Many pathogenic bacterial species produce spores, which may play an important role in their interaction with the host. Choo et al. show that the spores of Bacillus anthracis, the etiologic agent of anthrax, induce type I IFN signaling and disrupt host immune defense via TLR-stimulating RNA.
Abstract
The spores of pathogenic bacteria are involved in host entry and the initial encounter with the host immune system. How bacterial spores interact with host immunity, however, remains poorly understood. Here, we show that the spores of Bacillus anthracis (BA), the etiologic agent of anthrax, possess an intrinsic ability to induce host immune responses. This immunostimulatory activity is attributable to high amounts of RNA present in the spore surface layer. RNA-sensing TLRs, TLR7, and TLR13 in mice and their human counterparts, are responsible for detecting and triggering the host cell response to BA spores, whereas TLR2 mediates the sensing of vegetative BA. BA spores, but not vegetative BA, induce type I IFN (IFN-I) production. Although TLR signaling in itself affords protection against BA, spore RNA–induced IFN-I signaling is disruptive to BA clearance. Our study suggests a role for bacterial spore–associated RNA in microbial pathogenesis and illustrates a little known aspect of interactions between the host and spore-forming bacteria.
Introduction
Many bacterial species in the phylum Firmicutes produce spores and depend on this dormant yet durable form of life for their survival and dispersal in adverse environments (McKenney et al., 2013; Paredes-Sabja et al., 2014). The bacterial spore formers include pathogenic species of Bacillus and Clostridium, which enter the host in the spore form, and later germinate to become vegetative cells. Hence, it is conceivable that the first encounter of the host immune system with spore-forming pathogens involves stimulation of microbial sensors by spore-associated molecules and resultant induction of innate immune responses. Such interactions may also occur for commensal spore–forming bacteria (Browne et al., 2016), including those that actively modulate host immunity and physiology (Ivanov et al., 2009; Atarashi et al., 2013; Stefka et al., 2014; Yano et al., 2015). Notably, the spores of probiotic Bacillus subtilis (BS) strains have been shown to possess spore-intrinsic immunostimulatory activity and augment antigen-specific immune responses in mice (Huang et al., 2008; Foligné et al., 2012). This adjuvant-like property has been exploited to devise BS spore–based vaccines (Lee et al., 2010; Song et al., 2012). It remains unknown, however, which microbial sensing mechanisms serve to detect bacterial spores, and what effects spore-induced immune responses exert on bacterial pathogenesis and symbiosis.
Bacillus anthracis (BA) is a Gram-positive spore-forming bacterium and the causative agent of anthrax. Different forms of anthrax—cutaneous, inhalation, and gastrointestinal—develop depending on the site of spore entry (Dixon et al., 1999). BA has evolved a multitude of mechanisms to evade immune surveillance and commandeer host phagocytes as a medium for spore germination, bacillary replication, and dissemination within the infected host. Crucial to these capabilities are protein toxins that enzymatically perturb host cell signaling and an antiphagocytic poly-γ-d-glutamic acid capsule (Moayeri et al., 2015). These factors are produced by vegetative bacilli (VB) after their emergence from germinating spores, and therefore may not play a critical role during the very first stage of BA infection. Rather, host–spore interactions at the primary infection site are more likely to contribute to shaping the initial immune response in anthrax. This notion, despite its plausibility and potential importance, has yet to be substantiated by experimental evidence. Above all, it is unclear whether BA infection triggers host responses by means of spore-intrinsic immunostimulatory activity and, if so, which structural component of the spore serves as a direct agonist for host immune sensors.
Innate immunity not only affords immediate protection against invading microbes but also instructs adaptive immunity to select a specific arm of antimicrobial effector functions (Iwasaki and Medzhitov, 2015). This decision-making process is guided by the information that the immune system gathers during its first encounter with the microbial pathogen. In this regard, it is expected that cytokine production by and other accompanying signaling events from BA spore–exposed host phagocytes have a profound influence on subsequent phases of the immune response, such as the priming of spore antigen-specific T cells toward distinct effector subsets. The antimicrobial effector functions thus mobilized, however, may not be optimal for fighting the VB, whose cellular properties, niches within the host, and antigen repertoire differ from those of the spore. Based on this reasoning, we postulated that BA spore–induced immune responses might precede and forestall VB-directed responses, thereby interfering with the choice of effective antimicrobial strategies against vegetative BA cells and the toxins they produce.
In this study, we find that the two forms of BA, the VB and the spore, are recognized by distinct microbial sensors and induce differential immune responses. In contrast to TLR2-dependent recognition of vegetative BA, nucleic acid–sensing TLRs mediate BA spore–induced immune responses. Intriguingly, spore-associated RNA is identified as the immunostimulatory agent for TLR sensing. Upon recognition of the spore but not the VB, host phagocytes produce type I IFN (IFN-I), which functions to disrupt protective immune responses to BA. Collectively, our findings suggest that spore-associated RNA triggers early host responses to BA infection, yet the IFN-I signaling axis serves to misguide host immunity and impair antimicrobial defense against the vegetative form of BA at later stages of infection.
Results
Distinct mechanisms for sensing and responding to the vegetative and the spore form of BA
The Sterne strain of BA, commonly used in veterinary vaccination and laboratory studies, does not produce poly-d-glutamic acid that encapsulates vegetative cells. The genome sequence of Sterne (available from GenBank under accession no. AE017225.1), however, predicts that its spores retain all known structural elements found in the spores formed by capsule-producing BA strains (e.g., Ames). Using the Sterne strain, we compared the host immune response to vegetative BA versus BA spores. We first examined intracellular signaling in mouse macrophages exposed to the two forms of the bacterium. Both BA forms induced IκBα degradation, indicative of NF-κB activation, to comparable extents (Fig. 1 A). Phosphorylation, hence activation, of ERK, JNK, and p38 also occurred, but with greater magnitudes when exposed to VB (Fig. 1 A). Besides activating these intracellular signaling modules, BA exposure resulted in the production of cyclooxygenease-2 (COX-2) and inducible nitric oxide synthase (iNOS); intriguingly, VB and spores induced macrophages to produce disproportionate amounts of these enzymes, a greater amount of COX-2 by the former and iNOS by the latter (Fig. 1 B).
Figure 1.
BA VB and spores induce differential gene expression in macrophages via distinct microbial sensing mechanisms. (A–E) WT mouse macrophages (A, B, and D), macrophages from the indicated mice (C and E), or WT mouse plasmacytoid DCs (pDC; D) were exposed to VB or spores (Sp) of BA at moi of 3 (A–C and E) or at the indicated moi (D). Whole-cell lysates were prepared at the indicated time points (A) and 24 h (B) after exposure, and analyzed by immunoblotting. Relative mRNA amounts for VB/Sp-induced genes at the indicated time points (C) or 4 h (D and E) after exposure were determined by qPCR, and presented in color-coded arbitrary units (C) or plotted on a linear scale (D and E). ISG, IFN-stimulated gene. Data are representative of two experiments with similar results (A, B, D, and E) or from one experiment (C).
We extended our analysis of BA-exposed macrophages to the expression of cytokines, chemokines, and other immune response genes, and found that spores induced a wider repertoire of genes relative to VB (Fig. 1 C). Importantly, spores but not VB induced the IFN-I gene Ifnb1, as well as IFN-stimulated genes (ISGs), whose expression was elicited as a secondary response to IFN-I production (Fig. 1 C). Indeed, spore-induced expression of these ISGs was abrogated in macrophages lacking the IFN-I receptor subunit IFNAR1, an essential molecule mediating autocrine IFN-β action (Fig. 1 C). Plasmacytoid DCs are professional IFN-I–producing cells poised for robust induction of IFN-α members as well as IFN-β. Spore-induced IFN-I gene expression was also evident in plasmacytoid DCs (Fig. 1 D). Vegetative BA did not induce any members of IFN-I in macrophages and plasmacytoid DCs.
Vegetative BA–induced gene expression was nearly abolished in macrophages lacking TLR2 (Fig. 1 E), suggesting that the lipopeptide moiety of the cell wall was the major component recognized by the host immune system. In contrast, BA spore–induced gene expression remained intact in TLR2-deficient macrophages (Fig. 1 E). Therefore, BA spores appear to induce host macrophage responses via a microbial sensing mechanism different from that involved in detecting the vegetative form of BA.
We next examined the response of mice to infection initiated by footpad injection of the same cfu number of the vegetative and the spore form of BA. Given the filamentous structure of vegetative BA growing in multicellular chains, its enumeration by cfu numbers likely undercounted the bacilli that were actually present in the inoculum. Nevertheless, higher and more rapid mortality and greater bacterial burdens in the draining lymph nodes resulted from infection with spores than with VB (Fig. 2, A and B). Mice challenged with VB exhibited massive neutrophil infiltration and severe edema in the primary infection site, but these inflammatory reactions were much milder in spore-challenged mice (Fig. 2, C and D). Collectively, our findings suggested that BA spores interacted with host immunity in a manner different from, and were more effective at causing lethal infection than vegetative BA.
Figure 2.
Spore-initiated infection results in higher BA burdens and milder inflammatory responses than VB-initiated infection. (A) Mice were infected with BA (107 cfu per host) by footpad injection of VB or spores (Sp). Host survival (n = 10–15 mice/group) was monitored for 10 d. (B–D) Mice were infected with BA (2.5 × 106 cfu per host) by footpad injection of VB or Sp. Bacterial burdens in popliteal lymph nodes were determined (B) and neutrophil infiltration in footpad skin was analyzed by flow cytometry (C) 3 d after infection. Footpad swelling (n = 4 mice/group) was determined daily for 3 d (D). Data are representative of two experiments (A, B, and D) or four animals (C) with similar results. *, P < 0.05; **, P < 0.01; Log-rank test (A) and Student’s t test (B).
MyD88-dependent sensing of BA spores
A multitude of microbial sensors exist in mammals; these include TLRs, RIG1-like receptors (RLRs), Nod-like receptors (NLRs), C-type lectin receptors (CLRs), and cyclic GMP-AMP synthase (cGAS). These microbial sensors depend on distinct signaling adaptors for eliciting host cell responses to their cognate microbial-derived agonists: MyD88 and TRIF for TLRs; MAVS (also known as IPS1, VISA, and CARDIF) for RLRs; RIP2 for NLRs; CARD9 for CLRs; and STING for cGAS. Investigation of macrophages with a deletion or mutation of the gene encoding each signaling adaptor revealed that BA spore–induced gene expression depended on MyD88, whereas other adaptors were dispensable for this response (Fig. 3, A and B). The requirement of MyD88 in spore-induced gene expression was broad, encompassing all of the genes examined. MyD88 deficiency prevented spore-induced ERK, JNK, and p38 signaling (Fig. 3 C) and impaired NF-κB activation, as shown by greatly diminished nuclear translocation of the NF-κB proteins RelA and c-Rel (Fig. 3 D). Consistent with the broad effects of MyD88 deficiency on the induction of immune-response genes in macrophages, MyD88-KO mice exhibited increased susceptibility to BA spore–initiated infection compared with WT mice (Fig. 3, E and F).
Figure 3.
BA spore–induced signaling and gene expression in macrophages depend on MyD88, but not other signaling adaptors for microbial sensors. (A–D) Macrophages from the indicated mice were exposed to BA spores at moi of 3. Relative mRNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR and presented in color-coded arbitrary units (A). Whole-cell lysates (B and C), and cytoplasmic and nuclear extracts (D) were prepared at the indicated time points after exposure, and analyzed by immunoblotting. (E and F) WT and MyD88-KO mice were infected with BA (107 and 2.5 × 106 cfu per host; E and F, respectively) by footpad injection of spores. Host survival (n = 6–7 mice per group) was monitored for 10 d (E). Bacterial burdens in popliteal lymph nodes were determined 3 d after infection (F). Data are from one experiment (A and B) or representative of two experiments with similar results (C–F). **, P < 0.01; Log-rank test (E) and Student’s t test (F).
Detection of BA spores by endosomal nucleic acid-sensing TLRs
Our finding of MyD88 as the single most important adaptor for BA spore–induced host cell responses suggested that TLR signaling was crucial for sensing and responding to BA spores. We therefore examined macrophages with a single TLR deficiency to determine the specific TLR member that was responsible for the immune response to BA spores. This analysis used macrophages from mice with a deletion of the gene encoding TLR2, TLR3, TLR4, TLR5, TLR7, TLR8, TLR9, TLR11, TLR12, or TLR13. None of these single TLR deficiencies completely suppressed BA spore–induced expression of the cytokine gene Tnf and the chemokine gene Cxcl2 (Fig. 4 A), which we selected as readouts of TLR-mediated macrophage responses; this observation raised the possibility of redundancy among TLR members.
Figure 4.
BA spore–induced macrophage gene expression depends on endosomal nucleic acid–sensing TLRs. (A–C) Macrophages from the indicated mice (A and C) or WT mouse macrophages (B) were exposed to BA spores at moi of 3. Relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and plotted on a linear scale. Macrophages were pretreated with the following pharmacological agents 1 h before spore exposure (B): bafilomycin A1 (Bm), dynasore (Ds), z-FA-cmk (FA), and CA074-Me (CA). (D) Macrophages derived from control and UNC93B1-KO THP-1 cells were exposed to BA spores at moi of 3. Relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and then plotted on a linear scale. Data are representative of three experiments with similar results (A) or from one experiment (B–D).
A subset of mammalian TLRs is present in the endosomal compartment, undergoes functional maturation via endosomal proteolytic cleavage of their ectodomains, and encounters their ligands only after ligand uptake and delivery to the endosome. Therefore, the signaling function of these TLRs depends on endosomal acidity and proteinase activity as well as specific vesicle-trafficking events, and can be suppressed by small-molecule compounds that block endosomal proton transport (e.g., bafilomycin A1), cathepsin-dependent proteolysis (e.g., z-FA-cmk and CA074-Me), and dynamin-dependent endocytosis (e.g., dynasore). We found that Tnf and Cxcl2 induction by BA spores was attenuated in macrophages treated with these inhibitors (Fig. 4 B). Also, BA spore–induced gene expression was decreased or abolished in macrophages with a deletion or mutation of Unc93b1, a gene essential for the endosomal sorting of TLRs (Fig. 4 C). These results pointed to a role for endosomal TLRs in recognizing BA spores in mice and humans.
To examine whether the human immune system also detects BA spores in an UNC93B1-dependent manner, we investigated macrophages prepared by in vitro differentiation of human THP-1 cells with genetic ablation of UNC93B1 (Pelka et al., 2014). BA spores induced immune-response genes in control THP-1–derived macrophages, but far less potently in UNC93B1-deficient counterparts (Fig. 4 D), suggesting similar BA spore–sensing mechanisms in mice and humans.
Detection of BA spores by RNA-sensing TLRs
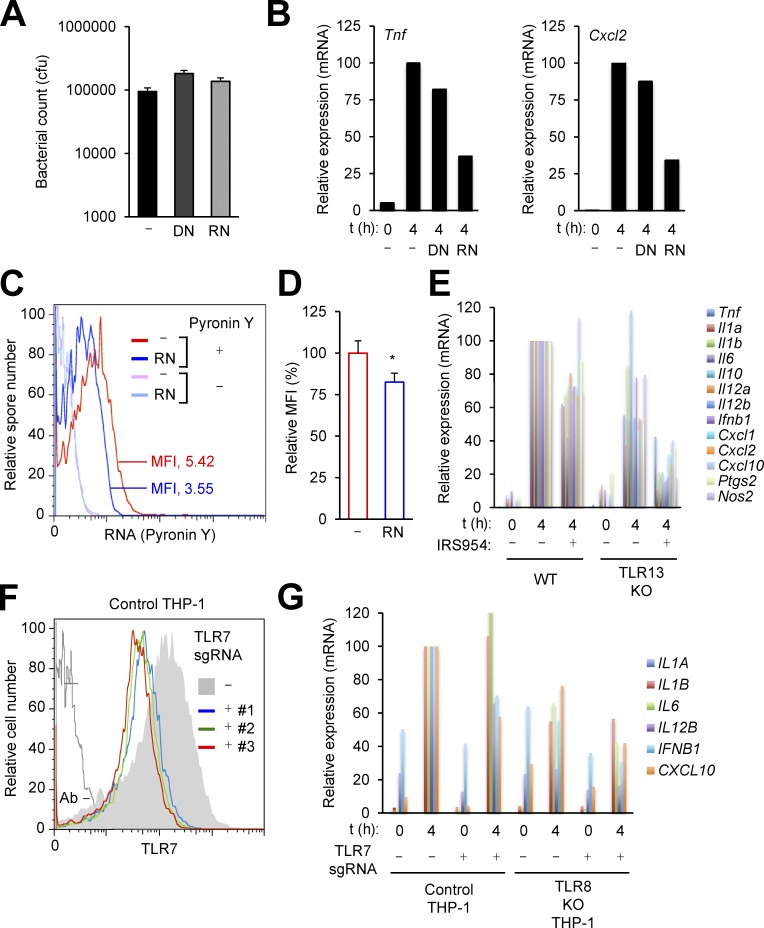
The endosomal TLRs include nucleic acid sensors that detect DNA (TLR9), double-stranded RNA (TLR3), and single-stranded RNA (TLR7, TLR8, and TLR13). To explore whether the immunostimulatory activity of BA spores is attributable to spore-associated nucleic acids, we tested its sensitivity to DNase and RNase treatment. Treatment with these enzymes did not reduce spore viability (Fig. 5 A). RNase-treated spores exhibited a substantially reduced ability to induce Tnf and Cxcl2 expression, whereas DNase treatment did not exert a comparable effect (Fig. 5 B), a finding that indicated a contribution of RNA to immune sensing of BA spores. The spores of Bacillus species have indeed been reported to contain varying amounts of RNA in addition to chromosomal and plasmid DNA (Doi and Igarashi, 1964; Matz et al., 1970; Jeng and Doi, 1974). BA spore–associated RNA was detectable by flow cytometry after spore staining with the RNA-labeling fluorescent dye Pyronin Y. RNase treatment of BA spores, which attenuated their immunostimulatory activity, resulted in only a moderate loss of their staining with Pyronin Y (a decrease in mean fluorescence intensity; Fig. 5, C and D). This indicated that the subpopulation of RNA responsible for immunostimulation by spores might be readily accessible to and destructible by RNase in a condition that did not permit the removal of the total spore RNA pool.
Figure 5.
BA spore–induced macrophage gene expression depends on RNA-sensing TLRs. (A–D) BA spores were treated with DNase I (DN) or RNase A (RN). Spore viability was determined after enzyme treatment (A). WT mouse macrophages were exposed to enzyme-treated BA spores at moi of 3; relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and plotted on a linear scale (B). Enzyme-treated spores were stained for RNA (Pyronin Y) and subjected to flow cytometry (C and D). MFI, mean fluorescence intensity. *, P < 0.05; Student’s t test. (E) Macrophages from WT and TLR13-KO mice were exposed to BA spores at moi of 3. Macrophages were pretreated with IRS954 1 h before exposure. Relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and plotted on a linear scale. (F and G) Control and TLR8-KO THP-1 cells were infected with lentiviruses expressing Cas9 alone (-) or TLR7-specific sgRNA (+; #1-#3). Macrophages derived from these cells were subjected to flow cytometry (F). Macrophages derived from the indicated cells were exposed to BA spores at moi of 3; relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and plotted on a linear scale (G). sgRNA #1 was used to disrupt the TLR7 gene (G). Data are from one experiment (A, B, and G) or representative of three experiments (C–E) or two experiments (F) with similar results.
It is noteworthy that although BA spore–induced Tnf and Cxcl2 expression was not completely abolished by a single deficiency of the RNA-sensing TLRs (TLR3, TLR7, TLR8, and TLR13), loss of TLR7 or TLR13 led to moderate decreases in the strength of their induction (Fig. 4 A). We suspected that TLR7 and TLR13 redundantly contributed to sensing BA spores, and sought to determine the effects of preventing the functioning of both TLRs. To this end, we examined gene expression in TLR13-deficient macrophages treated with IRS954, an oligonucleotide known to block TLR7 function. Macrophage gene induction by BA spores was abrogated by this combination of genetic ablation and pharmacological inhibition of TLR signaling (Fig. 5 E). IRS954 exerted only partial inhibitory effects on gene induction in WT macrophages. These findings pointed to the role of the RNA sensors TLR7 and TLR13 in the recognition of and immune response to the spore form of BA.
The human immune system does not possess a TLR13 homologue; instead, human TLR8 is known to serve as a functional analogue of TLR13 (Krüger et al., 2015). To determine the role of human TLR7 and TLR8 in detecting BA spores, we sought to generate THP-1 cells doubly deficient for the two receptors. To this end, we first examined the efficiency of TLR7 gene disruption in THP-1 cells by CRISPR/Cas9 genome editing. Infection of THP-1 cells with lentiviruses expressing TLR7 gene-specific single guide RNA (sgRNA) resulted in diminished TLR7 expression in a majority of macrophages derived from the infected cells (Fig. 5 F). Using this CRISPR/Cas9-based approach, we deleted the TLR7 gene from TLR8-deficient THP-1 cells that were established in a previous study (Krüger et al., 2015). Double deficiency of TLR7 and TLR8 greatly diminished BA spore–induced gene expression in THP-1–derived macrophages (Fig. 5 G). A single deficiency of either TLR also produced some effects but only moderately relative to the simultaneous deficiency of both. Collectively, our findings from the gene ablation experiments suggested TLR7 and TLR13 in mice and their counterparts in humans as innate immune sensors of BA spores.
Immunostimulatory activity of BA spore–derived RNA
Given the role of RNA-sensing TLRs in inducing immune responses to BA spores, we postulated that RNA integral to the spore structure was responsible for immunostimulation. We subjected BA spores to an RNA extraction and purification protocol, and obtained RNA adequate for in vitro macrophage stimulation. Treatment of macrophages with the RNA preparation resulted in the induction of Tnf and Cxcl2 expression, IκBα degradation, and ERK, JNK, and p38 phosphorylation (Fig. 6, A and B).
Figure 6.
BA spores harbor immunostimulatory RNA. (A and B) WT mouse macrophages were treated with RNA (1 and 0.25 µg/ml; A and B, respectively) isolated from BA spores. Relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and plotted on a linear scale (A). Whole-cell lysates were prepared at the indicated time points after exposure, and analyzed by immunoblotting (B). (C) BA spore RNA was analyzed by Bioanalyzer electrophoresis. The positions of bacterial rRNA and size markers are indicated at the top and bottom, respectively. (D and E) The composition of BA spore RNA was determined by RNA-Seq. The sequence reads from the long and small RNA libraries (D and E, respectively) were used to calculate RPKM values for each gene. RNA components with RPKM >20 (D) and 100 (E) are plotted. Red and blue columns correspond to the RNA components listed in Tables S1 and Table S2, respectively. * and **, columns representing degraded 23S and 16S RNA, respectively. (F) WT mouse macrophages were treated with the indicated synthetic and in vitro-transcribed RNA (1 µg/ml) isolated from BA spores. Relative RNA amounts for spore-induced genes 4 h after exposure were determined by qPCR, and plotted on a linear scale. Data are representative of two experiments with similar results (A, B, and F) or from one experiment (C–E).
BA spore–derived RNA was heterogeneous, and mostly ranged from 20–6,000 nt in size (Fig. 6 C). To verify the origin of BA spore–associated RNA (i.e., BA cells rather than artifactual sources such as contaminants from the culture medium) and determine its composition, we performed RNA sequencing (RNA-Seq) analysis. To this end, we divided total spore-derived RNA into two fractions according to size with a cut-off of 200 nt, and used each fraction for constructing a library for massively parallel sequencing. The sequence reads generated from the two libraries mapped to the BA genome with high rates (88 and 95.6% for the long and small RNA libraries, respectively), indicating BA cells as the origin of BA spore–associated RNA. The genome-aligned reads revealed overall heterogeneity in sequence composition, yet 23S and 16S rRNA were the two most abundant species in the spore-associated RNA pool (Fig. 6 D and Table S1). Other RNA species with a substantial share of the BA spore transcriptome included mRNA, as well as noncoding RNA such as tRNA (Fig. 6 E and Table S2). The profile of spore-associated mRNA represented a gene expression program associated with sporulation, many of the transcripts encoding structural components of the spore.
The occurrence of 23S rRNA and tRNA in the BA spore was consistent with the previous studies of these two bacterial RNA species serving as TLR13 and TLR7 agonists, respectively (Gehrig et al., 2012; Hidmark et al., 2012; Jöckel et al., 2012; Li and Chen, 2012; Oldenburg et al., 2012). The ability of bacterial 23S rRNA to trigger TLR13 signaling has been mapped to a segment in the domain V (Li and Chen, 2012; Oldenburg et al., 2012). This immunostimulatory sequence is preserved in BA 23S rRNA. A synthetic polyribonucleotide corresponding to this sequence (the nucleotide positions from 2063 to 2077) induced cytokine and chemokine gene expression in macrophages (Fig. 6 F). In vitro–transcribed BA valine and methionine tRNA, when used alone, did not show such immunostimulatory activity, but markedly enhanced 23S rRNA-induced gene expression (Fig. 6 F).
Enrichment of RNA in the exosporium of the BA spore
BA spores treated with an RNA-specific fluorescent labeling agent, SYTO RNASelect, or Pyronin Y, displayed intense staining in the spore periphery (Fig. 7, A and B). Peripheral RNA-specific staining was observed in the majority (>95%) of BA spores. The outermost layer of the BA spore, or the exosporium, consists of proteins including the glycoprotein BclA, which forms hair-like filaments that project from the base of the exosporium, therefore externally demarcating the spore (Stewart, 2015). The fluorescence signals representing spore-associated RNA co-localized with BclA-specific immunofluorescence (Fig. 7 B). We treated BA spores with sodium dodecyl sulfate and urea to remove exosporial proteins; this treatment almost completely eliminated peripheral RNA staining while sparing DNA and RNA in the spore core (Fig. 7 C). Similarly, spores of BA deficient in the gene cotE (cotEΔ), which do not form a stable exosporial layer, exhibited greatly reduced amounts of peripheral RNA but readily detectable core RNA (Fig. 7 D). This loss of exosporial RNA corresponded to a decrease in total spore RNA staining with Pyronin Y (Fig. 7, E and F). Infection with exosporium-deficient cotEΔ spores resulted in greater survival of and lower bacterial burden in host mice than infection with exosporium-intact counterparts (Fig. 7, G and H). It should be noted, however, that cotE deficiency did not reduce virulence in an earlier study (Giorno et al., 2007). The experiments in this study differed from ours in terms of the genetic background of both BA and the host, as well as the route of infection (Aimes, BALB/c, and intranasal instead of Sterne, C57BL/6, and subcutaneous); the effect of cotE deficiency may be nuanced by these differences.
Figure 7.
BA spore RNA is mainly localized to the exosporium. (A–F) Spores of the indicated BA strains were stained for RNA (CYTO RNASelect and Pyronin Y), DNA (Hoechst 33342), and BclA (fluorescently labeled antibody) as indicated. Fluorescence signals from the labeling agents were analyzed by microscopy (A–D) or flow cytometry (E and F). Where indicated (C), BA spores were treated with SDS and urea before staining to eliminate the proteinaceous surface layers. *, nonspecific signals. Bar, 1 µm. MFI, mean fluorescence intensity. (G and H) Mice were infected with the indicated BA strains (107 and 2.5 × 106 cfu per host; F and G, respectively) by footpad injection of spores. Host survival (n = 8 mice per group) was monitored for 10 d (G). Bacterial burdens in popliteal lymph nodes were determined 3 d after infection (H). Data are representative of three experiments (A, E, and F) or two experiments (B–D) with similar results, or from one experiment (G and H). *, P < 0.05; **, P < 0.01; Student’s t test (F and H) and Log-rank test (G).
BS spores naturally lack an exosporium layer (McKenney et al., 2013). Unlike BA spores, BS spores did not stain strongly for surface-localized RNA (Fig. 8, A–C). Correspondingly, we found that the RNA content of BS spores was far lower than that of BA spores; the amounts of total RNA extracted and purified from BS and BA spores were 0.02 µg versus 32 µg per 109 spores, respectively. Although BS spores were capable of inducing immune response genes in macrophages, they did not elicit strong ISG expression in macrophages (Fig. 8 D). BS spore–induced gene expression was dependent on MyD88 and TLR2, but independent of UNC93B1 (Fig. 8 E), suggesting that peptidoglycan, present in the spore cortex, contributed to immunostimulation by BS spores, whereas spore-associated nucleic acids exerted a negligible, if any, effect on it.
Figure 8.
BS spores do not depend on RNA for immunostimulation. (A–C) BA and BS spores were stained for RNA (Pyronin Y) and DNA (Hoechst 33342) as indicated. Fluorescence signals from the labeling agents were analyzed by microscopy (A) or flow cytometry (B and C). *, nonspecific signals. Bar, 1 µm. MFI, mean fluorescence intensity. **, P < 0.01; Student’s t test. (D and E) WT mouse macrophages (D) or macrophages from the indicated mice (E) were exposed to BA (D) or BS (D and E) spores at moi of 3. Relative mRNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and presented in color-coded arbitrary units (D) or plotted on a linear scale (E). ISG, IFN-stimulated gene. Data are from one experiment (A, D, and E) or representative of three experiments (B and C) with similar results.
Adverse effects of IFN-I signaling in BA infection
IFN-β and ISG expression was induced in macrophages after exposure to the spore form, but not the vegetative form, of BA (Fig. 1 C). IFN regulatory factor (IRF) family transcription factors play a pivotal role in IFN-β gene induction, but individual IRFs are differentially activated by distinct IFN-inducing stimuli and the signaling pathways they engage. The best characterized mechanisms of IFN-β induction are those that work in the context of signaling via TRIF, MAVS, and STING, and entail IRF3 activation (Liu et al., 2015). BA spore–induced IFN-β expression was, however, independent of IRF3, and instead required IRF5 and IRF7 (Fig. 9 A). These two IRFs have been shown to mediate IFN-I gene transcription in response to endosomal TLR-driven intracellular signaling (Schoenemeyer et al., 2005; Yasuda et al., 2007, 2013). Therefore, the requirement for IRF5 and IRF7 in BA spore–induced IFN-β expression was consistent with TLR sensing of spore-associated RNA.
Figure 9.
Type I interferon signaling is detrimental to host resistance to BA. (A) Macrophages from the indicated mice were exposed to BA spores at moi of 3. Relative RNA amounts for spore-induced genes at the indicated time points after exposure were determined by qPCR, and plotted on a linear scale. (B–E) The indicated mice were infected with BA (107 and 2.5 × 106 cfu per host; B and C–E, respectively) by footpad injection of spores. Host survival (n = 15–19 mice per group) was monitored for 10 d (B). Bacterial burdens in popliteal lymph nodes (C), footpad swelling (D), and serum concentrations of IL-1β (E) were determined 3 d after infection. Data are from one experiment (A and E) or representative of two experiments with similar results (B–D). *, P < 0.05; ***, P < 0.001; Log-rank test (B) and Student’s t test (C).
IFN-I pathway activation by BA spore–associated RNA might have evolved as a pathogenic mechanism for disrupting host immunity. To address how IFN-I signaling affected host defense against BA, we compared the response of WT and IFNα/βR-KO mice to spore-initiated infection. Deficiency in IFN-I signaling enhanced the survival of and reduced bacterial burden in BA-infected mice (Fig. 9, B and C). In contrast, edema formation in the primary infection site and the production of circulating IL-1β, both indicators of inflammatory reactions, in IFNα/βR-KO mice were similar to or moderately increased compared with those in WT mice (Fig. 9, D and E). Collectively, these findings demonstrated that BA spore–induced IFN-I signaling was dispensable for triggering immune responses and, rather, served to impair immune defense against BA.
DISCUSSION
The spores of BA and other pathogenic bacteria play a key role in the initiation of infection. Spore-forming bacteria also include commensal species that are known to exert a variety of effects on host immunity and metabolism. Evidence has recently emerged that points to bacterial spore-intrinsic immunostimulatory properties, and even inspired attempts to develop bacterial spore-based vaccines and therapeutics. Nevertheless, the current understanding of how bacterial spores interact with the host immune system, let alone its practical applications, has been formed mainly by empirical findings. We have shown that BA and BS spores both have the ability to activate host immunity, but their immunostimulatory properties are mediated by distinct molecules and produce dissimilar effects on host immune responses. Bacteria may have evolved diverse spore-intrinsic mechanisms for immunostimulation depending on the necessities determined by their mode of pathogenesis as well as by the architecture of their spores.
BA had long been studied as a human and veterinary pathogen even before the potential for the use of its spores in bioterrorism and biowarfare—and the actual 2001 attacks in the United States—raised the degree of public concern about anthrax (Schwartz, 2009). In fact, BA was an important model pathogen to pioneers of modern microbiology and immunology. A wealth of knowledge has since been generated regarding the role of the virulence factors produced by vegetative BA in anthrax pathogenesis. In contrast, much less is known about the contribution of spores to host–BA interactions. Of note, studies preceding ours provided some clues, showing that BA spores could induce cytokine expression in host cells and do so in a manner dependent on MyD88 (Basu et al., 2007; Glomski et al., 2007). In this study, we have identified RNA-sensing TLRs as crucial for detecting and inducing immune responses to BA spores. Also, we have determined the composition of BA spore–associated immunostimulatory RNA and its spatial distribution in the spore. Pathogen-sensing mechanisms are coupled to signaling pathways that instruct the immune system to deploy specific effector functions optimal for defense against the detected pathogens. Growing evidence suggests that among the multiple immune effector functions, those orchestrated by IL-1β and IL-17 afford effective protection against BA infection. These cytokines serve to mobilize neutrophil-mediated antimicrobial mechanisms against extracellular pathogens. Mice with a defect in inflammasome-dependent IL-1β production or IL-17 signaling, as well as mice depleted of neutrophils exhibited an increased susceptibility to BA infection (Moayeri et al., 2010; Terra et al., 2010; Garraud et al., 2012). Both spores and VB are likely to induce the immune system to deploy these BA-optimized effector functions. However, not all effector functions mobilized upon TLR sensing of BA spores seem to be beneficial to the host. We have shown that BA spore–induced IFN-I signaling exerts adverse effects on host immune defense. IFN-I signaling has indeed been demonstrated to interfere with key molecular events associated with the IL-1 and IL-17 signaling axes in various immunological settings (Guo et al., 2008; Shinohara et al., 2008; Guarda et al., 2011; Reboldi et al., 2014; Stock et al., 2014). We postulate that BA has evolved to use spore-associated RNA to activate IFN-I signaling and thereby evade host immunity.
Our study reveals BA spore–associated RNA as an inducer of host TLR signaling. Besides mediating host–BA interactions, RNA may serve other functions related to spore stability and germination. The exosporial enrichment of BA spore–associated RNA raises the question of whether other bacterial species forming spores with an exosporium use similar RNA-dependent mechanisms to activate host immune responses. In addition to BA, several other pathogenic Bacillus species, most notably B. cereus, an opportunistic human pathogen, and B. thuringensis, an insect pathogen, produce spores that possess an exosporium (Stewart, 2015). Furthermore, an exosporium-like layer is found in the spores of pathogenic Clostridium species such as C. difficile and C. botulinum (Stewart, 2015). The exosporial proteins of the pathogenic members of Bacillus show high degrees of conservation, but this phylogenetic similarity does not extend to the constituents of clostridial exosporium-like structures (Henriques and Moran, 2007). It remains to be seen how our findings with BA spores translate to spores of other bacterial species.
Materials and methods
Mice
Primary cell isolation and infection experiments were performed using mice from the following gene deletion and mutation lines: MyD88-KO (Myd88tm1.1Defr), TRIF-KO (Ticam1tm1Aki), MAVS-KO (Mavstm1Tsc), RIP2-KO (Ripk2tm1Flv), CARD9-KO (Card9tm1Xlin), STING gt (Tmem173gt), TLR2-KO (Tlr2tm1Kir), TLR3-KO (Tlr3tm1Flv), TLR4-KO (Tlr4tm1Aki), TLR5-KO (Tlr5tm1Flv), TLR7-KO (Tlr7tm1Aki), TLR8-KO (Tlr8tm1Vlcg), TLR9-KO (Tlr9tm1Aki), TLR11-KO (Tlr11tm1Gho), TLR12-KO (Tlr12tm1Gho), TLR13-KO (Tlr13tm1(KOMP)Vlcg), UNC93B1 3d (Unc93b13d), IFNα/βR-KO (Ifnar1tm1Agt), IRF3-KO (Irf3tm1Ttg), IRF5-KO (Irf5tm1Ttg), and IRF7-KO (Irf7tm1Ttg). The IRF5-KO mice used in this study did not carry the Dock2 mutation (Yasuda et al., 2013) that some IRF5-KO lines were found to harbor (Purtha et al., 2012). All mice were on a C57BL/6 background and maintained in a specific pathogen–free condition. C57BL/6 WT mice (The Jackson Laboratory) were used as controls. All animal experiments were conducted under protocols approved by the Institutional Animal Care and Use Committee of the Massachusetts General Hospital.
BA strains, spores, and VB
BA Sterne strain 7702 and BS strain 168 were used to prepare spores and VB as previously described (Lyons et al., 2004). In brief, BA or BS from a colony formed on Brain Heart Infusion agar was grown in 2X SG liquid medium with shaking at 37°C until mid-log phase, harvested by centrifugation, washed with and resuspended in phosphate-buffered saline, and used as VB. To induce sporulation, bacilli in 2X SG liquid medium were grown to stationary phase, and then diluted fivefold with water, and further grown for 40 h. The spores formed were harvested, washed, and resuspended as with VB, and heat-treated for 40 min at 68°C. The titers of spore and vegetative bacillus stocks were determined by plating serial dilutions on Luria-Bertani agar plates and counting colonies after 16 h of incubation at 37°C. In some experiments, spores were treated with DNase I (Promega) or RNase A (Sigma-Aldrich). A cotE-null BA strain, cotEΔ (Giorno et al., 2007), was used to prepare spores for microscopic analysis.
Cell culture and stimulation
Primary macrophages were prepared by culturing mouse bone marrow cells in DMEM with high glucose (Life Technologies) supplemented with FBS (10%), l-glutamine (2 mM), sodium pyruvate (1 mM), penicillin (100 U/ml), streptomycin (100 µg/ml), and recombinant mouse macrophage-colony stimulating factor (10 ng/ml; PeproTech) for 7 d. Plasmacytoid DCs were isolated from mouse spleen using the Plasmacytoid Dendritic Cell Isolation kit (Miltenyi Biotec). THP-1 cells were cultured in RPMI 1640 medium (Life Technologies) supplemented with FBS (10%), penicillin (100 U/ml), and streptomycin (100 µg/ml). THP-1–derived macrophages were prepared by treating THP-1 cells with phorbol 12-myristate 13-acetate (100 nM; Sigma-Aldrich) for 2 d. Cells to be stimulated with spores or VB were rinsed and maintained in medium lacking antibiotics. To suppress extracellular bacterial replication, gentamicin (50 µg/ml) was added to the culture medium 45 min after the cell exposure to spores or VB. In some experiments, macrophages were treated with the following chemical compounds: bafilomycin A1, z-FA-cmk, CA074-Me, and dynasore (all from EMD Millipore) and IRS954 (Enzo Life Sciences).
Lentiviral transduction for CRISPR/Cas9 genome editing
CRISPR/Cas9 lentiviral vectors were constructed by inserting sgRNA sequences into pLentiCRISPR v2 (a gift from F. Zhang; Addgene plasmid #52961; Sanjana et al., 2014). The TLR7-targeting sgRNA constructs were specific to the following target sequences: #1, 5′-GGTGAGGTTCGTGGTGTTCG-3′; #2, 5′-AGAGCCTTTTCCGGAGCTGG-3′; and #3, 5′-TGTGCACCTGTGATGCTGTG-3′. Lentiviral particles were generated by 293T cell transfection and quantified using the QuickTiter Lentivirus Titer kit (Cell Biolabs). THP-1 cells were infected with lentiviruses in the presence of polybrene (8 µg/ml) for 4 h, and subjected to selection with puromycin (2 µg/ml) 6 d after virus infection.
Infection and analysis of mice
The viable count (cfu) of the inoculum was determined immediately before mouse infection. A total of 2.5 × 106 or 107 cfu of BA spores or vegetative BA were injected into the left hind footpad of 8-wk-old mice. Survival of infected host mice is monitored for 10 d. To determine bacterial burdens, popliteal lymph nodes were isolated from infected mice and homogenized in 0.5 ml of 2.5% saponin; cfu assays were performed on serially diluted homogenates. The extent of footpad swelling was determined by measuring the change in thickness of the infected paw with a caliper (Thermo Fisher Scientific). For flow cytometry of tissue-infiltrating neutrophils, single-cell suspensions obtained from minced and collagenase-digested footpad skin were treated with Fc receptor-blocking anti-CD16/CD32 antibodies, stained with fluorescent dye–conjugated CD11b (12–0112; eBioscience) and Ly6G (551460; BD) antibodies, and analyzed using FACSCanto (BD) and FlowJo software (Tree Star). Serum IL-1β was assayed by ELISA (R&D Systems).
Extraction and analysis of host cell proteins
To prepare whole-cell lysates, 2 × 106 cells were resuspended in 0.2 ml of lysis buffer (20 mM Hepes-potassium, pH 7.6, 150 mM sodium chloride, 2 mM EDTA acid, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 10% glycerol, 25 mM β-glycerophosphate, and protease inhibitors), incubated for 5 min at 4°C, and centrifuged 5 min at 16,000 g. The supernatant was collected for use as the whole-cell lysate. To prepare cytoplasmic and nuclear fractions, 5 × 106 cells were resuspended in 0.5 ml of buffer L1 (50 mM Tris-chloride, pH 8.0, 2 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 25 mM β-glycerophosphate, and protease inhibitors), incubated for 5 min at 4°C, and centrifuged 5 min at 4,500 g. The supernatant was stored for use as the cytoplasmic fraction. The pelleted nuclei were rinsed with 0.5 ml of buffer L1, resuspended in 0.1 ml of lysis buffer, incubated for 5 min at 4°C, and centrifuged 5 min at 16,000 g. The supernatant was collected for use as the nuclear fraction. Immunoblot analysis was performed using antibodies to the following proteins: phosphorylated p38 (9211), phosphorylated JNK (9251), and phosphorylated ERK (9101; all from Cell Signaling Technology); iNOS (sc-651), IκBα (sc-371), RelA (sc-372), c-Rel (sc-71), and RelB (sc-226; all from Santa Cruz Biotechnology); COX-2 (160126; Cayman Chemical); IL-1α (AF-400-NA), and IL-1β (AF-401-NA; both from R&D Systems); RAC1 (05–389; EMD Millipore); and actin (A4700; Sigma-Aldrich).
Isolation and analysis of host and spore cell RNA
Total RNA was isolated from host cells with the TRIzol Reagent (Ambion), and used in cDNA synthesis with the SuperScript System (Life Technologies). Quantitative PCR was performed using the SYBR Green PCR Mater Mix (Applied Biosystems) and gene-specific primers (Table S3). Spore RNA was isolated using the FastRNA Blue kit (MP Biomedicals) and a TissueLyser homogenizer (QIAGEN). The size range of isolated spore RNA was determined by electrophoresis using a 2100 Bioanalyzer instrument (Agilent Technologies). For RNA-Seq analysis, sequencing libraries were constructed with the TruSeq RNA Library Preparation kit and the TruSeq Small RNA Library Preparation kit (Illumina), and subjected to sequencing with a HiSeq 2000/2500 instrument (Illumina). Trimmed sequence reads were mapped to the BA genome (NC_003997.3) using Bowtie; multiple mapping was allowed, as the majority of noncoding RNA genes exist in multiple copies. RPKM values were calculated using in-house script.
Synthesis of immunostimulatory RNA
The oligoribonucleotide corresponding to the nucleotide positions from 2063 to 2077 of BA 23S rRNA (5′-ACGGAAAGACCCCGU-3′) was chemically synthesized (Sigma-Aldrich). BA valine and methionine tRNA were prepared by in vitro transcription using the MEGAscript T7 Transcription kit (Ambion) and DNA templates (BA_5814 and BA_5781, respectively) generated by PCR; a T7 promoter sequence was added to the 5′ end of the DNA templates as a part of the PCR primers.
Fluorescent labeling, microscopy, and flow cytometry of spores
Spores on glass slides were fixed in methanol-acetic acid (3:1 [vol/vol]) or ethanol, and stained with the following fluorescent labeling agents individually or in combination: SYTO RNASelect (Life Technologies), Pyronin Y (Sigma-Aldrich), and Hoechst 33342 (Molecular Probes). For immunostaining, spores were sequentially treated with 1% BSA, 0.5% Tween 20, and an Alexa Fluor 488–conjugated anti-BclA antibody (EF12; Swiecki et al., 2006). Stained spores were analyzed by fluorescence microscopy using an Axio Observer.Z1 microscope (ZEISS). Pyronin Y– and Hoechst 33342–stained spores were also analyzed using FACSCanto and FlowJo software as in host cell analysis.
Statistical analysis
Data values are expressed as mean ± SEM or mean ± SD. P-values were obtained with the unpaired two-tailed Student’s t test and the Log-rank (Mantel-Cox) test.
Online supplemental material
Tables S1 and S2, available as an Excel file, show BA spore–associated RNA molecules identified by RNA-Seq. Table S3, available as an Excel file, lists oligonucleotide primers used in real-time qPCR.
Supplementary Material
Acknowledgments
We thank Shizuo Akira, Tadatsugu Taniguchi, Jürg Tschopp, Ann Marshak-Rothstein, and Regeneron for gene KO mice; Carsten Kirschning, Anne Krüger, and Veit Hornung for UNC93B1-KO and TLR8-KO THP-1 cells; Charles Turnbough, Jr. for the Alexa Fluor 488–conjugated anti-BclA antibody; and Ramnik Xavier, Arthur Friedlander, Jun Huh, and Aimee Shen for advice.
This study was supported by National Institutes of Health grants U54 AI057159 (NERCE BEID) and R01 AI070999 (J.M. Park). C. Kim and H. Chong are employees of Macrogen Inc.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BA
- Bacillus anthracis
- BS
- Bacillus subtilis
- CLR
- C-type lectin receptor
- COX-2
- cyclooxygenease-2
- IFN-I
- type I IFN
- IRF
- IFN regulatory factor
- ISG
- IFN-stimulated genes
- NLR
- Nod-like receptor
- RLR
- RIG1-like receptor
- RNA-Seq
- RNA sequencing
- VB
- vegetative bacillus/bacilli
- cGAS
- cyclic GMP-AMP synthase
- iNOS
- inducible nitric oxide synthase
- sgRNA
- single guide RNA
References
- Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., et al. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 500:232–236. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- Basu S., Kang T.J., Chen W.H., Fenton M.J., Baillie L., Hibbs S., and Cross A.S.. 2007. Role of Bacillus anthracis spore structures in macrophage cytokine responses. Infect. Immun. 75:2351–2358. 10.1128/IAI.01982-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D., Goulding D., and Lawley T.D.. 2016. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 533:543–546. 10.1038/nature17645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon T.C., Meselson M., Guillemin J., and Hanna P.C.. 1999. Anthrax. N. Engl. J. Med. 341:815–826. 10.1056/NEJM199909093411107 [DOI] [PubMed] [Google Scholar]
- Doi R.H., and Igarashi R.T.. 1964. Ribonucleic acids of Bacillus subtilis spores and sporulating cells. J. Bacteriol. 87:323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foligné B., Peys E., Vandenkerckhove J., Van Hemel J., Dewulf J., Breton J., and Pot B.. 2012. Spores from two distinct colony types of the strain Bacillus subtilis PB6 substantiate anti-inflammatory probiotic effects in mice. Clin. Nutr. 31:987–994. 10.1016/j.clnu.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Garraud K., Cleret A., Mathieu J., Fiole D., Gauthier Y., Quesnel-Hellmann A., and Tournier J.N.. 2012. Differential role of the interleukin-17 axis and neutrophils in resolution of inhalational anthrax. Infect. Immun. 80:131–142. 10.1128/IAI.05988-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig S., Eberle M.E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W.M., Erdmann V.A., Sprinzl M., et al. 2012. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209:225–233. 10.1084/jem.20111044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno R., Bozue J., Cote C., Wenzel T., Moody K.S., Mallozzi M., Ryan M., Wang R., Zielke R., Maddock J.R., et al. 2007. Morphogenesis of the Bacillus anthracis spore. J. Bacteriol. 189:691–705. 10.1128/JB.00921-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glomski I.J., Fritz J.H., Keppler S.J., Balloy V., Chignard M., Mock M., and Goossens P.L.. 2007. Murine splenocytes produce inflammatory cytokines in a MyD88-dependent response to Bacillus anthracis spores. Cell. Microbiol. 9:502–513. 10.1111/j.1462-5822.2006.00806.x [DOI] [PubMed] [Google Scholar]
- Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R.A., Romero P., and Tschopp J.. 2011. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 34:213–223. 10.1016/j.immuni.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Guo B., Chang E.Y., and Cheng G.. 2008. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J. Clin. Invest. 118:1680–1690. 10.1172/JCI33342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques A.O., and Moran C.P. Jr. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555–588. 10.1146/annurev.micro.61.080706.093224 [DOI] [PubMed] [Google Scholar]
- Hidmark A., von Saint Paul A., and Dalpke A.H.. 2012. Cutting edge: TLR13 is a receptor for bacterial RNA. J. Immunol. 189:2717–2721. 10.4049/jimmunol.1200898 [DOI] [PubMed] [Google Scholar]
- Huang J.M., La Ragione R.M., Nunez A., and Cutting S.M.. 2008. Immunostimulatory activity of Bacillus spores. FEMS Immunol. Med. Microbiol. 53:195–203. 10.1111/j.1574-695X.2008.00415.x [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., and Medzhitov R.. 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16:343–353. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng Y.H., and Doi R.H.. 1974. Messenger ribonucleic acid of dormant spores of Bacillus subtilis. J. Bacteriol. 119:514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöckel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H., Ehm G., Schnare M., Nain M., Kaufmann A., and Bauer S.. 2012. The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209:235–241. 10.1084/jem.20111075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger A., Oldenburg M., Chebrolu C., Beisser D., Kolter J., Sigmund A.M., Steinmann J., Schäfer S., Hochrein H., Rahmann S., et al. 2015. Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. EMBO Rep. 16:1656–1663. 10.15252/embr.201540861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Belitsky B.R., Brinker J.P., Kerstein K.O., Brown D.W., Clements J.D., Keusch G.T., Tzipori S., Sonenshein A.L., and Herrmann J.E.. 2010. Development of a Bacillus subtilis-based rotavirus vaccine. Clin. Vaccine Immunol. 17:1647–1655. 10.1128/CVI.00135-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., and Chen Z.J.. 2012. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. eLife. 1:e00102 10.7554/eLife.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., and Chen Z.J.. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347:aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Lyons C.R., Lovchik J., Hutt J., Lipscomb M.F., Wang E., Heninger S., Berliba L., and Garrison K.. 2004. Murine model of pulmonary anthrax: kinetics of dissemination, histopathology, and mouse strain susceptibility. Infect. Immun. 72:4801–4809. 10.1128/IAI.72.8.4801-4809.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz L.L., Beaman T.C., and Gerhardt P.. 1970. Chemical composition of exosporium from spores of Bacillus cereus. J. Bacteriol. 101:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney P.T., Driks A., and Eichenberger P.. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11:33–44. 10.1038/nrmicro2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M., Crown D., Newman Z.L., Okugawa S., Eckhaus M., Cataisson C., Liu S., Sastalla I., and Leppla S.H.. 2010. Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS Pathog. 6:e1001222 10.1371/journal.ppat.1001222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M., Leppla S.H., Vrentas C., Pomerantsev A.P., and Liu S.. 2015. Anthrax Pathogenesis. Annu. Rev. Microbiol. 69:185–208. 10.1146/annurev-micro-091014-104523 [DOI] [PubMed] [Google Scholar]
- Oldenburg M., Krüger A., Ferstl R., Kaufmann A., Nees G., Sigmund A., Bathke B., Lauterbach H., Suter M., Dreher S., et al. 2012. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 337:1111–1115. 10.1126/science.1220363 [DOI] [PubMed] [Google Scholar]
- Paredes-Sabja D., Shen A., and Sorg J.A.. 2014. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 22:406–416. 10.1016/j.tim.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K., Phulphagar K., Zimmermann J., Stahl R., Schmid-Burgk J.L., Schmidt T., Spille J.H., Labzin L.I., Agrawal S., Kandimalla E.R., et al. 2014. Cutting edge: the UNC93B1 tyrosine-based motif regulates trafficking and TLR responses via separate mechanisms. J. Immunol. 193:3257–3261. 10.4049/jimmunol.1301886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtha W.E., Swiecki M., Colonna M., Diamond M.S., and Bhattacharya D.. 2012. Spontaneous mutation of the Dock2 gene in Irf5−/− mice complicates interpretation of type I interferon production and antibody responses. Proc. Natl. Acad. Sci. USA. 109:E898–E904. 10.1073/pnas.1118155109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A., Dang E.V., McDonald J.G., Liang G., Russell D.W., and Cyster J.G.. 2014. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 345:679–684. 10.1126/science.1254790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenemeyer A., Barnes B.J., Mancl M.E., Latz E., Goutagny N., Pitha P.M., Fitzgerald K.A., and Golenbock D.T.. 2005. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 280:17005–17012. 10.1074/jbc.M412584200 [DOI] [PubMed] [Google Scholar]
- Schwartz M. 2009. Dr. Jekyll and Mr. Hyde: a short history of anthrax. Mol. Aspects Med. 30:347–355. 10.1016/j.mam.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Shinohara M.L., Kim J.H., Garcia V.A., and Cantor H.. 2008. Engagement of the type I interferon receptor on dendritic cells inhibits T helper 17 cell development: role of intracellular osteopontin. Immunity. 29:68–78. 10.1016/j.immuni.2008.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Hong H.A., Huang J.M., Colenutt C., Khang D.D., Nguyen T.V., Park S.M., Shim B.S., Song H.H., Cheon I.S., et al. 2012. Killed Bacillus subtilis spores as a mucosal adjuvant for an H5N1 vaccine. Vaccine. 30:3266–3277. 10.1016/j.vaccine.2012.03.016 [DOI] [PubMed] [Google Scholar]
- Stefka A.T., Feehley T., Tripathi P., Qiu J., McCoy K., Mazmanian S.K., Tjota M.Y., Seo G.Y., Cao S., Theriault B.R., et al. 2014. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA. 111:13145–13150. 10.1073/pnas.1412008111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.C. 2015. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol. Mol. Biol. Rev. 79:437–457. 10.1128/MMBR.00050-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A.T., Smith J.M., and Carbone F.R.. 2014. Type I IFN suppresses Cxcr2 driven neutrophil recruitment into the sensory ganglia during viral infection. J. Exp. Med. 211:751–759. 10.1084/jem.20132183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M.K., Lisanby M.W., Shu F., Turnbough C.L. Jr., and Kearney J.F.. 2006. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J. Immunol. 176:6076–6084. 10.4049/jimmunol.176.10.6076 [DOI] [PubMed] [Google Scholar]
- Terra J.K., Cote C.K., France B., Jenkins A.L., Bozue J.A., Welkos S.L., LeVine S.M., and Bradley K.A.. 2010. Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J. Immunol. 184:17–20. 10.4049/jimmunol.0903114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano J.M., Yu K., Donaldson G.P., Shastri G.G., Ann P., Ma L., Nagler C.R., Ismagilov R.F., Mazmanian S.K., and Hsiao E.Y.. 2015. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 161:264–276. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Richez C., Maciaszek J.W., Agrawal N., Akira S., Marshak-Rothstein A., and Rifkin I.R.. 2007. Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J. Immunol. 178:6876–6885. 10.4049/jimmunol.178.11.6876 [DOI] [PubMed] [Google Scholar]
- Yasuda K., Nündel K., Watkins A.A., Dhawan T., Bonegio R.G., Ubellacker J.M., Marshak-Rothstein A., and Rifkin I.R.. 2013. Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int. Immunol. 25:295–306. 10.1093/intimm/dxs114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.