Deeg et al. show a novel line of transgenic mice expressing restriction factor MxA exhibits robust resistance to influenza viruses of avian but not human origin. In vivo evasion of MxA is mediated by distinct amino acids in the nucleoprotein of human influenza viruses.
Abstract
Zoonotic transmission of influenza A viruses can give rise to devastating pandemics, but currently it is impossible to predict the pandemic potential of circulating avian influenza viruses. Here, we describe a new mouse model suitable for such risk assessment, based on the observation that the innate restriction factor MxA represents an effective species barrier that must be overcome by zoonotic viruses. Our mouse lacks functional endogenous Mx genes but instead carries the human MX1 locus as a transgene. Such transgenic mice were largely resistant to highly pathogenic avian H5 and H7 influenza A viruses, but were almost as susceptible to infection with influenza viruses of human origin as nontransgenic littermates. Influenza A viruses that successfully established stable lineages in humans have acquired adaptive mutations which allow partial MxA escape. Accordingly, an engineered avian H7N7 influenza virus carrying a nucleoprotein with signature mutations typically found in human virus isolates was more virulent in transgenic mice than parental virus, demonstrating that a few amino acid changes in the viral target protein can mediate escape from MxA restriction in vivo. Similar mutations probably need to be acquired by emerging influenza A viruses before they can spread in the human population.
Introduction
Influenza A viruses pose a constant threat to humans because zoonotic viral transmissions can cause severe disease and give rise to devastating pandemics (Watanabe et al., 2014; Yoon et al., 2014; Neumann and Kawaoka, 2015). Innate restriction factors represent effective species barriers that need to be overcome by viruses to invade the human host (Cauldwell et al., 2014; Iwasaki and Pillai, 2014). In mice, innate immune control of influenza viruses is mediated by the IFN-regulated Mx1 gene (Haller et al., 2015). In man, the MX1 ortholog (encoding MxA protein) may play a similar role, as it provides broad resistance to influenza and other viruses in cell culture systems (Pavlovic et al., 1990; Haller et al., 2015). However, formal genetic proof that MxA helps protect humans from influenza virus–induced disease has so far been missing, as none of the known minor MX1 gene variants has to date been linked to enhanced influenza virus susceptibility (Ciancanelli et al., 2016).
MxA belongs to the dynamin superfamily of multi-domain large GTPases, and the MxA coding sequence is highly conserved in humans and other primates (Mitchell et al., 2013; Haller et al., 2015). Sequence comparison revealed that loop L4 is the most divergent part of the otherwise conserved molecule, and mutational analysis showed that the L4 domain acts as an autonomous antiviral module that determines the antiviral specificities in different species (Mitchell et al., 2012; Patzina et al., 2014). L4 binds to the influenza viral nucleoprotein (Patzina et al., 2014), which leads to viral inhibition by a yet to be defined mechanism that requires GTP hydrolysis and oligomerization of MxA (Mitchell et al., 2013; Haller et al., 2015; Nigg and Pavlovic, 2015).
When major restriction factors are at work, viruses usually evolve to acquire mutations that help them to evade such host defense strategies (Simon et al., 2015). Recent studies suggest that MxA is targeting the nucleoprotein of influenza A viruses which encapsidates the viral genome, a process that is pivotal to virus replication (Dittmann et al., 2008; Zimmermann et al., 2011). Indeed, more recent studies indicate that influenza A viruses that successfully established stable lineages in humans acquired adaptive mutations in the nucleoprotein that allow partial MxA escape (Mänz et al., 2013; Götz et al., 2016). It remained unclear, however, whether these cell culture experiments accurately reflect the in vivo situation.
A suitable animal model is required to answer this question. Early attempts to constitutively express MxA cDNA in mice yielded disappointing results, as the transgene was not expressed well in the lungs (Pavlovic et al., 1995). We now generated a new mouse line that carries the complete human MX1 locus as a transgene and readily expresses MxA in response to IFN exposure in all major organs. Using this mouse model, we provide first in vivo evidence that a few surface-exposed amino acids on the nucleoprotein are indeed responsible for MxA evasion and play a key role in the adaptation of avian influenza A viruses to humans, as previously indicated by biochemical and cell culture experiments (Mänz et al., 2013; Götz et al., 2016). These results indicate that MxA is a decisive factor that confers influenza virus resistance in humans. They provide strong evidence that newly emerging influenza viruses need to acquire the ability to evade MxA restriction to spread efficiently in the human population.
Results and discussion
Cells from transgenic mice carrying the human Mx locus exhibit IFN-dependent resistance to influenza A virus
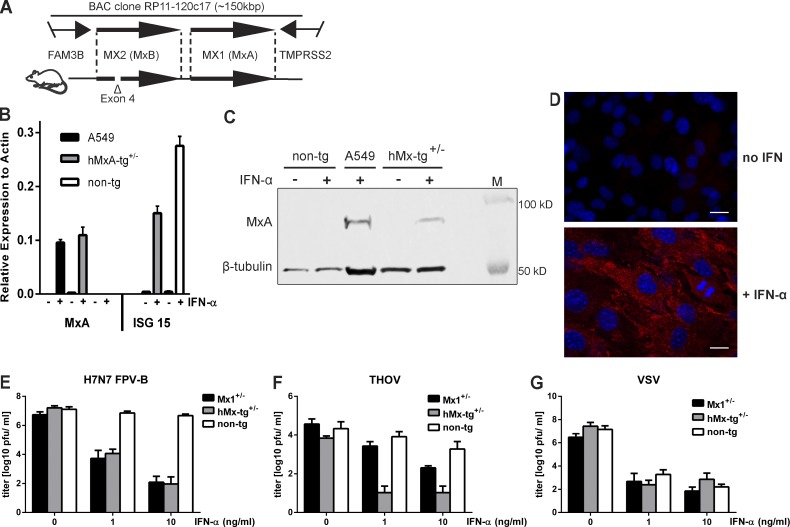
To assess the role of MxA in vivo, we developed a transgenic mouse that carries a bacterial artificial chromosome that includes the entire human MX locus (Fig. 1 A). Repeated backcrossing of founder animals to C57BL/6 mice lacking functional endogenous Mx genes (Staeheli et al., 1988) yielded a new transgenic mouse line, designated hMx-tg. cDNA sequencing revealed that the MX2 gene is defective in hMx-tg mice as a result of an accidental deletion that includes exon 4 (Fig. 1 A). Thus, MX1 is the only intact human gene in our hMx-tg mouse strain.
Figure 1.
Cells from transgenic mice carrying the human MX locus exhibit IFN-dependent resistance to influenza A virus and THOV. (A) Schematic drawing showing the fragment of human chromosome 21 present in BAC clone Rp11-120c17 (top) and transgenic mice (bottom). Note that the MX2 gene is defective in our transgenic mice as a result of a deletion that includes coding exon 4. (B and C) MEFs prepared from transgenic (hMx-tg+/−) or nontransgenic (non-tg) littermates were treated with 10 ng per ml of IFN-α for 18 h (+) or were left untreated (–) before analysis of the cellular MxA content by RT-qPCR (B) or Western blot (C). IFN-treated human A549 cells served as positive control. ISG15 served as IFN treatment control in MEFs. Western blots were simultaneously probed with an antibody recognizing β-tubulin to demonstrate similar loading of the gel. RT-qPCR ΔCT values were calculated relative to actin gene expression from technical triplicates; SEM is shown. (D) MEFs from hMx-tg+/− mice were treated with 10 ng per ml of IFN-α for 18 h before visualization of MxA expression (red) by indirect immunofluorescence. Untreated cells served as controls. Nuclei were stained with DAPI (blue). Bars, 10 µm. (E) MEFs from hMx-tg+/− and non-tg littermates were treated for 18 h with plain medium (0) or medium containing 1 or 10 ng per ml of IFN-α before infection (MOI, 0.01) with H7N7 avian influenza A virus strain FPV-B. Culture supernatants were harvested at 24 h after infection and virus titers were determined. Cells from mouse embryos carrying one functional allele of the endogenous Mx1 gene (Mx1+/−) were used as controls. Mean values with standard error of means of three independent experiments are shown. (F and G) MEFs from hMx-tg+/− and non-tg littermates were treated for 18 h with plain medium (0) or medium containing 1 or 10 ng per ml of IFN-α before infection (MOI = 0.1) with THOV (F) or VSV (G). Culture supernatants were harvested at 24 h (VSV) or 72 h (THOV) after infection and virus titers were determined. Mx1+/− MEFs were used as controls. Mean values with SEM of three independent experiments are shown.
Cultured mouse embryonic fibroblasts (MEFs) prepared from hMx-tg mice expressed the transgene when stimulated with IFN-α, but not under standard culture conditions, and maintained normal regulation of other IFN-stimulated genes such as ISG15 (Fig. 1 B). MxA protein levels in IFN-treated transgenic MEFs were slightly lower than in IFN-treated human A549 cells (Fig. 1 C). MxA accumulated in the cytoplasm of IFN-treated hMx-tg MEFs (Fig. 1 D) as expected from earlier studies of human cells (Staeheli and Haller, 1985). When pretreated with IFN-α and challenged with a mouse-adapted H7N7 influenza A virus (Israël, 1979), MEFs from hMx-tg mice but not from nontransgenic littermates exhibited a high degree of resistance that was comparable to cells from mice carrying one functional copy of the endogenous Mx1 resistance gene (Fig. 1 E). Furthermore, IFN-treated MEFs from hMx-tg but not from nontransgenic mice were highly resistant to Thogoto virus (THOV) known to be extremely sensitive to MxA (Kochs and Haller, 1999; Fig. 1 F). In contrast, IFN-induced resistance to vesicular stomatitis virus (VSV) was independent of either endogenous Mx1 or transgenically expressed MxA (Fig. 1 G), confirming other studies that additional IFN-induced restriction factors contribute to inhibition of VSV (Sadler and Williams, 2008).
hMx-tg mice are highly resistant to infection with avian influenza A viruses and THOV
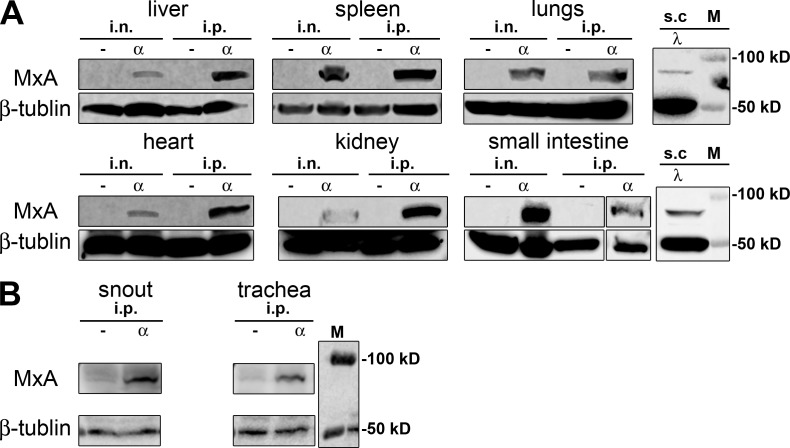
The transgene was expressed in all organs tested when the animals were treated with IFN-α by either the i.n. or the i.p. route (Fig. 2). Detectable levels of MxA were also induced by IFN-λ in lungs and small intestine, whereas transgene expression was not detected in organs of untreated mice (Fig. 2 A). We additionally checked MxA expression in the upper respiratory tract (trachea and snout) for relevance to influenza virus infection. As expected, MxA was induced upon IFN-α treatment in the upper respiratory tract (Fig. 2 B). These results demonstrated that our transgenic mice carry a copy of the human MX1 gene that is functional and expressed in response to type I and type III IFN, similar to the authentic gene in human tissue (Holzinger et al., 2007).
Figure 2.
IFN-induced expression of the MxA-encoding transgene in vivo. (A) hMx-tg mice were either left untreated (–) or treated with 1 µg per animal of either IFN-α (α) or IFN-λ (λ) by i.n., i.p., or s.c. routes 18 h before the indicated organs were collected and analyzed for MxA content by Western blot. Blots were simultaneously probed with an antibody recognizing β-tubulin to demonstrate similar loading of the gel. (B) Mice were left untreated (-) or were treated for 18 h with 1 µg of IFN-α per animal before homogenates from trachea and snouts were prepared. Western blot analysis was done as described for A.
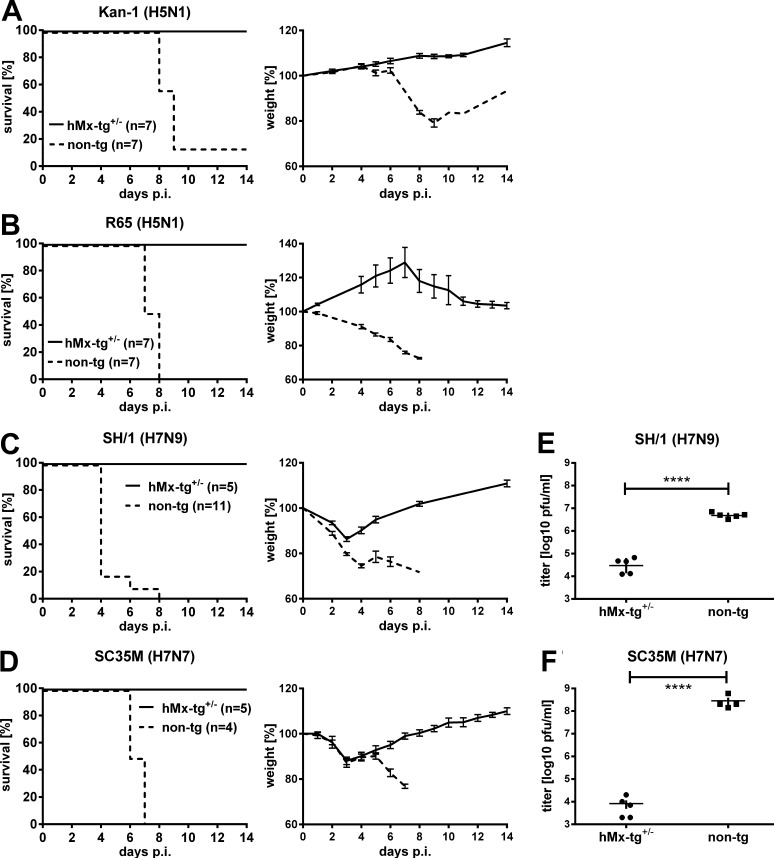
To assess influenza virus susceptibility, transgenic mice were challenged with 10–30 median lethal doses 50 (LD50) of highly pathogenic H5 and H7 influenza A viruses of avian origin. All transgenic mice survived such infections, whereas the nontransgenic littermates developed severe disease and most of them had to be sacrificed for animal welfare reasons before day 9 after infection (Fig. 3, A–D). Resistance of hMx-tg mice to these avian viruses was remarkably robust: all transgenic animals exhibited only very mild body weight losses and survived challenge infections with >104 LD50 (Table 1) of the highly pathogenic H5N1 influenza A virus strains A/KAN-1 (Fig. 3 A) and A/R65 (Fig. 3 B). Resistance of hMx-tg mice toward the H7N9 strain A/SH/1 (Fig. 3 C) and the H7N7 strain A/SC35M (Fig. 3 D) was similarly pronounced, and the LD50 values of these viruses in hMx-tg mice were ∼30-fold higher than in nontransgenic controls (Table 1). Enhanced survival of transgenic mice infected with SH/1 and SC35M correlated with strongly reduced virus titers in the lungs compared with nontransgenic controls (Fig. 3, E and F). Earlier work indicated that avian H7N9 influenza A viruses, which recently emerged in China, can partially evade restriction by MxA in cell culture (Riegger et al., 2015). In contrast, our infection studies with transgenic mice clearly demonstrated that MxA is able to control the A/SH1 human H7N9 isolate very efficiently in the context of the whole organism. Thus, the MxA restriction factor is protective against a broad range of avian influenza viruses and represents a veritable barrier that limits zoonotic virus transmissions.
Figure 3.
hMx-tg mice are highly resistant to infection with influenza A viruses of avian origin. Groups of hMx-tg+/− and non-tg mice were challenged by the i.n. route with 10–30 LD50 (values refer to non-tg mice) of (A) the avian influenza A virus strains KAN-1 (1.5 × 102 pfu; seven animals each), (B) R65 (2 × 102 pfu; seven animals each), (C) SH/1 (104 pfu; five hMx-tg+/− and eleven non-tg), or (D) SC35M (5 × 103 pfu; five hMx-tg+/− and four non-tg). Weight loss was monitored daily; animals were sacrificed and scored dead when weight loss reached 25% of the initial weight on the day of infection. (E and F) Other groups of transgenic and nontransgenic mice were challenged through the i.n. route with (E) SH/1 (104 pfu; five animals each) or (F) SC35M (5 × 103 pfu; five hMx-tg+/− and four non-tg). Viral titers in the lungs at day 5 after infection are shown. ****, P < 0.0001, Student’s t test.
Table 1. Approximate LD50 values of influenza A viruses of avian or human origin in C57BL/6 mice carrying functional endogenous Mx1 genes (Mx1), lacking functional endogenous Mx1 (non-tg) or carrying the human MxA transgene (hMx-tg).
| Influenza strain | LD50 [pfu] | ||
|---|---|---|---|
| hMx-tg | non-tg | Mx1 | |
| Avian | |||
| KAN-1 (H5N1) | >106 | 5 × 10° | >106 |
| R65 (H5N1) | 8 × 104 | 5 × 10° | 3 × 105 |
| SH/1 (H7N9) | 3 × 104 | 1 × 103 | >105 |
| SC35M (H7N7) | 2 × 104 | 8 × 102 | 1 × 105 |
| Human | |||
| PR8 (H1N1) | 2 × 103 | 1 × 103 | >106 |
| WSN (H1N1) | 2 × 103 | 5 × 102 | >4 × 105 |
| HK68 (H3N2) | 5 × 101 | 1 × 101 | 4 × 104 |
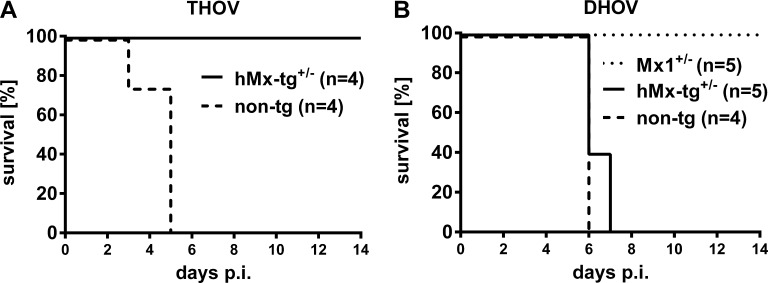
The Mx1 protein of mice confers a high degree of resistance to infection by members of the Thogotoviridae family, such as THOV and Dhori virus (DHOV), whereas human MxA selectively inhibits THOV (Frese et al., 1995; Haller et al., 1995). Our hMxA-tg mice exhibited robust resistance to challenges with THOV (Fig. 4 A), but were highly susceptible to DHOV (Fig. 4 B), demonstrating that the antiviral specificity of MxA is maintained in our transgenic mice. These findings are in agreement with earlier findings from studies with a transgenic mouse line that constitutively expressed low levels of MxA and confirmed that these two related viruses differ drastically with regard to MxA sensitivity (Thimme et al., 1995). Collectively, these results demonstrated that our transgenic mice behaved as predicted from previous studies with cultured human cells (Haller et al., 2015), and that they reproduced the innate anti-influenza immune response of humans.
Figure 4.
hMx-tg mice are resistant to infection with THOV but susceptible to DHOV. hMx-tg+/− and non-tg mice were challenged by i.p. infection with (A) 103 pfu of THOV (four animals each) or (B) 102 pfu of DHOV (five hMx-tg+/− and four non-tg). Survival was monitored for 14 d. Mice carrying one functional allele of the endogenous Mx1 gene (Mx1+/−, five animals) served as positive control for the DHOV challenge experiment. Animals were sacrificed and scored dead when weight loss reached the 25% limit.
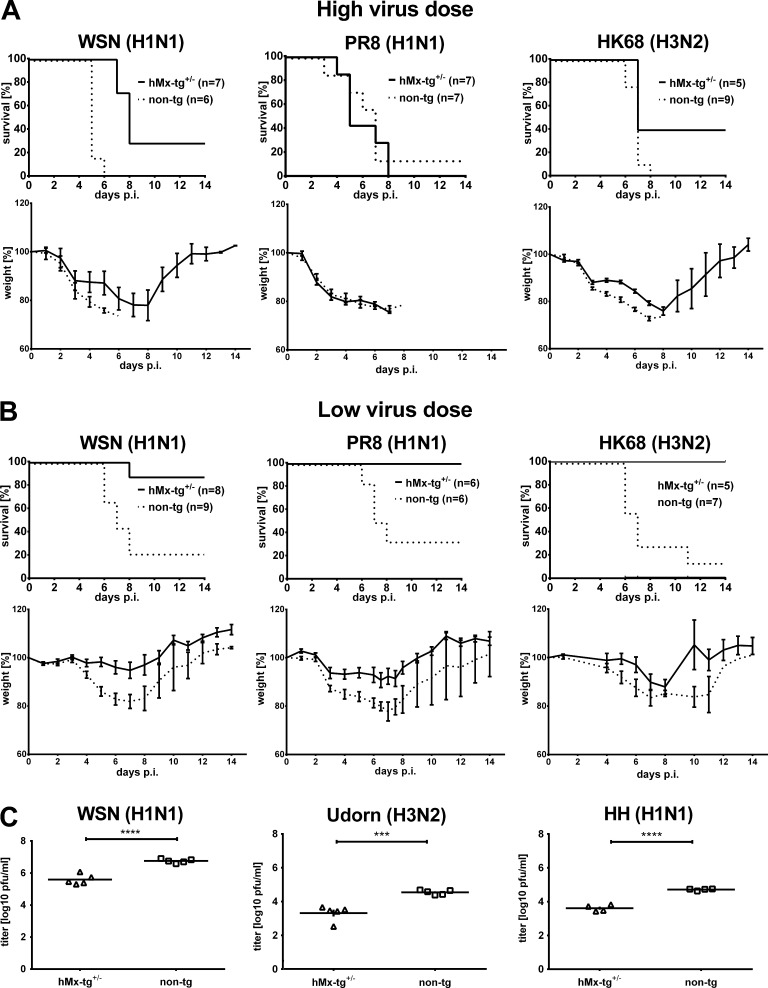
hMx-tg mice exhibit only moderate resistance to influenza A viruses of human origin
When challenged with ∼10 LD50 of mouse-adapted A/WSN (H1N1), A/PR8 (H1N1), or A/HK68 (H3N2), most of the transgenic mice developed severe disease symptoms with only slightly delayed kinetics compared with nontransgenic littermates (Fig. 5 A). These challenge conditions were not particularly stringent as control mice carrying a functional endogenous Mx1 gene survived such virus doses (Table 1) without developing severe disease symptoms. Nevertheless, the transgene conferred some protection which became obvious only when viral titers in the lungs were measured (Fig. 5 C) or when the challenge virus dose was reduced to ∼3 LD50 (Fig. 5 B). Under such nonstringent conditions, most of the transgenic mice lost <25% of their initial body weight and survived the infection, whereas >50% of nontransgenic littermates had to be sacrificed.
Figure 5.
hMx-tg mice exhibit only moderate resistance to influenza A viruses of human origin. (A) Groups of hMx-tg+/− and non-tg mice were challenged by the i.n. route with ∼10 LD50 (high dose; values refer to non-tg mice) of mouse-adapted influenza A virus strains of human origin, namely A/WSN (5 × 103 pfu; seven hMx-tg+/− and six non-tg), A/PR8 (104 pfu; seven animals each) or A/HK68 (100 pfu; five hMx-tg+/− and nine non-tg). (B) Other groups of transgenic and nontransgenic mice were challenged with 2–3 LD50 (low dose) of A/WSN (1.3 × 103 pfu; eight hMx-tg+/− and nine non-tg), A/PR8 (3 × 103 pfu; six animals each) or A/HK68 (30 pfu; five hMx-tg+/− and seven non-tg). Weight loss was monitored daily; animals were sacrificed and scored dead when weight loss reached the 25% limit. (C) Groups of hMx-tg+/− and non-tg mice were infected with 104 pfu of human influenza virus strains WSN, Udorn or HH. Viral titers in the lungs at day 4 after infection are shown. ***, P < 0.0005; ****, P < 0.0001, Student’s t test.
A similar picture emerged when influenza virus strains with no passage history in mice, such as A/Udorn/72 and A/HH/2009, were used for challenge experiments. These nonadapted viruses, which do not induce disease in mice replicated to significantly higher titer in the lungs of nontransgenic mice compared with transgenic animals, but the differences were moderate, as in the case of mouse-adapted WSN (Fig. 5 C), indicating that mouse adaptation does not alter the MxA resistance phenotype of human influenza virus isolates. It should be noted that influenza viruses of both avian and human origin were all restricted very well in mice that express endogenous Mx1 genes (Table 1), indicating that human influenza viruses evolved to specifically evade restriction by human MxA but not mouse Mx1. This finding is most likely caused by the fact that the mouse does not serve as a natural host for influenza viruses, and mouse Mx1 does therefore not exert any selective pressure on influenza virus evolution. In contrast, human MX1 and influenza virus genes are apparently genes in conflict in an evolutionary arms race (Mitchell et al., 2013). Accordingly, our in vivo findings clearly demonstrate that influenza A viruses of human origin are rather poorly restricted by MxA, confirming and extending previous results from experiments with human cell culture systems (Dittmann et al., 2008). Collectively, the present animal experiments provide first solid in vivo evidence that MxA is indeed a major influenza restriction factor in humans.
Avian influenza virus with adaptive mutations in the nucleoprotein gains virulence in hMx-tg mice
Partial escape of human influenza A viruses from restriction by MxA in a cell culture system was recently linked to three signature mutations in the viral nucleoprotein (Mänz et al., 2013). The vast majority of avian influenza viruses possess nucleoproteins with arginine at position 100, leucine at position 283, and phenylalanine at position 313, whereas most influenza A viruses of human origin possess nucleoproteins with isoleucine or valine at position 100, proline at position 283, and tyrosine or valine at position 313. Avian influenza A viruses with engineered nucleoproteins carrying all three human signature mutations are genetically instable (Mänz et al., 2013), suggesting that MxA escape has a substantial fitness cost. Due to this technical difficulty, conclusive experimental proof that these adaptive mutations play a decisive role in MxA evasion during viral infections was not available.
Recently, a variant of the avian H7N7 influenza A virus strain SC35M was described that carries all three human signature mutations (100V, 283P and 313Y) along with one additional glycine to aspartic acid mutation at position 16 in the nucleoprotein (Götz et al., 2016). This SC35M variant carrying four mutations in the nucleoprotein, designated SC35M-4x, was genetically stable and showed no detectable growth retardation in MDCK cells. Importantly, SC35M-4x replicated to higher titers than parental SC35M in human A549 cells constitutively expressing MxA (Götz et al., 2016), indicating that SC35M-4x may represent a suitable tool for testing the hypothesis that viruses carrying nucleoproteins with a human signature are far more virulent in the face of a functional MxA-driven innate immune response than viruses carrying nucleoproteins with avian signature.
In hMx-tg mice, SC35M-4x induced slightly more weight loss and replicated to significantly higher titers in the lungs of such animals than parental SC35M (Fig. 6 A). The difference in viral lung titers was most pronounced on day 5 after infection, indicating that SC35M-4x continued to replicate even when the transgene is expected to be expressed well in the infected lungs as a result of virus-induced IFN synthesis. To verify this exceptional property of SC35M-4x more directly and to exclude the possibility that SC35M-4x had simply outpaced the innate immune response, we preactivated MxA transgene expression by treating the animals with IFN-α one day before virus challenge. Under such experimental conditions, which are expected to result in high levels of MxA in the lung tissue within 18 h (Fig. 2), high doses of parental SC35M no longer caused detectable weight loss in our transgenic mice (Figs. 6, B and C). In contrast, SC35M-4x still induced pronounced weight loss (Fig. 6 B), and six of seven IFN-treated transgenic animals that received 106 pfu of SC35M-4x had to be sacrificed (Fig. 6 C). Titers of SC35M-4x in the lungs of IFN-treated transgenic mice were ∼10-fold higher than parental SC35M on day 3 after infection (Fig. 6 B). Of note, in IFN-treated mice lacking the MxA transgene, lung titers of SC35M and SC35M-4x did not differ significantly at day 3 after infection (Fig. 6 D), demonstrating that the IFN-mediated resistance in transgenic mice is strongly dependent on MxA and that enhanced virulence of SC35M-4x is caused bysuccessful evasion of MxA-mediated growth restriction by this virus.
Figure 6.
SC35M-4x exhibits enhanced MxA resistance in transgenic mice. (A) hMx-tg+/− mice were infected with 4 × 103 pfu of either parental SC35M-wt or SC35M-4x (six and five animals, respectively). Body weight changes with SEM and lung titers on days 3 and 5 after infection are shown. (B and C) hMx-tg+/− were pretreated with 10 µg of IFN-α by s.c. injection 1 d before infection with (B) 105 pfu (six animals with SC35M-wt and five animals with SC35M-4x) or (C) 106 pfu of either parental SC35M-wt or SC35M-4x (six animals SC35M-wt and five animals SC35M-4x). Kinetics of body weight changes, survival, and lung titers at day 3 (six animals SC35M-wt and five animals SC35M-4x) after infection are depicted. Weight loss was monitored daily and SEM depicted; animals were sacrificed and scored dead when body weight loss reached the 25% limit. (D) Groups (n = 5) of nontransgenic mice were infected with 103 pfu either parental SC35M-wt or SC35M-4x, and viral lung titers were determined on days 3 and 5 after infection. *, P < 0.05; ***, P < 0.0001.
Collectively, our new transgenic mouse shows fine-tuned expression of the human influenza virus restriction factor MxA in all major organs, including the upper and the lower respiratory tract. These mice thus represent the first small animal model that accurately mimics one important aspect of the innate immune response of humans toward influenza virus. Unlike all other currently available mouse models, our MxA-transgenic mouse can readily distinguish between MxA-sensitive influenza virus strains and virus strains which can evade MxA restriction and, consequently, possess a high pandemic potential in humans. Our MxA-transgenic mice will facilitate future basic research aimed at understanding the molecular details of influenza virus restriction by the innate immune system. For example, our current work revealed that H5N1 viruses are inhibited far more strongly by MxA than the H7 avian influenza viruses. We suspect that the high MxA susceptibility of H5 viruses is multifactorial, but further work is required to understand the molecular basis of this phenomenon. Our MxA-transgenic mouse might also be used to rapidly assess the pathogenic potential and overall fitness of emerging influenza A viruses in humans. Such functional analysis could complement current risk assessment strategies of emerging influenza viruses, including viral genome sequencing and screening for alterations in known viral virulence genes. Moreover, our transgenic mice might be useful in assessing the in vivo relevance of polymorphic MxA mutations that occur naturally in the human population. Such mutations could easily be introduced into the transgene of our mouse by CRISPR/Cas9 technology. Further, our transgenic mice might be used to analyze the in vivo importance of MxA cofactors.
Materials and methods
Transgenic mice
Human male bacterial artificial chromosome library clone RP11-120C17 (Cheung et al., 2001) was used to generate transgenic mice. This clone contains the complete human MX1 and MX2 genes, as well as fragments of the adjacent genes FAM3B and TMPRSS2. Microinjection of DNA into the pronucleus of fertilized eggs from superovulated B6.SJL females and transfer of manipulated eggs into pseudo-pregnant Swiss Webster female mice was performed by a commercial service (Taconic). Integration of the transgene into the genome of resulting offspring was verified by PCR using the following targeted primer sets: MX1, 5′-GCCTTTAATCAGGACATCACTGCTCTCATG-3′, 5′-CTGAACCCACAATATCTCTGAGGTATGCC-3′; FAM3B, 5′-TGAATTCCTTTCTGCAGACTTAGG-3′, 5′-GCAATTTGGGAGGATTGAGG-3′; TMPRSS2, 5′-GAGGTTAGGACTGCACTTGAGG-3′, 5′-TCCAGGCTTGTGGATCAGC-3′. Founder animals harboring the transgene were repeatedly backcrossed to C57BL/6 mice. Heterozygous males and females from backcrosses 5–12 (designated hMx-tg) were used for the experiments described in this study. Nontransgenic littermates served as controls.
Sequencing the MX gene transcripts
MEFs of hMx-tg and nontransgenic littermates were stimulated with human chimeric IFN-αB/D (10 ng/ml; Horisberger and de Staritzky, 1987) for 18 h. RNA was isolated using the Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer’s protocol and reverse transcribed into cDNA with the QuantiTect Reverse Transcription kit (QIAGEN). MX1 and MX2 genes were amplified by PCR. Products were purified with peqGOLD Cycle-Pure kit (PEQLAB) according to the manufacturer´s protocol before sequencing. Primers for MX1 were as follows: 5′-ATGGTTGTTTCCGAAGTGGAC-3′, 5′-TGTCAAGTGCCGGGGCCAGC-3′; primers for MX2 were as follows: 5′-ATGTCTAAGGCCCACAAGCCTTGGC-3′, 5′-TTCAGTGGATCTCTTTGCTGGAGAATTG-3′.
Viruses
For infection of mice, the following influenza A virus strains were used: A/Swan/Germany/R65/2006 (H5N1) designated R65, A/Thailand/1(KAN-1)/2004 (H5N1) designated KAN-1, A/Shanghai/1/2013 (H7N9) designated SH/1, A/Seal/Massachusetts/1/1980 (H7N7) designated SC35M, A/PR/8/1934 (H1N1) designated PR8, A/WSN/1933 (H1N1) designated WSN, A/Hong Kong/8/68 (H3N2) designated HK68, A/Udorn/72 (H3N2) designated Udorn, and A/HH/05/2009 (H1N1) designated HH. R65 and SH/1 are avian isolates with no passage history in mammals. The avian influenza virus strain KAN-1 was isolated from a diseased boy and has since been passaged in MDCK cells. SC35M represents a mouse-adapted variant of an avian H7N7 virus that was originally isolated from a diseased seal. PR8, WSN, and HK68 are mouse-adapted viruses of human origin, whereas the human virus isolates Udorn and HH have no passage history in mice. SC35M-4x differs from parental SC35M by four amino acid substitutions in the nucleoprotein, namely G16D, R100V, L293P, and F313Y (Götz et al., 2016). Two different THOVs were used for infection studies in mice: Thogoto virus strain SiAr126 lacking a functional ML gene (designated THOV; Albanese et al., 1972) and DHOV strain India1313/61 (designated DHOV; Anderson and Casals, 1973). The L cell–adapted variant of A/Fowl plague virus/Dobson (H7N7), designated FPV-B (Israël, 1979), and vesicular stomatitis virus serotype New Jersey (VSV) were used for infection of MEFs.
Cells
To generate MEFs, 14 d post coitum embryos of hMx-tg and nontransgenic littermates were mechanically and enzymatically homogenized before released cells were collected by centrifugation and resuspended in cell culture medium. Madin-Darby canine kidney epithelial cells (MDCK), human lung epithelial cells (A549), and baby hamster kidney cells (BHK-21) were originally obtained from the ATCC. All cells were cultivated in DMEM supplemented with 10% fetal bovine serum, 526 mg/liter glutamine, 20,000 U/liter penicillin and 40 mg/liter streptomycin at 37°C, 95% relative humidity and 5% CO2.
Immunofluorescence microscopy
MEFs were stimulated with human chimeric IFN-αB/D (Horisberger and de Staritzky, 1987; 10 ng per ml for 18 h) or were mock treated before fixing with 3% formaldehyde. Cells were permeabilized for 10 min using PBS containing 5% normal donkey serum and 0.1% Triton X-100 before they were incubated with a rabbit anti-Mx serum (Meier et al., 1990) in PBS containing 3% normal donkey serum at 4°C overnight. Goat α-rabbit Cy3 (Jackson ImmunoResearch Laboratories) served as a secondary antibody. Slides were mounted in DAPI-containing IS Mounting Medium (Dianova). Digital images were taken with an ApoTome fluorescence microscope (ZEISS) using AxioVision software.
Virus infection of cell cultures
MEFs were seeded, stimulated the next day with different doses of human chimeric IFN-αB/D (Horisberger and de Staritzky, 1987) for 18 h, and subsequently infected with a MOI of 0.1 of VSV or THOV for 24 and 72 h, respectively. Infections with the influenza A virus strain FPV-B were performed at a MOI of 0.01, and supernatants were harvested at 24 h after infection. Viral titers were determined by plaque assay.
IFN treatment of mice
Human chimeric IFN-αB/D (Horisberger and de Staritzky, 1987) or mouse IFN-λ (IL-28A; PeproTech) were applied i.n. in a volume of 40 µl to ketamin/xylazine-anesthetized animals. Where indicated, IFN was administered in 100 µl into the peritoneal cavity or in 120 µl s.c. on the back of the animals.
RT-qPCR
Reverse transcription was performed using the Revert Aid First Strand cDNA Synthesis kit (Fermentas) as recommended by the manufacturer. QuantiTect SYBR Green PCR kit and commercially available primer pairs (QIAGEN) were used for comparative analysis. ΔCT values were used to characterize expression of target genes compared with housekeeping genes.
Western blot
Protein samples from homogenized cells or tissues were incubated at 95°C in Lämmli buffer and loaded on SDS-PAGE gels. Proteins were blotted on a methanol-activated PVDF membrane (PerfectBlue Semi-Dry electro blotter; PEQLAB). Membranes were blocked in PBS containing 1% milk powder before incubation with primary antibodies (mouse anti-MxA; Flohr et al., 1999) and mouse anti–tubulin-β (Sigma-Aldrich) for 1 h, followed by incubation with horseradish peroxidase–coupled secondary antibody (goat anti–mouse IgG; Jackson ImmunoResearch Laboratories) for 1 h. Bound antibodies were visualized using Super Signal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Mouse infection experiments
All mice used in the study were bred locally in our facility and handled in accordance with guidelines of the Federation for Laboratory Animal Science Associations and the national animal welfare body. Animal experiments were performed in compliance with the German animal protection law and approved by the local animal welfare committee (Regierungspräsidium Freiburg; permit G-12/46). Virus stocks were diluted in PBS. Infection of ketamin/xylazine-anesthetized animals with influenza viruses was performed i.n. with 40 µl inoculum. For infection with THOVs, 100 µl of the virus dilutions were administered into the peritoneal cavity. Mice were monitored daily. If severe illness or weight loss of >25% occurred, mice were sacrificed by cervical dislocation. If required, organs were flash frozen in liquid nitrogen and stored at −80°C until further processing.
Plaque assay
Serial dilutions of cell culture supernatants or homogenized organ samples were placed on MDCK cells for 1 h at room temperature. After inoculum removal, cells were incubated with medium containing 1.5% Avicel for three days at 37°C. Cells were fixed with 4% formaldehyde and plaques were stained using crystal violet.
LD50 determination
The 50% median lethal dose (LD50) values were calculated as described previously (Riegger et al., 2015) by using three to five animals for each virus dilution.
Statistical analysis
Virus titration data of cell culture supernatants are from at least three independent experiments with three technical replicates. Geometric mean values of virus titers in lungs of at least four individual mice were determined. Where indicated, two tailed Student’s t test was used to determine if a difference in virus titers between two groups was significant. Values of P < 0.05 were considered to indicate a significant difference. Statistical analyses were conducted with GraphPad Prism v6.2.
Acknowledgments
We thank Annette Ohnemus for mouse genotyping and other technical support.
This study was supported by a grant from the German Research Foundation (DFG; SFB 1160, project 13) to P. Staeheli and M. Schwemmle. A fellowship from the German Academic Exchange Service (DAAD) supported E. Hassan.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- DHOV
- Dhori virus
- LD50
- lethal dose 50
- MEF
- mouse embryonic fibroblast
- THOV
- Thogoto virus
References
- Albanese M., Bruno-Smiraglia C., Di di Cuonzo G., Lavagnino A., and Srihongse S.. 1972. Isolation of Thogoto virus from Rhipicephalus bursa ticks in western Sicily. Acta Virol. 16:267. [PubMed] [Google Scholar]
- Anderson C.R., and Casals J.. 1973. Dhori virus, a new agent isolated from Hyalomma dromedarii in India. Indian J. Med. Res. 61:1416–1420. [PubMed] [Google Scholar]
- Cauldwell A.V., Long J.S., Moncorgé O., and Barclay W.S.. 2014. Viral determinants of influenza A virus host range. J. Gen. Virol. 95:1193–1210. 10.1099/vir.0.062836-0 [DOI] [PubMed] [Google Scholar]
- Cheung V.G., Nowak N., Jang W., Kirsch I.R., Zhao S., Chen X.N., Furey T.S., Kim U.J., Kuo W.L., Olivier M., et al. BAC Resource Consortium . 2001. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 409:953–958. 10.1038/35057192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M.J., Abel L., Zhang S.Y., and Casanova J.L.. 2016. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr. Opin. Immunol. 38:109–120. 10.1016/j.coi.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann J., Stertz S., Grimm D., Steel J., García-Sastre A., Haller O., and Kochs G.. 2008. Influenza A virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J. Virol. 82:3624–3631. 10.1128/JVI.01753-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohr F., Schneider-Schaulies S., Haller O., and Kochs G.. 1999. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett. 463:24–28. 10.1016/S0014-5793(99)01598-7 [DOI] [PubMed] [Google Scholar]
- Frese M., Kochs G., Meier-Dieter U., Siebler J., and Haller O.. 1995. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J. Virol. 69:3904–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz V., Magar L., Dornfeld D., Giese S., Pohlmann A., Höper D., Kong B.W., Jans D.A., Beer M., Haller O., and Schwemmle M.. 2016. Influenza A viruses escape from MxA restriction at the expense of efficient nuclear vRNP import. Sci. Rep. 6:23138 10.1038/srep23138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Frese M., Rost D., Nuttall P.A., and Kochs G.. 1995. Tick-borne thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J. Virol. 69:2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Staeheli P., Schwemmle M., and Kochs G.. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 23:154–163. 10.1016/j.tim.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Holzinger D., Jorns C., Stertz S., Boisson-Dupuis S., Thimme R., Weidmann M., Casanova J.L., Haller O., and Kochs G.. 2007. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J. Virol. 81:7776–7785. 10.1128/JVI.00546-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisberger M.A., and de Staritzky K.. 1987. A recombinant human interferon-alpha B/D hybrid with a broad host-range. J. Gen. Virol. 68:945–948. 10.1099/0022-1317-68-3-945 [DOI] [PubMed] [Google Scholar]
- Israël A. 1979. Preliminary characterization of the particles from productive and abortive infections of L cells by fowl plague virus. Ann. Microbiol. (Paris). 130B:85–100. [PubMed] [Google Scholar]
- Iwasaki A., and Pillai P.S.. 2014. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14:315–328. 10.1038/nri3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G., and Haller O.. 1999. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl. Acad. Sci. USA. 96:2082–2086. 10.1073/pnas.96.5.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mänz B., Dornfeld D., Götz V., Zell R., Zimmermann P., Haller O., Kochs G., and Schwemmle M.. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 9:e1003279 10.1371/journal.ppat.1003279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier E., Kunz G., Haller O., and Arnheiter H.. 1990. Activity of rat Mx proteins against a rhabdovirus. J. Virol. 64:6263–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.S., Patzina C., Emerman M., Haller O., Malik H.S., and Kochs G.. 2012. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe. 12:598–604. 10.1016/j.chom.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.S., Emerman M., and Malik H.S.. 2013. An evolutionary perspective on the broad antiviral specificity of MxA. Curr. Opin. Microbiol. 16:493–499. 10.1016/j.mib.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., and Kawaoka Y.. 2015. Transmission of influenza A viruses. Virology. 479-480:234–246. 10.1016/j.virol.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg P.E., and Pavlovic J.. 2015. Oligomerization and GTP-binding Requirements of MxA for Viral Target Recognition and Antiviral Activity against Influenza A Virus. J. Biol. Chem. 290:29893–29906. 10.1074/jbc.M115.681494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzina C., Haller O., and Kochs G.. 2014. Structural requirements for the antiviral activity of the human MxA protein against Thogoto and influenza A virus. J. Biol. Chem. 289:6020–6027. 10.1074/jbc.M113.543892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic J., Zürcher T., Haller O., and Staeheli P.. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64:3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic J., Arzet H.A., Hefti H.P., Frese M., Rost D., Ernst B., Kolb E., Staeheli P., and Haller O.. 1995. Enhanced virus resistance of transgenic mice expressing the human MxA protein. J. Virol. 69:4506–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegger D., Hai R., Dornfeld D., Mänz B., Leyva-Grado V., Sánchez-Aparicio M.T., Albrecht R.A., Palese P., Haller O., Schwemmle M., et al. . 2015. The nucleoprotein of newly emerged H7N9 influenza A virus harbors a unique motif conferring resistance to antiviral human MxA. J. Virol. 89:2241–2252. 10.1128/JVI.02406-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., and Williams B.R.. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568. 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V., Bloch N., and Landau N.R.. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 16:546–553. 10.1038/ni.3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeheli P., and Haller O.. 1985. Interferon-induced human protein with homology to protein Mx of influenza virus-resistant mice. Mol. Cell. Biol. 5:2150–2153. 10.1128/MCB.5.8.2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeheli P., Grob R., Meier E., Sutcliffe J.G., and Haller O.. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518–4523. 10.1128/MCB.8.10.4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimme R., Frese M., Kochs G., and Haller O.. 1995. Mx1 but not MxA confers resistance against tick-borne Dhori virus in mice. Virology. 211:296–301. 10.1006/viro.1995.1404 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Watanabe S., Maher E.A., Neumann G., and Kawaoka Y.. 2014. Pandemic potential of avian influenza A (H7N9) viruses. Trends Microbiol. 22:623–631. 10.1016/j.tim.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.W., Webby R.J., and Webster R.G.. 2014. Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol. 385:359–375. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Mänz B., Haller O., Schwemmle M., and Kochs G.. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J. Virol. 85:8133–8140. 10.1128/JVI.00712-11 [DOI] [PMC free article] [PubMed] [Google Scholar]