Abstract
The innate immune system comprises both a cellular and a humoral arm. Neutrophils are key effector cells of the immune and inflammatory responses and have emerged as a major source of humoral pattern recognition molecules (PRMs). These molecules, which include collectins, ficolins, and pentraxins, are specialized in the discrimination of self versus non-self and modified-self and share basic multifunctional properties including recognition and opsonisation of pathogens and apoptotic cells, activation and regulation of the complement cascade and tuning of inflammation. Neutrophils act as a reservoir of ready-made soluble PRMs, such as the long pentraxin PTX3, the peptidoglycan recognition protein PGRP-S, properdin and M-ficolin, which are stored in neutrophil granules and are involved in neutrophil effector functions. In addition, other soluble PRMs, such as members of the collectin family, are not expressed in neutrophils but can modulate neutrophil-dependent immune responses. Therefore, soluble PRMs are an essential part of the innate immune response and retain antibody-like effector functions. Here, we will review the expression and general function of soluble PRMs, focusing our attention on molecules involved in neutrophil effector functions.
Keywords: neutrophil, pattern-recognition receptor, innate immunity, pentraxin, ficolin
1. Introduction
The immune system of mammalians is composed by innate and the adaptive arms. The adaptive immune system is more recent in terms of evolution and its activation requires a set of specific receptors encoded by genes undergoing rearrangement. This system provides the basis for the immunological memory. The innate immune system constitutes the first line of defence against infections and is required for a correct activation of the adaptive immune response. The innate immune system, which is composed by a cellular and a humoral arm, uses a set of germline-encoded molecules involved in the discrimination of self versus non-self and modified-self [1, 2].
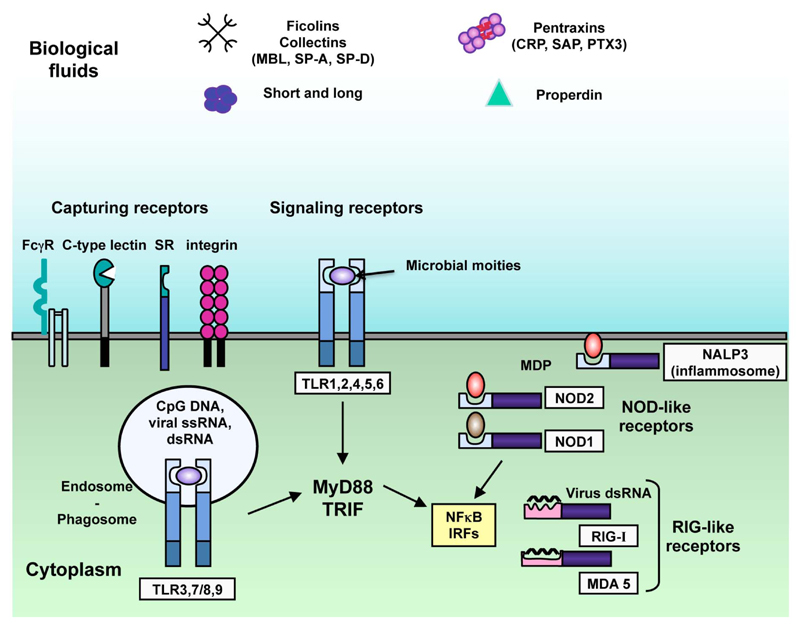
The innate immunity receptors have been called pattern recognition molecules (PRMs) since they recognize motifs expressed by microorganisms and called pathogen associated molecular patterns (PAMP). In addition, PRMs can recognize a set of motifs expressed by dying cells (i.e. apoptotic cell-associated motifs (ACAMP)) and a set of alarmins, such as endogenous molecules released by necrotic cells (e.g. HMGB1) [3]. Based on their localisation, PRMs have been divided between cell-associated PRMs and soluble PRMs (Fig. 1). Cell-associated PRMs include endocytic receptors, such as scavenger receptors, and signalling receptors, which can be both membrane-associated (e.g. Toll like receptors (TLRs)) or cytoplasmic molecules (e.g. RNA helicases, such as melanoma differentiation-associated gene 5 (MDA5) and retinoic acid-inducible gene I (RIG-I), and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs)) (Fig. 1) [3]. Fluid phase PRMs are heterogeneous in terms of structure, expression and specificity and include collectins, ficolins and pentraxins [1]. These molecules are essential in the activation, regulation and effector functions of innate and adaptive immunity and are considered functional ancestor of antibodies (Table 1) [1].
Figure 1. Cell-associated and soluble pattern recognition molecules.
Cell associated PRMs include endocytic receptors (e.g. scavenger receptors) and signalling receptors (TLRs) and can be found on the plasma membrane (e.g. TLRs, scavenger receptors, lectin receptors), in the cytoplasm (e.g. NOD-like receptors and RIG-like receptors) or in the endosomes (e.g. TLRs). TLRs recognize microbial moieties, RIG-like receptors recognize viral double-stranded RNA and NOD-like receptors recognize muramyl dipeptide (MDP), a subunit of bacterial peptidoglycan also recognized by the component of the inflammasome NALP3. Signalling receptors induce the activation of transcription factors, including NF-κB and IRFs. Soluble PRMs include collectins (MBL, SP-A, SP-D), ficolins, pentraxins (CRP, SAP, PTX3) and properdin.
Table 1.
Expression sites, ligands and activities of soluble pattern recognition molecules.
| Soluble pattern recognition molecules | Expression sites | Ligands | Activities on innate immunity and neutrophil-dependent immunity |
|---|---|---|---|
| Collectins (MBL, SP-A, SP-D) |
|
|
|
| Ficolins |
|
|
|
| Properdin |
|
|
|
| Long pentraxin PTX3 |
|
|
|
| Peptidoglycan-recognition proteins PGLYRP-1 |
|
|
|
Abbreviations: MBL, mannose-binding lectin; SP, surfactant protein; LPS, lipopolysaccharide; NETs, neutrophil-extracellular traps; GlcNAc, N-Acetylglucosamine; GalNAc, N-Acetylgalactosamine, ANCA, anti-neutrophil cytoplasmique antibody, Omp, Outer membrane protein; IαI, inter-α-trypsin inhibitor; TSG-6, TNF-α-induced protein 6; TREM, Triggering receptor expressed on myeloid cells
Neutrophils are essential innate immune cells and the life-threatening condition of people with neutropenia or with abnormalities in neutrophil functions underlines their role in immunity and defence against pathogens [4, 5]. In addition to their involvement during the acute phase of inflammation and to eliminate pathogens, neutrophils can produce cytokines, chemokines and express an important number of both cell-associated and soluble PRMs implicated in the activation and regulation of the innate and adaptive immune responses [4, 6–9]. Cell-associated PRMs in neutrophils include receptors on the plasma membrane, in particular all TLRs with the exception of TLRs 3 and 7 and receptors of the C-type lectin family, such as Dectin-1, CLEC2 and CLEC4E, and receptors found in the cytoplasmic compartment, in particular NOD-1, RIG1, MDA5 and the DNA sensor interferon-inducible protein 16 (IFI16) [8, 10, 11]. All these receptors are involved in the activation and modulation of neutrophil effector functions (e.g. phagocytosis, expression of cytokines and chemokines, production of antimicrobial peptides and reactive oxygen species, and formation of neutrophil extracellular traps (NETs) [8, 11].
Here we will review key soluble PRMs produced by neutrophils, describing their expression, their structures and their roles in immunity, inflammation and neutrophil-dependent responses.
2. Soluble pattern recognition molecules in neutrophils
2.1. Collectins
2.1.1. Structure and expression
Collectins are oligomeric proteins where subunits are composed by three identical polypeptide chains. The degree of multimerization varies among collectins and can significantly affect protein functions [12]. The protomer of each molecule consists of a globular C-terminal carbohydrate recognition domain (CRD) linked to a collagen-like region through an alpha-helical hydrophobic neck region composed by 24 to 28 amino acids and a N-terminal region composed by 7 to 28 amino acids [13]. The collagen-like region is composed by n repetitions of the triplet Gly-Xaa-Yaa (Xaa and Yaa are mostly proline or hydroxyproline) and is involved in the stability of the molecule and the formation of triple helices, which is also stabilized by the neck region. Multimerization of the triple polypeptide chains is supported by hydrophobic interactions and stabilized by interchain disulphide bonds [1]. Mannose-binding lectin (MBL) and surfactant protein (SP)-A are formed by octadecamers of six trimeric subunits and have a polarized bouquet-like structure, whereas SP-D, conglutinin and collectin (CL)-46 are formed by dodecamers of four trimeric subunits and have a cruciform-like structure [1, 12]. Based on structure and function similarities, the complement component C1q was related to this family.
To date, six secreted collectins have been reported: in addition to the well-characterized classical collectins MBL, SP-A and SP-D, CL-43 and CL-46 have been described only in bovidae [12]. MBL is mainly produced by the liver and found in serum whereas SP-A and SP-D are mainly expressed in the lungs by alveolar type II cells [14].
2.1.2. Role of collectins in innate immunity and in neutrophil-dependent immunity
Collectins have the capacity to interact with carbohydrates and lipids exposed on pathogen surfaces (i.e. bacteria, fungi, viruses and parasites). For instance, collectins interact with bacterial PAMP, such as lipopolysaccharide (LPS) from gram-negative bacteria, lipotechoic acid (LTA) from Bacillus subtilis and peptitoglycan from Staphylococcus aureus [12, 15]. The recognition of pathogens by collectins was shown to be protective for the host and to be associated with the induction of an appropriate immune response. For instance, collectins have opsonic activity, enhancing phagocytosis of pathogens, and the capacity to activate the lectin pathway of the complement system, leading to the formation of the membrane attack complex on microbial surfaces [14, 16].
Despite that the expression of collectins has not been reported in neutrophils, these molecules play a fundamental role in innate immunity and were involved in neutrophil-dependent immune response. For instance, MBL, together with MBL-associated serine proteases (MASP), activates the complement system and facilitates C3/C4-mediated phagocytosis of yeasts by neutrophils [17]. Interestingly, the deposition of C3 and C4 and opsonophagocytosis of Candida albicans by neutrophils mediated by MBL can be enhanced by the presence of the long pentraxin PTX3, likely due to a MBL-pentraxin heterocomplex on the pathogen surface and a cooperation between the lectin and classical complement pathways [18]. In addition, recent data have suggested that MBL can also amplify the phagocytosis of yeast by neutrophils in a mechanism coupled with a Dectin-1-triggered ROS production [19]. SP-A and SP-D were also shown to increase neutrophil phagocytosis of pathogens, through a mechanism that involved aggregation of microbes [20]. Interestingly, proteases produced by Staphylococcus aureus can degrade SP-A, leading to the abolishment of SP-A biological activity, including the promotion of S. aureus phagocytosis by neutrophils [21]. Finally, it has been suggested that SP-D can promote neutrophil-extracellular trap (NET)-mediated bacterial trapping [22]. NETs are an extracellular fibrillary network released by activated neutrophils composed by DNA, histones and decorated by a set of proteins from neutrophil granules (e.g. myeloperoxidase (MPO), neutrophil elastase (NE), PTX3) [8, 23]. These structures have the capacity to trap microbes and to favour their elimination [8]. Interestingly, SP-D, which is not produced by neutrophils, can bind simultaneously to carbohydrate ligands found on pathogen surfaces and to NET DNA fragments, leading to efficient trapping of agglutinated bacteria [22].
2.2. Ficolins
2.2.1. Structure and expression
Ficolins are lectin proteins identified in vertebrates with a general structure resembling to that of collectins [1]. Ficolins are oligomeric proteins assembled from a protomer with a collagen-like domain and a C-terminal fibrinogen-like domain involved in the recognition of pathogens. The oligomeric structure is supported by the crosslinking of three monomer subunits through hydrophobic interactions [24]. The globular organization of the fibrinogen-like domain is similar to the CRD of collectins [1]. To date, three members have been identified in human. M-ficolin (also called Ficolin-1), L-ficolin (also called Ficolin-2) and H-ficolin (also called Hakata antigen or Ficolin-3) [25]. L-ficolin is a serum protein mainly produced in the liver and for which the proposed structure is a dodecamers of 35-kDa subunits [26]. H-ficolin is also a serum protein mainly expressed in the liver but also by ciliated bronchial epithelial cells and type II alveolar epithelial cells [27]. An octadecameric structure consisting of a hexamer of trimers was proposed for H-ficolin [1]. M-ficolin was originally identified as a membrane protein on granulocytes and circulating monocytes [28]. Subsequently, M-ficolin was identified as secreted protein expressed in neutrophils, monocytes, macrophages and type II alveolar epithelial cells and found in serum [29].
Two ficolins (i.e. M-ficolin and L-ficolin) have been identified in mouse and pig and one ficolin has been identified in the horseshoe crab Tachypleus tridentatus [29].
2.2.2. Role of ficolins in innate immunity and in neutrophil-dependent immunity
Ficolins recognize carbohydrates through their fibrinogen-like domain. For instance, GlcNAc is recognized by L-ficolin, H-ficolin, M-ficolin and GalNAc is recognized by H-ficolin, and M-ficolin. Other microbial ligands interact with ficolins, such as lipoteichoic acid and β-(1,3)-D-glucan with L-ficolin and lipopolysaccharide with H-ficolin [27, 30, 31].
Ficolins belong to the humoral arm of innate immunity, playing a role in innate defence against invading pathogens, via mechanisms of opsonophagocytosis and the activation of the complement lectin pathway [27, 29]. For instance, L-ficolin interacts with fungi (e.g. Aspergillus fumigatus), gram-negative bacteria (e.g. Salmonella typhimurium and Pseudomonas aeruginosa), and gram-positive bacteria (e.g. S. aureus), leading to increased clearance of pathogens by phagocytes, including neutrophils [29, 32, 33]. L-ficolin was shown to interact also with Mycobacterium tuberculosis, reduce the infectivity in human lung cells and increase the elimination of the pathogen by opsonophagocytosis [34]. M-ficolin and H-ficolin can also interact with gram-negative bacteria (e.g. Salmonella minnesota, Escherichia spp) and gram-positive bacteria (e.g. S. aureus for M-ficolin and Aerococcus viridans for H-ficolin) [29]. In addition to bacteria, ficolins interact with viruses and can have anti-viral effects (e.g. inhibition of viral infectivity and hemagglutination) [29, 35, 36].
Neutrophil precursors (myelocytes, metamyelocytes, and band cells) express the mRNA for M-ficolin and, in accordance with the targeting-by-timing hypothesis (i.e. the time of biosynthesis determines the protein content into distinct neutrophil granules), the protein was found in gelatinase granules and in highly mobilizable gelatinase-poor granules [37, 38]. Therefore, M-ficolin is secreted by neutrophils after stimulation with PMA or fMLF and detected on the surface membrane [37, 39]. In addition, M-ficolin was identified in the phagosome skeleton of neutrophils [40]. Subsequently, the mechanism of binding of M-ficolin to the neutrophil surface has been related to its capacity to interact with the main membrane sialoprotein of neutrophils, leukosialin, also called CD43 [41]. Interestingly, the binding of M-ficolin to resting neutrophils was shown to trigger cell functions described with an anti-CD43 monoclonal antibody (i.e. cell polarization, aggregation and adhesion) and to induce the activation of complement, likely through the lectin pathway, resulting in C3d deposition on the cell membrane [41]. Mouse M-ficolin is also expressed and stored in immature granulocytes [42, 43]. Interestingly, native mouse M-ficolin isolated from neutrophil lysates or supernatants activates the complement lectin pathway [42]. As observed for the human orthologue, mouse L-ficolin interacts with A. fumigatus, leading to increased pathogen phagocytosis by neutrophils and modulation of neutrophil-associated inflammatory response [44].
2.3. Properdin
2.3.1. Structure and expression
Properdin is a highly positively charged glycoprotein found in the plasma. The molecule exists as dimers, trimers and tetramers, which are formed by a head-to-tail interaction of monomers [45, 46]. Each monomer is composed by 442 amino acids and contains a repetition of seven thrombospondin type I repeats involved in the stabilization of the interaction between properdin and ligands [47].
Properdin is a protein of the complement system. In contrast with most other complement molecules, which are produced in the liver, properdin has different production sites. For instance, primary monocytes, dendritic cells, T cells, mast cells, endothelial cells and adipocytes were shown to constitutively produce properdin [47, 48]. In addition, properdin is stored in neutrophil secondary granules and released upon stimulation by chemotactic and inflammatory agonists, such as TNF-α, C5a, CXCL8 or fMLF [49].
2.3.2. Role of Properdin in innate immunity and in neutrophil-dependent immunity
When discovered, properdin was first proposed to be an initiator of the alternative pathway of the complement system [50]. In contrast with the classical and lectin pathways, which are initiated following the recognition of structures by antibodies or specialised receptors, the alternative pathway is initiated in the fluid-phase by spontaneous hydrolysis of the central protein of the complement system, C3 to C3(H2O) [51, 52]. Then, C3(H2O) binds to factor B, leading to the formation of an unstable fluid phase C3 convertase C3(H2O)Bb which cleaves C3 into C3a and C3b and propagates further complement activation [52]. Properdin interacts with the formal C3 convertase C3bBb and extends considerably its half-life (5 to 10 fold), leading to an amplification loop of C3b formation and deposition on surfaces [47].
Properdin can bind to a variety of cell surfaces and acts as a pattern recognition molecule activating the complement system. Indeed, the binding of properdin to Bioacore sensor chips provides a focal point for the association of C3b with factor B on a surface and the formation of C3bBb [53]. In addition, another model proposed that properdin can serve as a stabilizer of preformed C3 convertase by interaction with C3b on target surfaces [52]. However, the presence of properdin might be not necessary for the activation of the alternative pathway on specific surfaces [52].
Properdin was proposed to interact with non-self ligands, such as zymosan, Neisseria gonorrhoeae, rabbit erythrocytes and some strains of E. coli, and modified-self ligands, such as apoptotic cells and necrotic cells [54, 55]. In line with the mechanisms described above, properdin might serve as a focal point for local amplification of the alternative pathway on microbes [47, 54, 55] or might be recruited on initially deposited C3b [56].
In human, properdin deficiency was associated with increased susceptibility to meningococcal disease [57]. Interestingly, administration of recombinant properdin in mice showed therapeutic activity against Streptococcus pneumoniae and Neisseria meningitidis infections, suggesting new opportunities for fighting infections caused by multidrug-resistant strains. [58]. It is important to note that properdin was also involved in complement-dependent autoimmune disorders [52]. For instance, properdin deficiency was associated with a protective effect in models of arthritis induced by K/BxN serum transfer and zymosan [59, 60].
As mentioned above, properdin is stored in neutrophil granules and released upon stimulation [49]. Interestingly, a tissue-specific deletion of properdin gene in myeloid lineage cells showed more than 95% reduction of properdin in plasma, suggesting that neutrophils might be the major source of circulating properdin [61]. Besides its role in protecting the host against infections, the expression of properdin by neutrophils was also related to neutrophil-mediated diseases (e.g. anti-neutrophil cytoplasmic antibody (ANCA)-mediated glomerulonephritis). In human, the binding of ANCA on neutrophils was speculated to induce the release of properdin and the amplification of the complement activation [62]. Consistently, neutrophil-secreted properdin can bind to activated neutrophils, leading to the stabilization of C3 convertase on neutrophil surface and the subsequent formation of C5 convertase [63]. Therefore, this mechanism allows the formation of C5a, which, in turn, induces further activation of neutrophils, creating an amplification loop of the inflammatory response [63]. Recently, properdin was also found on neutrophil-extracellular traps (NETs) induced by ANCA, suggesting that properdin might activate the alternative complement pathway on NETs and participate in the pathogenesis of ANCA-associated vasculitis [64].
2.4. Pentraxins
2.4.1. Structure and expression
Pentraxins constitute a superfamily of conserved multimeric proteins, characterized by the presence of a “pentraxin domain” in their carboxy-terminal, which is a conserved 8-amino acid long-sequence (HxCxS/TWxS, where x is any amino acid). Based on the primary structure of the protomer, pentraxins were divided in short pentraxins, that include C reactive protein (CRP) and serum amyloid P (SAP), and long pentraxins, that include the prototype long pentraxin PTX3 identified during the early 1990s [65–67]. Subsequently, other long pentraxins, including guinea pig apexin, neuronal pentraxin (NP) 1, NP2, neuronal pentraxin receptor (NPR), and PTX4 have been identified [68].
CRP and SAP are about 25-kDa proteins organized in five identical subunits arranged in a pentameric radial symmetry [1, 67]. CRP and SAP are produced by hepatocytes and constitute the main acute phase proteins in human and mouse, respectively [1]. CRP and SAP share functional properties, such as regulation of the complement system, recognition of pathogens, and interaction with Fcγ receptors (FcγR) resulting in phagocytosis of microorganisms and cytokine secretion [69].
The human and murine genes coding for PTX3 are localized on chromosome 3 and are organized in three exons coding for the leader signal peptide, the N-terminal domain and the C-terminal pentraxin domain, respectively [67]. Human and murine PTX3 gene promoters have potential binding sites for many inflammatory transcription factors, including Pu1, AP-1, NF-κB, Sp-1 and NF-IL-6 [70]. In addition, the PI3K/Akt axis and JNK were involved in PTX3 transcription [70]. Recently, it has been shown that PTX3 expression is regulated by an enhancer located in a region spanning the second exon and that methylation of this region is responsible of PTX3 silencing in cancer cells [71].
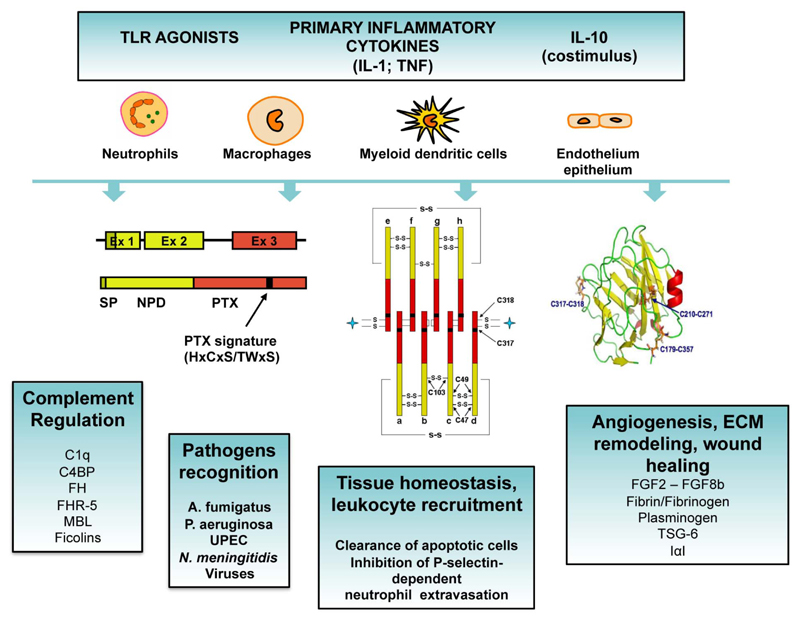
Inflammatory cytokines (e.g. TNF-α, IL-1β), TLR agonists (e.g. LPS), microbial moieties (e.g. KpOmpA) or pathogens (e.g. UPEC, A. fumigatus) induce the expression of PTX3 in various cell types, including dendritic cells, monocytes, macrophages, epithelial cells, endothelial cells, fibroblasts and adipocytes (Fig. 2) [70–74]. Neutrophils are a peculiar PTX3 producing cell type, since only myeloid precursors (promyelocytes and myelocytes/metamyelocytes) transcribe and synthetize PTX3, whereas mature cells store PTX3 in specific granules in a ready-to-use form, and do not express PTX3 mRNA [75]. In response to microorganisms or TLR agonists, or in conditions of tissue damage such as acute myocardial infarction [76], neutrophils rapidly release preformed PTX3, which in part associates to neutrophil extracellular traps (NETs) [75].
Figure 2. Expression, protein structure and roles of PTX3.
Inflammatory cytokines, TLR agonists and microbial moieties induce the expression of PTX3 in various cell types (i.e. dendritic cells, monocytes, macrophages, epithelial cells, endothelial cells, fibroblasts and adipocytes). Neutrophils store PTX3 in specific granules in a ready-to-use form. The gene coding for PTX3 is organized in three exons. The first two exons code for the signal peptide (SP) and the N-terminal domain of the protein (NTD), respectively, and the third exon codes for the pentraxin domain (PTX) containing the PTX signature. PTX3 has a quaternary structure with eight subunits, associated together to form an octamer by a network of disulfide bonds. The three-dimensional model of the pentraxin domain has been generated based on the crystallographic structures of CRP and SAP, showing that the pentraxin domain of PTX3 adopts a β-jelly roll topology. PTX3 plays a role in complement activation and regulation, pathogen recognition, leukocyte recruitment, recognition of apoptotic cells, angiogenesis, extracellular matrix (ECM) remodelling and wound healing.
The protein sequence is conserved among species with 82% identical amino acids in human and mouse. The protomer consists of 381 amino acids including a 17 amino acid-long signal peptide, a N-terminal domain unrelated to any known protein and a C-terminal pentraxin domain homologous to the short pentraxins CRP and SAP. The multimer has a complex quaternary structure characterized by two tetramers linked together by interchain bridges to form an octamer of 340 kDa folding into an elongated structure with a large and a small domain interconnected by a stalk region [77]. In addition, PTX3 has a single N-glycosylation site localized in the C-terminal domain at Asn220 and occupied by core-fucosylated and sialylated complex type oligosaccharides [78]. This glycosidic moiety modulates the interaction of PTX3 with other soluble PRMs, including C1q [78], factor H [79] and ficolin-1 [80], and is required for recognition of influenza virus [81] and engagement of P-selectin [82].
2.4.2. Role of PTX3 in innate immunity and in neutrophil-dependent immunity
PTX3 binds to a wide range of microorganisms, including fungi (e.g. A. fumigatus conidia, Paracoccidioides brasiliensis), bacteria (e.g. P. aeruginosa, Klebsiella pneumoniae, uropathogenic E. coli, Neisseria meningitides) and viruses (e.g. human and murine cytomegalovirus (CMV) and selected strains of influenza virus) [70]. Microbial ligands involved in these interactions are still poorly defined and only Outer membrane protein A of K. pneumoniae (KpOmpA) [83], selected meningococcal antigens of the outer membrane vesicles of N. meningitidis and viral hemagglutinin (HA) glycoprotein of influenza A virus have been identified so far [81, 84]. PTX3-deficient mice showed increased susceptibility to several fungal and bacterial infections, demonstrating the protective role of PTX3 in innate resistance to infections. For instance, PTX3-deficient mice were more susceptible to invasive pulmonary aspergillosis, and PTX3-deficient phagocytes showed defective phagocytosis and clearance of A. fumigatus conidia [75, 85, 86]. Neutrophil-associated PTX3 was essential for resistance against this pathogen, since adoptive transfer of PTX3 competent neutrophils was sufficient to rescue PTX3-deficient mice from the infection [75, 85, 86]. In humans, PTX3 allelic variants were associated with susceptibility to develop A. fumigatus infections in patients undergoing hematopoietic stem cell transplantation [87], or fungal infections in solid organ transplanted patients [88].
PTX3 was also involved in defence against P. aeruginosa and uropathogenic E. coli, playing a non-redundant role both in animal models and in humans [70]. Indeed, recombinant PTX3 had a potential therapeutic effect in a model of P. aeruginosa chronic lung infection that mimics the infection in cystic fibrosis patients by reducing lung colonization, proinflammatory cytokine levels and leukocyte recruitment in the airways [89]. The therapeutic effect was mediated by facilitated recognition and phagocytosis of pathogens by neutrophils through the interplay between complement and FcγRs [85, 89]. In the case of urinary tract infections, PTX3 produced by the uroepithelium and infiltrating myelomonocytic cells facilitated the phagocytosis of E. coli microbes by neutrophils, protecting mice from inflammatory responses and tissue damage [70]. PTX3 can also interact with M-ficolin, L-ficolin and MBL, and PTX3/L-ficolin and PTX3/MBL heterocomplexes promote the deposition of complement on the surface of pathogens [18, 90]. The relevance in humans of these data obtained in mice is supported by results showing that a specific PTX3 haplotype was associated with protection from P. aeruginosa colonization of cystic fibrosis patients [91] and PTX3 allelic variants were associated to the frequency of pyelonephritis [74]. In these infectious conditions, PTX3 contributed to dampen the inflammatory response reasonably by facilitating the elimination of microbes by phagocytes and controlling the microbial burden.
PTX3 also played a protective role in a model of meningococcal meningitis by amplifying the humoral responses to N. meningitides [84] and against certain strains of influenza A viruses (H1N1, H3N2, and H7N9) by neutralizing the virus [81, 92, 93]. In contrast, the interaction with arthritogenic alphaviruses (chikungunya virus and Ross River virus) through the PTX3 N-terminal domain facilitated viral entry and replication enhancing viral infectivity and prolonged disease [16].
In different models of sterile inflammatory responses, PTX3 was shown to play a protective role by limiting inflammation, including neutrophil infiltration. One of the mechanism underlying this effect is based on the binding of PTX3 to P-selectin and on the competition with the interaction between P-selectin and P-selectin glycoprotein ligand-1 (PSGL-1), leading to tuning of P-selectin-dependent neutrophil recruitment [82, 94, 95]. Thus, PTX3 rapidly released by neutrophils in inflammatory conditions activates a negative feed back loop, which prevents excessive neutrophil recruitment in tissues. A second mechanism depends on the common property of PTX3 and short pentraxins, to interact with different complement components (i.e C1q, Factor H, Factor H-related protein 5 (FHR-5), C4b-binding protein (C4BP)) and regulate the activation of the complement system acting at multiple levels [70]. For instance, the interaction between PTX3 and Factor H, FHR-5 and C4BP can favour the deposition of these negative complement regulators on PTX3-coated surfaces, such as apoptotic cells and extracellular matrix, limiting complement activation [79, 96, 97]. Moreover, PTX3 facilitates the deposition of M-ficolin on late apoptotic cells and their phagocytosis by macrophages [98]. More recently, it has been shown that PTX3 acted as an oncosuppressor by regulating tumor promoting complement-dependent cancer related inflammation and had a non-redundant role in tissue repair and remodelling, through the interaction with fibrin and plasminogen, facilitating plasminogen-dependent pericellular fibrinolysis [71, 73].
Neutrophil-derived PTX3 has finally a role in the safe removal of apoptotic cells, and in particular of apoptotic neutrophils. Indeed, it has been shown that PTX3 actively translocates from granules to the cell membrane and accumulates in blebs at the surface of apoptotic neutrophils, favouring the capture of late apoptotic neutrophils by macrophages [99]. However, PTX3 emerges as a novel antineutrophil cytoplasmic antibodies (ANCA) antigen, since anti-PTX3 autoantibodies were detected in the sera of ANCA-associated vasculitis patients [100].
2.5. Peptidoglycan recognition proteins
2.5.1. Structure and expression
Peptidoglycan recognition proteins (PGRPs) were originally discovered in haemolymph of Bombyx mori as peptidoglycan recognition molecules leading to the activation of an antibacterial host defence response [101]. PGRPs genes were subsequently found in other invertebrates, insects, and mammals [102]. In mammals, a family of four proteins was identified and called on the basis of transcript lengths (i.e. PGRP-S for short, PGRP-L for long and PGRP-Iα and PGRP-Iβ for intermadiate). Subsequently, the Human Genome Organization Gene Nomenclature Committee modified their names to PGLYRP (peptidoglycan recognition proteins)-1, 2, 3 and 4 [103]. The peptidoglycan recognition domain present on all PGLYRP is composed by ≈165 amino acids and is homologous to bacterial type 2 amidases, which catalyse the hydrolysis of amide bonds [102]. The short PGLYRP-1 is 196 and 182 amino acid long in human and mice, respectively and mostly composed by the peptidoglycan recognition domain [103]. Intermediate (i.e. PGLYRP-3 and PGLYRP-4) molecules have two peptidoglycan recognition domains and the long molecule PGLYRP-2 is composed by a C-terminal peptidoglycan recognition domain coupled to a non-conserved amino terminal sequence [102]. All mammalian PGRPs are secreted molecules and form homodimers. In addition, PGLYRP-3 and PGLYRP-4 can form disulphide-linked heterodimers [104, 105].
PGLYRP-1 is mainly expressed in bone marrow and stored in neutrophil granules and PGLYRP-3 and PGLYRP-4 are expressed in skin and mucous membrane, in particular in the mouth, throat, salivary glands, small and large intestines and stomach [106–108]. PGLYRP-2 is constitutively produced by the liver and secreted into the circulation [102, 109]. In addition, the expression of PGLYRP-2 was induced in keratinocytes and oral epithelial cells after stimulation by bacteria and cytokines [110, 111].
2.5.2. Role of PGRPs in innate immunity and in neutrophil-dependent immunity
All mammalian PGRPs have a bactericidal activity against non-pathogenic and pathogenic bacteria [106]. In addition to its bactericidal activity, PGLYRP-2 has an amidase activity that hydrolyses bacterial peptidoglycan [112, 113]. All mammalian PGRPs were involved in maintaining normal intestinal microflora and deficiencies in individual PGRP induced gastrointestinal dysbiosis [108, 114]. Therefore, mice deficient for PGLYRP-1, PGLYRP-2, PGLYRP-3 or PGLYRP-4 had increased susceptibility to experimental colitis induced by dextran sulphate sodium (DSS) [102, 108, 114]. PGRPs bind to peptidoglycan of Gram-positive bacteria cell wall, near the site of daughter cell separation, and bind to peptidoglyan of the outer cell membrane of Gram-negative bacteria [115]. PGRPs kill bacteria by mechanisms leading to arrest of the major biosynthetic processes, depolarization of the bacterial membrane and release of toxic hydroxyl radicals [115, 116].
As mentioned above, PGLYRP-1 is stored in neutrophil granules. In particular, PGLYRP-was identified in bovine, murine and human neutrophils. Interestingly, neutrophils isolated from PGLYRP-1 deficient mice are defective in intracellular killing of bacteria but not in phagocytosis, suggesting that PGLYRP-1 supports the antimicrobial activity of neutrophils. The defective killing activity was related to a low induction of oxidative burst in response to bacteria [117]. In addition, PGLYRP-1 has an additive and synergistic antimicrobial effect with lysozyme, an antimicrobial protein stored in neutrophil granules, and both molecules localize in NETs [118]. Most recently, PGLYRP-1 was proposed as a ligand for triggering receptor expressed on myeloid cells (TREM)-1, an orphan receptor expressed on monocytes, macrophages and neutrophils and involved in the proinflammatory response [119]. Soluble PGLYRP-1 secreted by stimulated-neutrophils binds to peptidoglycan and forms functional ligand complexes capable of activating adjacent TREM-1-expressing myeloid cells, creating an amplification loop of the response [119]. In addition multimeric presentation of PGLYRP-1 is sufficient to activate neutrophils, inducing the production of CXCL8 and the generation of superoxide radicals [119]. It is important to underline that other cells than neutrophils can express PGLYRP-1, such as eosinophils, and that its expression was induced in macrophages and epithelial cells upon stimulation [102, 120]. For instance, in a model of experimental asthma induced by intranasal instillation of house dust mite, the expression of PGLYRP-1 was induced in lung epithelial cells and macrophages. Therefore, the relevance of neutrophil-associated PGLYRP-1 in this model remains unknown [120].
3. Conclusion
Soluble PRMs belong to the humoral arm of innate immunity and share fundamental effector mechanisms, such as activation and regulation of the classical, alternative and lectin pathways of the complement system, opsonisation of pathogens, aggregation and neutralisation of viral particles and recognition of apoptotic cells. Consistently, PRMs participate in the fine-tuning of immune and inflammatory responses through the discrimination of self versus non-self and modified-self (Table 1). Evidence suggests that some fluid phase PRMs interact and act synergistically. For instance, the interaction of the long pentraxin PTX3 with MBL, M-ficolin and L-ficolin leads to the formation of heterocomplexes, which amplifies the recognition potential of non-self and modified-self, and the functional consequences [18, 90, 98].
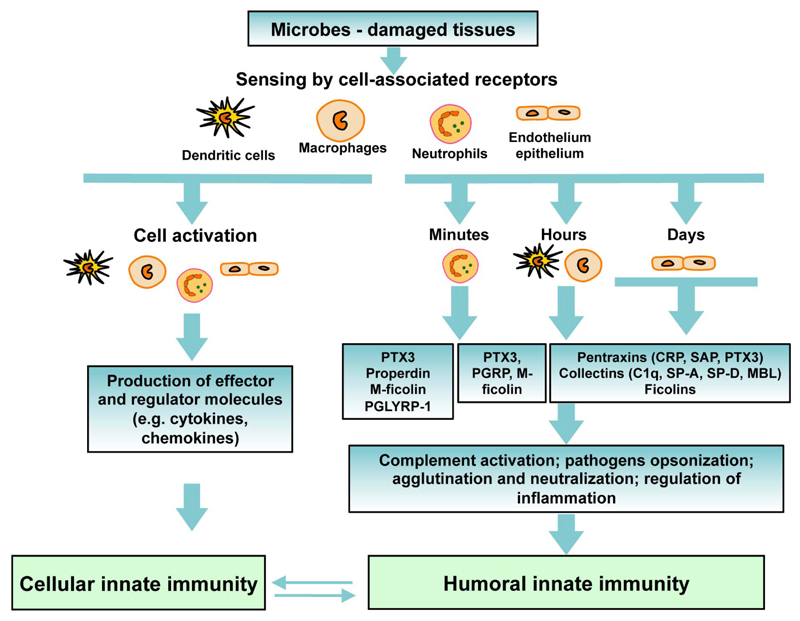
Importantly, the kinetic and source of production of PRMs ensure the continuous presence of these molecules both in the systemic circulation and tissues in response to infection and tissue damage. Indeed, epithelial tissues, including the liver or lung, support the expression of humoral PRMs and their presence in the systemic circulation (e.g. short pentraxins, ficolins) and in tissues (e.g. collectins in the lungs). Other cell types and in particular macrophages and dendritic cells produce PTX3, properdin, and M-ficolin, sustaining a rapid expression of these PRMs in a gene expression–dependent fashion. Finally, neutrophils act as a reservoir of ready-made PRMs (i.e. PTX3, M-ficolin, PGLYRP-1 and properdin) for rapid release in minutes in sites of tissue damage or microbial stimulation and represent the first source of soluble PRMs, thus providing an immediate availability of diverse antimicrobial and immunomodulatory molecules (Fig. 3).
Figure 3. Integration of the humoral innate immunity in the innate response.
The activation of innate immunity by cell-associated receptors induces the production of regulatory molecules (i.e cytokines) and soluble pattern recognition molecules. Soluble PRMs are expressed and secreted by a variety of cells, including myeloid cells, epithelial cells and endothelial cells, allowing the production of PRMs over time. These molecules share effector mechanisms, including complement activation and pathogen recognition. A cross-talk between the humoral and cellular arms of innate immunity participates in the regulation of the innate response.
Highlights.
Soluble pattern recognition molecules belong to the humoral arm of innate immunity
Soluble pattern recognition molecules share effector mechanisms
Neutrophils are a reservoir of ready-made soluble pattern recognition molecules
Soluble pattern recognition molecules are involved in neutrophil effector functions
Acknowledgements
The contribution of Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) (project FIRB RBAP11H2R9), Ministero della salute (Ricerca finalizzata), the Associazione Italiana Ricerca sul Cancro (AIRC and AIRC 5x1000), the European Research Council (ERC; to A. Montavoni), the European Commission (FP7-HEALTH-2011-ADITEC-280873), and the Fondazione CARIPLO (Project 2009-2582) is gratefully acknowledged.
Abbreviations
- ANCA
anti-neutrophil cytoplasmic antibody
- C4BP
C4b-binding protein
- CL
collectin
- CRD
C-terminal carbohydrate recognition domain
- CRP
C reactive protein
- FHR-5
Factor H-related protein 5
- LPS
lipopolysaccharide
- LTA
lipotechoic acid
- MBL
mannose-binding lectin
- MDA5
melanoma differentiation-associated gene 5
- MPO
myeloperoxidase
- NETs
neutrophil extracellular traps
- NLR
NOD-like receptors
- NOD
nucleotide-binding oligomerization domain
- PAMP
pathogen associated molecular patterns
- PGLYRP
peptidoglycan recognition proteins
- PGRPs
peptidoglycan recognition proteins
- PRMs
pattern recognition molecules
- PTX3
pentraxin 3
- RIG-I
retinoic acid-inducible gene I
- SAP
serum amyloid P
- SP
surfactant protein
- TLRs
Toll like receptors.
Footnotes
The authors declare no competing financial interests.
References
- [1].Bottazzi B, Doni A, Garlanda C, Mantovani A. An Integrated View of Humoral Innate Immunity: Pentraxins as a Paradigm. Annu Rev Immunol. 2010;28:157–183. doi: 10.1146/annurev-immunol-030409-101305. [DOI] [PubMed] [Google Scholar]
- [2].Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jeannin P, Jaillon S, Delneste Y. Pattern recognition receptors in the immune response against dying cells. Curr Opin Immunol. 2008;20:530–537. doi: 10.1016/j.coi.2008.04.013. [DOI] [PubMed] [Google Scholar]
- [4].Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- [5].Wirths S, Bugl S, Kopp HG. Neutrophil homeostasis and its regulation by danger signaling. Blood. 2014;123:3563–3566. doi: 10.1182/blood-2013-11-516260. [DOI] [PubMed] [Google Scholar]
- [6].Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- [7].Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- [8].Jaillon S, Galdiero MR, Del Prete D, Cassatella MA, Garlanda C, Mantovani A. Neutrophils in innate and adaptive immunity. Semin Immunopathol. 2013;35:377–394. doi: 10.1007/s00281-013-0374-8. [DOI] [PubMed] [Google Scholar]
- [9].Scapini P, Cassatella MA. Social networking of human neutrophils within the immune system. Blood. 2014;124:710–719. doi: 10.1182/blood-2014-03-453217. [DOI] [PubMed] [Google Scholar]
- [10].Zimmermann M, Arruda-Silva F, Bianchetto-Aguilera F, Finotti G, Calzetti F, Scapini P, et al. IFNalpha enhances the production of IL-6 by human neutrophils activated via TLR8. Sci Rep. 2016;6:19674. doi: 10.1038/srep19674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tamassia N, Cassatella MA. Cytoplasmic receptors recognizing nucleic acids and mediating immune functions in neutrophils. Curr Opin Pharmacol. 2013;13:547–554. doi: 10.1016/j.coph.2013.05.003. [DOI] [PubMed] [Google Scholar]
- [12].Gupta G, Surolia A. Collectins: sentinels of innate immunity. Bioessays. 2007;29:452–464. doi: 10.1002/bies.20573. [DOI] [PubMed] [Google Scholar]
- [13].Uemura T, Sano H, Katoh T, Nishitani C, Mitsuzawa H, Shimizu T, et al. Surfactant protein A without the interruption of Gly-X-Y repeats loses a kink of oligomeric structure and exhibits impaired phospholipid liposome aggregation ability. Biochemistry. 2006;45:14543–14551. doi: 10.1021/bi061338u. [DOI] [PubMed] [Google Scholar]
- [14].Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- [15].van de Wetering JK, van Eijk M, van Golde LM, Hartung T, van Strijp JA, Batenburg JJ. Characteristics of surfactant protein A and D binding to lipoteichoic acid and peptidoglycan, 2 major cell wall components of gram-positive bacteria. J Infect Dis. 2001;184:1143–1151. doi: 10.1086/323746. [DOI] [PubMed] [Google Scholar]
- [16].Foo SS, Reading PC, Jaillon S, Mantovani A, Mahalingam S. Pentraxins and Collectins: Friend or Foe during Pathogen Invasion? Trends Microbiol. 2015 doi: 10.1016/j.tim.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brouwer N, Dolman KM, van Houdt M, Sta M, Roos D, Kuijpers TW. Mannose-binding lectin (MBL) facilitates opsonophagocytosis of yeasts but not of bacteria despite MBL binding. J Immunol. 2008;180:4124–4132. doi: 10.4049/jimmunol.180.6.4124. [DOI] [PubMed] [Google Scholar]
- [18].Ma YJ, Doni A, Skjoedt MO, Honore C, Arendrup M, Mantovani A, et al. Heterocomplexes of mannose-binding lectin and the pentraxins PTX3 or serum amyloid P component trigger cross-activation of the complement system. J Biol Chem. 2011;286:3405–3417. doi: 10.1074/jbc.M110.190637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li D, Dong B, Tong Z, Wang Q, Liu W, Wang Y, et al. MBL-mediated opsonophagocytosis of Candida albicans by human neutrophils is coupled with intracellular Dectin-1-triggered ROS production. PLoS One. 2012;7:e50589. doi: 10.1371/journal.pone.0050589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hartshorn KL, Crouch E, White MR, Colamussi ML, Kakkanatt A, Tauber B, et al. Pulmonary surfactant proteins A and D enhance neutrophil uptake of bacteria. Am J Physiol. 1998;274:L958–969. doi: 10.1152/ajplung.1998.274.6.L958. [DOI] [PubMed] [Google Scholar]
- [21].Kantyka T, Pyrc K, Gruca M, Smagur J, Plaza K, Guzik K, et al. Staphylococcus aureus proteases degrade lung surfactant protein A potentially impairing innate immunity of the lung. J Innate Immun. 2013;5:251–260. doi: 10.1159/000345417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Douda DN, Jackson R, Grasemann H, Palaniyar N. Innate immune collectin surfactant protein D simultaneously binds both neutrophil extracellular traps and carbohydrate ligands and promotes bacterial trapping. J Immunol. 2011;187:1856–1865. doi: 10.4049/jimmunol.1004201. [DOI] [PubMed] [Google Scholar]
- [23].Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- [24].Endo Y, Matsushita M, Fujita T. Role of ficolin in innate immunity and its molecular basis. Immunobiology. 2007;212:371–379. doi: 10.1016/j.imbio.2006.11.014. [DOI] [PubMed] [Google Scholar]
- [25].Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- [26].Hummelshoj T, Thielens NM, Madsen HO, Arlaud GJ, Sim RB, Garred P. Molecular organization of human Ficolin-2. Mol Immunol. 2007;44:401–411. doi: 10.1016/j.molimm.2006.02.023. [DOI] [PubMed] [Google Scholar]
- [27].Matsushita M. Ficolins: complement-activating lectins involved in innate immunity. J Innate Immun. 2010;2:24–32. doi: 10.1159/000228160. [DOI] [PubMed] [Google Scholar]
- [28].Thielens NM. The double life of M-ficolin: what functions when circulating in serum and tethered to leukocyte surfaces? J Leukoc Biol. 2011;90:410–412. doi: 10.1189/jlb.0611281. [DOI] [PubMed] [Google Scholar]
- [29].Ren Y, Ding Q, Zhang X. Ficolins and infectious diseases. Virol Sin. 2014;29:25–32. doi: 10.1007/s12250-014-3421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ma YG, Cho MY, Zhao M, Park JW, Matsushita M, Fujita T, et al. Human mannose-binding lectin and L-ficolin function as specific pattern recognition proteins in the lectin activation pathway of complement. J Biol Chem. 2004;279:25307–25312. doi: 10.1074/jbc.M400701200. [DOI] [PubMed] [Google Scholar]
- [31].Lynch NJ, Roscher S, Hartung T, Morath S, Matsushita M, Maennel DN, et al. L-ficolin specifically binds to lipoteichoic acid, a cell wall constituent of Gram-positive bacteria, and activates the lectin pathway of complement. J Immunol. 2004;172:1198–1202. doi: 10.4049/jimmunol.172.2.1198. [DOI] [PubMed] [Google Scholar]
- [32].Matsushita M, Endo Y, Taira S, Sato Y, Fujita T, Ichikawa N, et al. A novel human serum lectin with collagen- and fibrinogen-like domains that functions as an opsonin. J Biol Chem. 1996;271:2448–2454. doi: 10.1074/jbc.271.5.2448. [DOI] [PubMed] [Google Scholar]
- [33].Bidula S, Sexton DW, Abdolrasouli A, Shah A, Reed A, Armstrong-James D, et al. The serum opsonin L-ficolin is detected in lungs of human transplant recipients following fungal infections and modulates inflammation and killing of Aspergillus fumigatus. J Infect Dis. 2015;212:234–246. doi: 10.1093/infdis/jiv027. [DOI] [PubMed] [Google Scholar]
- [34].Luo F, Sun X, Wang Y, Wang Q, Wu Y, Pan Q, et al. Ficolin-2 defends against virulent Mycobacteria tuberculosis infection in vivo, and its insufficiency is associated with infection in humans. PLoS One. 2013;8:e73859. doi: 10.1371/journal.pone.0073859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu J, Ali MA, Shi Y, Zhao Y, Luo F, Yu J, et al. Specifically binding of L-ficolin to N-glycans of HCV envelope glycoproteins E1 and E2 leads to complement activation. Cell Mol Immunol. 2009;6:235–244. doi: 10.1038/cmi.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Verma A, White M, Vathipadiekal V, Tripathi S, Mbianda J, Ieong M, et al. Human H-ficolin inhibits replication of seasonal and pandemic influenza A viruses. J Immunol. 2012;189:2478–2487. doi: 10.4049/jimmunol.1103786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rorvig S, Honore C, Larsson LI, Ohlsson S, Pedersen CC, Jacobsen LC, et al. Ficolin-1 is present in a highly mobilizable subset of human neutrophil granules and associates with the cell surface after stimulation with fMLP. J Leukoc Biol. 2009;86:1439–1449. doi: 10.1189/jlb.1008606. [DOI] [PubMed] [Google Scholar]
- [38].Rorvig S, Ostergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 2013;94:711–721. doi: 10.1189/jlb.1212619. [DOI] [PubMed] [Google Scholar]
- [39].Liu Y, Endo Y, Iwaki D, Nakata M, Matsushita M, Wada I, et al. Human M-ficolin is a secretory protein that activates the lectin complement pathway. J Immunol. 2005;175:3150–3156. doi: 10.4049/jimmunol.175.5.3150. [DOI] [PubMed] [Google Scholar]
- [40].Xu P, Crawford M, Way M, Godovac-Zimmermann J, Segal AW, Radulovic M. Subproteome analysis of the neutrophil cytoskeleton. Proteomics. 2009;9:2037–2049. doi: 10.1002/pmic.200800674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Moreno-Amaral AN, Gout E, Danella-Polli C, Tabarin F, Lesavre P, Pereira-da-Silva G, et al. M-ficolin and leukosialin (CD43): new partners in neutrophil adhesion. J Leukoc Biol. 2012;91:469–474. doi: 10.1189/jlb.0911460. [DOI] [PubMed] [Google Scholar]
- [42].Weber-Steffens D, Hunold K, Kurschner J, Martinez SG, Elumalai P, Schmidt D, et al. Immature mouse granulocytic myeloid cells are characterized by production of ficolin-B. Mol Immunol. 2013;56:488–496. doi: 10.1016/j.molimm.2013.06.015. [DOI] [PubMed] [Google Scholar]
- [43].Hunold K, Weber-Steffens D, Runza VL, Jensenius JC, Mannel DN. Functional analysis of mouse ficolin-B and detection in neutrophils. Immunobiology. 2012;217:982–985. doi: 10.1016/j.imbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- [44].Bidula S, Sexton DW, Schelenz S. Serum opsonin ficolin-A enhances host-fungal interactions and modulates cytokine expression from human monocyte-derived macrophages and neutrophils following Aspergillus fumigatus challenge. Med Microbiol Immunol. 2015 doi: 10.1007/s00430-015-0435-9. [DOI] [PubMed] [Google Scholar]
- [45].Smith CA, Pangburn MK, Vogel CW, Muller-Eberhard HJ. Molecular architecture of human properdin, a positive regulator of the alternative pathway of complement. J Biol Chem. 1984;259:4582–4588. [PubMed] [Google Scholar]
- [46].Pangburn MK. Analysis of the natural polymeric forms of human properdin and their functions in complement activation. J Immunol. 1989;142:202–207. [PubMed] [Google Scholar]
- [47].Cortes C, Ohtola JA, Saggu G, Ferreira VP. Local release of properdin in the cellular microenvironment: role in pattern recognition and amplification of the alternative pathway of complement. Front Immunol. 2012;3:412. doi: 10.3389/fimmu.2012.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li K, Fazekasova H, Wang N, Sagoo P, Peng Q, Khamri W, et al. Expression of complement components, receptors and regulators by human dendritic cells. Mol Immunol. 2011;48:1121–1127. doi: 10.1016/j.molimm.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wirthmueller U, Dewald B, Thelen M, Schafer MK, Stover C, Whaley K, et al. Properdin, a positive regulator of complement activation, is released from secondary granules of stimulated peripheral blood neutrophils. J Immunol. 1997;158:4444–4451. [PubMed] [Google Scholar]
- [50].Pillemer L, Blum L, Lepow IH, Ross OA, Todd EW, Wardlaw AC. The properdin system and immunity. I. Demonstration and isolation of a new serum protein, properdin, and its role in immune phenomena. Science. 1954;120:279–285. doi: 10.1126/science.120.3112.279. [DOI] [PubMed] [Google Scholar]
- [51].Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- [52].Lesher AM, Nilsson B, Song WC. Properdin in complement activation and tissue injury. Mol Immunol. 2013;56:191–198. doi: 10.1016/j.molimm.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hourcade DE. The role of properdin in the assembly of the alternative pathway C3 convertases of complement. J Biol Chem. 2006;281:2128–2132. doi: 10.1074/jbc.M508928200. [DOI] [PubMed] [Google Scholar]
- [54].Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179:2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- [55].Xu W, Berger SP, Trouw LA, de Boer HC, Schlagwein N, Mutsaers C, et al. Properdin binds to late apoptotic and necrotic cells independently of C3b and regulates alternative pathway complement activation. J Immunol. 2008;180:7613–7621. doi: 10.4049/jimmunol.180.11.7613. [DOI] [PubMed] [Google Scholar]
- [56].Harboe M, Garred P, Lindstad JK, Pharo A, Muller F, Stahl GL, et al. The role of properdin in zymosan- and Escherichia coli-induced complement activation. J Immunol. 2012;189:2606–2613. doi: 10.4049/jimmunol.1200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sjoholm AG, Braconier JH, Soderstrom C. Properdin deficiency in a family with fulminant meningococcal infections. Clin Exp Immunol. 1982;50:291–297. [PMC free article] [PubMed] [Google Scholar]
- [58].Ali YM, Hayat A, Saeed BM, Haleem KS, Alshamrani S, Kenawy HI, et al. Low-dose recombinant properdin provides substantial protection against Streptococcus pneumoniae and Neisseria meningitidis infection. Proc Natl Acad Sci U S A. 2014;111:5301–5306. doi: 10.1073/pnas.1401011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dimitrova P, Ivanovska N, Schwaeble W, Gyurkovska V, Stover C. The role of properdin in murine zymosan-induced arthritis. Mol Immunol. 2010;47:1458–1466. doi: 10.1016/j.molimm.2010.02.007. [DOI] [PubMed] [Google Scholar]
- [60].Kimura Y, Zhou L, Miwa T, Song WC. Genetic and therapeutic targeting of properdin in mice prevents complement-mediated tissue injury. J Clin Invest. 2010;120:3545–3554. doi: 10.1172/JCI41782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Miwa T, Sato S, Gullipalli D, Nangaku M, Song WC. Blocking properdin, the alternative pathway, and anaphylatoxin receptors ameliorates renal ischemia-reperfusion injury in decay-accelerating factor and CD59 double-knockout mice. J Immunol. 2013;190:3552–3559. doi: 10.4049/jimmunol.1202275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xiao H, Schreiber A, Heeringa P, Falk RJ, Jennette JC. Alternative complement pathway in the pathogenesis of disease mediated by anti-neutrophil cytoplasmic autoantibodies. Am J Pathol. 2007;170:52–64. doi: 10.2353/ajpath.2007.060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Camous L, Roumenina L, Bigot S, Brachemi S, Fremeaux-Bacchi V, Lesavre P, et al. Complement alternative pathway acts as a positive feedback amplification of neutrophil activation. Blood. 2011;117:1340–1349. doi: 10.1182/blood-2010-05-283564. [DOI] [PubMed] [Google Scholar]
- [64].Wang H, Wang C, Zhao MH, Chen M. Neutrophil extracellular traps can activate alternative complement pathways. Clin Exp Immunol. 2015;181:518–527. doi: 10.1111/cei.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Breviario F, d'Aniello EM, Golay J, Peri G, Bottazzi B, Bairoch A, et al. Interleukin-1-inducible genes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J Biol Chem. 1992;267:22190–22197. [PubMed] [Google Scholar]
- [66].Lee GW, Lee TH, Vilcek J. TSG-14, a tumor necrosis factor- and IL-1-inducible protein, is a novel member of the pentaxin family of acute phase proteins. J Immunol. 1993;150:1804–1812. [PubMed] [Google Scholar]
- [67].Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. 2005;23:337–366. doi: 10.1146/annurev.immunol.23.021704.115756. [DOI] [PubMed] [Google Scholar]
- [68].Martinez de la Torre Y, Fabbri M, Jaillon S, Bastone A, Nebuloni M, Vecchi A, et al. Evolution of the pentraxin family: the new entry PTX4. J Immunol. 2010;184:5055–5064. doi: 10.4049/jimmunol.0901672. [DOI] [PubMed] [Google Scholar]
- [69].Lu J, Marnell LL, Marjon KD, Mold C, Du Clos TW, Sun PD. Structural recognition and functional activation of FcgammaR by innate pentraxins. Nature. 2008;456:989–992. doi: 10.1038/nature07468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Jaillon S, Bonavita E, Gentile S, Rubino M, Laface I, Garlanda C, et al. The long pentraxin PTX3 as a key component of humoral innate immunity and a candidate diagnostic for inflammatory diseases. Int Arch Allergy Immunol. 2014;165:165–178. doi: 10.1159/000368778. [DOI] [PubMed] [Google Scholar]
- [71].Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160:700–714. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- [72].Salio M, Chimenti S, De Angelis N, Molla F, Maina V, Nebuloni M, et al. Cardioprotective function of the long pentraxin PTX3 in acute myocardial infarction. Circulation. 2008;117:1055–1064. doi: 10.1161/CIRCULATIONAHA.107.749234. [DOI] [PubMed] [Google Scholar]
- [73].Doni A, Musso T, Morone D, Bastone A, Zambelli V, Sironi M, et al. An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J Exp Med. 2015;212:905–925. doi: 10.1084/jem.20141268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jaillon S, Moalli F, Ragnarsdottir B, Bonavita E, Puthia M, Riva F, et al. The Humoral Pattern Recognition Molecule PTX3 Is a Key Component of Innate Immunity against Urinary Tract Infection. Immunity. 2014;40:621–632. doi: 10.1016/j.immuni.2014.02.015. [DOI] [PubMed] [Google Scholar]
- [75].Jaillon S, Peri G, Delneste Y, Fremaux I, Doni A, Moalli F, et al. The humoral pattern recognition receptor PTX3 is stored in neutrophil granules and localizes in extracellular traps. J Exp Med. 2007;204:793–804. doi: 10.1084/jem.20061301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Maugeri N, Rovere-Querini P, Slavich M, Coppi G, Doni A, Bottazzi B, et al. Early and transient release of leukocyte pentraxin 3 during acute myocardial infarction. J Immunol. 2011;187:970–979. doi: 10.4049/jimmunol.1100261. [DOI] [PubMed] [Google Scholar]
- [77].Inforzato A, Rivieccio V, Morreale AP, Bastone A, Salustri A, Scarchilli L, et al. Structural characterization of PTX3 disulfide bond network and its multimeric status in cumulus matrix organization. J Biol Chem. 2008;283:10147–10161. doi: 10.1074/jbc.M708535200. [DOI] [PubMed] [Google Scholar]
- [78].Inforzato A, Peri G, Doni A, Garlanda C, Mantovani A, Bastone A, et al. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry. 2006;45:11540–11551. doi: 10.1021/bi0607453. [DOI] [PubMed] [Google Scholar]
- [79].Deban L, Jarva H, Lehtinen MJ, Bottazzi B, Bastone A, Doni A, et al. Binding of the long pentraxin PTX3 to factor H: interacting domains and function in the regulation of complement activation. J Immunol. 2008;181:8433–8440. doi: 10.4049/jimmunol.181.12.8433. [DOI] [PubMed] [Google Scholar]
- [80].Gout E, Moriscot C, Doni A, Dumestre-Perard C, Lacroix M, Perard J, et al. M-ficolin interacts with the long pentraxin PTX3: a novel case of cross-talk between soluble pattern-recognition molecules. J Immunol. 2011;186:5815–5822. doi: 10.4049/jimmunol.1100180. [DOI] [PubMed] [Google Scholar]
- [81].Reading PC, Bozza S, Gilbertson B, Tate M, Moretti S, Job ER, et al. Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J Immunol. 2008;180:3391–3398. doi: 10.4049/jimmunol.180.5.3391. [DOI] [PubMed] [Google Scholar]
- [82].Deban L, Russo RC, Sironi M, Moalli F, Scanziani M, Zambelli V, et al. Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat Immunol. 2010;11:328–334. doi: 10.1038/ni.1854. [DOI] [PubMed] [Google Scholar]
- [83].Jeannin P, Bottazzi B, Sironi M, Doni A, Rusnati M, Presta M, et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity. 2005;22:551–560. doi: 10.1016/j.immuni.2005.03.008. [DOI] [PubMed] [Google Scholar]
- [84].Bottazzi B, Santini L, Savino S, Giuliani MM, Duenas Diez AI, Mancuso G, et al. Recognition of Neisseria meningitidis by the Long Pentraxin PTX3 and Its Role as an Endogenous Adjuvant. PLoS One. 2015;10:e0120807. doi: 10.1371/journal.pone.0120807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Moalli F, Doni A, Deban L, Zelante T, Zagarella S, Bottazzi B, et al. Role of complement and Fc{gamma} receptors in the protective activity of the long pentraxin PTX3 against Aspergillus fumigatus. Blood. 2010;116:5170–5180. doi: 10.1182/blood-2009-12-258376. [DOI] [PubMed] [Google Scholar]
- [86].Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 2002;420:182–186. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- [87].Cunha C, Aversa F, Lacerda J, Busca A, Kurzai O, Grube M, et al. Genetic deficiency of PTX3 and aspergillosis in stem cell transplantation. N Engl J Med. 2014;370:421–432. doi: 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- [88].Wojtowicz A, Lecompte TD, Bibert S, Manuel O, Rueger S, Berger C, et al. PTX3 Polymorphisms and Invasive Mold Infections After Solid Organ Transplant. Clin Infect Dis. 2015;61:619–622. doi: 10.1093/cid/civ386. [DOI] [PubMed] [Google Scholar]
- [89].Moalli F, Paroni M, Veliz Rodriguez T, Riva F, Polentarutti N, Bottazzi B, et al. The therapeutic potential of the humoral pattern recognition molecule PTX3 in chronic lung infection caused by Pseudomonas aeruginosa. J Immunol. 2011;186:5425–5434. doi: 10.4049/jimmunol.1002035. [DOI] [PubMed] [Google Scholar]
- [90].Ma YJ, Doni A, Hummelshoj T, Honore C, Bastone A, Mantovani A, et al. Synergy between ficolin-2 and pentraxin 3 boosts innate immune recognition and complement deposition. J Biol Chem. 2009;284:28263–28275. doi: 10.1074/jbc.M109.009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G, Mazza C, et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. 2010;11:665–670. doi: 10.1038/gene.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Job ER, Deng YM, Tate MD, Bottazzi B, Crouch EC, Dean MM, et al. Pandemic H1N1 influenza A viruses are resistant to the antiviral activities of innate immune proteins of the collectin and pentraxin superfamilies. J Immunol. 2010;185:4284–4291. doi: 10.4049/jimmunol.1001613. [DOI] [PubMed] [Google Scholar]
- [93].Job ER, Bottazzi B, Short KR, Deng YM, Mantovani A, Brooks AG, et al. A single amino acid substitution in the hemagglutinin of H3N2 subtype influenza A viruses is associated with resistance to the long pentraxin PTX3 and enhanced virulence in mice. J Immunol. 2014;192:271–281. doi: 10.4049/jimmunol.1301814. [DOI] [PubMed] [Google Scholar]
- [94].Han B, Haitsma JJ, Zhang Y, Bai X, Rubacha M, Keshavjee S, et al. Long pentraxin PTX3 deficiency worsens LPS-induced acute lung injury. Intensive Care Med. 2011 doi: 10.1007/s00134-010-2067-2. [DOI] [PubMed] [Google Scholar]
- [95].Lech M, Rommele C, Grobmayr R, Eka Susanti H, Kulkarni OP, Wang S, et al. Endogenous and exogenous pentraxin-3 limits postischemic acute and chronic kidney injury. Kidney Int. 2013;83:647–661. doi: 10.1038/ki.2012.463. [DOI] [PubMed] [Google Scholar]
- [96].Braunschweig A, Jozsi M. Human pentraxin 3 binds to the complement regulator c4b-binding protein. PLoS One. 2011;6:e23991. doi: 10.1371/journal.pone.0023991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Csincsi AI, Kopp A, Zoldi M, Banlaki Z, Uzonyi B, Hebecker M, et al. Factor H-related protein 5 interacts with pentraxin 3 and the extracellular matrix and modulates complement activation. J Immunol. 2015;194:4963–4973. doi: 10.4049/jimmunol.1403121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ma YJ, Doni A, Romani L, Jurgensen HJ, Behrendt N, Mantovani A, et al. Ficolin-1-PTX3 complex formation promotes clearance of altered self-cells and modulates IL-8 production. J Immunol. 2013;191:1324–1333. doi: 10.4049/jimmunol.1300382. [DOI] [PubMed] [Google Scholar]
- [99].Jaillon S, Jeannin P, Hamon Y, Fremaux I, Doni A, Bottazzi B, et al. Endogenous PTX3 translocates at the membrane of late apoptotic human neutrophils and is involved in their engulfment by macrophages. Cell Death Differ. 2009;16:465–474. doi: 10.1038/cdd.2008.173. [DOI] [PubMed] [Google Scholar]
- [100].Simon A, Subra JF, Guilpain P, Jeannin P, Pignon P, Blanchard S, et al. Detection of Anti-Pentraxin-3 Autoantibodies in ANCA-Associated Vasculitis. PLoS One. 2016;11:e0147091. doi: 10.1371/journal.pone.0147091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem. 1996;271:13854–13860. doi: 10.1074/jbc.271.23.13854. [DOI] [PubMed] [Google Scholar]
- [102].Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- [103].Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006;8:1059–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- [104].Royet J, Dziarski R. Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- [105].Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, et al. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J Biol Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- [106].Dziarski R, Gupta D. Review: Mammalian peptidoglycan recognition proteins (PGRPs) in innate immunity. Innate Immun. 2010;16:168–174. doi: 10.1177/1753425910366059. [DOI] [PubMed] [Google Scholar]
- [107].Mathur P, Murray B, Crowell T, Gardner H, Allaire N, Hsu YM, et al. Murine peptidoglycan recognition proteins PglyrpIalpha and PglyrpIbeta are encoded in the epidermal differentiation complex and are expressed in epidermal and hematopoietic tissues. Genomics. 2004;83:1151–1163. doi: 10.1016/j.ygeno.2004.01.003. [DOI] [PubMed] [Google Scholar]
- [108].Saha S, Jing X, Park SY, Wang S, Li X, Gupta D, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe. 2010;8:147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhang Y, van der Fits L, Voerman JS, Melief MJ, Laman JD, Wang M, et al. Identification of serum N-acetylmuramoyl-l-alanine amidase as liver peptidoglycan recognition protein 2. Biochim Biophys Acta. 2005;1752:34–46. doi: 10.1016/j.bbapap.2005.07.001. [DOI] [PubMed] [Google Scholar]
- [110].Li X, Wang S, Wang H, Gupta D. Differential expression of peptidoglycan recognition protein 2 in the skin and liver requires different transcription factors. J Biol Chem. 2006;281:20738–20748. doi: 10.1074/jbc.M601017200. [DOI] [PubMed] [Google Scholar]
- [111].Uehara A, Sugawara Y, Kurata S, Fujimoto Y, Fukase K, Kusumoto S, et al. Chemically synthesized pathogen-associated molecular patterns increase the expression of peptidoglycan recognition proteins via toll-like receptors, NOD1 and NOD2 in human oral epithelial cells. Cell Microbiol. 2005;7:675–686. doi: 10.1111/j.1462-5822.2004.00500.x. [DOI] [PubMed] [Google Scholar]
- [112].Wang ZM, Li X, Cocklin RR, Wang M, Fukase K, Inamura S, et al. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J Biol Chem. 2003;278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- [113].Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem Biophys Res Commun. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- [114].Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS One. 2016;11:e0146162. doi: 10.1371/journal.pone.0146162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kashyap DR, Wang M, Liu LH, Boons GJ, Gupta D, Dziarski R. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat Med. 2011;17:676–683. doi: 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kietzman C, Tuomanen E. PGRPs kill with an ancient weapon. Nat Med. 2011;17:665–666. doi: 10.1038/nm0611-665. [DOI] [PubMed] [Google Scholar]
- [117].Dziarski R, Platt KA, Gelius E, Steiner H, Gupta D. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood. 2003;102:689–697. doi: 10.1182/blood-2002-12-3853. [DOI] [PubMed] [Google Scholar]
- [118].Cho JH, Fraser IP, Fukase K, Kusumoto S, Fujimoto Y, Stahl GL, et al. Human peptidoglycan recognition protein S is an effector of neutrophil-mediated innate immunity. Blood. 2005;106:2551–2558. doi: 10.1182/blood-2005-02-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Read CB, Kuijper JL, Hjorth SA, Heipel MD, Tang X, Fleetwood AJ, et al. Cutting Edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J Immunol. 2015;194:1417–1421. doi: 10.4049/jimmunol.1402303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Park SY, Jing X, Gupta D, Dziarski R. Peptidoglycan recognition protein 1 enhances experimental asthma by promoting Th2 and Th17 and limiting regulatory T cell and plasmacytoid dendritic cell responses. J Immunol. 2013;190:3480–3492. doi: 10.4049/jimmunol.1202675. [DOI] [PMC free article] [PubMed] [Google Scholar]