Hepatocellular cancers arise in a background of liver damage and inflammation. Bakiri et al. describe the function of the transcription factor c-Fos/AP-1 using mouse models and human data. c-Fos affects cholesterol and bile acid metabolism and induces DNA damage and inflammation, thus promoting liver cancer.
Abstract
Human hepatocellular carcinomas (HCCs), which arise on a background of chronic liver damage and inflammation, express c-Fos, a component of the AP-1 transcription factor. Using mouse models, we show that hepatocyte-specific deletion of c-Fos protects against diethylnitrosamine (DEN)-induced HCCs, whereas liver-specific c-Fos expression leads to reversible premalignant hepatocyte transformation and enhanced DEN-carcinogenesis. c-Fos–expressing livers display necrotic foci, immune cell infiltration, and altered hepatocyte morphology. Furthermore, increased proliferation, dedifferentiation, activation of the DNA damage response, and gene signatures of aggressive HCCs are observed. Mechanistically, c-Fos decreases expression and activity of the nuclear receptor LXRα, leading to increased hepatic cholesterol and accumulation of toxic oxysterols and bile acids. The phenotypic consequences of c-Fos expression are partially ameliorated by the anti-inflammatory drug sulindac and largely prevented by statin treatment. An inverse correlation between c-FOS and the LXRα pathway was also observed in human HCC cell lines and datasets. These findings provide a novel link between chronic inflammation and metabolic pathways important in liver cancer.
Introduction
Primary liver cancer is the fifth and seventh most common cancer in men and women, respectively, and hepatocellular carcinoma (HCC), the most common histological subtype, accounts for 70–85% of cases (Jemal et al., 2011). HCC develops almost exclusively in the context of liver diseases associated with chronic inflammation. For this reason, liver cirrhosis, a consequence of many chronic liver diseases such as viral hepatitis, alcohol-induced hepatitis, or non–alcohol-induced hepatitis, is the main risk factor for HCC (Fattovich et al., 2004). Previous findings using genetically engineered mouse models (GEMMs) indicate that continuous rounds of hepatocyte injury, necrosis, inflammation-induced cell death, and subsequent compensatory proliferation are essential for liver cancer initiation and promotion (Farazi and DePinho, 2006). This is supported by the fact that during progression of chronic liver disease, HCC risk increases (Fattovich et al., 2004).
Although some improvement in the management of HCC has been achieved in the last 30 years, beneficial treatment is only possible at early stages by local ablative therapies, resection, or transplantation (Villanueva et al., 2013). Long-term prognosis after surgical resection of HCC remains poor, owing to the high rate of metastasis or de novo recurrence. Systemic chemotherapies and targeted therapies have failed in the treatment of HCC (Villanueva et al., 2013). Besides prevention and treatment of the causative liver diseases, early diagnosis, identification of high-risk patients, and prevention of malignant transformation are the most promising approaches. The critical step to identify biomarkers and develop effective preventive therapies is a better understanding of the mechanisms responsible for cancer initiation. The pathways most frequently involved are p53, Wnt/β-catenin, mTOR, TGF-β, Ras, Rb, HGF/c-Met, and IGF1. Transcription factors such as NF-κB, c-Myc, and AP-1 play an important role in HCC development (Liu et al., 2002; Wagner and Nebreda, 2009; Jain et al., 2010; He and Karin, 2011; Nault and Zucman-Rossi, 2011).
The AP-1 transcription factor is a dimeric complex composed of members of the Jun (c-Jun, JunB, JunD) and Fos (c-Fos, FosB, Fra1, Fra2) families of bZIP proteins. Components of AP-1 including c-Jun and c-Fos are important regulators of tumor development (Eferl and Wagner, 2003). In human HCC, both c-Jun and c-Fos are highly expressed, and genome-wide expression analysis of human HCCs revealed that AP-1 is at the center of an oncogenic signaling network in a subset of HCC with poor prognosis (Yuen et al., 2001; Liu et al., 2002; Lee et al., 2006). Mouse models for liver cancer have further established the importance of AP-1 in liver pathology and HCC (Bakiri and Wagner, 2013). In the diethylnitrosamine (DEN)-induced liver cancer model, mice with liver-specific inactivation of c-Jun have significantly fewer tumors because of p53-dependent apoptosis of tumor cells (Eferl et al., 2003). Consistently, inhibition of JNKs, which activate and stabilize c-Jun, leads to reduced proliferation and increased sensitization toward apoptosis in HCC models in vivo and in vitro (Sakurai et al., 2006; Hui et al., 2007; Wagner and Nebreda, 2009; Seki et al., 2012). c-Jun is specifically required for mouse liver tumorigenesis during cancer initiation, and we have shown that c-Jun promotes preneoplastic cell survival by regulating c-Fos– and SIRT6-dependent expression of survivin (Eferl et al., 2003; Min et al., 2012). The cancer-promoting function of c-Jun was also shown in mouse models for HBV- and HCV-related liver tumorigenesis (Machida et al., 2010; Trierweiler et al., 2016). Finally c-Jun promotes hepatocyte survival in experimental models of hepatitis, ER stress, and activated β-catenin–induced liver damage (Hasselblatt et al., 2007; Fuest et al., 2012; Trierweiler et al., 2012).
The role of c-Fos in HCC development is less well defined. Studies in human HCC cell lines indicate that c-Fos is important for cell migration (Fan et al., 2013), and ectopic expression of c-Fos in immortalized human hepatocytes increased cell proliferation (Güller et al., 2008). We recently documented how distinct AP-1 dimers regulate the expression of PPARγ in the liver in models of nonalcoholic fatty liver disease (Hasenfuss et al., 2014b), a condition associated with obesity and an established risk factor for HCC (Michelotti et al., 2013). Accumulation of cholesterol, especially free cholesterol and cholesterol derivatives such as oxysterols and bile acids (BAs), promotes hepatotoxicity, oxidative stress, inflammation, and even hepatocyte transformation (Tabas, 2002; Ikonen, 2006; Jusakul et al., 2011; Wang et al., 2013). The nuclear receptor and transcription factor LXRα (Nr1h3), which is activated by oxysterols, is essential for cholesterol homeostasis. In the liver, LXRα forms transcriptional heterodimers with retinoid X receptors (RXRs) and mediates cholesterol removal by promoting both cholesterol conversion to BAs and cholesterol excretion to the bile (Calkin and Tontonoz, 2012). As a result, Lxrα knockout mice display altered hepatic cholesterol and BA metabolism, with cholesterol accumulation and impaired hepatic function (Peet et al., 1998; Zhang et al., 2012). The BA receptor FXR (Nr1h4) is another important nuclear receptor and RXR dimerization partner essential for lipid and BA homeostasis (Calkin and Tontonoz, 2012). Mice lacking FXR have increased BAs, liver damage, inflammation, and late-onset liver tumors, all prevented by restoring BA homeostasis through BA-sequestering agents or intestinal FXR expression (Kim et al., 2007; Yang et al., 2007; Degirolamo et al., 2015).
In this study, we show that hepatocyte-specific expression of c-Fos leads to reversible premalignant transformation of hepatocytes, with mRNA signatures resembling human HCCs. Importantly, using the DEN experimental HCC model, we demonstrate that c-Fos is essential for hepatocyte transformation and HCC development. Mechanistically, c-Fos negatively regulates LXRα expression, which affects cholesterol and BA homeostasis.
Results
Hepatic c-Fos expression leads to premalignant transformation
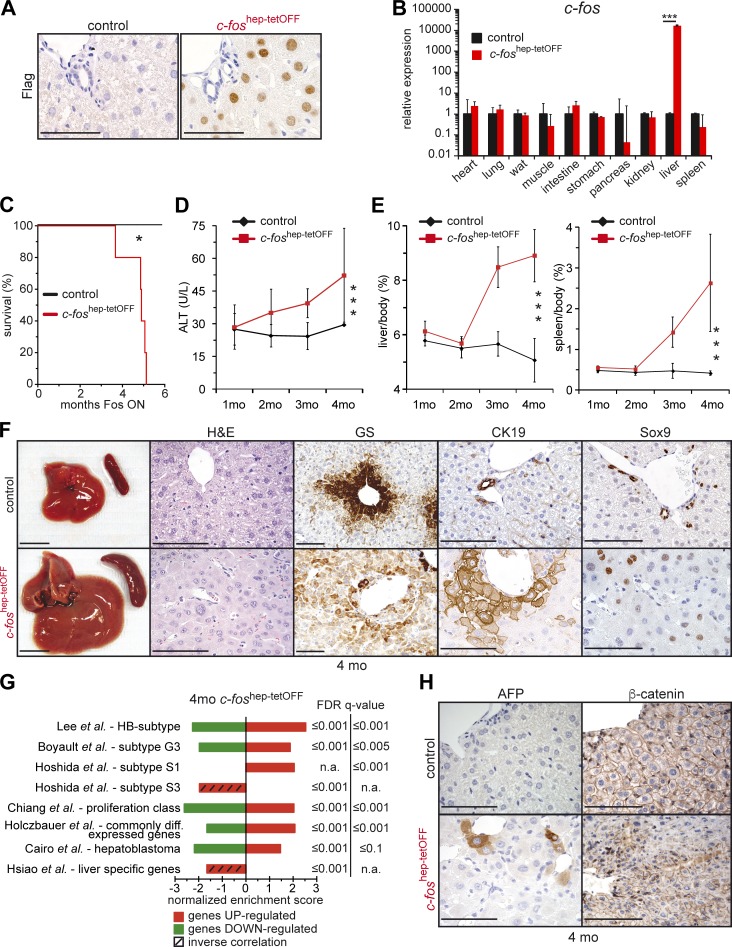
To investigate the role of c-Fos expression in liver physiology and carcinogenesis, a hepatocyte-specific doxycycline-switchable mouse model was used. LAP-tTA; col1a1:Tet-O-fosFlag double-transgenic mice are referred to as c-foshep-tetOFF mice, and single-transgenic littermates were used as controls throughout the study. Expression of c-Fos was switched on at the age of 3 wk by doxycycline removal. Although endogenous c-Fos is undetectable in control livers, hepatocyte-specific expression of Flag-tagged ectopic c-Fos was observed by immunohistochemical staining (IHC; Fig. 1 A), quantitative RT-PCR (qRT-PCR; Fig. 1 B), and Western blot (Fig. S1 A). No ectopic c-Fos mRNA was detected in any other tissue tested (Fig. S1 B). c-foshep-tetOFF mice had a median survival of 5 mo (Fig. 1 C). Therefore, changes in liver physiology were analyzed at up to 4 mo of c-Fos expression. The first histological observation was single-cell necrosis with surrounding immune cell infiltrates, mainly periportal, at 1 mo (Fig. S1 C). These lesions progressed over time to multiple necrotic foci at 3 and 4 mo (Fig. S1 C). Increased serum alanine aminotransferase (ALT), a marker for liver damage, was observed at 2 mo of c-Fos expression (Fig. 1 D), whereas elevated serum γ-glutamyl transferase and decreased albumin and blood urea nitrogen, all indicative of liver dysfunction, were observed at 4 mo (Fig. S1 D). Macroscopically, progressive hepatosplenomegaly was observed (Fig. 1 E), whereas body weight was not affected (Fig. S1 E). An altered growth pattern of hepatocytes was visible at 4 mo of c-Fos expression, with trabecular proliferation and double as well as poly nuclei (Fig. 1 F). In addition, although the controls showed the expected central vein localization of glutamine synthetase (GS), mutant livers displayed diffuse cytoplasmic staining, indicative of disturbed liver zonation (Fig. 1 F). Moreover, scattered positive staining for the dedifferentiation markers CK19 and Sox9 was apparent (Fig. 1 F) at as early as 2.5 mo of c-Fos expression (Fig. S1 F; see also Fig. 9 E). These results indicate that hepatocyte-specific c-Fos expression leads to altered hepatocyte morphology, necrosis, and a dedifferentiated phenotype indicative of premalignant transformation.
Figure 1.
Phenotypic consequences of hepatocyte-specific c-Fos expression. Ectopic expression of c-Fos was achieved in 3-wk-old mice by doxycycline removal. Mice were analyzed at 1, 2, 3, and 4 mo of c-Fos expression. (A) Representative Flag IHC in liver sections from c-foshep-tetOFF and control mice 2 mo after doxycycline removal.(B) qRT-PCR analyses of c-fos in different tissues. Bar graphs represent mean ± SD; n = 2/3; mean expression in controls set to 1; ***, P ≤ 0.001 by Student’s t test. (C) Kaplan–Meier curve (n = 5/cohort); 5 mo = median survival in mutants. *, P ≤ 0.05. (D and E) Serum ALT (D; n = 4; 12; 3; 9/3; 11; 3; 6) and liver/body and spleen/body weight ratio (E; n = 4; 5; 3; 10/3; 5; 3; 7) upon c-Fos expression. Plots represent mean ± SD. ***, P ≤ 0.001 by two-way ANOVA. (F) Representative liver pictures, H&E, and IHC for GS, CK19, and Sox9 in c-foshep-tetOFF and controls at 4 mo. (G) Normalized enrichment scores at 4 mo ectopic c-Fos expression relative to controls (RNA-seq, n = 3/cohort) compared with human HCC molecular classes and relevant gene signatures by GSEA. False discovery rate (FDR) q-values are indicated on the right side. n.a., not applicable, as the published gene signature was unidirectional (only enriched genes). Hatched bars highlight inverse correlations computed with up-regulated (red) or down-regulated (green) gene sets. (H) Representative IHC for AFP and β-catenin in c-foshep-tetOFF and controls at 4 mo. (A, F, and H) Bars: (F, left) 1 cm; (A, F [right], and H) 100 µm.
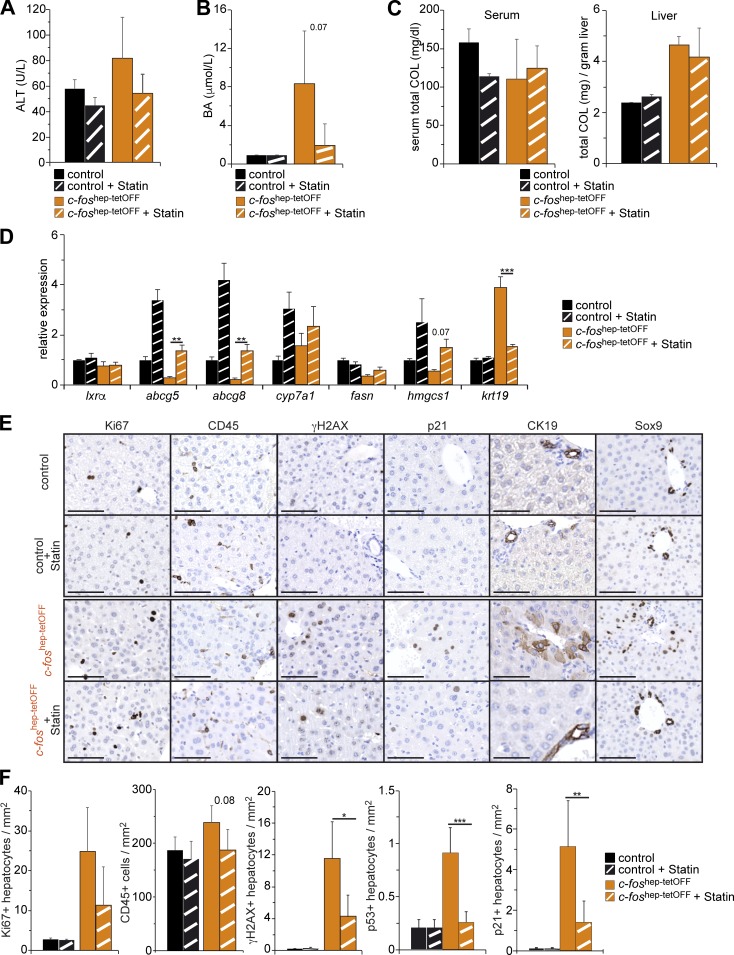
Figure 9.
Phenotypic consequences of statin treatment in c-foshep-tetOFF mice. (A–C) Serum ALT (A), serum BAs (B), and serum and liver total cholesterol (C) in untreated and statin-treated c-foshep-tetOFF and controls (n = 2; 2/4; 6). (D) qRT-PCR analyses in total liver tissue from untreated and statin-treated c-foshep-tetOFF and controls (n = 2; 2/3; 5, mean expression in untreated controls set to 1). (E) Representative IHC for Ki67, CD45, γH2AX, p21, CK19, and Sox9 in untreated and statin-treated c-foshep-tetOFF and controls. Bars, 100 µm. (F) Quantification of CD45-positive cells and Ki67-, γH2AX-, p53-, and p21-positive hepatocytes in liver sections of untreated and statin-treated c-foshep-tetOFF and controls (n = 2; 2/4; 6). Bar graphs represent mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 by Student’s t test.
Fos-expressing mouse livers exhibit molecular characteristics of human HCCs
Genome-wide transcription profiling using RNA-seq of liver mRNA at 4 mo of c-Fos expression was compared by gene set enrichment analysis (GSEA) with the well-established molecular classifications of human HCC (reviewed in Hoshida et al., 2010; van Malenstein et al., 2011; Pinyol et al., 2014). Notably, a significant positive correlation was observed with the hepatoblast (HB) subtype of human HCC with poor prognosis (Fig. 1 G), in which c-Fos/AP-1 was previously shown to be up-regulated and located in the center of an oncogenic signaling network (Lee et al., 2006). c-Fos–expressing gene expression profiles also significantly correlated with published HCC gene signatures, in particular the subtypes G3 (Boyault et al., 2007), S1 (Hoshida et al., 2009), the proliferative class (Chiang et al., 2008), and pediatric hepatoblastoma (Cairo et al., 2008; Fig. 1 G). These gene signatures are all characteristic of dedifferentiation, fetal liver–like gene expression, high proliferation, and aggressiveness (Hoshida et al., 2010; van Malenstein et al., 2011; Pinyol et al., 2014). We also observed a significantly positive correlation, with a 590-gene signature differentially expressed across mouse primary hepatic progenitor cells, HBs, and transformed adult hepatocytes (Holczbauer et al., 2013), whereas a significant inverse correlation was computed with signatures derived from healthy livers (Hsiao et al., 2001) or the well-differentiated and less aggressive S3 HCC subclass (Hoshida et al., 2009). A large fraction of these correlations could already be computed in RNA-seq profiles of livers expressing c-Fos at 2 mo (Fig. S1 G). These results imply that c-Fos expression leads to molecular characteristics of hepatocyte dedifferentiation and premalignant transformation. Consistent with dedifferentiation and premalignant transformation, α-fetoprotein (AFP) positivity and nuclear β-catenin were also observed (Fig. 1 H). Because c-Fos mutant mice showed dramatically reduced survival as a result of hepatic dysfunction, we speculate that c-Fos–expressing mice die before these premalignant cells progress to HCCs.
Liver carcinogenesis is enhanced by c-Fos expression
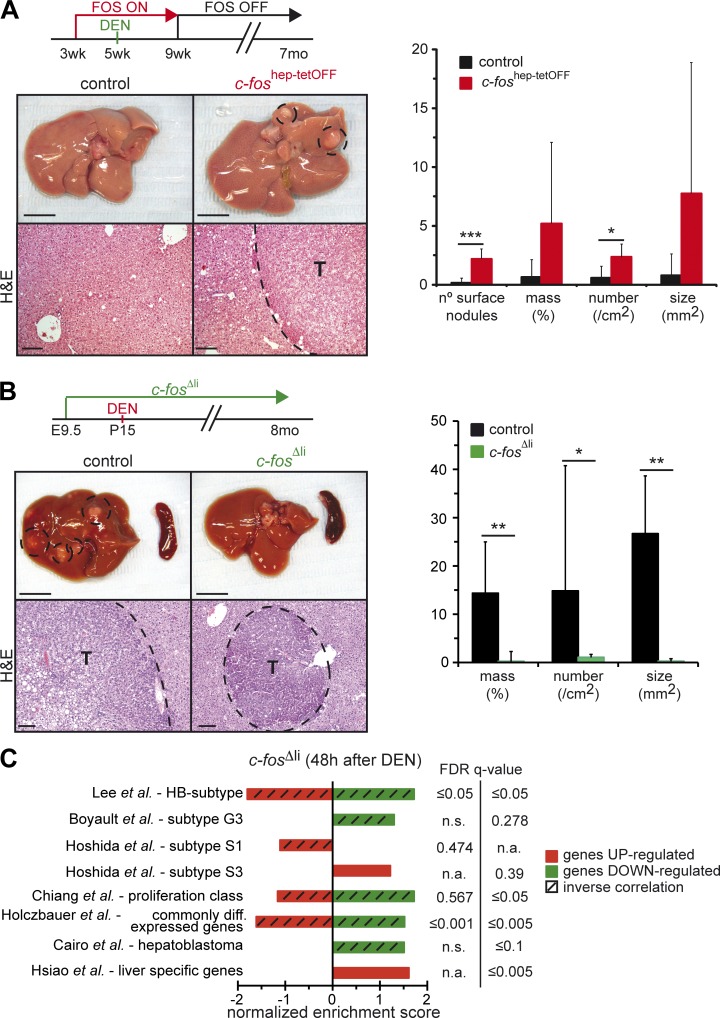
To circumvent the lethality induced by sustained c-Fos expression, the DEN-induced experimental carcinogenesis paradigm was applied to adult mice, with c-Fos expression restricted to tumor initiation (Fig. 2 A). In this setting, control mice very rarely developed HCCs (Bakiri and Wagner, 2013). However, when ectopic c-Fos expression was induced for a short period during DEN-induced tumor initiation, tumor development at 7 mo was significantly increased (Fig. 2 A). qRT-PCR analyses revealed increased c-Fos expression in tumor-bearing livers, whereas ectopic c-Fos was not expressed (Fig. S2 A). These data strongly imply a promoting function of c-Fos operating during the early events of malignant hepatocyte transformation and HCC development.
Figure 2.
c-Fos expression promotes and is essential for HCC development. (A) 5-wk-old c-foshep-tetOFF and controls were injected with 100 mg/kg DEN, while c-Fos expression was maintained from 3 to 9 wk. Representative liver pictures and histology 7 mo after DEN. (Right) Macroscopic and histological tumor quantification at 7 mo (n = 6/5). (B) 15-d-old c-fosΔli and c-fosf/f littermates were injected with 25 mg/kg DEN. Representative liver pictures and H&E 8 mo later.. (Right) Histological tumor quantification in c-fosΔli and controls 8 mo after DEN (n = 14/10). (A and B) Bars: (top) 1 cm; (bottom) 100 µm. T, tumor. Bar graphs represent mean ± SD; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 by Student’s t test. (C) Normalized enrichment scores 48 h after DEN injection in 8-wk-old c-fosΔli mice relative to controls (RNA-seq, n = 3/cohort) compared with human HCC molecular classes and relevant gene signatures by GSEA. False discovery rate (FDR) q-values are indicated on the right side. n.a., not applicable, as the published gene signature was unidirectional (with only enriched genes). n.s., not significant, as only one out of the two signatures published for the gene set was enriched in the GSEA. Hatched bars highlight inverse correlations computed with up-regulated (red) or down-regulated (green) gene sets. Note that the enrichment scores in c-fosΔli livers are inverse orientation, compared with the analysis of c-foshep-tetOFF livers presented in Fig. 1 G and Fig. S1 G.
DEN-induced HCC formation requires c-Fos expression
We next applied the DEN-induced mouse liver cancer model to mice with hepatocyte-specific knockout of c-Fos (c-fosΔli). Mice with conditional alleles (c-fosf/f) combined with the Alfp-Cre transgene were used to delete c-fos specifically in hepatocytes (Kellendonk et al., 2000; Fleischmann et al., 2003). DEN was injected in 2-wk-old pups to allow efficient tumor formation. Whereas all control mice developed multiple HCCs 8 mo later, c-fosΔli mice hardly developed any visible tumor (Fig. 2 B and Fig. S2 B). Histological analyses documented a significantly reduced number and size of foci in these mutants, which mainly displayed nodular proliferation and almost no HCC. PCR analyses of genomic DNA from liver and tumor tissue confirmed deletion of c-fos in c-fosΔli mice (Fig. S2 C). Moreover, DEN-induced DNA adducts were increased in c-fosΔli mice (Fig. S2 D), indicating that decreased tumor load in c-Fos mutants is not caused by decreased carcinogenic DEN metabolites.
Genome-wide transcription profiling using RNA-seq was performed using liver mRNA from c-fosΔli mice 48 h after DEN injection. Strikingly, several of the gene sets that correlated with Fos-overexpressing livers were also found enriched in the dataset from DEN-injected c-fosΔli mice but in the opposite direction. Most notably, whereas the S3-HCC subclass and healthy liver–specific genes were positively correlated with the c-fosΔli expression dataset, the HCC-HB subtype, the S1 and proliferative HCC subclasses, and the gene set characteristic of dedifferentiated/transformed hepatocytes were negatively correlated (Fig. 2 C). These data further confirm the association between c-Fos expression and malignant hepatocyte transformation during HCC and establish that c-Fos is not only sufficient to induce malignant transformation of hepatocytes, but is also required for HCC development in vivo.
Liver inflammation and genotoxic stress induced by c-Fos
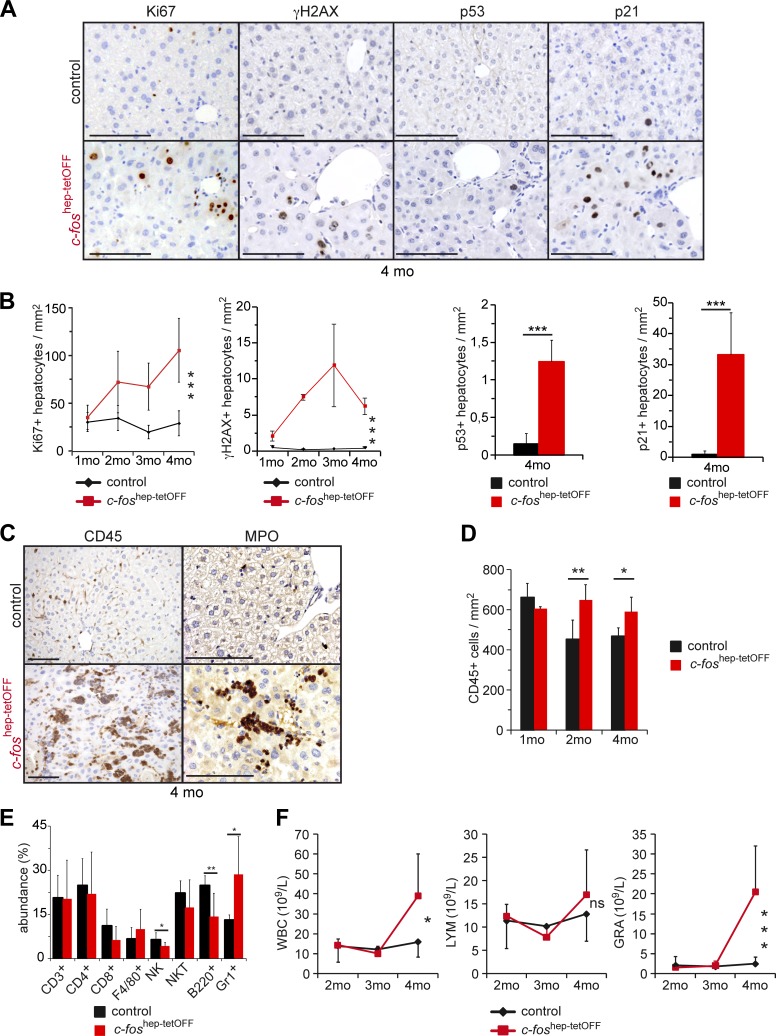
The c-Fos–induced hepatomegaly and premalignant transformation was accompanied by an increase in the number of Ki67-positive nuclei indicative of proliferation (Fig. 3, A and B). S139 phosphorylation of histone H2AX (γH2AX), a surrogate marker of DNA damage (Kinner et al., 2008), and increased p53- and p21-positive nuclei (Fig. 3, A and B) were also observed, indicating that the DNA damage response (DDR) was activated. Although necrosis was apparent, no obvious hepatocyte apoptosis was detected by cleaved caspase 3 IHC (not depicted). Significantly more immune cells were detected in the liver, in particular myeloperoxidase-positive granulocytes (Fig. 3, C and D). Flow cytometry analyses confirmed the accumulation of CD45-positive cells in the liver (Fig. S2 E) and the specific increase in Gr1+ cells at 2 mo, whereas NK and B cells were reduced (Fig. 3 E). Increased circulating leukocytes and granulocytes were also observed in the blood at later time points (Fig. 3 F), consistent with hepatic immune cell infiltration and increased spleen size. Several pro-inflammatory and pro-oncogenic pathways are activated in the livers of c-foshep-tetOFF mice, as indicated by increased phosphorylation of Stat3, Jnk1/2, and Akt (Fig. S2 F), likely as a response to increased inflammation and genotoxic stress. These data imply that c-Fos–induced liver inflammation, hepatocyte proliferation, and DDR activation are early events.
Figure 3.
c-Fos–dependent proliferation, DDR, and inflammation. (A) Representative IHC pictures for Ki67, S139 phosphorylation of histone H2AX (γH2AX), p53, and p21 in c-foshep-tetOFF and controls at 4 mo. (B) Quantification of Ki67- and γH2AX-positive hepatocytes at different time points (n = 4; 5; 3; 7/3; 5; 3; 6 and 4; 4; 2; 4/5; 5; 3; 7) and of p21- and p53-positive hepatocytes at 4 mo (n = 4/5). (C) Representative IHC pictures for CD45 and myeloperoxidase (MPO) at 4 mo. (D) Quantification of CD45-positive cells in liver sections (n = 3; 5; 4/3; 5; 4). (E) Immune cell subtypes in the liver at 2 mo (flow cytometry, n = 6/8). (F) Blood cell count of white blood cells (WBC), lymphocytes (LYM), and granulocytes (GRA) at the indicated time points of ectopic c-Fos expression (n = 4; 7; 6; 8/3; 6; 7; 6). Bars, 100 µm. Bar graphs and plots represent mean ± SD; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant by Student's t test or two-way ANOVA.
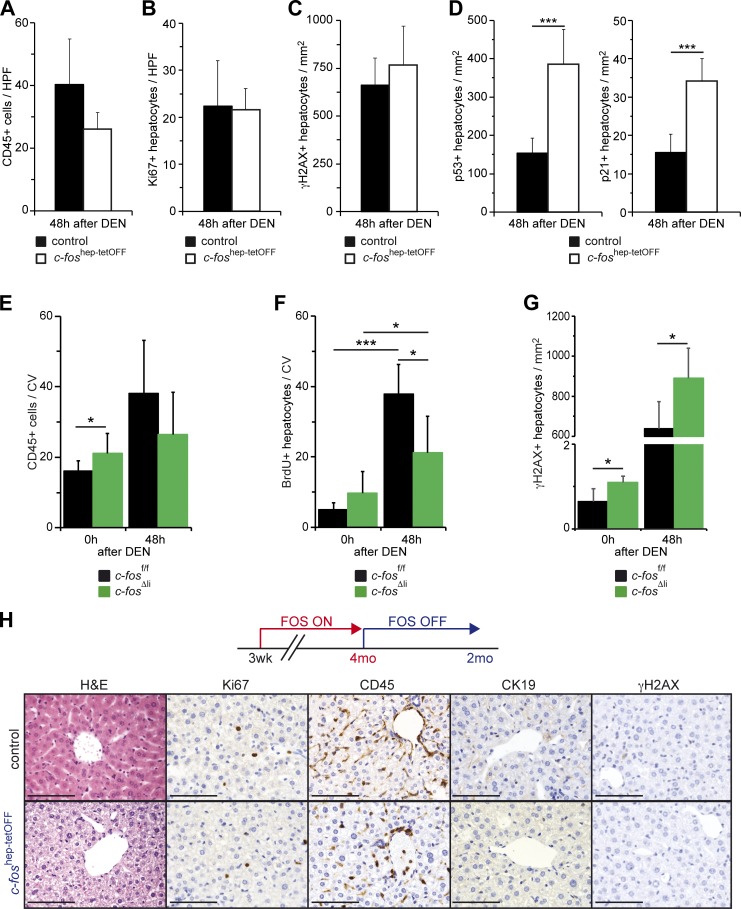
We next examined the early events occurring after DEN injection in the livers of c-Fos gain- and loss-of-function GEMMs. 8-wk-old c-foshep-tetOFF mice allowed to express c-Fos for 5 wk were injected with DEN and analyzed 48 h later. Similar to our observations when c-Fos was expressed for 1 mo in the absence of DEN (Figs. 1 and 3), serum ALT and hepatocyte apoptosis were not changed (Fig. S3 A). Short-term ectopic c-Fos expression combined with DEN appeared to have little effect on immune cell infiltration or proliferation (Fig. 4, A and B). However, despite comparable γH2AX counts, p53- and p21-positive nuclei were increased (Fig. 4, C and D), indicating that increased DDR likely contributes to tumorigenesis when c-Fos expression is combined with DEN.
Figure 4.
c-Fos–dependent early carcinogenic events and phenotype reversibility. (A–D) Quantification of CD45-positive cells (A; n = 5/5) and Ki67-positive (B; n = 5/5), γH2AX-positive (C; n = 4/5), and p53-positive (n = 4/5) and p21-positive (D; n = 4/5) hepatocytes in liver sections from 8-wk-old c-foshep-tetOFF and control mice 48 h after DEN. (E–G) Quantification of CD45-positive (E; n = 8; 6/8; 5) and BrdU-positive (F; n = 4; 7/3; 6) cells around the central vein (CV) and γH2AX-positive hepatocytes (G; n = 4/5) in liver sections from 8-wk-old c-fosΔli and control mice untreated (0) and 48 h after DEN. (A–G) Plots represent mean ± SD; *, P ≤ 0.05; ***, P ≤ 0.001 by Student’s t test. (H) Ectopic expression of c-Fos was allowed during 4 mo and then stopped by administration of doxycycline. Representative liver histology (H&E) and IHC (Ki67, CD45, CK19, and γH2AX) from c-foshep-tetOFF and controls 2 mo after switching off c-Fos expression. Bars, 100 µm.
Deletion of c-Fos in hepatocytes did not affect serum ALT or cell death under basal conditions or 48 h after DEN injection (Fig. S3 B), and c-Fos also had little impact on immune cell infiltration after DEN injection (Fig. 4 E). However, c-fosΔli mice showed increased γH2AX-positive nuclei and reduced compensatory proliferation after DEN-induced liver damage (Fig. 4, F and G), possibly accounting for decreased tumor load.
We next investigated whether c-Fos is necessary to maintain the premalignant phenotype and took advantage of the tetracycline control of c-Fos expression in c-foshep-tetOFF mice. c-Fos expression was induced for 4 mo, and the mice were subsequently put back on doxycycline to repress c-Fos expression. 2 mo later, c-Fos expression in c-foshep-tetOFF reverted mice was comparable with that of controls, and ectopic c-Fos was undetectable (Fig. S3 C). Furthermore, no hepatosplenomegaly was observed (Fig. S3 D), and serum ALT and blood cell counts were comparable with those of control mice (Fig. S3, E and F). Histological analyses confirmed normal liver morphology with no Ki67-, CK19-, or γH2AX-positive hepatocytes and no immune cell infiltration (Fig. 4 H and Fig. S3 G). The c-foshep-tetOFF reverted mice were viable and appeared healthy more than 6 mo after turning off c-Fos expression (not depicted). These results demonstrate that liver damage, inflammation, hepatocyte proliferation, and preneoplastic transformation induced by c-Fos expression are reversible and that c-Fos is essential to maintain the premalignant phenotype.
Hepatic metabolic pathways are affected by c-Fos
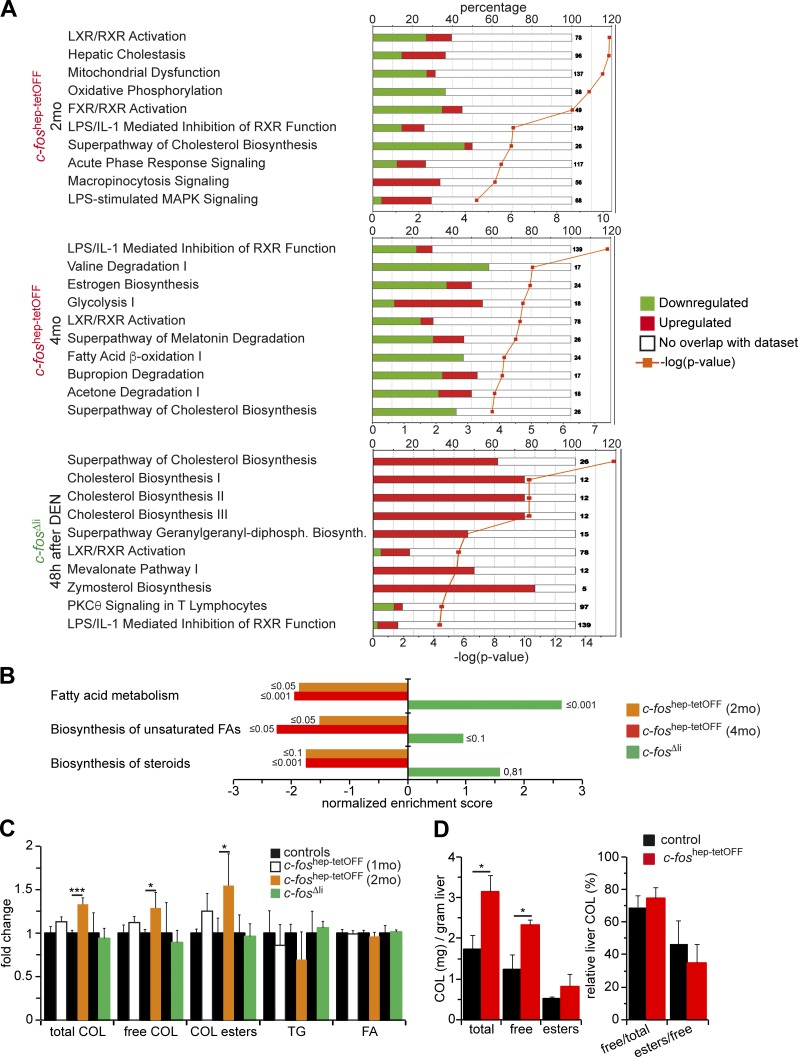
We next analyzed the three gene expression profiles obtained by RNA-seq using Ingenuity Pathway Analysis. The 10 top regulated pathways common to all datasets revealed a potential Fos-dependent regulation of metabolic pathways, particularly the ones connected to cholesterol and fatty acid (FA) metabolism. In particular, the LXR/RXR pathway was down-regulated upon c-Fos expression and up-regulated in c-fos–deficient mice (Fig. 5 A). GSEA confirmed that genes in cholesterol and FA biosynthesis were repressed in c-foshep-tetOFF mice and enriched in c-fos–deficient mice (Fig. 5 B).
Figure 5.
Metabolic pathways are regulated by c-Fos in the liver. (A) Ingenuity canonical pathway analyses. The 10 top affected canonical pathways in each condition are shown (n = 2/3/3/cohort). (B) Normalized enrichment scores for each indicated genotype and condition derived from GSEA and focused on lipid metabolism pathways (RNA-seq, n = 2; 3; 3/cohort). False discovery rate (FDR) q-values are indicated on each bar. (C) Relative change in liver cholesterol species, TGs, and FAs identified by NMR in c-foshep-tetOFF (1 and 2 mo of c-Fos expression; n = 3; 5/cohort) and c-fosΔli (48 h after DEN; n = 5/cohort) mice. Control groups were set to 1.(D) Cholesterol species in liver tissue of c-foshep-tetOFF and control mice at 4 mo of c-Fos expression measured by a colorimetric method (n = 7/6). (C and D) Bar graphs represent mean ± SD;*, P ≤ 0.05; ***, P ≤ 0.001 by Student’s t test.
Liver metabolites were next analyzed by nuclear magnetic resonance (NMR) spectroscopy. Increased cholesterol species were observed in the livers of c-foshep-tetOFF mice relative to controls, whereas triglycerides (TGs) and FAs were not affected (Fig. 5 C). Hepatic cholesterol accumulation in c-foshep-tetOFF mice was confirmed by a colorimetric assay, which also indicated that the ratio between cholesterol species was unaffected (Fig. 5 D). On the other hand, serum cholesterol was reduced at 4 mo of hepatic c-Fos expression (Fig. S4 A), with a decreased esterified/free cholesterol ratio (Fig. S4 B), and restored in the c-foshep-tetOFF reverted mice (Fig. S4 C). These results demonstrate that cholesterol accumulation in hepatocytes is a phenotypic consequence of increased c-Fos expression.
LXRα-mediated cholesterol accumulation in Fos-expressing livers
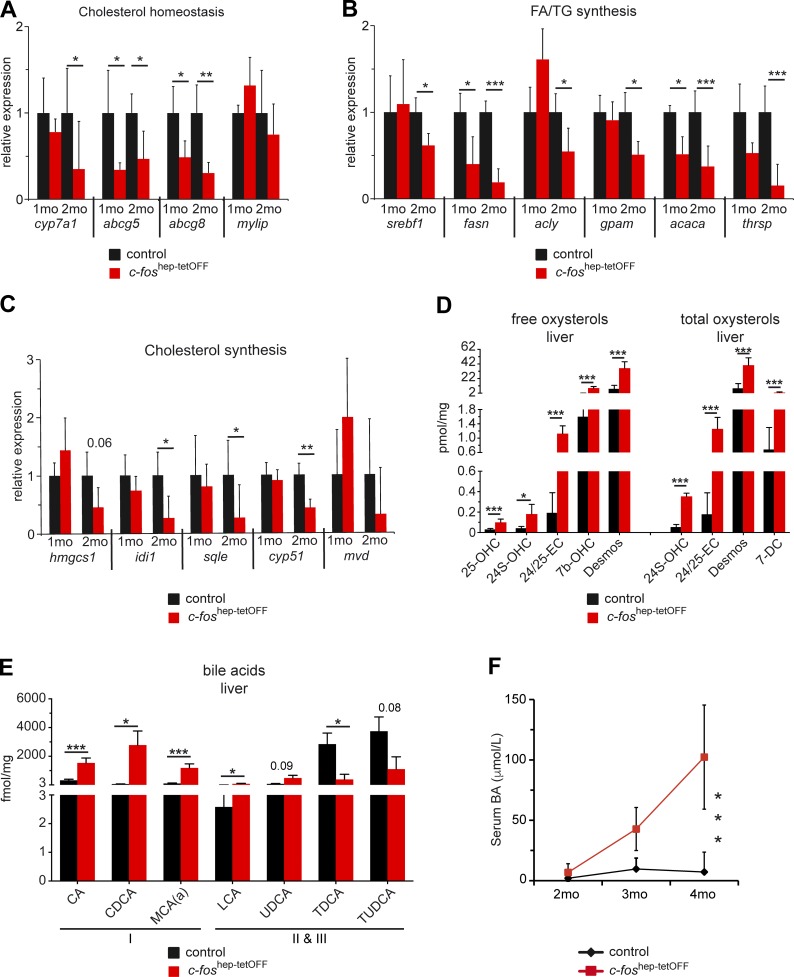
The nuclear receptor LXRα (Nr1h3) is a major regulator of cholesterol homeostasis responsible for cholesterol elimination from the cell (Calkin and Tontonoz, 2012). qRT-PCR analyses of genes involved in cholesterol homeostasis and FA and TG synthesis, including bona fide LXRα target genes, were performed. Several LXRα target genes such as abcg5, abcg8, fasn, and acaca were significantly down-regulated at 1 mo of c-Fos expression (Fig. 6, A and B). Consistent with the Ingenuity analysis, reduced expression of genes involved in cholesterol synthesis was observed at 2 mo (Fig. 6 C), indicating compensatory down-regulation. No significant changes in expression of LXRα target genes could be observed in c-fos–deficient or reverted livers (Fig. S4, D and E).
Figure 6.
Metabolic gene expression and cholesterol derivatives in c-foshep-tetOFF mice. (A–C) qRT-PCR analyses of genes involved in cholesterol homeostasis (A), FA and TG (B), and cholesterol (C) synthesis in liver of c-foshep-tetOFF and controls at 1 and 2 mo of c-Fos expression (n = 3/5); mean expression in each control group set to 1. (D and E) Free and total oxysterol (D) and BA (E) species in liver extracts from c-foshep-tetOFF at 4 mo of c-Fos expression (n = 5/cohort) determined and quantified by MS. Names of oxysterol and BA species are listed in Table S1. (F) Serum BAs (colorimetry) at the indicated time points of c-Fos expression (n = 9;3;12/11;4;8). Plots and bar graphs represent mean ± SD; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 by two-way ANOVA or Student’s t test.
Oxysterols, which are cholesterol metabolites and natural ligands for LXRα, were next analyzed by mass spectrometry. Total and free oxysterols were significantly increased in livers of c-foshep-tetOFF mice (Fig. 6 D, and Fig. S4 F, and Table S1), consistent with increased cholesterol and decreased expression of the Abcg5/Abcg8 transporter, which promotes cholesterol excretion from hepatocytes into bile, and further indicating that reduced activity of LXRα is not caused by lack of ligands. Cyp7a1, the rate-limiting enzyme for primary BA synthesis, is also modulated by LXRα in mice and was decreased at 2 mo of c-Fos expression (Fig. 6 A and Fig. S4 G). However, hepatotoxic primary BAs (CA, CDCA, and αMCA) were significantly increased in the liver (Fig. 6 E and Table S1), which could be explained by the overall increase in cholesterol. On the other hand, hepatoprotective taurine conjugates (TDCA and TUDCA) were reduced (Fig. 6 E), consistent with reduced mRNA expression of Baat and Scl27a5, two enzymes responsible for BA–amino acid conjugation (Fig. S4 G). BAs were also increased in serum of c-foshep-tetOFF mice (Fig. 6 F) reflecting both the hepatic increase in toxic BA species and liver damage. These results indicate that c-Fos expression affects LXRα-mediated control of cholesterol homeostasis, leading to compensatory reduced expression of genes involved in cholesterol synthesis and the accumulation of potentially toxic cholesterol species and derivatives in the liver.
Inhibition of LXRα expression by c-Fos
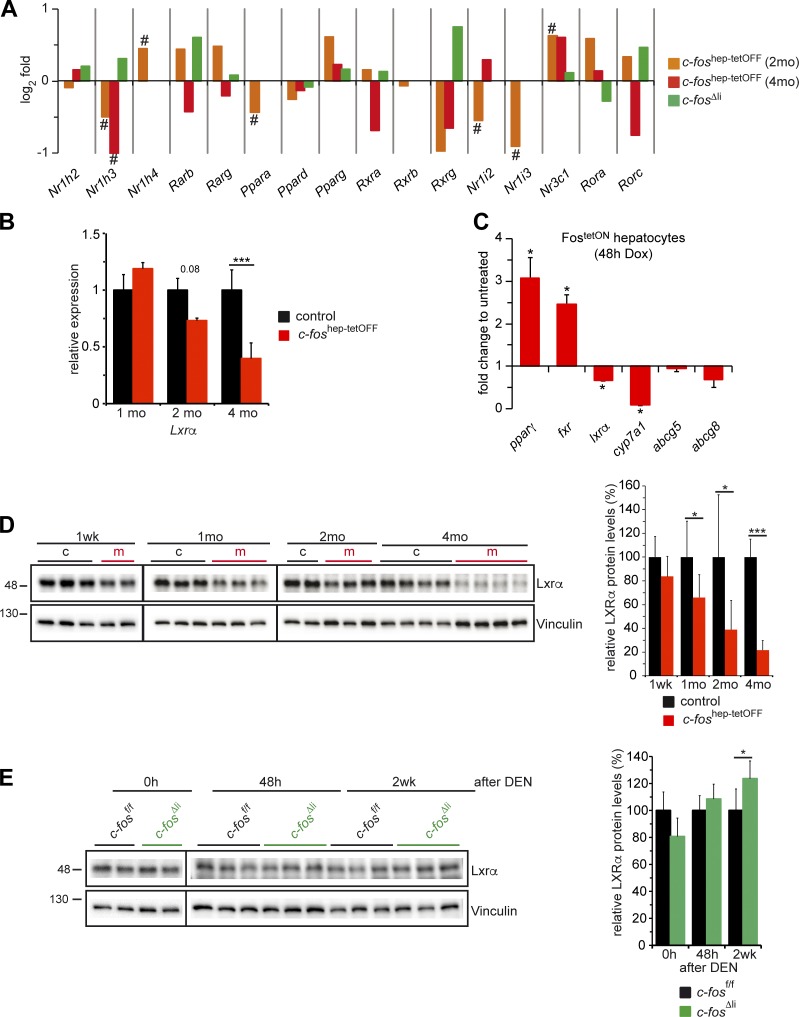
A notable down-regulation of hepatic LXRα mRNA was found in c-foshep-tetOFF mice (Fig. 7, A and B). This effect is likely cell autonomous to hepatocytes, as it was also observed in primary hepatocytes isolated from FostetON mice and induced to express c-Fos by addition of doxycycline to the culture medium (Fig. 7 C). Consistently, reduced mRNA of some LXRα target genes was also observed (Fig. 7 C). Although the mRNA of PPARγ, a direct c-Fos target gene (Hasenfuss et al., 2014b), was up-regulated in primary hepatocytes (Fig. 7 C) as well as in c-fos–expressing livers (Fig. 7 A and Fig. S4 H), mRNA expression of the BA receptor FXR (encoded by Nr1h4) was also found to be increased (Fig. 7 A and Fig. S4 H), indicating possible regulation by c-Fos or, as previously reported (Zhang et al., 2004), by PPARγ. No consistent changes in mRNA expression of other nuclear receptors, such as PPARα/δ or RXRs, were observed in RNA-seq or qRT-PCR analyses (Fig. 7 A and Fig. S4 H).
Figure 7.
Regulation of LXRα expression by c-Fos. (A) Relative expression of the indicated nuclear receptors, including Nr1h3 encoding for LXRα, by RNA-seq in c-foshep-tetOFF mice at 2 and 4 mo of ectopic c-Fos expression; (n = 2; 3/cohort) and in c-fosΔli mice 48 h after DEN (n = 3/cohort). Bar graphs represent mean fold changes (log2); # indicates significance after multiple testing corrections. (B) qRT-PCR analyses of Lxrα in total liver tissue of c-foshep-tetOFF and control mice at 1, 2, and 4 mo of c-Fos expression. Mean expression in controls set to 1; n = 5; 5; 7/5; 5; 7. (C) qRT-PCR analyses of FostetON primary hepatocytes (n = 4 mice/culture) induced to express c-Fos in vitro during 48 h. Expression in untreated cells set to 1. (D) Immunoblot analyses of total liver lysates from c-foshep-tetOFF at 1 wk and 1, 2, and 4 mo of c-Fos expression. (Right) Immunoblot quantification normalized to vinculin (n = 3; 7; 6; 12/2; 8; 9; 12). (E) LXRα immunoblot of liver lysates from c-fosΔli untreated (0 h), 48 h, and 2 wk after DEN injection. (Right) Immunoblot quantification normalized to vinculin (n = 2; 9; 5/2; 9; 5). Molecular mass is indicated in kilodaltons. (B–E) Bar graphs represent mean ± SD; *, P ≤ 0.05; ***, P ≤ 0.001 by Student’s t test.
Decreased Lxrα protein was already detectable at 1 mo in hepatic protein extracts from c-foshep-tetOFF mice (Fig. 7 D), consistent with reduced LXRα target genes and decreased LXRα pathway activity. LXRα protein expression was comparable to controls in c-fos–deficient livers under basal conditions. However, LXRα protein was significantly up-regulated in c-fos–deficient livers 2 wk after DEN-induced carcinogenesis (Fig. 7 E).
The Srebfs/Srebps transcription factors are also involved in cholesterol homeostasis and FA metabolism. GSEA revealed down-regulation of Srebf target genes (Horton et al., 2003) in c-fos–expressing livers, whereas these genes were induced in DEN-treated c-fos–deficient livers (Fig. S4 I). Although Srebf1 mRNA was reduced in c-fos–expressing livers at the 2-mo time point (Fig. 6 B), consistent with its reported transcriptional modulation by LXRα, increased cleaved (active) Srebf1 protein was detected, in particular at the late time point (Fig. S4 J). In the same samples, cleaved (active) Srebf2 protein was decreased, whereas Srebf2 mRNA appeared unchanged (Fig. S4, J and K). These results suggest that c-Fos–dependent alterations in LXR/RXR activity caused by decreased expression of LXRα are likely responsible for the observed metabolic changes.
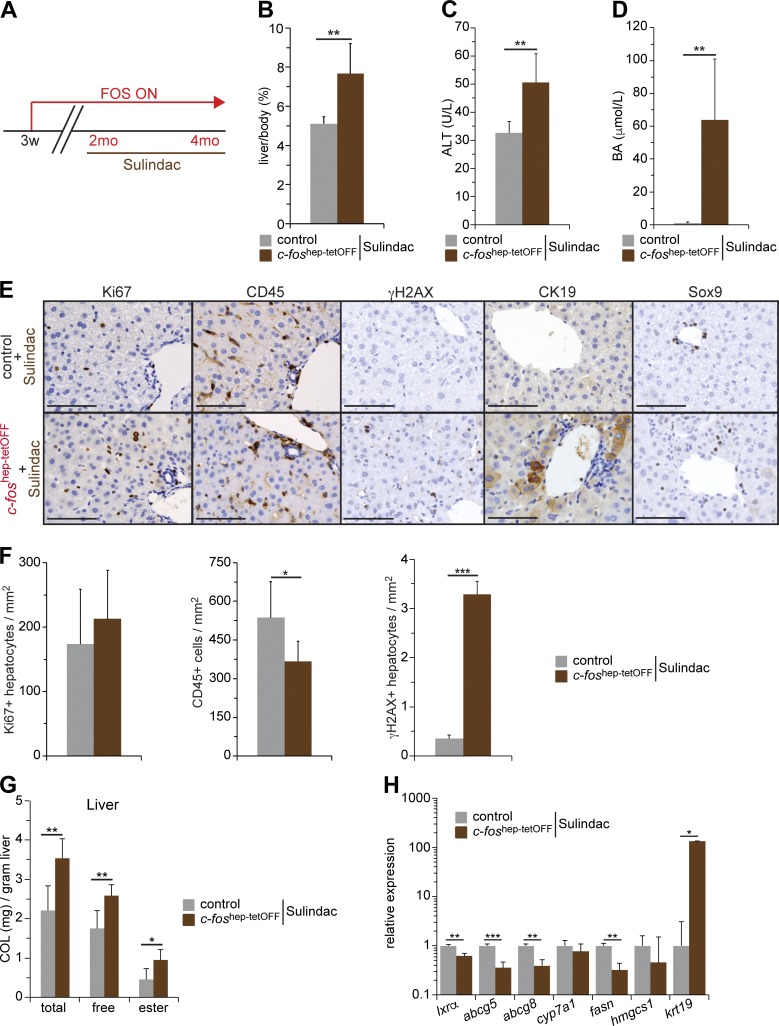
Inhibiting inflammation by sulindac treatment
The contribution of inflammation to the complex phenotypic changes induced by hepatic c-Fos expression was explored next. Control and c-foshep-tetOFF mice were treated with the COX1/2 inhibitor sulindac for 2 mo starting at 2 mo of c-Fos expression (Fig. 8 A). Sulindac had no effect on hepatic expression of total or ectopic c-Fos (Fig. S5 A). Increased liver size, serum ALT, and serum BA were still apparent in sulindac-treated mutant mice (Fig. 8, B and C). However, liver histology revealed normalized numbers of Ki67, reduced CD45-positive cells, and numerous hepatocytes positive for the dedifferentiation markers CK19 and Sox9 and γH2AX (Fig. 8, E and F). In addition, whereas serum cholesterol appeared to be normalized (Fig. S5 B), liver cholesterol species were still elevated in sulindac-treated mutant mice (Fig. 8 G). Consistently, mRNA expression of LXRα and most LXRα target genes was significantly reduced (Fig. 8 H).
Figure 8.
Phenotypic consequences of inhibiting inflammation in c-foshep-tetOFF mice. (A) Ectopic expression of c-Fos was allowed during 2 mo and then combined with sulindac for an additional 2 mo. (B–D) Liver/body weight (B), serum ALT (C), and serum BAs (D) in sulindac-treated c-foshep-tetOFF and controls (n = 6/6). (E) Representative IHC for Ki67, CD45, γH2AX, CK19, and Sox9 in sulindac-treated c-foshep-tetOFF and controls. Bars, 100 µm. (F) Quantification of CD45-positive cells and Ki67- and γH2AX-positive hepatocytes in liver sections of sulindac-treated c-foshep-tetOFF and controls (n = 6/6). (G) Liver cholesterol species in sulindac-treated c-foshep-tetOFF and controls (n = 6/6). (H) qRT-PCR analyses in total liver tissue from sulindac-treated c-foshep-tetOFF and controls (n = 6/6, mean expression in controls set to 1). Bar graphs represent mean ± SD; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.01 by Student’s t test.
These results suggest that hepatic inflammation contributes to sustaining hepatocyte proliferation upon c-Fos expression but has little contribution to c-Fos–dependent alterations in LXR/RXR activity, hepatic cholesterol accumulation, activation of DDR, and hepatocyte preneoplastic transformation.
Statin treatment partially reverses the c-Fos–dependent hepatic phenotype
The contribution of cholesterol and BA metabolism to the phenotype was next assessed using an inhibitor of the HMG-CoA reductase (HMGCR) atorvastatin (in short, statin). HMGCR catalyzes the conversion of HMG-CoA to mevalonate, an early step in cholesterol biosynthesis. Although cholesterol synthesis pathway activity was overall decreased (Figs. 5 A and 6 C), Hmgcr was up-regulated in the livers of c-foshep-tetOFF mice at 2 mo (Fig. S5 C), indicating that statin treatment might be effective. Additionally, besides decreasing circulating cholesterol and increasing intestinal excretion of cholesterol and BA (Parker et al., 2013), atorvastatin is reported to increase hepatic mRNA expression of Cyp7a1, Abcg5, and Abcg8 in wild-type mice (Fu et al., 2014). c-foshep-tetOFF mice were subjected to statin treatment for 2 wk, starting at 2 mo of c-Fos expression (Fig. S5 D). A smaller cohort of littermate controls was processed in parallel. No changes in total or ectopic c-Fos expression were observed in statin-treated mice (Fig. S5 E). Although statin treatment moderately affected serum ALT (Fig. 9 A), serum BAs were decreased to control levels in treated mutants (Fig. 9 B). Statin decreased circulating cholesterol and low-density lipoprotein (LDL) cholesterol in controls, as expected (Lawman et al., 2004; Parker et al., 2013), but did not further decrease these parameters in mutant mice (Fig. 9 C and Fig. S5 F). Consistent with a previous study (Parker et al., 2013), statin did not affect total hepatic cholesterol (Fig. 9 C). Importantly, statin increased hepatic mRNA expression of Abcg5, Abcg8, Cyp7a1, and Hmgcs1 in all treated groups, with the levels of Abcg5 and Abcg8 in statin-treated mutants reaching those of untreated controls (Fig. 9 D). Although hepatic Ki67- and CD45-positive cells were slightly diminished, no AFP-positive hepatocytes could be detected (Fig. S5 G), and significantly fewer hepatocytes expressed CK19 and Sox9 by qRT-PCR (Fig. 9 D) and IHC (Fig. 9 E) upon statin treatment. Furthermore, γH2AX-, p53-, and p21-positive hepatocytes were notably decreased in statin-treated c-foshep-tetOFF mice (Fig. 9, E and F; and Fig. S5 G).
These results suggest that the c-Fos–dependent DDR activation and preneoplastic transformation of hepatocytes are likely caused by decreased expression of the Abcg5/Abcg8 sterol transporter and LXR/RXR target genes and by the subsequent alterations in cholesterol and BA metabolism.
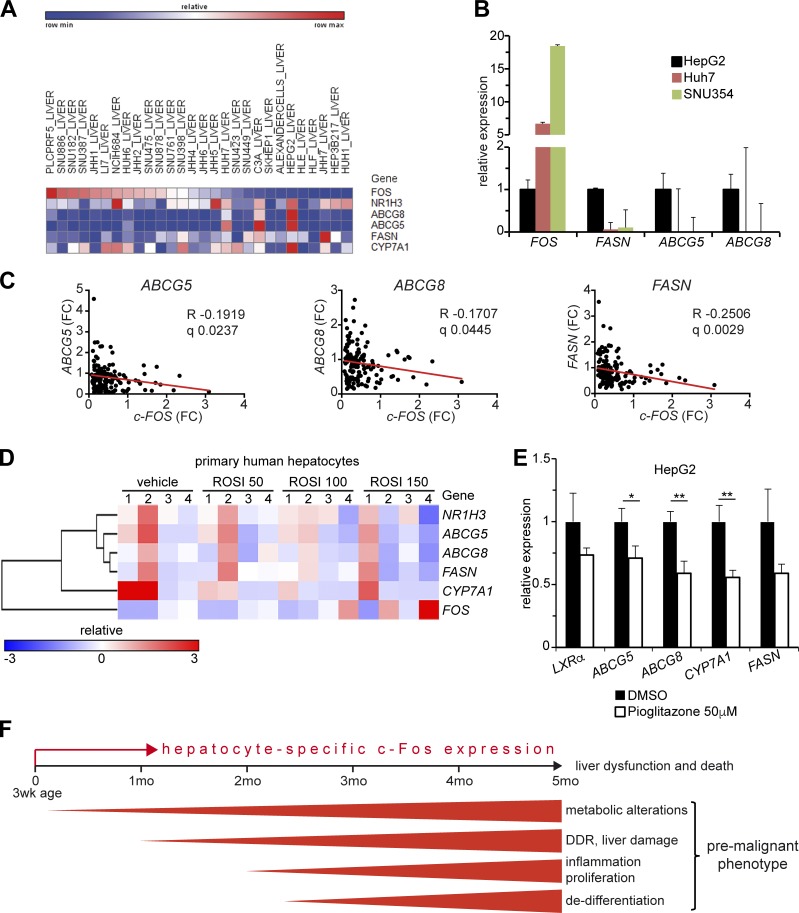
Pathway conservation in human liver cells
Using the Cancer Cell Line Encyclopedia of the Broad Institute, an inverse correlation of c-FOS expression with LXRα and its target genes was observed in human HCC cell lines (Fig. 10 A) and confirmed by qPCR analyses comparing the c-Fos low-expressing HepG2 to the high-expressing Huh7 and SNU354 HCC cell lines (Fig. 10 B). A significant negative correlation between c-FOS and LXRα target genes (FASN, ABCG8, ABCG5) was also computed in the gene expression dataset of the HB subtype of human HCC (Fig. 10 C). These correlations suggest that the conclusions drawn from analyzing GEMMs are likely relevant for human HCCs. We next attempted to define the connection between c-FOS and LXRα using human cells. PPARγ, a direct c-Fos/AP-1 target gene, is up-regulated in mouse hepatocytes expressing c-Fos (Hasenfuss et al., 2014b; Fig. 7 C and Fig. S4 H). In rat primary hepatocytes and mouse livers, activation of PPARα/γ was reported to decrease LXRα/RXR activity (Yoshikawa et al., 2003). This was also observed in primary human hepatocytes and the HepaRG hepatoma cell line (Rogue et al., 2011), in which treatment with PPARγ agonists notably decreased the expression of ABCG5, ABCG8, and CYP7A1 (Fig. 10 D). Decreased mRNA of ABCG5, ABCG8, and CYP7A1, and to a lesser extent, LXRα and FASN, was also observed in HepG2 cells treated with the PPARγ agonist pioglitazone (Fig. 10 E), whereas c-FOS, PPARγ, and FXR were unaffected and the PPARγ target gene FABP1 was up-regulated (Fig. S5 H). Increased PPARγ in c-Fos–expressing hepatocytes is therefore likely to be responsible for decreased LXRα expression and activity. Whether decreased LXRα expression and activity in liver cells occurs through direct binding of PPARγ to the LXRα promoter, as shown in human macrophages (Chinetti et al., 2001; Laffitte et al., 2001), or rather, as proposed by Yoshikawa et al. (2003), through competitive reduction of LXR/RXR dimers, remains to be addressed.
Figure 10.
Liver carcinogenesis by c-Fos in human HCC and GEMMs. (A) Correlation analyses of c-FOS, NR1H3 encoding for LXRα, and LXRα target genes in human HCC cell lines using the Broad Institute Cancer Cell Line Encyclopedia. (B) Relative mRNA expression of c-FOS and LXRα target genes in the indicated human HCC cell lines compared with HepG2. Bar graphs represent mean ± SD. (C) Correlation analyses of c-FOS and LXRα target genes in the HCC-HB subtype. (D) Correlation analyses of NR1H3 encoding for LXRα, LXRα target genes, and c-FOS in human primary hepatocytes from four adult donors treated with the PPARγ agonist rosiglitazone (ROSI) or vehicle (DMSO) for 24 h. Original data were deposited by Rogue et al. (2011) in the GEO databank under accession no. GSE27183. (E) Relative mRNA expression of LXRα and its target genes in HepG2 cells 24 h after treatment with the PPARγ agonist pioglitazone or vehicle (DMSO). Bar graphs represent mean ± SD; n = 3; DMSO-treated cells set to 1. *, P ≤ 0.05; **, P ≤ 0.01; by Student’s t test. (F) Schematic view of c-Fos functions in liver pathology: hepatocyte-specific c-Fos expression induced in 3-wk-old mice causes metabolic changes as early as 1 mo later, manifested by inhibition of Lxrα expression and LXR/RXR pathway activity, leading to liver damage visible by localized necrotic foci. Over time, these changes, together with inflammation, induce hepatocyte proliferation, dedifferentiation, and a premalignant phenotype. Within 4 mo of c-Fos expression, hepatic cholesterol, oxysterols, and BAs accumulate, which are likely responsible together with liver dysfunction for the death of the mutant mice. When c-Fos expression is combined in adults with a single dose of DEN treatment (GOF), HCCs develop 7 mo later; in contrast, the absence of c-Fos (LOF) prevents DEN-induced HCC development when the mutagen is applied at 2 wk of age.
Discussion
GEMMs are essential for advancing the molecular understanding of the basic mechanisms of liver diseases (Bakiri and Wagner, 2013). Here we show for the first time that hepatocyte-specific expression of c-Fos leads to liver inflammation, hepatocyte proliferation, DDR activation, and premalignant transformation, which depends on sustained c-Fos expression. When c-Fos expression is experimentally switched off, a complete regression of the phenotype is observed. However, when combined with a potent mutagen, such as DEN, to provide a second pro-oncogenic signal, c-Fos promotes HCC development. Importantly, carcinogenesis experiments using loss-of-function GEMMs show that c-Fos is not just sufficient, but essential for HCC development.
Mechanistically, hepatocyte-specific c-Fos expression leads to hepatic cholesterol accumulation, likely as a result of reduced expression and activity of the nuclear receptor LXRα, which is essential for cholesterol homeostasis (Peet et al., 1998; Calkin and Tontonoz, 2012; Zhang et al., 2012). Accumulation of cholesterol and toxic cholesterol derivatives (such as oxysterols and primary BAs), DDR activation, and subsequent inflammation and hepatocyte proliferation are the initiators of the premalignant phenotype in c-foshep-tetOFF mice (Fig. 10 F).
A significant correlation with several human HCC signatures characteristic of poorly differentiated, aggressive tumors, such as the S1 and G3 and the proliferative classes (Boyault et al., 2007; Chiang et al., 2008; Hoshida et al., 2009, 2010; van Malenstein et al., 2011; Pinyol et al., 2014), was identified using RNA-seq profiles of c-Fos–expressing murine livers. The correlation with the specific HB subtype of human HCCs with poor prognosis identified by Lee et al. (2006) is particularly compelling. Network-based analyses revealed high Fos/AP-1 activity in this subtype, and AP-1 was proposed as the main oncogenic driver (Lee et al., 2006). Our data experimentally support this hypothesis, as we observed not only a strong positive correlation with the HB subtype in livers from the c-foshep-tetOFF mice, but also a significant inverse correlation in the tumor-resistant, hepatocyte-specific c-Fos knockout livers. This inverse correlation pattern of Fos-expressing and Fos-deficient livers was also observed with the HCC classification signatures and signatures characteristic of dedifferentiated/transformed or healthy livers (Hsiao et al., 2001; Cairo et al., 2008; Holczbauer et al., 2013). Besides the molecular signatures, necrosis, inflammation, proliferation, and dedifferentiation were histologically observed, all consistent with premalignant transformation. c-foshep-tetOFF mice did not develop HCC, most likely because of their reduced lifespan. However, when c-Fos expression was restricted to the initiation stage of DEN-induced carcinogenesis, liver tumors efficiently developed, indicating that c-Fos is a potent inducer of hepatocyte transformation.
We previously reported that c-Jun promotes HCC by repressing c-Fos expression in hepatocytes, thus modulating a SIRT6/Survivin axis and initiating cancer cell survival (Min et al., 2012). In the current study, ectopic expression of c-Fos in c-Jun–proficient livers leads to hepatocyte proliferation, DDR activation, and premalignant transformation, whereas c-Fos inactivation conversely decreases DEN-induced hepatocyte proliferation and carcinogenesis. These new findings thus indicate that the functions and pathways controlled by c-Fos in liver carcinogenesis are multiple and stage, context, and c-Jun/AP-1 dependent.
c-Fos affects the LXR/RXR pathway controlling hepatic cholesterol. 1 mo of c-Fos expression already leads to reduced pathway activity, with a subsequent increase in total and free cholesterols and oxysterols. Oxysterols are LXRα natural ligands; thus, decreased LXRα activity is not caused by reduced ligand availability, but rather reduced LXRα mRNA and protein. Increased PPARγ in c-Fos–expressing hepatocytes likely contributes to decreased LXRα–RXR pathway activity, possibly through direct regulation of LXRα expression. Because RXR is required as an obligatory heterodimerization partner, competitive reduction of LXR/RXR dimers by PPARγ and FXR might also occur (Yoshikawa et al., 2003; Chan and Wells, 2009). Although systemic PPARγ activation is considered rather beneficial in HCC (Yu et al., 2010; Wu et al., 2012), ectopic expression of PPARγ in wild-type livers by adenoviral delivery enhances hepatocyte proliferation, increases liver size, and is deleterious in the context of Pten and Akt inactivation (Panasyuk et al., 2012). Defining the contribution of nuclear receptors such as PPARγ, RXR, and FXR in HCC downstream or independently of AP-1 certainly merits further investigation.
The c-Fos–dependent metabolic alterations are similar to those observed in Lxrα knockout mice (Peet et al., 1998; Zhang and Friedman, 2012). Lxrα mutants fed a high-cholesterol diet displayed further increased hepatic cholesterol and liver dysfunction, although no malignant transformation was reported. This is surprising because an anti-oncogenic function for LXRα in cancer cell lines has been documented (Mehrotra et al., 2011; Lo Sasso et al., 2013). Consistent with our observations in c-Fos–expressing livers, LXRα promoted the differentiation of human liver progenitor cells (Chen et al., 2014). In a subset of human HCC tissue samples in which oxysterols and cholesterol were elevated, LXRα mRNA and LXR target gene expression were found to be significantly reduced (Lu et al., 2013). This uncoupling between LXR activity and oxysterol accumulation, which is also apparent in the livers of c-foshep-tetOFF mice, was attributed to impaired sterol catabolism and efflux pathways, a metabolic adaptation of tumor cells to oxysterol-induced cytotoxicity (Lu et al., 2013). Consistently, we computed an inverse correlation between c-Fos and LXRα target genes in human HCC cell lines and in the HCC-HB subtype, indicating that our findings in mouse models are likely relevant to human HCC.
Cholesterol is essential for proliferating cells, but an overload of intracellular cholesterol, in diseases such as atherosclerosis and Niemann–Pick type C, is cytotoxic and affects plasma membrane, ER, lysosomes, and mitochondria (Tabas, 2002; Feng et al., 2003; Ikonen, 2006; Ibrahim et al., 2011). Cholesterol derivatives such as oxysterols and BAs are hepatotoxic, genotoxic, pro-oxidative, proinflammatory, and carcinogenic (Perez and Briz, 2009; Jusakul et al., 2011; Wang et al., 2013). Oxysterols and primary BAs are increased in c-Fos–expressing livers, whereas BA conjugation, which results in less toxic BA species (Perez and Briz, 2009), is decreased, and mRNA expression of the two enzymes catalyzing amino acid conjugation to BA is reduced. It is thus tempting to speculate that c-Fos/AP-1 might modulate the expression of enzymes implicated in BA conjugation and/or detoxification, similar to the other AP-1 protein Fra-1 (Hasenfuss et al., 2014a).
Accumulation of oxysterol and BAs, DDR, and liver damage combined with hepatic inflammation was evident in livers of c-foshep-tetOFF mice. Short-term treatment with atorvastatin largely prevented the deleterious consequences of c-Fos expression, in particular DDR activation and the early signs of preneoplastic transformation. Although long-term statin treatment of c-fos mutant mice might provide additional insights, these data strongly support our hypothesis, that deregulation of cholesterol metabolism caused by the LXRα–Abcg5/8 sterol transporter axis is crucial in the adverse sequence of events after increased c-Fos expression in hepatocytes. Preclinical and observational studies indicate that statins, widely used to treat hypercholesterolemia, might reduce HCC risk in humans (Björkhem-Bergman et al., 2014; Singh et al., 2014; Kim et al., 2017) and obese mice (Shimizu et al., 2011).
Inflammation is an important pathophysiological mechanism for HCC (Fattovich et al., 2004). Expression of c-Fos in hepatocytes causes liver inflammation, with granulocytes forming the bulk of recruited immune cells. The main trigger is likely the chronic damage to hepatocytes caused by accumulation of cholesterol, oxysterols, and primary BAs. Consistently, statin treatment decreased inflammatory infiltrates in the liver of c-foshep-tetOFF mice, whereas reducing hepatic inflammation using sulindac only modestly affected circulating cholesterol and LXR target gene expression and had little impact on DDR. In hepatocytes, inflammatory stress was reported to affect PPAR/LXR-controlled intracellular cholesterol homeostasis (Chen et al., 2012). We have documented that specific AP-1 dimers control the transcription of the nuclear receptor PPARγ during nutrient overload (Hasenfuss et al., 2014b). Here we show that c-Fos/AP-1 links modulation of LXRα nuclear receptor activity to inflammatory stress (Fig. 10 F). Under stress conditions, e.g., liver regeneration, these metabolic alterations might be necessary for hepatocyte proliferation. However, chronic liver damage, inflammation, and premalignant hepatocyte transformation predispose to HCC development. Interrupting this vicious cycle of chronic inflammation, metabolic alterations, and liver damage may constitute a novel targetable pathway for HCC prevention and treatment.
Materials and methods
Animal procedures
The Col1a1::TetOP-c-fos, LAP-tTA, c-fos floxed, and Alfp-Cre alleles are described elsewhere (Kistner et al., 1996; Kellendonk et al., 2000; Fleischmann et al., 2003; Briso et al., 2013). c-foshep-tetOFF and c-fosΔli mice were maintained on a C57BL/6 and mixed (C57BL/6 × 129sv) background, respectively, and housed in a specific pathogen–free facility accredited by the American Association for Laboratory Animal Care, with food and water ad libitum. Doxycycline (1 g/liter) was supplied in the drinking water (Sigma-Aldrich) or in food pellets (Research Diet). Mice were treated ad libitum with sulindac (Sigma-Aldrich) supplied in the drinking water (180 mg/L). Atorvastatin (TCI Chemicals) was dissolved at a final concentration of 20 mg/ml in cyclodextrin (5 mg/ml; Abmole Bioscience), and mice received daily oral gavage for 2 wk (100 mg/kg/d). 2-wk-old pups or 8-wk-old mice were injected intraperitoneally with 25 or 100 mg/kg DEN (Sigma-Aldrich), respectively. Mice were sacrificed 8 mo after DEN injection to monitor HCC development or earlier to analyze the acute effects of DEN. BrdU (Sigma-Aldrich) was supplied in sucrose-containing (1%) drinking water at a concentration of 1 mg/ml. Liver tumor detection by micro–computed tomography (micro-CT) was performed on anesthetized mice after intravenous injection of the iodinated contrast agent Iopamiro 300 (Bracco) using a small-animal micro-CT system (eXplore Vista PET/CT; GE Healthcare). In all experiments, sex-matched littermates were used as controls. All animal experiments were performed in accordance with institutional, national, and European guidelines for animals used in biomedical research and approved by the Spanish National Cancer Research Centre (CNIO) Institutional Animal Care and Use Committee and the CNIO–Instituto de Salud Carlos III Ethics Committee for Research and Animal Welfare.
Blood analyses
Blood was collected by submandibular vein or cardiac (experimental endpoint) puncture. Complete blood count was performed using a hematology analyzer Abacus JunVet (Diatron), and serum parameters were measured using a VetScan chemistry analyzer (Abaxis) or a Reflovet Plus blood chemistry analyzer (Scil Diagnostics) according to the manufacturer’s instructions. LDL cholesterol was calculated from total cholesterol, high-density lipoprotein cholesterol, and TGs using the Friedewald formula.
Detection of DNA adducts
10 µg liver DNA was treated with 0.4 M NaOH/10 mM EDTA for 10 min at 99°C. Afterward, DNA was dot-blotted on nylon membrane with 0.4 M NaOH. Membrane was rinsed in 2× SSC buffer and air-dried. Membrane was blocked with 5% skim milk powder in 1× TBS and 0.5% Tween. For detection of DNA adducts, membrane was incubated overnight with mAb to O6-ethyl-2-deoxyguanosine (EM 21; Squarix). The blots were incubated with anti–mouse secondary HRP-coupled antibodies (GE Healthcare) and developed using Luminata Western HRP Substrate (EMD Millipore) and Amersham ECL Hyperfilms (GE Healthcare). Loading control was performed using methylene blue staining.
Histology and immunohistochemistry
Tissue was fixed in 4% PFA and embedded in either paraffin or OCT. Hematoxylin and eosin (H&E) staining was performed according to standard procedures. For paraffin-embedded sections, antigen retrieval was performed using citrate buffer, pH 6.0, in a pressure cooker. PBS supplemented with 0.1% Triton X-100, 0.05% Tween, and 1.5% BSA and 10% serum compatible with the secondary antibody was used for antibody dilution. The following antibodies were used for IHC: Flag (Cell Signaling Technology), AFP (R&D Systems), CK19 (Developmental Studies Hybridoma Bank), MPO (Dako), Sox9 (Abcam), Phospho-Ser139-Histone H2AX (EMD Millipore), p53 (CNIO mAb unit), p21 (CNIO mAb unit), β-catenin (Cell Signaling Technology), CD45 (BD), Ki67 (Master Diagnostic), BrdU (AbD Serotec), GS (Sigma-Aldrich), and cleaved caspase 3 (Cell Signaling Technology) together with matching secondary antibodies from the Vectastain Elite ABC kits (Vector Laboratories). Counterstaining was performed using Carazzi’s hematoxylin (Panreac AppliChem). Quantification was performed on digital scans using Panoramic Viewer software (3DHISTECH).
Flow cytometry
Immune cells were isolated from the liver using two-step collagenase perfusion with liver perfusion medium (Gibco) and liver digest medium (Gibco) and filtered through a 100-µm cell strainer. Hepatocytes were removed by centrifugation at 50 g. After lysing red blood cells (Sigma-Aldrich), cells were incubated with FC-Block (BD) and the following antibodies against immune cell surface markers: F4/80-AF647, CD45-PerCP, and CD45R-APC-Cy7 (BioLegend); CD3-AF700 (eBioscience); and CD4-PE-Cy7, CD8-PE-Cy5, NK-1.1-PE, and Ly-6G/Ly-6C-PerCP-Cy5.5 (BD). Cells were fixed in 2% PFA. Data were acquired on a BD LSRII Fortessa and analyzed using FlowJo 9.5.3. Live cells were gated for CD45+, and at least 10,000 individual cells were collected. The different immune cell populations in the CD45+ population were gated as follows: T cells, CD3+, NK1.1−; NK cells, CD3−, NK1.1+; NKT cells, CD3+, NK1.1+; CD4+ T cells, CD4+, CD8−; CD8+ T cells, CD4−, CD8+; macrophages and monocytes, F4/80+; B cells, CD45R+ (B220+); and granulocytes, Gr1+ (Ly-6G/Ly-6C).
Protein isolation and Western blot
Tissue was disrupted using a Precellys device (Bertin Technologies) in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, and 0.1% SDS). Protein lysates were quantified using BCA protein assay reagent (Thermo Fisher Scientific). For Western blot analysis, 50 µg protein per sample was loaded. Membranes were blocked with 5% BSA or nonfat dry milk in TBS-T. The following primary antibodies were used: c-Fos (Santa Cruz Biotechnology, Inc.), p-AKT (Cell Signaling Technology), AKT (Cell Signaling Technology), p-Stat3 (Cell Signaling Technology), Stat3 (Cell Signaling Technology), p-JNK (Cell Signaling Technology), JNK (Cell Signaling Technology), Vinculin (Sigma-Aldrich), LXRa (R&D Systems), Srebf1 (Abcam), Srebf2 (Abcam), and Gapdh (Sigma-Aldrich). Blots were incubated with the appropriate secondary HRP-coupled antibody (GE Healthcare and Santa Cruz Biotechnology, Inc.) and developed using Luminata Western HRP Substrate (EMD Millipore) and Amersham ECL Hyperfilms (GE Healthcare) or a ChemiDoc XRS+ imaging system with Image Lab image acquisition and analysis software (Bio-Rad).
Cell culture
Primary mouse hepatocytes were isolated from adult mice and cultivated as previously described (Hasenfuss et al., 2014a), and doxycycline (Sigma-Aldrich) was dissolved in water and added to reach a final concentration of 1 µg/ml. Human HepG2, Huh7, and SNU354 cellular carcinoma cell lines were maintained in DMEM supplemented with 10% FBS. A final concentration of 50 µM of the PPARγ agonist pioglitazone (Sigma-Aldrich; dissolved in DMSO) was used to treat HepG2 cells during 24 h without obvious effect on cell viability.
RNA isolation and RT-PCR
Total RNA was isolated using TRI Reagent (Sigma-Aldrich), complementary DNA was synthesized using Ready-To-Go-You-Prime-First-Strand Beads (GE Healthcare), and RT-qPCR used GoTaq RT-qPCR Master Mix (Promega) and Eppendorf fluorescence thermocyclers, all according to manufacturers’ instructions. The 2ΔΔCT method was used to quantify amplified fragments. Expression levels were normalized using at least one housekeeping gene (gapdh and actin). Primer sequences are listed in Table S2.
RNA-seq and data analysis
Total RNA was isolated using TRI Reagent (Sigma-Aldrich), and RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies). Samples of RNA integrity score >8 were used for RNA-seq. RNA processing was performed as described in Illumina’s TruSeq RNA Sample Preparation V2 (Part #15026495 Rev B, February 2012). The resulting purified cDNA library was applied to an Illumina flow cell for cluster generation (TruSeq cluster generation kit v5) and sequenced on the Genome Analyzer IIx with SBS TruSeq v5 reagents according to the manufacturer’s protocols. RNA-seq read quality was checked with FastQC (Andrews, 2010). Fastq files (Cock et al., 2010) were randomly down-sampled to generate datasets with similar numbers of reads in all the samples.
The 40-nt single-end reads were aligned to the mouse genome (GRCm38/mm10) with TopHat-2.0.4 (Trapnell et al., 2012), using Bowtie 0.12.7 (Langmead et al., 2009) and Samtools 0.1.16 (Li et al., 2009), allowing two mismatches and five multihits. Transcript assembly and estimation of abundance were calculated with Cufflinks 1.3.0, using the mouse genome annotation dataset GRCm38/mm10 from the UCSC Genome Browser (Karolchik et al., 2014). Differential expression between genes in both conditions was calculated with Cuffdiff (Trapnell et al., 2012), and those genes with FPKM expression values lower than 0.05 in both conditions were excluded. GSEA was performed to test for relevant pathways in the data (Subramanian et al., 2005). Data are deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE81079.
Metabolite measurements
Liver samples were homogenized using chloroform/isopropanol/NP-40 (7:11:0.1). Chloroform was removed from the supernatant by vacuum centrifugation (GeneVac). Colorimetric cholesterol was measured using a BioVision kit (K603-100). NMR was performed at the Spectroscopy and Nuclear Magnetic Resonance Spectroscopy Unit. Oxysterols and BAs were measured by Biocrates Life Sciences AG (Innsbruck, Austria) using mass spectrometry. The sum of all oxysterols species resolved by liquid chromatography/tandem mass spectrometry (LC-MS/MS) as well as individual species was analyzed.
For determination of liver metabolite concentrations by NMR (Beckonert et al., 2007), dual-phase fractionation using extraction in methanol/water/chloroform (1:1:1) was used, according to a modified Salomon protocol for the simultaneous extraction of polar and apolar metabolites (Tyagi et al., 1996). In brief, 150–200 mg of tissue samples were disrupted in a Precellys 24 device using 3 ml of ice-cold methanol and transferred to centrifuge glass tubes (30-ml borosilicate glass tubes; Kimble-Chase), where equal volumes of methanol and water were sequentially added and vigorously mixed. Fractionation was accelerated by centrifugation at 5,000 g for 15 min at 4°C, after which the polar and apolar fractions were carefully separated and the latter dried by vacuum centrifugation (GeneVac). NMR samples of the apolar fractions were prepared by dissolving the dry extract in 500 µl of DCCl3 plus 300 µl of 20 mM deuterated EDTA, pH 6.0, in D2O/deuterated methanol (1:2) and transferred to 5-mm NMR tubes for measurement. High-resolution NMR spectra were registered on a Bruker Avance spectrometer operating at 16.4 T (proton Larmor frequency of 700 MHz) at 293 K using a TXI probe with pulsed field gradient capabilities and equipped with a BACS120 sample changer. 1D proton NMR spectra (1dH) of the apolar fraction of the liver lysates were recorded using a NOESY pulse sequence (noesygppr1d in Bruker nomenclature) using pulse field gradients during the mixing time of 10 ms and a 2-s recovery delay between consecutive scans. 256 scans were accumulated using a spectral width of 20 ppm centered at 6.37 ppm and with acquisition time of 1.6 s, resulting in an acquisition time of 16.5 min per sample. In addition, 2D sensitivity-enhanced 1H-13C heteronuclear single quantum coherence spectra (2dHC, hsqcetgpsisp2 in Bruker nomenclature; Schleucher et al., 1994) were recorded for each apolar extract sample (13C natural abundance) with 13C decoupling during the 60-ms acquisition time, using an indirect (13C) spectral width of 40 ppm centered at 68 ppm, with 35 indirect increments (complex points), 256 scans per increment, and 1.2-s recovery delay, resulting in a total acquisition time of 6.5 h per 2dHC spectrum. All 1dH free induction decays were processed with exponential multiplication (0.5 Hz line-broadening) before Fourier transformation and followed by baseline correction using Topspin2.1 (Bruker). The 2dHC spectra were processed with NMRPipe (Delaglio et al., 1995) using squared cosine window functions in both dimensions, and were visualized and analyzed using nmrViewJ (Johnson, 2004). 1H/13C chemical shifts were referenced to internal deuterated methanol (3.36/49.6 ppm). Metabolites in 1dH spectra were quantified from their signal/s integral/s, using integration regions of variable size that were manually defined to include all metabolite signals using AMIX3.8 software (Bruker). The relative levels of cholesterol and lipids were obtained from the maximum intensity of the most intense and best-resolved correlations (or combinations thereof) of the different chemical moieties for each metabolite (Vinaixa et al., 2010), as detected in the 2dHC spectra. For example, for assessing total cholesterol, the mean of four NMR signals corresponding to five methyl groups (C18, C19, C21, and C26 and C27, which overlap) was used. Reported metabolite concentrations are normalized to total extracted metabolites, as calculated from the relative sum of all metabolite signals in 1dH spectra, and thus accounting for metabolite level differences resulting from different mass of liver tissue and possible differences in extraction efficiency as described in Petruzzelli et al. (2014).
To extract metabolites from liver tissue for mass spectrometry, samples were homogenized using Precellys with 100% ethanol containing 0.01% butylated hydroxytoluene. For measuring metabolite concentrations, samples were centrifuged, and the supernatant was used for analysis. Biocrates Bile Acids kit validated for mouse plasma was used for BA quantification. A highly selective reversed-phase LC-MS/MS analysis method in negative ion multiple reaction monitoring (MRM) detection mode was applied to determine the concentrations of BAs. Samples were extracted via dried filter spot technique in 96-well plate format. Sample extracts were measured by electrospray ionization LC-MS/MS (SCIEX, Thermo Fisher Scientific, or Waters Corp.). For highly accurate quantification, seven-point external calibration curves and 10 stable isotope–labeled internal standards were applied. Data of BAs were quantified using the appropriate MS software (SCIEX, Analyst; Thermo Fisher Scientific, Xcalibur; and Waters, MassLynx), and the results were imported into Biocrates MetIDQ software for further analysis. Oxysterols, both free and esterified, were extracted from samples with methanol using a Biocrates kit filter plate. The plate was loaded with an internal standard mixture beforehand. The extract was then subjected to alkaline hydrolysis to release oxysterols from their respective esters. After neutralization, the metabolites were determined by ultra-high-performance LC-MS/MS with MRM in positive mode using a SCIEX API Qtrap 5500 mass spectrometer with electrospray ionization. The assay has been validated according to European Medicines Agency guidelines.
Statistics
Data are expressed as mean ± SD. Statistical significance was determined using two-tailed Student’s t test for all bar graphs and two-way ANOVA for all plots, except for Kaplan–Meier plots where Mantel–Cox log-rank was used. For all experiments, values of P < 0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows additional phenotypic consequences of hepatocyte-specific c-Fos expression. Fig. S2 shows DEN experiments using c-Fos loss- and gain-of-function mutant mice. Fig. S3 includes additional early DEN-induced events in c-Fos mutants and illustrates the reversibility of the c-Fos–induced phenotype. Fig. S4 shows additional liver metabolic pathways affected by c-Fos. Fig. S5 depicts data supporting the c-Fos–LXRα connection in mouse and human samples. Table S1 lists the full and abbreviated name of BAs and oxysterols species determined by mass spectrometry. Table S2 lists the specific primers used in this study.
Supplementary Material
Acknowledgments
We thank Drs. N. Djouder, M. Petruzzelli, R. Ricci, F.X Real, K.D. Bissig, and members of the Wagner laboratory for critical reading of the manuscript and valuable suggestions; Dr. H. Schönthaler for help with the bioinformatics analysis; V. Bermeo for technical help; and G. Luque, S. Leceta, and G. Medrano for assisting with mouse experiments.
The E.F. Wagner laboratory is supported by grants from the Spanish Ministry of Economy, Industry, and Competitiveness (BFU2012-40230 and SAF2015-70857, cofunded by the European Regional Development Fund), a European Research Council–advanced grant (ERC-FCK/2008/37), and Worldwide Cancer Research (13-0216). R. Hamacher was supported by the Deutsche Forschungsgemeinschaft (HA 6068/1-1), M.K. Thomsen by AUFF Nova, and S.C. Hasenfuss by a Boehringer Ingelheim Fonds PhD fellowship.
The authors declare no competing financial interests.
Author contributions: L. Bakiri and R. Hamacher designed and performed experiments, analyzed data, prepared figures, and wrote the manuscript. O. Graña analyzed RNA-seq and public microarray data, A. Guío-Carrión provided expert technical assistance, R. Campos-Olivas acquired and analyzed NMR data, L. Martinez analyzed flow cytometry data, M.K. Thomsen performed experiments with human cell lines, S.C. Hasenfuss performed experiments with primary hepatocytes and data mining, and H.P. Dienes performed pathological analysis on tissue sections. E.F. Wagner directed the study, approved the data, and wrote and edited the paper. All authors read and commented on the manuscript.
Footnotes
Abbreviations used:
- AFP
- α-fetoprotein
- ALT
- alanine aminotransferase
- BA
- bile acid
- DDR
- DNA damage response
- DEN
- diethylnitrosamine
- FA
- fatty acid
- GEMM
- genetically engineered mouse model
- GS
- glutamine synthetase
- GSEA
- gene set enrichment analysis
- HB
- hepatoblast
- HCC
- hepatocellular carcinoma
- IHC
- immunohistochemical staining
- NMR
- nuclear magnetic resonance
- qRT-PCR
- quantitative RT-PCR
- RXR
- retinoid X receptor
- TG
- triglyceride
References
- Andrews S.2010. FastQC a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed March 1, 2013)
- Bakiri L., and Wagner E.F.. 2013. Mouse models for liver cancer. Mol. Oncol. 7:206–223. 10.1016/j.molonc.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O., Keun H.C., Ebbels T.M., Bundy J., Holmes E., Lindon J.C., and Nicholson J.K.. 2007. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2:2692–2703. 10.1038/nprot.2007.376 [DOI] [PubMed] [Google Scholar]
- Björkhem-Bergman L., Backheden M., and Söderberg Löfdal K.. 2014. Statin treatment reduces the risk of hepatocellular carcinoma but not colon cancer—results from a nationwide case-control study in Sweden. Pharmacoepidemiol. Drug Saf. 23:1101–1106. 10.1002/pds.3685 [DOI] [PubMed] [Google Scholar]
- Boyault S., Rickman D.S., de Reyniès A., Balabaud C., Rebouissou S., Jeannot E., Hérault A., Saric J., Belghiti J., Franco D., et al. . 2007. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 45:42–52. 10.1002/hep.21467 [DOI] [PubMed] [Google Scholar]
- Briso E.M., Guinea-Viniegra J., Bakiri L., Rogon Z., Petzelbauer P., Eils R., Wolf R., Rincón M., Angel P., and Wagner E.F.. 2013. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes Dev. 27:1959–1973. 10.1101/gad.223339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo S., Armengol C., De Reyniès A., Wei Y., Thomas E., Renard C.A., Goga A., Balakrishnan A., Semeraro M., Gresh L., et al. . 2008. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 14:471–484. 10.1016/j.ccr.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Calkin A.C., and Tontonoz P.. 2012. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13:213–224. 10.1038/nrm3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan L.S., and Wells R.A.. 2009. Cross-talk between PPARs and the partners of RXR: A molecular perspective. PPAR Res. 2009:925309 10.1155/2009/925309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.T., Pernelle K., Tsai Y.H., Wu Y.H., Hsieh J.Y., Liao K.H., Guguen-Guillouzo C., and Wang H.W.. 2014. Liver X receptor α (LXRα/NR1H3) regulates differentiation of hepatocyte-like cells via reciprocal regulation of HNF4α. J. Hepatol. 61:1276–1286. 10.1016/j.jhep.2014.07.025 [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen Y., Zhao L., Chen Y., Mei M., Li Q., Huang A., Varghese Z., Moorhead J.F., and Ruan X.Z.. 2012. Inflammatory stress exacerbates hepatic cholesterol accumulation via disrupting cellular cholesterol export. J. Gastroenterol. Hepatol. 27:974–984. 10.1111/j.1440-1746.2011.06986.x [DOI] [PubMed] [Google Scholar]
- Chiang D.Y., Villanueva A., Hoshida Y., Peix J., Newell P., Minguez B., LeBlanc A.C., Donovan D.J., Thung S.N., Solé M., et al. . 2008. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 68:6779–6788. 10.1158/0008-5472.CAN-08-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G., Lestavel S., Bocher V., Remaley A.T., Neve B., Torra I.P., Teissier E., Minnich A., Jaye M., Duverger N., et al. . 2001. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 7:53–58. 10.1038/83348 [DOI] [PubMed] [Google Scholar]
- Cock P.J., Fields C.J., Goto N., Heuer M.L., and Rice P.M.. 2010. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 38:1767–1771. 10.1093/nar/gkp1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirolamo C., Modica S., Vacca M., Di Tullio G., Morgano A., D’Orazio A., Kannisto K., Parini P., and Moschetta A.. 2015. Prevention of spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice by intestinal-specific farnesoid X receptor reactivation. Hepatology. 61:161–170. 10.1002/hep.27274 [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., and Bax A.. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Eferl R., and Wagner E.F.. 2003. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer. 3:859–868. 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- Eferl R., Ricci R., Kenner L., Zenz R., David J.P., Rath M., and Wagner E.F.. 2003. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 112:181–192. 10.1016/S0092-8674(03)00042-4 [DOI] [PubMed] [Google Scholar]
- Fan Q., He M., Deng X., Wu W.K., Zhao L., Tang J., Wen G., Sun X., and Liu Y.. 2013. Derepression of c-Fos caused by microRNA-139 down-regulation contributes to the metastasis of human hepatocellular carcinoma. Cell Biochem. Funct. 31:319–324. 10.1002/cbf.2902 [DOI] [PubMed] [Google Scholar]
- Farazi P.A., and DePinho R.A.. 2006. Hepatocellular carcinoma pathogenesis: From genes to environment. Nat. Rev. Cancer. 6:674–687. 10.1038/nrc1934 [DOI] [PubMed] [Google Scholar]
- Fattovich G., Stroffolini T., Zagni I., and Donato F.. 2004. Hepatocellular carcinoma in cirrhosis: Incidence and risk factors. Gastroenterology. 127:S35–S50. 10.1053/j.gastro.2004.09.014 [DOI] [PubMed] [Google Scholar]
- Feng B., Yao P.M., Li Y., Devlin C.M., Zhang D., Harding H.P., Sweeney M., Rong J.X., Kuriakose G., Fisher E.A., et al. . 2003. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat. Cell Biol. 5:781–792. 10.1038/ncb1035 [DOI] [PubMed] [Google Scholar]
- Fleischmann A., Hvalby O., Jensen V., Strekalova T., Zacher C., Layer L.E., Kvello A., Reschke M., Spanagel R., Sprengel R., et al. . 2003. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J. Neurosci. 23:9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.D., Cui J.Y., and Klaassen C.D.. 2014. Atorvastatin induces bile acid-synthetic enzyme Cyp7a1 by suppressing FXR signaling in both liver and intestine in mice. J. Lipid Res. 55:2576–2586. 10.1194/jlr.M053124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuest M., Willim K., MacNelly S., Fellner N., Resch G.P., Blum H.E., and Hasselblatt P.. 2012. The transcription factor c-Jun protects against sustained hepatic endoplasmic reticulum stress thereby promoting hepatocyte survival. Hepatology. 55:408–418. 10.1002/hep.24699 [DOI] [PubMed] [Google Scholar]
- Güller M., Toualbi-Abed K., Legrand A., Michel L., Mauviel A., Bernuau D., and Daniel F.. 2008. c-Fos overexpression increases the proliferation of human hepatocytes by stabilizing nuclear Cyclin D1. World J. Gastroenterol. 14:6339–6346. 10.3748/wjg.14.6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenfuss S.C., Bakiri L., Thomsen M.K., Hamacher R., and Wagner E.F.. 2014a Activator Protein 1 transcription factor Fos-related antigen 1 (Fra-1) is dispensable for murine liver fibrosis, but modulates xenobiotic metabolism. Hepatology. 59:261–273. 10.1002/hep.26518 [DOI] [PubMed] [Google Scholar]
- Hasenfuss S.C., Bakiri L., Thomsen M.K., Williams E.G., Auwerx J., and Wagner E.F.. 2014b Regulation of steatohepatitis and PPARγ signaling by distinct AP-1 dimers. Cell Metab. 19:84–95. 10.1016/j.cmet.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt P., Rath M., Komnenovic V., Zatloukal K., and Wagner E.F.. 2007. Hepatocyte survival in acute hepatitis is due to c-Jun/AP-1-dependent expression of inducible nitric oxide synthase. Proc. Natl. Acad. Sci. USA. 104:17105–17110. 10.1073/pnas.0706272104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., and Karin M.. 2011. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 21:159–168. 10.1038/cr.2010.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holczbauer A., Factor V.M., Andersen J.B., Marquardt J.U., Kleiner D.E., Raggi C., Kitade M., Seo D., Akita H., Durkin M.E., and Thorgeirsson S.S.. 2013. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology. 145:221–231. 10.1053/j.gastro.2013.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., and Goldstein J.L.. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100:12027–12032. 10.1073/pnas.1534923100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y., Nijman S.M., Kobayashi M., Chan J.A., Brunet J.P., Chiang D.Y., Villanueva A., Newell P., Ikeda K., Hashimoto M., et al. . 2009. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69:7385–7392. 10.1158/0008-5472.CAN-09-1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y., Toffanin S., Lachenmayer A., Villanueva A., Minguez B., and Llovet J.M.. 2010. Molecular classification and novel targets in hepatocellular carcinoma: Recent advancements. Semin. Liver Dis. 30:035–051. 10.1055/s-0030-1247131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao L.L., Dangond F., Yoshida T., Hong R., Jensen R.V., Misra J., Dillon W., Lee K.F., Clark K.E., Haverty P., et al. . 2001. A compendium of gene expression in normal human tissues. Physiol. Genomics. 7:97–104. 10.1152/physiolgenomics.00040.2001 [DOI] [PubMed] [Google Scholar]
- Hui L., Bakiri L., Mairhorfer A., Schweifer N., Haslinger C., Kenner L., Komnenovic V., Scheuch H., Beug H., and Wagner E.F.. 2007. p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat. Genet. 39:741–749. 10.1038/ng2033 [DOI] [PubMed] [Google Scholar]
- Ibrahim S.H., Kohli R., and Gores G.J.. 2011. Mechanisms of lipotoxicity in NAFLD and clinical implications. J. Pediatr. Gastroenterol. Nutr. 53:131–140. 10.1097/MPG.0b013e31822578db [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. 2006. Mechanisms for cellular cholesterol transport: Defects and human disease. Physiol. Rev. 86:1237–1261. 10.1152/physrev.00022.2005 [DOI] [PubMed] [Google Scholar]
- Jain S., Singhal S., Lee P., and Xu R.. 2010. Molecular genetics of hepatocellular neoplasia. Am. J. Transl. Res. 2:105–118. [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Bray F., Center M.M., Ferlay J., Ward E., and Forman D.. 2011. Global cancer statistics. CA Cancer J. Clin. 61:69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- Johnson B.A. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278:313–352. [DOI] [PubMed] [Google Scholar]
- Jusakul A., Yongvanit P., Loilome W., Namwat N., and Kuver R.. 2011. Mechanisms of oxysterol-induced carcinogenesis. Lipids Health Dis. 10:44 10.1186/1476-511X-10-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. . 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42:D764–D770. 10.1093/nar/gkt1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C., Opherk C., Anlag K., Schütz G., and Tronche F.. 2000. Hepatocyte-specific expression of Cre recombinase. Genesis. 26:151–153. [DOI] [PubMed] [Google Scholar]
- Kim G., Jang S.Y., Han E., Lee Y.H., Park S.Y., Nam C.M., and Kang E.S.. 2017. Effect of statin on hepatocellular carcinoma in patients with type 2 diabetes: A nationwide nested case-control study. Int. J. Cancer. 140:798–806. 10.1002/ijc.30506 [DOI] [PubMed] [Google Scholar]
- Kim I., Morimura K., Shah Y., Yang Q., Ward J.M., and Gonzalez F.J.. 2007. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 28:940–946. 10.1093/carcin/bgl249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner A., Wu W., Staudt C., and Iliakis G.. 2008. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36:5678–5694. 10.1093/nar/gkn550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner A., Gossen M., Zimmermann F., Jerecic J., Ullmer C., Lübbert H., and Bujard H.. 1996. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl. Acad. Sci. USA. 93:10933–10938. 10.1073/pnas.93.20.10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte B.A., Joseph S.B., Walczak R., Pei L., Wilpitz D.C., Collins J.L., and Tontonoz P.. 2001. Autoregulation of the human liver X receptor alpha promoter. Mol. Cell. Biol. 21:7558–7568. 10.1128/MCB.21.22.7558-7568.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawman S., Mauri C., Jury E.C., Cook H.T., and Ehrenstein M.R.. 2004. Atorvastatin inhibits autoreactive B cell activation and delays lupus development in New Zealand black/white F1 mice. J. Immunol. 173:7641–7646. 10.4049/jimmunol.173.12.7641 [DOI] [PubMed] [Google Scholar]
- Lee J.S., Heo J., Libbrecht L., Chu I.S., Kaposi-Novak P., Calvisi D.F., Mikaelyan A., Roberts L.R., Demetris A.J., Sun Z., et al. . 2006. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 12:410–416. 10.1038/nm1377 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Kimmoun E., Legrand A., Sauvanet A., Degott C., Lardeux B., and Bernuau D.. 2002. Activation of NF-kappa B, AP-1 and STAT transcription factors is a frequent and early event in human hepatocellular carcinomas. J. Hepatol. 37:63–71. 10.1016/S0168-8278(02)00064-8 [DOI] [PubMed] [Google Scholar]
- Lo Sasso G., Bovenga F., Murzilli S., Salvatore L., Di Tullio G., Martelli N., D’Orazio A., Rainaldi S., Vacca M., Mangia A., et al. . 2013. Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology. 144:1497–1507.e13. 10.1053/j.gastro.2013.02.005 [DOI] [PubMed] [Google Scholar]
- Lu M., Hu X.H., Li Q., Xiong Y., Hu G.J., Xu J.J., Zhao X.N., Wei X.X., Chang C.C., Liu Y.K., et al. . 2013. A specific cholesterol metabolic pathway is established in a subset of HCCs for tumor growth. J. Mol. Cell Biol. 5:404–415. 10.1093/jmcb/mjt039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K., Tsukamoto H., Liu J.C., Han Y.P., Govindarajan S., Lai M.M., Akira S., and Ou J.H.. 2010. c-Jun mediates hepatitis C virus hepatocarcinogenesis through signal transducer and activator of transcription 3 and nitric oxide-dependent impairment of oxidative DNA repair. Hepatology. 52:480–492. 10.1002/hep.23697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra A., Kaul D., and Joshi K.. 2011. LXR-α selectively reprogrammes cancer cells to enter into apoptosis. Mol. Cell. Biochem. 349:41–55. 10.1007/s11010-010-0659-3 [DOI] [PubMed] [Google Scholar]
- Michelotti G.A., Machado M.V., and Diehl A.M.. 2013. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 10:656–665. 10.1038/nrgastro.2013.183 [DOI] [PubMed] [Google Scholar]
- Min L., Ji Y., Bakiri L., Qiu Z., Cen J., Chen X., Chen L., Scheuch H., Zheng H., Qin L., et al. . 2012. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat. Cell Biol. 14:1203–1211. 10.1038/ncb2590 [DOI] [PubMed] [Google Scholar]
- Nault J.C., and Zucman-Rossi J.. 2011. Genetics of hepatobiliary carcinogenesis. Semin. Liver Dis. 31:173–187. 10.1055/s-0031-1276646 [DOI] [PubMed] [Google Scholar]
- Panasyuk G., Espeillac C., Chauvin C., Pradelli L.A., Horie Y., Suzuki A., Annicotte J.S., Fajas L., Foretz M., Verdeguer F., et al. . 2012. PPARγ contributes to PKM2 and HK2 expression in fatty liver. Nat. Commun. 3:672 10.1038/ncomms1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R.A., Garcia R., Ryan C.S., Liu X., Shipkova P., Livanov V., Patel P., and Ho S.P.. 2013. Bile acid and sterol metabolism with combined HMG-CoA reductase and PCSK9 suppression. J. Lipid Res. 54:2400–2409. 10.1194/jlr.M038331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet D.J., Turley S.D., Ma W., Janowski B.A., Lobaccaro J.M., Hammer R.E., and Mangelsdorf D.J.. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 93:693–704. 10.1016/S0092-8674(00)81432-4 [DOI] [PubMed] [Google Scholar]
- Perez M.J., and Briz O.. 2009. Bile-acid-induced cell injury and protection. World J. Gastroenterol. 15:1677–1689. 10.3748/wjg.15.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzelli M., Schweiger M., Schreiber R., Campos-Olivas R., Tsoli M., Allen J., Swarbrick M., Rose-John S., Rincon M., Robertson G., et al. . 2014. A switch from white to brown fat increases energy expenditure in cancer-associated cachexia. Cell Metab. 20:433–447. 10.1016/j.cmet.2014.06.011 [DOI] [PubMed] [Google Scholar]
- Pinyol R., Nault J.C., Quetglas I.M., Zucman-Rossi J., and Llovet J.M.. 2014. Molecular profiling of liver tumors: Classification and clinical translation for decision making. Semin. Liver Dis. 34:363–375. 10.1055/s-0034-1394137 [DOI] [PubMed] [Google Scholar]
- Rogue A., Lambert C., Jossé R., Antherieu S., Spire C., Claude N., and Guillouzo A.. 2011. Comparative gene expression profiles induced by PPARγ and PPARα/γ agonists in human hepatocytes. PLoS One. 6:e18816 10.1371/journal.pone.0018816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T., Maeda S., Chang L., and Karin M.. 2006. Loss of hepatic NF-κB activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc. Natl. Acad. Sci. USA. 103:10544–10551. 10.1073/pnas.0603499103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleucher J., Schwendinger M., Sattler M., Schmidt P., Schedletzky O., Glaser S.J., Sørensen O.W., and Griesinger C.. 1994. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR. 4:301–306. 10.1007/BF00175254 [DOI] [PubMed] [Google Scholar]
- Seki E., Brenner D.A., and Karin M.. 2012. A liver full of JNK: Signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology. 143:307–320. 10.1053/j.gastro.2012.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Yasuda Y., Sakai H., Kubota M., Terakura D., Baba A., Ohno T., Kochi T., Tsurumi H., Tanaka T., and Moriwaki H.. 2011. Pitavastatin suppresses diethylnitrosamine-induced liver preneoplasms in male C57BL/KsJ-db/db obese mice. BMC Cancer. 11:281 10.1186/1471-2407-11-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Singh P.P., Roberts L.R., and Sanchez W.. 2014. Chemopreventive strategies in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 11:45–54. 10.1038/nrgastro.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I. 2002. Consequences of cellular cholesterol accumulation: Basic concepts and physiological implications. J. Clin. Invest. 110:905–911. 10.1172/JCI0216452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trierweiler C., Blum H.E., and Hasselblatt P.. 2012. The transcription factor c-Jun protects against liver damage following activated β-Catenin signaling. PLoS One. 7:e40638 10.1371/journal.pone.0040638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trierweiler C., Hockenjos B., Zatloukal K., Thimme R., Blum H.E., Wagner E.F., and Hasselblatt P.. 2016. The transcription factor c-JUN/AP-1 promotes HBV-related liver tumorigenesis in mice. Cell Death Differ. 23:576–582. 10.1038/cdd.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi R.K., Azrad A., Degani H., and Salomon Y.. 1996. Simultaneous extraction of cellular lipids and water-soluble metabolites: Evaluation by NMR spectroscopy. Magn. Reson. Med. 35:194–200. 10.1002/mrm.1910350210 [DOI] [PubMed] [Google Scholar]
- van Malenstein H., van Pelt J., and Verslype C.. 2011. Molecular classification of hepatocellular carcinoma anno 2011. Eur. J. Cancer. 47:1789–1797. 10.1016/j.ejca.2011.04.027 [DOI] [PubMed] [Google Scholar]
- Villanueva A., Hernandez-Gea V., and Llovet J.M.. 2013. Medical therapies for hepatocellular carcinoma: A critical view of the evidence. Nat. Rev. Gastroenterol. Hepatol. 10:34–42. 10.1038/nrgastro.2012.199 [DOI] [PubMed] [Google Scholar]
- Vinaixa M., Rodríguez M.A., Rull A., Beltrán R., Bladé C., Brezmes J., Cañellas N., Joven J., and Correig X.. 2010. Metabolomic assessment of the effect of dietary cholesterol in the progressive development of fatty liver disease. J. Proteome Res. 9:2527–2538. 10.1021/pr901203w [DOI] [PubMed] [Google Scholar]
- Wagner E.F., and Nebreda A.R.. 2009. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 9:537–549. 10.1038/nrc2694 [DOI] [PubMed] [Google Scholar]
- Wang X., Fu X., Van Ness C., Meng Z., Ma X., and Huang W.. 2013. Bile acid receptors and liver cancer. Curr. Pathobiol. Rep. 1:29–35. 10.1007/s40139-012-0003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.W., Farrell G.C., and Yu J.. 2012. Functional role of peroxisome-proliferator-activated receptor γ in hepatocellular carcinoma. J. Gastroenterol. Hepatol. 27:1665–1669. 10.1111/j.1440-1746.2012.07213.x [DOI] [PubMed] [Google Scholar]
- Yang F., Huang X., Yi T., Yen Y., Moore D.D., and Huang W.. 2007. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 67:863–867. 10.1158/0008-5472.CAN-06-1078 [DOI] [PubMed] [Google Scholar]
- Yoshikawa T., Ide T., Shimano H., Yahagi N., Amemiya-Kudo M., Matsuzaka T., Yatoh S., Kitamine T., Okazaki H., Tamura Y., et al. . 2003. Cross-talk between peroxisome proliferator-activated receptor (PPAR) alpha and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARs suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Mol. Endocrinol. 17:1240–1254. 10.1210/me.2002-0190 [DOI] [PubMed] [Google Scholar]
- Yu J., Shen B., Chu E.S., Teoh N., Cheung K.F., Wu C.W., Wang S., Lam C.N., Feng H., Zhao J., et al. . 2010. Inhibitory role of peroxisome proliferator-activated receptor gamma in hepatocarcinogenesis in mice and in vitro. Hepatology. 51:2008–2019. 10.1002/hep.23550 [DOI] [PubMed] [Google Scholar]
- Yuen M.F., Wu P.C., Lai V.C., Lau J.Y., and Lai C.L.. 2001. Expression of c-Myc, c-Fos, and c-jun in hepatocellular carcinoma. Cancer. 91:106–112. [DOI] [PubMed] [Google Scholar]
- Zhang D.Y., and Friedman S.L.. 2012. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 56:769–775. 10.1002/hep.25670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Castellani L.W., Sinal C.J., Gonzalez F.J., and Edwards P.A.. 2004. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 18:157–169. 10.1101/gad.1138104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Breevoort S.R., Angdisen J., Fu M., Schmidt D.R., Holmstrom S.R., Kliewer S.A., Mangelsdorf D.J., and Schulman I.G.. 2012. Liver LXRα expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J. Clin. Invest. 122:1688–1699. 10.1172/JCI59817 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.