Abstract
We introduce a general framework for estimation of inverse covariance, or precision, matrices from heterogeneous populations. The proposed framework uses a Laplacian shrinkage penalty to encourage similarity among estimates from disparate, but related, subpopulations, while allowing for differences among matrices. We propose an efficient alternating direction method of multipliers (ADMM) algorithm for parameter estimation, as well as its extension for faster computation in high dimensions by thresholding the empirical covariance matrix to identify the joint block diagonal structure in the estimated precision matrices. We establish both variable selection and norm consistency of the proposed estimator for distributions with exponential or polynomial tails. Further, to extend the applicability of the method to the settings with unknown populations structure, we propose a Laplacian penalty based on hierarchical clustering, and discuss conditions under which this data-driven choice results in consistent estimation of precision matrices in heterogenous populations. Extensive numerical studies and applications to gene expression data from subtypes of cancer with distinct clinical outcomes indicate the potential advantages of the proposed method over existing approaches.
Keywords and phrases: Graph Laplacian, graphical modeling, heterogeneous populations, hierarchical clustering, high-dimensional estimation, precision matrix, sparsity
1. Introduction
Estimation of large inverse covariance, or precision, matrices has received considerable attention in recent years. This interest is in part driven by the advent of high-dimensional data in many scientific areas, including high throughput omics measurements, functional magnetic resonance images (fMRI), and applications in finance and industry. Applications of various statistical methods in such settings require an estimate of the (inverse) covariance matrix. Examples include dimension reduction using principal component analysis (PCA), classification using linear or quadratic discriminant analysis (LDA/QDA), and discovering conditional independence relations in Gaussian graphical models (GGM).
In high-dimensional settings, where the data dimension p is often comparable or larger than the sample size n, regularized estimation procedures often result in more reliable estimates. Of particular interest is the use of sparsity inducing penalties, specifically the ℓ1 or lasso penalty [30], which encourages sparsity in off-diagonal elements of the precision matrix [7, 8, 33, 34]. Theoretical properties of ℓ1-penalized precision matrix estimation have been studied under both multivariate normality, as well as some relaxations of this assumption [4, 19, 25, 26].
Sparse estimation is particularly relevant in the setting of GGMs, where conditional independencies among variables correspond to zero off-diagonal elements of the precision matrix [14]. The majority of existing approaches for estimation of high-dimensional precision matrices, including those cited in the previous paragraph, assume that the observations are identically distributed, and correspond to a single population. However, data sets in many application areas include observations from several distinct subpopulations. For instance, gene expression measurements are often collected for both healthy subjects, as well as patients diagnosed with different subtypes of cancer. Despite increasing evidence for differences among genetic networks of cancer and healthy subjects [11, 27], the networks are also expected to share many common edges. Separate estimation of graphical models for each of the subpopulations would ignore the common structure of the precision matrices, and may thus be inefficient; this inefficiency can be particularly significant in high-dimensional low sample settings, where p ≫ n.
To address the need for estimation of graphical models in related subpopulations, few methods have been recently proposed for joint estimation of K precision matrices , k = 1, …, K [6, 9]. These methods extend the penalized maximum likelihood approach by combining the Gaussian likelihoods for the K subpopulations
| (1) |
Here, nk and are the number of observations and the sample covariance matrix for the kth subpopulation, respectively, is the total sample size and tr(·) and det(·) denote matrix trace and determinant.
To encourage similarity among estimated precision matrices, Guo et al. [9] modeled the (i, j)-element of Ω(k) as product of a common factor θij and group-specific parameters , i.e. . Identifiability of the estimates is ensured by assuming δij ≥ 0. A zero common factor δij = 0 induces sparsity across all subpopulations, whereas results in condition-specific sparsity for . This reparametrization results in a non-convex optimization problem based on the Gaussian likelihood with ℓ1-penalties ∑i≠j δij and . Danaher et al. [6] proposed two alternative estimators by adding an additional convex penalty to the graphical lasso objective function: either a fused lasso penalty (FGL), or a group lasso penalty (GGL). The fused lasso penalty has also been used by Kolar et al. [13], for joint estimation of multiple graphical models in multiple time points. The fused lasso penalty strongly encourages the values of to be similar across all subpopulations, both in values as well as sparsity patterns. On the other hand, the group lasso penalty results in similar estimates by shrinking all across subpopulations to zero if is small.
Despite their differences, methods of Guo et al. [9] and Danaher et al. [6] inherently assume that precision matrices in K subpopulations are equally similar to each other, in that they encourage and and and to be equally similar. However, when K > 2, some subpopulations are expected to be more similar to each other than others. For instance, it is expected that genetic networks of two subtypes of cancer be more similar to each other than to the network of normal cells. Similarly, differences among genetic networks of various strains of a virus or bacterium are expected to correspond to the evolutionary lineages of their phylogenetic trees. Unfortunately, existing methods for joint estimation of multiple graphical models ignore this heterogeneity in multiple subpopulations. Furthermore, existing methods assume subpopulation memberships are known, which limits their applicability in settings with complex but unknown population structures; an important example is estimation of genetic networks of cancer cells with unknown subtypes.
In this paper, we propose a general framework for joint estimation of multiple precision matrices by capturing the heterogeneity among subpopulations. In this framework, similarities among disparate subpopulations are presented using a subpopulation networkG(V, E, W), a weighted graph whose node set V is the set of subpopulations. The edges in E and the weights Wkk′ for (k, k′) ∈ E represent the degree of similarity between any two subpopulations k, k′. In the special case where Wkk′ = 1 for all k, k′, the subpopulation similarities are only captured by the structure of the graph G. An example of such a subpopulation network is the line graph corresponding to observations over multiple time points, which is used in estimation of time-varying graphical models [13]. As we will show in Section 2.3, other existing methods for joint estimation of multiple graphical models, e.g. proposals of Danaher et al. [6], can also be seen as special cases of this general framework.
Our proposed estimator is the solution to a convex optimization problem based on the Gaussian likelihood with both ℓ1 and graph Laplacian [15] penalties. The graph Laplacian has been used in other applications for incorporating a priori knowledge in classification [24], for principal component analysis on network data [28], and for penalized linear regression with correlated covariates [10, 15, 17, 18, 32, 37]. The Laplacian penalty encourages similarity among estimated precision matrices according to the subpopulation network G. The ℓ1-penalty, on the other hand, encourages sparsity in the estimated precision matrices. Together, these two penalties capture both unique patterns specific to each subpopulation, as well as common patterns shared among different subpopulations.
We first discuss the setting where G(V, E, W) is known from external information, e.g. known phylogenetic trees (Section 2), and later discuss the estimation of the subpopulation memberships and similarities using hierarchical clustering (Section 4). We propose an alternating methods of multipliers (ADMM) algorithm [3] for parameter estimation, as well as its extension for efficient computation in high dimensions by decomposing the problem into block-diagonal matrices. Although we use the Gaussian likelihood, our theoretical results also hold for non-Gaussian distributions. We establish model selection and norm consistency of the proposed estimator under different model assumptions (Section 3), with improved rates of convergence over existing methods based on penalized likelihood. We also establish the consistency of the proposed algorithm for the estimation of multiple precision matrices, in settings where the subpopulation network G or subpopulation memberships are unknown. To achieve this, we establish the consistency of hierarchical clustering in high dimensions, by generalizing recent results of Borysov et al. [1] to the setting of arbitrary covariance matrices, which is of independent interest.
The rest of the paper is organized as follows. In Section 2 we describe the formal setup of the problem and present our estimator. Theoretical properties of the proposed estimator are studied in Section 3, and Section 4 discusses the extension of the method to the setting where the subpopulation network is unknown. The ADMM algorithm for parameter estimation and its extension for efficient computation in high dimensions are presented in Section 5. Results of the numerical studies, using both simulated and real data examples, are presented in Section 6. Section 7 concludes the paper with a discussion. Technical proofs are collected in the Appendix.
2. Model and Estimator
2.1. Problem Setup
Consider K subpopulations with distributions ℘(k), k = 1, …, K. Let X(k) = (X(k),1, …, X(k),p)T ∈ ℝp be a random vector from the kth subpopulation with mean μk and the covariance matrix . Suppose that an observation comes from the kth subpopulation with probability πk > 0.
Our goal is to estimate the precision matrices , k = 1, …, K. To this end, we use the Gaussian log-likelihood based on the correlation matrix (see Rothman et al. [26]) as a working model for estimation of true , k = 1, …, K. Let , i = 1, …, nk, be independent and identically distributed (i.i.d.) copies from ℘(k), k = 1, …, K. We denote the correlation matrices and their inverse by , and , k = 1, …, K, respectively. The Gaussian log-likelihood based on the correlation matrix can then be written as
| (2) |
where , k = 1, …, K is the sample correlation matrix for subpopulation k.
Examining the derivative of (2), which consists of , k = 1, …, K, justifies its use as a working model for non-Gaussian data: the stationary points of (2) is , which gives a consistent estimate of . Thus we do not, in general, need to assume multivariate normality. However, in certain applications, for instance LDA/QDA and GGM, the resulting estimate is useful only if the data follows a multivariate normal distribution.
2.2. The Laplacian Shrinkage Estimator
Let Θ = (Θ(1), …, Θ(K)) and write , i, j = 1, …, p for a vector of (i, j)-elements across subpopulations. Our proposed estimator, Laplacian Shrinkage for Inverse Covariance matrices from Heterogeneous populations (LASICH), first estimates the inverse of the correlation matrices for each of the K subpopulations, and then transforms them into the estimator of inverse covariance matrices, as in Rothman et al. [26]. In particular, we first obtain the estimate Θ̂ of the true inverse correlation matrix by solving the following optimization problem
| (3) |
where Θ = ΘT enforces the symmetry of individual inverse correlation matrices, i.e. Θ(k) = (Θ(k))T, and Θ ≻ 0 requires that Θ(k) is positive definite for k = 1, …, K. The ℓ1-penalty in (3) encourages sparsity in estimated inverse correlation matrices. The graph Laplacian penalty, on the other hand, exploits the information in the subpopulation network G to encourage similarity among values of and . The tuning parameters ρn and ρnρ2 control the size of each penalty term.
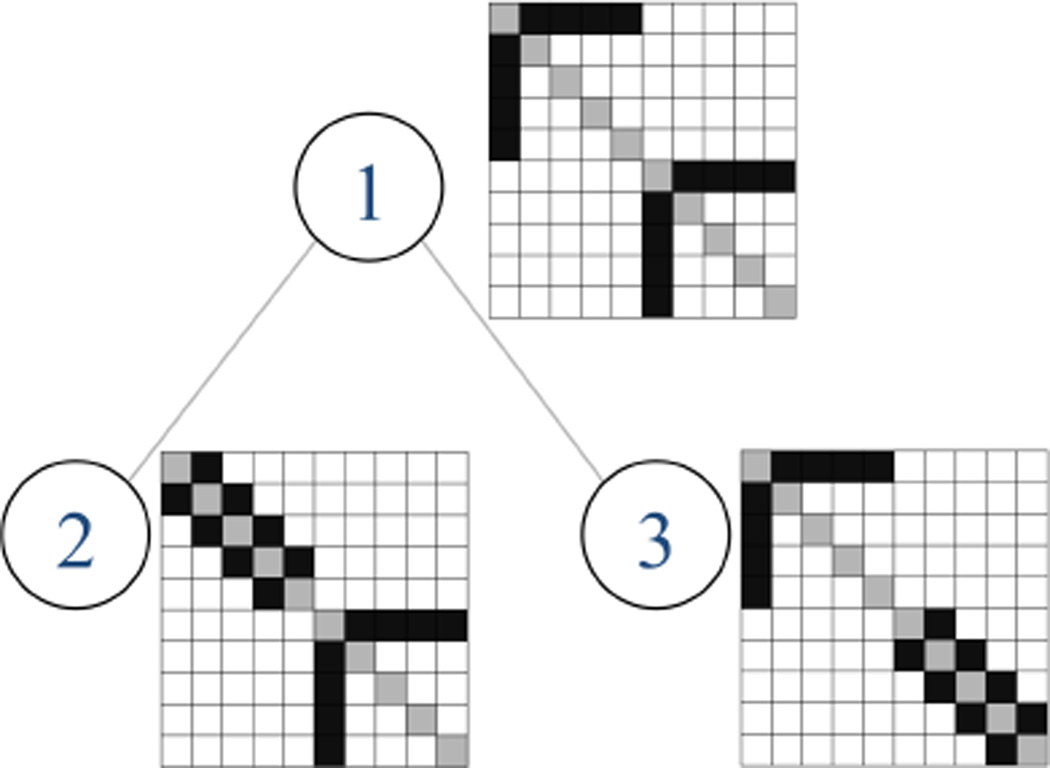
Figure 1 illustrates the motivation for the graph Laplacian penalty ‖Θij‖L in (3). The gray-scale images in the figure show the hypothetical sparsity patterns of precision matrices Θ(1), Θ(2), Θ(3) for three related subpopulations. Here, Θ(1) consists of two blocks with one “hub” node in each block; in Θ(2) and Θ(3) one of the blocks is changed into a “banded” structure. It can be seen that one of the two blocks in both Θ(2) and Θ(3) have a similar sparsity pattern as Θ(1). However, Θ(2) and Θ(3) are not similar. The subpopulation network G in this figure captures the relationship among precision matrices of the three subpopulations. Such complex relationships cannot be captured using the existing approaches, e.g. Danaher et al. [6], Guo et al. [9], which encourage all precision matrices to be equally similar to each other. More generally, G can be a weighted graph, G(V, E, W), whose nodes represent the subpopulations 1, …, K. The edge weights W : E → ℝ+ represent the similarity among pairs of subpopulations, with larger values of Wkk′ ≡ W (k, k′) > 0 corresponding to more similarity between precision matrices of subpopulations k and k′.
Fig 1.
Illustration of similarities in the sparsity patterns of precision matrices Ω(1), Ω(2) and Ω(3). Nonzero and zero off-diagonal entries are colored in black and white, respectively, while diagonal entires are colored in gray. The associated subpopulation network G reflects the similarities between precision matrices of subpopulations 1 and 2 and 1 and 3. The simulation experiments in Section 6.1 use a similar subpopulation network in a high-dimensional setting.
In this section, we assume that the weighted graph G is externally available, and defer the discussion of data-driven choices of G, based on hierarchical clustering, to Section 4. Given G, the (unnormalized) graph Laplacian penalty ‖Θij‖L is defined as
| (4) |
where Wkk′ = 0 if k and k′ are not connected. The Laplacian shrinkage penalty can be alternatively written as , where is the Laplacian matrix [5] of the subpopulation network G defined as
where dk = ∑k′≠kWkk′ is the degree of node k in G with Wkk′ = 0 if k and k′ are not connected. The Laplacian shrinkage penalty can also be defined in terms of the normalized graph Laplacian, I − D−1/2W D−1/2, where D = diag(d1, …, dK) is the diagonal degree matrix. The normalized Laplacian penalty,
which we also denote as ‖Θij‖L, imposes smaller shrinkage on coefficients associated with highly connected subpopulations. We henceforth primarily focus on the normalized penalty.
Given estimates of the inverse correlation matrices Θ̂(1), …, Θ̂(K) from (3), we obtain estimates of precision matrices Ω(k) by noting that Ω(k) = Ξ(k)Θ(k)Ξ(k), where Ξ(k) is the diagonal matrix of reciprocals of the standard deviations . Our estimator of precision matrices Ω is thus defined as
where with sample variance for the ith element in the kth subpopulation.
A number of alternative strategies can be used instead of the graph Laplacian penalty in (3). First, similarity among coefficients of precision matrices can also be imposed using a ridge-type penalty, . The main difference is that our penalty ‖Θij‖L discourages the inclusion of edges if they are very different across the K subpopulations. Another option is to use the graph trend filtering [31], which impose a fused lasso penalty over the subpopulation graph G. Finally, ignoring the weights Wkk′ in (4), the Laplacian shrinkage penalty resembles the Markov random field (MRF) prior used in Bayesian variable selection with structured covariates [16]. While our paper was under review, we became aware of the recent work by Peterson et al. [23], who utilize an MRF prior to develop a Bayesian framework for estimation of multiple Gaussian graphical models. This method assumes that edges between pairs of random variable are formed independently, and is hence more suited for Erdős-Rényi networks. Our penalized estimation framework can be seen as an alternative to using an MRF prior to estimate the precision matrices in a mixture of Gaussian distributions.
2.3. Connections to Other Estimators
To connect our proposed estimator to existing methods for joint estimation of multiple graphical models, we first give an alternative interpretation of the graph Laplacian penalty as a norm for a transformed version of . More specifically, consider the mapping gG : ℝK → ℝK defined based on the Laplacian matrix for graph G
if G has at least one edge. For a graph with no edges, define gG(Θij) = IK⊗Θij = diag(Θij), where IK is the K-identity matrix, and ⊗ denotes the Kronecker product. It can then be seen that the graph Laplacian penalty can be rewritten as
where ‖·‖F is the Frobenius norm.
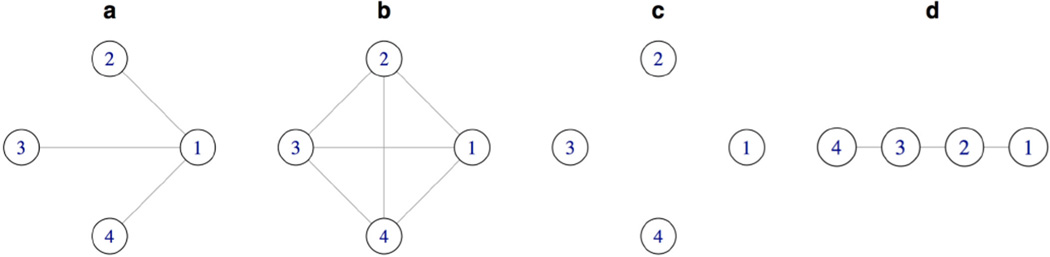
Using the above interpretation, other methods for joint estimation of multiple graphical models can be seen as penalties on transformations gG(Θij) corresponding to different graphs G. We illustrate this connection using the hypothetical subpopulation network shown in Figure 2a.
Fig 2.
Comparison of subpopulation networks used in the penalty for different methods for joint estimation of multiple precision matrices: a) the true network, modeled by LASICH; b) FGL; c) GGL & Guo et al; and d) estimation of time-varying networks (Kolar & Xing, 2009); see Section 2.3 for details.
Consider first the FGL penalty of Danaher et al. [6], applied to elements of the inverse correlation matrix . Let GC be a complete unweighted graph (Wkk′ = 1 ∀k ≠ k′), in which all node-pairs are connected to each other (Figure 2b). It is then easy to see that
where the factor of can be absorbed into the tuning parameter for the FGL penalty. A similar argument can also be applied to the GGL penalty of Danaher et al. [6], ‖Θij‖, by considering instead an empty graph Ge with no edges between nodes (Figure 2c). In this case, the mapping gG would give a diagonal matrix with elements , and hence ‖Θij‖ = ‖gGe(Θij)‖F.
Unlike proposals of Danaher et al. [6], the estimator of Guo et al. [9] is based on a non-convex penalty, and does not naturally fit into the above framework. However, Lemma 2 in Guo et al. [9] establishes a connection between the optimal solutions of the original optimization problem, with those obtained by considering a single penalty of the form . Similar to GGL, the connection with the method of Guo et al. [9] can be build based on the above alternative formulation, by considering again the empty graph Ge (Figure 2c), but instead the ‖·‖1,2 penalty, which is a member of the CAP family of penalties [36]. More specifically,
Using the above framework, it is also easy to see the connection between our proposed estimator and the proposal of Kolar et al. [13]: the total variation penalty in Kolar et al. [13] is closely related to FGL, with summation over differences in consecutive time points. It is therefore clear that the penalty of Kolar et al. [13] (up to constant multipliers) can be obtained by applying the graph Laplacian penalty defined for a line graph connecting the time points (Figure 2d).
The above discussion highlights the generality of the proposed estimator, and its connection to existing methods. In particular, while FGL and GGL/Guo et al. [9] consider extreme cases with isolated, or fully connected nodes, one can obtain more flexibility in estimation of multiple precision matrices by defining the penalty based on the known subpopulation network, e.g. based on phylogenetic trees or spatio-temporal similarities between fMRI samples. The clustering-based approach of Section 4 further extends the applicability of the proposed estimator to the settings where the subpopulation network in not known a priori. The simulation results in Section 6 show that the additional flexibility of the proposed estimator can result in significant improvements in estimation of multiple precision matrices, when K > 2. The above discussion also suggests that other variants of the proposed estimator can be defined, by considering other norms. We leave such extensions to future work.
3. Theoretical Properties
In this section, we establish norm and model selection consistency of the LASICH estimator. We consider a high-dimensional setting p ≫ nk, k = 1, …, K, where both n and p go to infinity. As mentioned in the Introduction, the normality assumption is not required for establishing these results. We instead require conditions on tails of random vectors X(k) for each k = 1, …, K. We consider two cases, exponential tails and polynomial tails, which both allow for distributions other than multivariate normal.
Condition 1 (Exponential Tails)
There exists a constant c1 ∈ (0, ∞) such that
Condition 2 (Polynomial Tails)
There exist constants c2, c3 > 0 and c4such that
Since we adopt the correlation-based Gaussian log-likelihood, we require the boundedness of the true variances to control the error between true and sample correlation matrices.
Condition 3 (Bounded variance)
There exist constants c5 > 0 andc6 < ∞ such that and .
Condition 4 (Sample size)
Let . Let
- (Exponential tails). It holds that
and log p/n → 0. - (Polynomial tails). Let where ρnis given in Lemma 1 in theAppendixand c7 > 0 be some constant. It holds that
Condition 4 determines the sufficient sample size n = Σk for consistent estimation of precision matrices Θ(1), …, Θ(K) in relation to, among other quantities, the number of variables p, the sparsity pattern s and the spectral norm of the Laplacian matrix ‖L‖2 of the subpopulation network G. While a general characterization of ‖L‖2 is difficult, investigating its value in special cases provides insight into the effect of the underlying population structure on the required sample size. Consider, for instance, two extreme cases: for a fully connected graph G associated with K subpopulations, ‖L‖2 = 1/(K − 1); for a minimally connected “line” graph, corresponding to e.g. multiple time points, ‖L‖2 = 2: with K = 5, 30% more samples are needed for the line graph, compared to a fully connected network. The above calculations match our intuition that fewer samples are needed to consistently estimate precision matrices of K subpopulations that share greater similarities. This, of course, makes sense, as information can be better shared when estimating parameters of similar subpopulations. Note that, here L represents the Laplacian matrix of the true subpopulation network capturing the underlying population structure. The above conditions thus do not provide any insight into the effect of misspecifying the relationship between subpopulations, i.e., when an incorrect L is used. This is indeed an important issue that garners additional investigation; see Zhao and Shojaie [37] for some insight in the context of inference for high dimensional regression. In Section 4, we will discuss a data-driven choice of L that results in consistent estimation of precision matrices.
Before presenting the asymptotic results, we introduce some additional notations. For a matrix , we denote the spectral norm ‖A‖2 = maxx∈ℝp,‖x‖=1‖Ax‖, and the element-wise ℓ∞-norm ‖A‖∞ = maxi,j |ai,j| where ‖x‖ is the Euclidean norm for a vector x. We also write the induced ℓ∞-norm ‖A‖∞/∞ = sup‖x‖∞=1‖Ax‖∞ where ‖x‖∞ = maxi |xi| for x = (x1, …, xp). For the ease of presentation, the results in this section are presented in asymptotic form; non-asymptotic results and proofs are deferred to the Appendix.
3.1. Consistency in Spectral Norm
Let , and . The following theorem establishes the rate of convergence of the LASICH estimator, in spectral norm, under either exponential or polynomial tail conditions (Condition 1 or 2). Convergence rates for LASICH in ℓ∞-and Frobenius norm are discussed in Section 3.3.
Theorem 1
Suppose Conditions 3 and 4 hold. Under Condition 1 or 2,
as n, p → ∞ where ρnis given in Lemma 1 in theAppendixwith γ = mink πk/2.
Theorem 1 is proved in the Appendix. The proof builds on tools from Negahban et al. [20]. However, our estimation procedure does not match their general framework: First, we do not penalize the diagonal elements of the inverse correlation matrices; our penalty is thus not a norm. Second, the Laplacian matrix is nonpositive definite. Thus, the Laplacian shrinkage penalty is not strictly convex. The results from Negahban et al. [20] are thus not directly applicable to our problem. To establish the estimation consistency, we first show, in Lemma 3, that the function r(·) = ‖·‖1 + ρ2‖·‖L is a seminorm, and is, moreover, convex and decomposable. We also characterize the subdifferential of this seminorm in Lemma 6, based on the spectral decomposition of the graph Laplacian L. The rest of the proof uses tools from Negahban et al. [20], Rothman et al. [26] and Ravikumar et al. [25], as well as new inequalities and concentration bounds. In particular, in Lemma 4 we establish a new ℓ∞ bound for the empirical covariance matrix for random variables with polynomial tails, which is used to established the consistency in the spectral norm under Condition 2.
The convergence rate in Theorem 1 compares favorably to several other methods based on penalized likelihood. Few results are currently available for estimation of multiple precision matrices. An exception is Guo et al. [9], who obtained a slower rate of convergence Op({(s + p) log p/n}1/2) under the normality assumption and based on a bound on the Frobenius norm. Our rates of convergence are comparable to the results of Rothman et al. [26] for spectral norm convergence of a single precision matrix, obtained under the normality assumption. Ravikumar et al. [25], on the other hand, assumed the irrepresentability condition to obtain the rate Op({min{s + p, d2} log p/n}1/2) and Op({min{s + p, d2}pτ/(c2+c3+1)/n}1/2), under exponential and polynomial tail conditions, respectively, where τ > 2 is some scalar. The rate in Theorem 1 is obtained without assuming the irrepresentability condition. In fact, our rates of convergence are faster than those of Ravikumar et al. [25] given the irrepresentability condition 5 (see Corollary 1). Cai et al. [4] obtained improved rates of convergence under both tail conditions for an estimator that is not found by minimizing the penalized likelihood objective function, and may be nonpositive definite. Finally, note that the results in [4, 25, 26] are for separate estimation of precision matrices and hold for the minimum sample size across subpopulations, minknk, whereas our results hold for the total samples size Σknk.
3.2. Model Selection Consistency
Let be the support of , and denote by d the maximum number of nonzero elements in any rows of , k = 1, …, K. Define the event
| (5) |
where sign(a) is 1 if a > 0, 0 if a = 0 and −1 if a < 0. We say that an estimator Ω̂ρn of Ω0 is model-selection consistent if .
We begin by discussing an irrepresentability condition for estimation of multiple graphical models. This restrictive condition is commonly assumed to establish model selection consistency of lasso-type estimators, and is known to be almost necessary [19, 35]. For the graphical lasso, Ravikumar et al. [25] showed that the irrepresentability condition amounts to a constraint on the correlation between entries of the Hessian matrix Γ = Ω−1 ⊗ Ω−1 in the set S corresponding to nonzero elements of Ω, and those outside this set. Our irrepresentability condition is motivated by that in Ravikumar et al. [25], however, we adjust the index set S to also account for covariances of “non-edge variables” that are correlated with each other. More specifically, the description of irrepresentability condition in Ravikumar et al. [25] involves ΓSS consisting only of elements σijσkl with (i, j) ∈ S and (k, l) ∈ S. However, σij ≠ 0 for (i, j) ∉ S is not taken into account by this definition. We thus adjust the index set S so that ΓSS also includes elements σijσkl if (i, k) ∈ S and (j, l) ∈ S. This definition is based on the crucial observations that Γ = Σ ⊗ Σ involves the covariance matrix Σ instead of the precision matrix Ω, and that some variables are correlated (i.e., σij ≠ 0) even though they may be conditionally independent (i.e., ωij = 0). Defining S(k) for k = 1, …, K as above, we assume the following condition.
Condition 5 (Irrepresentability condition)
The inverse of the correlation matrix satisfies the irrepresentability condition for S(k)with parameter α: (a) and are invertible, and (b) there exists some α ∈ (0, 1] such that
| (6) |
for k = 1, …, K where .
In addition to the irrepresentability condition, we require bounds on the magnitude of and their normalized difference.
Condition 6 (Lower bounds for the inverse correlation matrices)
There exists a constant c8 ∈ ℝ such that
Moreover, for Ω0,ij ≠ 0, LΩ0,ij ≠ 0 and there exists a constant c9 > 0 such that
The first lower bound in Condition 6 is the usual “min-beta” condition for model selection consistency of lasso-type estimators. The second lower bound, which is represented here for the normalized Laplacian penalty, is a mild condition which ensures estimates based on inverse correlation matrices can be mapped to precision matrices. For any pair of subpopulations k and k′ connected in G it requires that if the difference in (normalized) entries of the entires of the precision matrices are nonzero, the difference in (normalized) entries of inverse correlation matrices are bounded away from zero. In other words, the bound guarantees that Θ0,ij is not in the null space of L, whenever Ω0,ij is outside of the null space. This bound can be relaxed if we use a positive definite matrix Lε = L + εI for ε > 0 small.
Our last condition for establishing the model selection consistency concerns the minimum sample size and the tuning parameter for the graph Laplacian penalty. This condition is necessary to control the ℓ∞-bound of the error Θ̂ρn − Θ0, as in Ravikumar et al. [25]. Our minimum sample size requirement is related to the irrepresentability condition. Let κΓ be the maximum of the absolute column sums of the matrices {(Γ(k))−1}S(k)S(k), k = 1, …, K, and κΨ be the maximum of the absolute column sums of the matrices , k = 1, …, K. The minimum sample size in Ravikumar et al. [25] is also a function of the irrepresentability constant, in particular, their κΓ involves . There is, therefore, a subtle difference between our definition and theirs: in our definition, the matrix is first inverted and then partitioned, while in Ravikumar et al. [25], the matrix is first partitioned and then inverted. Corollary 2 establishes the model selection consistency under a weaker sample size requirement, by exploiting instead the control of the spectral norm in Theorem 1.
Condition 7 (Sample size and regularization parameters)
Let
- (Exponential tails). It holds
(Polynomial tails). It holds .
It holds that .
With these condition, we obtain
Theorem 2
Suppose that Conditions 3, 5, 6 and 7 hold. Under Condition 1 or 2, P(ℳ(Ω̂ρn, Ω0)) → 1 as n, p → ∞ where ρnis given in Lemma 1 in theAppendixwith γ = mink πk/2.
3.3. Additional Results
In this section, we establish norm and variable selection consistency of LASICH under alternative assumptions. Our first result gives better rates of convergence for consistency in the ℓ∞-, spectral and Frobenius norms, under the condition for model selection consistency. Our rates in Corollary 1 improve the previous results by Ravikumar et al. [25], and are comparable to that of Cai et al. [4] in the ℓ∞- and spectral norms under both tail conditions.
Corollary 1
Suppose the conditions in Theorem 2 hold. Then, under Condition 1 or 2,
Our next result in Corollary 2 establishes the model selection consistency under a weaker version of the irrepresentability condition (Condition 6). Aside from the difference in the index sets S(k), the form of the Condition 6 and the assumption of invertibility of are similar to those in Ravikumar et al. [25]. On the other hand, Ravikumar et al. [25] do not require invertibility of . However, their proof is based on an application of Brouwer’s fixed point theorem, which does not hold for the corresponding function (Eq. (70) in page 973) since it involves a matrix inverse, and is hence not continuous on its range. The additional inevitability assumption in Condition 6 is used to address this issue in Lemma 11. The condition can be relaxed if we assume an alternative scaling of the sample size stated in Condition 8 below instead of Condition 7.
Condition 8
Let . Suppose and
- (Exponential tails)
or - (Polynomial tails)
Corollary 2
Suppose that Conditions 3, 6 and 8 hold. Suppose also that Condition 5 holds without requiring the invertibility of . Then, under Condition 1 or 2, P(ℳ(Ω̂ρn, Ω0)) → 1 as n, p → ∞ where ρnis given in Lemma 1 in theAppendix with γ = mink πk/2.
4. Laplacian Shrinkage based on Hierarchical Clustering
Our proposed LASICH approach utilizes the information in the subpopulation network G. In practice, however, similarity between subpopulations may be difficult to ascertain or quantify. In this section, we present a modified LASICH framework, called HC-LASICH, which utilizes hierarchical clustering to learn the relationships among subpopulations. The information from hierarchical clustering is then used to define the weighted subpopulation network. Importantly, HC-LASICH can even be used in settings where the subpopulation membership is unavailable, for instance, to learn the genetic network of cancer patients, where cancer subtypes may be unknown.
We use hierarchical clustering with a complete, single or average linkage to estimate both the subpopulation memberships and the weighted subpopulation network G. Specifically, the length of a path between two subpopulations in the dendrogram is used as a measure of dissimilarity between two subpopulations; the weights for the subpopulation networks are simply defined by taking the inverse of these lengths. Throughout this section, we assume that the number of subpopulations K is known. While a number of methods have been proposed for estimating the number of subpopulations in hierarchical clustering (see e.g. Borysov et al. [1] and the references therein), the problem is beyond the scope of this paper.
Let I = (I(1), …, I(K)) be the subpopulation membership indicator such that I follows the multinomial distribution MultK (1, (π1, …, πK)) with parameter 1 and subpopulation membership probabilities (π1, …, πK) ∈ (0, 1)K. Note that I is missing and is to be estimated. Let Ii, i = 1, …, n be i.i.d. copies of I and be an estimated subpopulation indicator for the ith observation via hierarchical clustering. Based on the estimated subpopulation membership and subpopulation network Ĝ, we apply our method to obtain the estimator, HC-LASICH, . Interestingly, HC-LASICH enjoys the same theoretical properties as LASICH, under the normality assumption. To show this, we first establish the consistency of hierarchical clustering in high dimensions, which is of independent interest. Our result is motivated by the recent work of [1], who study the consistency of hierarchical clustering for independent normal variables X(k) ~ N(μ(k), σ(k)I); we establish similar results for multivariate normal distributions with arbitrary covariance structures. We make the following assumption.
Condition 9
For k, k′ = 1, …, K, let
where λ(k),jis the eigenvalues of Σ(k)with λ(k),1 ≤ λ(k),2 ≤ … ≤ λ(k),p, and the spectral decomposition of Σ(k) + Σ(k′)is . It holds that
for constants m and M.
Under the normality assumption, the following results shows that the probability of successful clustering converges to 1, as p, n → ∞.
Theorem 3
Suppose that that X(k), k = 1, …, K, is normally distributed. Under Condition 9,
| (7) |
as n, p → ∞.
To proof of Theorem 3 generalizes recent results of Borysov et al. [1] to the case of arbitrary covariance structures. A key component of the proof is a new bound on the ℓ2 norm of a multivariate normal random variable with arbitrary mean and covariance matrix established in Lemma 14. The proof of the lemma uses new concentration inequalities for high-dimensional problems in [2], and may be of independent interest.
Note that the consistent estimation of subpopulation memberships (7) implies that the estimated hierarchy among clusters also matches the true hierarchy. Thus, with successful clustering established in Theorem 3, theoretical properties of Ω̂HC, ρn naturally follow.
Theorem 4
Suppose that X(k), k = 1, …, K, is normally distributed and that Condition 9 holds. (i) Under the conditions of Theorem 1,
Suppose, moreover, that the conditions of Theorem 2 holds. Then
(ii) Under the conditions of Theorem 2,
5. Algorithms
We develop an alternating directions method of multipliers (ADMM) to efficiently solve the convex optimization problem (3).
Let , k = 1, … K. Define A = (A(1), …, A(K)), B = (B(1), …, B(K)), C = (C(1), …, C(K)), D = (D(1), …, D(K)), and where .
To facilitate the computation, we consider instead a perturbed graph Laplacian Lε = L + εI, where I is the identity matrix and ε > 0 is a small perturbation. The difference between solutions to the original and modified optimization problem is largely negligible for small ε; however, the positive definiteness of Lε results in more efficient computation. A similar idea was used in Guo et al. [9] and Rothman et al. [26] to avoid dividision by zero. The optimization problem (3) with L replaced by Lε can then be written as
| (8) |
Using Lagrange multipliers E = (EA, EB, EC)T, with with , k = 1, …, K, with , k = 1, …, K, and with , k = 1, …, K, the augmented Lagrangian in scaled form is given by
Here ϱ > 0 is a regularization parameter and is the square root of Lε with .
The proposed ADMM algorithm is as follows.
Step 0. Initialize A(k) = A(k),0, B(k) = B(k),0, C(k) = C(k),0, D(k) = D(k),0, and choose ϱ > 0. Select a scalar ϱ > 0.
- Step m. Given the (m − 1)th estimates,
-
–(Update A(k)) Find Am minimizing (see pages 46–47 of Boyd et al. [3] for details).
-
–(Update B(k)) Compute , where Sy(x) is x − y if x > y, is 0 if |x| ≤ y, and is x + y if x < −y.
-
–(Update C(k)) For (x)+ = max{x, 0}, compute
-
–(Update D(k)) Compute
-
–(Update EA) Compute .
-
–(Update EB) Compute ,
-
–(Update EC) Compute .
-
–
Repeat the iteration until the maximum of the errors , s(k),m = ϱ(D(k),m − D(k),m−1) in the Frobenius norm is less than a specified tolerance level.
The proposed ADMM algorithm facilitates the estimation of parameters of moderately large problems. However, parameter estimation in high dimensions can be computationally challenging. We next present a result that determines whether the solution to the optimization problem (3), for given values of tuning parameters ρn, ρ2, is block diagonal. (Note that this result is an exact statement about the solution to (3), and does not assume block sparsity of the true precision matrices; see Theorems 1 and 2 of Danaher et al. [6] for similar results.) More specifically, the condition in Proposition 1 provides a very fast check, based on the entries of the empirical correlation matrices , k = 1, …, K, to identify the block sparsity pattern in , k = 1, …, K after some permutation of the features.
Let UL = [u1 … uK] ∈ ℝK×K where u1, …, uK’s are eigenvectors of L corresponding to 0, λL,2, …, λL,K. Define as the diagonal matrix with diagonal elements 0, .
Proposition 1
The solution , k = 1, …, Kto the optimization problem(3)consists of the block diagonal matrices with the same block structure diag(Ω1, …, ΩB) among all groups if and only if for
| (9) |
and for all i, j such that the (i, j) element is outside the blocks.
The proof of the Proposition is similar to Theorems 1 of Danaher et al. [6] and is hence omitted. Condition 9 can be easily verified by applying quadratic programming to the left hand side of the inequality. The solution to (3) can then be equivalently found by solving the optimization problem separately for each of the blocks; this can result in significant computational advantages for moderate to large values of ρnρ2.
6. Numerical Results
6.1. Simulation Experiments
We compare our method with four existing methods, graphical lasso, the method of Guo et al. [9], FGL and GGL of Danaher et al. [6]. For graphical lasso, estimation was carried out separately for each group with the same regularization parameter.
Our simulation setting is motivated by estimation of gene networks for healthy subjects and patients with two similar diseases caused by inactivation of certain biological pathways. We consider K = 3 groups with sample sizes n = (50, 100, 50) and dimension p = 100. Data are generated from multivariate normal distributions , k = 1, 2, 3; all precision matrices are block diagonal with 4 blocks of equal size.
To create the precision matrices, we first generated a graph with 4 components of equal size, each as either an Erdős-Rényi or scale free graphs with 95 total edges. We randomly assigned Unif((−7, −5) ∪ (.5, .7)) values to nonzero entries of the corresponding adjacency matrix A and obtained a matrix Ã. We then added 0.1 to the diagonal of à to obtain a positive definite matrix . For each of subpopulations 2 and 3, we removed one of the components of the graph by setting the off diagonal entries of à to zero, and added a perturbation from Unif(−2, .2) to nonzero entries in Ã. Positive definite matrices and were obtained by adding 0.1 to the diagonal elements. All partial correlations ranges from .28 to .54 in the absolute values. A similar setting was considered in in Danaher et al. [6], where the graph included more components, but no perturbation was added. We consider two simulation settings, with known and unknown subpopulation network G.
6.1.1. Known subpopulation network G
In this case, we set μ(k) = 0, k = 1, 2, 3 and use the graph in Figure 1 as the subpopulation network.
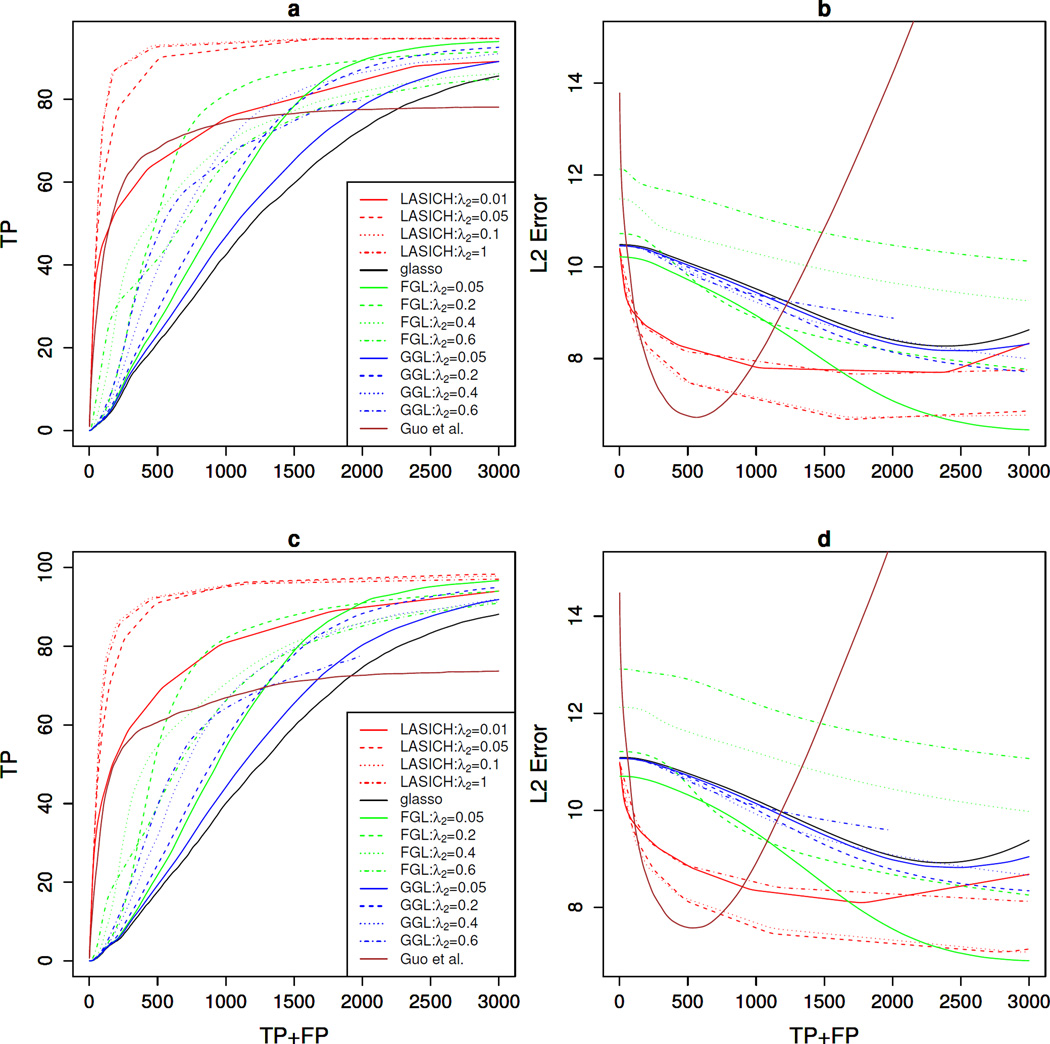
Figures 3a,c show the average number of true positive edges versus the average number of detected edges over 50 simulated data sets. Results for multiple choices of the second tuning parameter are presented for FGL, GGL and LASICH. It can be seen that in both cases, LASICH outperforms other methods, when using relatively large values of ρ2. Smaller values of ρ2, on the other hand, give similar results as other methods of joint estimation of multiple graphical models. These results indicate that, when the available subpopulation network is informative, the Laplacian shrinkage constraint can result in significant improvement in estimation of the underlying network.
Fig 3.
Simulation results for joint estimation of multiple precision matrices with known subpopulation memberships. Results show the average number of true positive edges (a & c) and estimation error, in Frobenius norm (b & d) over 50 data sets with n = 200 multivariate normal observations generated from a graphical model with p = 100 features; results in top row (a & b) are for an Erdős-Rényi graph and those in bottom row (c & d) are for a scale free (power-law) graph.
Figures 3b,d show the estimation error, in Frobenius norm, versus the number of detected edges. LASICH has larger errors when the estimated graphs have very few edges, but, its error decreases as the number of detected edges increase, eventually yielding smaller errors than other methods. The non-convex penalty of Guo et al. [9] performs well in terms of estimation error, although determining the appropriate range of tuning parameter for this method may be difficult.
6.1.2. Unknown subpopulation network G
In this case, the subpopulation memberships and the subpopulation network G are estimated based on hierarchical clustering. We randomly generated μ(1) from a multivariate normal distribution with a covariance matrix σ2I. For subpopulations 2 and 3, the elements of μ(1) corresponding to the empty components of the graph were set to zero to obtain μ(2) and μ(3). Hierarchical clustering with complete linkage was applied to data to obtain the dendrogram; we took inverse of distances in the dendrogram to obtain similarity weights used in the graph Laplacian.
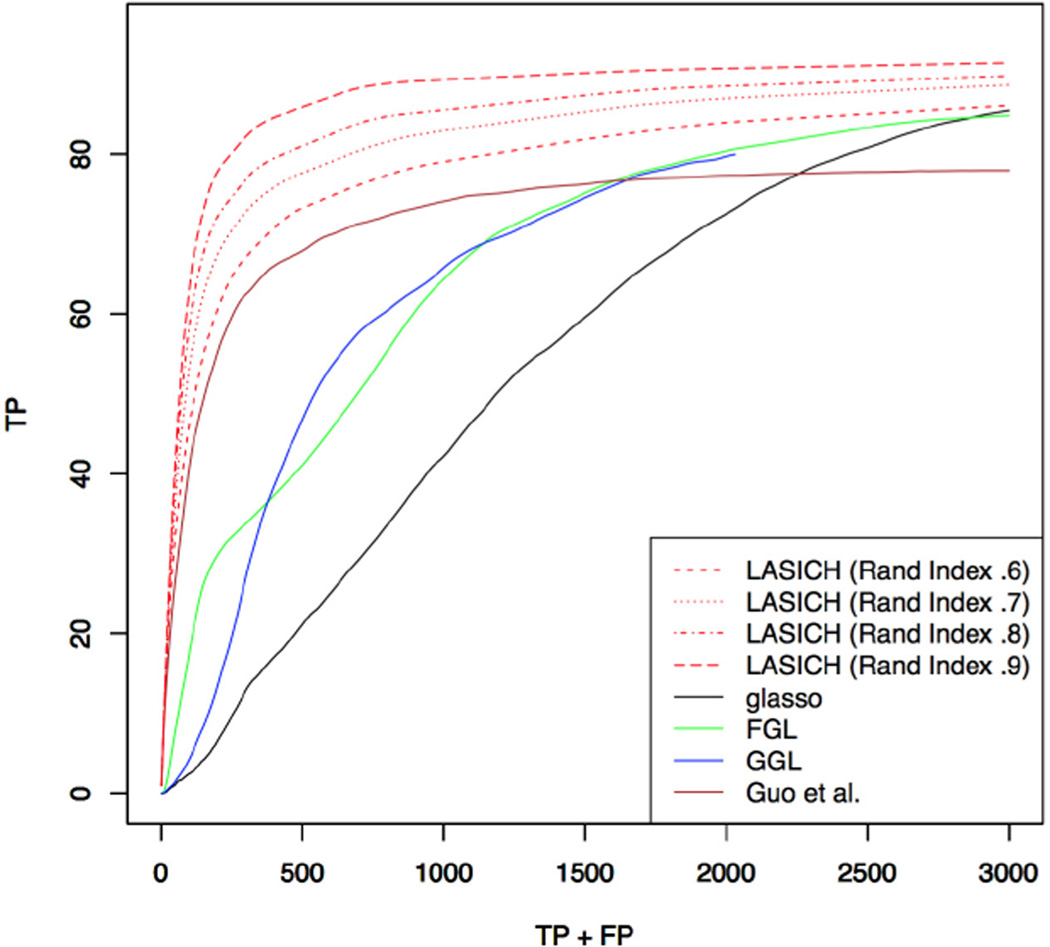
Figures 4 compares the performance of HC-LASICH, in terms of support recovery, to competing methods, in the setting where the subpopulation memberships and network are estimated from data (Section 4). Here the differences in subpopulation means μ(k,k′) are set up to evaluate the effect of clustering accuracy. The four settings considered correspond to average Rand indices of .6 .7, .8 and .9 across 50 data sets, respectively. Here the second tuning parameter for HC-LASICH, GGL and FGL is chosen according to the best performing model in Figure 3. As expected, changing the mean structure, and correspondingly the Rand index, does not affect the performance of other methods. The results indicate that, as long as features can be clustered in a meaningful way, HC-LASICH can result in improved support recovery. Data-adaptive choices of the tuning parameter corresponding to the Laplacian shrinkage penalty may result in further improvements in the performance of the HC-LASICH. However, we do not pursue such choices here.
Fig 4.
Simulation results for joint estimation of multiple precision matrices with unknown subpopulation memberships. Results show the average number of true positive edges over 50 data sets with n = 200 multivariate normal observations generated from a graphical model with over an Erdős-Rényi graph with p = 100 features. Results for HC-LASICH and FGL/GGL correspond to the best choice of the second tuning parameter among those in Figure 3a. The Rand indices for HC-LASICH are averages over 50 generated data sets.
6.2. Genetic Networks of Cancer Subtypes
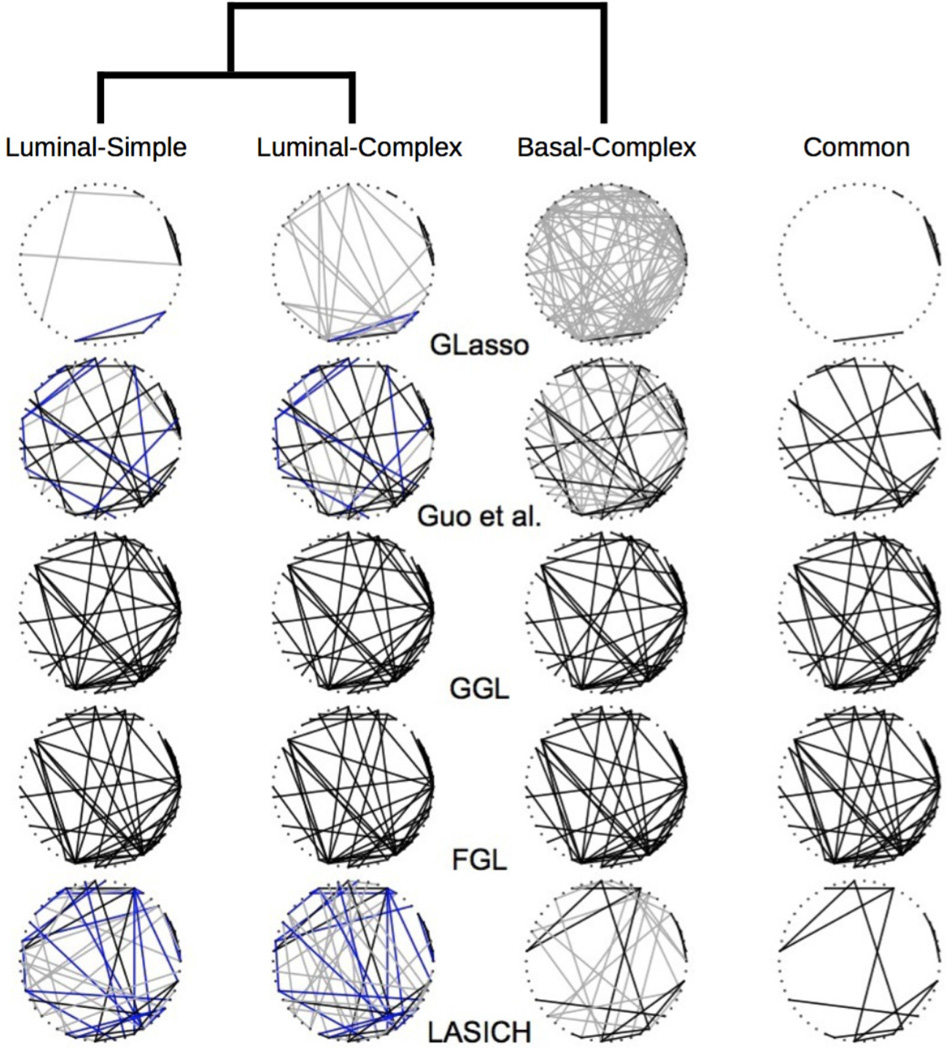
Breast cancer is heterogenous with multiple clinically verified subtypes [22]. Jönsson et al. [12] used copy number variation and gene expression measurements to identify new subtypes of breast cancer and showed that the identified subtypes have distinct clinical outcomes. The genetic networks of these different subtypes are expected to share similarities, but to also have unique features. Moreover, the similarities among the networks are expected to corroborate with the clustering of the subtypes based on their molecular profiles. We applied network estimation methods of Section 6.1 to a subset of the microarray gene expression data from Jönsson et al. [12], containing data for 218 patients classified into three previously known subtypes of breast cancer: 46 Luminal-simple, 105 Luminal-complex and 67 Basal-complex samples. For ease of presentation, we focused on 50 genes with largest variances. The hierarchical clustering results of Jönsson et al. [12], reproduced in Figure 5 for the above three subtypes, were used to identify the subpopulation membership; reciprocals of distances in the dendrogram were used to define similarities among subtypes used in the graph Laplacian penalty.
Fig 5.
Dendrogram of hierarchical clustering of three subtypes of breast cancer from Jönsson et al. (2010) along with estimated gene networks using graphical lasso (Glasso), method of Guo et al., FGL and GGL of Daneher et al. (2014) and LASICH. Blue edges are common to Luminal subtypes and black edges are shared by all three subtypes; condition specific edges are drawn in gray.
To facilitate the comparison, tuning parameters were selected such that the estimated networks of the three subtypes using each method contained a total of 150 edges. For methods with two tuning parameters, pairs of tuning parameters were determined using the Bayesian information criterion (BIC), as described in Guo et al. [9]. Estimated genetic networks of the three cancer subtypes are shown in Figure 5. For each method, edges common in all three subtypes, those common in Luminal subtypes and subtype specific edges are distinguished.
In this example, separate graphical lasso estimates and FGL/GGL estimates are two extremes. Estimated network topologies from graphical lasso vary from subtype to subtype, and common structures are obscured; this variability may be because similarities among subtypes are not incorporated in the estimation. In contrast, FGL and GGL give identical networks for all subtypes, perhaps because both methods encourage the estimated networks of all subtypes to be equally similar. Intermediate results are obtained using LASICH and the method of Guo et al. [9]. The main difference between these two methods is that Guo et al. [9] finds more edges common to all three subtypes, whereas LASICH finds more edges common to the Luminal subtypes. This difference is likely because LASICH prioritizes the similarity between the Luminal subtypes via graph Laplacian while the method of Guo et al. [9] does not distinguish between the three subtypes. The above example highlights the potential advantages of LASICH in providing network estimates that better corroborate with the known hierarchy of subpopulations.
7. Discussion
We introduced a flexible method for joint estimation of multiple precision matrices, called LASICH, which is particularly suited for settings where observations belong to three or more subpopulations. In the proposed method, the relationships among heterogenous subpopulations is captured by a weighted network, whose nodes correspond to subpopulations, and whose edges capture their similarities. As a result, LASICH can model complex relationships among subpopulations, defined, for example, based on hierarchical clustering of samples.
We established asymptotic properties of the proposed estimator in the setting where the relationship among subpopulations is externally defined. We also extended the method to the setting of unknown relationships among subpopulations, by showing that clusters estimated from the data can accurately capture the true relationships. The proposed method generalizes existing convex penalties for joint estimation of graphical models, and can be particularly advantageous in settings with multiple subpopulations.
A particularly appealing feature of the proposed extension of LASICH is that it can also be applied in settings where the subpopulation memberships are unknown. The latter setting is closely related to estimation of precision matrices for mixture of Gaussian distributions. Both approaches have limitations and drawbacks: on the one hand, the extension of LASICH to unknown subpopulation memberships requires certain assumptions on differences of population means (Section 4). On the other hand, estimation of precision matrices for mixture of Gaussians is computationally challenging, and known rates of convergence of parameter estimation in mixture distributions (e.g. in Städler et al. [29]) are considerably slower.
Throughout this paper we assumed that the number of subpopulations is known. Extensions of this method to estimation of graphical models in populations with an unknown number of subpopulations would be particularly interesting for analysis of genetic networks associated with heterogeneity in cancer samples, and are left for future research.
Acknowledgments
This work was partially supported by NSF grants DMS-1161565 & DMS-1561814 to AS.
Appendix
8. Appendix: Proofs and Technical Detials
We denote true inverse correlation matrices as and true correlation matrices as , where , and . The estimates of the population parameters are dented as , and . For a vector x = (x1, …, xp)T and J ⊂ {1, …, p}, we denote xJ = (xj, j ∈ J)T. For a matrix A, λk(A) is the kth smallest eigenvalue and A⃗ is the vectorization of A. For J ⊂ {(i, j) : i, j = 1, …, p} and A ∈ ℝp×p, A⃗J is a vector in ℝ|J| obtained by removing elements corresponding to (i, j) ∉ J from A⃗. A zero-filled matrix AJ ∈ ℝp×p is obtained from A by replacing aij by 0 for (i, j) ∉ J.
8.1. Consistency in Matrix Norms
Theorem 1 is a direct consequence of the following result.
Lemma 1
- Suppose that Condition 1 holds. Let γ ∈ (0, mink πk) be arbitrary. For
and , we have with probability (1 − 2K/p)(1 − 2K exp(−2n(mink πk − γ)2)) that - Suppose that Condition 2 holds with p ≤ c7nc2, c2, c3, c7 > 0. For ρn = C1Kδnsatisfying
and we have with probability (1 − 2K exp(−2n(mink πk − γ)2))ν nthat
where
and
Our proofs adopt several tools from Negahban et al. [20]. Note however that our penalty does not penalize the diagonal elements, and is hence a seminorm; thus, their results do not apply to our case. We first introduce several notations. To treat multiple precision matrices in a unified way, our parameter space is defined to be the set ℝ̃(pK)×(pK) of (pK) × (pK) symmetric block diagonal matrices, where the kth diagonal block is a p × p matrix corresponding to the precision matrix of subpopulation k. We write A ∈ ℝ̃(pK)×(pK) for a K-tuple of diagonal blocks A(k) ∈ ℝp×p. Note that for A, B ∈ ℝ̃(pK)×(pK), where 〈·, ·〉p is the trace inner product on ℝp×p. In this parameter space, we evaluate the following map from ℝ̃(pK)×(pK) to ℝ given by
where r : ℝ̃(pK)×(pK) ↦ ℝ is given by r(Θ) = ‖Θ‖1+ ρ2‖Θ‖L. This map provides information on the behavior of our criterion function in the neighborhood of Θ0. A similar map with a different penalty was studied in Rothman et al. [26]. A key observation is that f(0) = 0 and f(Δ̂n) ≤ 0 where Δ̂n = Θ̂ρn − Θ0.
The following lemma provides a non-asymptotic bound on the Frobenius norm of Δ (see Lemma 4 in Negahban et al. [21] for a similar lemma in a different context). Let be the union of the supports of . Define a model subspace and its orthocomplement under the trace inner product in ℝ̃(pK)×(pK). For , we write A = Aℳ + Aℳ⊥ where Aℳ and Aℳ⊥ are the projection of A into ℳ and ℳ⊥, in the Frobenius norm, respectively. In other words, the (i, j)-element of Aℳ is aij if (i, j) ∈ S and zero otherwise, and the (i, j)-element of Aℳ⊥ is aij if (i, j) ∉ S and zero otherwise. Note that Θ0 ∈ ℳ. Define the set 𝒞 = {Δ ∈ ℝ̃(pK)×(pK) : r(Δℳ⊥) ≤ 3r(Δℳ)}.
Lemma 2
Let ε > 0 be arbitrary. Suppose . Iff (Δ) > 0 for all elements Δ ∈ 𝒞 ⋂ {Δ ∈ ℝ̃(pK)×(pK) : ‖Δ‖F = ε} then ‖Δ̂n‖F ≤ ε.
Proof
We first show that Δ̂n ∈ 𝒞. We have by the convexity of −ℓ̃n(Θ) that
It follows from Lemma 3(iv) with our choice ρn that the right hand side of the inequality is further bounded below by −2−1 ρn (r(Δ̂n,ℳ) + r(Δ̂n,ℳ⊥)). Applying Lemma 3(iii), we obtain
or r(Δ̂n,ℳ⊥) ≤ 3r(Δ̂n,ℳ). This verifies Δ̂n ∈ 𝒞. Note that f, as a function of Δ is sum of two convex functions ℓn and r, and is hence convex. Thus, the rest of the proof follows exactly as Lemma 4 in Negahban et al. [21].
Lemma 3
Let Δ ∈ ℝ̃(pK)×(pK).
- The gradient of ℓ̃n(Θ0) is a block diagonal matrix given by
(10) - Let c > 0 be a constant. For ‖Δ‖F ≤ c and nk/n ≥ γ > 0 for all k and n,
(11) - The map r is a seminorm, convex, and decomposable with respect to (ℳ, ℳ⊥) in the sense that r(Θ1 + Θ2) = r(Θ1) + r(Θ2) for every Θ1 ∈ ℳ and Θ2 ∈ ℳ⊥. Moreover,
- For Δ ∈ ℝ̃(pK)×(pK),
(12) - For Θ ∈ ℝ̃(pK)×(pK),
Proof
The result follows by taking derivatives blockwise.
-
Rothman et al. [26] (page 500–502) showed thatSince ‖A‖2 ≤ ‖A‖F, nk/n ≥ γ and ‖Δ‖F ≤ c, this is further bounded below by
-
Because the graph Laplacian L is a positive semidefinite matrix, the triangle inequality r(Θ1 + Θ2) ≤ r(Θ1) + r(Θ2) holds. To see this let L = L̃L̃T be any Cholesky decomposition of L. ThenIt is clear that r(cΘ) = cr(Θ) for any constant c. Thus, given that r does not penalize the diagonal elements, it is a seminorm. The decomposability follows from the definition of r. The convexity follows from the same argument for the triangle inequality. Since Θ0 + Δ = Θ0 + Δℳ + Δℳ⊥, the triangle inequality and the decomposability of r yield
-
We show that, for A, B ∈ ℝ̃(pK)×(pK) with diag(B) = 0, 〈A, B〉 ≤ r(A)‖B‖∞. If A is a diagonal matrix (or if A = 0), the inequality trivially holds since 〈A, B〉 = 0. If not, r(A) ≠ 0 so that
Since the diagonal elements of ∇ℓ̃n(Θ0) are all zero, the result follows.
-
For s ≠ 0, we have
In the last inequality we used that , which follows by the concavity of the square root function. For s = 0, we trivially have . Combining these two cases yields the desired result.
Next, we obtain an upper bound for , which holds with high-probability assuming the tail conditions of the random vectors.
Lemma 4
Suppose that nk/n ≥ γ > 0 for all k and n.
- Suppose that Condition 1 holds. Then for n ≥ 6γ−1 log p we have
(13) - Suppose that Condition 2 holds with c2, c3 > 0 and p ≤ c7nc2. Then we have for
where(14)
with Suppose that Condition 3 holds and that P(‖Σ̂n − Σ0‖∞ ≥ bn) = o(1) and bn = o(1) as n → ∞. Then P(‖Ψ̂n − Ψ0‖∞ ≥ C1bn) = o(1).
Proof
This was proved by Ravikumar et al. [25].
-
Note thatWe have
where the first inequality follows from the triangle inequality. Note that(15) It follows from Bernstein’s inequality that(16) Now, for , νn,2 → 0 as p → ∞. Note that for this to hold it suffices to have
so that the power in the exponent is negative. This inequality reduces toWe can solve this by changing a quadratic equation for τ, since τ of our interest is positive. Combining (15) and (16) yields(17) LetProceeding as for ’s, we have
andThus, we have
and(18) (19) Note that , νn,1, νn,2, νn,3 → 0 as n, p → ∞ if log p/n → 0. Note also that and are on the set where nk/n ≥ γ.
For example, we have by Jensen’s inequality that -
Given that
whereinSince bn → 0, bn ≤ c5/2 for n sufficiently large by Condition 3. On the event ‖Σ̂n − Σ0‖∞ ≤ bn with n large, . Thus,It follows thatThus, we have
So far we have assumed nk/n ≥ γ in lemmas. We evaluate the probability of this event noting that nk ~ Binom(n, πk).
Lemma 5
Let ε > 0 such that γ ≡ mink πk − ε > 0. Then
| (20) |
Proof
We have by Hoeffding’s inequality that
Proof of Lemma 1
We apply Lemma 2 to obtain the non-asymptotic error bounds.
We first compute a lower bound for f(Δ). Suppose ε ≤ c. For Δ ∈ 𝒞 ∩ {Δ ∈ ℝ̃(pK)×(pK) : ‖Δ‖F = ε}, we have by Lemma 3(ii) and (iii) that
The assumption on ρn and Lemma 3(iii) and (iv) then yield
From this inequality and Lemma 3(v) we have
Viewing the right hand side of the above inequality as a quadratic equation in ‖Δ‖F, we have f(Δ) > 0 if
Thus, if we show that there exists a c0 > 0 such that εc0 ≤ c0, Lemma 2 yields that ‖Θ̂ρn − Θ0‖F ≤ εc0.
Consider the inequality (x + y)2z1/2 ≤ y where x, y, z ≥ 0. This inequality holds for (x, y, z) such that x = y and xz1/2 = 1/4. We apply the inequality above with x = λΘ, y = c, and solve xz ≤ 1/4 for n. (i) For , xz ≤ 1/4 yields
and (x + y)4z becomes
(ii) For ρn = C1Kδn, there is no closed form solution for n. Note that δn → 0 if log p/n → 0 so that xz ≤ 1/4 holds for n sufficiently large, given that .
Computing appropriate probabilities using Lemmas 4 and 5 completes the proof.
Proof of Theorem 1
The estimation error in the spectral norm can be bounded and evaluated in the same way as in the proof of Theorem 2 of Rothman et al. [26] together with Lemma 1.
8.2. Model Selection Consistency
Our proof is based on the primal-dual witness approach of Ravikumar et al. [25], with some modifications to overcome a difficulty in their proof when applying the fixed point theorem to a discontinuous function. First, we define the oracle estimator by
| (21) |
where indicates that for (i, j) ∉ S(k).
Lemma 6
- Let A ∈ ℝp×pbe a positive semidefinite matrix with eigenvalues 0 ≤ λ1 ≤ λ2 ≤ ⋯ ≤ λpand corresponding eigenvectors ui satisfying ui ⊥ uj, i ≠ j and ‖ui‖ = 1. The subdifferential of is
where U ∈ ℝp×phas ui as the ith columns and Λ1/2is the diagonal matrix with , i = 1, …, p, as diagonal elements. Furthermore, the subgradients are bounded above, i.e. -
Let A ∈ ℝp×pbe a positive semidefinite matrix and S = {Si} ⊂ {1, …, p}. Suppose ASS has eigenvalues 0 ≤ λ1,S ≤ λ2,S ≤ ⋯ ≤ λ|S|,Sand corresponding eigenvectors ui,S satisfying ui,S ⊥ uj,S, i ≠ j and ‖ui,S‖ = 1. Let gS : ℝ|S| → ℝpbe a map defined by gS(x) = y where yi = xSj for i = Sj for and yi = 0 for i ∉ S. The subdifferential equals to the subdifferential of given by
where US ∈ ℝ|S|×|S|has ui,S as the ith columns and is the diagonal matrix with , i = 1, …, |S|, as diagonal elements. For x with ASSx ≠ 0, there is a relationship between and at y = gS(x) given bySubgradients are bounded above:
Proof
-
For x with Ax ≠ 0, f(x) is differentiable and the subgradient of f at x is simply the matrix derivative. By definition, for x with Ax = 0, the subgradient υ of f at x satisfies the following inequality
for all y. Choosing y = 2x and y = 0 yield 0 ≥ 〈x, υ〉 and 0 ≥ − 〈x, υ〉, implying 〈x, υ〉 = 0. The inequality (22) reduces to , for any y. If Ay = 0, a similar argument implies that 〈y, υ〉 = 0. Hence υ ⊥ y for every y with Ay = 0.(22) Let j0 be the smallest index such that λj0 > 0. Because uj ’s form an orthonormal basis, any arbitrary vector y can be written as . Moreover, the null space of A is the span of u1, …, uj0−1. Thus, the subgradient υ can be written as . Thus, using the spectral decomposition of A as , we can write . On the other hand, . Thus, the inequality (22) further reduces toIt follows from the Cauchy-Schwartz inequality that the left hand side of the inequality is bounded from above;Thus,It is easy to see that this set is the image of the map UΛ1/2 on the closed ball of radius 1.
Given that ‖x‖∞ ≤ ‖x‖, to establish the bound in the ℓ∞-norm, we compute the bound in the Euclidean norm. We use the same notation as in (i). For x with Ax ≠ 0,But , because ‖UT x‖ = ‖x‖. For x with Ax = 0, for every y. Because of the form of the subdifferential and the fact that ‖U x‖ = ‖x‖, the result follows.
-
Let BS be a product of elementary matrices for row and column exchange such that BSgS(x) = (x, 0). Notice that and that since BS only rearranges elements of vectors and exchanges rows by multiplication from the left. Note also that ‖BS‖2 ≤ ‖BS‖∞/∞ = 1, since ‖C‖2 ≤ ‖C‖∞/∞ for C = CT and each row of BS has only one element with value 1. Because
the subdifferential of hA,S(x) follows from (ii). For x with ASSx ≠ x and y = gS(x), because of invertibility of BS. The relationship holds sinceAn ℓ∞-bound follows from (i) and the fact that .
Lemma 7
For sample correlation matrices and any ρn > 0, the convex problem(3)has a unique solution with , k = 1, …, K, characterized by
| (23) |
with and for every i ≠ j and k = 1, …, K. Moreover,
| (24) |
with for every i = 1, …, p, andk = 1, …, K.
For each (i, j) ∈ S, let . The convex problem(21)has a unique solution with characterized by
| (25) |
with and for every i ≠ j and k = 1, …, K. Moreover,
| (26) |
with for every i = 1, …, p, and k = 1, …, K.
Proof
A proof for the uniqueness of the solution is similar to the proof of Lemma 3 of Ravikumar et al. [25]. The rest is the KKT condition using Lemma 6.
We choose a pair Ũ = (Ũ1, Ũ2) of the subgradients of the first and second regularization terms evaluated at Θ̌ρn. For each (i, j) with Ω0,ij = 0 or with LΘ̌ρn,ij = 0, set
For (i, j) with , for all k = 1, …, K, set
For (i, j) with LΘ̌ρn,ij ≠ 0, Ω0,ij ≠ 0 but for some k′, set
and
if . Otherwise, let
Here, lk is the kth row of L.
The main idea of the proof is to show that (Θ̌ρn, Ũ) satisfies the optimality conditions of the original problem with probability tending to 1. In particular, we show the following equation, which holds by construction of Ũ1 and Ũ2, is in fact the KKT condition of the original problem (3):
| (27) |
To this end, we show that Ũ1 and Ũ2 are both subgradients of the original problem. We can then conclude that the oracle estimator in the restricted problem (21) is the solution to the original problem (3). Then it follows from the uniqueness of the solution that Θ̌ρn = Θ̂ρn.
Let , and .
Lemma 8
Suppose that max{‖Ξ(k)‖∞, R(k)(Δ̌(k))‖∞} ≤ αρn/8, and . Suppose moreover thatLΘ̌ρn,ij ≠ 0 for (i, j) ∈ S. Then for (i, j) ∈ (S(k))c.
Proof
We rewrite (27) to obtain
We further rewrite the above equation via vectorization;
We separate this equation into two equations depending on S(k);
| (28) |
where (Ũ⃗l)J ≡ Ũ⃗k,J, l = 1, 2. Here we used . Since is invertible, we solve the first equation to obtain
Substituting this expression into (28) yields
Taking the ℓ∞-norm yields
Here we used that ‖Ax‖∞ ≤ ‖A‖∞/∞ ≤ ‖A‖∞ and , and applied Lemma 6 to bound ‖Ũ⃗2, (S(k))c‖∞ and ‖Ũ⃗2, (S(k))‖∞ by . We also used by construction of Ũ1 and the assumption that . It follows by the assumption of the lemma that
Lemma 9 (Lemma 5 of Ravikumar et al. [25])
Suppose that ‖Δ‖∞ ≤ 1/(3κΨd) with
Then ‖H(k)‖∞/∞ ≤ 3/2 where , k = 1, …, K, andR(k)(Δ(k)) has representation with .
Lemma 10
Suppose with . Then ‖H(k)‖∞/∞ ≤ 2 where , k = 1, …, K, andR(k)(Δ(k)) has representation with .
Proof
Note that the Neumann series for a matrix (I − A)−1 converges if the operator norm of A is strictly less than 1, and that the ℓ∞-norm is bounded by the operator norm. A proof is similar to that of Lemma 5 of Ravikumar et al. [25] with the induced infinity norm ‖·‖∞/∞ replaced by the operator norm in appropriate inequalities.
The following lemma is similar to the statement of Lemma 6 of Ravikumar et al. [25].
Lemma 11
Suppose that
for k = 1, …, K. Suppose moreover that are invertible for k = 1, …, K. Then with probability ,
Proof
We apply Shauder’s fixed point theorem on the event mink πk/2 ≤ nk/n, which holds with probability by Lemma 5 with ε = mink πk/2. We first define the function fk and its domain 𝒟k to which the fixed point theorem applies. Let S̅(k) = S(k) ∪ {(i, i) : 1 ≤ i ≤ p}, and define
where 𝕊p×p is the space of symmetric p × p matrices. Then 𝒟k is a convex, compact subset of the set of 𝕊p×p.
Let , l = 1, 2, be zero-filled matrices whose (i, j)-element is in Lemma 7 if (i, j) ∈ S(k) and zero otherwise. Define the map gk on the set of invertible matrices in ℝp×p by . Note that is the KKT condition for the restricted problem (21). Let δ > 0 be a constant such that δ < min{1/2, 1/{10(4dr + 1)}}r and . Define a continuous function fk : 𝒟k ↦ 𝒟k as
where
Let . Then fk(A) = (f̃k(A))S(k) + A for A ∈ 𝒟k.
We now verify the conditions of Shauder’s fixed point theorem below. Once these conditions are established, the theorem yields that fk(A) = A. Since (fk(A))(S̅(k))c = A for any A ∈ 𝒟k, and hk(A) > 0, the solution A to fk(A) = A is determined by . Vectorizing this equation to obtain , it follows from the invertibility of that . By the uniqueness of the KKT condition, the solution is . Since A ∈ 𝒟k, and δ < r/2, we conclude .
In the following, we write A⃗ = vec(A) for a matrix A for notational convenience. For J ⊂ {(i, j) : i, j = 1, …, p}, vec(A)J should be understood as A⃗J.
The function fk is continuous on 𝒟k. To see this, note first that is positive definite for every A ∈ 𝒟k so that the inversion is continuous. Note also that all elements in the matrices involved with eigenvalues in hk(A) are uniformly bounded in 𝒟k, and hence the eigenvalues are also uniformly bounded.
To show that fk(A) ∈ 𝒟k, first we show that is positive semidefinite. This follows because for any x ∈ ℝp
To see this, note that if is positive, then the inequality easily follows. On the other hand, if λA < −1, we have
Lastly, if −1 ≤ λA < 0, we have
Next, we show that ‖fk(A)S̅(k)‖∞ ≤ r. Because diag(fk(A)) = diag(A), it suffices to show ‖fk(A)S(k)‖∞ ≤ r. Since ,
It then follows from Lemma 9 that
Thus, adding and subtracting yields
Vectorization and restriction on S(k) gives
| (29) |
where . Here we used hk(A) ≤ (1/4 + 1/2)/1 = 3/4. For the first term of the upper bound in (29), it follows by the inequality ‖Ax‖∞ ≤ ‖A‖∞/∞‖x‖∞ for A ∈ ℝp×p and x ∈ ℝp, Lemma 9 and the choice of δ satisfying that
For the second term, it follows by the assumption, the inequality that ‖Ax‖∞ ≤ ‖A‖∞/∞‖x‖∞ for A ∈ ℝp×p and x ∈ ℝp, and Lemma 6 that
Thus, we can further bound ‖vec((f̃k(A) + A)S(k))‖∞ by
| (30) |
Since
and δ ≤ r/2, a similar reasoning shows that
Thus, the inequality ‖B‖2 ≤ ‖B‖∞/∞ for B = BT implies that
Hence hk(A) ≥ 1/(8dr + 2) for every A ∈ 𝒟k.
Now (30) is further bounded by r:
Here we used the fact that δ ≤ r/{10(4dr + 1)} and 1/(8dr + 2) ≤ hk(A) < 1. Thus, ‖(fk(A))S(k)‖∞ ≤ r.
Since (fk(A))(S(k))c = 0 by definition, all the conditions for the fixed point theorem are established. This completes the proof.
We are now ready to prove Theorem 2. Note that Condition 7 implies that
Proof of Theorem 2
We prove that the oracle estimator Θ̌ρn satisfies (I) the model selection consistency and (II) the KKT conditions of the original problem (3) with (Θ̌ρn, Ũ1, Ũ2). The model selection consistency of Θ̂ρn = Θ̌ρn then follows by the uniqueness of the solution to the original problem. The following discussion is on the event that mink πk/2 ≤ nk/n, k = 1, …, K, and maxk‖Ξ(k)‖∞ ≤ α/8. Note that this event has probability approaching 1 by Lemmas 4 and 5.
First we obtain an ℓ∞-bound of the error of the oracle estimator. Note that by Condition 7 and the fact that α ∈ [0, 1)
Thus, it follows from Condition 7 that
Because is invertible by Condition 5, we can apply Lemma 11 to obtain with probability approaching 1.
As a consequence of the ℓ∞-bound, Θ̌ρn,ij ≠ 0 for (i, j) ∈ S, because by Conditions 6 and 7. This establishes the model selection consistency of the oracle estimator.
Next, we show that the Oracle estimator satisfies the KKT condition of the original problem (3). As the first step, we prove for every i, j, k with probability approaching 1. Since Θ̌ρn,ij ≠ 0 for (i, j) ∈ S with probability approaching 1, for (i, j) ∈ S(k) by construction. For (i, j) ∈ (S(k))c, we need to prove for every i, j, k. To this end, it suffices to verify that and apply Lemma 8. Applying Lemma 9 with and Condition 7 gives
Next, we prove that for every (i, j). For (i, j) with for all k = 1, …, K, . For (i, j) with Ω0,ij = 0, by Lemma 6. For (i, j) with Ω0,ij ≠ 0 and for some k′,
if LΘ̌ρn,ij ≠ 0. To see LΘ̌ρn,ij ≠ 0 holds with probability approaching 1, let (k, k′) ∈ S with k ≠ k′ such that . This pair (k, k′) exists by Condition 6 and the assumption LΘ0,ij ≠ 0. We assume without loss of generality . Since , it follows from Condition 7 that
Hence, or LΘ̌ρn,ij ≠ 0.
Finally, we show that Equation (27) for the KKT condition holds. For the (i, j)-element of the equation with Ω0, ij = 0, this equation hold by construction for every k = 1, …, K. For the (i, j)-element with for every k = 1, …, K, the equation holds for every k = 1, …, K, because it is the equation for the KKT condition of the corresponding element in a restricted problem (21). For (i, j)-element with Ω0, ij ≠ 0 and for some k′, note that Θ̌ρn,ij ≠ 0 with probability approaching 1 and that the rearrangement in Θij and corresponding exchange of rows and columns of L for each i, j does not change the original and restricted optimization problems (3) and (21). Thus, with the appropriate rearrangement of elements and exchange of rows and columns, with is in fact . Thus for such k the equation holds because of the corresponding KKT condition in the restricted problem (21). For other k, the equation holds by construction. We thus conclude the oracle estimator satisfies the KKT condition of the original problem (3). This completes the proof.
Proof of Corollary 1
In the proof of Theorem 2, the ℓ∞-bound of the error yields
Note that if one of two matrices A and B is diagonal, ‖AB‖∞ ≤ ‖A‖∞‖B‖∞. Thus, we can proceed in the same way as in the proof of Theorem 2 of Rothman et al. [26] to conclude that
The result follows from a similar argument to the proof of Corollary 3 in Ravikumar et al. [25].
Proof of Corollary 2
It follows from Condition 8 and Lemma 1 applied to Θ̌ρn that . Then we can apply Lemma 10 instead of Lemma 9. The rest is similar to the proof of Theorem 2.
Hierarchical Clustering
For simplicity, we prove Theorem 3 for the case of K = 2; the proof can be easily generalized to K > 2. Let X and Y be the random variable from the first and subpopulation, respectively. Suppose that X = (X1, …, Xp)T ~ N(μX, ΣX) with μX = (μ1,X, …, μp,X) and the spectral decomposition of ΣX where λ1,X, …, λp,X are the eigenvalues of ΣX and that Y ~ N(μY, ΣY) with μY = (μ1,Y, …, μp,Y) and the spectral decomposition of ΣY where λ1,Y, …, λp,Y are the eigenvalues of ΣY. Define Z = (X − Y) = (Z1, …, Zp)T ~ N(μZ, ΣZ) with μZ = (μ1,Z, …, μp,Z) and the spectral decomposition of ΣZ where λ1,Z, …, λp, Z are the eigenvalues of ΣZ. Let and . Then X̃ ~ N(μ̃X, ΛX), Ỹ ~ N(μ̃Y, ΛY) and Z̃ ~ N(μ̃Z, ΛZ), where
Let also
Lemma 12 (Lemma 1 of Borysov et al. [1])
Let W1, …, Wp be independent non-negative random variables with finite second moments. Let and . Then for any t > 0 P(S ≤ −t) ≤ exp(−t2/(2υ)).
The following lemma is an extension of Lemma 2 in Borysov et al. [1].
Lemma 13
Let . Then
Proof
Note that elements of X̃ are independent and that X̃j ~ N(μ̃j,X, λj,X). Thus, we have
Applying Lemma 12 with , since P (‖X‖2 < ap) = P (‖X̃‖2 < ap), we get
The following is an extension of Lemma 3 in Borysov et al. [1].
Lemma 14
Let . Then
Proof
By Markov’s inequality, for , we get
Since for all u ∈ (0, 1), −log(1 − u) − u ≤ u2/{2(1 − u)} (see page 28 of Boucheron et al. [2]), the above display is bounded above by
Using the following result from Boucheron et al. [2]
wherein , u > 0, we further obtain the upper bound
Taking γ ↓ 0, the upper bound becomes
Choosing t = ap, we have
Note that f(u) = (1 + 2u)1/2 ≤ u for u ≥ 0 because f′(0) = 1 and f′ is decreasing for u > 0. Thus, as p → ∞.
Proof of Theorem 3
For simplicity, we present the proof for the case of K = 2; the proof can be easily generalized to K > 2. Let n1 and n2 be the sample sizes for the first and second subpopulations, respectively. Define
for a fixed a > 0 satisfying the assumption. The intersection E1 ∩ E2 is contained in the event that the clustering performs in the way that two subpopulations are joined in the last step. The intersection E3 ∩ E4 ∩ E5 is also contained in E1 ∩ E2, or in other words, . Thus, it suffices to show that as n, p → ∞.
For and we have by Lemma 14 that
and that
for a satisfying a > 2 max{λ̄X, λ̄Y}.
Note that log nk/p → 0, k = 1, 2 as n1, n2, p → ∞. Moreover for x > 0. Thus, and as n1, n2, p → ∞. For , we have by Lemma 13 that
for . Given the assumption c10 ≤ λj,X ≤ c11, c10 ≤ λj,Y ≤ c11, max{|μj,X|, |μj,Y|} ≤ c11, j = 1, 2, …. Thus, we get as n1, n2, p → 1.
Since 2λ̄X − λp,X − λp,Y ≥ 2λ̄X − λ̄Z, and 2λ̄Y − λp,X − λp,Y ≥ 2λ̄Y − λ̄Z, the assumption that implies that there exists a such that a < μ̄Z̃ + λ̄Z and a > 2 max{λ̄X, λ̄Y}. This completes the proof.
Contributor Information
Takumi Saegusa, Department of Mathematics, University of Maryland, College Park, MD 20742 USA.
Ali Shojaie, Department of Biostatistics, University of Washington, Seattle, WA 98195 USA.
References
- 1.Borysov Petro, Hannig Jan, Marron JS. Asymptotics of hierarchical clustering for growing dimension. Journal of Multivariate Analysis. 2014;124:465–479. [Google Scholar]
- 2.Boucheron Stéphane, Lugosi Gábor, Massart Pascal. Concentration inequalities: A nonasymptotic theory of independence. Oxford University Press; 2013. [Google Scholar]
- 3.Boyd Stephen, Parikh Neal, Chu Eric, Peleato Borja, Eckstein Jonathan. Distributed optimization and statistical learning via the alternating direction method of multipliers. Foundations and Trends in Machine Learning. 2011;3(1):1–122. [Google Scholar]
- 4.Cai Tony, Liu Weidong, Luo Xi. A constrained ℓ1 minimization approach to sparse precision matrix estimation. J. Amer. Statist. Assoc. 2011;106(494):594–607. ISSN 0162-1459. [Google Scholar]
- 5.Chung Fan RK. Spectral graph theory. Vol. 92. American Mathematical Soc; 1997. [Google Scholar]
- 6.Danaher Patrick, Wang Pei, Witten Daniela M. The joint graphical lasso for inverse covariance estimation across multiple classes. Journal of the Royal Statistical Society: Series B (Statistical Methodology. 2014;76(2):373–397. doi: 10.1111/rssb.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.d’Aspremont Alexandre, Banerjee Onureena, Ghaoui Laurent El. First-order methods for sparse covariance selection. SIAM J. Matrix Anal. Appl. 2008;30(1):56–66. ISSN 0895-4798. [Google Scholar]
- 8.Friedman Jerome, Hastie Trevor, Tibshirani Robert. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2007;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Jian, Levina Elizaveta, Michailidis George, Zhu Ji. Joint estimation of multiple graphical models. Biometrika. 2011;98(1):1–15. doi: 10.1093/biomet/asq060. ISSN 0006-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Jian, Ma Shuangge, Li Hongzhe, Zhang Cun-Hui. The sparse Laplacian shrinkage estimator for high-dimensional regression. Ann. Statist. 2011;39(4):2021–2046. doi: 10.1214/11-aos897. ISSN 0090-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ideker Trey, Krogan Nevan J. Differential network biology. Molecular systems biology. 2012;8(1) doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jönsson Göran, Staaf Johan, Vallon-Christersson Johan, Ringnér Markus, Holm Karolina, Hegardt Cecilia, Gunnarsson Haukur, Fagerholm Rainer, Strand Carina, Agnarsson Bjarni A, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Research. 2010;12(3):1–14. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolar Mladen, Song Le, Xing Eric P. Sparsistent learning of varying-coefficient models with structural changes. Advances in Neural Information Processing Systems. 2009:1006–1014. [Google Scholar]
- 14.Lauritzen Steffen L. Graphical models. Oxford University Press; 1996. [Google Scholar]
- 15.Li Caiyan, Li Hongzhe. Variable selection and regression analysis for graph-structured covariates with an application to genomics. Ann. Appl. Stat. 2010;4(3):1498–1516. doi: 10.1214/10-AOAS332. ISSN 1932-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Fan, Zhang Nancy R. Bayesian variable selection in structured high-dimensional covariate spaces with applications in genomics. Journal of the American Statistical Association. 2010;105(491):1202–1214. [Google Scholar]
- 17.Liu F, Lozano AC, Chakraborty S, LI F. A graph laplacian prior for variable selection and grouping. Biometrika. 2011;98(1):1–31. [Google Scholar]
- 18.Liu Fei, Chakraborty Sounak, Li Fan, Liu Yan, Lozano Aurelie C, et al. Bayesian regularization via graph laplacian. Bayesian Analysis. 2014;9(2):449–474. [Google Scholar]
- 19.Meinshausen Nicolai, Bühlmann Peter. High-dimensional graphs and variable selection with the lasso. Ann. Statist. 2006;34(3):1436–1462. ISSN 0090-5364. [Google Scholar]
- 20.Negahban Sahand N, Ravikumar Pradeep, Wainwright Martin J, Yu Bin. A unified framework for high-dimensional analysis of m-estimators with decomposable regularizers. Stat. Sci. 2012;27(4):538–557. [Google Scholar]
- 21.Negahban Sahand N, Ravikumar Pradeep, Wainwright Martin J, Yu Bin. Supplementary material for “a unified framework for high-dimensional analysis of m-estimators with decomposable regularizers”. Stat. Sci. 2012 [Google Scholar]
- 22.Perou Charles M, Sørlie Therese, Eisen Michael B, van de Rijn Matt, Jeffrey Stefanie S, Rees Christian A, Pollack Jonathan R, Ross Douglas T, Johnsen Hilde, Akslen Lars A, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 23.Peterson Christine, Stingo Francesco C, Vannucci Marina. Bayesian inference of multiple gaussian graphical models. Journal of the American Statistical Association. 2015;110(509):159–174. doi: 10.1080/01621459.2014.896806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapaport Franck, Zinovyev Andrei, Dutreix Marie, Barillot Emmanuel, Vert Jean-Philippe. Classification of microarray data using gene networks. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravikumar Pradeep, Wainwright Martin J, Raskutti Garvesh, Yu Bin. High-dimensional covariance estimation by minimizing ℓ1-penalized log-determinant divergence. Electron. J. Stat. 2011;5:935–980. ISSN 1935-7524. [Google Scholar]
- 26.Rothman Adam J, Bickel Peter J, Levina Elizaveta, Zhu Ji. Sparse permutation invariant covariance estimation. Electron. J. Stat. 2008;2:494–515. ISSN 1935-7524. [Google Scholar]
- 27.Sedaghat Nafiseh, Saegusa Takumi, Randolph Timothy, Shojaie Ali. Comparative study of computational methods for reconstructing genetic networks of cancer-related pathways. Cancer Informatics. 2014;13(Suppl 2):55–66. 09. doi: 10.4137/CIN.S13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shojaie Ali, Michailidis George. Penalized principal component regression on graphs for analysis of subnetworks. In: Lafferty John D, Williams Christopher KI, Shawe-Taylor John, Zemel Richard S, Culotta Aron., editors. NIPS. Curran Associates, Inc; 2010. pp. 2155–2163. [Google Scholar]
- 29.Städler Nicolas, Bühlmann Peter, Van De Geer Sara. ℓ1-penalization for mixture regression models. Test. 2010;19(2):209–256. [Google Scholar]
- 30.Tibshiranit Robert. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996;58(1):267–288. [Google Scholar]
- 31.Wang Yu-Xiang, Sharpnack James, Smola Alex, Tibshirani Ryan J. Trend filtering on graphs. arXiv preprint arXiv:1410.7690. 2014 [Google Scholar]
- 32.Weinberger Kilian Q, Sha Fei, Zhu Qihui, Saul Lawrence K. Graph laplacian regularization for large-scale semidefinite programming. Advances in neural information processing systems (NIPS) 2006:1489–1496. [Google Scholar]
- 33.Yuan Ming. High dimensional inverse covariance matrix estimation via linear programming. J. Mach. Learn. Res. 2010;11:2261–2286. ISSN 1532-4435. [Google Scholar]
- 34.Yuan Ming, Lin Yi. Model selection and estimation in the Gaussian graphical model. Biometrika. 2007;94(1):19–35. ISSN 0006-3444. [Google Scholar]
- 35.Zhao Peng, Yu Bin. On model selection consistency of lasso. The Journal of Machine Learning Research. 2006;7:2541–2563. [Google Scholar]
- 36.Zhao Peng, Rocha Guilherme, Yu Bin. The composite absolute penalties family for grouped and hierarchical variable selection’. Annals of Statistics. 2009;37(6A):3468–3497. [Google Scholar]
- 37.Zhao Sen, Shojaie Ali. A significance test for graph-constrained estimation. Biometrics (forthcoming) 2015 doi: 10.1111/biom.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]