Fu et al. demonstrate that reactive oxygen species (ROS)–sensing molecule misshapen/NIK-related kinase 1 (MINK1) can specifically suppress Th 17 cell differentiation through direct phosphorylation of SMAD2. This study provides the mechanism of how ROS limit inflammatory response and unveils the potential health risk of antioxidant supplementation.
Abstract
T helper type 17 cells (Th17 cells) are major contributors to many autoimmune diseases. In this study, we demonstrate that the germinal center kinase family member MINK1 (misshapen/NIK-related kinase 1) negatively regulates Th17 cell differentiation. The suppressive effect of MINK1 on induction of Th17 cells is mediated by the inhibition of SMAD2 activation through direct phosphorylation of SMAD2 at the T324 residue. The importance of MINK1 to Th17 cell differentiation was strengthened in the animal model of experimental autoimmune encephalomyelitis (EAE). Moreover, we show that the reactive oxygen species (ROS) scavenger N-acetyl cysteine boosts Th17 cell differentiation in a MINK1-dependent manner and exacerbates the severity of EAE. Thus, we have not only established MINK1 as a critical regulator of Th17 cell differentiation, but also clarified that accumulation of ROS may limit the generation of Th17 cells. The contribution of MINK1 to ROS-regulated Th17 cell differentiation may suggest an important mechanism for the development of autoimmune diseases influenced by antioxidant dietary supplements.
Introduction
T helper type 17 cells (Th17 cells) have been identified and characterized as a distinct T cell lineage that maintains mucosal tissue homeostasis and contributes to the host defense against extracellular pathogens (Harrington et al., 2005; Park et al., 2005; Korn et al., 2009; Song et al., 2016). Importantly, dysregulated Th17 cell activity has been linked to a predisposition to the pathogenesis of human autoimmune diseases and relevant mouse models (Langrish et al., 2005; Reynolds et al., 2010; Wang et al., 2015). Transforming growth factor β (TGF-β), in combination with IL-6, has been well established as the most important instructive stimulus for the differentiation of Th17 cells (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006a). Ablation of TGF-β signaling in T cells prevents the induction of Th17 cells and the initiation of experimental autoimmune encephalomyelitis (EAE; Veldhoen et al., 2006b). However, it has recently been reported that stimuli other than cytokines, including high salt, lipid metabolites, and redox status, can also influence Th17 cell differentiation (Kleinewietfeld et al., 2013; Wang et al., 2015; Ravindran et al., 2016). It is currently unknown whether these stimuli work through signal cross talk with cytokine-induced pathways, including the TGF-β–receptor-regulated SMAD (TGF-β–R-SMAD) axis.
Misshapen (Msn)/NIK-related kinase 1 (MINK1) is a serine-threonine kinase that belongs to the germinal center kinase IV family (Dan et al., 2000). MINK1 has been reported to mediate oncogene-induced ovarian epithelial cell growth arrest via a mechanism involving reactive oxygen species (ROS) and MAPK p38 activation (Nicke et al., 2005). It has also been suggested that MINK1 may mediate thymocyte selection in bone marrow chimeras through activation of JNK and Bim phosphorylation (McCarty et al., 2005). Recent studies have demonstrated that increased oxidative metabolism and the production of ROS inhibit the survival and proliferation of Th17 cells (Won et al., 2013; Gerriets et al., 2015). Thus, MINK1 may act as an intermediate to bridge the ROS effect to Th17 induction. Intriguingly, besides functioning as an upstream kinase for MAPK pathways, MINK1 can also serve as an Msn kinase phosphorylating R-SMADs (Kaneko et al., 2011). Therefore, it is of interest to determine whether MINK1 is a vital player in ROS/redox balance–influenced Th17 cell induction and to elucidate the signaling pathways associated with MINK1.
Here, we evaluated the role of MINK1 in Th17 cell differentiation in a mouse EAE model. Upon induction of EAE, Mink1−/− mice exhibited increased Th17 cell differentiation and disease severity. Direct phosphorylation at residue T324 in the α-helix 1 region and inactivation of SMAD2 were found to be the major cellular mechanism by which MINK1 modulated Th17 cell differentiation. Moreover, the ROS scavenger N-acetyl cysteine (NAC) markedly boosted the induction of Th17 cells in a MINK1-dependent manner both in vitro and in vivo. In sum, MINK1 is a suppressor in the redox status–regulated differentiation of Th17 cells.
Results
Increased numbers of Th17 cells in the periphery of Mink1−/− mice
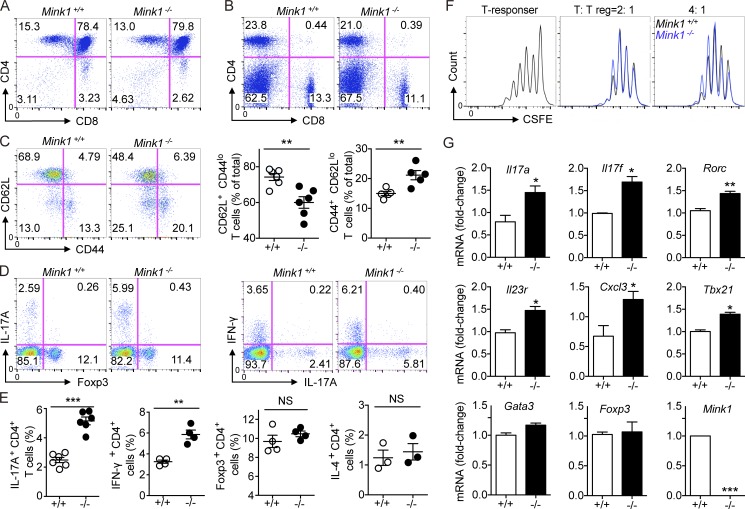
First, we analyzed the T cell compartment in Mink1−/− mice. The numbers and ratios of major thymocyte subsets were comparable with those in Mink1+/+ littermates (WT), suggesting a minimal role of MINK1 in thymocyte development (Fig. 1 A and not depicted). Although in vitro studies have documented a Ras-dependent apoptotic pathway that is mediated by MINK1 (Nicke et al., 2005), additional analysis has suggested a functional redundancy in Ras-dependent negative selection (Kortum et al., 2012) that may be independent of MINK1. In the periphery, the numbers of splenocytes and CD4+ T cells in Mink1−/− mice were normal, except for a slightly reduced number of CD8+ T cells (Fig. 1 B and not depicted). However, we found a marked increase in memory-like (CD44hiCD62Llo) T cells as well as a reduction of naive T cells in Mink1−/− CD4+ and CD8+ T cells (Fig. 1 C and not depicted). Then, we enumerated the effector T cell subsets in the periphery of both Mink1−/− and WT mice. Upon stimulation with PMA and ionomycin, MINK1 deficiency led to a two-to-three–times increase of Th17 (IL-17A+CD4+) and Th1 (IFN-γ+CD4+) cells, compared with WT cells (Fig. 1, D and E). Meanwhile, the percentage of Th2 (IL-4+CD4+) cells was not markedly changed (not depicted and Fig. 1 E).
Figure 1.
Loss of MINK1 in T cells results in the accumulation of Th17 cells in vivo. (A) Surface staining of CD4 and CD8 on Mink1−/− and WT thymocytes. Numbers in or adjacent to outlined areas (or in quadrants) indicate the percentages of cells in each throughout. (B) Splenocytes from Mink1−/− and WT mice stained for CD4 and CD8. Numbers in quadrants indicate the percentages of cells in each throughout. (C, left) Splenocytes from Mink1−/− and WT mice were stained for CD4, CD44, and CD62L and analyzed by flow cytometry. The gated CD4+ T cells were analyzed for CD44 and CD62L expression. Numbers in quadrants indicate the percentages of cells in each throughout. (Right) Percentages of naive (CD4+CD62L+) and memory (CD4+CD44+) T cells in the spleen of Mink1−/− and WT mice. (D) Splenocytes from Mink1−/− and WT mice were stimulated ex vivo with PMA + ionomycin for 5 h and analyzed for IL-17A–, IFN-γ–, and Foxp3-expressing CD4+ T cells by flow cytometry. The data shown were gated on CD4+ splenocytes, and numbers in quadrants indicate the percentages of cells in each throughout. (E) Percentages of splenic IL-17A+, IFN-γ+, IL-4+, and Foxp3+ CD4+ T cells in Mink1−/− and WT mice. (F) Suppression of CFSE-labeled CD4+ T cells by Mink1−/− and WT T reg cells, presented as CFSE dilution in responding T cells cultured at a ratio of 2:1 or 4:1 with T reg cells. (G) Real-time PCR analysis of the indicated genes’ expression in purified Mink1−/− and WT peripheral CD4+ T cells. Error bars show mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. n = 3–6 in each group; Student’s t test. Data are representative of three experiments.
Intriguingly, the frequency and number of Foxp3+ regulatory T cells (T reg cells) did not change in Mink1−/− mice in both spleen and LN T cells (Fig. 1, D and E; and not depicted). Mink1−/− T reg cells were able to suppress CD4+ T cell proliferation in vitro with similar efficiency as WT T reg cells (Fig. 1 F). We also compared the expression of Th cell signature genes in CD4+ T cells from Mink1−/− and WT mice. We found that the expression of Th17 lineage–specific genes (Il17a, Il17f, and Rorc) and Th1 signature Tbx21 was significantly increased in the Mink1−/− T cells, whereas Foxp3 and Gata3 gene expression were unchanged (Fig. 1 G). Together, these data suggest that MINK1 deficiency may favor T cell differentiation toward Th17 and Th1 cell lineages.
Changes of Th17, Th1, and T reg cells by MINK1 deficiency were further checked in the CD4+CD44+ T cell population. When Mink1−/− cells were compared with WT cells, the frequency of Th17 cells was three to four times higher (Fig. 2 A), whereas the frequencies of Th1 and T reg cells were comparable (Fig. 2 A). The data indicate that the increased frequency of Th17 cells in the Mink1−/− mice was not caused by the changed ratio of effector/memory T cells and suggest a specific regulatory role of MINK1 in Th17 cell differentiation. This is consistent with the observations in the lamina propria (LP) where an increased frequency of IL-17A+CD4+ but not Foxp3+CD4+ and IFN-γ+CD4+ T cells was detected in the small intestine of Mink1−/− mice (Fig. 2 B). Because γδ T cells are another major source of IL-17 production (Martin et al., 2009), we checked IL-17A production in γδ T cells from Mink1−/− mice. IL-17A–producing γδT cells were more prevalent in Mink1−/− mice than in WT controls (Fig. 2 C). Currently, we do not know whether it is a γδT-intrinsic effect.
Figure 2.
MINK1 suppress Th17 cell differentiation through a cell-intrinsic mechanism. (A, left) Splenocytes from Mink1−/− and WT mice were stimulated with PMA + ionomycin for 5 h, and the CD44+CD4+ population was gated and then analyzed for IL-17A+, IFN-γ+, and Foxp3+ cells as indicated. Numbers in quadrants indicate the percentages of cells in each throughout. (Right) Summary of IL-17A+, IFN-γ+, and Foxp3+ cells from the CD44+CD4+ population of Mink1−/− and WT mice. (B, left) FACS analysis of LP CD4+ T cells in the small intestine for IL-17A, Foxp3, and IFN-γ expression. (Right) Summary of CD4+IL-17A+, CD4+Foxp3+ T, and CD4+IFN-γ+ cells in Mink1−/− and WT small intestine. (C) Splenic γδT cells derived from Mink1−/− or WT animals were stimulated with PMA + ionomycin for 5 h and stained for IL-17A production. (D) Rag1−/− mice were reconstituted with mixed bone marrow cells from B6.SJL (CD45.1+) and Mink1−/− (CD45.2+) mice or from B6.SJL (CD45.1+) and WT (CD45.2+) mice at a 1:1 ratio. Total splenocytes in the recipient mice were analyzed 8 wk later for IL-17A+CD4+ and Foxp3+CD4+ T cells. The data shown were gated on CD4+CD45.1+ or CD4+CD45.2+ (Mink1−/− or WT) populations. (E, top) Total splenocytes in the recipient mice as in Fig. 2 D were analyzed for CD62L and CD44 expression. (Bottom) Percentages of naive (CD4+CD62L+) and memory (CD4+CD44+) T cells in the spleen of host mice. The numbers in the flow cytometry graphs show the percentages of the gated populations. Error bars show mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. n = 3–6 in each group; Student’s t test. Data are representative of three (A and B) or two (C–E) experiments.
MINK1 suppresses in vivo Th17 cell differentiation through a cell-intrinsic mechanism
To test whether the defects in Th17 cell differentiation in the Mink1−/− mice were intrinsic to T cells, we reconstituted irradiated Rag1−/− mice with Mink1−/− (CD45.2+) and WT congenic (CD45.1+) bone marrow cells mixed at a 1:1 ratio and analyzed the phenotypes of donor T cells 8 wk later. The frequency of the Mink1−/− IL-17A+ CD4+ T cells in the spleen was consistently higher than their WT counterparts derived from the cotransferred congenic WT bone marrow cells in the same host (Fig. 2 D). Once more, the numbers of Foxp3-expressing T reg cells were comparable with those in controls (Fig. 2 D). The peripheral Mink1–/– T cells from those Rag1−/− mice also recapitulated the enhanced memory phenotypes of CD4+ T cells from the parental Mink1−/− mice, suggesting an increased spontaneous activation of these cells caused by dysregulation of Th17 cell differentiation (Fig. 2 E). Together, these data demonstrate that MINK1 suppresses Th17 cell lineage differentiation in vivo through a T cell–intrinsic mechanism.
MINK1 specifically suppresses Th17 cell differentiation in vitro
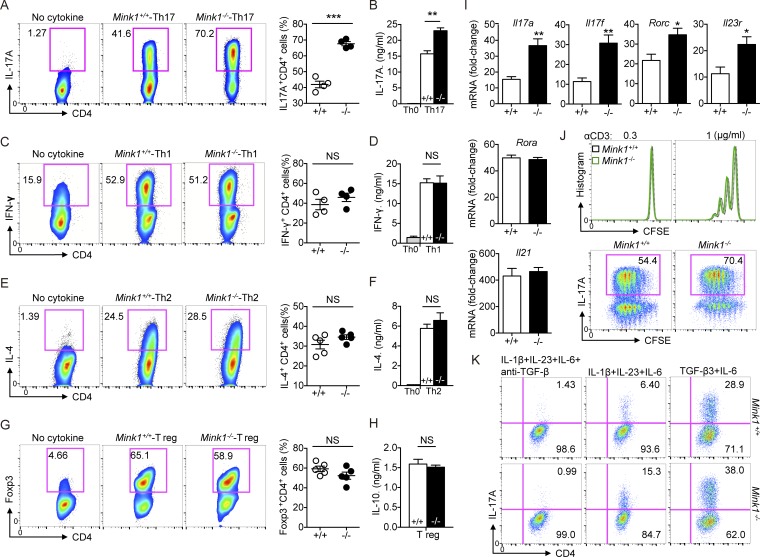
To further elucidate the contribution of MINK1 to Th17 cell differentiation, we polarized-sorted naive CD4+ T cells into Th17 cells through the use of TGF-β1 and IL-6 and compared the efficiency of Mink1−/− and WT naive T cells. The frequency of IL-17–producing cells generated from Mink1−/− T cell cultures was much greater than that of WT T cells (Fig. 3 A), and this was correlated with enhanced IL-17A secretion into culture supernatants (Fig. 3 B). Th1 and Th2 cell differentiation was not notably affected upon MINK1 deficiency (Fig. 3, C–F). The enhancement of Th17 cells was not accompanied by induced T reg cell (iT reg cell) dysfunction in Mink1−/− T cells, as naive T cells from Mink1−/− and WT mice expressed comparable levels of Foxp3 and IL-10 when activated under neutral (Th0) or iT reg cell–polarizing conditions (Fig. 3, G and H). We also found that expression of the key signatures of Th17 cells, such as Il17a, Il17f, Rorc, and Il23r, but not Il21 and Rora, were elevated in Mink1−/− T cells under the Th17 condition (TGF-β1 + IL-6; Fig. 3 I). Furthermore, CFSE dilution showed parallel proliferation capacity of Mink1−/− and WT T cells cultured under TCR stimulation or Th17 conditions (Fig. 3 J). In sum, these results indicate that MINK1 plays a negative role in Th17 cell differentiation.
Figure 3.
MINK1 suppresses Th17 cell polarization in vitro. (A, left) Naive CD4+ T cells (CD4+CD62LhiCD44loCD25−) from Mink1−/− and WT mice were differentiated into Th17 cells with 10 ng/ml IL-6 and 3 ng/ml TGF-β1. On day 5, the differentiated cells were stimulated with PMA + ionomycin for 5 h. The frequencies of IL-17A+ cells were determined by flow cytometry. (Right) Percentages of IL-17A+CD4+ T cells generated from in vitro polarization. (B) On day 5 after in vitro polarization, the supernatants of cell cultures were collected, and the secreted amounts of IL-17A were measured by ELISA. (C–H) Naive CD4+ T cells from Mink1−/− and WT mice were polarized under Th1 (C), Th2 (E), and iT reg (G) cell conditions and analyzed by flow cytometry 5 d after differentiation. Cytokines secreted into the cell culture supernatant (D, F, and H) were measured by ELISA. (I) Naive CD4+ T cells from Mink1−/− and WT mice were differentiated into Th17 cells with 10 ng/ml IL-6 and 3 ng/ml TGF-β for 5 d. The indicated genes’ expressions were analyzed by real-time PCR. (J, top) CFSE staining of naive T cells after 3 d of stimulation in the presence of plate-bound anti-CD3 antibody. (Bottom) Representative intracellular staining of IL-17A against CFSE on day 5 as in Fig. 3 A. (K) Naive CD4+ T cells from Mink1−/− and WT mice were differentiated into Th17 cells with the indicated cytokines. Induction of IL-17A+ cells was analyzed 5 d after differentiation. When indicated, the TGF-β antibody was added throughout the culture. The numbers in the flow cytometry graphs show the percentages of the gated populations. Error bars show mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. n = 3–6 in each group; Student’s t test. Data are representative of three experiments.
Cytokine combinations of IL-1β + IL-6 + IL-23 or TGF-β3 + IL-6 have recently been implicated in inducing pathogenic Th17 cells (Lee et al., 2012; Zhou et al., 2015). Thus, we examined the in vitro differentiation of naive Mink1−/− CD4+ T cells through the use of these two combinations. We found that the Th17 cell differentiation from naive Mink1−/− T cells was augmented in both conditions when compared with WT T cells (Fig. 3 K). A neutralizing antibody against TGF-β could block this Th17 cell differentiation in both Mink1−/− and WT T cells (Fig. 3 K).
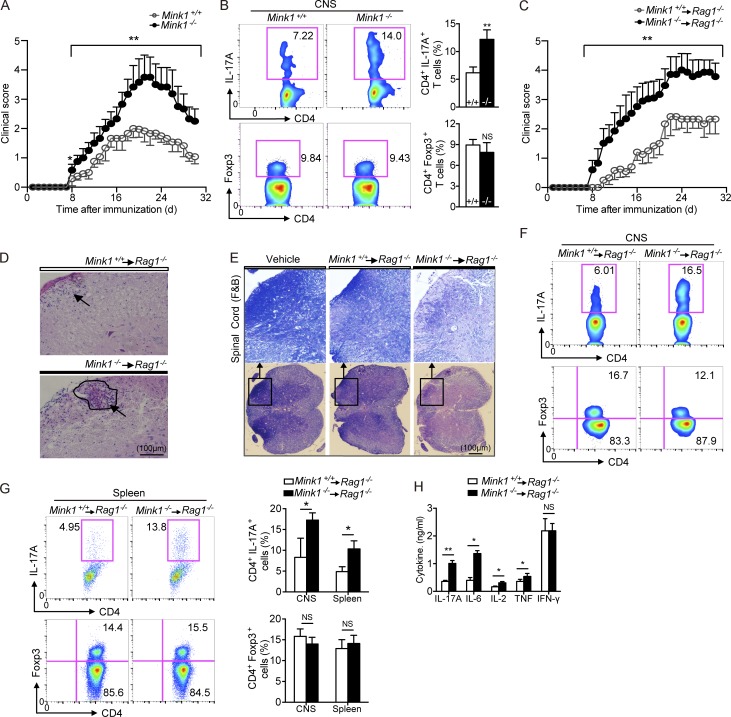
MINK1 deficiency leads to increased severity of EAE
To determine whether the altered number of Th17 cells caused by MINK1 deficiency impacts the development of Th17-dependent inflammatory diseases, we immunized Mink1−/− and WT mice with myelin oligodendrocyte glycoprotein peptide 35–55 (MOG35–55) to induce EAE. Mink1−/− mice exhibited significantly increased disease severity (Fig. 4 A). Histopathological analysis revealed more prominent demyelination in both the central nervous system (CNS) and spinal cord of MOG35–55-immunized Mink1−/− mice, as well as more numerous inflammatory foci (not depicted). Consistently, CD4+ T cells that infiltrated the CNS produced more IL-17A in Mink1−/− mice (Fig. 4 B). When we restimulated T cells isolated from mice immunized with MOG35–55 in vitro, the production of IL-17A by Mink1−/− T cells was considerably greater than that by WT T cells, whereas IFN-γ production was unaffected (not depicted). To delineate the intrinsic role of MINK1 in T cells, we sorted naive T cells (CD44loCD62L+) from Mink1−/− and WT mice, transferred these cells into Rag1−/− mice, and then monitored them for the development of EAE after MOG35–55 immunization. In contrast to the transfer of WT T cells, which resulted in mild disease, transfer of Mink1−/−-naive T cells resulted in EAE with high disease incidence, severity, and mortality (Fig. 4 C and not depicted). Histological examination showed significantly more mononuclear cell infiltration and demyelination in the spinal cord of hosts with Mink1−/− cells (Fig. 4, D and E). This was accompanied by elevated IL-17A production in both the CNS and the spleen, whereas T reg cells numbers appeared to be comparable (Fig. 4, F and G). In recall response to MOG35–55, the amount of IL-17A, IL-2, IL-6, and TNF secretion increased in mice with Mink1−/− T cells, whereas the IFN-γ levels were unchanged, as compared with WT mice (Fig. 4 H). These findings indicate that the primary determinant of the IL-17–dependent EAE phenotype in Mink1−/− mice is T cell intrinsic.
Figure 4.
T cell–intrinsic effects of MINK1 deficiency in EAE induction and Th17 cell accumulation. (A) Mean clinical scores for EAE in Mink1−/− and WT mice after being immunized with MOG35–55 + CFA plus pertussis toxin. (B, left) 18 d after EAE induction (at the peak of disease), CD4+ T cells were gated from the isolated leukocytes in the CNS and further analyzed for the frequencies of IL-17A+ and Foxp3+ cells. (Right) Summary of CNS IL-17A+CD4+ and Foxp3+CD4+ cells in Mink1−/− and WT EAE mice. (C) Mean clinical scores for EAE in Rag1−/− recipients of Mink1−/− and WT naive CD4+ T cells. (D and E) Representative histology of the spinal cord (hematoxylin and eosin in D and Luxol fast blue [F&B] in E) of Rag1−/− mice after EAE induction (day 32). (D) Arrowheads indicate inflammatory infiltration. (E) Outlines indicate demyelination by loss of normal blue staining of myelin. (F) On day 18 after EAE induction of Rag1−/− hosts, CD4+ T cells were gated from leukocytes isolated from the CNS and analyzed for the frequencies of IL-17A+ and Foxp3+ cells. (G, left) On day 18 after EAE induction of Rag1−/− hosts, spleen CD4+ T cells were analyzed for IL-17A+ and Foxp3+ expression. (Right) Summary of CNS and splenic IL-17A+CD4+ and Foxp3+CD4+ cells in Rag1−/− EAE mice. (H) Splenocytes isolated from EAE Rag1−/− mice (day 20) were restimulated in vitro with 5 µg/ml MOG peptide for 3 d, and then, the indicated cytokine production in the culture supernatants was assessed by ELISA. The numbers in the flow cytometry graphs show the percentages of the gated populations. Error bars show mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; Student’s t test. Data are representative six pairs of mice from two independent experiments.
MINK1 inhibits SMAD2 activation through direct phosphorylation of SMAD2-T324 residue
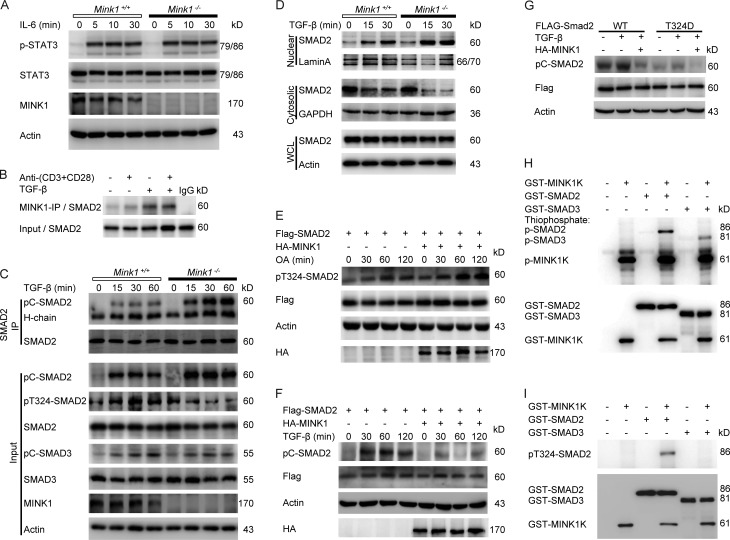
Then, we examined the pathways whereby MINK1 regulates Th17 cell differentiation. IL-6 signaling is required for the differentiation of Th17 cells, and the activation of STAT3 is a vital component of the Th17 cell–inducing mechanism. However, Western blot analysis showed no difference in STAT3 activation between Mink1−/− and WT CD4+ T cells stimulated with IL-6 (Fig. 5 A), suggesting that MINK1 does not play a major role in the modulation of early signaling events downstream of the IL-6 receptor. A previous study demonstrated a negative regulatory role of the Msn family kinase in MAD (mother against decapentaplegic; Drosophila SMAD) activation through a built-in phosphorylation of MAD in Drosophila. The Msn phosphorylated site of MAD (T312) is conserved in mouse SMAD2 (T324; Kaneko et al., 2011). Therefore, we proposed that MINK1 regulates Th17 cell differentiation through phosphorylation of SMAD2. Initially, we investigated whether MINK1 interacts with SMAD2 using coimmunoprecipitation assays. Stimulation with TGF-β induced an interaction between endogenous MINK1 and SMAD2 in both mouse primary T cells and human Jurkat T cells (Fig. 5 B and not depicted). Then, we compared the activation (C-terminal phosphorylation) of SMAD2 in Mink1−/− CD4+ T cells and their WT counterparts. Activation by anti-CD3 and anti-CD28 antibodies along with TGF-β treatment led to rapid C-terminal phosphorylation and nuclear translocation of SMAD2 in CD4+ T cells. These were markedly enhanced in Mink1−/− T cells, accompanied by diminished levels of T324 phosphorylation, suggesting an inhibitory role of MINK1 in SMAD2 activation through T324 phosphorylation (Fig. 5, C and D). However, SMAD3 phosphorylation was comparable in Mink1−/− and WT T cells (Fig. 5 C), indicating that MINK1 may selectively regulate the physiological responses to TGF-β mediated by SMAD2.
Figure 5.
MINK1 inhibits SMAD2 activation via direct phosphorylation of the T324 residue in the SMAD2 α-helix 1 region. (A) Purified CD4+ T cells were stimulated for the indicated times with 50 ng/ml IL-6. Phosphorylated and total STAT3 proteins were detected in Western blot assays. (B) Mouse primary CD4+ T cells were stimulated with TGF-β, anti–CD3 + CD28 antibody, or both for 30 min and then lysed and immunoprecipitated (IP) with anti-MINK1 and probed with anti-SMAD2 in the immunoblot analysis. (C, top) After anti–CD3 + CD28 activation (5 µg/ml for 30 min), Mink1−/− and WT CD4+ T cells were stimulated with 10 ng/ml TGF-β as indicated, whole-cell lysates were immunoprecipitated with anti-SMAD2, and bound proteins were probed as indicated. (Bottom) The cell lysates were probed with the indicated antibodies in the immunoblots. (D) Mouse primary CD4+ T cells were treated as in Fig. 5 C. Both the nuclear and cytoplasmic fractions were collected, and the SMAD2 levels were examined. The nuclear-localized protein lamin A and cytoplasmic-localized protein GAPDH demonstrate separation of the fractions and proper sample loading. WCL, whole-cell lysate. (E and F) Flag-tagged SMAD2 was overexpressed with or without hemagglutinin (HA)-tagged MINK1 in 293T cells and treated with either the serine/threonine phosphatase inhibitor okadaic acid (OA; 125 nM; E) or 2 ng/ml TGF-β (F) for the indicated times and then lysed and immunoblotted with the indicated antibodies. (G) 293T cells were transfected with either WT or SMAD2-mutant T324D with or without MINK1 and treated with TGF-β. Cell lysates were immunoblotted with the indicated antibodies. (H) In vitro kinase assays were performed with purified recombinant SMAD2 or SMAD3 (GST tagged) as substrates in the presence of ATP-γ-S. The reaction products were immunoblotted with anti-thiophosphate ester antibody. Recombinant MINK1 kinase domain (GST-MINK1K) purified from E. coli directly phosphorylated SMAD2. (I) In vitro kinase assay. The recombinant MINK1 kinase domain directly phosphorylated SMAD2 at the T324 residue. The reaction products were immunoblotted with anti-pT324–SMAD2 antibody. Data are representative of three independent experiments.
To confirm that MINK1 is capable of promoting T324 phosphorylation, we coexpressed MINK1 with SMAD2 in human 293T cells and found that T324 phosphorylation of SMAD2 was greatly enhanced when coexpressed with MINK1 (Fig. 5 E); In contrast, the C-terminal phosphorylation of SMAD2 was markedly decreased when MINK1 was transiently expressed (Fig. 5 F). In addition, mutation of the T324 into aspartic acid (D), which mimics the phosphorylated threonine (Thr), almost completely abolished the C-terminal phosphorylation of SMAD2 in response to TGF-β (Fig. 5 G). Moreover, in vitro kinase assays with glutathione S-transferase (GST) fusion of the kinase domain of MINK1 produced in Escherichia coli showed direct phosphorylation of the SMAD2-T324 residue by MINK1 (Fig. 5, H and I). Collectively, these data demonstrate that MINK1 inhibits SMAD2 activation through direct phosphorylation of the T324 residue.
Th17 cell differentiation is regulated by phosphorylation of the SMAD2-T324 residue
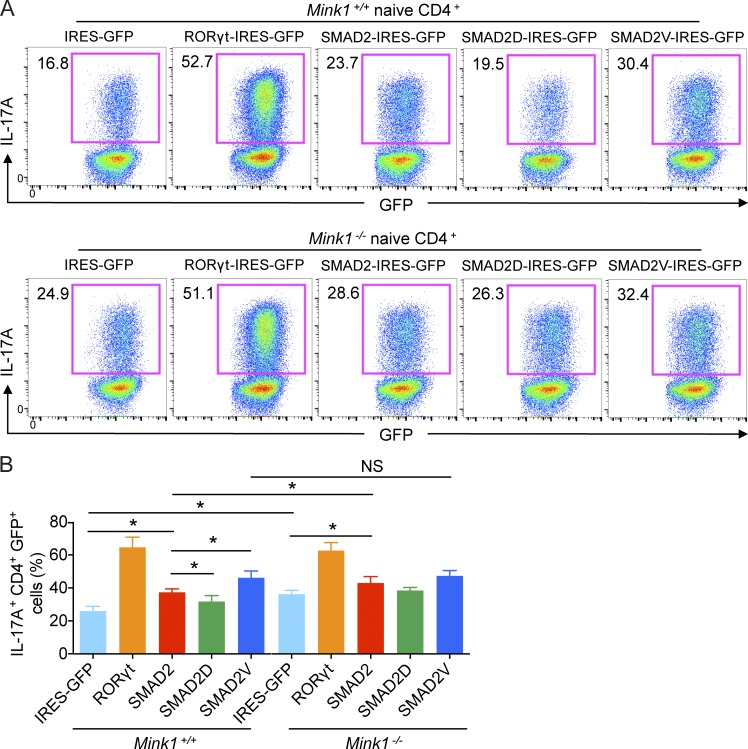
To directly test the contribution of SMAD2-T324 phosphorylation during Th17 cell differentiation, naive T cells from Mink1−/− or WT mice were infected with retroviruses that expressed WT SMAD2 or SMAD2-T324D/V mutants (i.e., mutation of T324 to phosphomimetic aspartic or a nonphosphorylatable valine; Kaneko et al., 2011) and differentiated them into Th17 cells in vitro. Transcription factor RORγt overexpression was used as a positive control (Fig. 6, A and B). As previously reported (Martinez et al., 2010; Chang et al., 2011), expression of WT SMAD2 enhanced Th17 cell differentiation compared with the expression of the empty vector (Fig. 6, A and B). More importantly, SMAD2-T324V mutants were much more potent in inducing Th17 cell differentiation than WT SMAD2 (30 vs. 23%), whereas T324D mutants had the opposite effect (19 vs. 23%; Fig. 6, A and B), indicating that phosphorylation of SMAD2 at α-helix 1 inhibits Th17 cell differentiation. In Mink1−/− T cells, overexpression of WT SMAD2 by retrovirus also induced more IL-17A–expressing cells than the vector, and this was higher than in infected WT T cells (Fig. 6, A and B). However, SMAD2-T324V, which could not be phosphorylated by MINK1, induced similar proportions of IL-17A–expressing cells from both Mink1−/− and WT T cells (Fig. 6, A and B). Together with the results from the biochemical studies described above, our data suggest that MINK1 negatively controls SMAD2-dependent Th17 cell differentiation through phosphorylation of SMAD2 at T324.
Figure 6.
MINK1 mediates Th17 cell differentiation by mediating SMAD2-α–helix 1 phosphorylation. (A) Sorted Mink1−/− and WT naive T cells were polarized under Th17 cell conditions as described in Fig. 3 A, except that the cells were first infected with the indicated retrovirus, and the cytokines were added 24 h later. The infection was repeated once at 48 h. 5 d later, the cells were analyzed for IL-17A+ cells after restimulation with PMA + ionomycin for 5 h. The infection efficiency was determined by GFP expression. The numbers in the graphs show the percentages of the gated populations. (B) Summary of retrovirus-infected Th17 cells as described in Fig. 6 A. Error bars show mean ± SD. *, P ≤ 0.05. n = 3 in each group; Student’s t test. Data are representative of two independent experiments. IRES, internal ribosomal entry site.
NAC potentiates Th17 cell differentiation in vitro in a MINK1-dependent manner
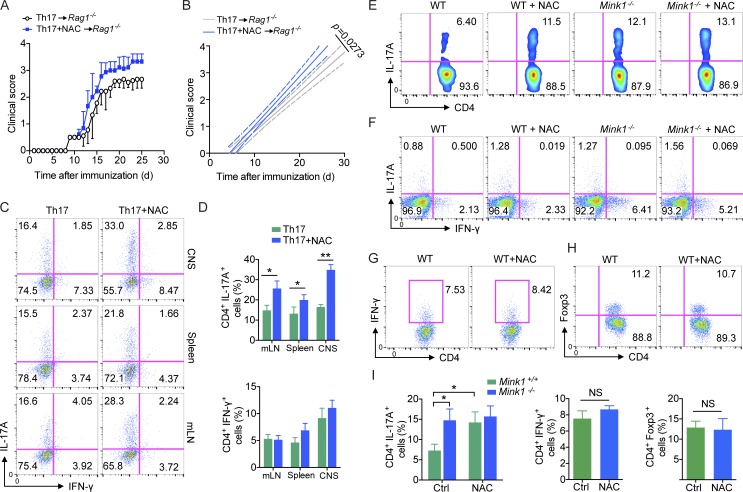
MINK1 has been reported to be activated by ROS (Nicke et al., 2005), and ROS have recently been implicated in regulating Th17 cell differentiation, although their effect is quite controversial (Zhi et al., 2012; Lu et al., 2013; Won et al., 2013; Gerriets et al., 2015). Considering the increased Th17 phenotype in Mink1−/− mice, we raised the question of whether that MINK1 mediates the suppression of Th17 cell differentiation reflects the effects of ROS. In 293T cells, MINK1 kinase activity was mildly enhanced by addition of H2O2 and significantly reduced by addition of NAC to cell cultures (Fig. 7, A and B). Pretreatment of cells with NAC also inhibited H2O2-induced MINK1 activity (Fig. 7, A and B). In primary mouse CD4+ T cells, MINK1 was fully activated by TCR stimulation in a ROS-dependent manner, as the kinase activity of MINK1 could not be further elevated by H2O2 treatment but was critically compromised by treatment with the antioxidant NAC (Fig. 7 C). Also, the kinase activity of MINK1 was profoundly reduced in Th17 conditions by NAC treatment (Fig. 7 D). Furthermore, we investigated the differentiation of Th17 cells under Th17 cell–polarizing conditions in the presence of NAC and confirmed increased numbers of IL-17A+ cells with additional 0.5-mM NAC (Fig. 7 E). Notably, NAC-induced increase in Th17 cell development did not occur in MINK1-deficient T cells (Fig. 7 E). In contrast, NAC treatment did not affect iT reg cell differentiation or T reg cell suppressive activity (Fig. 7, F and G). Consistently, treatment of CD4+ T cells with TGF-β plus NAC resulted in stronger C-terminal phosphorylation and weaker T324 phosphorylation of SMAD2 than TGF-β alone (Fig. 7 H). Importantly, the phosphorylation levels of both were not affected by NAC treatment in Mink1−/− CD4+ T cells (Fig. 7 H). Thus, our data indicated that NAC boosts Th17 cell differentiation in vitro in a MINK1-dependent manner.
Figure 7.
NAC inhibits MINK1 activity and promotes Th17 cell differentiation in vitro in a MINK1-dependent manner. (A) Human 293T cells were transfected with a hemagglutinin-tagged MINK1 expression construct and treated with 0.2 mM H2O2, 2 mM NAC, or H2O2 after pretreatment with NAC, as indicated, for 30 min. MINK1 was immunoprecipitated from the lysates with anti-MINK1. In vitro kinase assays were performed with myelin basic protein (MBP) as substrate in the presence of ATP-γ-S. The reaction products were immunoblotted with anti-thiophosphate ester antibody. Rel., relative. (B) MINK1 was immunoprecipitated as in Fig. 7 A, and MINK1 kinase activity was assessed with a luminescence-based assay. (C) MINK1 kinase activity assays in CD4+ T cells treated with H2O2 or NAC. The kinase activity was assessed with luminescence. (D) Naive CD4+ T cells were polarized into Th17 cells with the indicated concentrations of NAC in culture for 12 h. MINK1 was immunoprecipitated, and in vitro kinase assays were performed with myelin basic protein as the substrate in the presence of ATP-γ-S. The reaction products were immunoblotted with anti-thiophosphate ester antibody. (E, left) Naive CD4+ T cells were polarized in vitro under Th17 conditions (TGF-β1 + IL-6) in the presence (NAC) or absence (Vehicle) of 0.5 mM NAC and analyzed for IL-17A by flow cytometry. (Right) IL-17A secretion in the culture supernatant measured by ELISA. (F) Foxp3 production by Mink1−/− and WT naive T cells stimulated under iT reg cell conditions in the presence (NAC) or absence (Vehicle) of 0.5 mM NAC was analyzed by flow cytometry. (E and F) The numbers in the flow cytometry graphs show the percentages of the gated populations. (G) Suppression of CFSE-labeled CD4+ T cells by NAC-intervened T reg cells, presented as CFSE dilution in responding T cells cultured at a ratio of 2:1 with T reg cells. (H) After anti–CD3 + CD28 activation (5 µg/ml for 30 min), Mink1−/− and WT mice primary CD4+ T cells were stimulated with 10 ng/ml TGF-β and 0.5 mM NAC, as indicated, and the cell lysates were probed with the indicated antibodies in the immunoblots. Error bars show mean ± SD. ***, P ≤ 0.001. n = 3–5 in each group; Student’s t test. Data are representative of three independent experiments.
NAC induces Th17 cells in vivo and exacerbates EAE through a T cell–intrinsic mechanism
To determine whether increased Th17 cells upon NAC treatment could result in an enhanced pathogenicity in autoimmune disease, we differentiated MOG35–55-immunized CD4+ T cells (day 8) cells into Th17 cells with or without NAC treatment in vitro and then transferred these cells to Rag1−/− mice and induced EAE. Mice that received NAC-treated cells developed more severe disease compared with those that received control cells (Fig. 8, A and B). We observed a dramatically higher frequency of Th17 cells in the CNS, spleen, and mesenteric LN of Rag1−/− mice that received NAC-treated cells than in those of controls (Fig. 8, C and D). The percentage of IFN-γ–producing T cells in these organs was not altered in mice that received NAC-treated cells (Fig. 8, C and D), suggesting that NAC treatment directly affected the pathogenicity of antigen-specific T cells through promoting Th17 cell differentiation.
Figure 8.
Dietary supplementation of NAC potentiates Th17 cell differentiation in vivo. (A) On day 8 after immunized with MOG35–55 + CFA, WT CD4+ T cells were sorted and differentiated with 10 ng/ml IL-6 and 3 ng/ml TGF-β1 in the presence or absence of 0.5 mM NAC and transferred into Rag1−/− mice. 1 d later, the recipient mice were immunized with MOG35–55 plus pertussis toxin to induce EAE. Clinical scores were recorded and calculated each day for the indicated times. (B) Linear regression curves of Fig. 8 A. Dashed lines indicate the 95% confidence intervals of the regression lines. (C) IL-17A and IFN-γ production by CD4+ T cells isolated from the indicated organs of Rag1−/− mice in Fig. 8 A at the peak of disease after immunization with MOG35–55 plus pertussis toxin. mLN, mesenteric LN. (D) Quantification of CD4+IL-17A+ and CD4+IFN-γ+ T cells from the indicated organs of EAE animals at the peak of disease in Fig. 8 A. (E) Representative intracellular staining of IL-17A+ CD4+ T cells from the small intestine of Mink1−/− and WT mice treated with NAC or regular drinking water for 2 mo. (F) Mesenteric LN IL-17A+CD4+ and IFN-γ+CD4+ cell frequencies of Mink1−/− and WT mice treated with either NAC or regular drinking water. (G and H) Small intestine LP IFN-γ+CD4+ (G) or Foxp3+CD4+ (H) cell frequencies of unimmunized WT mice either untreated or treated with NAC. (I) Percentages of small intestine LP IL-17A+CD4+, IFN-γ+CD4+, and Foxp3+CD4+ cell frequencies of NAC-treated or untreated mice. Ctrl, control. The numbers in the flow cytometry graphs show the percentages of the gated populations. Error bars show mean ± SD. *, P ≤ 0.05; **, P ≤ 0.01; Student’s t test. Data are representative of six (A–D) or three (E–I) mice in each group from two independent experiments.
Previous studies have shown that different components of common dietary supplements and the gut microbiota can overtly affect the percentage of effector T cells in the gut (Berer et al., 2011; Atarashi et al., 2015). In addition, recent data indicate that molecules associated with ROS homeostasis can influence Th17 cell responses and inflammation (Gerriets et al., 2015; Ravindran et al., 2016). To further understand the effect of NAC on Th17 cell generation in vivo, we added NAC into the drinking water of Mink1−/− and WT mice. After 2 mo on NAC, nonimmunized WT mice showed a marked increase in the frequency of Th17 cells in the LP and mesenteric LNs (Fig. 8, E, F, and I), whereas there was no change in IFN-γ+ and Foxp3+ cells (Fig. 8, G–I). Importantly, the Th17 cell frequency was not affected in the gut of MINK1-deficient mice that had received NAC (Fig. 8, E and I).
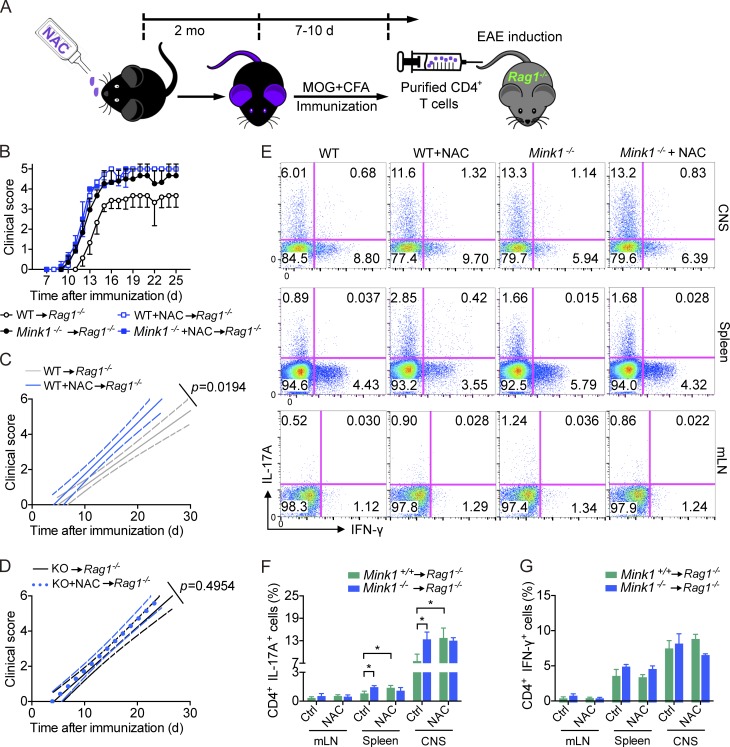
Finally, to understand better whether NAC supplementation would affect the development of EAE in vivo, we immunized NAC-treated mice with MOG35–55 to induce EAE. To our surprise, NAC supplementation did not significantly affect the severity of EAE as measured by clinical scores during the course of disease (not depicted). We suspect NAC may affect cells other than T cells. It is well known that NAC could reduce tissue injury during inflammation (Sayin et al., 2014; Schieber and Chandel, 2014). To focus the effect of NAC on T cells, we purified CD4+ T cells from these mice, transferred them into Rag1−/− hosts, and then examined their ability to induce EAE (Fig. 9 A). Mice that received T cells from NAC-treated mice developed more severe disease than those that received T cells from control mice. Again, the severity of disease was increased in mice receiving MINK1-deficient T cells, which, however, was not further elevated by NAC treatment (Fig. 9, B–D). Consistently, Th17 cell numbers in the CNS of Rag1−/− mice that received NAC-treated T cells were notably higher than in controls (Fig. 9, E–G). We also found a higher frequency of Th17 cells in the lymphoid organs of mice with T cell transfer from NAC-treated mice at the peak of disease (Fig. 9, E–G). The data presented here indicate that dietary supplementation with the antioxidant NAC potentiates Th17 cell generation in vivo in a MINK1-dependent manner and therefore has the potential to increase the risk of promoting autoimmune inflammatory diseases.
Figure 9.
NAC supplementation results in elevated potentiality of T cells in inducing EAE. (A) Schematic illustration of adoptive transfer experiments shown in Fig. 9 B. (B) Mink1−/− and WT mice either untreated (vehicle only) or treated with NAC were immunized with MOG35–55 + CFA. On day 8 after immunization, CD4+ T cells were sorted and transferred into Rag1−/− mice. 1 d later, the recipient mice were immunized with MOG35–55 plus pertussis toxin to induce EAE. Clinical scores were recorded and calculated each day for the indicated times. (C and D) Linear regression curves of Fig. 9 B. Dashed (thinner) lines indicate the 95% confidence intervals of the regression lines. (E) IL-17A and IFN-γ production by CD4+ T cells isolated from indicated organs of Rag1−/− mice in Fig. 9 B at the peak of disease after immunization with MOG35–55 plus pertussis toxin. The numbers in the graphs show the percentages of the gated populations. (F and G) Quantification of CD4+IL-17A+ and CD4+IFN-γ+ T cells from the indicated organs of EAE animals (at the peak of diseases) in Fig. 9 B. Error bars show mean ± SD. *, P ≤ 0.05; Student’s t test. Data are representative of nine mice in each group from two independent experiments. Ctrl, control; mLN, mesenteric LN.
Discussion
Here, we reveal that MINK1 suppresses Th17 cell differentiation. Activated downstream of ROS, MINK1 phosphorylated SMAD2 at T324 and inhibited the activation of SMAD2. This regulation was further demonstrated in vivo as more severe EAE disease in Mink1−/− mice. These findings suggest cross talk between redox status and cytokine-induced signals during Th17 cell differentiation.
Although MINK1 had previously been established as a regulator in thymocyte development, this study is the first to unveil its contribution to Th17 cell differentiation. MINK1 (MAP4K6) is a kinase that belongs to the MAP4K family, several members of which have been implicated in T cell activation and differentiation. For instance, loss of HGK (HPK1/GCK-like kinase; MAP4K4) in T cells enhances Th17 cell differentiation in adipose tissue (Chuang et al., 2014). GLK (GCK-like kinase; MAP4K3) deficiency impairs T cell–mediated immune responses in mice (Chuang et al., 2011). HPK1 (MAP4K1) has been identified as a negative regulator of TCR signaling (Shui et al., 2007). Interestingly, despite their original identification as regulators of MAPK pathways, these MAP4Ks modulate T cell development and differentiation in both a MAPK-dependent (HPK1) and a MAPK-independent (HGK and GLK) manner (Shui et al., 2007; Chuang et al., 2011, 2014). The suppression of Th17 cell differentiation by MINK1 is clearly independent of MAPKs, as indicated by unaltered TCR-induced activation of Erk, p38, and JNK in Mink1−/− CD4+ T cells (unpublished data).
SMAD proteins comprise the canonical signaling pathway downstream of TGF-β receptor activation (Derynck and Zhang, 2003). Regulated SMAD proteins, especially SMAD2 and SMAD3, have been reported to be essential in both Th17 and T reg cell differentiation (Tone et al., 2008; Martinez et al., 2010; Yoon et al., 2015). MINK1 was found to specifically target the differentiation of Th17 cells, but not iT reg cells, via selective phosphorylation of SMAD2, which dampened its activity. The cell-type preference of MINK1 is in keeping with findings that SMAD2 is dominant in Th17 cell differentiation (Malhotra et al., 2010; Martinez et al., 2010; Yoon et al., 2015). Despite bearing almost homologous amino acid sequences, SMAD2 and SMAD3 do differ in certain residues, including those that can be specifically modified (Wrighton et al., 2009; Kaneko et al., 2011; Bruce and Sapkota, 2012). This feature was exemplified in our experiments that showed that SMAD3 was not phosphorylated by MINK1 and that the activity of SMAD3 was not changed by MINK1 deficiency. The unaffected iT reg differentiation in MINK1-deficient cells may largely reflect the unaltered SMAD3 activity. Serine residue phosphorylation is critical for the activation of SMAD proteins, and this is best indicated by studies on regulation of Th17 and T reg cell differentiation by phosphorylation of C-terminal SXS motifs and the linker region on SMAD2 and SMAD3 (Chang et al., 2011; Yoon et al., 2015). In accord with a previous study in Drosophila showing that MAD activation can be regulated by the conserved threonine residue T324 in the α-helix 1 region (Kaneko et al., 2011), we have provided evidence that MINK1 regulates the activation of SMAD2 through direct phosphorylation of this residue during Th17 cell differentiation. To the best of our knowledge, this is the first report of the physiological/pathophysiological significance of this regulation, which is involved in the differentiation of a crucially important T cell subtype in the context of autoimmune disease. The T324 residue of SMAD2 also exists in SMAD1 (T312), which can be activated by TGF-β3 (Lee et al., 2012). Considering the augmented in vitro Th17 cell differentiation in Mink1−/− T cells under the TGF-β3 + IL-6 condition (Fig. 3 K), our supposition does not exclude the possibility that the EAE phenotype (Fig. 4) of Mink1−/− mice is a functional redundancy of both SMADs’ activation.
It is well known that intracellular ROS are elevated during T cell activation as a result of increased glycolytic flux after TCR engagement (Sena et al., 2013). Because of their destructive roles in cells and tissues upon release, ROS are traditionally viewed as strong mediators of inflammation (Sena et al., 2013; Schieber and Chandel, 2014). However, ROS were recently identified as possible negative regulators of the T cell–mediated inflammatory responses (Hultqvist et al., 2009). Consistent with the findings that T cells with low ROS production owing to genetic mutation are more potent in inducing arthritis (Olofsson et al., 2002; Hultqvist et al., 2004; Gerriets et al., 2015), we observed elevated levels of Th17 cell differentiation upon the inhibition of ROS by NAC. Thus, the current study not only explained the negative regulatory role of ROS in the Th17 cell–mediated autoimmune inflammatory disease model, but also established MINK1 as a key molecule executing the effects of ROS on the differentiation of Th17 cells. Although the mechanism by which ROS activate MINK1 remains to be elucidated, our findings suggest that a cautious approach is required when antioxidants are used to counteract the side effects of inflammation and in the treatment of inflammatory-related diseases, such as tumors, aging, and metabolic disorders, particularly when Th17 cells and autoimmune disease such as multiple sclerosis are considered.
Materials and methods
Mice, EAE induction, and adoptive transfer
Mink1−/− mice were generated by homologous recombination-mediated gene targeting as previously described on a mixed B6 × 129 background and backcrossed to B6 for at least eight times (Yue et al., 2016). C57BL/6(B6) and Rag1−/− mice were purchased from the Model Animal Research Center of Nanjing University, and CD45.1 (B6.SJL) mice were purchased from The Jackson Laboratory. All mice were housed in Zhejiang University Laboratory Animal Center. The experimental protocols were approved by the Review Committee of Zhejiang University School of Medicine and were in compliance with institutional guidelines.
EAE was induced as described previously (Lu et al., 2007). In brief, male mice age 6–8 wk were immunized with 200 mg MOG35–55 (MEVGWYRSPFSRVVHLYRNGK; Sangon) in an equal amount of complete Freund’s adjuvant (Chondrex, Inc.) and received 200 ng pertussis toxin (List Biochemicals) intravenously on days 0 and 2 after induction. Clinical evaluation was performed daily using a five-point scale: 0, no clinical signs; 1, limp tail; 2, limp tail, impaired righting reflex, and paresis of one limb; 3, hind-limb paralysis; 4, hind-limb and fore-limb paralysis; and 5, moribund. NAC (616-91-1; ≥99% purity; Sigma-Aldrich) was administered in the drinking water at 1 g/L. The NAC water was replaced every 3 d.
Bone marrow cells were transplanted as described previously (Wang et al., 2012). In brief, bone marrow cells were isolated from Mink1−/−, WT, or CD45.1 mice, and 5 × 106 cells were injected intravenously into Rag1−/− mice, which had received sublethal irradiation (400 rad) 1 d before. Chimeric mice were analyzed 6–8 wk after transplantation.
For EAE induction in Rag1−/− mice, naive T cells or CD4+ T cells from WT and Mink1−/− mice were sorted by an AutoMacs separator (STEMCELL Technologies) and intravenously transferred into Rag1−/− mice at 7 × 106 per mouse for naive T cell transfer, 1 × 106 per mouse for CD4+ T cell transfer, and 2 × 105 per mouse for differentiated Th17 cell transfer. 2 d later, the recipient mice were immunized with MOG35–55 as described in the second paragraph of this section.
Antibodies, recombinant cytokines, and reagents
The following monoclonal antibodies and reagents were used: for surface staining, anti-CD4 (GK1.5), anti-CD8a (53-6.7), anti-CD25 (PC61), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-CD45.1 (A20), anti-CD45.2 (104), and anti–TCR-β chain (H57-597; all from BioLegend); for intracellular (cytoplasmic and nuclear) staining, anti–IL-17A (TC11-18H10.1), anti–IL-4 (11B11), anti–IFN-γ (XMG1.2; from BioLegend), anti-Foxp3 (FJK-16s), and anti-RORγt (B2D; from eBioscience); and for T cell stimulation, anti-CD3 (145-2c11) and anti-CD28 (37.51; from BioLegend). Recombinant mouse TGF-β1, IL-6, IL-4, IL-12, IL-1β, and IL-23 were from R&D Systems and recombinant mouse IL-2 was from PeproTech. Neutralizing anti–IFN-γ (XMG1.2), anti–IL-4 (11B11), and anti–IL-12 (C17.8) were from BioLegend. CFSE was from Invitrogen.
In vitro T cell differentiation
Sorted naive CD4+ T cells (CD4+CD62L+CD44−) were cultured on irradiated (30 Gy) CD90.2lo splenocytes with 1 µg/ml anti-CD3 and 2 µg/ml anti-CD28 at a ratio of 1:5. The naive cells were cultured at 106/ml in RPMI 1640 medium supplemented with 10% FBS, sodium pyruvate, Hepes, penicillin/streptomycin, gentamicin sulfate, and 2-mercaptoethanol. The following cytokines were added to generate each subset: Th1, 10 ng/ml IL-12 and 10 µg/ml anti–IL-4; Th2, 50 ng/ml IL-4 and 10 µg/ml anti–IFN-γ; and Th17, 10 ng/ml IL-6, 3 ng/ml TGF-β1, 3 ng/ml TGF-β3, 20 ng/ml IL-1β, 20 ng/ml IL-23, 10 µg/ml anti–IFN-γ, and 10 µg/ml anti–IL-4. When indicated, 5 µg/ml TGF-β antibody (1D11; R&D Systems) was added into the culture. For T reg cells, 3 ng/ml TGF-β, 10 µg/ml anti–IFN-γ, and 10 µg/ml anti–IL-4 were used. On day 3 after stimulation, cells were split 1:2 and replated with 50 U/ml IL-2 alone for an additional 2 d before analysis.
In vitro suppression assays
Mouse CD4+ T cells were negatively selected from the spleen and LNs of mice by using an AutoMacs separator (STEMCELL Technologies). Sorted CD4+ CD25+ T reg cells were added to CD4+ T cells at a final concentration of 3 × 105 cells per 500 µl and stimulated for 72 h with anti-CD3.
Isolation of LP lymphocytes
Mice were sacrificed, and the intestines were removed and placed in ice-cold PBS. After removal of residual mesenteric fat tissue, Peyer’s patches were carefully excised, and the intestine was opened longitudinally. Then, the intestine was thoroughly washed in ice-cold PBS and cut into 1–2-cm pieces. The pieces were incubated in 15 ml of 5 mM EDTA in 10% FBS/RPMI medium for 15–20 min at 37°C with slow rotation at 100 rpm. After incubation, the epithelial cell layer, containing the intraepithelial lymphocytes, was removed by intensive vortexing and passaged through a 100-µm cell strainer. Then, the pieces were washed in RPMI medium and placed in 10 ml of digestion solution containing 4% fetal calf serum, 1 mg/ml collagenase II (Sigma-Aldrich), and 50 µg/ml DNase I (Sigma-Aldrich). Digestion was performed by incubating the pieces at 37°C in a water bath for 20 min with rotation at 120 rpm. After the initial 20 min, the solution was vortexed intensely and passed through a 100-µm cell strainer. The pieces were collected and placed into fresh digestion solution, and the procedure was repeated. Supernatants from both digestions (or from the EDTA treatment for intraepithelial lymphocyte isolation) from a single small intestine were combined, washed once in cold FACS buffer, resuspended in 5 ml of the 40% fraction of a 40:80 Percoll gradient, and overlaid on 2.5 ml of the 80% fraction in a 15-ml Falcon tube. Percoll gradient separation was performed by centrifugation for 20 min at 2,500 rpm at room temperature. LP lymphocytes were collected at the interphase of the Percoll gradient, washed once, and resuspended in FACS buffer or T cell medium. The cells were used immediately for experiments.
Retroviral infection
Retroviruses were produced by transfecting Plat-E cells with pMX-IRES-GFP, pMX-WT SMAD2 IRES-GFP, and pMX-SMAD2-T324D/V IRES-GFP. Fresh viruses from the supernatants were collected 3 d later and used to infect T cells. To infect T cells, naive T cells were first stimulated with anti-CD3 and anti-CD28 antibodies. At the 24- and 36-h time points, activated T cells (0.5 × 106) were spun infected for 1 h at 2,000 rpm with 500 µl of viral supernatant in the presence of 10 µg/ml polybrene and incubated at 37°C for one additional hour before being removed from the viral supernatant and resuspended in the indicated T cell differentiation medium.
Flow cytometry and cytokine detection
Mice were perfused with PBS before brains were harvested. Brains were pretreated with 2 µg/ml collagenase D (Roche) and 1 µg/ml DNase I (Sigma-Aldrich), and total cells were isolated by cell straining (70 µm for spleen and mesenteric LN and 100 µm for brain). Brain homogenates were separated into neuronal and leukocyte populations by discontinuous density gradient centrifugation using Percoll (Pharmacia). For surface staining, cells were stained with the respective antibodies for 20 min in PBS containing 2% FBS before analysis. For intracellular staining, cells were stimulated for 4–5 h with 50 ng/ml PMA and 500 ng/ml ionomycin (both from Beyotime) in the presence of brefeldin A (BioLegend), fixed, made permeable (Fix/Perm; eBioscience) according to the manufacturer’s instructions, and stained with the respective antibodies for 20–30 min for intracellular cytokine detection. Before fixation, cells were stained with Fixable Viability Dye (eBioscience) to exclude dead cells. Data were acquired on a FACSCalibur cell analyzer (BD) and analyzed with FlowJo software (Tree Star). Culture supernatants were measured by ELISA for secretion of IL-17A, IL-6, IL-2, TNF, and IFN-γ (eBioscience) according to the manufacturer’s instructions.
Quantitative RT-PCR
Cells were stimulated as indicated, and RNA was extracted using RNAiso-Plus (9109; Takara Bio Inc.). The following primers were used in the study. Il17a: forward, 5′-TCCAGAAGGCCCTCAGACTA-3′ and reverse, 5′-TCAGGACCAGGATCTCTTGC-3′; Il17f: forward, 5′-CCCCATGGGATTACAACATCAC-3′ and reverse, 5′-CATTGATGCAGCCTGAGTGTCT-3′; Il23r: forward, 5′-AACATGACATGCACCTGGAA-3′ and reverse, 5′-TCCATGCCTAGGGAATTGAC-3′; Cxcl3: forward, 5′-GTTGTGGCCAGTGAGCTG-3′ and reverse, 5′-CAAGCTCTGGATGGTCTCAA-3′; Tbx21: forward, 5′-GCCAGGGAACCGGTTATATG-3′ and reverse, 5′-GACGATCATCTGGGTCACAT-3′; Rorc: forward, 5′-TGAGGCCATTCAGTATGTGG-3′ and reverse, 5′-ACACCACCGTATTTGCCTTC-3′; Il21: forward, 5′-ATGCAGCTTTTGCCTGTTTT-3′ and reverse, 5′-TGGGTGTCCTTTTCTCATACG-3′; Rora: forward, 5′-GTGGAGACAAATCGTCAGGAAT-3′ and reverse, 5′-TGGTCCGATCAATCAAACAGTTC-3′; Foxp3: forward, 5′-CCCATCCCCAGGAGTCTTG-3′ and reverse, 5′-ACCATGACTAGGGGCACTGTA-3′; Gata3: forward, 5′-AAGCTCAGTATCCGCTGACG-3′ and reverse, 5′-GTTTCCGTAGTAGGACGGGAC-3′; and Mink1: forward, 5′-CCACCTACTATGGGGCCTTTA-3′ and reverse, 5′-AGCACCGCAGAACTCCATC-3′. SYBR Premix Ex Taq II and a 7500 Fast Real-Time PCR system were used for real-time PCR, and data were analyzed with 7500 software (v2.0.1; Applied Biosystems). Results were normalized to β-actin expression.
Western blotting and immunoprecipitation
2–5 × 106 cells were lysed in whole-cell extract buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% NP-40 [NDB0385; Life-Biotech], 1 mM EDTA, 1 mM Na3VO4, 10% glycerol, 1 mM PMSF, and protease inhibitors) to obtain whole-cell lysates. Proteins were separated by SDS-PAGE (6–10%) followed by transfer to nitrocellulose membrane. To block nonspecific binding, membranes were incubated with 5% BSA in TBST (0.5 M NaCl, Tris-HCl, pH 7.5, and 0.05% [vol/vol] Tween 20) for 60 min and washed twice with TBST. Proteins of interest were detected by incubating the membranes overnight at 4°C in 5% BSA/TBST with the indicated antibodies (listed in this section), washed three times with TBST for 10 min, and incubated with horseradish peroxidase–conjugated anti–rabbit or anti–mouse antibody (A0208 and A0216; Beyotime). Bound antibody was detected using Immobilon Western chemiluminescent HRP substrate (Cyanagen). Antibodies were used for Western blot analysis as follows. The pT324-SMAD2 antibody was raised against the peptide NRNA [pT] TVEMTRRHIGC, and the sera were first absorbed by beads conjugated with the nonphosphopeptide and then affinity purified using the phosphopeptide. Anti-MINK1 (NBP1-22990; Novus Biologicals), anti–pC-SMAD2 (3101; Cell Signaling Technology), anti-SMAD2 (3103; Cell Signaling Technology), anti-SMAD3 (9523; Cell Signaling Technology), anti–pC-SMAD3 (9520; Cell Signaling Technology), anti–lamin A (613501; BioLegend), anti–β-actin (LK2002; LKtag), anti-GAPDH (EM1101; HuaAn Biotechnology), anti-Flag (MA191878; Thermo Fisher Scientific), and anti-HA (0906-1; HuaAn Biotechnology) were used.
For immunoprecipitation, cell lysates were prepared as described in the previous paragraph, and proteins were immunoprecipitated by incubation of lysates with 1–10 µg antibody overnight at 4°C and pulldown of antibody–protein precipitates with SureBeads Protein A (or protein G) magnetic beads (Bio-Rad Laboratories). The presence of immunocomplexed proteins was determined by Western blot analysis.
Cell fractionation and in vitro kinase activity assay
3 × 107 mice CD4+ T cells were treated with TGF-β at the time periods indicated in Fig. 5, harvested with fractionation buffer (BSP001; Sangon Biotech) for 20 min on ice, and then fractionated by centrifugation. The fractions were analyzed on SDS-PAGE and by subsequent Western blot analysis.
Cell lysates were prepared, and MINK1 was immunoprecipitated from the lysates with anti-MINK1 antibody as described in the Mice, EAE induction, and adoptive transfer section of Materials and methods. The immunoprecipitate was washed extensively, and MINK1 kinase activity was assessed with a luminescence-based assay (MINK1 kinase ADP-Glo kinase assay; V8001; Promega) and an anti-thiophosphate antibody (ab92570; Abcam) according to the manufacturers’ instructions. Purified GST-MINK1 kinase domain (Sigma-Aldrich) was used for in vitro kinase reactions at 10 µg/ml in a final volume of 25 µl.
Plasmids
Mouse MINK1 cDNA was cloned into the BamHI and XhoI sites of the pcDNA 3.1 expression vector and SMAD2 at the EcoRI and XhoI sites. GST-tagged SMAD2 or SMAD3 was constructed by subcloning of the appropriate cDNA into the pGEX-4T-1 GST expression vector (GE Healthcare).
Statistical analysis
Statistical analysis was performed using Prism (GraphPad Software). The data were analyzed by the paired sign Student’s t test or Student’s t test. All p-values ≤0.05 were considered statistically significant.
Acknowledgments
The authors thank Drs. Bin Li and Chen Dong for helpful discussion and A. Alison for assistance with manuscript editing. We are grateful to Xinhui Song, Youfa Zhu, Shuangshuang Liu, Jiajia Wang, and Yingying Huang from the core facilities (Zhejiang University School of Medicine) for technical assistance in histology and FACS analysis.
This work was supported by grants from the National Natural Science Foundation of China (31530019, 31325009, and 31270927 to L. Lu), the National Basic Research Program of China (973 Program; 2012CB945004 and 2011CB944100 to L. Lu), the Zhejiang Provincial Natural Science Foundation of China (LR14H100001 to D. Wang), and the Program of Introducing Talents of Discipline to Universities (B13026 to L. Lu). L. Lu also received funding from the Specialized Research Fund for the Doctoral Program of Higher Education (20130101110004) and the Zhejiang University K.P. Chao High Technology Development Foundation.
The authors declare no competing financial interests.
Author contributions: G. Fu, Q. Xu, Y. Ke, H. Hu, J. Wang, X. Cao, D. Wang, H. Cantor, and L. Lu designed the research. G. Fu, Q. Xu, Y. Qiu, X. Jin, and T. Xu performed the research. G. Fu, Q. Xu, and L. Lu analyzed the data. G. Fu, D. Wang, H. Cantor, and L. Lu wrote the paper.
Footnotes
Abbreviations used:
- CNS
- central nervous system
- EAE
- experimental autoimmune encephalomyelitis
- GST
- glutathione S-transferase
- LP
- lamina propria
- MAD
- mother against decapentaplegic
- MINK1
- Msn/NIK-related kinase 1
- MOG
- myelin oligodendrocyte glycoprotein peptide
- Msn
- misshapen
- NAC
- N-acetyl cysteine
- R-SMAD
- receptor-regulated SMAD
- ROS
- reactive oxygen species
References
- Atarashi K., Tanoue T., Ando M., Kamada N., Nagano Y., Narushima S., Suda W., Imaoka A., Setoyama H., Nagamori T., et al. . 2015. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 163:367–380. 10.1016/j.cell.2015.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K., Mues M., Koutrolos M., Rasbi Z.A., Boziki M., Johner C., Wekerle H., and Krishnamoorthy G.. 2011. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 479:538–541. 10.1038/nature10554 [DOI] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Bruce D.L., and Sapkota G.P.. 2012. Phosphatases in SMAD regulation. FEBS Lett. 586:1897–1905. 10.1016/j.febslet.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Chang X., Liu F., Wang X., Lin A., Zhao H., and Su B.. 2011. The kinases MEKK2 and MEKK3 regulate transforming growth factor-β-mediated helper T cell differentiation. Immunity. 34:201–212. 10.1016/j.immuni.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H.C., Lan J.L., Chen D.Y., Yang C.Y., Chen Y.M., Li J.P., Huang C.Y., Liu P.E., Wang X., and Tan T.H.. 2011. The kinase GLK controls autoimmunity and NF-κB signaling by activating the kinase PKC-θ in T cells. Nat. Immunol. 12:1113–1118. 10.1038/ni.2121 [DOI] [PubMed] [Google Scholar]
- Chuang H.C., Sheu W.H., Lin Y.T., Tsai C.Y., Yang C.Y., Cheng Y.J., Huang P.Y., Li J.P., Chiu L.L., Wang X., et al. . 2014. HGK/MAP4K4 deficiency induces TRAF2 stabilization and Th17 differentiation leading to insulin resistance. Nat. Commun. 5:4602 10.1038/ncomms5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I., Watanabe N.M., Kobayashi T., Yamashita-Suzuki K., Fukagaya Y., Kajikawa E., Kimura W.K., Nakashima T.M., Matsumoto K., Ninomiya-Tsuji J., and Kusumi A.. 2000. Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett. 469:19–23. 10.1016/S0014-5793(00)01247-3 [DOI] [PubMed] [Google Scholar]
- Derynck R., and Zhang Y.E.. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 425:577–584. 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E., et al. . 2015. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125:194–207. 10.1172/JCI76012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.E., Hatton R.D., Mangan P.R., Turner H., Murphy T.L., Murphy K.M., and Weaver C.T.. 2005. Interleukin 17–producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6:1123–1132. 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- Hultqvist M., Olofsson P., Holmberg J., Bäckström B.T., Tordsson J., and Holmdahl R.. 2004. Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc. Natl. Acad. Sci. USA. 101:12646–12651. 10.1073/pnas.0403831101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultqvist M., Olsson L.M., Gelderman K.A., and Holmdahl R.. 2009. The protective role of ROS in autoimmune disease. Trends Immunol. 30:201–208. 10.1016/j.it.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Kaneko S., Chen X., Lu P., Yao X., Wright T.G., Rajurkar M., Kariya K., Mao J., Ip Y.T., and Xu L.. 2011. Smad inhibition by the Ste20 kinase Misshapen. Proc. Natl. Acad. Sci. USA. 108:11127–11132. 10.1073/pnas.1104128108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinewietfeld M., Manzel A., Titze J., Kvakan H., Yosef N., Linker R.A., Muller D.N., and Hafler D.A.. 2013. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 496:518–522. 10.1038/nature11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Kortum R.L., Sommers C.L., Pinski J.M., Alexander C.P., Merrill R.K., Li W., Love P.E., and Samelson L.E.. 2012. Deconstructing Ras signaling in the thymus. Mol. Cell. Biol. 32:2748–2759. 10.1128/MCB.00317-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langrish C.L., Chen Y., Blumenschein W.M., Mattson J., Basham B., Sedgwick J.D., McClanahan T., Kastelein R.A., and Cua D.J.. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240. 10.1084/jem.20041257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Awasthi A., Yosef N., Quintana F.J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D.A., et al. . 2012. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13:991–999. 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Ikizawa K., Hu D., Werneck M.B., Wucherpfennig K.W., and Cantor H.. 2007. Regulation of activated CD4+ T cells by NK cells via the Qa-1–NKG2A inhibitory pathway. Immunity. 26:593–604. 10.1016/j.immuni.2007.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Chen B., Song J.-H., Zhen T., Wang B.-Y., Li X., Liu P., Yang X., Zhang Q.L., Xi X.D., et al. . 2013. Eriocalyxin B ameliorates experimental autoimmune encephalomyelitis by suppressing Th1 and Th17 cells. Proc. Natl. Acad. Sci. USA. 110:2258–2263. 10.1073/pnas.1222426110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra N., Robertson E., and Kang J.. 2010. SMAD2 is essential for TGFβ-mediated Th17 cell generation. J. Biol. Chem. 285:29044–29048. 10.1074/jbc.C110.156745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-β induces development of the TH17 lineage. Nature. 441:231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Martin B., Hirota K., Cua D.J., Stockinger B., and Veldhoen M.. 2009. Interleukin-17-producing γδ T cells selectively expand in response to pathogen products and environmental signals. Immunity. 31:321–330. 10.1016/j.immuni.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Martinez G.J., Zhang Z., Reynolds J.M., Tanaka S., Chung Y., Liu T., Robertson E., Lin X., Feng X.H., and Dong C.. 2010. Smad2 positively regulates the generation of Th17 cells. J. Biol. Chem. 285:29039–29043. 10.1074/jbc.C110.155820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty N., Paust S., Ikizawa K., Dan I., Li X., and Cantor H.. 2005. Signaling by the kinase MINK is essential in the negative selection of autoreactive thymocytes. Nat. Immunol. 6:65–72. 10.1038/ni1145 [DOI] [PubMed] [Google Scholar]
- Nicke B., Bastien J., Khanna S.J., Warne P.H., Cowling V., Cook S.J., Peters G., Delpuech O., Schulze A., Berns K., et al. . 2005. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol. Cell. 20:673–685. 10.1016/j.molcel.2005.10.038 [DOI] [PubMed] [Google Scholar]
- Olofsson P., Holmberg J., Tordsson J., Lu S., Akerström B., and Holmdahl R.. 2002. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet. 33:25–32. 10.1038/ng1058 [DOI] [PubMed] [Google Scholar]
- Park H., Li Z., Yang X.O., Chang S.H., Nurieva R., Wang Y.H., Wang Y., Hood L., Zhu Z., Tian Q., and Dong C.. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141. 10.1038/ni1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran R., Loebbermann J., Nakaya H.I., Khan N., Ma H., Gama L., Machiah D.K., Lawson B., Hakimpour P., Wang Y.C., et al. . 2016. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature. 531:523–527. 10.1038/nature17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J.M., Pappu B.P., Peng J., Martinez G.J., Zhang Y., Chung Y., Ma L., Yang X.O., Nurieva R.I., Tian Q., and Dong C.. 2010. Toll-like receptor 2 signaling in CD4+ T lymphocytes promotes T helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity. 32:692–702. 10.1016/j.immuni.2010.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin V.I., Ibrahim M.X., Larsson E., Nilsson J.A., Lindahl P., and Bergo M.O.. 2014. Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 6:221ra15 10.1126/scitranslmed.3007653 [DOI] [PubMed] [Google Scholar]
- Schieber M., and Chandel N.S.. 2014. ROS function in redox signaling and oxidative stress. Curr. Biol. 24:R453–R462. 10.1016/j.cub.2014.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena L.A., Li S., Jairaman A., Prakriya M., Ezponda T., Hildeman D.A., Wang C.R., Schumacker P.T., Licht J.D., Perlman H., et al. . 2013. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 38:225–236. 10.1016/j.immuni.2012.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui J.W., Boomer J.S., Han J., Xu J., Dement G.A., Zhou G., and Tan T.H.. 2007. Hematopoietic progenitor kinase 1 negatively regulates T cell receptor signaling and T cell–mediated immune responses. Nat. Immunol. 8:84–91. 10.1038/ni1416 [DOI] [PubMed] [Google Scholar]
- Song X., He X., Li X., and Qian Y.. 2016. The roles and functional mechanisms of interleukin-17 family cytokines in mucosal immunity. Cell. Mol. Immunol. 13:418–431. 10.1038/cmi.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., and Tone M.. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202. 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R.J., Atkins C.J., Locksley R.M., and Stockinger B.. 2006a TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R.J., Flavell R.A., and Stockinger B.. 2006b Signals mediated by transforming growth factor-β initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7:1151–1156. 10.1038/ni1391 [DOI] [PubMed] [Google Scholar]
- Wang C., Yosef N., Gaublomme J., Wu C., Lee Y., Clish C.B., Kaminski J., Xiao S., Meyer Zu Horste G., Pawlak M., et al. . 2015. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell. 163:1413–1427. 10.1016/j.cell.2015.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zheng M., Lei L., Ji J., Yao Y., Qiu Y., Ma L., Lou J., Ouyang C., Zhang X., et al. . 2012. Tespa1 is involved in late thymocyte development through the regulation of TCR-mediated signaling. Nat. Immunol. 13:560–568. 10.1038/ni.2301 [DOI] [PubMed] [Google Scholar]
- Won H.Y., Jang E.J., Lee K., Oh S., Kim H.K., Woo H.A., Kang S.W., Yu D.Y., Rhee S.G., and Hwang E.S.. 2013. Ablation of peroxiredoxin II attenuates experimental colitis by increasing FoxO1-induced Foxp3+ regulatory T cells. J. Immunol. 191:4029–4037. 10.4049/jimmunol.1203247 [DOI] [PubMed] [Google Scholar]
- Wrighton K.H., Lin X., and Feng X.H.. 2009. Phospho-control of TGF-β superfamily signaling. Cell Res. 19:8–20. 10.1038/cr.2008.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.-H., Sudo K., Kuroda M., Kato M., Lee I.-K., Han J.S., Nakae S., Imamura T., Kim J., Ju J.H., et al. . 2015. Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat. Commun. 6:7600 10.1038/ncomms8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M., Luo D., Yu S., Liu P., Zhou Q., Hu M., Liu Y., Wang S., Huang Q., Niu Y., et al. . 2016. Misshapen/NIK-related kinase (MINK1) is involved in platelet function, hemostasis, and thrombus formation. Blood. 127:927–937. 10.1182/blood-2015-07-659185 [DOI] [PubMed] [Google Scholar]
- Zhi L., Ustyugova I.V., Chen X., Zhang Q., and Wu M.X.. 2012. Enhanced Th17 differentiation and aggravated arthritis in IEX-1–deficient mice by mitochondrial reactive oxygen species-mediated signaling. J. Immunol. 189:1639–1647. 10.4049/jimmunol.1200528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Tang J., Cao H., Fan H., and Li B.. 2015. Tissue resident regulatory T cells: novel therapeutic targets for human disease. Cell. Mol. Immunol. 12:543–552. 10.1038/cmi.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]