Abstract
The Neuronal Ceroid Lipofuscinoses (NCLs), also known as Batten disease, result from mutations in over a dozen genes. Although, adults are susceptible, the NCLs are frequently classified as pediatric neurodegenerative diseases due to their greater pediatric prevalence. Initial clinical presentation usually consists of either seizures or retinopathy but develops to encompass both in conjunction with declining motor and cognitive function. The NCLs result in premature death due to the absence of curative therapies. Nevertheless, preclinical and clinical trials exist for various therapies. However, the genotypes of NCL animal models determine which therapeutic approaches can be assessed. Mutations of the CLN2 gene encoding a soluble lysosomal enzyme, tripeptidyl peptidase 1 (TPP1), cause late infantile NCL/CLN2 disease. The genotype of the original mouse model of CLN2 disease, Cln2-/-, excludes mutation guided therapies like antisense oligonucleotides and nonsense suppression. Therefore, the purpose of this study was to develop a model of CLN2 disease that allows for the assessment of all therapeutic approaches. Nonsense mutations in CLN2 disease are frequent, the most common being CLN2R208X. Thus, we created a mouse model that carries a mutation equivalent to the human p.R208X mutation. Molecular assessment of Cln2R207X/R207X tissues determined significant reduction in Cln2 transcript abundance and TPP1 enzyme activity. This reduction leads to the development of neurological impairment (e.g. tremors) and neuropathology (e.g. astrocytosis). Collectively, these assessments indicate that the Cln2R207X/R207X mouse is a valid CLN2 disease model which can be used for the preclinical evaluation of all therapeutic approaches including mutation guided therapies.
Introduction
Children across the globe are affected by any one of more than 40 different lysosomal storage disorders (LSDs); many of them resulting from deficiencies in either a lysosomal membrane protein or lysosomal enzyme [1, 2]. These deficiencies affect normal lysosomal function resulting in an accumulation of various lysosomal substrates. LSDs are commonly categorized according to the constituents of this accumulated material thus resulting in generalized LSD categories; for example the mucopolysaccharidoses accumulate mucopolysaccharides [1, 2]. All LSDs affect multiple physiological systems however, most appear to illicit a significant effect on the central nervous system (CNS) [3–5].
The Neuronal Ceroid Lipofuscinoses (NCLs) are a special category of LSD and accumulate a storage material referred to as ceroid lipofuscin [6–8]. Most commonly, NCL onset begins in childhood however in rarer occasions onset can occur in adulthood. Symptomatic hallmarks of the NCLs consist of but are not limited to epilepsy, tremors, retinopathy, gross and fine motor deterioration, and cognitive decline. Given the large neurological manifestations and predominance in childhood onset, the NCLs are considered to be the most common pediatric neurodegenerative disease [9–12]. Unfortunately, the NCLs are not limited to a single gene or genetic mutation. Currently, several mutations have been identified in more than 13 different genes; all resulting in a specific form of NCL [12–15]. Generally, these NCL forms are based on disease onset and mutated gene. The most common altered genes are CLN1, CLN2, and CLN3 causing classic Infantile Neuronal Ceroid Lipofuscinosis (cINCL), classic Late Infantile Neuronal Ceroid Lipofuscinosis (cLINCL), and classic Juvenile Neuronal Ceroid Lipofuscinosis (cJNCL) respectively [9, 16, 17].
cLINCL, also referred to as CLN2 disease, stems from a deficiency in the lysosomal enzyme tripeptidyl peptidase 1 (TPP1). This deficiency leads to a disease onset ranging from 2 to 4 years of age with initial clinical symptoms consisting of developmental delay and epilepsy [9, 16, 18]. Like all other NCLs, cLINCL results in premature death. To combat this premature death, numerous therapeutic approaches have been hypothesized and assessed [19, 20]. Experimental and clinical trials for cLINCL focus on gene therapy and enzyme replacement therapy. However, the assessment of some therapeutic approaches (i.e. antisense oligonucleotides and nonsense suppression compounds) is limited by the genotypes of available cLINCL animal models; for example, the genotype of the commonly used Cln2-/- mouse model does not permit the assessment of significantly relevant mutation-targeted therapies. Therefore, the purpose of this study was to generate an alternative cLINCL animal model that would allow for the assessment of all therapeutic approaches including nonsense suppression therapies. The most commonly annotated CLN2 mutations are c.509-G>C and p.R208X [21, 22]. Since one of the two most common mutations is a nonsense mutation, we generated a mouse model that carries the equivalent p.R208X mutation. Amino acid sequence for murine TPP1 is shorter than human TPP1 by 1 amino acid which occurs prior to the mutation; thus, the new cLINCL animal model is referred to as Cln2R207X/R207X. To be considered a valid model of cLINCL, Cln2R207X/R207X mice were vigorously assessed using various molecular, histological, and behavioral assessments. Results indicate that the Cln2R207X mutation leads to a significant reduction in both Cln2 transcript level and TPP1 enzyme activity. Consequences of these reductions consist of hallmark histopathological changes (i.e. substrate accumulation and astrocytosis) and neurological deficits (i.e. declining motor skills and tremors).
Materials and methods
Ethics statement
All animal research carried out in this manuscript followed National Institute of Health (NIH) and Sanford Research Institutional Animal Care and Use Committee (IACUC) guidelines. The animal protocol for this study was reviewed and approved by the Sanford Research IACUC.
Animals
Mice heterozygous for the Cln2 c.619C>T mutation in exon 6 (Cln2 p.R207X) and the neo cassette, Cln2R207X;neo/+, were generated and obtained from Applied Stem Cell, Inc. Generation of the Cln2R207X;neo/+mice was similar to the previously reported Cln1R151X;neo/+ mice [23]. Conversely, the neo cassette in the Cln2R207X;neo/+mice were flanked with FRT-sites. Cln2R207X;neo/+mice were natural bred to produce Cln2R207X;neo/R207X;neo. Cln2R207X;neo/R207X;neo mice were crossed with FLPo-10 mice (The Jackson Laboratory) to produce Cln2R207X/+ mice. Flippase-mediated excision of the neo cassette was assessed using polymerase chain reaction (Forward primer: 5’-GCTATTCGGCTATGACTGGG–3’; Reverse primer: 5’–CCCGGTAGAATTCCGATCAT-3’). Heterozygous Cln2R207X/+ mice were bred to produce colonies of Cln2R207X/R207X and Cln2+/+. Confirmation genotypic screening was performed using a quantitative real-time polymerase chain reaction assay (Forward primer: 5’- TGCCTGAGCCCACATCTTTG-3’; Reverse primer: 5’-TGGGAGTGACTCCGTCTGT-3’; Reporter 1(VIC): 5’- CAGGTTGTATCGCTGACG-3’; Reporter 2(FAM): 5’- CAGGTTGTATCACTGAC-3’) from Thermo Fisher Scientific. Running conditions for the qRT-PCR assay consisted of: 1) 95°C for 15 min. (1 cycle) and 2) 95°C for 1 sec. and 60°C for 1 min. (40 cycles). Genomic sequencing confirmation was conducted through Eurofins Genomics (Forward primer: 5’-CTTGCTGGCTGAGTGTCCGGT-3’; Reverse primer: 5’-CTACCAGCTTCCCCAGGCCTTG-3’). Both the Cln2R207X/R207X and Cln2+/+ colonies were on a mixed genetic background (C57BL/6x129S6/SvEv). All animals were housed, cared for, and used under the guidelines set by the NIH and the direction of the Sanford Research IACUC. Mice were housed in individually vented microisolator cages (four or five mice/cage) with ad libitum access to food and water. Mice were fed with the Teklad Global 2918 diet (Harlan Laboratories, Indianapolis, IN), and their drinking water was tap water.
Cln2R207X/R207X mice have a severe disease phenotype, twitching starts at 90 days of age and die within a couple months after twitching onset. Therefore, from 90 days of age, Cln2R207X/R207X mice were monitored daily and were euthanized by carbon dioxide when debilitating phenotypes (i.e. immobility, difficulties in feeding) were observed. The care for the Cln2R207X/R207X mice followed our animal protocol approved by the Sanford Research IACUC.
Protein sample preparation
Under the NIH and Sanford Research IACUC guidelines, 1-month-old Cln2R207X/R207X (n = 3) and Cln2+/+ (n = 3) mice were euthanized using a carbon dioxide chamber. Following euthanasia, animals were flushed with 10 ml of sterile phosphate buffered saline solution via cardiac perfusion. Tissues including cerebral cortex, cerebellum, liver, spleen, and kidney were harvested and flash frozen using dry ice. Tissues were stored at -80°C. Upon their removal from -80°C, tissue sections were removed using a sterile blade, placed into 2mL tubes containing zirconium beads and 500 μL lysis buffer [50 mM Tris-HCl pH 7.4; 150 mM NaCl; 0.2% Triton X-100; 0.3% NP-40; 0.1 mM PMSF; 1x HALT Protease Inhibitor Cocktail (Thermo Fisher Scientific)], and then homogenized using a Bertin Technologies Precellys 24 homogenizer. Samples were then transferred to clean 1.5-ml microcentrifuge tubes and pulse sonicated using a Branson Sonifier 250. Following sonication, samples were incubated on ice for 30 min and then centrifuged at 12,000g for 10min. Sample supernatants were then transferred to clean pre-chilled 1.5mL microcentrifuge tubes and stored at -80°C. Total protein concentrations were quantified using the Pierce 660 nm Protein Assay (Thermo Fisher Scientific).
TPP1 enzyme activity assay
Using previously described methods, the aforementioned Cln2R207X/R207X and Cln2+/+ total protein samples were evaluated for TPP1 enzyme activity [24, 25]. The assays were conducted in a black, flat-bottom 96-well plate. 10 μg of total protein were loaded into each well. Each sample was done in quadruplicates. 40 μl of 250 μM Ala-Ala-Phe-7-amido-4-methylcoumarin [10 mM Ala-Ala-Phe AMC (Sigma Aldrich) diluted in substrate buffer (150 mM NaCl; 0.1% Triton X-100; 100 mM sodium acetate; pH 4.0)] was added to each well. The plate was incubated in the dark at 37°C for 1h. 200 μl of stop buffer [0.1 M monochloroacetic acid; 0.13 M NaOH; 0.1 M acetic acid; pH 4.3] was added to each well. The plate was gently shaken and fluorescence was read using a BioTek Cytation 3 (Excite 380 nm/Emit 460 nm). Background fluorescence was accounted and removed from each sample. Relative fluorescence was calculated as a percentage of wild type fluorescence.
PPT1 enzyme activity assay
Using previously described methods, the aforementioned Cln2R207X/R207X and Cln2+/+ total protein samples were evaluated for PPT1 enzyme activity [25, 26]. The assays were conducted in a black, flat-bottom 96-well plate. 10 μg of total protein were loaded into each well. Each sample was done in quadruplicates. 20 μl of PPT1 substrate [0.64 mM 4-methylumbelliferyl-6-thiopalmitoyl-β-glucoside (Santa Cruz); 15 mM dithiothreitol with 2.3 mg/ml bovine serum albumin and 0.09% sodium azide; 0.2 M disodium phosphate with 0.1 M citric acid buffer pH 4.0; 0.945 mg/mL β-Glucosidase from almonds (Sigma Aldrich)] was added to each well. The plate was incubated in the dark at 37°C for 1h. 200 μl of stop buffer [0.5 M sodium bicarbonate with 0.5 M sodium carbonate pH 10.7; 0.025% Triton X-100] was added to each well. The plate was gently shaken and fluorescence was read using a BioTek Cytation 3 (Excite 355 nm/Emit 460 nm). Background fluorescence was accounted and removed from each sample. Relative fluorescence was calculated as a percentage of wild type fluorescence.
Sphingomyelinase activity assay
Sphingomyelinase activity was assessed using the Amplex Red Sphingomyelinase Assay Kit (Thermo Fisher Scientific). The assay was performed according to the kits protocol. 10 μg of total protein was assessed for each sample and done in triplicates. Fluorescence was read using a BioTek Cytation 3 (Excite 545 nm/Emit 590 nm). Background fluorescence was accounted and removed from each sample. Relative fluorescence was calculated as a percentage of wild type fluorescence.
RNA sample preparation
Under the NIH and Sanford Research IACUC guidelines, 1-month-old Cln2R207X/R207X (n = 3) and Cln2+/+ (n = 3) mice were euthanized using a carbon dioxide chamber. Following euthanasia, animals were flushed with 10 ml of sterile phosphate buffered saline solution via cardiac perfusion. Tissues including cerebral cortex, cerebellum, liver, spleen, and kidney were harvested and flash frozen using dry ice. Tissues were stored at -80°C. Upon their removal from -80°C, tissue sections were removed using a sterile blade and placed into RNAse free tubes. Maxwell 16 LEV simplyRNA tissue kits and a Maxwell 16 instrument (Promega) were utilized for RNA extractions. Samples were processed according to the Maxwell 16 LEV simplyRNA tissue kit. RNA samples were then assessed and quantified using the BioTek Epoch Microplate Spectrophotometer; all samples had concentrations between 200–1000 ng/μl and an A260/A280 > 2.
Reverse transcription
Using the GoScript Reverse Transcription System (Promega), cDNA synthesis was performed on 1 μg of total RNA from each sample. Reactions were run in an Applied Biosystems Veriti Thermal Cycler with the following conditions: 25°C for 5 min., 42°C for 60 min., 70°C for 15 min., and a 4°C hold. Samples were stored at -20°C.
Quantitative real-time polymerase chain reaction
Prior to their use, cDNA samples were diluted 1:20 using nuclease free water. Quantitative real-time polymerase chain reactions were carried out using the Absolute Blue QPCR Mix (Thermo Fisher Scientific). Cln2 expression was measured using the Tpp1 TaqMan Gene Expression Assay from Thermo Fisher Scientific (Mm00487016_m1). In addition, Gapdh expression was measured using the Gapdh TaqMan Gene Expression Assay from Thermo Fisher Scientific (Mm99999915_g1). All reactions were run in a Stratagene Mx3005P qPCR machine (Agilent Technologies). Reaction conditions consisted of the following: 1) 95°C for 15 min. (1 cycle) and 2) 95°C for 15 sec.; 60°C for 1 min. (40 cycles). ΔΔCT was used to calculate relative fold expression.
Behavioral testing
All behavioral tests were performed on male mice as they became available in our animal colony. The same cohorts of Cln2R207X/R207X (n = 20) and Cln2+/+ (n = 14) mice were tested at 1 and 3 months of age. Prior to performing any behavioral tests, the mice were acclimated to the environment for at least 20 min. Behavioral testing consisted of the following four day scenario: Day 1) Modified vertical pole test and force-plate actimeter, Day 2) Rotarod, Day 3) Light/Dark box, and Day 4) Hanging wire.
Modified vertical pole test
Vertical pole testing using a threaded metal rod was carried out in two phases: 1) Climbing downward and 2) Turning downward; according to our published method [23, 27]. First, the mouse was placed at the top of the pole facing downward, and was given a 60-s window to descend the pole. The time to descend the pole was recorded and tracked for 5 consecutive trials. After the 5th climbing down trial, the same mouse was placed towards the top of the pole facing upwards and timed to determine how long it takes to turn around and face downward. The mouse was given 4 consecutive trials and a 60-s window per trial. If the mouse fell from the pole during the climbing down or turning downward trials the mouse was given a time of 60 sec.
Force-plate actimeter
Mice were placed into a force-plate actimeter (BASi, West Lafayette, IN) and allowed to roam for 20 min. All tracked information was processed and analyzed using FPAanalysis software version 1.10.01 (BASi, West Lafayette, IN).
Rotarod test
An accelerating rotarod test was administered as we described previously [23, 27]. Rota Rod Rotamex-5 machines (Columbus Instruments, Columbus, OH) were used for animal testing. Testing parameters consisted of the following: 1) start speed: 0 rpm; end speed: 48 rpm, 2) acceleration occurred in increments of 0.2 rpm per second, and 3) total allotted test time: 240 s. Mice were trained on the rotarod in 3 consecutive trial runs. After a 1.5-h resting period, mice were tested on the rotarod in 3 test trials each consisting of three consecutive runs, with 15 minutes of rest between trials. Latency to fall was calculated as an average of the 9 test runs.
Light/Dark box test
Anxiety was measured in 3-month-old Cln2R207X/R207X and Cln2+/+ mice via the light/dark box test [28, 29]. Mice were placed into light/dark boxes (Stoelting Co., Wood Dale, IL) and allowed to roam freely for 20 min. Mouse movement was tracked and recorded using ANY-maze equipment and software (Stoelting Co., Wood Dale, IL). The recorded data was used to determine the amount of time spent in the dark and the amount of time spent in the light.
Hanging wire test
Strength in addition to motor coordination was assessed using a modified hanging wire test [29, 30]. Only 3-month-old Cln2R207X/R207X and Cln2+/+ mice were tested. To conduct this test, mice were placed in the center of a food rack which was subsequently inverted and suspended approximately 18 inches above a cage containing bedding. Mice were timed to see how long they could remain suspended; 60 s was used as the cut-off time. Each mouse was tested 5 times and the average time suspended was calculated.
Histological preparation
Under the NIH and Sanford Research IACUC guidelines, 3-month-old Cln2R207X/R207X and Cln2+/+ mice were euthanized using a carbon dioxide chamber. Following euthanasia, animals were flushed with 10 ml of sterile phosphate buffered saline solution via cardiac perfusion. PBS perfusion was followed by a 10 ml perfusion of 4% paraformaldehyde in PBS. Tissues including the cerebrum and cerebellum were harvested and placed into individual scintillation vials containing a solution of 4% paraformaldehyde in PBS. After 24 h, the 4% paraformaldehyde solution was replaced with PBS containing 0.02% sodium azide and stored at 4°C. Tissues were removed from storage and embedded into 3% low melting point agarose. Samples were sliced into 50μm sections using a Leica VT1000S vibratome and serially divided 1:6.
Immunohistochemistry
Serial sections of Cln2R207X/R207X (n = 5) and Cln2+/+ (n = 4) cerebrums were immunostained for mitochondrial ATP synthase subunit c (subunit c), glial fibrillary acidic protein (GFAP), and ionized calcium binding adapter molecule 1 (Iba1). Serial sections of Cln2R207X/R207X (n = 5) and Cln2+/+ (n = 4) cerebella were immunostained for mitochondrial ATP synthase subunit c (subunit c). All immunostainings were done on floating tissue sections and conducted in a 24-well plate. Incubations lasting an hour or more were done on a plate rocker. Tissue sections were blocked in blocking buffer [PBS; 3% bovine serum albumin; 5% goat serum; 0.2% Triton X-100; 0.02% sodium azide] for 1 h at room temperature. Blocking buffer was removed and sections were incubated with the primary antibody diluted in blocking buffer [anti-subunit c 1:100 (Abcam: ab181243); anti-GFAP 1:250 (Dako: 70334); anti-Iba1 1:250 (Biocare Medical: CP290 A,B)] at room temperature overnight. Primary antibody was removed and slices were washed three times in PBS. The secondary antibody [anti-rabbit IgG coupled to Alexa Fluor 568 (Thermo Fisher Scientific: z25306)] diluted 1:1500 and 4’,6-Diamidino-2-Phenylindole (DAPI) (Thermo Fisher Scientific: D1306) diluted 1:1000 all in PBS was added to each sample. The plates were covered in aluminum foil and incubated at room temperature for 1 h. The secondary antibody and DAPI were removed and slices were washed three times in PBS. Floating sections were then transferred to labeled glass slides, mounted using DAKO mounting medium, and covered with cover glass. Slides were allowed to dry at room temperature and then stored at 4°C.
Double staining for co-localization was conducted using the same protocol. Anti-mitochondrial ATP synthase subunit c 1:100 (Abcam: ab181243) and anti-lysosomal associated membrane protein 1 (LAMP-1) 1:1000 (Santa Cruz: sc-20011) diluted in blocking buffer were used as the primary antibodies. The secondary antibodies were: anti-rabbit IgG coupled to Alexa Flour 568 (Thermo Fisher Scientific: z25306) 1:1500 and anti-mouse IgG coupled to Alexa Fluor 488 (Thermo Fisher Scientific: z25002) 1:1500.
Image acquisition
All slides were viewed and images collected using a Nikon Eclipse Ni-E microscope equipped with Photometrics CoolSNAP MYO CCD camera. Cerebral and cerebellar sections were viewed and imaged using a 20X-objective. Cortical regions of cerebral sections were visually divided into superficial and deep cortical layers. Four images were collected per superficial and deep cortical layers per cerebral section. This approach was used for all cerebral immunostainings. Sections double immunostained for co-localization were viewed and imaged using an oil immersion 100X objective.
Image analysis
Images were analyzed and processed with the Cellomics Scan (Thermo Fisher Scientific) software using the Spot Detector BioAssay (Version 4). Subunit c positive staining was measured in a region of interest created based on the DAPI signal and expanded by 10 pixels using a threshold based analysis. Individual cells were then identified and counted based on the nuclei (DAPI), subunit c signal intensity was then measured in the region of interest and positive puncta were counted and measured for size. Averages of 6000 cells were counted per group. In GFAP analysis, cells stained positive for GFAP were used to measure the total staining intensity, the total area of GFAP staining, and the average area of a GFAP positive cell. In Iba1 analysis, cells stained positive for Iba1 were used to measure the total staining intensity, the average area of a Iba1 positive cell, and the average staining intensity per cell.
Statistical analysis
All data was graphed and statistically analyzed using GraphPad PRISM Version 5.04 software.
Results
CLN2 mutations
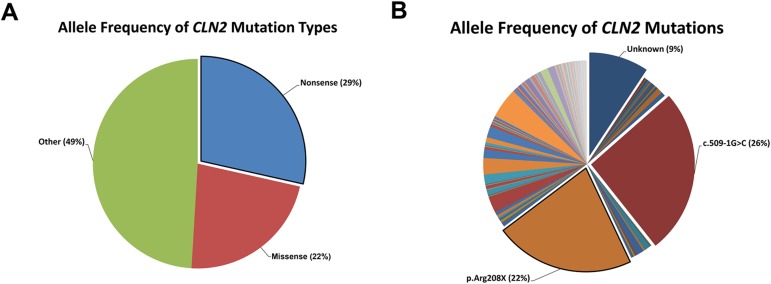
To establish mutation type frequencies (e.g. missense, nonsense) in CLN2 disease patients, the University College London’s online NCL mutation database was assessed for recorded CLN2 mutations [22]. Missense and nonsense mutations account for more than 50% of the recorded mutation allele frequencies; of which missense mutations contribute 22% and nonsense mutations contribute 29% (Fig 1A). Using the same set of data, we determined that the most common nonsense mutation in CLN2 disease patients is p.Arg208X; which appears to occur with an allele frequency of 22% (Fig 1B). This correlates with previous reports on the occurrence of p.Arg208X [21, 31]. The allele frequency of CLN2 p.Arg208X directed us to generate the murine equivalent, Cln2R207X.
Fig 1. Frequency of CLN2 disease-associated mutations.
(A) Allele frequency of CLN2 disease mutations demonstrates a predominance of nonsense (29%) and missense (22%) mutations. (B) The most common CLN2 disease mutations consists of either the intronic transversion c.509-1G>C that results in altered transcript splicing or the exonic transition c.622C>T that results in the p.R208X nonsense mutation.
Cln2R207X/R207X mouse model generation
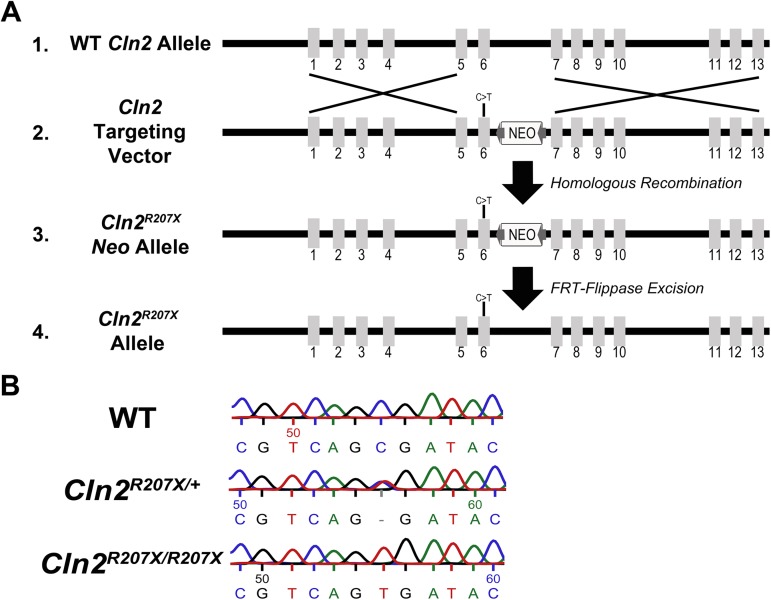
Complete overview of the Cln2R207X/R207X mouse model generation is represented in Fig 2A. Applied StemCell, Inc. was outsourced to generate our initial Cln2R207XNeo/+ mouse founders. To create these founders, a Cln2 targeting vector carrying a C to T transition in exon 6 and a Neomycin selection cassette flanked with FRT sites between exon 6 and exon 7, all within Cln2, was placed into mouse embryonic stem cells. Through homologous recombination the targeting vector inserted into the embryonic stem cell genome. Cln2R207XNeo/+ mouse founders were then bred to create Cln2R207XNeo/R207XNeo. The Neomycin selection cassette was removed via FRT-Flippase mediated excision by breeding Cln2R207XNeo/R207XNeo mice with FLPo-10 mice obtained from Jackson Laboratory. The Cln2R207X/+ mice obtained following Neomycin cassette removal were then used to generate our Cln2+/+ and Cln2R207X/R207X colonies. Sequencing results confirmed preservation of the point mutation (Fig 2B).
Fig 2. Genetic design of the CLN2 disease mouse model, Cln2R207X/R207X.
(A) A Cln2 targeting vector carrying the c.619C>T mutation (Line 2), also known as p.R207X mutation, was electroporated into C57BL/6x129S6/SvEv embryonic stem cells. Homologous recombination between wildtype Cln2 (Line 1) and the targeting vector (Line 2) resulted in the generation of a Cln2Neo;R207X allele (Line 3). Removal of the Neo cassette via FRT-flippase mediated excision (FRT sites represented by grey arrows flanking the Neo cassette) generated the Cln2R207X allele (Line 4) resulting in Cln2+/R207X mice. Cln2+/R207X mice were bred to create Cln2R207X/R207X mice. (B) Cln2 sequencing results confirm the retention of the mutation following breeding.
Cln2R207X transcript level and TPP1 enzyme activity in Cln2R207X/R207X mice
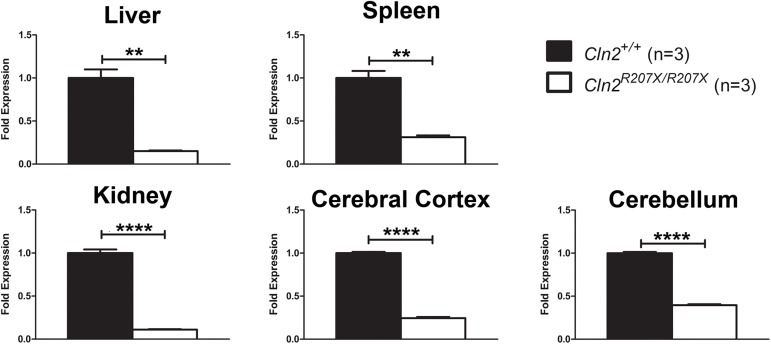
Transcripts containing nonsense mutations/premature termination codons (PTCs) are targeted for degradation via the nonsense mediated decay (NMD) pathway; thus minimalizing the abundance of PTC-containing transcripts and reducing the production of truncated and potentially harmful proteins. To assess NMD in Cln2R207X/R207X mice, Cln2 transcript levels were measured from liver, spleen, kidney, cerebral cortex, and cerebellum via quantitative real-time PCR (qRT-PCR). Cln2R207X/R207X mice demonstrated a 60–90% reduction in Cln2 transcript levels when compared to Cln2+/+ mice (Fig 3).
Fig 3. Ubiquitously reduced Cln2 transcript abundance in Cln2R207X/R207X mice.
Quantitative real-time PCR was used to measure endogenous levels of the Cln2 transcript from five different tissues obtained from 1-month-old Cln2+/+ (n = 3) and Cln2R207X/R207X (n = 3) mice. Three technical replicates were performed using the three biological samples obtained from Cln2+/+ and Cln2R207X/R207X mice. Cln2 transcript levels were normalized to Gapdh expression. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (**p < 0.01 and **** p < 0.0001).
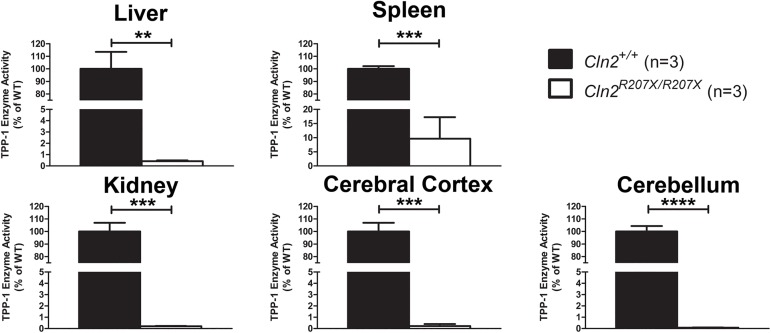
Transcripts containing PTCs that manage to escape NMD lead to the translation of truncated proteins; the functionality of these proteins is dependent on the location of the truncation. Therefore, in combination with NMD-mediated Cln2 transcript reduction, the activity of TPP1, the enzyme encoded by Cln2, should be significantly reduced in Cln2R207X/R207X mice. Using a previously established TPP1 fluorogenic enzyme activity assay, TPP1 activity was evaluated in Cln2R207X/R207X mice (Fig 4). TPP1 activity was significantly reduced in five different tissues when compared to Cln2+/+ mice; the overall reduction consisted of at least 90%. To ensure that lysosomal dysfunction was solely due to reduced TPP1 activity, similar methods (fluorogenic enzyme activity assays) were used to assess the activity of palmitoyl-protein thioesterase 1 (PPT1), the lysosomal enzyme associated with cINCL, and sphingomyelinase, the lysosomal enzyme associated with Niemann-Pick disease. Overall, Cln2R207X/R207X mice were found to have similar PPT1 and sphingomyelinase activity when compared to Cln2+/+ mice (S1 Fig). The only difference was found in the liver: a significant decrease in PPT1 activity in Cln2R207X/R207X mice.
Fig 4. Decreased TPP1 activity in various Cln2R207X/R207X mouse tissues.
Fluorogenic TPP1 enzyme activity assays were used to measure endogenous levels of TPP1 activity from five different tissues obtained from 1-month-old Cln2+/+ (n = 3) and Cln2R207X/R207X (n = 3) mice. Four technical replicates were performed using the three biological samples obtained from Cln2+/+ and Cln2R207X/R207X mice. Cln2R207X/R207X TPP1 activity was normalized to Cln2+/+ TPP1 activity. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (** p < 0.01, ***p < 0.001, and **** p < 0.0001).
Behavioral assessment of Cln2R207X/R207X mice
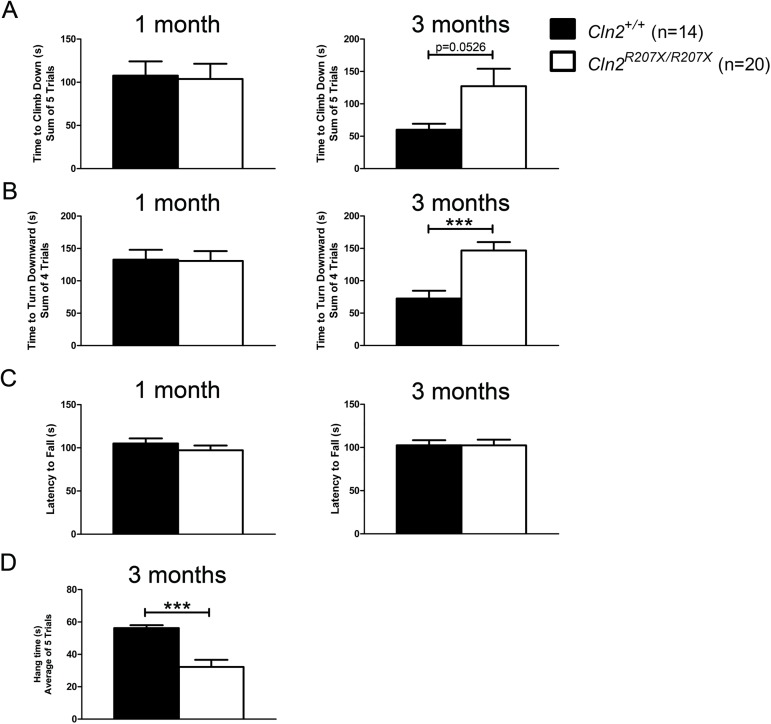
Cln2+/+ and Cln2R207X/R207X male mice were compared in a battery of behavioral tests at 1 and 3 months of age. Older mice were not tested because premature death in Cln2R207X/R207X mice started to appear after 3 months of age and progressively worsened; afflicting most mice by at least 6 months of age (Fig 5). To test for motor deficits, a modified vertical pole test was conducted as previously described [32]. First, the ability of a mouse to descend the pole was evaluated in 5 consecutive trials, and then the ability of the same mouse to turn around on the pole was tested in 4 consecutive trials. At 1 month of age, both Cln2+/+ and Cln2R207X/R207X mice descended the vertical pole in similar time (Fig 6A). At 3 months, Cln2R207X/R207X mice strongly trended towards taking a longer time to descend but the difference between Cln2+/+ and Cln2R207X/R207X mice was shy of statistical significance (Fig 6A). In terms of their ability to completely turn around on the vertical pole, while no differences were detected at 1 month, Cln2R207X/R207X mice at 3 months displayed significant difficulties (Fig 6B). Motor coordination and balance was also tested in an accelerating rotarod test (0-48rpm in 240 sec). At 1 and 3 months, the rotarod performance of Cln2+/+ and Cln2R207X/R207X mice was similar (Fig 6C). Given that 3-month-old Cln2R207X/R207X mice started to display some motor deficits, overall strength was evaluated using the hanging wire test. At 3 months of age, Cln2R207X/R207X mice showed significant impediments in their ability to hang from a wire rack (Fig 6D). To examine if anxiety is a contributing factor to the aforementioned behaviors, the anxiety level of 3-month-old Cln2+/+ and Cln2R207X/R207X mice were evaluated using the light/dark box test. No difference in anxiety between Cln2+/+ and Cln2R207X/R207X mice was found (S2 Fig).
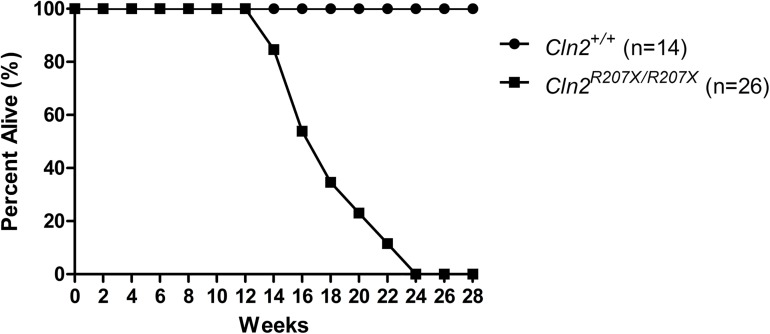
Fig 5. Significantly reduced lifespan in Cln2R207X/R207X mice.
Cln2+/+ (n = 14) and Cln2R207X/R207X (n = 26) mice were plotted on a survival curve and tracked overtime. Cln2R207X/R207X mice died between 3 and 6 months of age.
Fig 6. Altered motor skills in 3-month-old Cln2R207X/R207X mice.
A panel of behavioral tests including modified vertical pole (A and B), rotarod (C), and modified hanging wire (D) were used to assess motor skills in 1 and 3-month-old Cln2+/+ (n = 14) and Cln2R207X/R207X (n = 20) mice. The modified hanging wire was only evaluated at 3 months of age. (A) Climb down time as assessed by the vertical pole showed no differences at 1 month of age between Cln2+/+ and Cln2R207X/R207X but at 3 months of age Cln2R207X/R207X mice trended towards an increased time. (B) Time to turn downward, also assessed by the vertical pole, showed no differences at 1 month of age between Cln2+/+ and Cln2R207X/R207X. By 3 months of age however, Cln2R207X/R207X mice demonstrated significant difficulties turning around. (C) Cln2+/+ and Cln2R207X/R207X mice exhibited no differences in their latency to fall from an accelerating rotarod at both 1 and 3 months of age. (D) 3-month-old Cln2R207X/R207X mice have significant difficulties hanging from a wire rack. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (***p < 0.001).
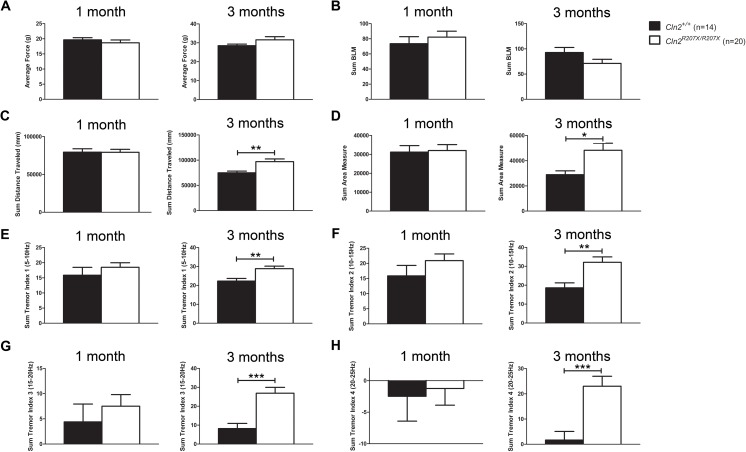
To test multiple behavioral parameters in freely-moving Cln2+/+ and Cln2R207X/R207X mice a force-plate actimeter was used (BASi, West Lafayette, IN; Ossowska et al., 2014, Neuropharmacology 83: 28–35). This device tracks the movement of mice by tracing the changes in the force applied to the plate. Several parameters including distance traveled, area covered, number of left and right turns, focused stereotypes, low mobility bouts, and tremor index are measured. The average force, closely related to the weight of the mice, did not differ between Cln2+/+ and Cln2R207X/R207X mice at 1 and 3 months (Fig 7A). Cln2+/+ and Cln2R207X/R207X mice also had similar bouts of low mobility at both 1 and 3 months (Fig 7B). Tracings of the mice indicated that at 3 months, Cln2R207X/R207X mice traveled a greater distance and covered a larger surface area (Fig 7C and 7D). These potential indicators of hyperactivity correlated with observations made during the handling of the mice. Through continuous tracking of the applied force, tremors or seizures can be detected and measured by calculating a tremor index score. Tremor indices for this study covered tremor frequencies ranging from 5–10 Hz, 10–15 Hz, 15–20 Hz, and 20–25 Hz. At 1 month of age, Cln2+/+ and Cln2R207X/R207X mice had similar tremor index scores for all frequency ranges (Fig 7E–7H). At 3 months of age, however, Cln2R207X/R207X mice showed a marked increase in all four tremor indices (Fig 7E–7H). Observationally, the tremor became more severe and debilitating after 3 months of age.
Fig 7. Cln2R207X/R207X mice display characteristics of hyperactivity and develop tremors by 3 months of age.
Various behavioral characteristics of 1 and 3-month-old Cln2+/+ (n = 14) and Cln2R207X/R207X (n = 20) mice were evaluated using a force-plate actimeter. (A) Average force increased from 1 to 3 months of age but did not differ between Cln2+/+ and Cln2R207X/R207X at either time points. (B) Cln2+/+ and Cln2R207X/R207X mice had similar bouts of low mobility at both 1 and 3 months. (C,D) By 3 months of age, Cln2R207X/R207X mice display signs of hyperactivity such as significantly increased distance traveled and area coverage. (E-H) Software analysis of the force-plate actimeter data permits the evaluation of tremors by calculating a tremor index score. Tremor index scores are separated according to tremor frequencies such as, 5–10 Hz (E), 10–15 Hz (F), 15–20 Hz (G) and 20–25 Hz (H). As Cln2R207X/R207X mice aged from 1 to 3 months, tremor index scores increased significantly. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (*p < 0.05, **p < 0.01, and ***p < 0.001).
Histological evaluation of Cln2R207X/R207X mice
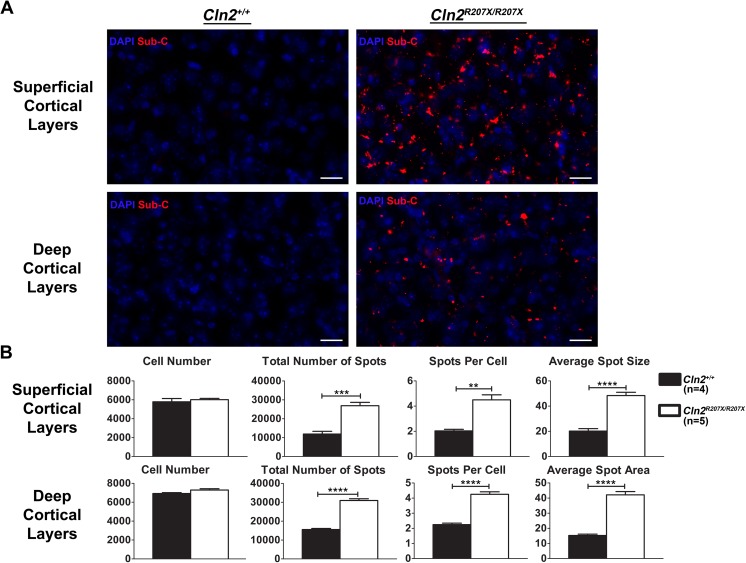
Based on the age of onset for behavioral anomalies in the Cln2R207X/R207X mice, 3-month-old Cln2R207X/R207X cerebrums and cerebellums were evaluated for NCL associated neuropathological hallmarks. Generally, these include lysosomal storage material accumulation, astrocytosis, and activated microglia [11, 17, 33–39]. Due to the natural fluorescence of NCL-associated lysosomal storage material, intracellular autofluorescence is usually measured. Three-month-old Cln2R207X/R207X cerebrum and cerebellum did not demonstrate autofluorescent storage material (data not shown). Nevertheless, a principal component of cLINCL storage material is mitochondrial ATP synthase subunit c, a substrate of TPP1 [40–43]. Using immunohistochemistry, Cln2+/+ and Cln2R207X/R207X brain sections were evaluated for mitochondrial ATP synthase subunit c accumulation. The cerebral cortex of Cln2R207X/R207X mice had a pronounced accumulation of mitochondrial ATP synthase subunit c throughout superficial and deep cortical layers; whereas, Cln2+/+ brain sections had little immunoreactivity (Fig 8A). Quantitative analysis of the immunoreactive puncta throughout the Cln2R207X/R207X cerebral cortex in comparison to the Cln2+/+ cortex revealed an increased frequency per tissue (superficial layers: 2.26 fold increase, p = 0.0004; deep layers: 1.98 fold increase, p < 0.0001), increased frequency per cell (superficial layers: 2.21 fold increase, p = 0.0011; deep layers: 1.89 fold increase, p < 0.0001), and a greater size (superficial layers: 2.4 fold increase, p < 0.0001; deep layers: 2.67 fold increase, p < 0.0001) (Fig 8B). Likewise, the cerebellum of Cln2R207X/R207X mice had profound mitochondrial ATP synthase subunit c accumulation throughout cerebellar regions, whereas the cerebellum of Cln2+/+ mice had little to none (S3 Fig). To confirm the subcellular location of the ATP synthase subunit c puncta, cerebral cortical sections from Cln2R207X/R207X mice were immunostained for LAMP-1, a lysosomal membrane protein, and for the mitochondrial ATP synthase subunit c. A significant portion of mitochondrial ATP synthase subunit c co-localized with LAMP-1 (S4 Fig).
Fig 8. Lysosomal accumulation of mitochondrial ATP synthase subunit c in Cln2R207X/R207X mice.
Subunit c accumulation was detected by immunofluorescent staining. (A) Images of superficial and deep cortical layers demonstrate diffuse and pronounced accumulation of mitochondrial ATP synthase subunit c in 3-month-old Cln2R207X/R207X mice. (B) Images from 3-month-old Cln2+/+ (n = 4) and Cln2R207X/R207X (n = 5) mice were blindly collected and analyzed for differences in cell number, total number of immunoreactive puncta, number of puncta per cell, and average punctum area. Cln2R207X/R207X mice have significantly increased total number of puncta, puncta per cell, and punctum size. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (**p < 0.01, ***p < 0.001, and **** p < 0.0001).
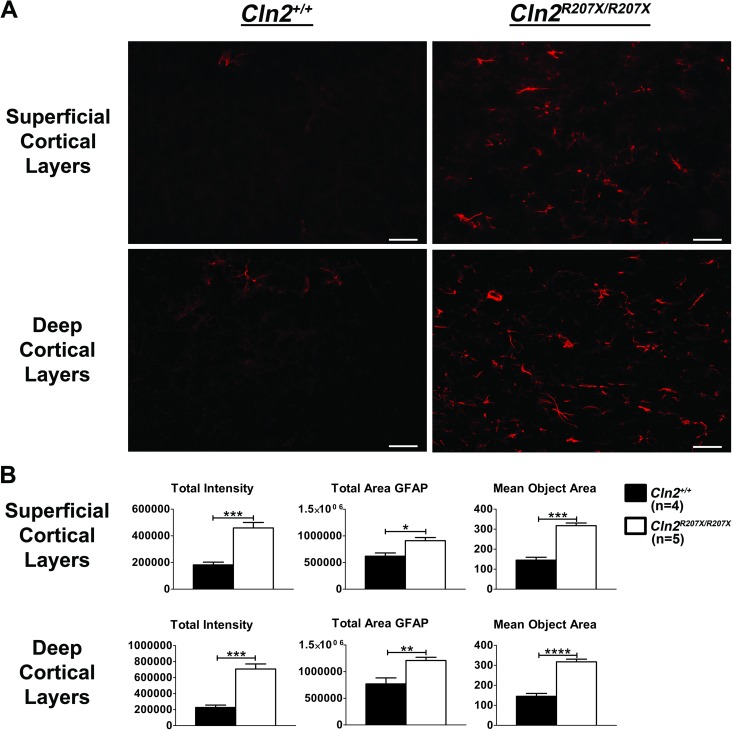
Astrocytosis is a common pathological finding in neurodegenerative diseases [44–47]. Most commonly, the immunostaining intensity of glial fibrillary acidic protein (GFAP) and astrocyte morphology are used to evaluate astrocytosis; even within the NCLs [11, 17, 33–36, 38, 48–51]. Therefore, 3-month-old Cln2+/+ and Cln2R207X/R207X cerebral sections were immunostained for GFAP. Superficial and deep cortical layers in sections from Cln2R207X/R207X mice demonstrated significantly increased GFAP immunostaining as well as hypertrophy of astrocytes when compared to sections from Cln2+/+ mice (Fig 9A). Superficial cortical layers of Cln2R207X/R207X mice were found to have a 2.52 fold increase in GFAP intensity (p = 0.0008) and a 2.04 fold increase in mean astrocyte area (p = 0.0002), whereas deep cortical layers of Cln2R207X/R207X mice showed a 3.12 fold increase in GFAP intensity (p = 0.0004) and a 2.19 fold increase in mean astrocyte area (p < 0.0001) (Fig 9B). An additional pathological feature of neurodegenerative diseases is activated microglia. Microglial activation in the brain can be evaluated using immunostaining for the microglial markers, Iba1, F4/80, and CD68 [52–56]. The morphology of microglial cells dramatically changes during activation: the resting cells lose their processes and become more rounded [52, 53, 57]. Cerebral sections from 3-month-old Cln2+/+ and Cln2R207X/R207X mice were immunostained for Iba1, and microglial activation was evaluated based on staining intensity and cell morphology. No detectable differences were identified in total Iba1 immunostaining intensity, average microglial Iba1 intensity, and average microglia size (S5 Fig). Although microglial activation in Cln2R207X/R207X mice was not present at 3 months, microglial activation during later periods of disease progression cannot be ruled out. However, for the purposes of this project we were interested in brain pathology prior to the sudden decline in the survival curve.
Fig 9. Diffuse cortical astrocytosis in Cln2R207X/R207X mice.
(A) Images of superficial and deep cortical layers from 3-month-old Cln2+/+ and Cln2R207X/R207X mice demonstrate increased GFAP immunostaining and enlarged astrocytes. (B) Images from 3-month-old Cln2+/+ (n = 4) and Cln2R207X/R207X (n = 5) mice were blindly collected and analyzed for GFAP immunostaining intensity, GFAP staining area, and astrocyte area. Cln2R207X/R207X mice have significantly increased GFAP immunostaining and astrocyte size relative to Cln2+/+. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (*p < 0.05, **p < 0.01, ***p < 0.001, and **** p < 0.0001).
Collectively, cerebral and cerebellar lysosomal accumulation of mitochondrial ATP synthase subunit c and cerebral astrocytosis indicate disease pathology at the onset of both behavioral phenotypes and decline in survival. However, these pathological findings do not identify a clear cause of premature death. To help identify potential causes of death and peripheral pathology, tissues (i.e. spleen, lung, heart, eye, and brain) from 4-month-old Cln2+/+ and Cln2R207X/R207X mice were sectioned, hematoxylin/eosin stained, and sent to a veterinary pathologist for examination. The veterinary pathologist was blinded from all identifying information. According to the pathology report, the brain was the only tissue with identifiable abnormalities which consisted of hypereosinophilic granules and sporadic neurodegeneration (S6 Fig). Consequently, no definitive causes of death were identified.
Discussion
Current preclinical therapeutic studies for CLN2 disease utilize the Cln2-/- mouse model which carries an uncommon missense mutation [37]. Cln2-/- mice are not suitable to test therapies such as read-through compounds, nonsense mediated decay inhibitors, and clinically relevant mutation targeting antisense oligonucleotides. Therefore, the primary purpose of this study was to generate an alternative model of CLN2 disease that allows for the evaluation of all therapies including those previously excluded.
The new model was chosen to have the most common nonsense mutation that occurs in CLN2 disease patients, p.R207X. To be considered a model of CLN2 disease, the Cln2R207X/R207X mouse was rigorously characterized to confirm the presence of CLN2 disease-associated anomalies. Molecular and biochemical characterization confirmed that the presence of the p.R207X mutation targeted Cln2 transcripts for the governing nonsense mediated decay pathway; this reduction in combination with TPP1 truncation resulted in a significant decrease in ubiquitous TPP1 activity. These alterations in TPP1 activity are similar to what is seen in CLN2 disease patients [25, 58, 59]. Loss of TPP1 activity results in lysosomal dysfunction. Thus, the lysosomes in CLN2 disease patients and models accumulate autofluorescent storage material which consists of various components including mitochondrial ATPase synthase subunit c [40, 60–62]. Histological assessment of Cln2R207X/R207X cerebral and cerebellar sections did not show autofluorescent storage material at the same time point as symptomatic onset (3 months). Nevertheless, similar sections strongly demonstrated the accumulation of mitochondrial ATPase synthase subunit c. Co-localization of mitochondrial ATPase synthase subunit c and LAMP-1 indicates lysosomal accumulation of non-autofluorescent material prior to the accumulation of autofluorescent components. Secondary pathological findings such as astrocytosis in 3-month-old Cln2R207X/R207X cerebral sections confirm previous findings of NCL. Overall, both biochemical and histological assessments of Cln2R207X/R207X mice demonstrate similar findings to the Cln2-/- mouse model and CLN2 disease patients.
Severe deficiency in TPP1 activity and neuropathological abnormalities lead CLN2 disease patients to develop incapacitating neurological deficits and ultimately death [9, 16]. Behavioral characterization of Cln2R207X/R207X mice identified motor and strength deficits starting at 3 months of age. Unfortunately most Cln2R207X/R207X mice did not survive to be tested for motor coordination at either 4 or 5 months, those that could be tested failed immediately. In addition to motor and strength deficits, Cln2R207X/R207X mice at 3 months of age developed a tremor that progressively worsened and became significantly debilitating. This behavioral anomaly is being characterized as a tremor based on presentation; although it cannot be ruled out as seizure activity without further specialized testing. Overall, these behavioral abnormalities contribute to a significant shortening of the Cln2R207X/R207X mouse lifespan. We tried to test visual acuity (e.g. visual cliff and mouse optometry system) in 1 and 3 month old wildtype and mutant mice. However, the hyperactivity of the mutant mice prohibited adequate assessment of visual acuity. Cln2R207X/R207X mice will be given to an expert in NCL vision loss and retinal pathology to investigate visual changes in these new Cln2 mutant mice.
Collectively, the biochemical, histological, and behavioral characterization of Cln2R207X/R207X mice clearly demonstrates the successful generation of a new CLN2 disease model. Unlike previous mouse models, this model carries a common disease causing CLN2 mutation. In addition, this model allows for the assessment of all therapies. Preclinical studies investigating the use of read through compounds and nonsense mediated decay inhibitors have shown positive outcomes in other disease models including lysosomal storage disorders [23, 25, 63–69]. Prior to the development of our Cln2R207X/R207X mouse model, preclinical studies into these therapies could not be evaluated for CLN2 disease. The generation of this new CLN2 disease model advances preclinical studies into CLN2 disease therapies.
Supporting information
Fluorogenic enzyme activity assays for sphingomyelinase (A) and PPT1 (B) were used to measure endogenous activity in five different tissues from 1-month-old Cln2+/+ (n = 3) and Cln2R207X/R207X (n = 3) mice. In the sphingomyelinase activity assay, three technical replicates were performed using the three biological samples obtained from Cln2+/+ and Cln2R207X/R207X mice; whereas, the PPT1 activity assays used four technical replicates from the three biological samples. Cln2R207X/R207X sphingomyelinase and PPT1 activity was normalized to Cln2+/+ activity levels. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (*p < 0.05).
(TIF)
The light/dark box test was used to assess for anxiety in 3-month-old Cln2+/+ (n = 14) and Cln2R207X/R207X (n = 20) mice. Both cohorts spent similar amounts of time in the dark. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test.
(TIF)
Images of the molecular (ML) and granular (GL) cerebellar layers demonstrate accumulation of mitochondrial ATP synthase subunit c in Cln2R207X/R207X mice when compared to Cln2+/+ controls. Pronounced accumulation is present in the Purkinje cell layer (at the border of ML and GL) in Cln2R207X/R207X mice.
(TIF)
Cln2R207X/R207X cerebral sections immunostained with anti-subunit c (red) and anti-LAMP-1 (green) reveal co-localization.
(TIF)
(A) Superficial and deep cortical layers from 3-month-old Cln2+/+ and Cln2R207X/R207X mice show minimal differences in immunostaining for the microglial marker, Iba1. (B) Images from 3-month-old Cln2+/+ (n = 4) and Cln2R207X/R207X (n = 5) mice were blindly collected and analyzed for Iba1 immunostaining total intensity, average intensity per Iba1 positive microglia, and average microglia size. Columns and bars represent mean ± SEM. Statistical significance was assessed using an unpaired t-test.
(TIF)
Tissues from Cln2+/+ (n = 3) and Cln2R207X/R207X (n = 3) mice were sectioned, hematoxylin/eosin stained, and evaluated by a veterinary pathologist under blinded conditions. The pathology report solely identified cerebral cellular hypereosinophilic inclusions (arrows) and signs of neurodegeneration (arrow heads) in Cln2R207X/R207X mice (A-C) when compared to Cln2+/+ controls (D).
(TIF)
(PDF)
Acknowledgments
Thank you to David K. Meyerholz, DVM, PhD, from the University of Iowa for the veterinary pathology examination. Lastly, we specially thank Katherine White from Sanford Research for providing advice and support on immunohistochemistry, image collection, and image analysis.
Abbreviations
- LSD
lysosomal storage disorder
- cINCL
classic infantile neuronal ceroid lipofuscinosis
- cLINCL
classic late infantile neuronal ceroid lipofuscinosis
- cJNCL
classic juvenile neuronal ceroid lipofuscinosis
- TPP1
tripeptidyl-peptidase 1
- PPT1
palmitoyl protein thioesterase 1
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- Iba1
ionized calcium binding adapter molecule 1
- DAPI
4’,6-Diamidino-2-Phenylindole
Data Availability
All data is contained within the paper and supplementary material.
Funding Statement
This work was supported by Sanford Health. The equipment used from the molecular, histology, and microscope cores at Sanford Research are funded by National Institutes of Health program grants P20GM103620 and P20GM103548 awarded to Sanford Research's Children's Health Research Center and Cancer Biology Research Center respectively.
References
- 1.Wang RY, Bodamer OA, Watson MS, Wilcox WR. Lysosomal storage diseases: diagnostic confirmation and management of presymptomatic individuals. Genetics in medicine: official journal of the American College of Medical Genetics. 2011;13(5):457–84. Epub 2011/04/20. [DOI] [PubMed] [Google Scholar]
- 2.Meikle PJ, Hopwood JJ, Clague AE, Carey WF. Prevalence of lysosomal storage disorders. JAMA. 1999;281(3):249–54. Epub 1999/01/26. [DOI] [PubMed] [Google Scholar]
- 3.Cheng SH. Gene Therapy for the Neurological Manifestations in Lysosomal Storage Disorders. Journal of Lipid Research. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballabio A, Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim Biophys Acta. 2009;1793(4):684–96. doi: 10.1016/j.bbamcr.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 5.Jeyakumar M, Dwek RA, Butters TD, Platt FM. Storage solutions: treating lysosomal disorders of the brain. Nat Rev Neurosci. 2005;6(9):713–25. doi: 10.1038/nrn1725 [DOI] [PubMed] [Google Scholar]
- 6.Mole SE. The genetic spectrum of human neuronal ceroid-lipofuscinoses. Brain Pathol. 2004;14(1):70–6. Epub 2004/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jalanko A, Braulke T. Neuronal ceroid lipofuscinoses. Biochim Biophys Acta. 2009;1793(4):697–709. Epub 2008/12/17. doi: 10.1016/j.bbamcr.2008.11.004 [DOI] [PubMed] [Google Scholar]
- 8.Haltia M, Goebel HH. The neuronal ceroid-lipofuscinoses: a historical introduction. Biochim Biophys Acta. 2013;1832(11):1795–800. doi: 10.1016/j.bbadis.2012.08.012 [DOI] [PubMed] [Google Scholar]
- 9.Mink JW, Augustine EF, Adams HR, Marshall FJ, Kwon JM. Classification and natural history of the neuronal ceroid lipofuscinoses. Journal of child neurology. 2013;28(9):1101–5. Epub 2013/07/11. PubMed Central PMCID: PMC3979348. doi: 10.1177/0883073813494268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siintola E, Lehesjoki AE, Mole SE. Molecular genetics of the NCLs—status and perspectives. Biochim Biophys Acta. 2006;1762(10):857–64. Epub 2006/07/11. doi: 10.1016/j.bbadis.2006.05.006 [DOI] [PubMed] [Google Scholar]
- 11.Mole SE, Williams RE, Goebel HH. The neuronal ceroid lipofuscinoses (Batten disease). 2nd ed. Oxford: Oxford University Press; 2011. xxx, 444 p. p. [Google Scholar]
- 12.Schulz A, Kohlschutter A, Mink J, Simonati A, Williams R. NCL diseases—clinical perspectives. Biochim Biophys Acta. 2013;1832(11):1801–6. PubMed Central PMCID: PMC4631127. doi: 10.1016/j.bbadis.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mole SE, Cotman SL. Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim Biophys Acta. 2015;1852(10 Pt B):2237–41. PubMed Central PMCID: PMC4567481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams RE, Mole SE. New nomenclature and classification scheme for the neuronal ceroid lipofuscinoses. Neurology. 2012;79(2):183–91. doi: 10.1212/WNL.0b013e31825f0547 [DOI] [PubMed] [Google Scholar]
- 15.Warrier V, Vieira M, Mole SE. Genetic basis and phenotypic correlations of the neuronal ceroid lipofusinoses. Biochim Biophys Acta. 2013;1832(11):1827–30. doi: 10.1016/j.bbadis.2013.03.017 [DOI] [PubMed] [Google Scholar]
- 16.Chabrol B, Caillaud C, Minassian B. Neuronal ceroid lipofuscinoses. Handbook of clinical neurology. 2013;113:1701–6. Epub 2013/04/30. doi: 10.1016/B978-0-444-59565-2.00038-1 [DOI] [PubMed] [Google Scholar]
- 17.Cooper JD. Progress towards understanding the neurobiology of Batten disease or neuronal ceroid lipofuscinosis. Curr Opin Neurol. 2003;16(2):121–8. Epub 2003/03/20. doi: 10.1097/01.wco.0000063762.15877.9b [DOI] [PubMed] [Google Scholar]
- 18.Mole SE, Williams RE, Goebel HH. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 2005;6(3):107–26. Epub 2005/06/21. doi: 10.1007/s10048-005-0218-3 [DOI] [PubMed] [Google Scholar]
- 19.Geraets RD, Koh S, Hastings ML, Kielian T, Pearce DA, Weimer JM. Moving towards effective therapeutic strategies for Neuronal Ceroid Lipofuscinosis. Orphanet journal of rare diseases. 2016;11:40 PubMed Central PMCID: PMCPMC4833901. doi: 10.1186/s13023-016-0414-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neverman NJ, Best HL, Hofmann SL, Hughes SM. Experimental therapies in the neuronal ceroid lipofuscinoses. Biochim Biophys Acta. 2015;1852(10 Pt B):2292–300. [DOI] [PubMed] [Google Scholar]
- 21.Kousi M, Lehesjoki AE, Mole SE. Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum Mutat. 2012;33(1):42–63. doi: 10.1002/humu.21624 [DOI] [PubMed] [Google Scholar]
- 22.Mole SE. NCL Resource 2014 [updated 05/10/2014]. Available from: http://www.ucl.ac.uk/ncl/mutation.shtml.
- 23.Miller JN, Kovacs AD, Pearce DA. The novel Cln1R151X mouse model of infantile neuronal ceroid lipofuscinosis (INCL) for testing nonsense suppression therapy. Hum Mol Genet. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohar I, Lin L, Lobel P. Enzyme-based diagnosis of classical late infantile neuronal ceroid lipofuscinosis: comparison of tripeptidyl peptidase I and pepstatin-insensitive protease assays. Clin Chem. 2000;46(7):1005–8. Epub 2000/07/15. [PubMed] [Google Scholar]
- 25.Miller JN, Chan CH, Pearce DA. The role of nonsense-mediated decay in neuronal ceroid lipofuscinosis. Hum Mol Genet. 2013;22(13):2723–34. doi: 10.1093/hmg/ddt120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Diggelen OP, Keulemans JL, Winchester B, Hofman IL, Vanhanen SL, Santavuori P, et al. A rapid fluorogenic palmitoyl-protein thioesterase assay: pre- and postnatal diagnosis of INCL. Mol Genet Metab. 1999;66(4):240–4. Epub 1999/04/07. doi: 10.1006/mgme.1999.2809 [DOI] [PubMed] [Google Scholar]
- 27.Kovacs AD, Pearce DA. Location- and sex-specific differences in weight and motor coordination in two commonly used mouse strains. Sci Rep. 2013;3:2116 Epub 2013/07/03. PubMed Central PMCID: PMC3698490. doi: 10.1038/srep02116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourin M, Hascoet M. The mouse light/dark box test. Eur J Pharmacol. 2003;463(1–3):55–65. [DOI] [PubMed] [Google Scholar]
- 29.Crawley JN. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 1999;835(1):18–26. Epub 1999/08/17. [DOI] [PubMed] [Google Scholar]
- 30.Watzlawik JO, Kahoud RJ, O'Toole RJ, White KA, Ogden AR, Painter MM, et al. Abbreviated exposure to hypoxia is sufficient to induce CNS dysmyelination, modulate spinal motor neuron composition, and impair motor development in neonatal mice. PLoS One. 2015;10(5):e0128007 PubMed Central PMCID: PMCPMC4447462. doi: 10.1371/journal.pone.0128007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks DA, Muller VJ, Hopwood JJ. Stop-codon read-through for patients affected by a lysosomal storage disorder. Trends Mol Med. 2006;12(8):367–73. doi: 10.1016/j.molmed.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 32.Kovacs AD, Pearce DA. Finding the most appropriate mouse model of juvenile CLN3 (Batten) disease for therapeutic studies: the importance of genetic background and gender. Dis Model Mech. 2015;8(4):351–61. PubMed Central PMCID: PMC4381334. doi: 10.1242/dmm.018804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haltia M. The neuronal ceroid-lipofuscinoses. J Neuropathol Exp Neurol. 2003;62(1):1–13. Epub 2003/01/17. [DOI] [PubMed] [Google Scholar]
- 34.Haltia M. The neuronal ceroid-lipofuscinoses: from past to present. Biochim Biophys Acta. 2006;1762(10):850–6. Epub 2006/08/16. doi: 10.1016/j.bbadis.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 35.Cooper JD, Russell C, Mitchison HM. Progress towards understanding disease mechanisms in small vertebrate models of neuronal ceroid lipofuscinosis. Biochim Biophys Acta. 2006;1762(10):873–89. Epub 2006/10/07. doi: 10.1016/j.bbadis.2006.08.002 [DOI] [PubMed] [Google Scholar]
- 36.Mitchison HM, Lim MJ, Cooper JD. Selectivity and types of cell death in the neuronal ceroid lipofuscinoses. Brain Pathol. 2004;14(1):86–96. Epub 2004/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sleat David E., Wiseman Jennifer A., El-Banna Mukarram, Kim Kwi-Hye, Mao Qinwen, Price Sandy, Macauley RLS Shannon L., Shen Michael M., Zhao Qi, Passini Marco A., Davidson Beverly L., Stewart aPL Gregory R.. A Mouse Model of Classical Late-Infantile Neuronal Ceroid Lipofuscinosis Based on Targeted Disruption of the CLN2 Gene Results in a Loss of Tripeptidyl-Peptidase I Activity and Progressive Neurodegeneration. Neurobiology of Disease. 2004;24(41):9117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shacka JJ. Mouse models of neuronal ceroid lipofuscinoses: useful pre-clinical tools to delineate disease pathophysiology and validate therapeutics. Brain Res Bull. 2012;88(1):43–57. doi: 10.1016/j.brainresbull.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 39.Lull ME, Block ML. Microglial activation and chronic neurodegeneration. Neurotherapeutics. 2010;7(4):354–65. PubMed Central PMCID: PMCPMC2951017. doi: 10.1016/j.nurt.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer DN, Fearnley IM, Walker JE, Hall NA, Lake BD, Wolfe LS, et al. Mitochondrial ATP synthase subunit c storage in the ceroid-lipofuscinoses (Batten disease). Am J Med Genet. 1992;42(4):561–7. Epub 1992/02/15. doi: 10.1002/ajmg.1320420428 [DOI] [PubMed] [Google Scholar]
- 41.Tian Y, Sohar I, Taylor JW, Lobel P. Determination of the substrate specificity of tripeptidyl-peptidase I using combinatorial peptide libraries and development of improved fluorogenic substrates. J Biol Chem. 2006;281(10):6559–72. Epub 2005/12/13. doi: 10.1074/jbc.M507336200 [DOI] [PubMed] [Google Scholar]
- 42.Ezaki J, Takeda-Ezaki M, Kominami E. Tripeptidyl peptidase I, the late infantile neuronal ceroid lipofuscinosis gene product, initiates the lysosomal degradation of subunit c of ATP synthase. J Biochem. 2000;128(3):509–16. Epub 2000/08/31. [DOI] [PubMed] [Google Scholar]
- 43.Ezaki J, Tanida I, Kanehagi N, Kominami E. A lysosomal proteinase, the late infantile neuronal ceroid lipofuscinosis gene (CLN2) product, is essential for degradation of a hydrophobic protein, the subunit c of ATP synthase. J Neurochem. 1999;72(6):2573–82. Epub 1999/06/01. [DOI] [PubMed] [Google Scholar]
- 44.Maragakis NJ, Rothstein JD. Mechanisms of Disease: astrocytes in neurodegenerative disease. Nat Clin Pract Neurol. 2006;2(12):679–89. doi: 10.1038/ncpneuro0355 [DOI] [PubMed] [Google Scholar]
- 45.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4(1):49–60. doi: 10.1038/nrn1007 [DOI] [PubMed] [Google Scholar]
- 46.Heneka MT, Rodriguez JJ, Verkhratsky A. Neuroglia in neurodegeneration. Brain Res Rev. 2010;63(1–2):189–211. doi: 10.1016/j.brainresrev.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Hu X, Qian L, O'Callaghan JP, Hong JS. Astrogliosis in CNS pathologies: is there a role for microglia? Mol Neurobiol. 2010;41(2–3):232–41. PubMed Central PMCID: PMCPMC3629545. doi: 10.1007/s12035-010-8098-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000). Neurochem Res. 2000;25(9–10):1439–51. [DOI] [PubMed] [Google Scholar]
- 49.Macauley SL, Pekny M, Sands MS. The role of attenuated astrocyte activation in infantile neuronal ceroid lipofuscinosis. J Neurosci. 2011;31(43):15575–85. PubMed Central PMCID: PMC3218425. doi: 10.1523/JNEUROSCI.3579-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pontikis CC, Cella CV, Parihar N, Lim MJ, Chakrabarti S, Mitchison HM, et al. Late onset neurodegeneration in the Cln3-/- mouse model of juvenile neuronal ceroid lipofuscinosis is preceded by low level glial activation. Brain Res. 2004;1023(2):231–42. Epub 2004/09/18. doi: 10.1016/j.brainres.2004.07.030 [DOI] [PubMed] [Google Scholar]
- 51.Pontikis CC, Cotman SL, MacDonald ME, Cooper JD. Thalamocortical neuron loss and localized astrocytosis in the Cln3Deltaex7/8 knock-in mouse model of Batten disease. Neurobiol Dis. 2005;20(3):823–36. Epub 2005/07/12. doi: 10.1016/j.nbd.2005.05.018 [DOI] [PubMed] [Google Scholar]
- 52.Boche D, Perry VH, Nicoll JA. Review: activation patterns of microglia and their identification in the human brain. Neuropathol Appl Neurobiol. 2013;39(1):3–18. doi: 10.1111/nan.12011 [DOI] [PubMed] [Google Scholar]
- 53.Graeber MB, Streit WJ. Microglia: biology and pathology. Acta Neuropathol. 2010;119(1):89–105. doi: 10.1007/s00401-009-0622-0 [DOI] [PubMed] [Google Scholar]
- 54.Macauley SL, Wozniak DF, Kielar C, Tan Y, Cooper JD, Sands MS. Cerebellar pathology and motor deficits in the palmitoyl protein thioesterase 1-deficient mouse. Exp Neurol. 2009;217(1):124–35. Epub 2009/05/07. PubMed Central PMCID: PMC2679857. doi: 10.1016/j.expneurol.2009.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kielar C, Maddox L, Bible E, Pontikis CC, Macauley SL, Griffey MA, et al. Successive neuron loss in the thalamus and cortex in a mouse model of infantile neuronal ceroid lipofuscinosis. Neurobiol Dis. 2007;25(1):150–62. Epub 2006/10/19. PubMed Central PMCID: PMC1866219. doi: 10.1016/j.nbd.2006.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyynela J, Cooper JD, Khan MN, Shemilts SJ, Haltia M. Hippocampal pathology in the human neuronal ceroid-lipofuscinoses: distinct patterns of storage deposition, neurodegeneration and glial activation. Brain Pathol. 2004;14(4):349–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bilbo SD, Schwarz JM. Early-life programming of later-life brain and behavior: a critical role for the immune system. Front Behav Neurosci. 2009;3:14 PubMed Central PMCID: PMCPMC2737431. doi: 10.3389/neuro.08.014.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sleat DE, Sohar I, Gin RM, Lobel P. Aminoglycoside-mediated suppression of nonsense mutations in late infantile neuronal ceroid lipofuscinosis. Eur J Paediatr Neurol. 2001;5 Suppl A:57–62. [DOI] [PubMed] [Google Scholar]
- 59.Sleat DE, Donnelly RJ, Lackland H, Liu CG, Sohar I, Pullarkat RK, et al. Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science. 1997;277(5333):1802–5. [DOI] [PubMed] [Google Scholar]
- 60.Williams RE, Aberg L, Autti T, Goebel HH, Kohlschutter A, Lonnqvist T. Diagnosis of the neuronal ceroid lipofuscinoses: an update. Biochim Biophys Acta. 2006;1762(10):865–72. Epub 2006/08/26. doi: 10.1016/j.bbadis.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 61.Sleat DE, El-Banna M, Sohar I, Kim KH, Dobrenis K, Walkley SU, et al. Residual levels of tripeptidyl-peptidase I activity dramatically ameliorate disease in late-infantile neuronal ceroid lipofuscinosis. Mol Genet Metab. 2008;94(2):222–33. Epub 2008/03/18. PubMed Central PMCID: PMC2467442. doi: 10.1016/j.ymgme.2008.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahmood F, Fu S, Cooke J, Wilson SW, Cooper JD, Russell C. A zebrafish model of CLN2 disease is deficient in tripeptidyl peptidase 1 and displays progressive neurodegeneration accompanied by a reduction in proliferation. Brain. 2013;136(Pt 5):1488–507. doi: 10.1093/brain/awt043 [DOI] [PubMed] [Google Scholar]
- 63.Keeling KM, Brooks DA, Hopwood JJ, Li P, Thompson JN, Bedwell DM. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of alpha-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum Mol Genet. 2001;10(3):291–9. [DOI] [PubMed] [Google Scholar]
- 64.Keeling KM, Wang D, Dai Y, Murugesan S, Chenna B, Clark J, et al. Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One. 2013;8(4):e60478 PubMed Central PMCID: PMC3622682. doi: 10.1371/journal.pone.0060478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D, Belakhov V, Kandasamy J, Baasov T, Li SC, Li YT, et al. The designer aminoglycoside NB84 significantly reduces glycosaminoglycan accumulation associated with MPS I-H in the Idua-W392X mouse. Mol Genet Metab. 2012;105(1):116–25. PubMed Central PMCID: PMC3253910. doi: 10.1016/j.ymgme.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gunn G, Dai Y, Du M, Belakhov V, Kandasamy J, Schoeb TR, et al. Long-term nonsense suppression therapy moderates MPS I-H disease progression. Mol Genet Metab. 2014;111(3):374–81. PubMed Central PMCID: PMCPMC3943726. doi: 10.1016/j.ymgme.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–94. doi: 10.1146/annurev-genom-091212-153527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baradaran-Heravi A, Balgi AD, Zimmerman C, Choi K, Shidmoossavee FS, Tan JS, et al. Novel small molecules potentiate premature termination codon readthrough by aminoglycosides. Nucleic Acids Res. 2016;44(14):6583–98. PubMed Central PMCID: PMCPMC5001621. doi: 10.1093/nar/gkw638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagel-Wolfrum K, Moller F, Penner I, Baasov T, Wolfrum U. Targeting Nonsense Mutations in Diseases with Translational Read-Through-Inducing Drugs (TRIDs). BioDrugs. 2016;30(2):49–74. doi: 10.1007/s40259-016-0157-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorogenic enzyme activity assays for sphingomyelinase (A) and PPT1 (B) were used to measure endogenous activity in five different tissues from 1-month-old Cln2+/+ (n = 3) and Cln2R207X/R207X (n = 3) mice. In the sphingomyelinase activity assay, three technical replicates were performed using the three biological samples obtained from Cln2+/+ and Cln2R207X/R207X mice; whereas, the PPT1 activity assays used four technical replicates from the three biological samples. Cln2R207X/R207X sphingomyelinase and PPT1 activity was normalized to Cln2+/+ activity levels. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test (*p < 0.05).
(TIF)
The light/dark box test was used to assess for anxiety in 3-month-old Cln2+/+ (n = 14) and Cln2R207X/R207X (n = 20) mice. Both cohorts spent similar amounts of time in the dark. Columns and bars represent mean ± SEM. Statistical significance was determined using an unpaired t-test.
(TIF)
Images of the molecular (ML) and granular (GL) cerebellar layers demonstrate accumulation of mitochondrial ATP synthase subunit c in Cln2R207X/R207X mice when compared to Cln2+/+ controls. Pronounced accumulation is present in the Purkinje cell layer (at the border of ML and GL) in Cln2R207X/R207X mice.
(TIF)
Cln2R207X/R207X cerebral sections immunostained with anti-subunit c (red) and anti-LAMP-1 (green) reveal co-localization.
(TIF)
(A) Superficial and deep cortical layers from 3-month-old Cln2+/+ and Cln2R207X/R207X mice show minimal differences in immunostaining for the microglial marker, Iba1. (B) Images from 3-month-old Cln2+/+ (n = 4) and Cln2R207X/R207X (n = 5) mice were blindly collected and analyzed for Iba1 immunostaining total intensity, average intensity per Iba1 positive microglia, and average microglia size. Columns and bars represent mean ± SEM. Statistical significance was assessed using an unpaired t-test.
(TIF)
Tissues from Cln2+/+ (n = 3) and Cln2R207X/R207X (n = 3) mice were sectioned, hematoxylin/eosin stained, and evaluated by a veterinary pathologist under blinded conditions. The pathology report solely identified cerebral cellular hypereosinophilic inclusions (arrows) and signs of neurodegeneration (arrow heads) in Cln2R207X/R207X mice (A-C) when compared to Cln2+/+ controls (D).
(TIF)
(PDF)
Data Availability Statement
All data is contained within the paper and supplementary material.