Abstract
The steroidogenic acute regulatory protein (StAR) governs the rate-limiting step in steroidogenesis, and its expression varies depending on the needs of the specific tissue. Tight control of steroid production is essential for multiple processes involved in reproduction, including follicular development, ovulation, and endometrial synchronization. Recently, there has been a growing interest in the role of noncoding RNAs in the regulation of reproduction. Here we demonstrate that StAR is a novel target of the microRNA let-7, which itself is regulated by the long noncoding RNA (lncRNA) H19. Using human and murine cell lines, we show that overexpression of H19 stimulates StAR expression by antagonizing let-7, which inhibits StAR at the post-transcriptional level. Our results uncover a novel mechanism underlying the regulation of StAR expression and represent the first example of lncRNA-mediated control of the rate-limiting step of steroidogenesis. This work thus adds to the body of literature describing the multiple roles in oncogenesis, cellular growth, glucose metabolism, and now regulation of steroidogenesis, of this complex lncRNA.
Precis: Overexpression of the lncRNA H19 stimulates StAR expression by antagonizing let-7 (which itself inhibits StAR at the posttranscriptional level). These results uncover a novel mechanism underlying the regulation of StAR expression and represent the first example of lncRNA-mediated control of the rate-limiting step of steroidogenesis.
Tight spatiotemporal control of steroid production is essential for multiple processes involved in reproduction and fertility, including appropriate follicular development, ovulation, and the development of a synchronous endometrial lining for implantation. Production of the steroid hormones estradiol and progesterone, which regulate the menstrual cycle, is governed initially via expression of steroidogenic acute regulatory protein (StAR), whose expression permits translocation of the steroid hormone substrate cholesterol from the outer to the inner mitochondrial membrane (1). StAR expression, like other menstrual cycle changes, is itself cyclic in nature. During the early phase of the menstrual cycle, the presence of StAR allows for follicular production of androgens (a substrate for follicular estrogen production). At the time of luteinization, StAR expression undergoes a dramatic upregulation (2, 3), promoting conversion to a progesterone-dominant microenvironment, which allows endometrial decidualization and support of a growing pregnancy.
Recently there has been a growing interest in the role of noncoding RNAs in the regulation of reproduction. Of particular interest is the role of the long noncoding RNA (lncRNA) H19, which belongs to a highly conserved imprinted gene cluster that also contains the reciprocally imprinted gene for insulin growth factor 2 (Igf2) and is integral for growth and development. It encodes a predominantly cytoplasmic, 2.6-kb–long RNA that has no known protein product. It is abundantly expressed during fetal life and repressed after birth except in a few adult organs, including skeletal and cardiac muscle, mammary gland, uterus, and ovaries (4–6).
We have recently discovered that H19 acts as a molecular “sponge” for the let-7 family of microRNAs (miRNAs); that is, H19 is able to bind to and negatively regulate let-7’s activity (7). Let-7 regulates gene expression via translational repression and mRNA degradation and plays critical roles in development, tumorigenesis, glucose metabolism, and endometrial development, and a decrease in let-7 bioavailability through this “sponge” mechanism results in increased expression of let-7 targets (8–13). In addition to its known roles in development and glucose metabolism, other wide-ranging roles for let-7, including in follicular and oocyte development, have been proposed (14–16).
Although the physiological significance of the H19/let-7 axis in adult human reproduction organs remains largely undefined, the presence of H19 in hormone-sensitive tissues, including breast, uterus, and ovary, suggests that the H19/let-7 axis may play a role as an effector of gene function in these tissues. Adding to this possibility, a cyclic pattern to H19 expression (similar to that of the menstrual cycle) has been observed. Arial et al. (5) reported a peak in H19 endometrial expression during the late secretory phase of the menstrual cycle (as well as after exogenous hormonal therapy) and found that ovarian expression was highest in postovulatory corpora lutea and atretic follicles. Using human granulosa cells as a model system, we show here that StAR is a novel target of let-7–mediated inhibition and that the expression of StAR is directly stimulated by the H19/let-7 axis.
Materials and Methods
Materials
The antibodies for StAR (rabbit monoclonal, #8449; Cell Signaling, Danvers, MA), β-tubulin (rabbit polyclonal, ab6046; Abcam, Cambridge, UK), and β-actin (mouse monoclonal, ab8226; Abcam) were purchased (Table 1). Human let-7 miRNA mimic hsa-let-7b (Let-7b, AM17100/PM11050) and PremiR negative control (AM17110; miCon,) were purchased as well. Primers for H19, StAR, β-tubulin, and β-actin were purchased from the Yale University W.M. Keck Oligonucleotide Synthesis Facility (New Haven, CT). The human H19 expression vector pH19 was previously described (7). The mouse H19 expression vector pmH19 was created by cloning a full-length mouse H19 (NR_001592) sequence into a pCMV6-based expression vector (BlueHeron Gene Synthesis Company, Bothell, WA).
Table 1.
Antibodies Used in This Study
| Antigen Sequence (if known) | Name of Antibody | Manufacturer, Catalog #, and/or Name of Individual Providing the Antibody | Species Raised in; Monoclonal or Polyclonal | Dilution Used |
|---|---|---|---|---|
| Arg272 of human StAR | StAR | Cell Signaling, 8449 | rabbit monoclonal | 1:500 |
| Human β-actin aa-100 | beta-actin | AbCam, 8226 | mouse monoclonal | 1:6000 |
Bioinformatics analysis
Let-7 binding sites in the StAR gene in human, mouse, monkey, and rat were predicted using a Web-based program RNAhybrid. RNAhybrid predicts microRNA (miRNA) targets by calculating the minimum free energy of hybridization between target RNA and miRNA sequences to predict miRNA/target duplexes (17).
Plasmids
Reporters were constructed based on the psiCHECK2TM vector. PsiCHECK2-let-7 4x (plasmid 20930; Addgene) was previously described (18). PsiCHECK2-STAR was created by inserting a 60-nt long PCR fragment from human StAR (nt 1467 relative to the transcriptional start site of human StAR) into psiCHECK2-let-7 4x [at the 3′-untranslated region (UTR) of Rluc, in place of 4x], opened with XhoI and NotI. This fragment was predicted to bind let-7b at position 1467. The forward and reverse primers (forward 5′x and reverse 5′x) were used, and the 4x let-7 binding sites were removed prior to the ligation. Renilla luciferase activity is used as an indicator of the effect of the artificial 3′UTR on translation efficiency. The psiCHECK2™ vector also contains a constitutively expressed firefly luciferase gene, which serves as an internal control to normalize transfection efficiency (19).
Cell culture and transfection
Human KGN cells were a gift from Maria Lalioti, PhD (Biogen, Weston, MA). Human embryonic kidney (HEK)293 (ATCC CRL-1573) cells were obtained from the ATCC Global Resource Center, Manassas, VA). Mouse Leydig cells (ATCC CRL-2065) were a gift from Jean-Ju Chung, PhD (Yale School of Medicine, New Haven, CT), and all cells were cultured using standard protocols provided by the ATCC. Cells were maintained in 100-mm culture dishes in DMEM-F12 with 10% fetal bovine serum (heat inactivated), 1% l-glutamine, and 1% penicillin-streptomycin.
Cells were transfected in a 48-well plate scale. To prepare plasmid transfection solution, 0.2 μg H19 plasmid DNA per well was mixed with 25 μl OPTI‐MEM per well by gentle pipetting. In parallel, 2 μl of lipofectamine 2000 per well was mixed with 25 μl of OPTI-MEM per well. After 5 minutes of incubation at room temperature, the two solutions were mixed by gentle pipetting and incubated for 20 minutes at room temperature to allow plasmid/lipid complexes to form. At the end of incubation, the 50 μl/well transfection solution was used to resuspend the cell pellet (5 × 105 cells/well). After incubation at room temperature for 10 minutes, regular growth medium was added at a ratio of 1:3, and the cell suspension was transferred to the culture plate. After 24-hour incubation at 37°C in 5% CO2, the medium was replaced with fresh growth medium. RNA and protein were extracted at the indicated time points after transfection. For let-7 mimic transfection, 1 pmol of control miRNA (miCon) or let-7b mimic was mixed with 50 μl of OPTI-MEM. In parallel, 0.5 μl of Lipofectamine 2000 was mixed with 50 μl of OPTI-MEM. After 5 minutes of incubation, the two solutions were mixed and incubated at room temperature for 20 minutes. The resulting 100 μl of transfection cocktail was added to MLTC-1 cells prewashed with OPTI-MEM. Cells were harvested at 12 hours post-transfection, and RNA levels were determined by quantitative reverse transcription–polymerase chain reaction (qRT-PCR). Results are representative of three independent transfection experiments.
Luciferase assays
The assays were carried out in a 48-well plate scale as previously described (20) with minor modifications. Briefly, 10 ng of the indicated luciferase reporter plus 140 ng of empty vector were transfected into HEK293 cells (2 × 105 cells/well), together with control miRNA or let-7b miRNAs at final concentrations of 8, 16, and 32 pM. Each concentration was run in triplicate. Luciferase activities were measured 18 hours post-transfection using Promega Dual-Luciferase Reporter Assay System (E1960) (Madison, WI) according to the manufacturer’s protocol. Renilla luciferase activity was normalized against Firefly luciferase activities and presented as percentage of inhibition.
Western blot, reverse transcription, and real-time PCR
qRT-PCR and Western blot analyses were carried out as previously described (7). Briefly, cDNA was synthesized using a Bio-Rad iSCRIPT kit (1725122) in a 20-μl reaction containing 0.5 μg of total RNA. qPCR was performed in a 25-μl reaction containing 0.5 to 1.5 μl of cDNA using iQSYBRGreen (Bio-Rad) in a Bio-Rad iCycler. PCR was performed by initial denaturation at 95°C for 5 minutes, followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle values of each sample were used in the post-PCR data analysis, and the ΔΔ threshold cycle method was used to calculate mRNA levels. The PCR primers for the indicated human and mouse genes are listed here. β-Tubulin was used as a housekeeping gene.
Human H19 forward: 5′-GCACCTTGGACATCTGGAGT
Human H19 reverse: 5′-TTCTTTCCAGCCCTAGCTCA
Human StAR forward: 5′-GGCATCCTTAGCAACCAAGA
Human StAR reverse: 5′-TCTCCTTGACATTGGGGTTC
Mouse StAR forward: 5′-TTGGGCATACTCAACAACCA
Mouse StAR reverse: 5′-GAAACACCTTGCCCACATCT
Human/mouse beta-tubulin forward: 5′-CGTGTTCGGCCAG AGTGGTGC
Human beta-tubulin reverse: 5′-GGGTGAGGGCATGACGCTGAA
To measure protein levels, cells were harvested, and cell pellets were directly lysed in 3 volumes of 2X SDS-sample buffer by heating at 95°C for 5 minutes with occasional vortexing to break chromosomal DNA. Cell lysates (5 to 10 μl/well) were resolved on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by Western blot analysis using StAR primary antibody (1:500) and goat anti-rabbit secondary antibody (1:6000).
Statistical analysis
All data are presented as mean ± SD. Data were analyzed using 2-tailed Student t test. P values at 0.05 or less were considered significant.
Results
H19 stimulates StAR expression
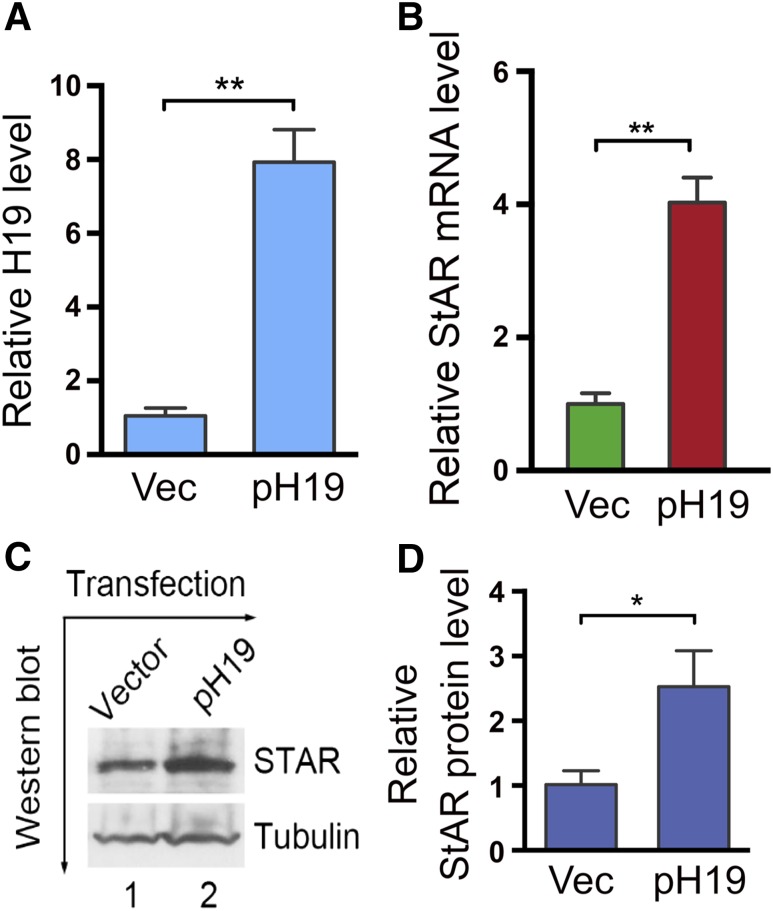
StAR expression is dramatically upregulated at the time of luteinization (2, 3), which coincides with the peak expression of H19 during the late secretory phase of the menstrual cycle (5). This suggests that there might be a regulatory relationship between StAR and H19. To test this possibility, we overexpressed H19 by transfecting a human full-length H19-expression vector pH19 (7) in KGN cells and examined effects on StAR expression. KGN cells are a human ovarian granulosa cell tumor line that express a low but detectable level of endogenous H19 (21). Transfection of pH19 increased H19 by ∼8-fold [Fig. 1(A)]. H19 overexpression also increased StAR expression by ∼4-fold [Fig. 1(B)]. Consistent with the mRNA level change, the StAR protein also increased by ∼2.3-fold [Fig. 1(C) and 1(D)]. Together, these results suggest that H19 positively regulates StAR expression.
Figure 1.
StAR expression increases in response to H19 overexpression. (A, B) KGN cells were transfected with empty vector (Vec) or with pH19. RNAs were isolated 48 h post-transfection and analyzed by qRT-PCR. Numbers are mean ± SD (n = 3). **P < 0.01. (C, D) KGN cells were transfected with vector or pH19. Proteins were extracted and analyzed by Western blotting. β-Tubulin was used as a loading control. Results are representative of three independent transfection experiments. Western blot quantifications (D) were performed using ImageJ. Numbers are mean ± SD (n = 3). *P < 0.05.
StAR mRNA contains let-7-binding sites at its 3′UTR
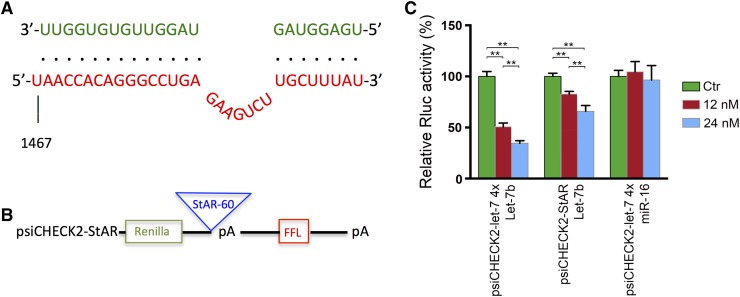
Because H19 stimulates gene expression by antagonizing let-7 activity (7, 10–12), we speculated that StAR might be a target of let-7 inhibition. Indeed, our bioinformatic analysis predicted a let-7b binding site in the 3′UTR of StAR mRNA [Fig. 2(A)]. To demonstrate that the predicted let-7 binding sites are functional, we created a reporter gene construct, psiCHECK2-StAR, containing a 60-nt long fragment from human StAR at the 3′-UTR of a Renilla luciferase gene, along with a downstream constitutively expressed firefly luciferase gene as an internal control and a polyA signal to reduce background [Fig. 2(B)]. psiCHECK2-let7-4x, which harbors four copies of let7 binding sites in the 3′-UTR of the Renilla luciferase gene, was used as a positive control. We transfected psiCHECK2-StAR and psiCHECK2-let7-4x into HEK293 cells together with increasing amounts of let-7. As expected, cotransfected let-7 inhibited Rluc expression in a dose-dependent manner in the HEK293 cells containing the positive control psiCHECK2–let7-4x. Additionally, the Rluc activity decreased in response to let-7 in the HEK293 cells containing psiCHECK2-StAR in a dose-dependent manner [Fig. 2(C)], supporting the functionality of the predicted let-7b binding site at the 3′UTR of StAR.
Figure 2.
Human StAR mRNA contains a functional let-7 binding site at its 3′-UTR. (A) Bioinformatics predicted 1 binding site for let-7b at position1467 in the 3′UTR of human StAR. Partial sequences of StAR (in red) and sequence of let-7b (in green) are shown. Base-paired interactions between the microRNA seed region and the target mRNA are indicated by dots. Numbers are in nucleotides relative to the transcriptional start site of StAR. (B) psiCHECK2-StAR, the let-7 reporter construct, containing a 60-nt long fragment from human StAR at the 3′-UTR of a Renilla luciferase gene, along with a downstream constitutively expressed firefly luciferase (FFL) gene and a polyA (pA) signal to reduce background. psiCHECK2-let7-4x, which harbors 4 copies of let7 binding sites in the 3′-UTR of the Renilla luciferase gene, was used as a positive control. (C) Indicated reporter constructs were individually transfected into HEK293 cells, together with control miRNA (Ctr), let-7b, or miR-16, at a final concentration of 12 or 24 nM. Luciferase activities were measured 18 h post-transfection. Results are representative of three independent transfection experiments. Numbers are mean ± SD (n = 3). **P < 0.01.
H19-mediated regulation of StAR expression is conserved between human and mouse
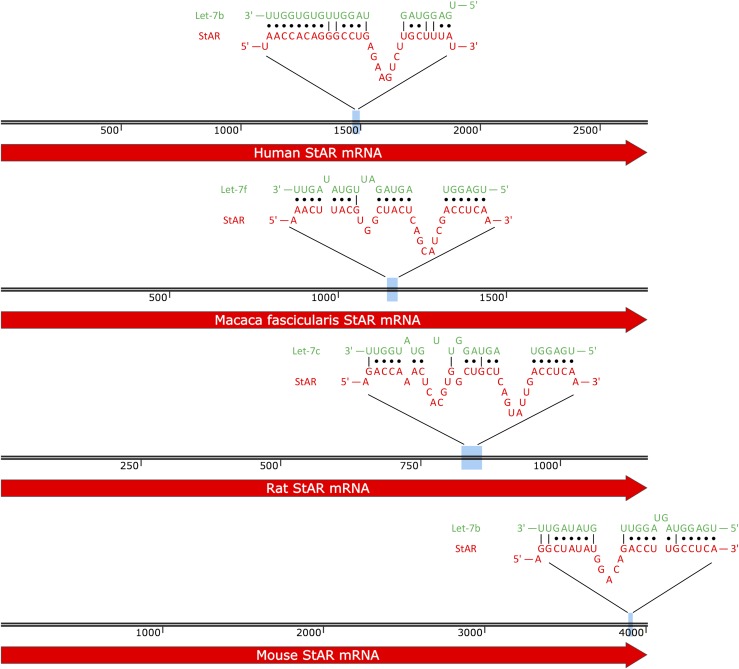
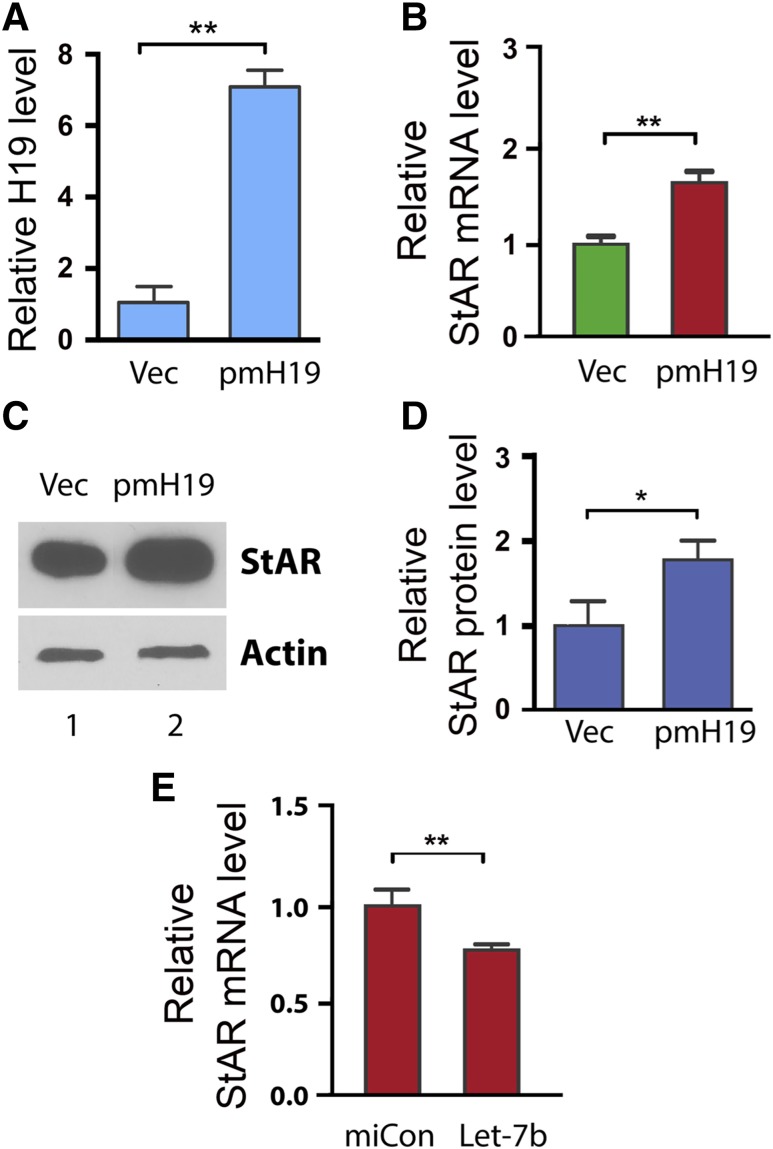
Our bioinformatics analysis predicted let-7 binding sites in StAR mRNA of other species including mouse, rat, and monkey (Fig. 3). To test whether the H19-mediated regulation of StAR is conserved, we examined the effect of H19 overexpression on StAR expression in a mouse steroidogenic cell line MLTC-1 (22). We transfected pmH19 (which expresses a mouse version of H19) or empty vector into MLTC-1 cells, followed by analysis of expression of StAR 48 hours later (Fig. 4). Consistent with our observation in the human KGN cells (Fig. 1), H19 overexpression resulted in increased expression of StAR at both the mRNA and protein levels [Fig. 3(B–D)].
Figure 3.
StAR mRNA let-7 binding sites are conserved across species. Our bioinformatics analysis predicted let-7 binding sites in StAR mRNA of other species including monkey, rat, and mouse. The human StAR binding site, along with conserved StAR binding sites, is presented.
Figure 4.
H19-mediated regulation of StAR is conserved in other steroidogenic cells. To test whether the H19-mediated regulation of StAR is conserved, we examined the effect of H19 overexpression on StAR expression in the mouse Leydig cell line MLTC-1. (A, B) MTLC-1 cells were transfected with empty vector (Vec) or with pH19. RNAs were isolated 48 h post-transfection and analyzed by qRT-PCR for H19 and StAR expression. Numbers are mean ± SD (n = 3). **P < 0.01. (C, D) StAR protein was extracted and analyzed by Western blotting. β-Tubulin was used as a loading control. Results are representative of 3 independent transfection experiments. Western blot quantifications (D) were performed using ImageJ. Numbers are mean ± SD (n = 3). *P < 0.05. (E) To confirm the functionality of the predicted let-7b binding site in the mouse StAR mRNA, we tested the effect of let-7b on inhibition of endogenous StAR expression. Thus, let-7b (or miRNA negative control miCon) was transfected into MLTC-1 cells. RNAs were extracted 12 h later, and StAR mRNA levels were determined by qRT-PCR. Let-7 transfection led to decreased StAR expression, supporting StAR being a target of let-7b inhibition in the mouse cells.
To confirm the functionality of the predicted let-7b binding site in the mouse StAR mRNA, we tested the effect of let-7b on inhibition of endogenous StAR expression. Thus, let-7b (or miRNA negative control miCon) was transfected into MLTC-1 cells. RNAs were extracted 12 hours later, and StAR mRNA levels were determined by qRT-PCR. Transfection of let-7 led to decreased StAR expression, supporting StAR being a target of let-7b inhibition in the mouse cells [Fig. 3(E)].
Discussion
We provide evidence that overexpression of the lncRNA H19 promotes expression of StAR, a protein that controls the rate-limiting step in steroidogenesis. We also show that StAR contains at least one functional let-7 binding site at its 3′UTR and is likely a novel target of let-7. Thus, we identify the H19/let-7 axis as a regulatory mechanism for StAR expression.
Steroid hormones are essential for maintenance of salt balance, carbohydrate metabolism, and reproductive capacity, and steroidogenic cells must move large amounts of cholesterol from the outer mitochondrial membrane to the inner membrane. In luteinized granulosa cells, this cholesterol is then converted to pregnenolone and ultimately to progesterone (23). The initial transport of cholesterol across the mitochondrial membrane is the rate-limiting step in steroidogenesis and requires StAR (1). StAR expression is low in preovulatory cells of the developing follicle; the midcycle LH surge then causes a dramatic upregulation of StAR expression within the dominant follicle. StAR expression is most abundant, therefore, in theca-lutein and granulosa-lutein cells during the early and midluteal phases, declining significantly toward the end of the luteal phase of the menstrual cycle. This upregulation allows the corpus luteum to produce substantial amounts of progesterone to support the potential growing pregnancy (2).
The cyclic pattern of H19 expression described by Arial et al. (5) (weak H19 expression in primordial, primary, and secondary follicles and increasing expression in antral follicles, especially in thecal cells, continuing until the corpus albicans state) closely mimics StAR’s expression pattern, lending credence to the possibility that H19 may be involved in the regulation of StAR. Moreover, noncoding RNA–mediated regulation of steroidogenic enzymes has been previously reported. For example, the miRNA miR-133b was recently shown to promote StAR expression and estradiol synthesis in mouse primary granulosa cells via downregulation of the transcriptional repressor Foxl2 (24). Although little is known about the ability of lncRNAs to directly regulate steroidogenic enzymes, our results show lncRNA-mediated mechanism of steroidogenic regulation.
Our bioinformatic analysis shows that the predicted let-7 binding sites on StAR are conserved. These binding sites, like all miRNA-mRNA binding sites, are predicted based on the seed sequence of miRNAs, which is usually located at positions 2 through 7 from the 5′ miRNA end. If the seed sequence is perfectly complimentary to the target mRNA, binding will occur even if the rest of the miRNA-mRNA sequence does not perfectly match. Thus, because the seed sequences listed in the manuscript are complementary, binding is expected to occur. Many miRNA prediction tools allow a limited number of G-U wobble pairs (the allowance of a G pairing with a U instead of a C) and insertions or deletions in other regions (25). Thus, this proposed mechanism may well hold true for all species.
These results suggest the possibility of a broader role for the H19/let-7 axis not only as a downstream effector of changes in steroid hormone levels but also as part of a larger feedback loop triggered by alterations in steroid homeostasis. There is a growing body of literature linking H19 expression to the presence of steroid receptors and linking H19 transcriptional regulation to hormonal stimulation (6). H19 expression has been linked to different stages of the estrous cycle (6) and to varying stages of follicular development (5) and has been noted to be specifically high during puberty and pregnancy, all processes that are hormonally driven (5, 6). The significance of this expression is unknown, but it is worth noting that the expression of StAR is also under tight spatiotemporal control in the ovary, with increased expression noted in antral and luteinizing follicles as well as corpora lutea (the same sites where expression of H19 has been observed) (26). H19 may thus not only be under the control of steroid hormones but also may act in a feedback loop with let-7 to regulate those same hormones and maintain steroid homeostasis. It has been reported that let-7 can target H19 for degradation in certain conditions, such as acute hyperinsulinemia, and that this “double-negative” feedback loop between H19 and let-7 contributes to glucose regulation (12). Thus, it is feasible that this let-7–mediated regulation of H19 is applicable to other physiologic systems as well. Moreover, let-7 is a downstream target of the RNA binding protein Lin28, which has a wide variety of target genes and is involved in multiple biological processes. Lin28 has recently been identified in testicular Leydig cells; further work could explore further the regulation of the H19/let-7 axis by RNA binding proteins (e.g., Lin28) in steroidogenic tissues (e.g., Leydig cells) (27). Moreover, there is a single report in the literature that demonstrates a direct effect of let-7 as a negative regulator of aromatase expression, suggesting that let-7 may also have direct steroidogenic effects (in addition to a possible role in H19 regulation and cholesterol homeostasis); these observations warrant further exploration as well (28).
Regarding the possibility of a broader role for the H19/let-7 axis in cholesterol homeostasis, the literature does suggest a role for let-7 in cholesterol metabolism: levels of let-7g display a female-specific elevation in individuals with metabolic syndrome, and elevated let-7g is significantly associated with low high-density lipoprotein in female patients (29). Thus, it could be predicted that the H19/let-7 axis might have a role in cholesterol metabolism; however, further study is required to provide a definitive answer to this question.
In conclusion, these results identify StAR as a novel target of let-7 and suggest a role for the H19/let-7 axis in the regulation of steroidogenesis via direct effect on StAR. Although further work must be done to investigate the significance of these findings in in vivo physiological systems, this work contributes to the growing body of literature describing the multiple and varied roles of this complex lncRNA.
Acknowledgments
We thank Yingqun Huang, Maria Lalioti, and Joshua Johnson for providing materials and assistance with experimental planning.
Acknowledgments
This work was supported by Grant K12 HD000849, awarded to the Reproductive Scientist Development Program (RSDP) by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the American Society for Reproductive Medicine as part of the RSDP, the Bennack-Polan Foundation grant, and the National Institutes of Health Loan Repayment Program.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- HEK
- human embryonic kidney
- lncRNA
- long noncoding RNA
- miRNA
- microRNA
- qRT-PCR
- quantitative reverse transcription–polymerase chain reaction
- StAR
- steroidogenic acute regulatory protein
- UTR
- untranslated region
References
- 1.Miller WL. Disorders in the initial steps of steroid hormone synthesis [published online ahead of print 6 March 2016]. J Steroid Biochem Mol Biol. 2016. [DOI] [PubMed] [Google Scholar]
- 2.Devoto L, Kohen P, Vega M, Castro O, González RR, Retamales I, Carvallo P, Christenson LK, Strauss JF et al. Control of human luteal steroidogenesis. Mol Cell Endocrinol. 2002;186(2):137–141. [DOI] [PubMed] [Google Scholar]
- 3.Kiriakidou M, McAllister JM, Sugawara T, Strauss JF III. Expression of steroidogenic acute regulatory protein (StAR) in the human ovary. J Clin Endocrinol Metab. 1996;81(11):4122–4128. [DOI] [PubMed] [Google Scholar]
- 4.Dugimont T, Curgy JJ, Wernert N, Delobelle A, Raes MB, Joubel A, Stehelin D, Coll J The H19 gene is expressed within both epithelial and stromal components of human invasive adenocarcinomas. Biol Cell. 1995;85(2-3):117–124. [DOI] [PubMed] [Google Scholar]
- 5.Ariel I, Weinstein D, Voutilainen R, Schneider T, Lustig-Yariv O, de Groot N, Hochberg A. Genomic imprinting and the endometrial cycle: the expression of the imprinted gene H19 in the human female reproductive organs. Diagn Mol Pathol. 1997;6(1):17–25. [DOI] [PubMed] [Google Scholar]
- 6.Adriaenssens E, Lottin S, Dugimont T, Fauquette W, Coll J, Dupouy JP, Boilly B, Curgy JJ. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene. 1999;18(31):4460–4473. [DOI] [PubMed] [Google Scholar]
- 7.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19(6):586–593. [DOI] [PubMed] [Google Scholar]
- 9.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 long non-coding RNA in cancer initiation, progression and metastasis: a proposed unifying theory. Mol Cancer. 2015;14(1):184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghazal S, McKinnon B, Zhou J, Mueller M, Men Y, Yang L, Mueller M, Flannery C, Huang Y, Taylor HS. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med. 2015;7(8):996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, Yang Y, Liu N, Zhao X, Santin AD, Taylor H, Huang Y. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34(23):3076–3084. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, Yang L, Mueller M, Zhou XB, Dandolo L, Szendroedi J, Roden M, Flannery C, Taylor H, Carmichael GG, Shulman GI, Huang Y. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014;42(22):13799–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci USA. 2013; 110(51):20693–20698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Zhou Y, Peng S, Wu L, Lin HY, Wang S, Wang H. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction. 2012;144(2):235–244. [DOI] [PubMed] [Google Scholar]
- 15.Ahn HW, Morin RD, Zhao H, Harris RA, Coarfa C, Chen ZJ, Milosavljevic A, Marra MA, Rajkovic A. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol Hum Reprod. 2010;16(7):463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles JR, McDaneld TG, Wiedmann RT, Cushman RA, Echternkamp SE, Vallet JL, Smith TP. MicroRNA expression profile in bovine cumulus-oocyte complexes: possible role of let-7 and miR-106a in the development of bovine oocytes. Anim Reprod Sci. 2012;130(1-2):16–26. [DOI] [PubMed] [Google Scholar]
- 17.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34(1):58–67. [DOI] [PubMed] [Google Scholar]
- 19.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132(23):7976–7981. [DOI] [PubMed] [Google Scholar]
- 20.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38(4):1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havelock JC, Rainey WE, Carr BR Ovarian granulosa cell lines. Mol Cell Endocrinol. 2004;228(1-2):67–78. [DOI] [PubMed] [Google Scholar]
- 22.Rebois RV. Establishment of gonadotropin-responsive murine leydig tumor cell line. J Cell Biol. 1982;94(1):70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YJ, Hsiao PW, Lee MT, Mason JI, Ke FC, Hwang JJ. Interplay of PI3K and cAMP/PKA signaling, and rapamycin-hypersensitivity in TGFbeta1 enhancement of FSH-stimulated steroidogenesis in rat ovarian granulosa cells. J Endocrinol. 2007;192(2):405–419. [DOI] [PubMed] [Google Scholar]
- 24.Dai A, Sun H, Fang T, Zhang Q, Wu S, Jiang Y, Ding L, Yan G, Hu Y. MicroRNA-133b stimulates ovarian estradiol synthesis by targeting Foxl2. FEBS Lett. 2013;587(15):2474–2482. [DOI] [PubMed] [Google Scholar]
- 25.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollack SE, Furth EE, Kallen CB, Arakane F, Kiriakidou M, Kozarsky KF, Strauss JF III. Localization of the steroidogenic acute regulatory protein in human tissues. J Clin Endocrinol Metab. 1997;82(12):4243–4251. [DOI] [PubMed] [Google Scholar]
- 27.Gaytan F, Sangiao-Alvarellos S, Manfredi-Lozano M, García-Galiano D, Ruiz-Pino F, Romero-Ruiz A, León S, Morales C, Cordido F, Pinilla L, Tena-Sempere M. Distinct expression patterns predict differential roles of the miRNA-binding proteins, Lin28 and Lin28b, in the mouse testis: studies during postnatal development and in a model of hypogonadotropic hypogonadism. Endocrinology. 2013;154(3):1321–1336. [DOI] [PubMed] [Google Scholar]
- 28.Shibahara Y, Miki Y, Onodera Y, Hata S, Chan MS, Yiu CC, Loo TY, Nakamura Y, Akahira J, Ishida T, Abe K, Hirakawa H, Chow LW, Suzuki T, Ouchi N, Sasano H. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J Pathol. 2012;227(3):357–366. [DOI] [PubMed] [Google Scholar]
- 29.Wang YT, Tsai PC, Liao YC, Hsu CY, Juo SH Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci. 2013;20:72. [DOI] [PMC free article] [PubMed] [Google Scholar]