Abstract
Humoral fluid phase pattern recognition molecules (PRMs) are a key component of the activation and regulation of innate immunity. Humoral PRMs are diverse. We focused on the long pentraxin PTX3 as a paradigmatic example of fluid phase PRMs. PTX3 acts as a functional ancestor of antibodies and plays a non-redundant role in resistance against selected microbes in mouse and man and in the regulation of inflammation. This molecule interacts with complement components, thus modulating complement activation. In particular PTX3 regulates complement-driven macrophage-mediated tumor progression, acting as an extrinsic oncosuppressor in preclinical models and selected human tumors. Evidence collected over the years suggests that PTX3 is a biomarker and potential therapeutic agent in humans, and pave the way to translation of this molecule into the clinic.
Keywords: pentraxins, PTX3, inflammation, opsonisation, complement activation, cancer-related inflammation
Introduction
The innate immune response is the first line of defense to invading microbes and tissue damage and is activated upon sensing of microbes and tissue injury through humoral and cell-associated pattern recognition molecules (PRMs). This response includes the production of inflammatory cytokines, the activation of the acute phase response, and leukocyte recruitment and polarization (1, 2). Innate responses have the general significance of defense and orchestration of tissue repair, but are also potentially involved in the pathogenesis of certain diseases.
The humoral arm of innate immunity includes the classic short pentraxins C reactive protein (CRP) and serum amyloid P component (SAP), the long pentraxin PTX3, complement recognition molecules such as C1q and ficolins, and collectins (3). Humoral PRMs are biochemically heterogeneous, but share the common property to behave as antibody-like molecules, recognizing microbial moieties, having opsonic activity and activating and regulating the complement cascade (3, 4). Furthermore, several humoral PRMs interact with extracellular matrix components and are involved in tissue remodeling (5). They are rapidly expressed in infectious or injury conditions in different cell types and tissues and with different kinetics. The liver is the main source of short pentraxins and sustains their presence in the systemic circulation. In contrast, macrophages, dendritic cells and endothelial cells locally and rapidly transcribe and produce PTX3 upon inflammatory stimulation (4), and neutrophils rapidly release it from intracellular granules at sites of tissue damage or microbial stimulation (6). The different kinetics and cell origin of short pentraxins and PTX3 provide the continuous presence of these molecules in the systemic circulation as well as within tissues during inflammatory conditions (3).
The pentraxin concentration in the circulation or in tissues rapidly increases in inflammatory conditions and in general correlates with the severity of the clinical condition inducing their expression. These results raised the question whether they are simple markers and innocent bystanders, or players in the pathogenesis of the disease (7–9). CRP is a widely used biomarker of inflammation and tissue damage in humans, but the identification of its actual role in inflammation has been precluded by the low conservation between mouse and human and the lack of appropriate gene targeted animal models (10, 11). In contrast, the high conservation between mouse and human PTX3 and Ptx3-/- mice have allowed defining the functional role of PTX3 in innate immunity and inflammation.
From these studies, PTX3 emerges as a non-redundant humoral PRM involved in recognizing and opsonizing microbes for facilitated phagocytosis, a regulator of inflammatory responses and a player of tissue remodelling (5, 12–14). Studies in animal models of fungal, bacterial and viral infections and the evidence that human PTX3 genetic variants are linked to susceptibility to specific infections indicate that PTX3 plays a protective role in the resistance to microbes and in modulating inflammatory responses associated with tissue damage (14–17). However, in specific contexts, PTX3 may contribute to the pathogenesis of the disease. In particular, PTX3 has been shown to increase inflammation and tissue damage (16, 18, 19), and its interaction with collectins and ficolins may increase complement activation (20). In addition, PTX3 can be exploited by specific pathogens to enter the cell (21). By modulating complement-driven inflammation, PTX3 has been shown to act as an oncosuppressor gene in mice and selected human tumors (12). By interacting with provisional matrix components, it has been shown to orchestrate wound healing, fibrin-rich inflammatory matrix remodelling and tissue repair (5). Finally, PTX3 has complex effects on blood vessels, by modulating the effect of angiogenic growth factors of the FGF family and affecting tumor-associated angiogenesis, (22, 23), and by regulating the vessel wall tone (24).
These studies indicate that depending on the disease context, cellular source and levels of protein released, PTX3 can exert dual roles in inflammation and infections, and may contribute to mechanisms of pathogenesis of specific diseases. Here we will summarize molecular and functional properties of PTX3 in microbial defense, regulation of inflammation and tissue remodeling and repair, and discuss its yin-yang role in immunopathology and potential use as disease marker and candidate prophylactic and therapeutic agent in infectious disorders.
Key structural features
The human PTX3 is a homo-multimeric glycoprotein, whose protomer subunits comprise 381 amino acids, including a 17 residue long leader peptide (Fig. 1A). The primary sequence of this long pentraxin is highly conserved among animal species (where the human and murine proteins share 92% of conserved amino acids), suggesting a strong evolutionary pressure to maintain its structure/function relationships. Analogous to other long-pentraxins, the PTX3 protomer contains a unique N-terminal region (residues 18-178 of the preprotein) and a C-terminal domain (amino acids 179–381) that is homologous to the short pentraxins CRP and SAP (25).
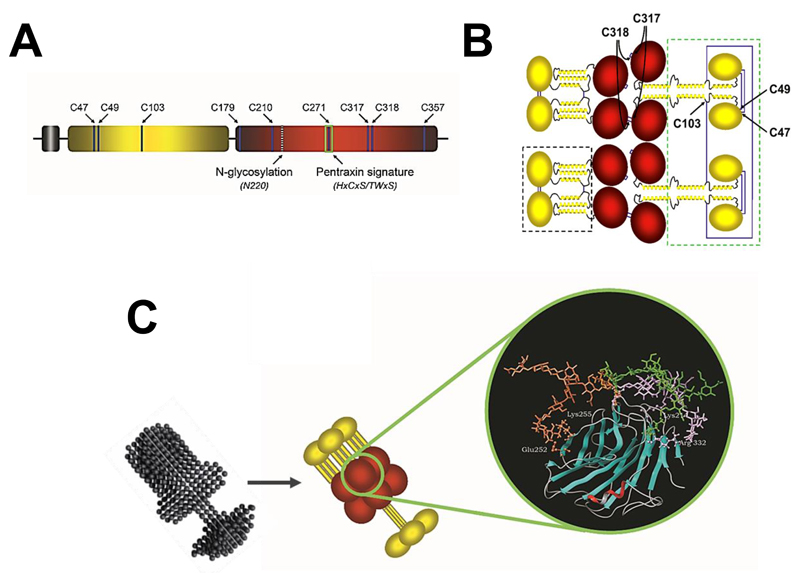
Figure 1. Model of the PTX3 protein.
A, schematic representation of the human PTX3 protomer: the leader peptide is in grey, the N- and C-terminal domains are in yellow and red, respectively. Shown is the relative position of Cys residues, the N-glycosylation site at Asn220, and the pentraxin signature motif. B, the mature protein is composed of eight protomer subunits held together by disulfide bonds. The N-terminal domain (in yellow) comprises an intrinsically disordered N-terminal segment (here represented by ovals) followed by three α-helices, which are predicted to form coiled-coils, and is believed to adopt two different conformations, either extended tetramers (green box) or compact dimers (black box). These are brought together by disulfide bonds formed by cysteine residues of the C-terminal domain (in red). Cysteines forming inter-chain disulfide bonds are indicated. C, the protein folds into asymmetric octamers with two differently sized domains linked by a short stalk. A SAXS model of PTX3 is shown, that is compared with a schematic drawing of the protein. The C-terminal domain of PTX3 (a 3D model of which that is based on the crystal structure of CRP is shown in the blow-up) is N-linked to complex type oligosaccharides, here represented by a core monofucosylated and disialylated biantennary glycan. Molecular dynamics indicates that glycans have different conformations (orange, green, and purple) and contact amino acids at the protein surface (ball-and-stick) through terminal sialic acid residues.
The N-terminal region has no sequence similarity to any protein of known structure. However, based on secondary structure predictions and circular dichroism analyses [(26) and Inforzato et al, unpublished], this portion of the protein most likely folds into four consecutive α-helices, three of which (amino acids 78-97, 109-135, and 144-170) are probably involved in the formation of coiled-coil supersecondary structures (23). Furthermore, in silico predictions point to the N-terminal end of this domain (amino acids 18-54) as an intrinsically disordered sequence; this region likely gains structure upon interaction of the PTX3 protein with its ligands, thus possibly contributing structural and functional versatility to this pentraxin (see below and Fig. 1B).
As anticipated above, the C-terminal domain of PTX3 is homologous to the short pentraxins, with up to 57% similarity (25). This allowed generating three-dimensional models of the C-terminal domain based on the crystallographic structures of CRP (PDBID:1b09) and SAP (PDBID:1sac) (27–29). In particular, the CRP-derived model shows the C-terminal portion of PTX3 to adopt a β-jelly roll topology, similar to that found in legume lectins (Fig. 1C). This structure is stabilized by three disulfide bonds, as supported by biochemical evidences from our own work (30). Cys210 and Cys271 are involved in an intrachain bond that is highly conserved amongst pentraxins. An additional intrachain disulfide links together Cys179 and Cys357 at the N- and C-ends of the pentraxin domain, thus limiting the flexibility of these terminal regions. The two remaining cysteine residues (i.e., Cys317 and Cys318) have been described to form both intra- and inter-chain disulfide linkages that, along with inter-chain disulfide bonds made by cysteine residues of the N-terminal domain, support the quaternary structure of the PTX3 protein (see below) (31). Most importantly, the amino acid residues that line the calcium-binding pocket in both CRP and SAP are missing in the pentraxin domain of PTX3, which might explain some differences in the binding properties of these pentraxins. As an example, PTX3 binds C1q in a calcium-independent fashion, as opposed to CRP and SAP that require this divalent cation for their interaction with C1q (27). In addition, the pentraxin domain of PTX3 lacks the amino acids that form the inter-protomer interface in the CRP and SAP 5-mers and 10-mers. Indeed, recombinant constructs of the PTX3 C-terminal domain do not establish stable intermolecular interactions and are monomeric in solution (30).
The modular nature of the PTX3 protomer provides this long pentraxin with the necessary structural versatility to support its interaction with a number of diverse ligands, and in this way mediate its biological activities. For example, among the PTX3 ligands, fibroblast growth factor 2 (FGF2), inter-α-inhibitor (IαI), TNF-α–induced protein 6 (TNFAIP6 or TSG-6), myeloid differentiation protein 2 (MD-2), and conidia of A. fumigatus each bind to the N-terminal region of the protein (23, 32–35); C1q and P-selectin interact with the pentraxin-like domain (25, 36, 37), whereas both domains have been implicated in the interaction of PTX3 with complement Factor H (38) (see below).
A single N-glycosylation site has been identified in the C-terminal domain of PTX3 at Asn220. This is fully occupied by complex type oligosaccharides, mainly fucosylated and sialylated biantennary sugars with a minor fraction of tri- and tetraantennary glycans. Based on three-dimensional models of the glycosylated C-terminal domain, we have proposed that the PTX3 oligosaccharides make contacts to polar and basic amino acids at the protein surface (i.e., Lys214, Glu252, Lys255, and Arg332) mainly through terminal sialic acid residues (Fig. 1C). These interactions are lost when sialic acid is removed. In addition, upon desialylation protein sites potentially relevant to ligands recognition become accessible, and might be involved in local modifications of the PTX3 tertiary/quaternary structure (28). Interestingly, we have found that the relative content of bi-, tri-, and tetraantennary oligosaccharides and the level of sialylation vary to a great extent amongst PTX3 isolates from different cellular sources. This suggests that the glycosylation pattern of this long pentraxin might change depending on cell type and inducing stimuli (28). This might have important functional implications given that the glycosidic moiety of PTX3 has been implicated in a number of biological activities. In this regard, we have reported that the glycosylation status of PTX3 modulates the protein interaction with C1q and factor H, recognition component of the classical complement pathway and major soluble inhibitor of the complement system, respectively (28, 38). In addition, the sialylated glycans of PTX3 act as major determinants in the interaction of PTX3 with selected influenza A virus strains. PTX3 could thus inhibit virus-induced hemagglutination and neutralize virus infectivity (39). Most importantly, the N-linked glycosidic moiety of PTX3 is essential for its binding to P-selectin. Thereby, PTX3 inhibits leukocyte rolling and extravasation in animal models of acute lung injury and pleurisy (36). PTX3 can therefore communicate with a range of diverse ligands through a common glycan code, and changes in the glycosylation status of the protein might represent a strategy to fine tune the biological activities of this long pentraxin (40).
In addition to the multidomain organization, the human PTX3 protein shows a complex quaternary structure with protomer subunits assembled into high order oligomers stabilized by disulfide bonds [Fig. 1B and (25)]. Mass spectrometry and site-directed mutagenesis analysis of the recombinant human protein indicate that PTX3 is made of covalent octamers (i.e. with a molecular mass of 340 kDa), where cysteine residues at positions 47, 49, and 103 in the N-terminal region form three inter-chain disulfides holding four protein subunits into a tetrameric arrangement. Two tetramers are linked together to form an octamer by additional inter-chain bridges involving the C-terminal residues Cys317 and Cys318 [see above and (31)]. A low-resolution model of the wild type full length PTX3 molecule has been generated based on data from Electron Microscopy (EM) and Small Angle X-ray Scattering (SAXS). According to the model, eight subunits of the protein fold into an elongated structure with a large and a small domain interconnected by a stalk region [Fig. 1C and (30)]. This oligomerization state and the asymmetric shape of the molecule make PTX3 unique amongst pentraxins. Indeed, it displays pseudo 4-fold symmetry along its longitudinal axis, in sharp contrast to the typical pentameric arrangement of the classical short pentraxins. The only other pentraxin that forms an octamer is SAP from Limulus polyphemus, which, however, has been reported to fold into a doubly stacked octameric ring (41).
Protein quaternary structure has different roles in the ligand binding properties of PTX3. For instance, we have shown that the PTX3 octamer contains two FGF2 binding sites, and tetramers of its N-terminal domain act as the functional units in recognition and inhibition of this angiogenic factor (30). However, we have also reported that it is the dimers of the N-terminal domain that mediate the binding of PTX3 to both IαI and TSG-6. Based on these evidences, we have suggested that the octameric structure of PTX3 provides multiple binding sites for each of these ligands, thus acting as a nodal molecule in cross-linking hyaluronic acid in the extracellular matrix (42, 43). Therefore, structural complexity and modular nature of the PTX3 protein probably explain the rather broad spectrum of ligands of this long pentraxin and the diversity of its biological functions as compared to the short pentraxins.
Mechanisms of anti-microbial resistance
A major function of PTX3 is in anti-microbial resistance. PTX3 exerts its anti-microbial effects through different mechanisms, including opsonization and promotion of phagocytosis, regulation of complement activity, and interaction with anti-microbial proteins.
PTX3 as an opsonin
PTX3 retains the opsonic properties of pentraxins, being able to bind to selected microbes and to enhance the phagocytic activity of macrophages and neutrophils (Fig. 2). The first evidence of PTX3 opsonic activity has been reported by Garlanda et al. (44), who demonstrated PTX3-assisted phagocytosis of conidia from Aspergillus fumigatus by murine alveolar macrophages. PTX3 binds to conidia directly, and PTX3-opsonized conidia are ingested by alveolar macrophages more efficiently than non-opsonized conidia. In addition, macrophages from ptx3-/- mice show a weaker phagocytic ability compared to macrophages from wild-type mice, a defect rescued by the administration of exogenous PTX3. In agreement, a higher in vivo susceptibility to invasive pulmonary aspergillosis (IPA) has been observed in ptx3-/- mice (44).
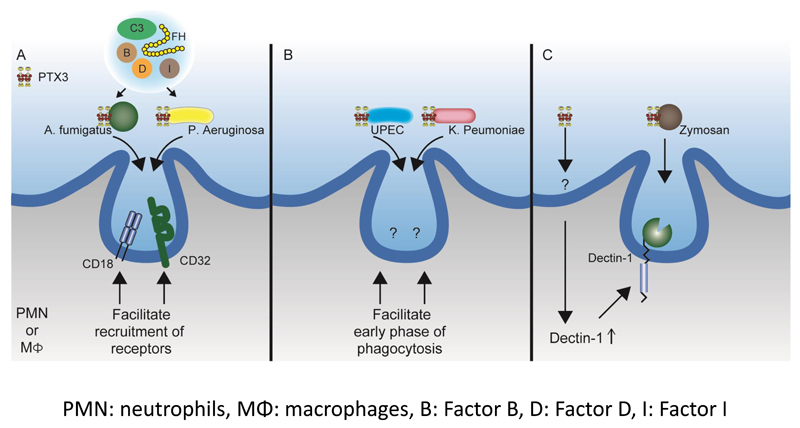
Figure 2. Opsonic activity of PTX3 with various pathways.
PTX3 binds to several microbes including A. fumigatus, P. aeruginosa, UPEC, K. pneumoniae and zymosan. Upon binding, neutrophils or macrophages display enhanced phagocytosis through several pathways; (A) Phagocytosis of PTX3-opsonized A. fumigatus and P. aeruginosa is exerted through FcγRIIa/CD32 and CD11b/CD18, together with alternative complement components. PTX3 opsonization also increases the recruitment of CD32 and CD18 in the phagocytic cup. (B) PTX3 opsonization facilitates the early phase of the phagocytosis of UPEC and K. pneumoniae through unknown mechanisms. (C) PTX3-opsonized zymosan is phagocyted through dectin-1. PTX3 itself has the ability to increase dectin-1 expression through unknown pathway, which creates a feedback loop in the dectin-1-mediated phagocytosis.
The enhancement of phagocytic activity by PTX3 is abolished when heat-inactivated serum is used and is not affected by IgG-depletion, suggesting the involvement of complement components in the process. To characterize the involvement of complement in PTX3-mediated amplification of phagocytosis, a series of experiments were conducted with commercially available sera depleted of specific complement components (34). Lack of C1q, the first component of the classical pathway of complement activation, or of C4, which is implicated in both the classical and the lectin pathways, did not affect PTX3 amplification. On the contrary PTX3-mediated amplification of phagocytosis was completely abrogated in the presence of Factor B- or complement Factor 3 (C3)-depleted serum (34), revealing the main involvement of the alternative complement pathway in the pro-phagocytic activity exerted by PTX3. More significantly, reconstitution of the alternative complement pathway with purified complement components (C3, Factor B, Factor H, Factor D, Factor I) is sufficient to promote the opsonophagocytic effect of PTX3 (Fig. 2A).
Fcγ receptor (FcγR) IIa (CD32) and complement receptor 3 (CD11b/CD18) are required for the PTX3-mediated phagocytic activity (34). Experiments with integrin-blocking antibodies, using FACS and confocal analysis, indicate that activation, internalization and recruitment to the phagocytic cup of CD11b/CD18, as well as CD11b-dependent phagocytosis, are increased in the presence of PTX3-opsonized conidia. Confocal studies also reveal that conidia opsonization by PTX3 enhances co-localization of CD11b and CD32 in the phagocytic cup as compared to non-opsonized conidia. Along the same line, CD11b recruitment to the phagocytic cup is defective in ptx3-/- mice. These results suggest that PTX3 facilitates the internalization of conidia through recruitment and activation of CD11b and CD32 in the phagocytic cup (Fig. 2A). PTX3 has been also reported to interact with CD32, which has been proposed as a potential cellular receptor for this pentraxin (45).
Neutrophils, with their storage of preformed PTX3 that can be promptly released upon microbial stimulation, are major players in the PTX3 opsonic activity (6). In this regard, neutrophils from ptx3-/- mice exhibited lower opsonic activity and less recruitment of CD11b to the phagocytic cup than wild-type neutrophils. In vivo studies revealed that the PTX3-mediated opsonic enhancement in neutrophils was lost in C3- and Fcγ-deficient mice (34).
A similar opsonic effect of PTX3 has been reported for other microbes and microbial moieties, such as Paracoccidioides brasiliensis, Pseudomonas aeruginosa, Klebsiella pneumoniae, uropathogenic Escherichia coli and Saccharomyces cerevisiae [Zymosan, (14, 18, 46, 47)]. PTX3 exerts a protective role in a murine model of P. aeruginosa infection, as shown by an increased susceptibility of ptx3-/- mice to Pseudomonas infection (44). In the course of assessing the therapeutic potential of PTX3 in chronic lung infections by P aeruginosa, Moalli et al. showed that PTX3 directly bound to P aeruginosa, acting as an opsonin and promoting phagocytosis of the opsonized bacteria (47). Similarly to what was described for conidia, this process was C3 and FcγRIIa dependent and C1q independent (Fig. 2A). Opsonization by PTX3 also facilitates the phagocytosis process in other microbial infections. PTX3 exerts a protective role against urinary tract infections (UTI) caused by uropathogenic Escherichia coli (UPEC). Jaillon et al. showed that PTX3 directly binds to UPEC and enhances UPEC phagocytosis leading to accelerated phagosome maturation by neutrophils (14) (Fig. 2B).
Soares et al. analyzed the role of PTX3 in infections caused by K. pneumoniae taking advantage of transgenic mice overexpressing the PTX3 protein (18). In contrast to what has been described until now, a detrimental or protective role of PTX3 was observed, depending on the microbial burden. In the case of a lower inoculum, PTX3-transgenic mice are protected from K. pneumoniae infection. Under such condition, neutrophils from PTX3-transgenic mice show faster phagocytic process (Fig. 2B). On the contrary, in the case of a high inoculum, PTX3 overexpression was associated with higher production of proinflammatory mediators (i.e. nitric oxide and TNF-α), leading to faster lethality and inability of the murine host to deal with the pulmonary infection.
Macrophages from PTX3-transgenic mice display a higher phagocytic activity for both zymosan and P. brasiliensis, irrespective of opsonization, while the addition of exogenous PTX3 enhances the phagocytic ability of macrophages from wild-type mice (46). Macrophages from PTX3-transgenic mice express consistently higher levels of dectin-1, a main receptor of β-glucan, in response to zymosan. Blockade of dectin-1 results in the inhibition of zymosan phagocytosis. These data suggest the existence of a PTX3-dectin-1 feedback loop involved in the phagocytosis of zymosan (Fig. 2C). Also in this case, a direct binding of PTX3 to both zymosan and P. brasiliensis was confirmed.
Thus PTX3, by controlling the production of proinflammatory mediators and acting both as an opsonin and an inducer of dectin-1 expression, may favor or disfavor handling by the murine host of different infectious agents.
Regulation of complement activation
The pentraxins CRP, PTX3 and SAP have all been found to interact with molecules of the complement system, indicating that they can use complement for their effector functions (Fig. 3). The interaction of CRP with complement has been investigated more extensively than that of PTX3. Therefore here CRP will also be analyzed as a reference molecule in the family.
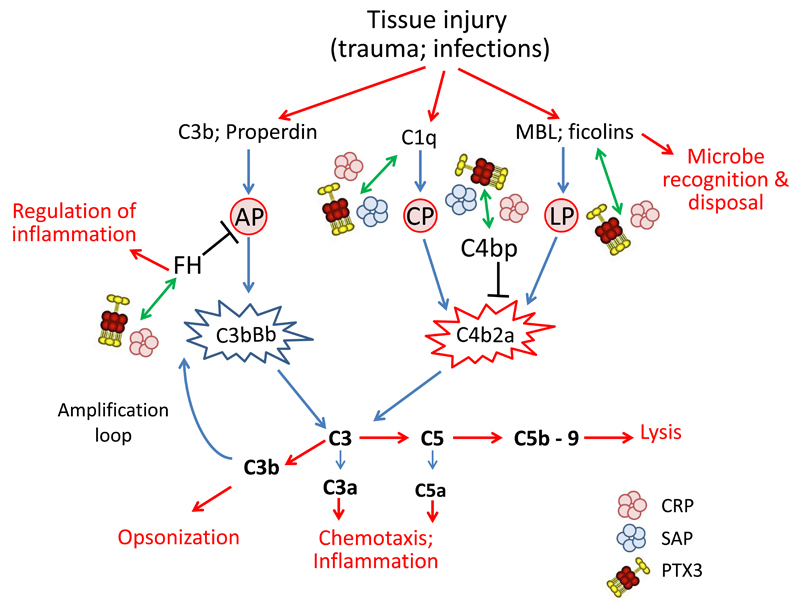
Figure 3. Schematic representation of pentraxins involvement in regulation of complement activity.
PTX3, CRP and SAP have been found to interact with molecules of the complement system. Complement activation is triggered by the interaction of microbes with recognition molecules. The lectin pathway (LP) is activated by interaction of MBL and ficolines with carbohydrates on microbial surface. Specific antibodies can bind to microorganisms forming binding sites for C1q and activate the classical complement pathway (CP). Finally C3 and properdin, once immobilized on a surface such as a microbial cell wall, can activate the alternative pathway (AP). Activation of all the three pathways lead to a cascade of events resulting in opsonization, leukocyte recruitment and cell lysis, activities essential for microbial removal. The first step is the formation of the C3 convertase (C4b2a for CP and LP and C3bBb for AP) that triggers hydrolysis of C3 with formation of C3a, C3b and C5a, and with subsequent formation of C5b-9, the membrane attack complex leading to cell lysis. PTX3, CRP and SAP they all bind C1q, regulating the classical complement pathway. PTX3 and CRP also participate to the regulation of the lectin pathway through interaction with MBL and ficolines. In addition pentraxins can recognize regulators of complement activation, in particular FH, the most important regulator of the alternative pathway, and C4bp, involved in the regulation of the C3 convertase C4b2a, common to the classical and lectin pathways.
In the acute phase response the levels of CRP can increase from levels below 1 µg/ml up to 500 µg/ml (48). No complement protein increases its levels during the acute phase response in a similar way, although levels of e.g. C3 can increase by about 50%. Therefore, the complement system can dynamically use CRP for directing complement activity into areas of tissue damage or against infectious agents when needed. This activity, indeed, is similar to antibodies, except that, for new antibodies to develop, much more time is needed in a de novo situation. CRP levels reach their maximum at 24-48 hours, whereas antibody development takes 1-2 weeks. Pre-existing antibodies or natural antibodies of IgM class, however, can be recruited much faster. Natural antibodies often have similar specificities as pentraxins. As an example, they can bind to certain phospholipids, like phosphatidylcholine or phosphatidylethanolamine. The difference is that CRP needs calcium for the interaction, while the pentameric IgM does not. Common to both is that they use multiple binding sites to increase the avidity towards target particles.
The complement system can thus use pentraxins as one group of sensor molecules for rapid recognition of patterns on targets that need to be cleared away before healing can start. Antibodies are another group and collectins yet another one to initiate complement activation. Also collectins exploit multiple binding sites to increase their overall binding avidities to physiologically meaningful levels (49). Pentraxins have five plate-like binding subunits in the pentraxin domains (50). The short pentraxins CRP and SAP are composed exclusively of the pentameric plates, whereas PTX3 and the other long pentraxins can have different quaternary conformations (Fig. 1). CRP can use all different parts of the molecule for its interactions. The bottom part of the plate binds in a calcium-dependent manner to surface phospholipids, while the top part and the sides are used for interactions with the complement proteins (see below) (51).
Functionally the interactions of pentraxins with targets and with complement lead to opsonization of foreign microbes or endogenous waste products. Bound molecules, mostly C1q and iC3b in the complement system, are recognized by specific receptors of phagocytes, macrophages, dendritic cells and neutrophils. These usually ingest the target particles in a relatively silent, non-inflammatory fashion. Recognition of iC3b-coated particles by macrophages via the integrin receptor CD11b/CD18 leads to synthesis of immunosuppressive IL-10 and TGF-beta cytokines (52). Upon contact with a larger number of microbes or in a more threatening situation, the level of complement activation is stronger and alarms a wider inflammatory response via the generation of the C5a anaphylatoxin and cell activation by the membrane attack complex of complement (MAC). MAC can lyse many gram-negative bacteria. Especially for protection against meningococcal and gonococcal infections (gram-negative Neisserias) complement lysis is essential. In alternative and terminal pathway complement deficiencies, the risk for neisserial infections can increase up to 1000-fold (53).
Following binding to their target, pentraxins can recruit the first component of complement C1q to the same site. The ability to bind C1q and activate the classical pathway has been described for CRP (54), SAP (55) and PTX3 (56). Since at least two different kinds of receptors exist for C1q, one for the globular domain and another for the collagenous part, the interactions can signal the opsonic event and lead to phagocytosis. C1q is not the only ligand for C1q receptors. Also many other collectins, like mannan-binding lectin (MBL) and ficolins 1-3 can bind to the C1q receptors (57). Collectins are so called because the subunits of the molecules contain a collagen-like triple-helical trunk and often a sugar binding globular lectin domain. Some of the collectins have been shown to bind PTX3 (58, 59). Direct interaction with FcγR, as described above, has also been reported for PTX3 and CRP (45, 60). By binding via collectins to the C1q receptors, pentraxins could act as “clean-up” molecules to assist in the removal of materials from injured or apoptotic cells. C1q can bind to many components of injured tissue itself, as well. Since specificity differences between C1q and pentraxins exist, the overall pattern recognition repertoire is wide. While C1q binding has broad specificity, that of pentraxins is more limited and often dependent on divalent cations, mostly Ca2+ ions.
In systemic lupus erythematosus (SLE), which is a disease characterized by a clearance problem and secondary autoimmunity, the CRP response is often compromised (61). Also, deficiencies of C1q and subsequent classical pathway complement components predispose to SLE (62). In rare cases of C1q deficiency, SLE is always severe and involves dysfunction of kidneys. In mice, SAP has been suggested to adopt the role equivalent to human CRP (63). SAP binds to chromatin in a calcium-dependent manner and prevents anti-DNA autoantibody formation. In SAP knock-out mice, accordingly, increased levels of antibodies against DNA have been detected (64). PTX3 also plays a role in SLE, as suggested by the results obtained crossing the autoimmune mouse strain B6lpr with ptx3-/- mice (65). In fact, besides an impaired clearance of apoptotic cells by ptx3-/- peritoneal macrophages, lack of PTX3 in B6lpr mice aggravated autoimmune lung disease (65). A common property of pentraxins thus appears to participate in the clearance of intra- or extracellular chromatin. This function is assisted by the interaction with the complement system.
Pentraxins can activate the classical pathway of the complement cascade. Each triplet of the globular domains of C1q binds into the center of the upper side of a pentraxin pentamer (66). It is thus likely, as with antibodies, that the C1q binding is strong enough to promote complement activation, once pentraxins are bound to targets with a sufficient density. Thus, for example, CRP pentamers would bind in a calcium-dependent manner to a surface with phospholipids exposed from the inner membrane leaflet and recruit C1q molecules. This could occur e.g. to apoptotic cells or to nonviable cells in an ischemic lesion, but not to neighboring sufficiently viable cells. Thereby the target area would become demarcated and labeled for later removal by opsonophagocytosis. Importantly, following recruitment of C1q or of other collectins, the phagocytes can directly recognize the bound ligands with C1q receptors.
In the classical pathway, after binding and a conformational change in C1q, the serine esterases C1r and C1s in the C1 complex (C1qr2s2) become activated by proteolytic cleavage. Activated C1s cleaves C4 and C2 to generate the classical pathway C3 convertase C4b2a. The C4b2a convertase formation and stability are controlled by the classical pathway inhibitor C4b binding protein, C4bp. The pentraxins CRP (67), PTX3 (68) (Deban et al, unpublished) and SAP (69) all have been shown to bind C4bp suggesting an important point of control in classical pathway complement activation. For example, in alcohol-induced liver damage the productions of both SAP and C4bp were reduced, whereas those of the classical pathway components were increased. This suggests an alcohol-induced decreased ability of tissue clearance and role for SAP and complement in the development of liver cirrhosis (70). A delicate balance thus exists between activities favoring complement activation and control at sites of tissue damage. This is understandable, because at the same time as the targets are being recognized and prepared for clearance, the neighboring normal tissue needs to be protected. By promoting cleavage of C4b by Factor I (C4b/C3b inactivator) the pentraxin-bound C4bp can irreversibly inactivate the C4b2a convertase. The binding of other collectins, like ficolins or MBL, to pentraxins may lead to similar consequences as binding to C1q. This is because complement activation via the classical and lectin pathways converges into the same C4b2a convertase and involves the same downstream molecules.. Indeed, both pathways ultimately lead to C3 conversion, which can launch the amplification cascade of the alternative complement pathway.
The alternative pathway of complement can become activated directly upon contact with a foreign, non-self surface, or following activation by any other means, including the classical and lectin pathways. After covalent binding of C3b to the target surface, Factor B will bind to it and generate the alternative pathway C3 convertase C3bBb, where B is cleaved to Bb by another serine esterase, Factor D (71). Alternative pathway can amplify efficiently its own activation because the C3bBb convertase will activate more C3 molecules to C3b and these will in turn become subunits of new C3bBb convertases. By this, the alternative pathway amplification system can coat the whole target, e.g. bacteria or yeast, with C3b molecules very fast, within minutes. The “explosive potential” of complement activation poses a risk to healthy normal tissues both at the constitutive as well as at an induced level.
Our own cell surfaces are, under normal circumstances, well protected against complement attack by specific membrane regulators, like CD35 (C3b/C4b receptor or complement receptor type 1, CR1), CD46 (membrane cofactor protein, MCP), CD55 (decay accelerating factor, DAF) and CD59 (protectin) (72). In addition, the alternative pathway can in a robust manner discriminate between self and non-self structures by using only 2 molecules, C3b and Factor H (73). After an initial random deposition of C3b to a surface, Factor H will bind to it if the surface is coated with sialic acid, glycosaminoglycans or other polyanions (74). Factor H bound to the C3b-polyanion complex acts as a cofactor for the C4b/C3b inactivator enzyme, Factor I. Thereby the C3b molecules become inactivated by Factor I and can no longer participate in the formation of the C3bBb convertase enzymes. The importance of Factor H activities in protecting self surfaces is illustrated by the severe diseases that follow if recognition fails (75). The first example is atypical hemolytic uremic syndrome (aHUS), where mutations in Factor H fail to recognize host cell surface-associated sialic acid (76).
During acute tissue damage or upon infection the need to carefully target complement activation and to regulate excessive activation locally needs to be organized at an “ad hoc” basis. This is where the pentraxins are in charge. Most importantly, viable tissues at local sites need to be protected from potentially detrimental effects of the amplification cascade. The need for protection and regulated activity extends also to the coagulation cascade, because often conditions related to tissue injury increase the pro-coagulant activity of vascular endothelial cells, platelets and blood cells. The increased pro-complement and pro-coagulant activities are typical for aHUS and other so called thrombotic microangiopathies (TMAs) (77). PTX3 is produced by both endothelial cells and blood leukocytes.
While the pentraxins are able to activate the classical or lectin pathways of complement, their role in the alternative pathway appears to limit activation and target it to only sites where needed. Both CRP and PTX3 have been found to bind Factor H (38, 78). Binding of the elongated Factor H molecule to CRP seems to occur to areas distinct from the surface interaction sites (at the bottom side of CRP) and C1q binding site (in the middle of CRP pentamer). Thus, the likeliest interaction area is at the external sides of CRP monomers (79, 80). CRP has multiple binding sites on Factor H, a well characterized site on domain 7 (78), a secondary site on domains 8-11 (78) and a site at the most C-terminal domains (80), where also important sites for C3d/C3b and the surface polyanions exist (81–84). Also PTX3 binds to the two “hot spot” domains 7 and 19-20 of Factor H (38). Interestingly, a common polymorphic amino acid variant in domain 7 (Tyr402His) has been found to influence binding of CRP (79, 85) but not of PTX3 to Factor H (38). This finding may be relevant for the pathogenesis of the most common cause of blindness in the Western world, age-related macular degeneration (AMD). AMD is linked to the Tyr402His polymorphism in Factor H (86, 87), with a risk factor up to 9 in case the person is homozygous for the variant (88). In addition to CRP binding, the Tyr402His variant has been found to influence glycosaminoglycan binding locally in the retina (89).
All the pentraxins CRP, PTX3 and SAP interact with both activating (C1q) and regulating (Factor H, C4bp) complement components (Fig. 3). But, how could these activities be viewed from a physiological point of view? The pentraxins, despite differences among themselves, seem all to be involved in the acute phase response or response to injury. Therefore, they could be considered as a task force or an acting arm for the complement system in emergent situations. Once tissue injury takes place, e.g. because of trauma or infection, the synthesis of acute phase proteins increases remarkably in response to cytokines, IL-1β and IL-6. CRP, PTX3 and SAP can bind to injured cells. Initial binding may be diffuse and follow a gradient towards the center of an ischemic or necrotic area. However, at the end, a demarcation line needs to be created between cells to be cleared away and cells to be saved. The activation of complement via C1q binding will generate an initial level of inflammation and stimulate formation of C3 convertase enzymes. Activation, however, needs to be limited in area and in time. Upon binding of C4bp, and especially of Factor H, the amplification of complement remains controlled. Binding of Factor H can help in a sharper demarcation between nonviable and viable cells or tissue structures by preventing activation by normal cells. At sites, where CRP or PTX3 have become bound, Factor H is recruited to limit excess complement activation, C5a production and MAC formation. Pentraxin-directed binding of Factor H promotes inactivation of C3b to iC3b thereby generating high affinity ligands for the macrophage, dendritic cell and neutrophil receptor CD11b/CD18, which is an integrin, complement receptor type 3 (CR3). This will aid the phagocytosis of opsonized targets, nonviable cells or remnants of cells (membrane fragments, cytoskeletal structures, nuclei and other intracellular organelles) in a directed fashion. The general role of pentraxins together with complement would thus be a directed removal of tissue debris and other waste in a noninflammatory fashion. In clinical situations, cooperation between pentraxins and complement may occur e.g. in rheumatoid arthritis, reactive arthritis, atherosclerosis and ischemic heart disease.
The targets for pentraxins would not only be endogenous tissue components but selected microbes, as well. PTX3 has been found to bind e.g. to the fungus A. fumigatus and the gram-negative bacteria P. aeruginosa and K. pneumoniae. CRP was originally described as a protein that binds to pneumococci (90). Thus, pentraxins could participate in the recognition of invading microbes and their removal by direct or complement-mediated phagocytosis..
Many pathogenic microbes efficiently use Factor H for their own protection against complement-mediated opsonophagocytosis (91). This is possible by the expression of microbial Factor H binding proteins that seem to be specific for each pathogen. It has, for example, been shown that many types of pneumococci express PspC-family proteins that bind Factor H (92). Interestingly, those pneumococci that lack Factor H binding proteins can recruit Factor H to their surfaces with the help of CRP (92, 93). Direct interactions between PTX3 and pneumococci have not yet been thoroughly addressed but it has been observed that high levels of PTX3 associated with a fatal disease in bacteremic patients (94). This could indicate misuse of PTX3 by the bacteria, as well. Clinically, it indicates that, like CRP, also PTX3 can potentially be used as a marker for serious bacterial infections. Thus, while we have developed efficient means to combat bacterial infections with pentraxins and complement, the microbes also have devised ways to counteract the attack.
Interaction with anti-microbial proteins
As already outlined in this review, the capacity of PTX3 to interact with several ligands supports the multi-functionality of this long pentraxin (3). Proteomic analysis of PTX3 complexes from the blood of septic patients confirmed the interaction of PTX3 with a series of complement components and extracellular matrix proteins (95). This analysis also revealed that PTX3 forms complexes with some of the bactericidal proteins associated with neutrophil extracellular traps (NETs), namely azurocidin 1 (AZU1) and myeloperoxidase (MPO). A calcium-dependent and high affinity binding (Kd 22-7.6 nM) has been confirmed between AZU1 and PTX3, that involves in particular the N-terminal PTX3 domain. NETs are mesh-like structures, composed of DNA, histones and microbicidal proteins, produced by neutrophils through a distinct apoptosis pathway called NETosis (96–98). Taking into account the fact that PTX3 is one of the NETs components (6, 99), this observation implies that the interplay of PTX3 and NETs proteins might have some host-protective roles against sepsis. Co-localization of AZU1-PTX3 and MPO-PTX3 in NETs has been observed (95, 100). AZU1 has broad bactericidal activity towards Gram-positive and Gram-negative bacteria and Candida albicans, likely due to its cationic and hydrophobic nature (101, 102). The major bactericidal activity of MPO is to catalyze the production of hypochlorous acid (HOCl), which is toxic to microbes, in the presence of hydrogen peroxide (H2O2) and chloride ions (103). This evidence strongly suggests an involvement of PTX3 in these bactericidal activities. It is, however, yet to be understood whether and how PTX3 modulates AZU1- and/or MPO-mediated bacterial killing.
Extracellular histones have emerged as new players that affect mortality for sepsis (104). In addition to cytotoxicity on endothelial cells, in vivo analysis revealed that they help the recruitment of neutrophils and contribute to intra-alveolar hemorrhage, vascular thrombosis, platelet aggregation and thrombocytopenia (104, 105). Thus, they are regarded as new targets for both diagnosis and treatment of sepsis. The proteomic analysis described above identified extracellular histones as new ligands of PTX3 (95), and a direct interaction between PTX3 and histone H1 has been reported (25, 26). In addition, the PTX3-histones interaction has novel functional implication in the fatal sepsis (26). In this respect, in vitro studies showed that PTX3 suppresses the extracellular histones-mediated cytotoxicity on endothelial cells. Of note, PTX3-histones interaction induced their aggregation through the loss of secondary structure. In vivo studies revealed that the administration of PTX3 suppresses the mortality caused by histones infusion in a murine model of sepsis. The N-terminal domain of PTX3 recapitulates the full length protein’s activity both in vitro and in vivo. Taken together, this study indicates that PTX3 might participate in the dampening of extracellular histones-mediated cytotoxicity in sepsis. Therefore, circulating levels of PTX3-histone complexes might reflect the severity of sepsis. Thus, an understanding of the molecular mechanism underlying PTX3-histones aggregation might pave the ground to new therapeutic strategies for sepsis targeting extracellular histones.
Exploitation by microbes
Although the results presented above demonstrated the protective role of PTX3 during microbial invasion, in specific contexts PTX3 may promote immunopathology. As already mentioned, in a model of K. pneumoniae infection, PTX3 overexpression played a dual role depending on the bacterial load. PTX3 overexpressing transgenic mice showed increased mortality, reduced neutrophil infiltration in lungs and increased bacterial dissemination compared to wild type animals upon infection with high bacterial load (18). In contrast, PTX3 overexpression conferred protection to lower K. pneumoniae pulmonary inocula, by favoring a moderately increased inflammatory response in the lungs and bacterial phagocytosis (18). Accordingly, PTX3 was shown to interact with the outer membrane protein A of K. pneumoniae (KpOmpA), and amplify the TLR2-dependent inflammatory response induced upon KpOmpA recognition by scavenger receptors LOX-1 and SREC-I (106). These results highlight the importance of balanced inflammatory reponses in innate resistance to K. pneumoniae and in the prevention of immunopathology (Fig. 2).
PTX3 has been proposed to play a role in defense against some viruses, such as human and murine cytomegalovirus (CMV) and influenza A virus [IAV, (39, 107)]. The protective role of PTX3 was due to the binding to human and murine CMV, which reduced viral entry into DCs and played a protective role against primary CMV infection and reactivation in mice (107). PTX3 also recognized specific strains of the H3N2 subtype IAV via the interaction between the sialic acid residue on its glycosidic moiety and hemagglutinin (HA) and neuraminidase glycoproteins present on the viral envelope, acting as a “receptor decoy” for the virus and preventing viral spread and infection by specific IAV strains (39). In contrast, PTX3 did not show anti-viral activities towards seasonal or pandemic H1N1 IAV or other H3N2 strains due to aminoacid substitutions in the viral HA sequence, which abolished the interaction with PTX3 and its viral neutralizing activity (108, 109).
In contrast with these data, the interaction of PTX3 with arthritogenic alphaviruses, namely with chikungunya virus (CHIKV) and Ross River virus (RRV), was shown to promote the early viral entry and replication in host cells (21, 110). During the acute phase of alphavirus infection, the expression of PTX3 increased both in patients and experimental animals, and was associated with an enhanced viral load and severity of the disease. Furthermore, PTX3-deficiency was associated with delayed disease progression and fast recovery, reduced inflammatory responses and viral replication (21). Even if the cellular receptors involved in the entrance of alphaviruses are still poorly defined, the aggregates formed between RRV and PTX3 may promote more efficient multivalent binding to cell surface receptor/s for RRV, thereby promoting enhanced receptor-mediated endocytosis and viral entry. Alternatively, PTX3 may opsonize RRV and promote its uptake via putative cell surface receptors for PTX3. The role of PTX3 in the pathogenesis of these viral infections is in line with recent evidence showing that alphavirus-induced diseases can be exacerbated by over-expression of C3 and MBL (111). It also highlights the potential contribution of humoral innate immunity and complement in the pathogenesis of alphaviral disease.
Regulation of inflammation and tissue damage
Given the production of PTX3 by myeloid cells in response to primary inflammatory cytokines, it is expected that the protein, similarly to the other members of the pentraxin family, plays a role in the regulation of inflammation. As outlined above, PTX3 regulates complement activation and, in some context, it can directly affect the production of proinflammatory mediators, as shown for instance in the model of infection with K. pneumoniae in PTX3-overexpressing mice. In addition, PTX3 amplification of the proinflammatory response induced by Kp-OmpA is complement dependent and is abrogated by treatment with complement inhibitors (112).
PTX3 can affect cell recruitment at inflamed sites through the interaction with the adhesion molecule P-selectin. A dose-dependent and saturable binding of PTX3 to P-selectin, but not to E- and L-selectin, has been observed, an interaction essentially mediated by the sialilated N-linked glycosidic moiety of PTX3 (36). Intra-vital microscopy of thrombin-stimulated mesenteric venules, a model of P-selectin dependent rolling, shows that ptx3-/- mice had significantly more rolling interactions than wild-type mice, and PTX3 administration to wild type vessels significantly and consistently attenuated the frequency of rolling interactions. This suggests that, through the binding with P-selectin, PTX3 limits early neutrophil recruitment in response to inflammatory stimuli. In agreement, ptx3-/- mice show greater neutrophil recruitment in a model of pleurisy, in acute lung injury and in ischemia/reperfusion-induced kidney damage (36, 113). Based on these data, PTX3 has been proposed as a negative feedback mechanism regulating PMN recruitment in inflamed tissues.
Recent observations demonstrate that PTX3 plays a non-redundant role in the normal wound healing processes (5). Different models of tissue damage were analyzed in ptx3-/- mice, including skin wound healing, sterile liver and lung injury, arterial thrombosis. In all cases lack of PTX3 is associated with increased fibrin deposition and persistence, and higher clot formation. Ptx3-/- macrophages show a defective pericellular fibrinolysis in vitro, and defective directional migration in the provisional fibrin-rich inflammatory matrix in vivo. The phenotype of ptx3-/- mice is attributed to the interaction of PTX3 with fibrin and plasminogen at acidic pH, an interaction essentially mediated by the PTX3 N-terminal domain. Administration of inhibitors of fibrin deposition and platelet activation rescued the phenotype of ptx3-/- mice, thus demonstrating that alteration in the fibrinolytic response to injury is responsible for the defective wound healing. The results obtained so far suggest that, by interacting with the provisional matrix protein fibrin, PTX3 contributes to the orchestration of tissue repair and remodeling. The acidic pH, which occurs during tissue injury and repair (114), is likely to act as a switch on signal that set PTX3 in a tissue repair mode and the pH dependency of PTX3 binding to fibrin and plasminogen ensures that the interaction does not occur in the circulation but rather at sites of tissue repair.
A role for PTX3 is described in different models of ischemia and reperfusion. In a model of post-ischemic renal injury, ptx3-/- mice showed increased tissue damage and administration of recombinant protein exerted a protective effect, with enhanced recovery, suppression of glomerulal sclerosis and inhibition of interstitial fibrosis (113, 115). Interaction between PTX3 and P-selectin also plays a role in this model. In fact, the increased post-ischemic leukocyte recruitment observed after renal injury in ptx3-/- mice was completely abrogated by treatment with a neutralizing antibody against P-selectin (113). On the contrary, in a model of intestinal ischemia and reperfusion, the absence of PTX3 was associated with lower inflammation and lethality (19).
PTX3 is also associated with inflammation in acute myocardial infarction (AMI). In humans as well as in mice, PTX3 is rapidly produced during acute myocardial ischemia and ptx3-/- mice have higher no-reflow area, increased neutrophil infiltration, decreased number of capillaries, and increased number of apoptotic cardiomyocytes. In addition higher C3 deposition was observed in lesional tissue (116). Higher aortic lesions and a more pronounced inflammatory profile are observed in a model of atherosclerosis in mice deficient for apolipoprotein E and PTX3 (117). In addition, in a murine model of myocarditis, induced by infection with coxsackievirus B3, PTX3-deficiency is associated with increased heart injury and cardiomyocyte apoptosis (118). The mechanisms of the protective role exerted by PTX3 are still debated, it is known that PTX3 does not exert a direct antiviral effect, but rather could facilitate the clearance of dead or dying cells. As a matter of fact, PTX3 binds apoptotic cells, enhancing C1q binding and complement-mediated clearance of the apoptotic debris (37, 119). In a murine model of seizure-induced degeneration, a reduced number of dying neurons (apoptotic and/or necrotic) was observed in wild type compared to ptx3-/- mice, suggesting that PTX3, through the binding with apoptotic cells, could exert a protective role in damaged tissues (120). Several studies have shown that efficient apoptosis inhibits the inflammatory response and reduces the development of atherosclerosis, therefore the capacity of PTX3 to affect engulfment of apoptotic cells could likely represent an additional mechanism of regulation of inflammation.
In a model of cerebral ischemia induced by transient middle cerebral artery occlusion, PTX3 deficiency was associated with impaired glial scar formation and alterations in scar-associated extracellular matrix production. Ptx3-/- mice also have defective resolution of brain edema, suggesting that PTX3 might support integrity of the brain blood barrier (121). Other examples of dual functions of PTX3 emerged recently. In a model of ventilator-induced lung injury (122), PTX3 overexpression resulted in increased inflammatory response and PTX3 has been found able to induce endothelial dysfunction inhibiting vasorelaxation induced by acetylcholine. Overall it emerged that PTX3 can affect vascular resistance, inducing morphological changes in endothelial cells (24). Thus, in the context of reperfusion and injury, PTX3 could exert dual opposite roles, being protective or deleterious depending on tissue district.
It is generally accepted that inflammation, whether sustained by infections or inflammatory conditions of diverse origins, plays an essential role in tumor development. Thus it is expected that PTX3, as regulator if inflammation, could play a role in cancer. The role of PTX3 in tumor development was analyzed taking advantage of ptx3-/- mice in different models of chemical carcinogenesis. PTX3 deficiency causes increased susceptibility to mesenchymal and epithelial carcinogenesis in the models of 3-Methylcholanthrene (3-MCA)-induced carcinogenesis and 7,12-dimethylbenz [α] anthracene/terephtalic acid (DMBA/TPA)-induced skin carcinogenesis (12). PTX3-deficiency is associated with an exacerbated inflammation, as revealed by enhanced levels of tumor-associated macrophages, higher production of pro-inflammatory cytokines, increased DNA damage, higher angiogenesis and C3 deposition. In this context, increased inflammation is potentially a cause of genetic instability, as suggested by increased Trp53 mutations and oxidative DNA damage (123). PTX3 regulation of C3 deposition on tumor cells occurs through recruitment of the negative complement regulator FH, suggesting that unleashed complement activation observed in Ptx3-/- mice plays a crucial role in promotion of an inflammatory, pro-tumoral microenvironment.
In humans, PTX3 expression is increased in different cancers, including glioma, lung cancer, liposarcoma, prostate and pancreatic carcinoma, breast cancer (124–128). PTX3 gene polymorphisms are associated with circulating levels of the protein and risk to develop hepatocellular carcinoma in subjects infected with hepatitis C virus (129). In a cohort of ovarian cancer patients, gene expression profile evidences PTX3 gene expression in a stromal signature associated to poor prognosis (130).
Epigenetic regulation of PTX3 expression has been characterized in human cancer. In esophageal squamous cell carcinoma, PTX3 promoter is hypermethylated, with consequent reduction of PTX3 expression (131). A similar hypermethylation of PTX3 promoter is reported for colorectal cancer (CRC; File: GSM801957 in the Epigenomics database http://www.ncbi.nlm.nih.gov/epigenomics). In a series of mesenchymal and epithelial cancers, as leyomiosarcomas, desmoid tumors, CRC, the PTX3 promoter and a regulatory region are highly methylated, in contrast with normal mesenchymal or epithelial tissues (12). The analysis of these regulatory regions of PTX3 gene indicates that methylation progressively increases from normal colon epithelium, to adenomas and to CRC, inducing in parallel PTX3 gene silencing (12). Taken together these data suggest that PTX3 acts as an extrinsic oncosuppressor gene in mouse and man, representing an example of the connection between inflammation and cancer perceived in the last years (132–135).
Beside the role as oncosuppressor, few reports outline a pro-tumoral role of PTX3. In gastric cancer and head and neck tumors, PTX3 promotes tumor cell migration and invasion (136–138), while in human glioma the protein sustains tumor cell proliferation (139). In addition, in gastric cancer PTX3 silencing suppresses cancer related inflammation. These contrasting results suggest that further studies will be necessary to define the real significance of PTX3 in the regulation of tumor-associated inflammation.
More in general, PTX3 could exert opposite roles in different contexts, being essentially protective against bacterial infections, where it exerts antibody-like functions, promoting complement activation and opsonization. On the contrary, following tissue damage, protective or deleterious roles are described, depending on the tissue district and the overall inflammation.
PTX3 as diagnostic/prognostic marker during inflammation and infection
PTX3 is an acute phase protein and its plasmatic levels increase rapidly (peak at 6-8 h) from a basal value of approximately 2 ng/ml in healthy subjects, to as much as hundreds of nanograms in inflammatory conditions. The production of PTX3 by different cell types guarantees a local effect of the molecule, this representing a main difference with the cognate protein CRP, produced essentially systemically by the liver. Led by the production induced by pro-inflammatory cytokines and TLR engagement, ongoing efforts have the purpose to investigate whether PTX3 could represent a novel marker of infectious or inflammatory diseases, complementary to CRP.
A first study published in 2000 reported a rapid increase of PTX3 plasma levels in patients with acute myocardial infarction (AMI), and described for the first time the correlation between PTX3 levels and severity of disease, proposing that PTX3 could be a predictor of mortality (140). This initial observation was extended in a large cohort of patients with AMI, where it emerged as the only independent predictor of mortality within 3 months from the acute event (141). In addition, PTX3 levels were associated with adverse cardiovascular outcomes and risk of cardiac events in patients with heart failure (142–145). In addition, multiple studies have reported increased PTX3 plasma levels in chronic kidney disease and hemodialysis patients, where it appears to be a promising biomarker of disease (146–151). From the analysis of large cohorts of patients emerged that high systemic PTX3 levels are associated with increased risk of cardiovascular morbidity and mortality (147, 148, 152–154), providing further insights into the role of inflammation in these pathological conditions.
In the context of infections, similar results on PTX3 association with disease severity are reported. In a small cohort of 101 consecutive critically ill patients admitted to the Intensive Care Unit (ICU) with systemic inflammatory response syndrome (SIRS), sepsis, or septic shock, PTX3 plasma levels were elevated, with a gradient from SIRS to septic shock, reflecting the severity of disease and acting as predictor of mortality (155). Several studies confirmed this original observation. A Danish study on 261 consecutive patients admitted to ICU for SIRS reported higher levels of PTX3 associated with the development of sepsis, severe sepsis and septic shock. Higher PTX3 levels correlated with SAPS2 score and patients with high PTX3 levels at admission have higher 90 day mortality than patients with lower levels (156). In addition, a study on more than 500 patients demonstrates that a high PTX3 level on hospital admission predicts severe sepsis and case fatality in patients with suspected infection (157). In 132 adult patients admitted to ICU with positive blood culture for the most common causative organisms in community-acquired bacteremia, (Staphylococcus. aureus, Streptococcus. pneumoniae, b-hemolytic streptococci or E. coli) maximum PTX3 values on days 1–4 were observed in nonsurvivors, while in this cohort CRP levels were not predictive of mortality (94). In a prospective study on 90 patients admitted to three general intensive care units for severe sepsis or septic shock, persisting high levels of circulating PTX3 over the first days from sepsis onset are associated with mortality (158). A “sepsis-like” syndrome is observed in the absence of infections after cardiopulmonary resuscitation. The systemic inflammatory response observed after a condition of total body ischemia and reperfusion is characterized by deregulated cytokines production, presence of endotoxin in plasma and coagulation abnormalities. In these patients PTX3 plasma levels are markedly elevated and are associated with higher risk of multiple organ dysfunction syndrome (159). Overall these data indicate that measurement of PTX3 in the course of a systemic inflammatory response may improve patient risk assessment and thus be useful in guiding subsequent therapeutic interventions.
Several studies address the role of PTX3 as marker in infections. PTX3 plasma levels were investigated in patients with Dengue virus infections, pulmonary tuberculosis (TB), leptospirosis, meningococcal disease (160–163). In patients suffering from Dengue shock syndrome, PTX3 levels were higher compared to levels found in patients with Dengue fever and Dengue hemorrhagic fever, indicating that PTX3 is a marker of infection better than CRP, that is not associated to disease severity (161). In a group of 220 newly diagnosed TB patients, PTX3 levels were higher than levels in healthy household controls and community controls. At the end of the therapeutic protocol, subject responsive to therapy presented a significant reduction of PTX3 levels, while in patients with treatment failure, PTX3 levels increased further. In addition plasma PTX3 levels increased in five previously healthy controls developing TB during the follow-up (160), suggesting that measurement of this protein may help the monitoring of disease activity and efficacy of therapy. High levels of PTX3 are associated to severity of disease and mortality in patients with severe leptospirosis (163). The list of infections showing an increase in PTX3 levels includes pulmonary aspergillosis, where levels in pediatric patients can monitor the response to antifungal therapy (164). Local production of PTX3 can be measured in bronchoalveolar lavage (BAL) where it predicts pneumonia in critically ill, intubated patients (165); and in urine of patients with urinary tract infections caused by uropathogenic Escherichia coli (14), where it correlates with disease severity. High PTX3 levels observed in patients with necrotizing soft tissue infections (NSTI) at time of admission were associated with septic shock, renal replacement therapy, amputation and risk of death (166).
Several studies have analyzed single-nucleotide polymorphisms (SNPs) in the PTX3 gene. Among the 22 SNPs spanning approximately 25 kb on chromosome 3, three SNPs resulted to be associated with susceptibility to infections. Two of the three SNPs are located in the non-coding region and one, exonic, causes a single amino acid substitution in position 48 (D/A). A first study by Olesen et al., reported the association of a particular PTX3 haplotype with the risk of TB in West Africa (167). In cystic fibrosis patients, PTX3 haplotype frequencies were significantly different between subjects with P. aeruginosa colonization, as compared with non-colonized patients (168). PTX3 variants are associated with susceptibility to invasive aspergillosis in patients undergoing hematopoietic stem-cell transplantation (17), to fungal infections in solid organ transplanted patients (169), to urinary tract infections (14), to meningococcal disease (Sprong, Barbati and Bottazzi, personal communication). Analysis of the molecular consequences of the different haplotypes suggested that the one determining higher susceptibility to invasive aspergillosis is associated with lower stability of PTX3 mRNA, thus determining lower levels of the protein (17, 170). This observation pave the way to a possible prophylactic and therapeutic use of PTX3 in infectious disordes.
Concluding remarks
The humoral arm of innate immunity is generally depicted as a collection of weird molecules belonging to different molecular classes (ficolins; collectins; complement components) (3). Pentraxins and PTX3 in particular are part of this complex system of fluid phase PRMs. Fluid phase PRMs interact and synergize in microbial recognition and disposal as illustrated by PTX3 and ficolins and MBL (59, 171, 172). In general, one can view fluid phase PRMs as evolutionarily ancient, antibody-like molecules. As antibodies do, PTX3 and in general fluid phase PRMs have a regulatory function on inflammation. Given the recent discovery that PTX3 acts as an extrinsic oncosuppressor in murine and selected human tumors, the role of humoral innate immunity in tumor progression needs to be investigated. The new vistas on humoral innate immunity and PTX3 in particular may pave the way to diagnostic and therapeutic translational efforts (17, 173, 174).
Acknowledgements
The financial support of Ministero della Salute (RF-2011-02348358), Cluster Alisei (MEDINTECH CTN01_00177_962865), the European Commission (FP7-HEALTH-2011-ADITEC-N°280873), the European Research Council (ERC – N° 669415 to AM), and the Italian Association for Cancer Research (AIRC, IG and 5x1000) is gratefully acknowledged. AI is recipient of a Young Investigator Grant from Ministero della Salute (GR-2011-02349539). SM received financial support from the Academy of Finland, The Sigrid Jusélius Foundation, The Stockmann Foundation, Signe and Ane Gyllenberg Foundation and Helsinki University Hospital Grants (EVO), Finland.
References
- 1.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 2.Garlanda C, Riva F, Bonavita E, Gentile S, Mantovani A. Decoys and Regulatory “Receptors” of the IL-1/Toll-Like Receptor Superfamily. Front Immunol. 2013;4:180. doi: 10.3389/fimmu.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottazzi B, Doni A, Garlanda C, Mantovani A. An Integrated View of Humoral Innate Immunity: Pentraxins as a Paradigm. Annu Rev Immunol. 2010;28:157–183. doi: 10.1146/annurev-immunol-030409-101305. [DOI] [PubMed] [Google Scholar]
- 4.Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. 2005;23:337–366. doi: 10.1146/annurev.immunol.23.021704.115756. [DOI] [PubMed] [Google Scholar]
- 5.Doni A, et al. An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J Exp Med. 2015;212:905–925. doi: 10.1084/jem.20141268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaillon S, et al. The humoral pattern recognition receptor PTX3 is stored in neutrophil granules and localizes in extracellular traps. J Exp Med. 2007;204:793–804. doi: 10.1084/jem.20061301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casas JP, Shah T, Hingorani AD, Danesh J, Pepys MB. C-reactive protein and coronary heart disease: a critical review. J Intern Med. 2008;264:295–314. doi: 10.1111/j.1365-2796.2008.02015.x. [DOI] [PubMed] [Google Scholar]
- 8.Wensley F, et al. Association between C reactive protein and coronary heart disease: mendelian randomisation analysis based on individual participant data. BMJ. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norata GD, Garlanda C, Catapano AL. The long pentraxin PTX3: a modulator of the immunoinflammatory response in atherosclerosis and cardiovascular diseases. Trends Cardiovasc Med. 2010;20:35–40. doi: 10.1016/j.tcm.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Hirschfield GM, et al. Transgenic human C-reactive protein is not proatherogenic in apolipoprotein E-deficient mice. Proc Natl Acad Sci U S A. 2005;102:8309–8314. doi: 10.1073/pnas.0503202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepys MB, et al. Targeting C-reactive protein for the treatment of cardiovascular disease. Nature. 2006;440:1217–1221. doi: 10.1038/nature04672. [DOI] [PubMed] [Google Scholar]
- 12.Bonavita E, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160:700–714. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Bottazzi B, et al. Recognition of Neisseria meningitidis by the long pentraxin PTX3 and its role as an endogenous adjuvant. PloS one. 2015;10:e0120807. doi: 10.1371/journal.pone.0120807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaillon S, et al. The humoral pattern recognition molecule PTX3 is a key component of innate immunity against urinary tract infection. Immunity. 2014;40:621–632. doi: 10.1016/j.immuni.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Garlanda C, Jaillon S, Doni A, Bottazzi B, Mantovani A. PTX3, a humoral pattern recognition molecule at the interface between microbe and matrix recognition. Curr Opin Immunol. 2016;38:39–44. doi: 10.1016/j.coi.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bottazzi B, et al. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J Hepatol. 2016;64:1416–1427. doi: 10.1016/j.jhep.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunha C, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med. 2014;370:421–432. doi: 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- 18.Soares AC, et al. Dual function of the long pentraxin PTX3 in resistance against pulmonary infection with Klebsiella pneumoniae in transgenic mice. Microbes Infect. 2006;8:1321–1329. doi: 10.1016/j.micinf.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Souza DG, et al. The long pentraxin PTX3 is crucial for tissue inflammation after intestinal ischemia and reperfusion in mice. Am J Pathol. 2009;174:1309–1318. doi: 10.2353/ajpath.2009.080240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inforzato A, et al. PTX3 as a paradigm for the interaction of pentraxins with the Complement system. Semin Immunol. 2013 doi: 10.1016/j.smim.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Foo SS, et al. Role of pentraxin 3 in shaping arthritogenic alphaviral disease: from enhanced viral replication to immunomodulation. PLoS pathogens. 2015;11:e1004649. doi: 10.1371/journal.ppat.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronca R, et al. Long-Pentraxin 3 Derivative as a Small-Molecule FGF Trap for Cancer Therapy. Cancer Cell. 2015;28:225–239. doi: 10.1016/j.ccell.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Presta M, Camozzi M, Salvatori G, Rusnati M. Role of the soluble pattern recognition receptor PTX3 in vascular biology. J Cell Mol Med. 2007;11:723–738. doi: 10.1111/j.1582-4934.2007.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrizzo A, et al. Pentraxin 3 Induces Vascular Endothelial Dysfunction Through a P-selectin/Matrix Metalloproteinase-1 Pathway. Circulation. 2015;131:1495–1505. doi: 10.1161/CIRCULATIONAHA.114.014822. [DOI] [PubMed] [Google Scholar]
- 25.Bottazzi B, et al. Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J Biol Chem. 1997;272:32817–32823. doi: 10.1074/jbc.272.52.32817. [DOI] [PubMed] [Google Scholar]
- 26.Daigo K, et al. Protective effect of the long pentraxin PTX3 against histone-mediated endothelial cell cytotoxicity in sepsis. Sci Signal. 2014;7:ra88. doi: 10.1126/scisignal.2005522. [DOI] [PubMed] [Google Scholar]
- 27.Goodman AR, Cardozo T, Abagyan R, Altmeyer A, Wisniewski HG, Vilcek J. Long pentraxins: an emerging group of proteins with diverse functions. Cytokine Growth Factor Rev. 1996;7:191–202. doi: 10.1016/1359-6101(96)00019-6. [DOI] [PubMed] [Google Scholar]
- 28.Inforzato A, et al. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry (Mosc) 2006;45:11540–11551. doi: 10.1021/bi0607453. [DOI] [PubMed] [Google Scholar]
- 29.Introna M, et al. Cloning of mouse ptx3, a new member of the pentraxin gene family expressed at extrahepatic sites. Blood. 1996;87:1862–1872. [PubMed] [Google Scholar]
- 30.Inforzato A, et al. The angiogenic inhibitor long pentraxin PTX3 forms an asymmetric octamer with two binding sites for FGF2. J Biol Chem. 2010;285:17681–17692. doi: 10.1074/jbc.M109.085639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inforzato A, et al. Structural characterization of PTX3 disulfide bond network and its multimeric status in cumulus matrix organization. J Biol Chem. 2008;283:10147–10161. doi: 10.1074/jbc.M708535200. [DOI] [PubMed] [Google Scholar]
- 32.Bozza S, et al. PTX3 binds MD-2 and promotes TRIF-dependent immune protection in aspergillosis. J Immunol. 2014;193:2340–2348. doi: 10.4049/jimmunol.1400814. [DOI] [PubMed] [Google Scholar]
- 33.Leali D, et al. Long pentraxin 3/tumor necrosis factor-stimulated gene-6 interaction: a biological rheostat for fibroblast growth factor 2-mediated angiogenesis. Arterioscler Thromb Vasc Biol. 2012;32:696–703. doi: 10.1161/ATVBAHA.111.243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moalli F, et al. Role of complement and Fc{gamma} receptors in the protective activity of the long pentraxin PTX3 against Aspergillus fumigatus. Blood. 2010;116:5170–5180. doi: 10.1182/blood-2009-12-258376. [DOI] [PubMed] [Google Scholar]
- 35.Scarchilli L, et al. PTX3 interacts with inter-alpha-trypsin inhibitor: implications for hyaluronan organization and cumulus oophorus expansion. J Biol Chem. 2007;282:30161–30170. doi: 10.1074/jbc.M703738200. [DOI] [PubMed] [Google Scholar]
- 36.Deban L, et al. Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat Immunol. 2010;11:328–334. doi: 10.1038/ni.1854. [DOI] [PubMed] [Google Scholar]
- 37.Nauta AJ, Daha MR, van Kooten C, Roos A. Recognition and clearance of apoptotic cells: a role for complement and pentraxins. Trends Immunol. 2003;24:148–154. doi: 10.1016/s1471-4906(03)00030-9. [DOI] [PubMed] [Google Scholar]
- 38.Deban L, et al. Binding of the long pentraxin PTX3 to Factor H: Interacting domains and function in the regulation of complement activation. J Immunology. 2008;181:8433–8440. doi: 10.4049/jimmunol.181.12.8433. [DOI] [PubMed] [Google Scholar]
- 39.Reading PC, et al. Antiviral Activity of the Long Chain Pentraxin PTX3 against Influenza Viruses. J Immunol. 2008;180:3391–3398. doi: 10.4049/jimmunol.180.5.3391. [DOI] [PubMed] [Google Scholar]
- 40.Inforzato A, Reading PC, Barbati E, Bottazzi B, Garlanda C, Mantovani A. The “sweet” side of a long pentraxin: how glycosylation affects PTX3 functions in innate immunity and inflammation. Front Immunol. 2013;3 doi: 10.3389/fimmu.2012.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrive AK, Metcalfe AM, Cartwright JR, Greenhough TJ. C-reactive protein and SAP-like pentraxin are both present in Limulus polyphemus haemolymph: crystal structure of Limulus SAP. J Mol Biol. 1999;290:997–1008. doi: 10.1006/jmbi.1999.2956. [DOI] [PubMed] [Google Scholar]
- 42.Ievoli E, et al. Implication of the oligomeric state of the N-terminal PTX3 domain in cumulus matrix assembly. Matrix Biol. 2011;30:330–337. doi: 10.1016/j.matbio.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baranova NS, et al. Incorporation of pentraxin 3 into hyaluronan matrices is tightly regulated and promotes matrix cross-linking. J Biol Chem. 2014;289:30481–30498. doi: 10.1074/jbc.M114.568154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garlanda C, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 2002;420:182–186. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- 45.Lu J, Marnell LL, Marjon KD, Mold C, Du Clos TW, Sun PD. Structural recognition and functional activation of FcgammaR by innate pentraxins. Nature. 2008;456:989–992. doi: 10.1038/nature07468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diniz SN, et al. PTX3 function as an opsonin for the dectin-1-dependent internalization of zymosan by macrophages. J Leukoc Biol. 2004;75:649–656. doi: 10.1189/jlb.0803371. [DOI] [PubMed] [Google Scholar]
- 47.Moalli F, et al. The therapeutic potential of the humoral pattern recognition molecule PTX3 in chronic lung infection caused by Pseudomonas aeruginosa. J Immunol. 2011;186:5425–5434. doi: 10.4049/jimmunol.1002035. [DOI] [PubMed] [Google Scholar]
- 48.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiel S, Gadjeva M. Humoral pattern recognition molecules: mannan-binding lectin and ficolins. Adv Exp Med Biol. 2009;653:58–73. doi: 10.1007/978-1-4419-0901-5_5. [DOI] [PubMed] [Google Scholar]
- 50.Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999;7:169–177. doi: 10.1016/S0969-2126(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 51.Oliveira EB, Gotschlich C, Liu TY. Primary structure of human C-reactive protein. J Biol Chem. 1979;254:489–502. [PubMed] [Google Scholar]
- 52.Amarilyo G, et al. iC3b-opsonized apoptotic cells mediate a distinct anti-inflammatory response and transcriptional NF-kappaB-dependent blockade. Eur J Immunol. 2010;40:699–709. doi: 10.1002/eji.200838951. [DOI] [PubMed] [Google Scholar]
- 53.Skattum L, van Deuren M, van der Poll T, Truedsson L. Complement deficiency states and associated infections. Mol Immunol. 2011;48:1643–1655. doi: 10.1016/j.molimm.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Jiang HX, Siegel JN, Gewurz H. Binding and complement activation by C-reactive protein via the collagen-like region of C1q and inhibition of these reactions by monoclonal antibodies to C-reactive protein and C1q. J Immunol. 1991;146:2324–2330. [PubMed] [Google Scholar]
- 55.Bristow CL, Boackle RJ. Evidence for the binding of human serum amyloid P component to Clq and Fab gamma. Mol Immunol. 1986;23:1045–1052. doi: 10.1016/0161-5890(86)90003-9. [DOI] [PubMed] [Google Scholar]
- 56.Nauta AJ, et al. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur J Immunol. 2003;33:465–473. doi: 10.1002/immu.200310022. [DOI] [PubMed] [Google Scholar]
- 57.Holmskov U, Malhotra R, Sim RB, Jensenius JC. Collectins: collagenous C-type lectins of the innate immune defense system. Immunol Today. 1994;15:67–74. doi: 10.1016/0167-5699(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 58.Doni A, Garlanda C, Bottazzi B, Meri S, Garred P, Mantovani A. Interactions of the humoral pattern recognition molecule PTX3 with the complement system. Immunobiology. 2012;217:1122–1128. doi: 10.1016/j.imbio.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Ma YJ, et al. Heterocomplexes of mannose-binding lectin and the pentraxins PTX3 or serum amyloid P component trigger cross-activation of the complement system. J Biol Chem. 2011;286:3405–3417. doi: 10.1074/jbc.M110.190637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu J, Marjon KD, Mold C, Du Clos TW, Sun PD. Pentraxins and Fc receptors. Immunol Rev. 2012;250:230–238. doi: 10.1111/j.1600-065X.2012.01162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maury CP, Helve T, Sjoblom C. Serum beta 2-microglobulin, sialic acid, and C-reactive protein in systemic lupus erythematosus. Rheumatol Int. 1982;2:145–149. doi: 10.1007/BF00286135. [DOI] [PubMed] [Google Scholar]
- 62.Walport MJ. Complement and systemic lupus erythematosus. Arthritis Res. 2002;4(Suppl 3):S279–293. doi: 10.1186/ar586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pepys MB, Baltz M, Gomer K, Davies AJ, Doenhoff M. Serum amyloid P-component is an acute-phase reactant in the mouse. Nature. 1979;278:259–261. doi: 10.1038/278259a0. [DOI] [PubMed] [Google Scholar]
- 64.Bickerstaff MC, et al. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. 1999;5:694–697. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- 65.Lech M, et al. Lack of the long pentraxin PTX3 promotes autoimmune lung disease but not glomerulonephritis in murine systemic lupus erythematosus. PloS one. 2011;6:e20118. doi: 10.1371/journal.pone.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaboriaud C, Ling WL, Thielens NM, Bally I, Rossi V. Deciphering the fine details of c1 assembly and activation mechanisms: “mission impossible”? Front Immunol. 2014;5:565. doi: 10.3389/fimmu.2014.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sjoberg AP, Trouw LA, McGrath FD, Hack CE, Blom AM. Regulation of complement activation by C-reactive protein: targeting of the inhibitory activity of C4b-binding protein. J Immunol. 2006;176:7612–7620. doi: 10.4049/jimmunol.176.12.7612. [DOI] [PubMed] [Google Scholar]
- 68.Braunschweig A, Jozsi M. Human pentraxin 3 binds to the complement regulator c4b-binding protein. PloS one. 2011;6:e23991. doi: 10.1371/journal.pone.0023991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwalbe RA, Dahlback B, Nelsestuen GL. Independent association of serum amyloid P component, protein S, and complement C4b with complement C4b-binding protein and subsequent association of the complex with membranes. J Biol Chem. 1990;265:21749–21757. [PubMed] [Google Scholar]
- 70.Bykov I, Junnikkala S, Pekna M, Lindros KO, Meri S. Effect of chronic ethanol consumption on the expression of complement components and acute-phase proteins in liver. Clin Immunol. 2007;124:213–220. doi: 10.1016/j.clim.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Pangburn MK, Muller-Eberhard HJ. The alternative pathway of complement. Springer Semin Immunopathol. 1984;7:163–192. doi: 10.1007/BF01893019. [DOI] [PubMed] [Google Scholar]
- 72.Morgan BP, Meri S. Membrane proteins that protect against complement lysis. Springer Semin Immunopathol. 1994;15:369–396. doi: 10.1007/BF01837366. [DOI] [PubMed] [Google Scholar]
- 73.Meri S. Self-nonself discrimination by the complement system. FEBS Lett. 2016 Jul 9; doi: 10.1002/1873-3468.12284. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 74.Meri S, Pangburn MK. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci U S A. 1990;87:3982–3986. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kavanagh D, Goodship T. Genetics and complement in atypical HUS. Pediatr Nephrol. 2010;25:2431–2442. doi: 10.1007/s00467-010-1555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hyvarinen S, Meri S, Jokiranta TS. Disturbed sialic acid recognition on endothelial cells and platelets in complement attack causes atypical hemolytic uremic syndrome. Blood. 2016;127:2701–2710. doi: 10.1182/blood-2015-11-680009. [DOI] [PubMed] [Google Scholar]
- 77.Meri S. Complement activation in diseases presenting with thrombotic microangiopathy. Eur J Intern Med. 2013;24:496–502. doi: 10.1016/j.ejim.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 78.Jarva H, Jokiranta TS, Hellwage J, Zipfel PF, Meri S. Regulation of complement activation by C-reactive protein: targeting the complement inhibitory activity of factor H by an interaction with short consensus repeat domains 7 and 8-11. J Immunol. 1999;163:3957–3962. [PubMed] [Google Scholar]
- 79.Laine M, et al. Y402H polymorphism of complement factor H affects binding affinity to C-reactive protein. J Immunol. 2007;178:3831–3836. doi: 10.4049/jimmunol.178.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okemefuna AI, Nan R, Miller A, Gor J, Perkins SJ. Complement factor H binds at two independent sites to C-reactive protein in acute phase concentrations. J Biol Chem. 2010;285:1053–1065. doi: 10.1074/jbc.M109.044529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hellwage J, et al. Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J Immunol. 2002;169:6935–6944. doi: 10.4049/jimmunol.169.12.6935. [DOI] [PubMed] [Google Scholar]
- 82.Jokiranta TS, Hellwage J, Koistinen V, Zipfel PF, Meri S. Each of the three binding sites on complement factor H interacts with a distinct site on C3b. J Biol Chem. 2000;275:27657–27662. doi: 10.1074/jbc.M002903200. [DOI] [PubMed] [Google Scholar]
- 83.Kajander T, et al. Dual interaction of factor H with C3d and glycosaminoglycans in host-nonhost discrimination by complement. Proc Natl Acad Sci U S A. 2011;108:2897–2902. doi: 10.1073/pnas.1017087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morgan HP, et al. Structural basis for engagement by complement factor H of C3b on a self surface. Nat Struct Mol Biol. 2011;18:463–470. doi: 10.1038/nsmb.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sjoberg AP, et al. The factor H variant associated with age-related macular degeneration (His-384) and the non-disease-associated form bind differentially to C-reactive protein, fibromodulin, DNA, and necrotic cells. J Biol Chem. 2007;282:10894–10900. doi: 10.1074/jbc.M610256200. [DOI] [PubMed] [Google Scholar]
- 86.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seitsonen S, et al. Analysis of variants in the complement factor H, the elongation of very long chain fatty acids-like 4 and the hemicentin 1 genes of age-related macular degeneration in the Finnish population. Mol Vis. 2006;12:796–801. [PubMed] [Google Scholar]
- 89.Clark SJ, et al. Tissue-specific host recognition by complement factor H is mediated by differential activities of its glycosaminoglycan-binding regions. J Immunol. 2013;190:2049–2057. doi: 10.4049/jimmunol.1201751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macleod CM, Avery OT. The Occurrence during Acute Infections of a Protein Not Normally Present in the Blood : Iii. Immunological Properties of the C-Reactive Protein and Its Differentiation from Normal Blood Proteins. J Exp Med. 1941;73:191–200. doi: 10.1084/jem.73.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meri T, et al. Microbes bind complement inhibitor factor H via a common site. PLoS Pathog. 2013;9:e1003308. doi: 10.1371/journal.ppat.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jarva H, Janulczyk R, Hellwage J, Zipfel PF, Bjorck L, Meri S. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded Hic protein that binds to short consensus repeats 8-11 of factor H. J Immunol. 2002;168:1886–1894. doi: 10.4049/jimmunol.168.4.1886. [DOI] [PubMed] [Google Scholar]
- 93.Mold C, Kingzette M, Gewurz H. C-reactive protein inhibits pneumococcal activation of the alternative pathway by increasing the interaction between factor H and C3b. J Immunol. 1984;133:882–885. [PubMed] [Google Scholar]
- 94.Huttunen R, et al. High plasma level of long pentraxin 3 (PTX3) is associated with fatal disease in bacteremic patients: a prospective cohort study. PloS one. 2011;6:e17653. doi: 10.1371/journal.pone.0017653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Daigo K, et al. The proteomic profile of circulating pentraxin 3 (PTX3) complex in sepsis demonstrates the interaction with azurocidin 1 and other components of neutrophil extracellular traps. Mol Cell Proteomics. 2012;11:M111 015073. doi: 10.1074/mcp.M111.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 97.Kaplan MJ, Radic M. Neutrophil extracellular traps: double-edged swords of innate immunity. J Immunol. 2012;189:2689–2695. doi: 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Urban CF, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Savchenko AS, et al. Long pentraxin 3 (PTX3) expression and release by neutrophils in vitro and in ulcerative colitis. Pathol Int. 2011;61:290–297. doi: 10.1111/j.1440-1827.2011.02651.x. [DOI] [PubMed] [Google Scholar]
- 100.Razvina O, et al. Differential expression of pentraxin 3 in neutrophils. Exp Mol Pathol. 2015;98:33–40. doi: 10.1016/j.yexmp.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 101.Linder A, Christensson B, Herwald H, Bjorck L, Akesson P. Heparin-binding protein: an early marker of circulatory failure in sepsis. Clin Infect Dis. 2009;49:1044–1050. doi: 10.1086/605563. [DOI] [PubMed] [Google Scholar]
- 102.Soehnlein O, Lindbom L. Neutrophil-derived azurocidin alarms the immune system. J Leukoc Biol. 2009;85:344–351. doi: 10.1189/jlb.0808495. [DOI] [PubMed] [Google Scholar]
- 103.Klebanoff SJ, Kettle AJ, Rosen H, Winterbourn CC, Nauseef WM. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J Leukoc Biol. 2013;93:185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu J, et al. Extracellular histones are major mediators of death in sepsis. Nat Med. 2009;15:1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fuchs TA, Bhandari AA, Wagner DD. Histones induce rapid and profound thrombocytopenia in mice. Blood. 2011;118:3708–3714. doi: 10.1182/blood-2011-01-332676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jeannin P, et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity. 2005;22:551–560. doi: 10.1016/j.immuni.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 107.Bozza S, et al. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood. 2006;108:3387–3396. doi: 10.1182/blood-2006-03-009266. [DOI] [PubMed] [Google Scholar]
- 108.Job ER, et al. A single amino acid substitution in the hemagglutinin of H3N2 subtype influenza A viruses is associated with resistance to the long pentraxin PTX3 and enhanced virulence in mice. J Immunol. 2014;192:271–281. doi: 10.4049/jimmunol.1301814. [DOI] [PubMed] [Google Scholar]
- 109.Job ER, et al. Pandemic H1N1 influenza A viruses are resistant to the antiviral activities of innate immune proteins of the collectin and pentraxin superfamilies. J Immunol. 2010;185:4284–4291. doi: 10.4049/jimmunol.1001613. [DOI] [PubMed] [Google Scholar]
- 110.Foo SS, Reading PC, Jaillon S, Mantovani A, Mahalingam S. Pentraxins and Collectins: Friend or Foe during Pathogen Invasion? Trends Microbiol. 2015;23:799–811. doi: 10.1016/j.tim.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gunn BM, et al. Mannose binding lectin is required for alphavirus-induced arthritis/myositis. PLoS Pathog. 2012;8:e1002586. doi: 10.1371/journal.ppat.1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cotena A, et al. Complement Dependent Amplification of the Innate Response to a Cognate Microbial Ligand by the Long Pentraxin PTX3. J Immunol. 2007;179:6311–6317. doi: 10.4049/jimmunol.179.9.6311. [DOI] [PubMed] [Google Scholar]
- 113.Lech M, et al. Endogenous and exogenous pentraxin-3 limits postischemic acute and chronic kidney injury. Kidney Int. 2013;83:647–661. doi: 10.1038/ki.2012.463. [DOI] [PubMed] [Google Scholar]
- 114.Schneider LA, Korber A, Grabbe S, Dissemond J. Influence of pH on wound-healing: a new perspective for wound-therapy? Arch Dermatol Res. 2007;298:413–420. doi: 10.1007/s00403-006-0713-x. [DOI] [PubMed] [Google Scholar]
- 115.Xiao Y, Yang N, Zhang Q, Wang Y, Yang S, Liu Z. Pentraxin 3 inhibits acute renal injury-induced interstitial fibrosis through suppression of IL-6/Stat3 pathway. Inflammation. 2014;37:1895–1901. doi: 10.1007/s10753-014-9921-2. [DOI] [PubMed] [Google Scholar]
- 116.Salio M, et al. Cardioprotective function of the long pentraxin PTX3 in acute myocardial infarction. Circulation. 2008;117:1055–1064. doi: 10.1161/CIRCULATIONAHA.107.749234. [DOI] [PubMed] [Google Scholar]
- 117.Norata GD, et al. Deficiency of the long pentraxin PTX3 promotes vascular inflammation and atherosclerosis. Circulation. 2009;120:699–708. doi: 10.1161/CIRCULATIONAHA.108.806547. [DOI] [PubMed] [Google Scholar]
- 118.Paeschke A, et al. The immunoproteasome controls the availability of the cardioprotective pattern recognition molecule Pentraxin3. Eur J Immunol. 2016;46:619–633. doi: 10.1002/eji.201545892. [DOI] [PubMed] [Google Scholar]
- 119.Rovere-Querini P, et al. Plasma and tissue expression of the long pentraxin 3 during normal pregnancy and preeclampsia. Obstet Gynecol. 2006;108:148–155. doi: 10.1097/01.AOG.0000224607.46622.bc. [DOI] [PubMed] [Google Scholar]
- 120.Ravizza T, et al. Dynamic induction of the long pentraxin PTX3 in the CNS after limbic seizures: evidence for a protective role in seizure-induced neurodegeneration. Neuroscience. 2001;105:43–53. doi: 10.1016/s0306-4522(01)00177-4. [DOI] [PubMed] [Google Scholar]
- 121.Rodriguez-Grande B, et al. The acute-phase protein PTX3 is an essential mediator of glial scar formation and resolution of brain edema after ischemic injury. J Cereb Blood Flow Metab. 2014;34:480–488. doi: 10.1038/jcbfm.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Real JM, et al. Pentraxin 3 accelerates lung injury in high tidal volume ventilation in mice. Mol Immunol. 2012;51:82–90. doi: 10.1016/j.molimm.2012.02.113. [DOI] [PubMed] [Google Scholar]
- 123.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 124.Choi B, et al. Elevated Pentraxin 3 in bone metastatic breast cancer is correlated with osteolytic function. Oncotarget. 2014;5:481–492. doi: 10.18632/oncotarget.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Diamandis EP, Goodglick L, Planque C, Thornquist MD. Pentraxin-3 Is a Novel Biomarker of Lung Carcinoma. Clin Cancer Res. 2011;17:2395–2399. doi: 10.1158/1078-0432.CCR-10-3024. [DOI] [PubMed] [Google Scholar]
- 126.Germano G, et al. Antitumor and anti-inflammatory effects of trabectedin on human myxoid liposarcoma cells. Cancer Res. 2010;70:2235–2244. doi: 10.1158/0008-5472.CAN-09-2335. [DOI] [PubMed] [Google Scholar]
- 127.Infante M, et al. Prognostic and diagnostic potential of local and circulating levels of pentraxin 3 in lung cancer patients. Int J Cancer. 2016;138:983–991. doi: 10.1002/ijc.29822. [DOI] [PubMed] [Google Scholar]
- 128.Stallone G, et al. Pentraxin 3: a novel biomarker for predicting progression from prostatic inflammation to prostate cancer. Cancer Res. 2014;74:4230–4238. doi: 10.1158/0008-5472.CAN-14-0369. [DOI] [PubMed] [Google Scholar]
- 129.Carmo RF, Aroucha D, Vasconcelos LR, Pereira LM, Moura P, Cavalcanti MS. Genetic variation in PTX3 and plasma levels associated with hepatocellular carcinoma in patients with HCV. J Viral Hepat. 2016;23:116–122. doi: 10.1111/jvh.12472. [DOI] [PubMed] [Google Scholar]
- 130.Tothill RW, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 131.Wang JX, He YL, Zhu ST, Yang S, Zhang ST. Aberrant methylation of the 3q25 tumor suppressor gene PTX3 in human esophageal squamous cell carcinoma. World J Gastroenterol. 2011;17:4225–4230. doi: 10.3748/wjg.v17.i37.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 134.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 135.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol. 2013;35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chang WC, et al. PTX3 gene activation in EGF-induced head and neck cancer cell metastasis. Oncotarget. 2015;6:7741–7757. doi: 10.18632/oncotarget.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Choi B, et al. Pentraxin-3 Silencing Suppresses Gastric Cancer-related Inflammation by Inhibiting Chemotactic Migration of Macrophages. Anticancer Res. 2015;35:2663–2668. [PubMed] [Google Scholar]
- 138.Kondo S, et al. Clinical impact of pentraxin family expression on prognosis of pancreatic carcinoma. Br J Cancer. 2013;109:739–746. doi: 10.1038/bjc.2013.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tung J-N, et al. Inhibition of pentraxin 3 in glioma cells impairs proliferation and invasion in vitro and in vivo. J Neurooncol. 2016:1–9. doi: 10.1007/s11060-016-2168-z. [DOI] [PubMed] [Google Scholar]
- 140.Peri G, et al. PTX3, A prototypical long pentraxin, is an early indicator of acute myocardial infarction in humans. Circulation. 2000;102:636–641. doi: 10.1161/01.cir.102.6.636. [DOI] [PubMed] [Google Scholar]
- 141.Latini R, et al. Prognostic significance of the long pentraxin PTX3 in acute myocardial infarction. Circulation. 2004;110:2349–2354. doi: 10.1161/01.CIR.0000145167.30987.2E. [DOI] [PubMed] [Google Scholar]
- 142.Suzuki S, et al. Pentraxin 3, a new marker for vascular inflammation, predicts adverse clinical outcomes in patients with heart failure. Am Heart J. 2008;155:75–81. doi: 10.1016/j.ahj.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 143.Matsubara J, et al. Pentraxin 3 is a new inflammatory marker correlated with left ventricular diastolic dysfunction and heart failure with normal ejection fraction. J Am Coll Cardiol. 2011;57:861–869. doi: 10.1016/j.jacc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 144.Latini R, et al. Pentraxin-3 in chronic heart failure: the CORONA and GISSI-HF trials. Eur J Heart Fail. 2012;14:992–999. doi: 10.1093/eurjhf/hfs092. [DOI] [PubMed] [Google Scholar]
- 145.Matsubara J, et al. Incremental prognostic significance of the elevated levels of pentraxin 3 in patients with heart failure with normal left ventricular ejection fraction. J Am Heart Assoc. 2014;3:e000928. doi: 10.1161/JAHA.114.000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Suliman ME, et al. Novel links between the long pentraxin 3, endothelial dysfunction, and albuminuria in early and advanced chronic kidney disease. Clin J Am Soc Nephrol. 2008;3:976–985. doi: 10.2215/CJN.03960907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suliman ME, et al. The long pentraxin PTX-3 in prevalent hemodialysis patients: associations with comorbidities and mortality. QJM. 2008;101:397–405. doi: 10.1093/qjmed/hcn019. [DOI] [PubMed] [Google Scholar]
- 148.Tong MC, C J, Qureshi AR, Anderstam B, Heimburger O, Barany P, Axelsson J, Alverstrand A, Stenvinkel P, Lindholm B, Suliman M. Plasma Pentraxin 3 in chronic kidney disease patients: association with renal function, protein-energy wasting, cardiovascular disease and mortality. Clin J Am Soc Nephrol. 2007;2:889–897. doi: 10.2215/CJN.00870207. [DOI] [PubMed] [Google Scholar]
- 149.Sjoberg B, et al. Association between levels of pentraxin 3 and incidence of chronic kidney disease in the elderly. J Intern Med. 2016;279:173–179. doi: 10.1111/joim.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sjoberg B, Qureshi AR, Anderstam B, Alvestrand A, Barany P. Pentraxin 3, a sensitive early marker of hemodialysis-induced inflammation. Blood Purif. 2012;34:290–297. doi: 10.1159/000342630. [DOI] [PubMed] [Google Scholar]
- 151.Sjoberg B, Snaedal S, Stenvinkel P, Qureshi AR, Heimburger O, Barany P. Three-month variation of plasma pentraxin 3 compared with C-reactive protein, albumin and homocysteine levels in haemodialysis patients. Clinical kidney journal. 2014;7:373–379. doi: 10.1093/ckj/sfu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jenny NS, Arnold AM, Kuller LH, Tracy RP, Psaty BM. Associations of pentraxin 3 with cardiovascular disease and all-cause death: the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 2009;29:594–599. doi: 10.1161/ATVBAHA.108.178947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jenny NS, Blumenthal RS, Kronmal RA, Rotter JI, Siscovick DS, Psaty BM. Associations of pentraxin 3 with cardiovascular disease: the multi-ethnic study of atherosclerosis. J Thromb Haemost. 2014;12:999–1005. doi: 10.1111/jth.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dubin R, Li Y, Ix JH, Shlipak MG, Whooley M, Peralta CA. Associations of pentraxin-3 with cardiovascular events, incident heart failure, and mortality among persons with coronary heart disease: data from the Heart and Soul Study. Am Heart J. 2012;163:274–279. doi: 10.1016/j.ahj.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Muller B, et al. Circulating levels of the long pentraxin PTX3 correlate with severity of infection in critically ill patients. Crit Care Med. 2001;29:1404–1407. doi: 10.1097/00003246-200107000-00017. [DOI] [PubMed] [Google Scholar]
- 156.Bastrup-Birk S, Skjoedt MO, Munthe-Fog L, Strom JJ, Ma YJ, Garred P. Pentraxin-3 serum levels are associated with disease severity and mortality in patients with systemic inflammatory response syndrome. PloS one. 2013;8:e73119. doi: 10.1371/journal.pone.0073119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Uusitalo-Seppala R, et al. Pentraxin 3 (PTX3) is associated with severe sepsis and fatal disease in emergency room patients with suspected infection: a prospective cohort study. PloS one. 2013;8:e53661. doi: 10.1371/journal.pone.0053661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mauri T, et al. Persisting high levels of plasma pentraxin 3 over the first days after severe sepsis and septic shock onset are associated with mortality. Intens Care Med. 2010;36:621–629. doi: 10.1007/s00134-010-1752-5. [DOI] [PubMed] [Google Scholar]
- 159.Ristagno G, et al. Elevations of inflammatory markers PTX3 and sST2 after resuscitation from cardiac arrest are associated with multiple organ dysfunction syndrome and early death. Clin Chem Lab Med. 2015;53:1847–1857. doi: 10.1515/cclm-2014-1271. [DOI] [PubMed] [Google Scholar]
- 160.Azzurri A, et al. IFN-gamma-inducible protein 10 and pentraxin 3 plasma levels are tools for monitoring inflammation and disease activity in Mycobacterium tuberculosis infection. Microbes Infect. 2005;7:1–8. doi: 10.1016/j.micinf.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 161.Mairuhu AT, et al. Elevated plasma levels of the long pentraxin, pentraxin 3, in severe dengue virus infections. J Med Virol. 2005;76:547–552. doi: 10.1002/jmv.20397. [DOI] [PubMed] [Google Scholar]
- 162.Sprong T, et al. Ptx3 and C-Reactive Protein in Severe Meningococcal Disease. Shock. 2009;31:28–32. doi: 10.1097/SHK.0b013e31817fd543. [DOI] [PubMed] [Google Scholar]
- 163.Wagenaar JF, et al. Long pentraxin PTX3 is associated with mortality and disease severity in severe Leptospirosis. J Infect. 2009;58:425–432. doi: 10.1016/j.jinf.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 164.Biagi E, et al. PTX3 as a potential novel tool for the diagnosis and monitoring of pulmonary fungal infections in immuno-compromised pediatric patients. J Pediatr Hematol Oncol. 2008;30:881–885. doi: 10.1097/MPH.0b013e318180bc1d. [DOI] [PubMed] [Google Scholar]
- 165.Mauri T, et al. Alveolar pentraxin 3 as an early marker of microbiologically confirmed pneumonia: a threshold-finding prospective observational study. Crit Care. 2014;18:562. doi: 10.1186/s13054-014-0562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hansen MB, Rasmussen LS, Garred P, Bidstrup D, Madsen MB, Hyldegaard O. Pentraxin-3 as a marker of disease severity and risk of death in patients with necrotizing soft tissue infections: a nationwide, prospective, observational study. Crit Care. 2016;20:1–11. doi: 10.1186/s13054-016-1210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Olesen R, et al. DC-SIGN (CD209), Pentraxin 3 and Vitamin D Receptor gene variants associate with pulmonary tubercolosis risk in West-Africans. Genes Immun. 2007;8:456–467. doi: 10.1038/sj.gene.6364410. [DOI] [PubMed] [Google Scholar]
- 168.Chiarini M, et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. 2010;11:665–670. doi: 10.1038/gene.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wojtowicz A, et al. PTX3 Polymorphisms and Invasive Mold Infections After Solid Organ Transplant. Clin Infect Dis. 2015;61:619–622. doi: 10.1093/cid/civ386. [DOI] [PubMed] [Google Scholar]
- 170.Barbati E, et al. Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and PTX3 plasma levels. PloS one. 2012;7:e53030. doi: 10.1371/journal.pone.0053030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ma YJ, et al. Synergy between ficolin-2 and pentraxin 3 boosts innate immune recognition and complement deposition. J Biol Chem. 2009;284:28263–28275. doi: 10.1074/jbc.M109.009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gout E, et al. M-ficolin interacts with the long pentraxin PTX3: a novel case of cross-talk between soluble pattern-recognition molecules. J Immunol. 2011;186:5815–5822. doi: 10.4049/jimmunol.1100180. [DOI] [PubMed] [Google Scholar]
- 173.van den Blink B, et al. Recombinant human pentraxin-2 therapy in patients with idiopathic pulmonary fibrosis: safety, pharmacokinetics and exploratory efficacy. Eur Respir J. 2016;47:889–897. doi: 10.1183/13993003.00850-2015. [DOI] [PubMed] [Google Scholar]
- 174.Dillingh MR, et al. Recombinant human serum amyloid P in healthy volunteers and patients with pulmonary fibrosis. Pulm Pharmacol Ther. 2013;26:672–676. doi: 10.1016/j.pupt.2013.01.008. [DOI] [PubMed] [Google Scholar]