Abstract
Long before bacteria infected humans, they infected amoebas, which remain a potentially important reservoir for human disease. Diverse soil amoebas including Dictyostelium and Acanthamoeba can host intracellular bacteria. Though the internal environment of free-living amoebas is similar in many ways to that of mammalian macrophages, they differ in a number of important ways, including temperature. A new study in PLOS Biology by Taylor-Mulneix et al. demonstrates that Bordetella bronchiseptica has two different gene suites that are activated depending on whether the bacterium finds itself in a hot mammalian or cool amoeba host environment. This study specifically shows that B. bronchiseptica not only inhabits amoebas but can persist and multiply through the social stage of an amoeba host, Dictyostelium discoideum.
Environmental amoebas came before animals as hosts to bacteria
The bacteria that most concern us are those that make us sick, but we are sometimes so preoccupied with our battle with them that we forget they have been waging a much longer war. More than a billion (109) years before the first animals, bacteria were evolving strategies first to resist being killed by protozoan predators and then to actually infect their former predators [1]. These strategies are likely to have laid the groundwork for the later evolution of animal–bacteria interactions, so understanding how they function provides an essential context for understanding modern-day bacterial pathogens in humans. This is particularly true for the bacteria that invade animals through macrophages [2]. Further, environmental amoebas are still ubiquitous in modern soil and water, so they may act as important reservoirs from which emerging human diseases can arise [3]. Many amoebas, including Acanthamoeba castellanii, D. discoideum, Hartmannella vermiformis, and Naegleria gruberi, have been found to harbor bacteria [4]. Bacteria that can defeat amoebas’ defenses gain a refuge in which to proliferate, where they are protected from hostile external conditions by their unwitting hosts [5–8].
It is worth pointing out that amoebas do not fall into a monophyletic group but instead share a life form and a diet based on phagocytosis. The bacteria that can evade amoeba defenses are called amoeba-resistant bacteria [3,4]. In these amoebas, resistant bacteria can survive, proliferate, and be protected in adverse situations, particularly when the host amoeba forms a hardy cyst with the bacteria inside.
Glossary
Amoeba-resistant bacteria: Bacteria that have evolved to resist being killed by free-living amoebas.
Bacterial secretion system: The mechanisms by which bacterial pathogens evolved to export various virulence factors across the phospholipid membrane and cell envelope.
Ejectosome: A peripheral cellular organelle responsible for ejecting cytosolic bacteria from the cell without lysing that cell.
Fruiting body: A multicellular structure on which spore-producing structures are borne.
Free-living amoebas: Widely distributed protozoa that have the ability to alter their shape and feed on bacteria, algae, fungi, and small organic particles.
Lysosome: A membrane-bound organelle that contains hydrolytic enzymes that can break down biomolecules.
Phagocytosis: The process by which a cell engulfs a solid particle to form an internal compartment known as a phagosome.
Phagosome: A vacuole formed around a particle engulfed by phagocytosis.
Symbiosis: A relationship between individuals of different species that live closely together.
Two-component regulatory system: One kind of mechanism of signal transduction that allows organisms to sense and respond to a changing environment.
Spore: A unit of sexual or asexual reproduction that is able to disperse and survive in unfavorable conditions.
Virulence factor: Molecules produced by pathogens that can increase their fitness in interactions with the host.
Survival strategies of intracellular bacteria within amoebas
Entry of bacteria into amoebas is simple because amoebas eat bacteria. Amoebas normally engulf food bacteria by phagocytosis and kill them inside the phagosome, where ingested bacteria are confronted with acidification, oxidative stress, nutrient deprivation, and various antimicrobial small molecules [2] [9,10]. Amoeba grazing has been suggested to be one of the major forces shaping bacterial abundance and diversity [11]. However, some bacteria have developed strategies to survive phagocytosis by amoebas and are able to exploit host cell resources. Bacteria like Legionella pneumophila that remain in the vacuole of macrophages in humans are perhaps the best-studied bacteria that infect humans and amoebas, but they are by no means the only ones (Table 1) [12,13].
Table 1. List of human pathogens that are found in free-living amoebas.
These bacteria are isolated from various amoeba hosts and have different lifestyles [8,14–16]. They have evolved sophisticated ways to export various virulence factors across their bacterial inner and sometimes outer membrane (in gram-negative bacteria), as well as through the host plasma membrane or phagosomal membrane, by using diverse secretion systems [17,18].
| Bacteria | Amoeba hosts | Location in amoebas | Bacterial secretion systems known to be present | Human diseases |
|---|---|---|---|---|
| β proteobacteria | ||||
| Burkholderia cepacia | Acanthamoeba | Extracellular | Type III secretion system; type VI secretion system | Pneumonia |
| Bu. pseudomallei | Acanthamoeba | Extracellular | Type III secretion system; type VI secretion system | Melioidosis |
| Burkholderia spp. | Dictyostelium | Facultative intracellular | Unknown | Unknown |
| γ proteobacteria | ||||
| Coxiella burnetii | Acanthamoeba | Obligate intracellular | Dot/Icm type IVB secretion system | Q fever |
| Escherichia coli O157 | Acanthamoeba | Extracellular | Type III secretion system; Tat secretion pathway | Hemorrhagic diarrhea; kidney failure |
| Francisella tularensis | Acanthamoeba | Facultative intracellular | Type VI secretion system | Tularemia |
| L. pneumophila | Various amoebas | Facultative intracellular | Type II secretion system; type IV secretion system; Tat secretion pathway | Legionnaires disease |
| L. anisa | Acanthamoeba | Facultative intracellular | Unknown | Pontiac fever; Legionnaires disease |
| Pseudomonas aeruginosa | Acanthamoeba | Extracellular | Tat secretion pathway; Type VI secretion system | Infect human cells |
| Vibrio cholerae | Acanthamoeba, Naegleria | Extracellular | Type I secretion system; type II secretion system; type VI secretion system | Cholera |
| ɛ proteobacteria | ||||
| Helicobacter pylori | Acanthamoeba | Facultative intracellular | Type IV secretion system | Asymptomatic disease |
| Chlamydia | ||||
| Chlamydophila pneumoniae | Acanthamoeba | Obligate intracellular | Type III secretion system | Pneumonia |
| Neochlamydia hartmanellae | Hartmannella | Obligate intracellular | Type III secretion system | Infect human cells |
| Parachlamydia acanthamoebae | Acanthamoeba | Obligate intracellular | Type III secretion system | Infect human cells |
| Simkania negevensis | Acanthamoeba | Obligate intracellular | Type III secretion system | Chronic obstructive pulmonary disease |
| Bacilli | ||||
| Listeria monocytogenes | Acanthamoeba | Facultative intracellular | Type VII secretion system | Listeriosis |
| Bacillus anthracis | Acanthamoeba | Obligate intracellular | Type IV secretion system | Anthrax |
| Actinobacteria | ||||
| Mycobacterium leprae | Acanthamoeba | Obligate intracellular | Type VII secretion system | Leprosy |
| M. avium | Acanthamoeba | Facultative intracellular | Type VII secretion system | Mycobacterium avium-intracellulare infection |
| M. marinum | Acanthamoeba | Facultative intracellular | Type VII secretion system | Opportunistic infections; aquarium granuloma |
| M. ulcerans | Acanthamoeba | Facultative intracellular | Type VII secretion system | Buruli ulcer |
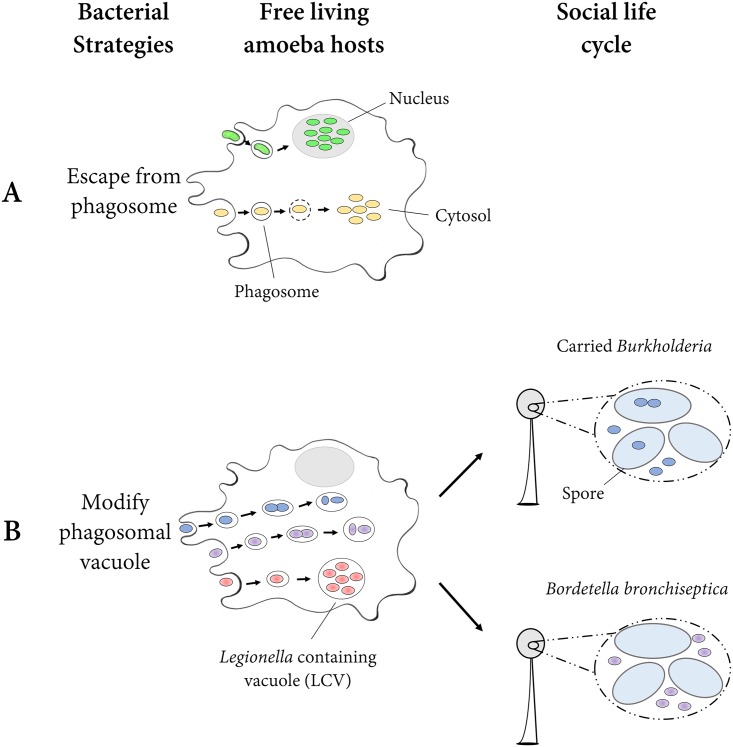
The most obvious strategy to avoid being killed by the amoeba host is to escape from its vacuole into the cytosol of the amoeba (Fig 1A). For example, M. marinum and M. tuberculosis have evolved this ability (Fig 1A, yellow). This process requires the mycobacterial type VII secretion system ESX-1 [12]. In addition, both M. marinum and M. tuberculosis can eject from the cell through an F-actin structure called an ejectosome and then spread from cell to cell [19,20].
Fig 1. Diagram of survival strategies of intracellular bacteria within amoebas.
The figure represents two general strategies that intracellular bacteria deploy to survive within amoebas. They can escape from the phagosome (Fig 1A) or stay within the phagosomal vacuole but modify it (Fig 1B). Green, intranuclear bacteria; yellow, bacteria that escape into the cytosol; blue, carried Burkholderia; purple, B. bronchiseptica; red, L. pneumophila.
In general, the cytosol is considered to be favorable for bacterial growth because it provides nutrients and is isolated from the host immune system [21]. Therefore, it is an ideal place for bacteria to thrive after escaping from the phagosome. Some intracellular pathogens can invade a more unusual intracellular niche: the eukaryotic nucleus (Fig 1A, green). This has been reported in the free-living amoebas—for example, a bacterium called strain Pn in Chlamydiae was found in nuclei of N. clarki [22]. A bacterium tentatively called “Candidatus Nucleicultrix amoepiphila” and distantly related to the Rickettsiales was found in nuclei of Hartmannella sp. [23]. Apparently, these two evolved the intranuclear habit independently.
The second strategy bacteria employ is to stay within the phagosomal vacuole but to subvert its antimicrobial mechanisms (Fig 1B). These subversion techniques include preventing phagosome-lysosome fusion, modulating phagosomal pH, damaging phagosomal membranes, and/or quenching oxidative bursts [5]. Intracellular pathogens use a combination of these approaches. For instance, L. pneumophila (Fig 1B, red) has evolved a complex system that allows the bacteria to hijack the phagocytic vacuole [24]. It evades the endocytic pathway and the subsequent phagosome-lysosome fusion, delays its acidification, and establishes a safe intracellular niche called a Legionella-containing vacuole (LCV), which allows intracellular replication [24,25]. Further studies suggest that L. pneumophila uses the Icm/Dot type IV secretion system (T4SS) and the Lsp type II secretion system (T2SS) to avoid death and to exploit host resources [24,26]. Other bacteria use similar strategies [12].
The well-studied amoeba D. discoideum adds another wrinkle to the story of amoeba–bacteria interactions. This social amoeba in the Amoebozoa and others in its family behave like other soil amoebas most of the time, eating bacteria and dividing by binary fission. But when they cease to find sufficient food bacteria, the amoebas aggregate by the tens of thousands into a multicellular slug that moves towards heat and light [27]. Ultimately, this slug forms a fruiting body in which about 20% of cells (formerly independent amoebas) die to form a sturdy stalk, and the remaining cells form hardy spores atop the stalk, where they are more likely to be transported [27,28].
Bacteria can exploit this amoeba [29,30]. Some bacteria can also remain inside the spores through the social cycle. Burkhoderia near fungorum is one such bacterium (Fig 1B). In fact, this and other strains of Burkholderia so change the phagosome machinery that D. discoideum infected with them can also carry food bacteria, which would otherwise be digested (Fig 1B, blue) [31–34]. These amoeba clones are called farmers because they can seed and harvest their crops in new environments [34].
Overall, the majority of intracellular pathogens of amoebas occupy phagosomal vacuoles, while only some are able to escape the phagosome [5]. This is possibly because specialized mechanisms are needed to escape from the phagosome [5,21]. There is no clear relationship between the type of survival strategies and whether the microbe is an obligate or facultative intracellular pathogen [5].
Interactions between B. bronchiseptica and amoebas
We began this piece by noting that amoebas antedated animals on the planet by more than a billion years. If bacteria began their infectious lives in soil and water, then we expect those lineages to be more ancient than those from animals. There is a comprehensive and recent study on this topic for B. bronchiseptica, which is a bacterium in the gram-negative Betaproteobacteria [35]. It causes respiratory infections in some species of mammals and is closely related to B. pertussis, which causes whooping cough in humans, accounting for about 89,000 deaths worldwide in 2008, according to the World Health Organization.
Soumana et al. constructed a phylogeny of Bordetella strains collected from environmental sources and from animals [36]. To do this, Soumana et al. searched the National Center for Biotechnology Information (NCBI) database for 16s ribosomal RNA sequence matches to several species of Bordetella and tied what they found to the sequence sources [36]. A neighbor-joining tree based on the 16S rRNA sequences indicated that environmental isolates were basal, as predicted [36].
This is not the only interesting thing about Bordetella. Most studies of amoeba–bacteria interactions take advantage of the similarities between amoebas and macrophages that are attributable to both having phagocytic activity [12,24]. While there are powerful advantages to using amoebas instead of animals as experimental hosts for bacteria, environmental amoebas generally live at much cooler temperatures (~21°C) than macrophages inside the human body (~37°C).
B. bronchiseptica has a two-component signal transduction system called BvgAS that regulates two distinct phases, the virulent Bvg+ phase and the avirulent Bvg− phase [37]. These systems operate differently at low and high temperatures [35]. At a higher temperature, virulence in the mammal host is regulated by Bvg+, which controls expression of over 100 genes [35]. At cooler temperatures, an equally large set of genes is activated in the Bvg− state. The latter genes allow growth at lower nutrient concentrations and turn on flagellar movement [35]. It turns out that the Bvg− state is what allows B. bronchiseptica to survive inside soil amoebas, including in the lab amoeba D. discoideum [35].
B. bronchiseptica remained present and alive after an hour when added to a culture of D. discoideum with the antibiotic gentamicin. By contrast, B. bronchiseptica could not survive an hour in the absence of D. discoideum with the same antibiotic. A standard food bacterium given to D. discoideum (namely, Klebsiella pneumoniae) was not present after an hour in either case, while the B. bronchiseptica bacteria were protected inside the amoebas. This result was confirmed with a similar experiment allowing B. bronchiseptica to invade another amoeba species distantly related to D. discoideum, A. castellanii.
When D. discoideum went through the social stage, B. bronchiseptica came right along, though outside the spores, which made it vulnerable at this stage to antibiotics (Fig 1B, purple). Not only did B. bronchiseptica bacteria survive in the fruiting bodies, but when the fruiting bodies were diluted 10-fold and replated on a new lawn of food, B. bronchiseptica proliferated right along with D. discoideum. This success of proliferation and survival in amoebas is due to the expression of the Bvg− system, something the authors demonstrated by showing how many fewer cells of a clone locked in the Bvg+ stage proliferated compared to either wild type or a clone locked in the Bvg− stage [35]. The authors further demonstrated that after passaging through spores of D. discoideum, the B. bronchiseptica were able to infect mouse respiratory tracts [35].
Bordetella is an ancient genus of bacteria that probably attacked environmental amoebas first but now also causes respiratory illness in mammals; this genus includes B. pertussis, which attacks only humans and is unable to survive in the environment [36].
Nevertheless, questions remain. Is B. bronchiseptica found in wild strains of D. discoideum or other species of Dictyostelium? Do other bacteria that invade both amoebas and animals have different sets of genes to adapt to both? Furthermore, a comprehensive survey of bacteria found in wild amoebas awaits future studies. Perhaps most insightful will be further discoveries of bacterial sequences in sequenced amoeba genomes.
Conclusions
As McFall-Ngai and coauthors so nicely put it, animals evolved in a world that already contained billions of bacteria, archaea, and amoebas [38]. Thus, it is no surprise that some bacterial pathogens of humans and other mammals not only came from ancestors that attacked amoebas but often retained that ability over evolutionary time. These new and exciting results tell the detailed story of how a bacterium can exploit the social cycle of an amoeba and completely change the virulence genes it deploys according to whether it is attacking a hot mammal or a chilly amoeba. This example is likely to be only the first of many careful studies that reveal exactly how bacteria pull off these tricks.
Acknowledgments
We thank Tyler Larsen and David Queller for extremely helpful comments.
Abbreviations
- LCV
Legionella-containing vacuole
- NCBI
National Center for Biotechnology Information
- T2SS
type II secretion system
- T4SS
type IV secretion system
Funding Statement
The authors received no specific funding for this work.
Footnotes
Provenance: Commissioned; externally peer reviewed.
References
- 1.Brüssow H (2007) Bacteria between protists and phages: from antipredation strategies to the evolution of pathogenicity. Molecular microbiology 65: 583–589. 10.1111/j.1365-2958.2007.05826.x [DOI] [PubMed] [Google Scholar]
- 2.Haas A (2007) The phagosome: compartment with a license to kill. Traffic 8: 311–330. 10.1111/j.1600-0854.2006.00531.x [DOI] [PubMed] [Google Scholar]
- 3.Scheid P (2014) Relevance of free-living amoebae as hosts for phylogenetically diverse microorganisms. Parasitology research 113: 2407–2414. 10.1007/s00436-014-3932-7 [DOI] [PubMed] [Google Scholar]
- 4.Denoncourt AM, Paquet VE, Charette SJ (2014) Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Frontiers in microbiology 5: 240 10.3389/fmicb.2014.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A (2008) Evolution of intracellular pathogens. Annu Rev Microbiol 62: 19–33. 10.1146/annurev.micro.61.080706.093305 [DOI] [PubMed] [Google Scholar]
- 6.Guimaraes AJ, Gomes KX, Cortines JR, Peralta JM, Peralta RHS (2016) Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiological Research 193: 30–38. 10.1016/j.micres.2016.08.001 [DOI] [PubMed] [Google Scholar]
- 7.Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S (2007) Environmental predators as models for bacterial pathogenesis. Environmental microbiology 9: 563–575. 10.1111/j.1462-2920.2007.01238.x [DOI] [PubMed] [Google Scholar]
- 8.Greub G, Raoult D (2004) Microorganisms resistant to free-living amoebae. Clinical microbiology reviews 17: 413–433. 10.1128/CMR.17.2.413-433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosson P, Lima WC (2014) Intracellular killing of bacteria: is Dictyostelium a model macrophage or an alien? Cellular Microbiology 16: 816–823. 10.1111/cmi.12291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.German N, Doyscher D, Rensing C (2013) Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiology 8: 1257–1264. 10.2217/fmb.13.100 [DOI] [PubMed] [Google Scholar]
- 11.Matz C, Kjelleberg S (2005) Off the hook–how bacteria survive protozoan grazing. Trends in microbiology 13: 302–307. 10.1016/j.tim.2005.05.009 [DOI] [PubMed] [Google Scholar]
- 12.Steinert M. Pathogen–host interactions in Dictyostelium, Legionella, Mycobacterium and other pathogens; 2011. Elsevier; pp. 70–76. [DOI] [PubMed] [Google Scholar]
- 13.Tosetti N, Croxatto A, Greub G (2014) Amoebae as a tool to isolate new bacterial species, to discover new virulence factors and to study the host–pathogen interactions. Microbial pathogenesis 77: 125–130. 10.1016/j.micpath.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 14.Tosetti N, Croxatto A, Greub G (2014) Amoebae as a tool to isolate new bacterial species, to discover new virulence factors and to study the host-pathogen interactions. Microbial Pathogenesis 77: 125–130. 10.1016/j.micpath.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 15.Bozzaro S, Eichinger L (2011) The Professional Phagocyte Dictyostelium discoideum as a Model Host for Bacterial Pathogens. Current Drug Targets 12: 942–954. 10.2174/138945011795677782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinert M (2011) Pathogen-host interactions in Dictyostelium, Legionella, Mycobacterium and other pathogens. Seminars in Cell & Developmental Biology 22: 70–76. [DOI] [PubMed] [Google Scholar]
- 17.Green ER, Mecsas J (2016) Bacterial Secretion Systems: An Overview. Microbiology Spectrum 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, et al. (2015) Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nature Reviews Microbiology 13: 343–359. 10.1038/nrmicro3456 [DOI] [PubMed] [Google Scholar]
- 19.Hagedorn M, Rohde KH, Russell DG, Soldati T (2009) Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323: 1729–1733. 10.1126/science.1169381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, et al. (2015) The autophagic machinery ensures nonlytic transmission of mycobacteria. Proceedings of the National Academy of Sciences of the United States of America 112: E687–E692. 10.1073/pnas.1423318112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray K, Marteyn B, Sansonetti PJ, Tang CM (2009) Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nature Reviews Microbiology 7: 333–340. 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- 22.Schulz F, Horn M (2015) Intranuclear bacteria: inside the cellular control center of eukaryotes. Trends in Cell Biology 25: 339–346. 10.1016/j.tcb.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 23.Schulz F, Lagkouvardos I, Wascher F, Aistleitner K, Kostanjsek R, et al. (2014) Life in an unusual intracellular niche: a bacterial symbiont infecting the nucleus of amoebae. ISME Journal 8: 1634–1644. 10.1038/ismej.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann C, Harrison CF, Hilbi H (2014) The natural alternative: protozoa as cellular models for Legionella infection. Cellular Microbiology 16: 15–26. 10.1111/cmi.12235 [DOI] [PubMed] [Google Scholar]
- 25.Escoll P, Rolando M, Gomez-Valero L, Buchrieser C (2013) From amoeba to macrophages: exploring the molecular mechanisms of Legionella pneumophila infection in both hosts. Curr Top Microbiol Immunol 376: 1–34. 10.1007/82_2013_351 [DOI] [PubMed] [Google Scholar]
- 26.Hubber A, Kubori T, Nagai H (2014) Modulation of the Ubiquitination Machinery by Legionella. Molecular Mechanisms in Legionella Pathogenesis 376: 227–247. [DOI] [PubMed] [Google Scholar]
- 27.Kessin RH (2001) Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge UK: Cambridge University Press; 308 p. [Google Scholar]
- 28.Smith J, Queller DC, Strassmann JE (2014) Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC evolutionary biology 14: 105 10.1186/1471-2148-14-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagele S, Kohler R, Merkert H, Schleicher M, Hacker J, et al. (2000) Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol 2: 165–171. [DOI] [PubMed] [Google Scholar]
- 30.Bozzaro S, Bucci C, Steinert M (2008) Phagocytosis and host–pathogen interactions in Dictyostelium with a look at macrophages. International review of cell and molecular biology 271: 253–300. 10.1016/S1937-6448(08)01206-9 [DOI] [PubMed] [Google Scholar]
- 31.DiSalvo S, Haselkorn TS, Bashir U, Jimenez D, Brock DA, et al. (2015) Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. Proceedings of the National Academy of Sciences 112: E5029–E5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stallforth P, Brock DA, Cantley AM, Tian X, Queller DC, et al. (2013) A bacterial symbiont is converted from an inedible producer of beneficial molecules into food by a single mutation in the gacA gene. Proceedings of the National Academy of Sciences 110: 14528–14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brock D, Read S, Bozchenko A, Queller DC, Strassmann JE (2013) Social amoeba farmers carry bacterial weapons to protect and privatize their crops. Nature Communications 4: 2385 10.1038/ncomms3385 [DOI] [PubMed] [Google Scholar]
- 34.Brock DA, Douglas TE, Queller DC, Strassmann JE (2011) Primitive agriculture in a social amoeba. Nature 469: 393–396. 10.1038/nature09668 [DOI] [PubMed] [Google Scholar]
- 35.Taylor-Mulneix DL, Bendor L, Linz B, Rivera I, Ryman VE, Dewan KK, et al. Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLoS Biol. 2017;15: e2000420 10.1371/journal.pbio.2000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soumana IH, Linz B, Harvill ET (2017) Environmental Origin of the Genus Bordetella. Frontiers in Microbiology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuk MH, Harvill ET, Miller JF (1998) The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Molecular Microbiology 28: 945–959. [DOI] [PubMed] [Google Scholar]
- 38.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, et al. (2013) Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences 110: 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]