Biechele T, Wei Z, Morrone S, Moon RT, Wang L, Xu W (2011) Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol 18: 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN (2010) Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem 285: 9172–9179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X et al (2011) Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev Cell 21: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu ML, Ahn VE, Choi HJ, Daniels DL, Nusse R, Weis WI (2013) Structural studies of Wnts and identification of an LRP6 binding site. Structure 21: 1235–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C (2001) LDL‐receptor‐related protein 6 is a receptor for Dickkopf proteins. Nature 411: 321–325 [DOI] [PubMed] [Google Scholar]
- 22. Gong Y, Slee RB, Fukai N, Rawadi G, Roman‐Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D et al (2001) LDL receptor‐related protein 5 (LRP5) affects bone accrual and eye development. Cell 107: 513–523 [DOI] [PubMed] [Google Scholar]
- 23. Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B et al (2002) A mutation in the LDL receptor‐related protein 5 gene results in the autosomal dominant high‐bone‐mass trait. Am J Hum Genet 70: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML (2005) Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass‐associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol 25: 4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams BO, Insogna KL (2009) Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling in bone. J Bone Miner Res 24: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mani A, Radhakrishnan J, Wang H, Mani MA, Nelson‐Williams C, Carew KS, Mane S, Najmabadi H, Wu D, Lifton RP (2007) LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 315: 1278–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De‐Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A et al (2007) Common genetic variation within the low‐density lipoprotein receptor‐related protein 6 and late‐onset Alzheimer's disease. Proc Natl Acad Sci USA 104: 9434–9439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Culi J, Mann RS (2003) Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila . Cell 112: 343–354 [DOI] [PubMed] [Google Scholar]
- 29. La Sala G, Marazziti D, Di Pietro C, Golini E, Matteoni R, Tocchini‐Valentini GP (2015) Modulation of Dhh signaling and altered Sertoli cell function in mice lacking the GPR37‐prosaposin receptor. FASEB J 29: 2059–2069 [DOI] [PubMed] [Google Scholar]
- 30. Marazziti D, Di Pietro C, Golini E, Mandillo S, Matteoni R, Tocchini‐Valentini GP (2009) Macroautophagy of the GPR37 orphan receptor and Parkinson disease‐associated neurodegeneration. Autophagy 5: 741–742 [DOI] [PubMed] [Google Scholar]
- 31. Marazziti D, Gallo A, Golini E, Matteoni R, Tocchini‐Valentini GP (1998) Molecular cloning and chromosomal localization of the mouse Gpr37 gene encoding an orphan G‐protein‐coupled peptide receptor expressed in brain and testis. Genomics 53: 315–324 [DOI] [PubMed] [Google Scholar]
- 32. Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R (2001) An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell 105: 891–902 [DOI] [PubMed] [Google Scholar]
- 33. Marazziti D, Di Pietro C, Golini E, Mandillo S, Matteoni R, Tocchini‐Valentini GP (2009) Induction of macroautophagy by overexpression of the Parkinson's disease‐associated GPR37 receptor. FASEB J 23: 1978–1987 [DOI] [PubMed] [Google Scholar]
- 34. Meyer RC, Giddens MM, Schaefer SA, Hall RA (2013) GPR37 and GPR37L1 are receptors for the neuroprotective and glioprotective factors prosaptide and prosaposin. Proc Natl Acad Sci USA 110: 9529–9534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang HJ, Vainshtein A, Maik‐Rachline G, Peles E (2016) G protein‐coupled receptor 37 is a negative regulator of oligodendrocyte differentiation and myelination. Nat Commun 7: 10884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cruciat CM, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, Boutros M, Niehrs C (2010) Requirement of prorenin receptor and vacuolar H+‐ATPase‐mediated acidification for Wnt signaling. Science 327: 459–463 [DOI] [PubMed] [Google Scholar]
- 37. Li Y, Lu W, He X, Bu G (2006) Modulation of LRP6‐mediated Wnt signaling by molecular chaperone Mesd. FEBS Lett 580: 5423–5428 [DOI] [PubMed] [Google Scholar]
- 38. Feldman M, van der Goot FG (2009) Novel ubiquitin‐dependent quality control in the endoplasmic reticulum. Trends Cell Biol 19: 357–363 [DOI] [PubMed] [Google Scholar]
- 39. Lemus L, Goder V (2014) Regulation of endoplasmic reticulum‐associated protein degradation (ERAD) by ubiquitin. Cells 3: 824–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dinter A, Berger EG (1998) Golgi‐disturbing agents. Histochem Cell Biol 109: 571–590 [DOI] [PubMed] [Google Scholar]
- 41. Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11: 381–389 [DOI] [PubMed] [Google Scholar]
- 42. Edkins AL (2015) CHIP: a co‐chaperone for degradation by the proteasome. Subcell Biochem 78: 219–242 [DOI] [PubMed] [Google Scholar]
- 43. Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R (2002) CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell 10: 55–67 [DOI] [PubMed] [Google Scholar]
- 44. McDonough H, Patterson C (2003) CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marazziti D, Di Pietro C, Golini E, Mandillo S, La Sala G, Matteoni R, Tocchini‐Valentini GP (2013) Precocious cerebellum development and improved motor functions in mice lacking the astrocyte cilium‐, patched 1‐associated Gpr37 l1 receptor. Proc Natl Acad Sci USA 110: 16486–16491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gotz M, Huttner WB (2005) The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788 [DOI] [PubMed] [Google Scholar]
- 47. Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y (2004) The Wnt/beta‐catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131: 2791–2801 [DOI] [PubMed] [Google Scholar]
- 48. Kalani MY, Cheshier SH, Cord BJ, Bababeygy SR, Vogel H, Weissman IL, Palmer TD, Nusse R (2008) Wnt‐mediated self‐renewal of neural stem/progenitor cells. Proc Natl Acad Sci USA 105: 16970–16975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K (2005) Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol 282: 397–410 [DOI] [PubMed] [Google Scholar]
- 50. Yu JM, Kim JH, Song GS, Jung JS (2006) Increase in proliferation and differentiation of neural progenitor cells isolated from postnatal and adult mice brain by Wnt‐3a and Wnt‐5a. Mol Cell Biochem 288: 17–28 [DOI] [PubMed] [Google Scholar]
- 51. Seib DR, Corsini NS, Ellwanger K, Plaas C, Mateos A, Pitzer C, Niehrs C, Celikel T, Martin‐Villalba A (2013) Loss of Dickkopf‐1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell 12: 204–214 [DOI] [PubMed] [Google Scholar]
- 52. Marazziti D, Golini E, Mandillo S, Magrelli A, Witke W, Matteoni R, Tocchini‐Valentini GP (2004) Altered dopamine signaling and MPTP resistance in mice lacking the Parkinson's disease‐associated GPR37/parkin‐associated endothelin‐like receptor. Proc Natl Acad Sci USA 101: 10189–10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mandillo S, Golini E, Marazziti D, Di Pietro C, Matteoni R, Tocchini‐Valentini GP (2013) Mice lacking the Parkinson's related GPR37/PAEL receptor show non‐motor behavioral phenotypes: age and gender effect. Genes Brain Behav 12: 465–477 [DOI] [PubMed] [Google Scholar]
- 54. Lopes JP, Morato X, Souza C, Pinhal C, Machado NJ, Canas PM, Silva HB, Stagljar I, Gandia J, Fernandez‐Duenas V et al (2015) The role of parkinson's disease‐associated receptor GPR37 in the hippocampus: functional interplay with the adenosinergic system. J Neurochem 134: 135–146 [DOI] [PubMed] [Google Scholar]
- 55. Marazziti D, Mandillo S, Di Pietro C, Golini E, Matteoni R, Tocchini‐Valentini GP (2007) GPR37 associates with the dopamine transporter to modulate dopamine uptake and behavioral responses to dopaminergic drugs. Proc Natl Acad Sci USA 104: 9846–9851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kelly OG, Pinson KI, Skarnes WC (2004) The Wnt co‐receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development 131: 2803–2815 [DOI] [PubMed] [Google Scholar]
- 57. Mattila SO, Tuusa JT, Petaja‐Repo UE (2016) The Parkinson's‐disease‐associated receptor GPR37 undergoes metalloproteinase‐mediated N‐terminal cleavage and ectodomain shedding. J Cell Sci 129: 1366–1377 [DOI] [PubMed] [Google Scholar]
- 58. Williams DB (2006) Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci 119: 615–623 [DOI] [PubMed] [Google Scholar]
- 59. Liu CC, Pearson C, Bu G (2009) Cooperative folding and ligand‐binding properties of LRP6 beta‐propeller domains. J Biol Chem 284: 15299–15307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J (2007) The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol 8: 319–330 [DOI] [PubMed] [Google Scholar]
- 61. Haines JL, Schnetz‐Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, Olson L, Kenealy SJ, Hauser M, Gilbert JR et al (2006) Functional candidate genes in age‐related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci 47: 329–335 [DOI] [PubMed] [Google Scholar]
- 62. van Meurs JB, Rivadeneira F, Jhamai M, Hugens W, Hofman A, van Leeuwen JP, Pols HA, Uitterlinden AG (2006) Common genetic variation of the low‐density lipoprotein receptor‐related protein 5 and 6 genes determines fracture risk in elderly white men. J Bone Miner Res 21: 141–150 [DOI] [PubMed] [Google Scholar]
- 63. Walker LC, Levine H III, Mattson MP, Jucker M (2006) Inducible proteopathies. Trends Neurosci 29: 438–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Inestrosa NC, Arenas E (2010) Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci 11: 77–86 [DOI] [PubMed] [Google Scholar]
- 65. Lucas FR, Salinas PC (1997) WNT‐7a induces axonal remodeling and increases synapsin I levels in cerebellar neurons. Dev Biol 192: 31–44 [DOI] [PubMed] [Google Scholar]
- 66. Davidson G, Shen J, Huang YL, Su Y, Karaulanov E, Bartscherer K, Hassler C, Stannek P, Boutros M, Niehrs C (2009) Cell cycle control of wnt receptor activation. Dev Cell 17: 788–799 [DOI] [PubMed] [Google Scholar]
- 67. Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM (2001) The Hsc70 co‐chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3: 100–105 [DOI] [PubMed] [Google Scholar]
- 68. Jiang X, Charlat O, Zamponi R, Yang Y, Cong F (2015) Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol Cell 58: 522–533 [DOI] [PubMed] [Google Scholar]
- 69. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dehairs J, Talebi A, Cherifi Y, Swinnen JV (2016) CRISP‐ID: decoding CRISPR mediated indels by Sanger sequencing. Sci Rep 6: 28973 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


