Kitada et al. demonstrate that transcription factor BATF2 induced by IFN-γ in macrophages and dendritic cells prevents Th17-mediated multiorgan pathology through suppression of IL-23 production during T. cruzi infection.
Abstract
Inappropriate IL-17 responses are implicated in chronic tissue inflammation. IL-23 contributes to Trypanosoma cruzi–specific IL-17 production, but the molecular mechanisms underlying regulation of the IL-23–IL-17 axis during T. cruzi infection are poorly understood. Here, we demonstrate a novel function of BATF2 as a negative regulator of Il23a in innate immune cells. IL-17, but not IFN-γ, was more highly produced by CD4+ T cells from spleens and livers of T. cruzi–infected Batf2−/− mice than by those of wild-type mice. In this context, Batf2−/− mice showed severe multiorgan pathology despite reduced parasite burden. T. cruzi–induced IL-23 production was increased in Batf2−/− innate immune cells. The T. cruzi–induced enhanced Th17 response was abrogated in Batf2−/−Il23a−/− mice. The interaction of BATF2 with c-JUN prevented c-JUN–ATF-2 complex formation, inhibiting Il23a expression. These results demonstrate that IFN-γ–inducible BATF2 in innate immune cells controls Th17-mediated immunopathology by suppressing IL-23 production during T. cruzi infection.
Introduction
Chagas disease, characterized by chronic cardiomyopathy, is caused by infection with the intracellular protozoan parasite Trypanosoma cruzi (Bonney et al., 2015). The TLR-MyD88/TRIF adaptor molecule pathways initiate both innate and adaptive immune responses, and thus contribute to the resistance of the host to intracellular protozoan parasites such as T. cruzi (Gazzinelli and Denkers, 2006; Kayama and Takeda, 2010; Kawai and Akira, 2011; Rodrigues et al., 2012). Recently, several studies have demonstrated the mechanisms underlying the TLR-independent host defense against T. cruzi infection, such as TLR-independent T-helper 1 (Th1) responses (Kayama et al., 2009).
Th17 cells, which produce IL-17A, IL-17F, IL-22, and GM-CSF, play essential roles in the immune response to infection (Miyazaki et al., 2010; McGeachy and McSorley, 2012; Bermejo et al., 2013). Th17 cell differentiation is induced by IL-6 and TGF-β, and IL-23 mediates the enhanced production of Th17 cell–related cytokines (Gaffen et al., 2014). IL-17A–producing CD4+ T cells are greatly increased in mice infected with T. cruzi (da Matta Guedes et al., 2010), and a lack of IL-17A is linked to the aggravation of the parasite burden and the failure of various organs after T. cruzi infection (da Matta Guedes et al., 2010; Miyazaki et al., 2010). These findings indicate that adequate IL-17 responses are required for host protection, but that enhanced IL-17 production can cause tissue immunopathology during infection with intracellular protozoan parasites. Several studies have demonstrated that the Th17 response, particularly induction of Th17 cell development, is tightly regulated by several mechanisms during the course of parasite infection. For example, the IL-27/WSX-1 signaling pathway plays an important role in the negative regulation of IL-17 production by CD4+ T cells during T. cruzi and Toxoplasma gondii infection (Stumhofer et al., 2006; Yoshimura et al., 2006), and the T cell-intrinsic transcription factor T-bet, encoded by Tbx21, contributes to the inhibition of T. cruzi–specific Th17 cell differentiation (Guo et al., 2009). IL-12p35 also down-regulates Th17 responses to T. cruzi (Cobb and Smeltz, 2012). However, it remains unclear how Th17 cell responses are controlled after induction.
IL-23, a heterodimer of the IL-23p19 and IL-12p40 subunits, is essential for the generation of pathogenic Th17 cells (McGeachy et al., 2009; Gaffen et al., 2014). In experimental autoimmune myocarditis (EAM), IL-23 functions as a key effector molecule by promoting the production of IL-17 by lymphocytes (Rangachari et al., 2006). IL-23 is also implicated in the development of colitis by stimulating the accumulation of Th17 cells (Ahern et al., 2010). In contrast, another study has shown that induction of histone modification by IFN-γ was linked to the suppression of Il23a expression, which encodes IL-23p19, in intestinal CD11b+ Mφs, thus preventing colitis (Sheikh et al., 2010). The IFN-γ released by CD8+ T cells also suppresses the development of EAM through inhibition of IL-17 production (Rangachari et al., 2006). These findings indicate that the IL-23–Th17 axis is tightly regulated by IFN-γ in a variety of contexts. T. cruzi infection induces IL-23 production by host immune cells, and antigen-specific Th17 responses are then promoted (Cobb et al., 2010; Erdmann et al., 2013). However, whether the IL-23–Th17 axis is controlled by IFN-γ–dependent mechanisms during T. cruzi infection remains unclear.
Transcription factor BATF2 was initially identified as an AP-1 inhibitor (Su et al., 2008). In cancer cells, BATF2 suppresses the expression of AP-1–dependent genes through its interaction with c-JUN (Su et al., 2008). In contrast, BATF2 functions as a transcriptional activator in BM-derived macrophages (Mφs [BMMφs]) by interacting with IRF1 in response to IFN-γ and the TLR4 ligand LPS (Roy et al., 2015). BATF2 also compensates CD103+ DC development in Batf3−/− mice (Tussiwand et al., 2012). We previously demonstrated that BATF2 is normally induced in Myd88−/−Trif−/− BM-derived DCs (BMDCs) during T. cruzi infection (Kayama et al., 2009). However, the roles of IFN-γ–inducible BATF2 in T. cruzi–infected innate immune cells are poorly understood.
In this study, we demonstrate the mechanism by which BATF2 regulates T. cruzi–specific IL-17 production by CD4+ T cells. IFN-γ–inducible BATF2 inhibits Il23a expression in DCs and Mφs during T. cruzi infection by disrupting the formation of the c-JUN–ATF-2 complex by directly binding to c-JUN, thus preventing Th17-mediated tissue damage during T. cruzi infection. Therefore, BATF2-mediated modulation of the IL-23–Th17 axis is involved in host resistance to T. cruzi infection.
Results
IFN-γ–induced expression of BATF2
We previously demonstrated the TLR-independent expression of 4933430F08Rik, encoding BATF2, in T. cruzi–infected BMDCs (Kayama et al., 2009). To assess the physiological role of BATF2 during T. cruzi infection, we generated Batf2−/− mice with gene targeting (Fig. S1, A and B). Stimulation with LPS plus IFN-γ robustly induced BATF2 mRNA in wild-type BMMφs, but not Batf2−/− BMMφs (Fig. S1 C). In wild-type BMMφs, IFN-γ, but not LPS, induced high levels of Batf2 expression (Fig. S1 D). The expression of the mRNAs of other BATF family members, such as BATF and BATF3, was not induced by IFN-γ in BMMφs (Fig. S1 E). Moreover, BATF2 deficiency did not lead to the compensatory up-regulation of Batf and Batf3 expression, as previously described (Roy et al., 2015; Fig. S1 E). A subset of TLR-independent genes shown to be expressed in Myd88−/−Trif−/− BMDCs during T. cruzi infection (Kayama et al., 2009) was normally induced in Batf2−/− BMMφs stimulated with IFN-γ (Fig. S1 F). These results indicate that BATF2 is not involved in the expression of a subset of IFN-γ–inducible genes.
To determine whether BATF2 is responsible for immune cell development, we analyzed the composition of both lymphocytes and innate myeloid cells in the spleens of wild-type and Batf2−/− mice (Fig. S1 G). The frequencies and total cell numbers of the CD11c+ CD103+, CD11b+ Ly-6C+, CD11b+ Ly-6G+, CD3+ CD4+, CD3+ CD8+, and B220+ CD19+ cell populations were not altered in the Batf2−/− mice, demonstrating that the development of neither innate nor adaptive immune cells was affected in Batf2−/− mice.
Increased production of IL-17, but not IFN-γ, by CD4+ T cells in T. cruzi–infected Batf2−/− mice
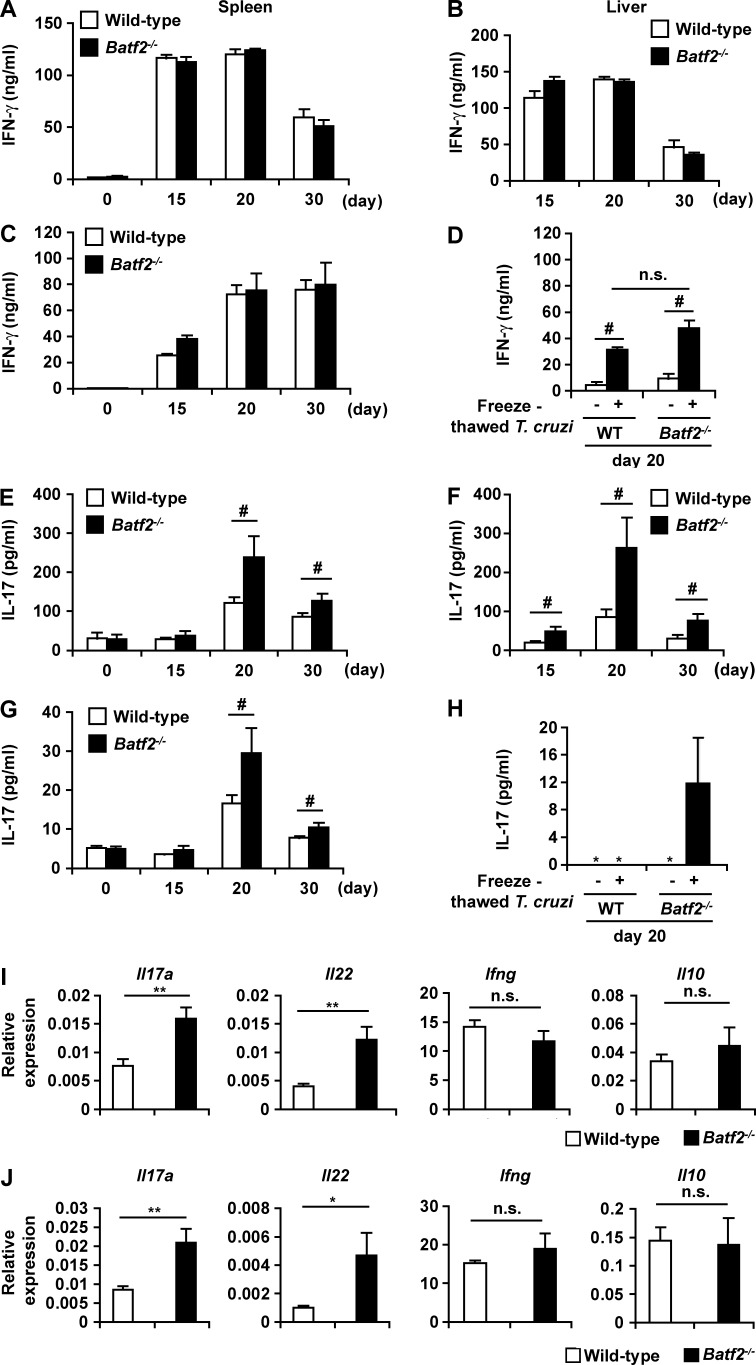
The TLR-independent production of IFN-γ by innate immune cells is implicated in driving Th1 responses during T. cruzi infection (Kayama et al., 2009). Because BATF2 was shown to be induced by a TLR-independent IFN-γ–dependent mechanism, we analyzed whether BATF2 is responsible for the induction of Th1 responses during T. cruzi infection. Wild-type and Batf2−/− mice were intraperitoneally infected with T. cruzi trypomastigotes. CD4+ T cells were isolated from their spleens 0, 15, 20, and 30 d after infection, whereas liver CD4+ T cells were isolated 15, 20, and 30 d after infection because there were fewer of these cells in the uninfected mice. The isolated CD4+ T cells were stimulated with anti-CD3 antibody (Fig. 1 A). The spleen CD4+ T cells of T. cruzi–infected wild-type mice showed considerably higher production of IFN-γ at 15, 20, and 30 d of infection than the uninfected mice, indicating induction of an intensive Th1 response. IFN-γ production by the spleen CD4+ T cells of T. cruzi–infected Batf2−/− mice was similar to that of wild-type mice. Furthermore, no difference in IFN-γ production was observed in liver CD4+ T cells of the wild-type and Batf2−/− mice after T. cruzi infection (Fig. 1 B). We next analyzed antigen-specific Th1 responses after infection by stimulating CD4+ T cells with freeze-thawed T. cruzi in the presence of antigen-presenting cells (Fig. 1, C and D). IFN-γ production by the Batf2−/− CD4+ T cells from the spleen and liver at any time point of infection was similar to that by wild-type CD4+ T cells, indicating that the Th1 responses during T. cruzi infection were not impaired in Batf2−/− mice.
Figure 1.
Increased production of IL-17 in CD4+ T cells in T. cruzi–infectedBatf2−/− mice. (A–H) CD4+ T cells were isolated from the spleens (left) and livers (right) of wild-type (WT) and Batf2−/− mice at 0 d (n = 5 per group), 15 d (n = 6 per group), 20 d (n = 8 per group), and 30 d (n = 5 per group) of T. cruzi infection. Isolated CD4+ T cells were stimulated with anti-CD3 antibody (A, B, E, and F) or heat-killed T. cruzi (C, D, G, and H) and analyzed for IFN-γ and IL-17 with ELISA. Graphs show mean values ± SEM. *, not detected; #, P < 0.05; n.s., not significant. (I and J) Expression of Il17a, Il22, Il10, and Ifng in CD4+ T cells was isolated from spleens (I) and livers (J) of wild-type (n = 5) and Batf2−/− (n = 4) mice after T. cruzi infection for 20 d (mean values ± SEM). *, P < 0.05; **, P < 0.01; n.s., not significant.
Recent studies have shown that IL-17 is responsible for host resistance to T. cruzi infection (da Matta Guedes et al., 2010; Miyazaki et al., 2010; Bermejo et al., 2013). Therefore, we examined IL-17 production by CD4+ T cells in the spleens and livers of T. cruzi–infected mice. More IL-17 was produced by CD4+ T cells from the spleens of T. cruzi–infected wild-type mice stimulated with anti-CD3 antibody than from uninfected mice (Fig. 1 E). These findings indicate that T. cruzi infection causes the induction of Th17 immune responses. At 20- and 30-d time points of infection, IL-17 production by CD4+ T cells stimulated with anti-CD3 antibody was considerably higher in the spleens of T. cruzi–infected Batf2−/− mice than in those of wild-type mice (Fig. 1 E). In the liver, IL-17 production by anti-CD3 antibody–stimulated Batf2−/− CD4+ T cells was greatly increased at 15, 20, and 30 d of infection compared with wild-type cells (Fig. 1 F). Furthermore, antigen-specific IL-17 production was markedly higher after T. cruzi infection in Batf2−/− spleen and liver CD4+ T cells than in the corresponding wild-type cells (Fig. 1, G and H). Intracellular staining analysis also showed that production of IL-17, but not IFN-γ and IL-10, was increased in CD4+ T cells from the spleens and livers of T. cruzi–infected Batf2−/− mice compared with those from wild-type mice, as indicated by increased mean fluorescence intensity of IL-17 (Fig. S2, A, B, D, and E). In addition, a higher frequency of IFN-γ+ IL-17+ CD4+ T cells was observed in the spleens, but not livers, of Batf2−/− mice (Fig. S2, C and F). Expression of Th17-related genes, such as Il17 and Il22, was markedly higher in CD4+ T cells from the spleens and livers of T. cruzi–infected Batf2−/− mice than in the corresponding cells from wild-type mice (Fig. 1, I and J), whereas expression of Ifng and Il10 in Batf2−/− CD4+ T cells was similar to that in wild-type cells. These findings indicate that BATF2 modulates Th17 responses during T. cruzi infection.
Increased multiorgan pathology in T. cruzi–infected Batf2−/− mice
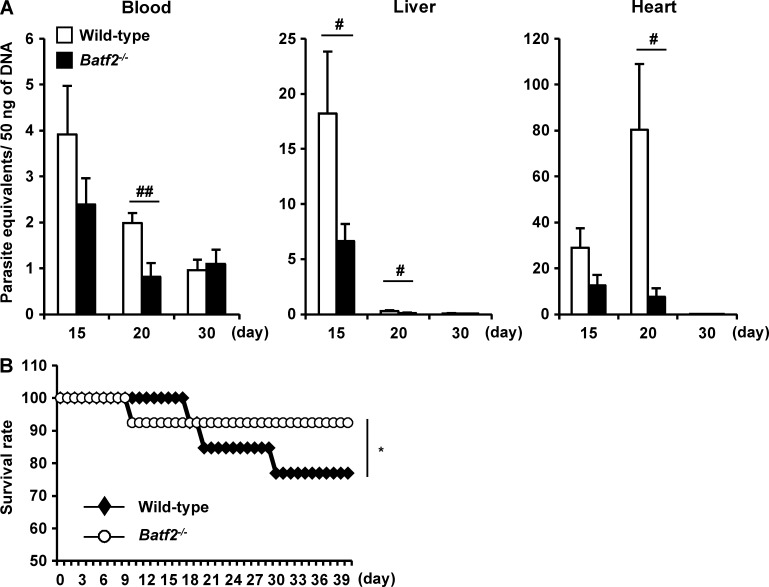
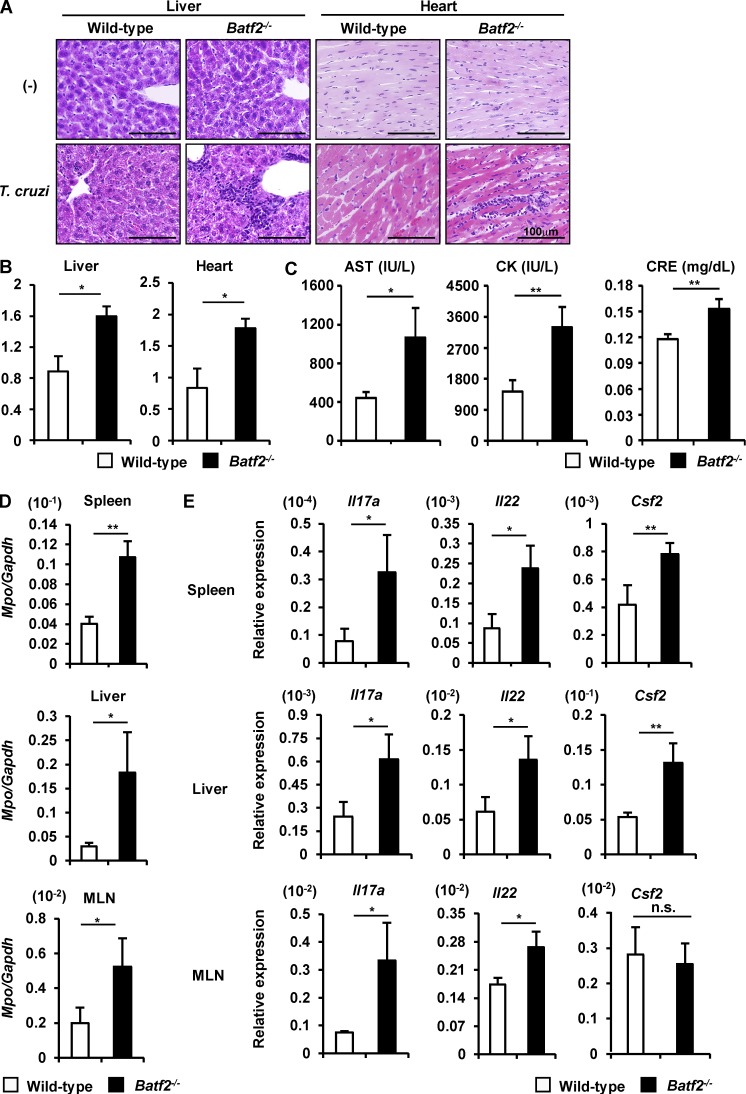
We next assessed parasitemia levels in the blood, livers, and hearts of wild-type and Batf2−/− mice after infection, using quantitative PCR (qPCR) assay to measure the amounts of T. cruzi DNA (Fig. 2 A). In the blood and livers of T. cruzi–infected wild-type mice, the parasite number increased as early as 15 d and then gradually decreased. In T. cruzi–infected Batf2−/− mice, the parasite number was reduced, particularly at 20 d of infection, compared with wild-type mice. In the hearts of T. cruzi–infected wild-type mice, the parasite number increased slowly and reached peak levels at 20 d. The parasite number in the hearts was dramatically reduced in Batf2−/− mice. In accordance with the reduced parasite burden, Batf2−/− mice showed higher survival rates compared with wild-type mice after T. cruzi infection (Fig. 2 B). We also analyzed pathological changes (Fig. 3, A and B) in the livers and hearts of T. cruzi–infected wild-type and Batf2−/− mice. There was greater hepatic inflammation and myocarditis in Batf2−/− mice after T. cruzi infection than in wild-type mice. We then measured the concentrations of markers for tissue damage: aspartate aminotransferase (AST) in the plasma, which is indicative of liver damage; creatinine kinase (CK) in the plasma, indicative of heart failure; and creatinine (CRE) in the serum, indicative of renal failure (Miyazaki et al., 2010; Fig. 3 C). AST, CK, and CRE were markedly higher in T. cruzi–infected Batf2−/− mice than in wild-type mice. The expression of Mpo, which encodes myeloperoxidase, a tissue-damaging factor produced by neutrophils, was elevated in the spleens, livers, and mesenteric LNs (MLNs) of T. cruzi–infected Batf2−/− mice (Fig. 3 D). We also analyzed the expression of Th17-related genes, such as Il17a, Il22, and Csf2, in the spleens, livers, and MLNs of wild-type and Batf2−/− mice after T. cruzi infection (Fig. 3 E). Expression of these Th17-related genes was extremely enhanced in all tissues of Batf2−/− mice compared with those of wild-type mice. Thus, enhanced Th17 responses correlated with heightened immune activation and tissue damage, despite reduced parasite numbers and mortality, in T. cruzi–infected Batf2−/− mice.
Figure 2.
Lower levels of parasitemia in T. cruzi–infected Batf2−/− mice. (A) Parasite equivalents in the livers, hearts, and blood of wild-type and Batf2−/− mice 15, 20, and 30 d after infection (n ≥ 5). #, P < 0.05; ##, P < 0.009. Data are mean values ± SEM. (B) Mortality of wild-type and Batf2−/− mice after T. cruzi infection. Line graphs represent data pooled from three independent experiments (n = 13). *, P = 0.036.
Figure 3.
Severe multiorgan pathology in T. cruzi–infected Batf2−/− mice. (A and B) Representative histopathology (A) and histopathological score (B) of wild-type (n = 8) and Batf2−/− (n = 9) mice 49 d after infection. Bars, 100 µm. Graphs show mean values ± SD. *, P < 0.01. (C) Concentrations of AST and CK in the plasma and CRE in tge sera of T. cruzi–infected wild-type (n ≥ 14) and Batf2−/− (n ≥ 19) mice. *, P < 0.05; **, P < 0.01. Graphs are mean values ± SEM. (D) Mpo expression in tissues of wild-type and Batf2−/− mice after T. cruzi infection (n = 3 per group). *, P < 0.05; **, P < 0.02. Data are mean values ± SEM. (E) Expression of Th17-related genes (n = 4 per group). *, P < 0.05; **, P < 0.02; n.s., not significant. All graphs show mean values ± SEM.
Increased LPS-induced IL-23 production by Batf2−/− innate immune cells
To determine the mechanisms by which BATF2 modulates Th17 responses during T. cruzi infection, we first examined whether T cell-intrinsic BATF2 is involved in either the generation of Th17 cells or the production of IL-17 by CD4+ T cells. Naive CD4+ T cells isolated from the spleens of wild-type and Batf2−/− mice were cultured under Th0, Th1, or Th17 conditions for 4 d and then analyzed for IFN-γ and IL-17 production with intracellular staining and ELISA. The frequency of IL-17–producing cells among Batf2−/− CD4+ T cells cultured under Th17 conditions was similar to that in wild-type cells (Fig. S3 A). Moreover, IFN-γ–producing Batf2−/− CD4+ T cells maintained under Th1 conditions were induced to levels similar to those in wild-type cells. There was no significant difference in the secretion of IL-17 or IFN-γ by wild-type and Batf2−/− CD4+ T cells stimulated with anti-CD3 antibody (Fig. S3 B), indicating that T cell–intrinsic BATF2 does not affect the differentiation of Th1 and Th17 cells or their cytokine production.
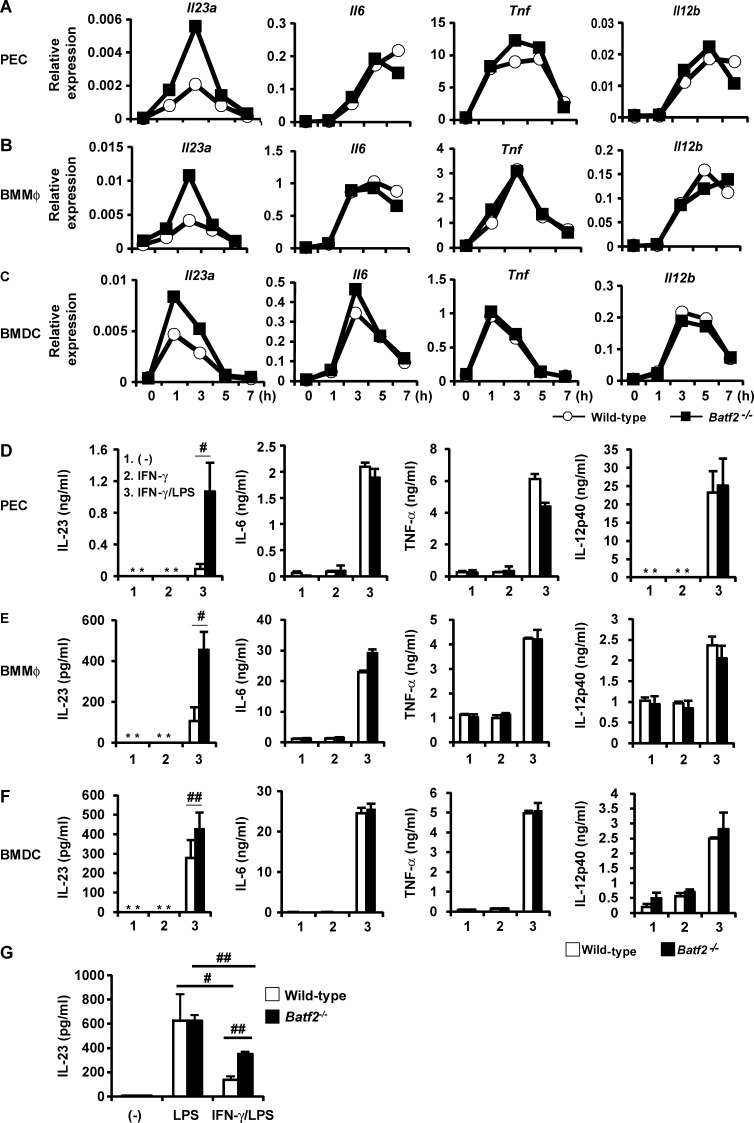
Accumulating evidence has shown that the Th17 responses are tightly controlled by the innate immune cells (Gaffen et al., 2014). Therefore, to determine the functions of BATF2 in innate immune responses, we comprehensively analyzed the overall gene expression patterns in wild-type and Batf2−/− BMMφs. RNA-seq analysis showed that 98 genes are up-regulated in Batf2−/− BMMφs stimulated with LPS and IFN-γ relative to their expression in wild-type cells (Table S1). Among these genes, we focused on Il23a, which encodes IL-23p19, because IL-23 promotes the expression of Il17a in Th17 cells (Gaffen et al., 2014). To confirm the results of the RNA-seq analysis, we examined LPS-induced expression of Il23a, Il6, Tnf, and Il12b in IFN-γ–pretreated innate immune cells prepared from wild-type and Batf2−/− mice, using real-time RT-PCR. The expression of Il23a, but not Il6, Tnf, or Il12b, was dramatically higher in Batf2−/− Mφs, such as peritoneal Mφs (PECs) and BMMφs, than in wild-type cells (Fig. 4, A and B). Batf2−/− BMDCs also showed higher expression of Il23a than wild-type BMDCs (Fig. 4 C). Because an increase in Il23a expression was observed in Batf2−/− innate immune cells, we next analyzed the LPS-induced production of proinflammatory cytokines in wild-type and Batf2−/− innate immune cells pretreated with IFN-γ. Consistent with the mRNA expression patterns, the production of IL-23 by IFN-γ–pretreated Batf2−/− PECs, BMMφs, and BMDCs was markedly higher than that by wild-type cells, whereas IL-6, TNF, and IL-12p40 were normally produced by the Batf2−/− cells (Fig. 4, D–F). We also analyzed LPS-induced cytokine production by IFN-γ–untreated innate immune cells. IFN-γ–untreated Batf2−/− and wild-type PECs, BMMφs, and BMDCs produced similar amounts of IL-23 as well as IL-6, TNF, and IL-12p40 in response to LPS (Fig. S4, A–C). LPS-induced Il23a expression is negatively regulated by an IFN-γ–dependent mechanism in Mφs (Sheikh et al., 2010). Therefore, we evaluated the LPS-induced production of IL-23 at the mRNA and protein levels in wild-type and Batf2−/− BMMφs pretreated with or without IFN-γ (Fig. 4 G and Fig. S4 D). LPS stimulation robustly induced IL-23 expression in the wild-type BMMφs, whereas IFN-γ pretreatment dramatically reduced this LPS-induced IL-23 expression. In the Batf2−/− BMMφs not pretreated with IFN-γ, the expression of IL-23 in response to LPS was similar to that in wild-type BMMφs, but the IFN-γ–induced reduction in IL-23 expression was lower in Batf2−/− cells stimulated with LPS than in stimulated wild-type cells. To investigate whether the newly synthesized proteins induced by IFN-γ are involved in the inhibition of LPS-induced Il23a expression in innate immune cells, BMMφs were pretreated with IFN-γ in the presence or absence of cycloheximide (CHX), an inhibitor of protein synthesis, and then stimulated with LPS (Fig. S4 E). Compared with control cells, LPS-induced Il23a expression was greatly increased in CHX-treated cells. Collectively, these findings suggest that IFN-γ–induced BATF2 contributes to the suppression of the LPS-dependent expression of Il23a, but not Il6, Tnf, or Il12b, in innate immune cells.
Figure 4.
Increased IL-23 production by Batf2−/− innate immune cells stimulated with LPS after IFN-γ treatment. (A–C) Expression of Il23a, Il6, Tnf, and Il12b in wild-type and Batf2−/− PECs (A), BMMφs (B), and BMDCs (C). All data are representative of at least two independent experiments. (D–F) Production of IL-23, IL-6, IL-12p40, and TNF by wild-type and Batf2−/− PECs (D), BMMφs (E), and BMDCs (F). Results are mean ± SEM from four independent experiments. *, not detected; #, P < 0.03; ##, P < 0.05. (G) IL-23 production by wild-type and Batf2−/− BMMφs. Data are mean ± SEM from three independent experiments. #, P < 0.05; ##, P < 0.001.
BATF2-mediated suppression of IL-23 production by innate immune cells during T. cruzi infection
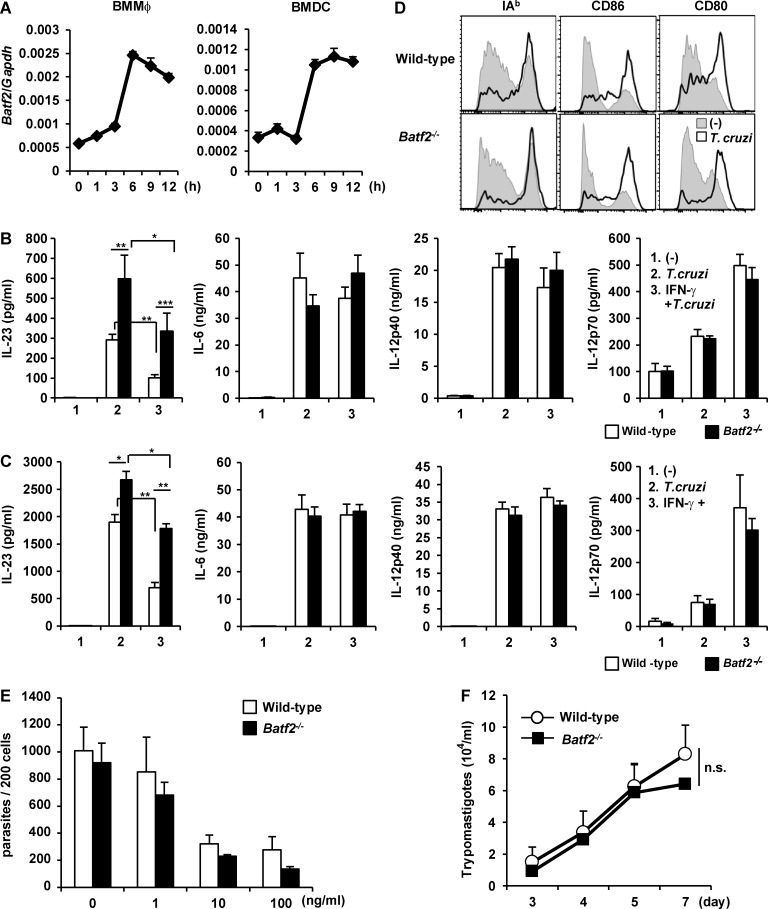
In the next experiment, we analyzed the effect of BATF2 on T. cruzi–induced IL-23 production. Batf2 expression was highest 6 h after T. cruzi infection, and this was maintained in both BMMφs and BMDCs even after 12 h of infection (Fig. 5 A). BMMφs and BMDCs prepared from wild-type and Batf2−/− mice were pretreated with or without IFN-γ for 4 h, exhaustively washed, and then infected with T. cruzi. At 24 h after infection, the culture supernatants were analyzed for IL-23, IL-6, IL-12p40, and IL-12p70 with ELISA (Fig. 5, B and C). Even in the absence of IFN-γ pretreatment, Batf2−/− cells showed higher levels of T. cruzi–mediated IL-23 production than wild-type cells. The enhanced T. cruzi–dependent response was specific to IL-23, because IL-6, IL-12p40, and IL-12p70 production and expression of I-Ab, CD80, and CD86 were normally increased in T. cruzi–infected Batf2−/− BMDCs (Fig. 5, C and D). IFN-γ pretreatment caused reduced production of IL-23, but not IL-6, IL-12p40, and IL-12p70, in T. cruzi–infected wild-type cells (Fig. 5, B and C). However, only a partial reduction in IL-23 production was observed in Batf2−/− cells. To investigate the involvement of BATF2 in the replication of T. cruzi within innate immune cells, BMMφs prepared from wild-type and Batf2−/− mice were pretreated with or without IFN-γ and infected with T. cruzi. At 6 h after infection, cells were thoroughly washed and changed to fresh media. At 72 h of culture, intracellular T. cruzi numbers in the wild-type BMMφs were decreased in an IFN-γ dose–dependent manner (Fig. 5 E). The numbers of intracellular parasites in Batf2−/− BMMφs were similarly reduced as IFN-γ concentration increased. Moreover, the numbers of trypomastigotes desorbed into the culture supernatant of the Batf2−/− BMMφs were similar to those of wild-type cells at 3, 4, 5, and 7 d after T. cruzi infection (Fig. 5 F). These findings indicate that BATF2 is required for the down-regulation of T. cruzi–induced IL-23 production in innate immune cells, but not for the killing of T. cruzi.
Figure 5.
Increased IL-23 production by T. cruzi–infected Batf2−/− innate immune cells. (A) Expression of Batf2 in BMMφs and BMDCs after T. cruzi infection. All data are representative of two independent experiments (mean values ± SD). (B and C) Production of IL-23, IL-12p40, IL-12p70, and IL-6 by T. cruzi–infected wild-type and Batf2−/− BMMφs (B) and BMDCs (C). Data are mean ± SEM from four independent experiments. *, P < 0.05; **, P < 0.03; ***, P < 0.01. (D) Surface expression of I-Ab, CD86, and CD80 on BMDCs after T. cruzi infection for 36 h. All data are representative of three independent experiments. (E) Parasite number per 200 BMMφs prepared from wild-type and Batf2−/− mice. (F) Numbers of trypomastigotes in the culture supernatants. (E and F) Graphs show the mean values ± SEM of three independent experiments. n.s., not significant.
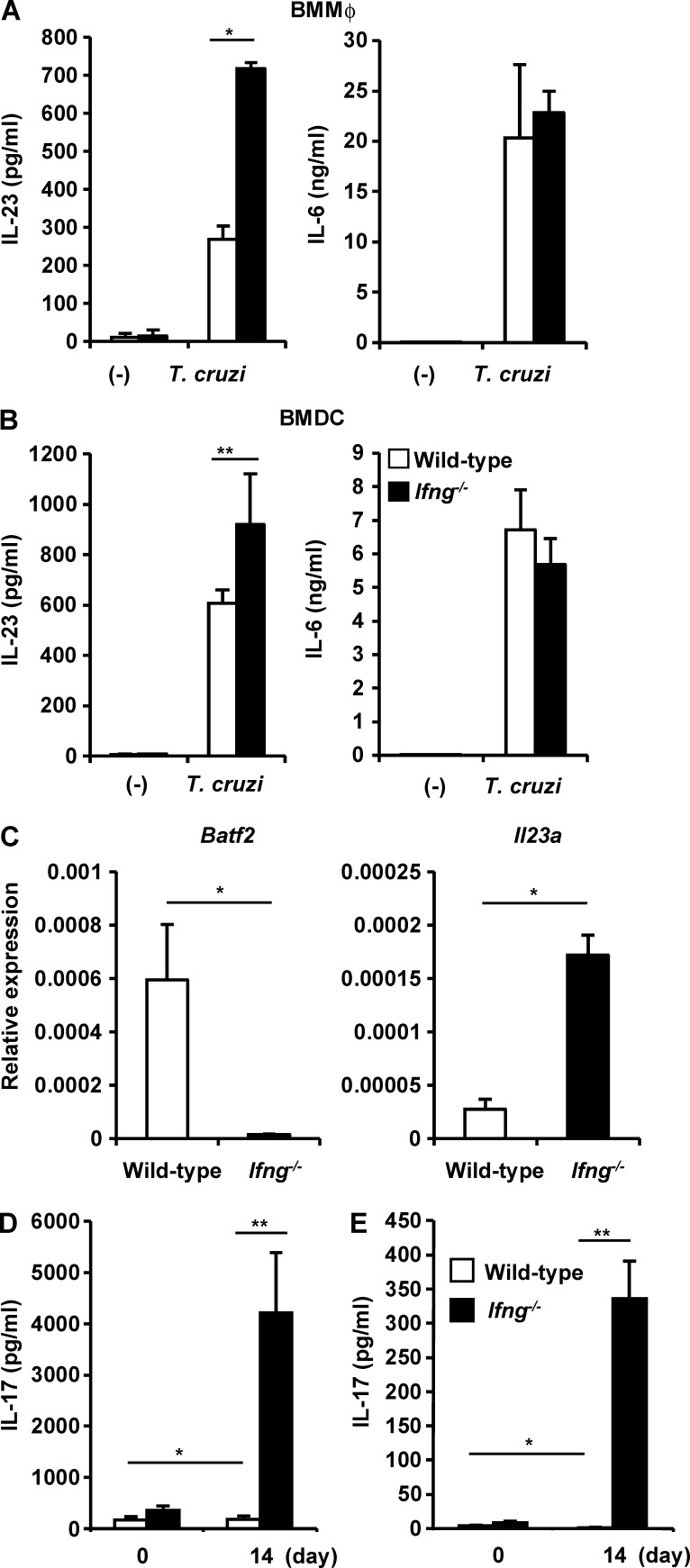
We then analyzed whether IFN-γ is responsible for the suppression of IL-23 production by T. cruzi–infected innate immune cells. BMMφs and BMDCs prepared from wild-type and Ifng−/− mice were infected with T. cruzi and cultured for 24 h. The culture supernatants were then analyzed for IL-23 and IL-6 (Fig. 6, A and B). Ifng−/− BMMφs showed markedly greater production of IL-23, but not IL-6, than wild-type cells. Ifng−/− BMDCs also released higher levels of IL-23 after T. cruzi infection than wild-type cells. CD11b+ F4/80+ Mφs isolated from the spleens of wild-type and Ifng−/− mice 14 d after T. cruzi infection were analyzed for the expression of Batf2 and Il23a (Fig. 6 C). In the CD11b+ F4/80+ Mφs from the spleens of Ifng−/− mice, Batf2 expression was severely reduced compared with that in the wild-type cells. However, the expression of Il23a was higher in the Ifng−/− Mφs than in the wild-type Mφs. IL-17 production by CD4+ T cells isolated from the spleens of wild-type and Ifng−/− mice infected with or without T. cruzi was evaluated (Fig. 6, D and E). IL-17 production by CD4+ T cells stimulated with anti-CD3 antibody was markedly higher in Ifng−/− mice at 14 d after infection than in wild-type mice (Fig. 6 D). Antigen-specific IL-17 production was also significantly higher in CD4+ T cells from Ifng−/− mice than in those from wild-type mice (Fig. 6 E). These findings demonstrate that IFN-γ mediates the suppression of Il23a expression in innate immune cells and the down-regulation of IL-17 production by CD4+ T cells during T. cruzi infection. Because Mφs IL-23 production and T cell IL-17 production were similarly increased in Ifng−/− and Batf2−/− mice, IFN-γ–induced BATF2 expression is supposed to be responsible for the suppression of the Th17 responses during T. cruzi infection.
Figure 6.
Increased expression of Il23a in Mφs and Th17 responses in T. cruzi–infected Ifng−/− mice. (A and B) Production of IL-23 and IL-6 by BMMφs (A) and BMDCs (B). Graphs show mean values ± SEM of four independent experiments. *, P < 0.003; **, P < 0.05. (C) Expression of Batf2 and Il23a in CD11b+ F4/80+ Mφs from the spleens of wild-type and Ifng−/− mice 14 d after infection (n = 3 per group). Mean values ± SD. *, P < 0.01. (D and E) IL-17 production by CD4+ T cells from the spleens of T. cruzi–infected mice (n = 4) or uninfected mice (n = 3) stimulated with anti-CD3 antibody (D) or freeze-thawed T. cruzi (E). Mean values ± SEM. *, P < 0.05; **, P < 0.001.
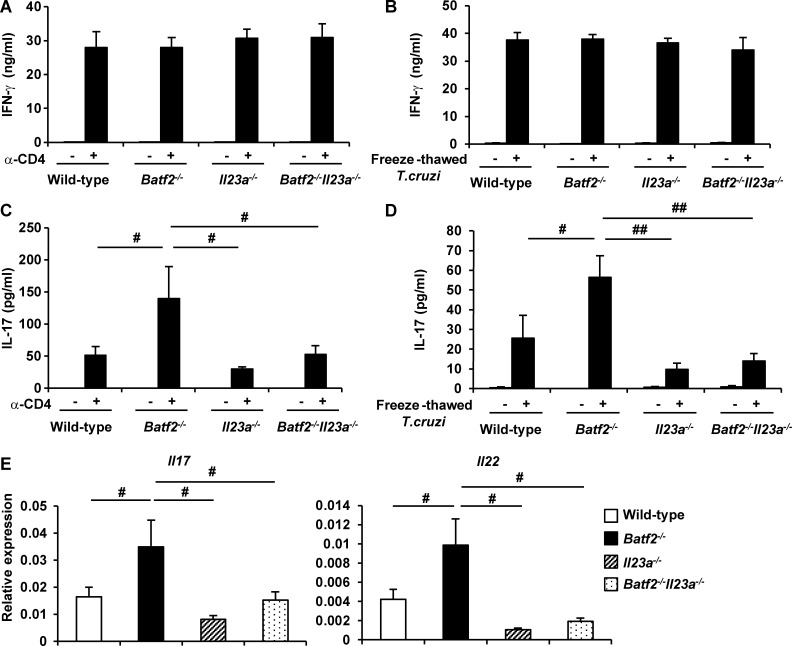
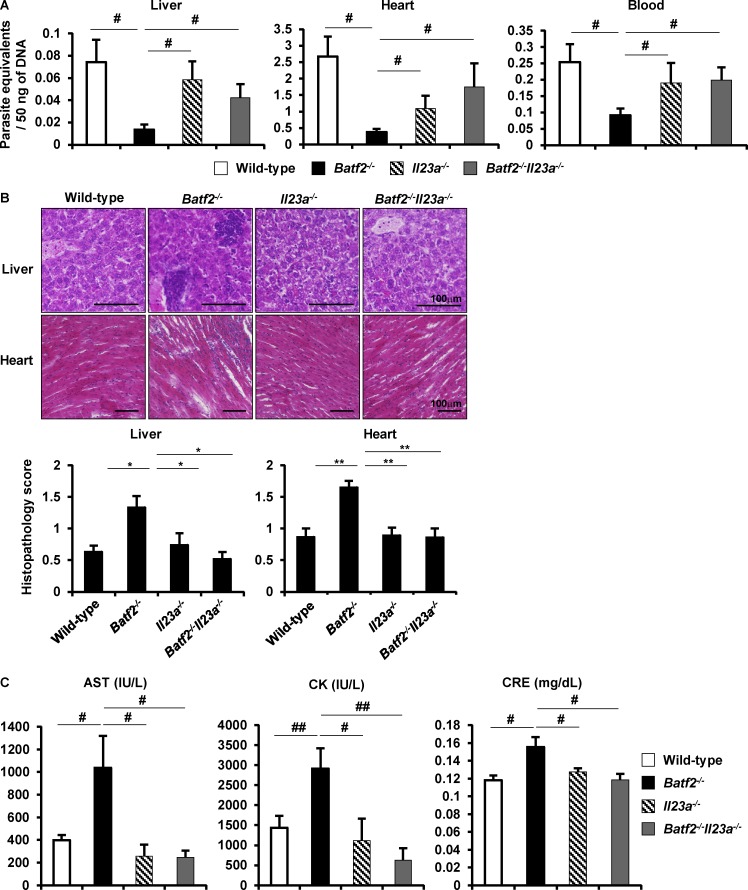
To further determine whether the higher levels of IL-23 production in innate immune cells are involved in the enhanced Th17 responses in T. cruzi–infected Batf2−/− mice, we generated Il23a−/− mice by the Cas9/CRISPR-mediated genome editing (Fig. S5 A). BMMφs prepared from Il23a−/− mice did not produce IL-23 in response to LPS (Fig. S5 B), indicating the successful generation of Il23a−/− mice by the genome editing method. Il23a−/− mice were used for generation of Batf2−/−Il23a−/− mice. Wild-type, Batf2−/−, Il23a−/−, and Batf2−/−Il23a−/− mice were infected i.p. with T. cruzi. CD4+ T cells isolated from their spleens 20 d after infection were analyzed for production of IFN-γ and IL-17 (Fig. 7, A–D). There was no difference in IFN-γ production by CD4+ T cells stimulated with anti-CD3 antibody (Fig. 7 A) or with freeze-thawed T. cruzi in the presence of antigen-presenting cells (Fig. 7 B) between wild-type, Batf2−/−, Il23a−/−, and Batf2−/−Il23a−/− mice. In contrast, introduction of Il23a deficiency into Batf2−/− mice drastically reduced IL-17 production by CD4+ T cells stimulated with anti-CD3 antibody (Fig. 7 C). IL-17 production by Batf2−/−Il23a−/− CD4+ T cells stimulated with freeze-thawed T. cruzi was also dramatically reduced compared with that by Batf2−/− CD4+ T cells (Fig. 7 D). In addition, expression of Il17 and Il22 was markedly reduced in liver CD4+ T cells from T. cruzi–infected Batf2−/−Il23a−/− mice compared with those in Batf2−/− mice (Fig. 7 E). Furthermore, Batf2−/−Il23a−/− mice exhibited increased parasite load in the livers, hearts, and blood compared with Batf2−/− mice (Fig. 8 A), but they showed reduced tissue damage after T. cruzi infection, as determined by lower pathological score and reduced serum or plasma concentration of AST, CK, and CRE (Fig. 8, B and C). These findings indicate that increased IL-23 production is responsible for the enhanced Th17 responses and immunopathology in T. cruzi–infected Batf2−/− mice.
Figure 7.
Reduced CD4+ T cell IL-17 production in T. cruzi–infected Batf2−/−Il23a−/− mice. (A–D) Wild-type (n = 18), Batf2−/− (n = 12), Il23a−/− (n = 9), and Batf2−/−Il23a−/− (n = 9) mice were infected with T. cruzi for 20 d. CD4+ T cells were isolated from the spleen of T. cruzi–infected mice and stimulated with anti-CD3 antibody (A and C) or freeze-thawed T. cruzi in the presence of antigen-presenting cells (B and D) for 24 h. The culture supernatants were analyzed for IFN-γ (A and B) and IL-17 (C and D) by ELISA. #, P < 0.045; ##, P < 0.006. (E) Expression of Il17a and Il22 in CD4+ T cells from the livers of T. cruzi–infected mice. #, P < 0.05. (A–E) Graphs show mean values ± SEM.
Figure 8.
IL-23–mediated immunopathology in Batf2−/− mice during T. cruzi infection. (A) Parasite equivalents in the livers, hearts, and blood of T. cruzi–infected wild-type (n = 18), Batf2−/− (n = 12), Il23a−/− (n = 9), and Batf2−/−Il23a−/− (n = 9) mice (mean values ± SEM). #, P < 0.05. (B) Representative sections (top) and pathological score (bottom) in T. cruzi–infected mice. Data are mean values ± SEM. Bars, 100 µm. *, P < 0.009; **, P < 0.006. (C) Concentrations of AST and CK in the plasma and CRE in the sera of T. cruzi–infected mice. Data are mean ± SEM. #, P < 0.05; ##, P < 0.01.
Inhibition of Il23a expression by BATF2 through its interaction with c-JUN
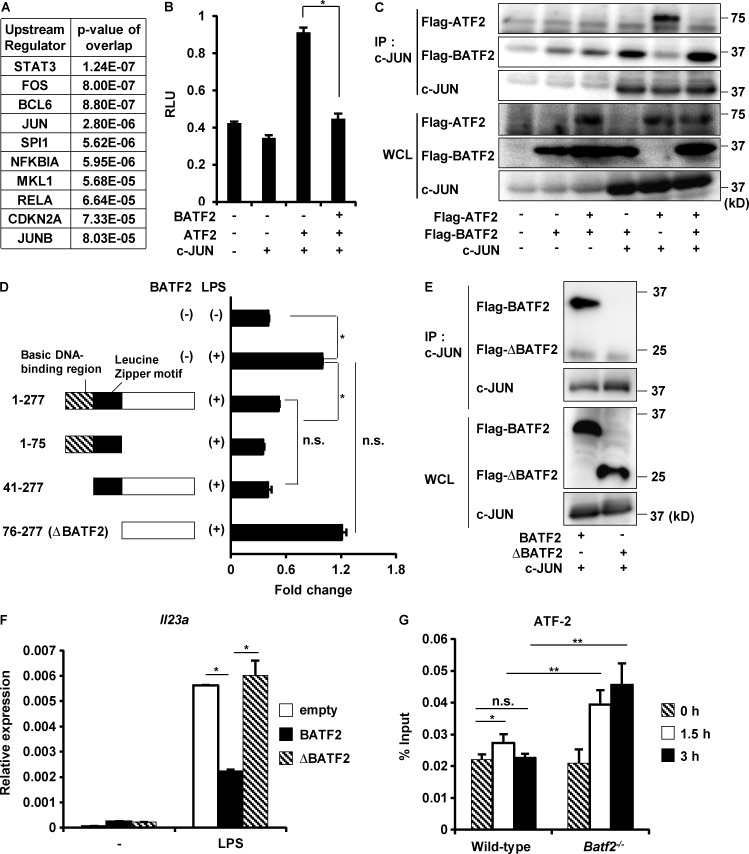
We next investigated how BATF2 selectively down-regulates the expression of Il23a in innate immune cells. Because BATF2 does not contain a transcriptional activation domain, we suspected that it interacts with other transcription factors to regulate downstream genes. Upstream Regulator Analysis within Ingenuity Pathway Analysis (IPA) showed the indicated transcription factors as significantly enriched upstream regulators of the genes up-regulated in Batf2−/− BMMφs shown in Table S1 (Fig. 9 A). IPA was also used to determine the protein–protein interaction networks associated with BATF2. Among the molecules predicted to be upstream regulators of BATF2-dependent genes (Fig. 9 A), IPA showed that BATF2 binds to c-JUN, JUNB, and RELA/NF-κBp65 (Fig. S5 C). Therefore, we analyzed the interactions between BATF2 and these molecules with coimmunoprecipitation assays. HEK293 cells were cotransfected with Flag-BATF2 expression vector and c-JUN or JUNB expression vectors. TLR4-expressing HEK293 cells were transfected with Flag-BATF2 expression vector and stimulated with LPS to analyze the interaction between BATF2 and NF-κBp65. The cell lysates were immunoprecipitated with anti-Flag antibody, and immunoblotting was performed with the indicated antibodies (Fig. S5, D–F). Among the three transcription factors, c-JUN and JUNB, but not NF-κBp65, interacted with BATF2. It has been shown that the heterodimer ATF-2/c-JUN, but not ATF-2/JUNB, drives the activity of the Il23a promoter (Liu et al., 2009). Therefore, we analyzed whether BATF2 negatively regulates c-JUN–ATF-2 heterodimer-mediated activation of the Il23a promoter (Fig. 9 B). HEK293 cells were transfected with a pLuc vector containing the Il23a promoter region with or without BATF2, c-JUN, and ATF-2 expression vectors, and luciferase activity in the cell lysates was measured. The introduction of c-JUN and ATF-2 induced activation of the Il23a promoter. However, the expression of BATF2 inhibited the c-JUN–ATF-2 heterodimer–induced activity of Il23a promoter. To further clarify how BATF2 down-regulates c-JUN–ATF-2 heterodimer–dependent expression of Il23a, HEK293 cells were transfected with the Flag-BATF2 expression vector, with or without the c-JUN and ATF-2 expression vectors, and the cell lysates were immunoprecipitated with anti–c-JUN antibody. Immunoblotting was then performed with the indicated antibodies (Fig. 9 C). c-JUN interacted individually with BATF2 and ATF-2. However, in the presence of BATF2, c-JUN was associated with BATF2, but not with ATF-2. BATF2 lacking the leucine zipper motif (ΔBATF2), which contains the protein–protein interaction domain, was unable to suppress the LPS-induced Il23a promoter activity (Fig. 9 D). ΔBATF2 also did not interact with c-JUN (Fig. 9 E). When Batf2−/− BMMφs were transfected with a vector expressing full-length BATF2 or ΔBATF2, the full-length protein, but not ΔBATF2, suppressed LPS-induced Il23a expression (Fig. 9 F). We also investigated the effect of BATF2-mediated disruption of c-JUN/ATF2 heterodimer formation on the transcription of Il23a. In addition to innate immune cells, BATF2 was robustly induced by IFN-γ in MEFs (Fig. S5 G). Furthermore, expression of Il23a was higher in Batf2−/− MEFs stimulated with LPS after IFN-γ treatment than in wild-type MEFs (Fig. S5 H). In this context, chromatin immunoprecipitation (ChIP) analysis showed that only a little ATF-2 was instantaneously recruited at the AP-1–binding site in the Il23a promoter in IFN-γ–pretreated wild-type MEFs in response to LPS, whereas NF-κBp65 recruitment to the Il23a promoter was markedly increased, as previously described (Mise-Omata et al., 2007; Mühlbauer et al., 2008; Fig. 9 G and Fig. S5 I). In contrast, Batf2−/− MEFs showed increased levels of ATF2 and prolonged ATF-2 binding to the Il23a promoter after LPS stimulation (Fig. 9 G). These findings indicate that IFN-γ–inducible BATF2 inhibits the recruitment of the c-JUN–ATF-2 heterodimer to the Il23a promoter by preventing the formation of the c-JUN–ATF-2 complex when it directly interacts with c-JUN, leading to the suppression of Il23a expression.
Figure 9.
BATF2 down-regulates Il23a expression through its interaction with c-JUN. (A) Enrichment analysis of the upstream regulators of genes up-regulated in Batf2−/− BMMφs. (B) Luciferase activity of Il23a promoter. Data are representative of three independent experiments. Graph show mean values ± SD of triplicate wells. *, P < 0.001. RLU, relative light unit. (C) Coimmunoprecipitation (IP) assay with anti–c-JUN antibody for immunoprecipitation and the indicated antibodies for immunoblotting. WCL, whole cell lysate. (D) Luciferase activity of Il23a promoter in response to LPS. Data are representative of three independent experiments. Mean values ± SD for triplicate wells are shown. *, P < 0.01; n.s., not significant. (E) Coimmunoprecipitation assay with anti–c-JUN antibody for immunoprecipitation and the indicated antibodies for immunoblotting. (F) Batf2−/− BMMφs were transfected with the indicated vectors by nucleofection and, after 18 h, stimulated with LPS for 1 h, and their expression of Il23a was analyzed. Data are representative of three independent experiments. Graphs show mean values ± SD of triplicate measurements. *, P < 0.01. (G) ChIP assay for the Il23a promoter of AP-1–binding site was performed with anti–ATF-2 antibody using wild-type and Batf2−/− MEFs. Means ± SD of triplicate PCRs on identical samples. *, P < 0.03; **, P < 0.005; n.s., not significant. These values are representative of three independent experiments.
Discussion
In this study, we have demonstrated the mechanism underlying regulation of the IL-23–Th17 pathway by BATF2 during T. cruzi infection. IFN-γ–inducible BATF2 in innate immune cells suppressed IL-23 production by preventing the formation of the c-JUN–ATF-2 heterodimer, resulting in the inhibition of excessive Th17 responses during T. cruzi infection.
Compared with wild-type mice, T. cruzi–infected Batf2−/− mice showed lower mortality despite the development of severe immunopathology. Previous studies have shown that parasite-infected Il27ra−/− mice produced high amounts of IFN-γ and IL-17, with high mortality rates (Hamano et al., 2003; Villarino et al., 2003; Stumhofer et al., 2006; Findlay et al., 2010). However, treatment with anti–IFN-γ antibody alone dramatically reduced mortality (Hamano et al., 2003), indicating that excessive IL-17 is not implicated in the enhanced host lethality during T. cruzi infection. Therefore, increased IL-17 production in Batf2−/− mice might be associated with reduced parasite burden leading to low mortality, while inducing the severe immunopathology. In Batf2−/−Il23a−/− mice, parasite number was partially reduced compared with that in wild-type mice, suggesting the possibility that BATF2-dependent genes other than Il23a also contribute to regulation of the parasite burden.
IL-17 promotes recruitment and activation of neutrophils (Kolaczkowska and Kubes, 2013). In accordance with the enhanced Th17 responses, a neutrophil-produced tissue damage factor, myeloperoxidase (Kolaczkowska and Kubes, 2013), was highly expressed in tissues of T. cruzi–infected Batf2−/− mice, suggesting that increased IL-23 production in Batf2−/− mice leads to neutrophil-related local tissue damage through enhanced IL-17 production. In the liver, Il23a is expressed in Kupffer cells and CD11c+ DCs (unpublished data), indicating that BATF2 might negatively regulate Il23a expression in these cells and thereby suppress immunopathological Th17 responses.
Multiple studies have identified several IL-17–producing cells besides Th17 cells, including invariant natural killer T cells, type 3 innate lymphoid cells, activated B cells, and innate γδ T cells (Cua and Tato, 2010; Bermejo et al., 2013). To determine whether CD4+ T cell–derived IL-17 is responsible for the multiorgan pathology in Batf2−/− mice, we attempted to deplete CD4+ T cells in Batf2−/− mice by intraperitoneally injecting with monoclonal anti-CD4 antibody after T. cruzi infection. However, effective depletion of CD4+ T cells in the livers and spleens could not be achieved, although CD4+ T cells in the peripheral blood severely decreased (unpublished data). Therefore, it would be an important future issue to characterize the IL-17–producing cells in detail during T. cruzi infection and to determine whether BATF2 controls IL-17 production in these cells by down-regulating Il23a expression in innate myeloid cells.
In the present study, Ifng−/− BMMφs and BMDCs showed higher T. cruzi–mediated IL-23 production than wild-type cells, indicating that BATF2, which is induced by IFN-γ autocrine signaling in innate immune cells, is responsible for the suppression of IL-23 production after T. cruzi infection. We also found that IFN-γ pretreatment leads to severely reduced IL-23 production in wild-type innate immune cells infected with T. cruzi, suggesting that BATF2 expression in innate myeloid cells is enhanced by potent IFN-γ producers in vivo, thereby restricting IL-23 production. However, in Batf2−/− innate immune cells, IFN-γ-mediated reduction of IL-23 production was partially observed during T. cruzi infection and LPS stimulation. In this regard, it has been shown that IFN-γ depresses the recruitment of NF-κBp65 to the Il23a promoter by inducing histone modification in the early phase of the LPS responses (Sheikh et al., 2010). Therefore, it appears that IFN-γ suppresses Il23a expression in two ways during T. cruzi infection: IFN-γ induces both histone modification–mediated and BATF2-mediated reduction of Il23a expression. In T. cruzi–infected Ifng−/− mice, production of IL-17 by CD4+ T cells was markedly increased, as previously described (Cobb et al., 2010). IFN-γ–inducible T-bet is implicated in the suppression of Th17 cell differentiation by preventing the activation of RORγt (Lazarevic et al., 2011). Furthermore, IFN-γ acts on the histone modification of Th17-related transcription factors, including RORγt, STAT3, and STAT1 (Liu et al., 2015). In this study, we characterized a novel mechanism by which IFN-γ–inducible BATF2 contributes to the attenuation of the Th17 responses by down-regulating IL-23 production.
A previous study reported that BATF2 is responsible for the induction of Tnf and Il12b expression by interacting with IRF1 in LPS-stimulated BMMφs induced by L929 cell–conditioned medium (Roy et al., 2015). However, we observed no changes in Tnf or Il12b expression in IFN-γ plus LPS–stimulated Batf2−/− BMMφs, which were induced by GM-CSF. LPS-induced expression of Tnf, Ccl5, and Nos2 was decreased in the absence of BATF2 when we prepared BMMφs using L929 cell–conditioned medium (unpublished data); however, these genes were normally induced in GM-CSF–induced Batf2−/− BMMφs (unpublished data). Thus, the discrepancy between the two studies may be explained by the polarization of Mφs because L929 cell–conditioned medium, containing high levels of M-CSF–induced BMMφs, is skewed to the M2Mφ phenotype, whereas GM-CSF-induced BMMφs is skewed to the M1Mφ phenotype. Therefore, BATF2 might regulate its downstream genes via Mφ type–specific mechanisms.
IL-27 suppresses tissue inflammation by inhibiting effector T cell responses and inducing IL-10–producing Tr1 cells during parasite infection (Hamano et al., 2003; Stumhofer et al., 2006; Hall et al., 2012). IL-27 production was slightly decreased in Batf2−/− BMMφs infected with T. cruzi (unpublished data). A modest reduction of IL-27 production in Batf2−/− BMMφs is not thought to be a major cause of the increased Th17 responses in Batf2−/− mice for the following reasons: (a) T. cruzi–infected Batf2−/− mice showed normal Th1 responses and IL-10–producing CD4+ T cell development; and (b) T. cruzi–infected Batf2−/−Il23a−/− mice showed decreased IL-17 production by CD4+ T cells. IL-27 suppresses Th17 responses during the chronic phase of T. gondii infection (Stumhofer et al., 2006). Thus, it would be an important future issue to analyze the effects of BATF2-dependent regulation of IL-27 production on the chronic phase of T. cruzi infection.
In the present study, we have demonstrated that IFN-γ–inducible BATF2 exerts its immunoregulatory function through the suppression of Il23a by interacting directly with c-JUN in innate immune cells, thereby preventing immunopathological Th17 responses during T. cruzi infection. BATF2 is also broadly expressed in nonhematopoietic cells, such as vascular cells (Murphy et al., 2013; Chmielewski et al., 2014) and fibroblasts, as shown in the current study. Therefore, it would be interesting to evaluate the precise role of BATF2 during T. cruzi infection using innate myeloid cell–specific BATF2-deficient mice in a future study. In addition, enhanced intestinal IL-23 production and Th17 responses, which were associated with disturbed intestinal homeostasis, were observed in Batf2−/− mice (unpublished data). Thus, BATF2-mediated regulation of Th17 responses is not specific to T. cruzi infection, but is applicable to several models. Analyses of the involvement of BATF2 in other infectious models and Th17-mediated immune disorder models would be an interesting future issue.
Materials and methods
Mice
C57BL/6 mice were purchased from Japan SLC. Ifng−/− mice were generated as described previously (Tagawa et al., 1997). All mice were maintained under specific pathogen–free conditions. All animal experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee of Osaka University.
Generation of Batf2−/− and Il23a−/− mice
To generate Batf2−/− mice, the targeted vector was constructed by replacing exon 3 of Batf2 with a neomycin resistance gene cassette, and a gene encoding herpes simplex virus thymidine kinase driven by a phosphoglycerate kinase promoter was inserted into the genomic fragment for negative selection. After embryonic day 14, embryonic stem cells were transfected with the targeted vector, and colonies doubly resistance to G418 and ganciclovir were selected and screened with PCR and Southern blot analysis. Homologous recombinants were microinjected into blastocysts of female C57BL/6 mice, and the heterozygous F1 progeny were intercrossed to obtain Batf2-deficient mice. Batf2-deficient mice and their wild-type littermates from these intercrosses were confirmed with Southern blot analysis and real-time RT-PCR analysis and were used for subsequent experiments. Batf2-deficient mice were backcrossed to C57BL/6 mice for at least six generations, and Batf2-deficient mice and their wild-type littermates from the intercrosses of heterozygous mice were used for the experiments.
To generate Il23a−/− mice by the Cas9/CRISPR system, Il23a-gAS1-F (5′-CACCGCAGAGCAGTAATAATGCTA-3′) and Il23a-gAS1-R (5′-AAACTAGCATTATTACTGCTCTGC-3′) oligonucleotides and Il23a-gAS2-F (5′-CACCCTGGAACGCACATGCACCAG-3′) and Il23a-gAS2-R (5′-AAACCTGGTGCATGTGCGTTCCAG-3′; Thermo Fisher Scientific) were annealed and inserted into the BbsI restriction site in the px330 vector (plasmid 42230; Addgene) by Ligation high v. 2 (Toyobo). This plasmid was designated as px330-Il23a-gAS1 and px330-Il23a-gAS2, respectively. The px330-Il23a-gAS1 together with px330-Il23a-gAS2 vectors were injected into the pronuclei of one-cell-stage embryos from B6D2F1 mice. The eggs were cultivated in kSOM overnight and then transferred into the oviducts of pseudo-pregnant females.
Reagents
The Ca2+ ionophore A23187, PMA, LPS (O55: B5), and an anti-Flag antibody (clone M1) were purchased from Sigma-Aldrich. Anti-mouse c-JUN (clone H-79), JUNB (clone N-17), and NF-kBp65 (clone C-20) antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-ATF2 antibody (clone 20F1) was purchased from Cell Signaling Technology.
Preparation of Mφs and DCs
To prepare BMDCs and BMMφs, BM cells were prepared from mouse femurs and tibias, passed through a nylon mesh, and cultured in RPMI 1640 medium containing 10% FBS, 100 µM 2-mercaptoethanol, and 10 ng/ml GM-CSF (PeproTech). After 6 d, nonadherent cells were used as BMDCs, and adherent cells were used as BMMφs in the experiments. To isolate PECs, mice were injected i.p. with 2 ml of 4% thioglycolate medium (Sigma-Aldrich), and cells in the peritoneal exudate were isolated from the peritoneal cavity 3 d after injection. The cells were incubated for 2 h and then washed three times with HBSS. The remaining adherent cells were used as PECs in the experiments.
Parasite and experimental infection
The T. cruzi Tulahuen strain was maintained in vitro in LLC-MK2 cells, with passage every 3 d. For the in vitro experiments, 5 × 105 Mφs or DCs were infected with 2.5 × 106 T. cruzi. For the in vivo experiments, the mice were injected i.p. with 5 × 102 T. cruzi.
Flow cytometry
The following antibodies were purchased from BD: anti–mouse-CD16/32 (2.4.G2), APC-conjugated anti-CD11c (clone HL3), PE-conjugated anti-CD103 (clone M290), PE-Cy7–conjugated anti–Ly-6C (clone AL-21), FITC-conjugated anti–Ly-6G (clone 1A8), biotin-conjugated anti-IAb (AF6-120.1), PE-conjugated anti-CD80 (B7-1), and streptavidin-APC antibodies. Pacific blue–conjugated anti-CD11b (clone M1/70), Percp-Cy5.5–conjugated anti-CD4 (clone CK1.5), APC-conjugated anti-CD8 (clone 53–6.7), PE-conjugated anti-CD19 (clone 6D5), APC-conjugated anti–IL-17A (TC11-18H10.1), APC-conjugated anti-F4/80 (BM8), FITC-conjugated anti–IFN-γ (clone XMG1.2), and PE-conjugated anti–IL-10 (JES5-16E3) antibodies were purchased from BioLegend. FITC-conjugated anti-CD86 antibody (clone GL1) was purchased from eBioscience. Flow cytometric analysis was performed with a FACSCanto II flow cytometer (BD) with FlowJo software (Tree Star). CD11b+ F4/80+ Mφs from the spleens of T. cruzi–infected mice were isolated with a FACSAria flow cytometer (BD). The instrumental compensation was set in each experiment using single-color, two-color, or four-color stained samples.
Real-time RT-PCR
CD4+ T cells isolated from the spleens and livers after T. cruzi infection for 20 d were stimulated with 50 ng/ml PMA and 5 µM calcium ionophore A23187 for 2 h, and total RNA was isolated. PECs, BMMφs, and BMDCs were pretreated with IFN-γ for 4 h and stimulated with LPS for the indicated periods, and total RNA was isolated. Total RNA was isolated using the RNeasy Mini kit (QIAGEN), and 1–2 µg of RNA was reverse transcribed with Moloney Murine Leukemia Virus Reverse transcription (Promega) and random primers (Toyobo) after treatment with RQ1 DNase I (Promega). Real-time RT-PCR was performed on a Step One Plus Real-Time PCR System (Applied Biosystems) using GoTaq qPCR Master Mix (Promega). All values were normalized to the expression of Gapdh, encoding glyceraldhyde-3-phosphate dehydrogenase, and the fold difference in expression relative to that of Gapdh is shown. The amplification conditions were as follows: 50°C (2 min), 95°C (10 min), and 40 cycles of 95°C (15 s) and 60°C (60 s). The following primer sets were used: Gapdh, 5′-CCTCGTCCCGTAGACAAAATG-3′ and 5′-TCTCCACTTTGCCACTGCAA-3′; Batf, 5′-AGACACAGAAAGCCGACACC-3′ and 5′-TCAGCACTGATGTGAAGTAC-3′; Batf2, 5′-GCCCAGCGCAGCCGGCAGAA-3′ and 5′-CCAGCTCAGTCTGCAAGGCCT-3′; Batf3, 5′-AAGGACGATGACAGGAAAG-3′ and 5′-CTCTCGTGCTCCTCGTGGAG-3′; Cd86, 5′-TACGGAAGCACCCACGATGG-3′ and 5′-AATAAGCTTGCGTCTCCACG-3′; Fgl2, 5′-AGTGTCCCAGCCAAGAACAC-3′ and 5′-GATCAGGGGTAACTCTGTAG-3′; Gbp7, 5′-TGAGAGAGCAAGAGGTGAAGCTG-3′ and 5′-ACTTACCAGAACCAGGCACTAC-3′; Gbp8, 5′-CCACAATGAAACATCTGTCCGTGAACC-3′ and 5′-CCAGAGGGAAACCGTGATTCTGTC-3′; Ifng, 5′-TCAAGTGGCATAGATGTGGAAGAA-3′ and 5′-TGGCTCTGCAGGATTTTCATG-3′; Irf1, 5′-CCGAAGACCTTATGAAGCTCTTTG-3′ and 5′-GCAAGTATCCCTTGCCATCG-3′; Il10, 5′-CTGGGTGAGAAGCTGAAGACC-3′ and 5′-CATGGCCTTGTAGACACCTTG-3′; Il12b, 5′-GGTTTGCCATCGTTTGCTGG-3′ and 5′-CATCTTCTTCAGGCGTGTCAC-3′; Il12rb1, 5′-CCTGCTTTGCCAGCTAGGCG-3′ and 5′-CCTGGGTCCTAAGGGTGAGC-3′; Il17a, 5′-GGACTCTCCACCGCAATGA-3′ and 5′-GGCACTGAGCTTCCCAGATC-3′; Il6, 5′-CTGCAAGAGACTTCCATCCAGTT-3′ and 5′-AAGTAGGGAAGGCCGTGGTT-3′; Il22, 5′-TGATGGCTGTCCTGCAGAAA-3′ and 5′-CAGCTGGCGGCAAAG-3′; Il23a, 5′-CCAGCAGCTCTCTCGGAATCT-3′ and 5′-ACTGCTGACTAGAACTCAGGC-3′; Irgm, 5′-AATAAAACCAGAGAGCCTCA-3′ and 5′-TAATGGGTCTCTGCCATGTT-3′; Mpo, 5′-ATCACGGCCTCCCAGGATACAATG-3′ and 5′-ACCGCCCATCCAGATGTCAAT-3′; Stat1, 5′-GCGTGCAGTGAGTGAGTGAG-3′ and 5′-CAACACAAGCAATCACCACA-3′; Tnfa 5′-TCCAGGCGGTGCCTATGT-3′ and 5′-CACCCCGAAGTTCAGTAGACAGA-3′; and Csf2, 5′-TCGAGCAGGGTCTACGGGGC-3′ and 5′-TCCGTTTCCGGAGTTGGGGG-3′.
Cytokine analysis
CD4+ T cells were isolated from the spleens and livers of mice on the indicated days after T. cruzi infection with MACS technology (Miltenyi Biotec) and stimulated with anti-CD3 antibody or freeze-thawed T. cruzi in the presence of antigen-presenting cells in a 1:1:1 ratio for 24 h. PECs, BMMφs, and BMDCs were pretreated with 10 ng/ml IFN-γ and stimulated with 100 ng/ml LPS for 24 h. BMMφs and BMDCs pretreated with or without 10 ng/ml IFN-γ for 4 h were thoroughly washed and infected with T. cruzi in a ratio of 1:5 and cultured for 24 h. The concentrations of IL-17, IFN-γ, IL-23, TNF, IL-12p40, IL-12p70, and IL-6 in the culture supernatants were measured with the Cytometric Bead Array kit (BD).
Intracellular cytokine staining
CD4+ T cell intracellular IFN-γ, IL-10, and IL-17A expression was analyzed with a Cytofix/Cytoperm Plus kit with GolgiStop (BD), according to the manufacturer’s instructions. In brief, CD4+ T cells were incubated with 50 ng/ml PMA, 5 µM calcium ionophore A23187, and GolgiStop in complete RPMI 1640 at 37°C for 4 h. Surface staining was performed with anti-CD4 antibody for 20 min at 4°C, and intracellular cytokine staining was performed with anti–IL-17A, anti–IL-10, and anti–IFN-γ antibodies for 20 min.
T. cruzi killing assay
Wild-type and Batf2−/− BMMφs were pretreated with or without the indicated concentration of IFN-γ for 18 h, rigorously washed, and then infected with T. cruzi for 6 h. After washing, the cells were cultured for 72 h and fixed. After staining with Diff-Quik (Sysmex), the intracellular parasites were counted.
Luciferase assay
HEK293 cells were transfected with the indicated expression plasmids together with the reporter plasmid Il23a–Luc and the internal control plasmid phRG-TK. After 12 h, luciferase activities of the whole cell lysates were measured with the Dual-Luciferase Reporter Assay System (Promega) and a Lumat LB 9507 luminometer (Berthold Technologies). TLR4-expressing HEK293 cells were transfected with the plasmids. After 12 h, the cells were stimulated with 100 ng/ml LPS for 24 h, lysed, and then measured with the Dual-Luciferase Reporter Assay System.
Transfection into BMMφs
Batf2−/− BMMφs were transfected with pcDNA3 containing the full-length form of BATF2 or the mutant form which the leucine zipper motif had been deleted, using nucleofection (Nucleofector kit for Mouse Macrophages; Lonza). After 12 h, the cells were stimulated with 100 ng/ml LPS for 3 h, and then the expression of Il23a was analyzed.
RNA-seq analysis
Wild-type and Batf2−/− BMMφs pretreated with 10 ng/ml IFN-γ for 4 h were stimulated with (designated WT_3 and KO_3, respectively) or without (designated WT_0 and KO_0, respectively) 100 ng/ml LPS for 3 h. Total RNAs were extracted from the cells with RNeasy Mini kit (QIAGEN), according to the manufacturer’s protocol. The libraries were constructed and sequenced at Kazusa DNA Research Institute (Chiba, Japan) using a SureSelect Strand-Specific RNA Library Prep kit (Agilent Technologies), according to the manufacturer’s instructions. Sequencing was performed on an Illumina HiSeq 2000 sequencer with a TruSeq Rapid SBS kit v3 HS (Illumina) in the 50-base single-end mode. Illumina Casava 1.8.2 software was used for base calling. The low-quality regions of the raw reads were trimmed with Btrim. The sequenced reads were mapped to the mouse reference genome sequence (mm9) using TopHat v2.0.12. The fragments per kilobase of exons per million mapped fragments (FPKMs) were calculated using Cufflinks v2.1.1. Among the calculated genes with a normalized FPKM value >1.0 in WT_3, 98 genes were up-regulated >1.5-fold from WT_0 to WT_3 and >2.0-fold from WT_3 to KO_3 (Table S1). These results have been deposited in the National Center Biotechnology Information Gene Expression Omnibus database (GSE81724).
Quantitation of T. cruzi in tissues
The parasite load in the tissue was analyzed as previously described (Cencig et al., 2011; Caldas et al., 2012). In brief, concentrations of DNA purified from the liver, heart, and blood of T. cruzi–infected mice were adjusted to 25 ng/µl. Each PCR reaction was performed with 50 ng of genomic tissue DNA. The amount of tissue in each PCR was normalized to mouse TNF DNA (Cencig et al., 2011). T. cruzi standard curves were generated as previously described (Caldas et al., 2012). qPCR was performed on the Step One Plus Real-Time PCR System (Applied Biosystems) using GoTaq qPCR Master Mix (Promega). The amplification conditions were as follows: 95°C for 10 min, 40 cycles of 94°C for 15 s, and 64.3°C for 1 min. The following primer sets were used: T. cruzi specific primers, 5′-CGAGCTGTTGCCCACACGGGTGCT-3′ and 5′-CCTCCAAGCAGCGGATAGTTCAGG-3′ (Cencig et al., 2011); and Tnf primers, 5′-TCCCTCTCATCAGTTCTATGGCCCA-3′ and 5′-CAGCAAGCATCTATGCACTTAGACCCC-3′ (Caldas et al., 2012).
Histopathological analysis
The livers and hearts collected from the mice 49 d after T. cruzi infection were fixed in 4% PFA. Paraffin-embedded 5-µm sections mounted on glass slides were stained with hematoxylin and eosin (H&E) and examined for histopathology with inflammatory cell infiltrates using light microcopy. Each section of the hearts (Leon et al., 2003) and livers (Barreto-de-Albuquerque et al., 2015) was evaluated for inflammation scores as previously described.
ChIP assay
The ChIP assay was performed as previously described, with modifications (Kayama et al., 2008). In brief, MEFs were stimulated with 100 ng/ml LPS for the indicated periods after treatment with 10 ng/ml IFN-γ for 5 h. The chromatin was cross-linked with 1% formaldehyde at room temperature for 10 min. The MEFs were washed with PBS, scraped, and then centrifuged at 800 g. The pellet was resuspended in SDS buffer supplemented with protease inhibitor. The chromatin was sonicated 15 times with 30 s pulses, centrifuged at 1,300 g to remove any debris, diluted fivefold with ChIP dilution buffer, and precleared with salmon sperm DNA/protein A agarose (EMD Millipore). The diluted chromatin was immunoprecipitated at 4°C overnight. The immune complexes were adsorbed onto salmon sperm DNA/protein A agarose beads, and washed once each with low-salt buffer, high-salt buffer, and LiCl buffer, and then twice with TE buffer. The immune complexes were extracted in elution buffer and incubated for 4 h at 65°C. The DNA was then extracted by incubating it in proteinase K buffer for 1 h at 45°C. The purified DNA was used in qPCR, using 10% of each input as the standard. The Il23a promoter-specific primer was designed to include the NF-κB–binding site or AP-1–binding site. The primer sequences were as follows: NF-κB–binding site, 5′-TGGCCTCACTCTGACGTCCA-3′ and 5′-GTAACACCCAGGGAAGCAGC-3′; AP-1–binding site, 5′-AATCCCACCTGCTCTGAGTC-3′ and 5′-GTAGTCCGACTTCTAAGCGG-3′.
Statistical analysis
Differences between the control and experimental groups were evaluated with Student’s t test. Differences of P < 0.05 were considered to be significant.
Online supplemental material
Fig. S1 shows the targeting construct for Batf2−/− mice, expression patterns of IFN-γ–dependent genes, and innate immune cell population of wild-type and Batf2−/− mice. Fig. S2 shows flow cytometric dot plots of cytokine-producing CD4+ T cells of T. cruzi–infected mice. Fig. S3 displays production of IFN-γ and IL-17 in wild-type and Batf2−/− naive CD4+ T cells cultured under Th0, Th1, and Th17 conditions. Fig. S4 shows cytokine production by wild-type and Batf2−/− PECs, BMMφs, and BMDCs. Fig. S5 shows the Cas9/gRNA-targeting site for Il23a and the analysis of BATF2-binding partners. Table S1, included in a separate Excel file, lists genes upregulated in response to LPS in Batf2−/− BMMφs pretreated with IFN-γ.
Supplementary Material
Acknowledgments
We thank S. Nishioka, T. Kondo, and Y. Magota for their technical assistance; S. Pareek for critical reading of the manuscript; and C. Hidaka for secretarial assistance.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Agency for Medical Research and Development (grant 15gm011002h0006; to K. Takeda) and by the Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care and the Kurata Memorial Hitachi Science and Technology Foundation (to H. Kayama).
The authors declare no competing financial interests.
Author contributions: S. Kitada, H. Kayama, R. Koga, and M. Kobayashi designed and performed the experiments, analyzed the data, and revised the manuscript. D. Okuzaki designed the experiments, analyzed data, and supervised the RNA-seq analysis. A. Kumanogoh and M. Ikawa supervised the generation of Il23a−/− mice by the Cas9/CRISPR system. Y. Arima and M. Murakami analyzed the Ifng−/− mice. K. Takeda supervised the study, designed the experiments, analyzed the data, and wrote the manuscript.
Footnotes
Abbreviations used:
- AST
- aspartate aminotransferase
- BMMφ
- BM-derived Mφ
- ChIP
- chromatin immunoprecipitation
- CK
- creatinine kinase
- CRE
- creatinine
- Mφ
- macrophage
- PEC
- peritoneal Mφ
- qPCR
- quantitative PCR
References
- Ahern P.P., Schiering C., Buonocore S., McGeachy M.J., Cua D.J., Maloy K.J., and Powrie F.. 2010. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 33:279–288. 10.1016/j.immuni.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-de-Albuquerque J., Silva-dos-Santos D., Pérez A.R., Berbert L.R., de Santana-van-Vliet E., Farias-de-Oliveira D.A., Moreira O.C., Roggero E., de Carvalho-Pinto C.E., Jurberg J., et al. . 2015. Trypanosoma cruzi infection through the oral route promotes a severe infection in mice: New disease form from an old infection? PLoS Negl. Trop. Dis. 9:e0003849 10.1371/journal.pntd.0003849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo D.A., Jackson S.W., Gorosito-Serran M., Acosta-Rodriguez E.V., Amezcua-Vesely M.C., Sather B.D., Singh A.K., Khim S., Mucci J., Liggitt D., et al. . 2013. Trypanosoma cruzi trans-sialidase initiates a program independent of the transcription factors RORγt and Ahr that leads to IL-17 production by activated B cells. Nat. Immunol. 14:514–522. 10.1038/ni.2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney K.M., Taylor J.M., Thorp E.B., Epting C.L., and Engman D.M.. 2015. Depletion of regulatory T cells decreases cardiac parasitosis and inflammation in experimental Chagas disease. Parasitol. Res. 114:1167–1178. 10.1007/s00436-014-4300-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas S., Caldas I.S., Diniz L.F., Lima W.G., Oliveira R.P., Cecílio A.B., Ribeiro I., Talvani A., and Bahia M.T.. 2012. Real-time PCR strategy for parasite quantification in blood and tissue samples of experimental Trypanosoma cruzi infection. Acta Trop. 123:170–177. 10.1016/j.actatropica.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Cencig S., Coltel N., Truyens C., and Carlier Y.. 2011. Parasitic loads in tissues of mice infected with Trypanosoma cruzi and treated with AmBisome. PLoS Negl. Trop. Dis. 5:e1216 10.1371/journal.pntd.0001216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewski S., Olejnik A., Sikorski K., Pelisek J., Błaszczyk K., Aoqui C., Nowicka H., Zernecke A., Heemann U., Wesoly J., et al. . 2014. STAT1-dependent signal integration between IFNγ and TLR4 in vascular cells reflect pro-atherogenic responses in human atherosclerosis. PLoS One. 9:e113318 10.1371/journal.pone.0113318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb D., and Smeltz R.B.. 2012. Regulation of proinflammatory Th17 responses during Trypanosoma cruzi infection by IL-12 family cytokines. J. Immunol. 188:3766–3773. 10.4049/jimmunol.1103478 [DOI] [PubMed] [Google Scholar]
- Cobb D., Hambright D., and Smeltz R.B.. 2010. T-bet-independent effects of IL-12 family cytokines on regulation of Th17 responses to experimental T. cruzi infection. J. Leukoc. Biol. 88:965–971. 10.1189/jlb.0410238 [DOI] [PubMed] [Google Scholar]
- Cua D.J., and Tato C.M.. 2010. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 10:479–489. 10.1038/nri2800 [DOI] [PubMed] [Google Scholar]
- da Matta Guedes P.M., Gutierrez F.R., Maia F.L., Milanezi C.M., Silva G.K., Pavanelli W.R., and Silva J.S.. 2010. IL-17 produced during Trypanosoma cruzi infection plays a central role in regulating parasite-induced myocarditis. PLoS Negl. Trop. Dis. 4:e604 10.1371/journal.pntd.0000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann H., Roßnagel C., Böhme J., Iwakura Y., Jacobs T., Schaible U.E., and Hölscher C.. 2013. IL-17A promotes macrophage effector mechanisms against Trypanosoma cruzi by trapping parasites in the endolysosomal compartment. Immunobiology. 218:910–923. 10.1016/j.imbio.2012.10.005 [DOI] [PubMed] [Google Scholar]
- Findlay E.G., Greig R., Stumhofer J.S., Hafalla J.C., de Souza J.B., Saris C.J., Hunter C.A., Riley E.M., and Couper K.N.. 2010. Essential role for IL-27 receptor signaling in prevention of Th1-mediated immunopathology during malaria infection. J. Immunol. 185:2482–2492. 10.4049/jimmunol.0904019 [DOI] [PubMed] [Google Scholar]
- Gaffen S.L., Jain R., Garg A.V., and Cua D.J.. 2014. The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 14:585–600. 10.1038/nri3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R.T., and Denkers E.Y.. 2006. Protozoan encounters with Toll-like receptor signalling pathways: Implications for host parasitism. Nat. Rev. Immunol. 6:895–906. 10.1038/nri1978 [DOI] [PubMed] [Google Scholar]
- Guo S., Cobb D., and Smeltz R.B.. 2009. T-bet inhibits the in vivo differentiation of parasite-specific CD4+ Th17 cells in a T cell-intrinsic manner. J. Immunol. 182:6179–6186. 10.4049/jimmunol.0803821 [DOI] [PubMed] [Google Scholar]
- Hall A.O., Beiting D.P., Tato C., John B., Oldenhove G., Lombana C.G., Pritchard G.H., Silver J.S., Bouladoux N., Stumhofer J.S., et al. . 2012. The cytokines interleukin 27 and interferon-γ promote distinct Treg cell populations required to limit infection-induced pathology. Immunity. 37:511–523. 10.1016/j.immuni.2012.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano S., Himeno K., Miyazaki Y., Ishii K., Yamanaka A., Takeda A., Zhang M., Hisaeda H., Mak T.W., Yoshimura A., and Yoshida H.. 2003. WSX-1 is required for resistance to Trypanosoma cruzi infection by regulation of proinflammatory cytokine production. Immunity. 19:657–667. 10.1016/S1074-7613(03)00298-X [DOI] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 34:637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Kayama H., and Takeda K.. 2010. The innate immune response to Trypanosoma cruzi infection. Microbes Infect. 12:511–517. 10.1016/j.micinf.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Kayama H., Ramirez-Carrozzi V.R., Yamamoto M., Mizutani T., Kuwata H., Iba H., Matsumoto M., Honda K., Smale S.T., and Takeda K.. 2008. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J. Biol. Chem. 283:12468–12477 (published errata appear in J. Biol. Chem. 2015. 290:4815 and 22446). 10.1074/jbc.M709965200 [DOI] [PubMed] [Google Scholar]
- Kayama H., Koga R., Atarashi K., Okuyama M., Kimura T., Mak T.W., Uematsu S., Akira S., Takayanagi H., Honda K., et al. . 2009. NFATc1 mediates Toll-like receptor-independent innate immune responses during Trypanosoma cruzi infection. PLoS Pathog. 5:e1000514 10.1371/journal.ppat.1000514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E., and Kubes P.. 2013. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13:159–175. 10.1038/nri3399 [DOI] [PubMed] [Google Scholar]
- Lazarevic V., Chen X., Shim J.H., Hwang E.S., Jang E., Bolm A.N., Oukka M., Kuchroo V.K., and Glimcher L.H.. 2011. T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12:96–104. 10.1038/ni.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon J.S., Wang K., and Engman D.M.. 2003. Captopril ameliorates myocarditis in acute experimental Chagas disease. Circulation. 107:2264–2269. 10.1161/01.CIR.0000062690.79456.D0 [DOI] [PubMed] [Google Scholar]
- Liu W., Ouyang X., Yang J., Liu J., Li Q., Gu Y., Fukata M., Lin T., He J.C., Abreu M., et al. . 2009. AP-1 activated by Toll-like receptors regulates expression of IL-23 p19. J. Biol. Chem. 284:24006–24016. 10.1074/jbc.M109.025528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ren S., Qu X., Ge C., Cheng K., and Zhao R.C.. 2015. Mesenchymal stem cells inhibit Th17 cells differentiation via IFN-γ-mediated SOCS3 activation. Immunol. Res. 61:219–229. 10.1007/s12026-014-8612-2 [DOI] [PubMed] [Google Scholar]
- McGeachy M.J., and McSorley S.J.. 2012. Microbial-induced Th17: Superhero or supervillain? J. Immunol. 189:3285–3291. 10.4049/jimmunol.1201834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O’Shea J.J., and Cua D.J.. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10:314–324. 10.1038/ni.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise-Omata S., Kuroda E., Niikura J., Yamashita U., Obata Y., and Doi T.S.. 2007. A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J. Immunol. 179:6596–6603. 10.4049/jimmunol.179.10.6596 [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Hamano S., Wang S., Shimanoe Y., Iwakura Y., and Yoshida H.. 2010. IL-17 is necessary for host protection against acute-phase Trypanosoma cruzi infection. J. Immunol. 185:1150–1157. 10.4049/jimmunol.0900047 [DOI] [PubMed] [Google Scholar]
- Mühlbauer M., Chilton P.M., Mitchell T.C., and Jobin C.. 2008. Impaired Bcl3 up-regulation leads to enhanced lipopolysaccharide-induced interleukin (IL)-23P19 gene expression in IL-10(-/-) mice. J. Biol. Chem. 283:14182–14189. 10.1074/jbc.M709029200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.L., Tussiwand R., and Murphy K.M.. 2013. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 13:499–509. 10.1038/nri3470 [DOI] [PubMed] [Google Scholar]
- Rangachari M., Mauermann N., Marty R.R., Dirnhofer S., Kurrer M.O., Komnenovic V., Penninger J.M., and Eriksson U.. 2006. T-bet negatively regulates autoimmune myocarditis by suppressing local production of interleukin 17. J. Exp. Med. 203:2009–2019. 10.1084/jem.20052222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M.M., Oliveira A.C., and Bellio M.. 2012. The immune response to Trypanosoma cruzi: Role of Toll-like receptors and perspectives for vaccine development. J. Parasitol. Res. 2012:507874 10.1155/2012/507874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Guler R., Parihar S.P., Schmeier S., Kaczkowski B., Nishimura H., Shin J.W., Negishi Y., Ozturk M., Hurdayal R., et al. . 2015. Batf2/Irf1 induces inflammatory responses in classically activated macrophages, lipopolysaccharides, and mycobacterial infection. J. Immunol. 194:6035–6044. 10.4049/jimmunol.1402521 [DOI] [PubMed] [Google Scholar]
- Sheikh S.Z., Matsuoka K., Kobayashi T., Li F., Rubinas T., and Plevy S.E.. 2010. Cutting edge: IFN-γ is a negative regulator of IL-23 in murine macrophages and experimental colitis. J. Immunol. 184:4069–4073. 10.4049/jimmunol.0903600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumhofer J.S., Laurence A., Wilson E.H., Huang E., Tato C.M., Johnson L.M., Villarino A.V., Huang Q., Yoshimura A., Sehy D., et al. . 2006. Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat. Immunol. 7:937–945. 10.1038/ni1376 [DOI] [PubMed] [Google Scholar]
- Su Z.Z., Lee S.G., Emdad L., Lebdeva I.V., Gupta P., Valerie K., Sarkar D., and Fisher P.B.. 2008. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN). Proc. Natl. Acad. Sci. USA. 105:20906–20911. 10.1073/pnas.0807975106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa Y., Sekikawa K., and Iwakura Y.. 1997. Suppression of concanavalin A-induced hepatitis in IFN-gamma(-/-) mice, but not in TNF-alpha(-/-) mice: Role for IFN-gamma in activating apoptosis of hepatocytes. J. Immunol. 159:1418–1428. [PubMed] [Google Scholar]
- Tussiwand R., Lee W.L., Murphy T.L., Mashayekhi M., Kc W., Albring J.C., Satpathy A.T., Rotondo J.A., Edelson B.T., Kretzer N.M., et al. . 2012. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 490:502–507. 10.1038/nature11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A., Hibbert L., Lieberman L., Wilson E., Mak T., Yoshida H., Kastelein R.A., Saris C., and Hunter C.A.. 2003. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity. 19:645–655. 10.1016/S1074-7613(03)00300-5 [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Takeda A., Hamano S., Miyazaki Y., Kinjyo I., Ishibashi T., Yoshimura A., and Yoshida H.. 2006. Two-sided roles of IL-27: Induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. J. Immunol. 177:5377–5385. 10.4049/jimmunol.177.8.5377 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.