Macrophages are important for tissue function, and adapt phenotypically to each tissue by factors produced locally. A-Gonzalez et al. now show that phagocytosis of unwanted cells additionally contributes to imprinting macrophage heterogeneity, thus promoting tissue homeostasis.
Abstract
Tissue-resident macrophages display varying phenotypic and functional properties that are largely specified by their local environment. One of these functions, phagocytosis, mediates the natural disposal of billions of cells, but its mechanisms and consequences within living tissues are poorly defined. Using a parabiosis-based strategy, we identified and isolated macrophages from multiple tissues as they phagocytosed blood-borne cellular material. Phagocytosis was circadianally regulated and mediated by distinct repertoires of receptors, opsonins, and transcription factors in macrophages from each tissue. Although the tissue of residence defined the core signature of macrophages, phagocytosis imprinted a distinct antiinflammatory profile. Phagocytic macrophages expressed CD206, displayed blunted expression of Il1b, and supported tissue homeostasis. Thus, phagocytosis is a source of macrophage heterogeneity that acts together with tissue-derived factors to preserve homeostasis.
Introduction
Most tissues are populated by macrophages, a cell type endowed with both immune and nonimmune tasks that preserve tissue integrity and proper function. Within different tissues, macrophages acquire unique properties that reflect distinct developmental origins, dynamics, and phenotypic attributes (Davies et al., 2013). These tissue-restricted features of macrophages are now known to be driven by the expression of defined repertoires of transcription factors that integrate signals emanating from the environment and trigger specific genetic programs (Lavin et al., 2014). Genetic deficiency in some of these factors can lead to striking phenotypes, such as the disappearance of discrete subsets of macrophages in certain organs and altered homeostasis in affected tissues (A-Gonzalez et al., 2013; Haldar et al., 2014; Rosas et al., 2014; Schneider et al., 2014). Phenotypic and functional heterogeneity of resident macrophages is therefore dictated by specific molecular cues present in each tissue.
A defining functional property of macrophages is their capacity to phagocytose unwanted material during development (Henson and Hume, 2006; Gordon, 2016), pathogenic infections, or tissue injury, thereby contributing to the resolution of inflammation (Dockrell et al., 2001; Kosmider et al., 2012; Martin et al., 2012), or through normal tissue function (Arandjelovic and Ravichandran, 2015). In most of these cases, the eliminated material consists of cells that become irreversibly damaged as a consequence of external aggression or programmed cell death, as is the case for neutrophils and other blood cells that display a short lifespan (Pillay et al., 2010; Tak et al., 2013). Indeed, billions of cells are eliminated every day from a healthy organism in a way that does not trigger damage but is instead part of a homeostatic clearance program that involves engulfment by macrophages and other cells types (Arandjelovic and Ravichandran, 2015; Gordon, 2016).
The importance of phagocytosis is illustrated in the elimination of developing spermatocytes in the testes, epithelial cells in the lungs or intestine, or circulating cells in the spleen and liver (Suratt et al., 2001; Dong et al., 2016; Grabiec and Hussell, 2016; Nakahashi-Oda et al., 2016). The efficiency of this clearance process is ensured by a series of soluble or cell-bound molecules that mediate the attraction, recognition, and engulfment of the target cells by macrophages (Hochreiter-Hufford and Ravichandran, 2013). Opsonins like Annexin A1 or milk-fat globule EGF factor 8 (Mfge8) bind phosphatidylserine or other marks of apoptotic cells (Hanayama et al., 2004; Dalli et al., 2012), and are recognized directly or indirectly by receptors on the surface of macrophages, including T cell immunoglobulin- and mucin-domain–containing molecule 4 (Tim4; Miyanishi et al., 2007) or the receptor tyrosine kinase Mer (Scott et al., 2001). Uptake of the cellular material can finally trigger activation of nuclear receptors such as LXRα and β or PPARδ and γ (A-Gonzalez et al., 2009; Roszer et al., 2011). These receptors amplify cell clearance by promoting the transcription of genes involved in the phagocytic cascade, while at the same time repressing the production of inflammatory mediators, with the overall outcome of efficient and immunologically silent removal of the cells (Fadok et al., 1998; Huynh et al., 2002; Notley et al., 2015). The importance of this process is further emphasized by the development of chronic inflammatory disease in mice lacking one or various genes involved in phagocytosis (Scott et al., 2001; Hanayama et al., 2004; A-Gonzalez et al., 2009; Mukundan et al., 2009; Nagata et al., 2010; Roszer et al., 2011). Physiological cell clearance can therefore influence the transcriptional profile of macrophages even in the steady state.
Despite the well-established connection between cell removal and changes in transcriptional profile, it remains unexplored whether and how phagocytosis contributes to the functional specialization of macrophages in intact tissues. Here, we have developed a strategy that allows identification and purification of macrophages that are actively engulfing cellular material, and show that macrophages residing in different tissues use distinct molecular mediators to remove unwanted cellular debris. We find that although phagocytic and nonphagocytic macrophages coexist and share a similar transcriptional profile in each tissue, phagocytosis imprints an antiinflammatory phenotype that is only partially preserved across different tissues. We propose that, by feeding cellular material to different tissues, the circulatory system contributes to the functional heterogeneity of macrophages.
Results
Identification of active phagocytosis by tissue-resident macrophages in the steady state
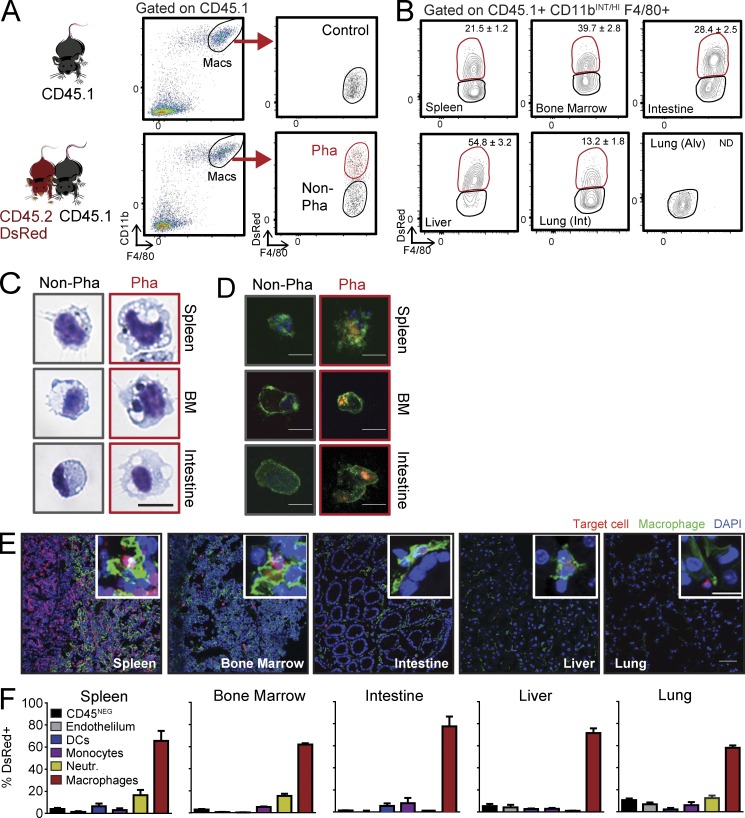
To identify macrophages that are actively phagocytic within intact tissues, we conjoined the circulation of wild-type CD45.2 DsRedTg mice and wild-type nonfluorescent CD45.1 mice by parabiosis for over 8 wk. Because circulating leukocytes naturally extravasate into tissues in the steady state (Scheiermann et al., 2015), this strategy allowed exchange of fluorescently tagged target cells (leukocytes and platelets, but not erythrocytes; Fig. S1 A) that could be spontaneously taken up by tissue-resident macrophages with minimal manipulation. We used flow cytometry to unambiguously define the populations of tissue-resident phagocytic macrophages from the CD45.1 partner that acquired fluorescence upon engulfment of DsRed+ cell content (Fig. 1 A), which was stable for at least 6 h in most tissues (Fig. S1 B). Using this strategy, we were able to identify and quantify phagocytic macrophages during homeostasis in many tissues, including the bone marrow, spleen, intestine, and liver, as well as the interstitial space of the lungs (Fig. 1 B and Fig. S1 C). We failed, however, to detect phagocytic cells among alveolar macrophages, which is consistent with the notion that circulating cells do not infiltrate alveoli of healthy mice. Cytometric identification and isolation of macrophages based on this strategy revealed that phagocytic macrophages were bigger and more complex than their nonphagocytic counterparts, a morphological feature that correlated with the presence of enlarged phagolysosomes in those cells (Fig. 1 C), the presence of what appear to be remnants of ingested cells or large cellular fragments (Fig. S1 D), as well as with internalized DsRed+ material (Fig. 1 D). Direct visualization of macrophages in tissues of the nonfluorescent CD45.1 partner revealed many DsRed+ cells from the fluorescent partner had infiltrated tissues and, in many instances, appeared to be in the proximity of or even inside resident macrophages (Fig. 1 E). Importantly, macrophages were the predominant population engulfing DsRed fluorescence in every analyzed tissue (Fig. 1 F), indicating specificity in the uptake of material from the parabiotic partner.
Figure 1.
Identification and characterization of phagocytic macrophages in healthy tissues. (A) Experimental strategy used for the identification of tissue macrophages that engulf blood-borne DsRed+ cells from their parabiotic partners by flow cytometry. Labeling of the differential CD45.1 allele and gain of DsRed identifies phagocytic versus nonphagocytic cells. (B) Representative density plots indicating the frequencies of phagocytic macrophages in different tissues from WT: DsRedTg parabionts; n = 9 pairs from three independent experiments. (C) Giemsa staining showing sorted phagocytic and nonphagocytic macrophages in spleen, bone marrow, and intestine. Bar, 10 µm. (D) Immunofluorescence staining of sorted CD45.1 macrophages from the same tissues showing ingested DsRed+ material from the parabiotic partner (green, F/80 or CD11b; blue, DAPI; red, DsRed). Bars, 10 µm. n = 10 pairs from four independent experiments. (E) Representative immunofluorescence images of tissues with resident macrophages (F4/80, green) and partner-derived target cells (red), some of which are inside macrophages (insets). Bars: 50 µm; (insets) 10 µm. n = 4 pairs. (F) Percentage of cells subsets that uptake DsRed+ material through the parabiotic exchange, within different tissues. Bars show mean ± SEM. CD45NEG cells are also CD31NEG; Neutr., neutrophils; n = 5 pairs from two independent experiments.
We considered the possibility that, besides intact cells, extracellular vesicles derived from fluorescent circulating cells (including exosomes and larger microvesicles; Colombo et al., 2014) could be mediating the transfer of fluorescence into macrophages through a nonphagocytic process. We found that only microvesicles, but not exosomes, present in the circulation of DsRed+ mice displayed low levels of fluorescence by flow cytometry (Fig. S2 A) and fluorimetry analysis (unpublished data). Intravenous transfer of exosomes or microvesicles purified from the blood of DsRed+ mice revealed that exosomes did not transfer fluorescence into tissue macrophages in vivo. Interestingly, a small fraction of liver macrophages took up DsRed+ microvesicles (Fig. S2 B), suggesting that both cells and cellular fragments contribute to label the phagocytic population in our parabiosis method. These findings, together with the observation that the fluorescence signal derived from the partner’s circulation was preferentially taken up by tissue macrophages (Fig. 1 F), indicated that our system allows detection and quantification of macrophages that are actively phagocytosing cellular material cleared from the circulation.
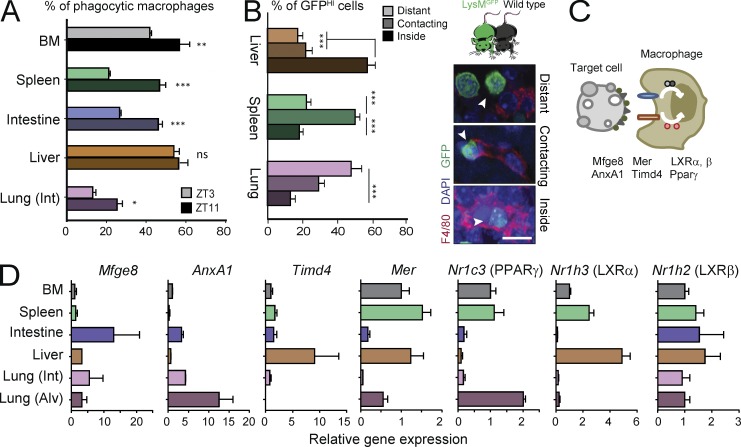
We and others have reported oscillations of leukocyte levels in blood that follow circadian patterns of clearance from blood (Scheiermann et al., 2012; Casanova-Acebes et al., 2013; Nguyen et al., 2013). We speculated that if extravasated leukocytes were bona fide target cells for tissue macrophages, their fluctuating levels in tissues would result in circadian patterns of phagocytosis. When we analyzed the frequency of phagocytic macrophages at different times of the day, we found increased percentages of phagocytic macrophages in the lung, spleen, bone marrow, and intestine at the beginning of the animal’s active phase (zeitgeber time or ZT11; Fig. 2 A and Fig. S2 C). This time coincides with the peak of leukocyte egress from blood (Casanova-Acebes et al., 2013), further supporting that phagocytic macrophages originate by uptake of circulating leukocytes. Interestingly, this rhythmic pattern of phagocytosis was conspicuously absent in hepatic macrophages (Fig. 2 A). The difference in the frequency of phagocytic macrophages at these times was not likely to be caused by rapid degradation of the fluorescent protein, as the signal was stable in macrophages at 37°C from explanted tissues for at least 6 h (Fig. S1 B). Interestingly, rapid degradation of fluorescence was only observed in the liver (Fig. S1 B), which may explain the lack of rhythmicity in this particular organ.
Figure 2.
Tissue-specific dynamics and expression of phagocytic mediators. (A) Frequency of phagocytic macrophages across different tissues in the morning (ZT3) or evening (ZT11). n = 6 per group from two independent experiments. (B) Distribution of partner-derived GFPHI neutrophils from WT: Lyz2GFP parabionts, relative to host macrophages in liver, spleen, and interstitial space of lungs. Data are from 12–46 tissue regions from two to three mice. Micrographs at right show examples of distant, contacting, and engulfed neutrophils from the same parabiotic pairs. (C) Scheme showing molecules associated with phagocytosis of target cells by macrophages. (D) Expression of genes related to phagocytosis in macrophages from bone marrow, spleen, liver, lung (interstitial or alveolar), and large intestine. All bars show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by unpaired Student’s t test (A) or one-way ANOVA analysis with Bonferroni correction (B and D). Int and Alv. refer to the interstitial and alveolar space of the lungs, respectively. n = 3 mice from one experiment.
The circadian pattern of phagocytosis suggested that circulating cells with a short lifespan and diurnal migratory patterns (e.g., neutrophils; Casanova-Acebes et al., 2013) might be preferred targets for tissue macrophages. Consistent with this possibility, mice set in parabiosis with partners that lacked neutrophils (using DsRedTg; Mrp8CreMcl1f/f mice; Dzhagalov et al., 2007) showed decreased frequency of phagocytic macrophages in most tissues, whereas exposure to DsRedTg neutrophilic partners (DsRedTg; P- and E-selectin doubly deficient or PEdKO mice; Frenette et al., 1996) resulted in more phagocytic macrophages in tissues (Fig. S2 D). In agreement with the lack of temporal patterns in the liver, the frequency of phagocytic macrophages in this tissue did not correlate with the numbers of neutrophils (Fig. S2 D). Thus neutrophils cleared from the circulation, but possibly other circulating leukocyte subsets as well, enter tissues and “feed” resident macrophages.
Apoptosis is thought to be the main form of cell death involved in homeostatic cellular turnover (Nagata et al., 2010). We therefore asked whether apoptosis of targets cells preceded their uptake by macrophages. Using TUNEL and Annexin V staining, however, we found little evidence of apoptosis among neutrophils cleared into tissues in the parabiosis model (Fig. S3, A and B). This correlated with the absence of labeling of tissue macrophages when we transferred DsRed+ apoptotic thymocytes into wild-type mice (Fig. S3 C), suggesting that, during homeostasis, apoptosis alone is not sufficient for phagocytosis of blood-borne leukocytes. Future studies will be needed to define whether other forms of cell death are required for phagocytosis in tissues.
Heterogeneous expression of phagocytic mediators
Phagocytosis of cells is a step-wise process in which target and phagocytic cells exchange signals and move toward each other, thereby facilitating their encounter and engulfment of the target cells in the relatively large space of a tissue (Medina and Ravichandran, 2016). By examining various tissues of the phagocytic mouse, we found that the position of partner-derived cells relative to macrophages varied depending on the tissue: in the interstitial space of lungs, target cells were closest to macrophages; in the spleen, contacts between potential target cells and macrophages were more frequent; and the liver featured the highest frequency of target cells that had been engulfed by macrophages (Fig. 2 B). This observation suggested that the repertoire of find-me signals, opsonins, phagocytic receptors or downstream transcription factors for effective phagocytosis might be tissue specific. In addition, because tissue-resident macrophages display a marked heterogeneity in the expression of receptors and other phagocytic molecules (Gautier et al., 2012), we predicted that differential expression of genes related to phagocytosis could be influencing this tissue-specific phagocytic activity. We thus analyzed the expression of genes encoding proteins implicated in different stages of phagocytosis of apoptotic cells (Fig. 2 C). We found diverse patterns of expression of Timd4, Mer, Mfge8, AnxA1, Nr1h3 (encoding for LXRα), Nr1h2 (LXRβ), and Nr1c3 (PPARγ) among the different tissues (Fig. 2 D). For example, expression of LXRα was particularly high in macrophages of the bone marrow, liver, and spleen; expression of Annexin A1 and PPARγ were highest in alveolar macrophages (Damazo et al., 2011; Schneider et al., 2014), and that of Timd4 dominated in the liver. In contrast, expression of LXRβ was homogeneous across all tissues (Fig. 2 D).
Thus, we identify a subset of resident macrophages within tissues whose active phagocytic activity is timed with the extravasation of circulating target cells, and that display heterogeneous expression of phagocytic molecules.
Resident macrophages use unique and shared pathways for phagocytosis among different tissues
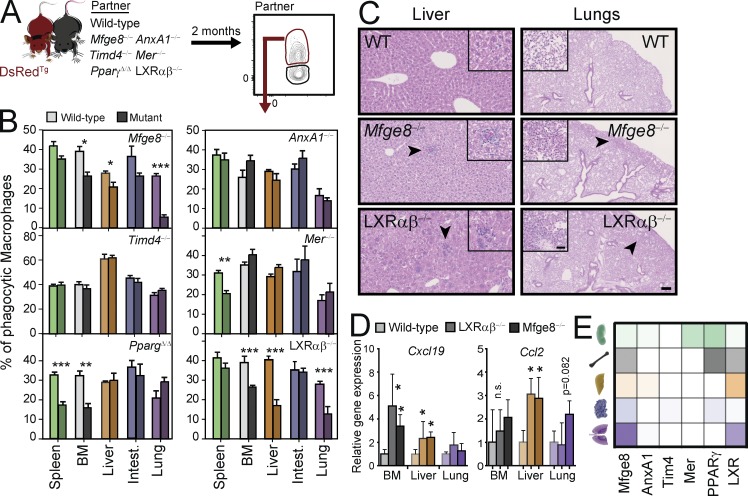
Circulating cells are believed to infiltrate tissues and to die by apoptosis (Nagata et al., 2010). To investigate which of the phagocytosis-associated molecules might be involved in the homeostatic engulfment of infiltrating cells across different tissues, we quantified the frequency of phagocytic macrophages in mice deficient in molecules involved in the various stages of phagocytosis of apoptotic cells (i.e., efferocytosis; Hochreiter-Hufford and Ravichandran, 2013). We set up parabiotic pairs of DsRedTg mice with mice deficient in the PtdSer-recognition receptors Tim4 or Mer, with mice deficient in the opsonins Mfge8 or AnxA1, and with mice deficient in the nuclear receptors LXRαβ or PPARγ (Fig. 3 A). We failed to find significant alterations in the frequency of phagocytic macrophages in Timd4−/− and Mer−/− in most of the analyzed tissues. Only the splenic red pulp macrophages in Mer−/− animals showed a reduction in the frequency of phagocytic macrophages (Fig. 3 B). Similarly, the opsonin AnxA1 was dispensable for the uptake of circulating cells in all analyzed tissues. In contrast, deletion of PPARγ (Mx1CREPpargf/f mice treated with poly I/C; referred to as PpargΔ/Δ mice) in adult mice resulted in deficient phagocytosis by bone marrow and spleen red pulp macrophages. Mfge8−/− and LXRαβ−/− mice showed reduced frequency of phagocytic macrophages in bone marrow, liver and the lung parenchyma but not in the red pulp of the spleen (Fig. 3 B). To exclude confounding effects caused by the absence of these genes in other cells, we examined phagocytosis in transplantation chimeras of wild-type and those mutants that displayed tissue-restricted deficiencies (Mfge8, Mer, PPARγ, and LXRαβ; Fig. S4). This approach, which allowed analysis of mutant and wild-type macrophages coexisting in the same environment, confirmed the findings in the global mutant mice (Fig. 3 B and Fig. S4), and demonstrated that phagocytic deficiencies were intrinsic to macrophages.
Figure 3.
Heterogeneous use of phagocytic mediators by tissue macrophages. Experimental scheme (A) and frequency (B) of phagocytic macrophages in different tissues of parabionts of DsRedTg mice with wild-type partners or partners deficient in various genes related to phagocytosis. n = 5–9 pairs from two independent experiments. (C) Histological sections of livers and lungs from the indicating mutants or control mice showing leukocyte infiltrates in Mfge8−/− and LXRαβ−/− mice (insets). Bars: 100 µm: (insets) 25 µm. (D) Expression of the inflammatory chemokines Cxcl19 and Ccl2 in bone marrow, liver, and lungs of LXRαβ−/−, Mfge8−/−, and control mice; n = 3 mice per genotype. (E) Functional heat map of phagocytic mediators according to their contribution across five different tissues, as determined in (B). n = 4 animals per group. All bars show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by unpaired Student’s t test. Lung refers to macrophages in the interstitial space of lungs.
To validate these findings and to investigate how phagocytic macrophages may contribute to tissue homeostasis, we analyzed the tissues that presented defective phagocytosis in the different mutants. We found leukocyte infiltrates in the livers of Mfge8−/− and LXRαβ−/− mice that formed discrete aggregates in the parenchyma (Fig. 3 C), and similar aggregates were visible in the lungs of those mice (Fig. 3 C). We could not observe, in contrast, major histological alterations in the bone marrow of PpargΔ/Δ, Mfge8−/−, and LXRαβ−/− mice or in the red pulp of the spleen of PpargΔ/Δ and Mer−/− mice (Fig. S5, A–C). Cytometric analyses, however, revealed increased numbers of neutrophils and monocytes in the bone marrow of PpargΔ/Δ and LXRαβ−/− mice compared with their WT counterparts (Fig. S5 B). We did not find changes in Mfge8−/− mice (unpublished data), which suggests that this molecule may overlap with other pathways for clearance. Similarly, despite normal splenic architecture we found increased numbers of neutrophils and monocytes in the spleens of PpargΔ/Δ, but not Mer−/− mice (Fig. S5 D). In contrast, although we could not directly assess engulfment of erythrocytes in our model, we found that mutants with defective phagocytosis in the spleen (i.e., Mer and Pparg mutants; Fig. 3 B) had normal numbers of circulating erythrocytes (unpublished data), suggesting no major defects in erythrocyte clearance in these mice.
Collectively, these results suggested that phagocytosis of circulating cells contributes to tissue homeostasis by preventing accumulation of inflammatory leukocytes in tissues. Leukocyte accumulation could originate from either local proliferation and/or inflammatory infiltration of affected tissues. Staining with the proliferation marker Ki67 in affected tissues of several mutants, however, ruled out a significant effect of local proliferation (Fig. S5. E and F). In contrast, expression analyses of whole tissues revealed increased expression of the genes encoding for the inflammatory chemokines CXCL19 and CCL2 in the livers of Mfge8−/− and LXRαβ−/− mice, and increased expression of CXCL19 in the bone marrow (Fig. 3 D). These data indicated that multiple molecular pathways required for efficient phagocytosis are active in the steady state, and revealed heterogeneity in how macrophages from different tissues engulfed unwanted cells (Fig. 3 E).
Distinct transcriptional signatures of tissue-resident phagocytic macrophages
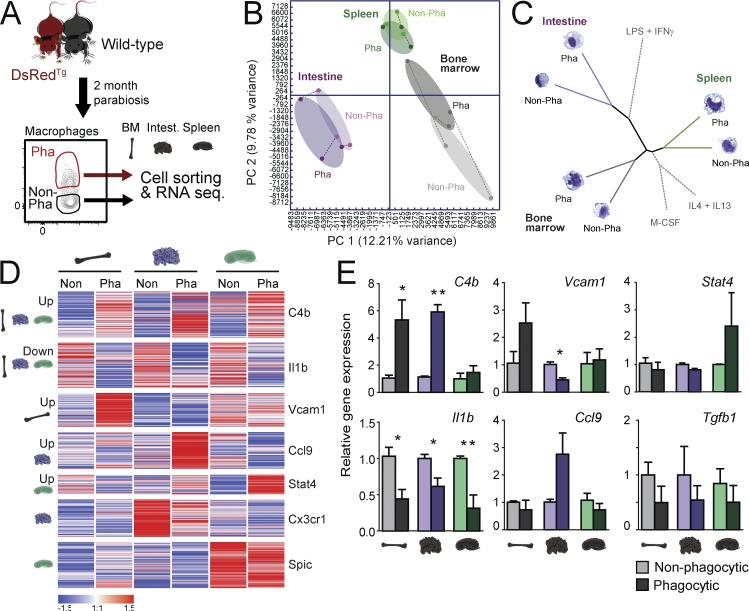
We next sought to define the impact that phagocytosis has on the transcriptional signature of macrophages residing in different tissues. We focused on splenic red pulp, large intestine, and bone marrow macrophages, of which we could isolate sufficient cells for analyses. We purified macrophages from these tissues by cytometric-sorting from parabiotic pairs of DsRedTg and wild-type partners into phagocytic and nonphagocytic subsets for transcriptomic analyses (Fig. 4 A). Principal component analysis of all genes expressed by the six populations under analysis revealed greater distance between intestinal macrophages compared with spleen and bone marrow macrophages. Importantly, these analyses could discriminate between phagocytic and nonphagocytic cells in all three tissues (Fig. 4 B). Macrophages, however, clustered best by their tissue of origin than by their phagocytic activity (Fig. 4 B) and, when we integrated our data with published signatures of cultured macrophages classically defined as M0 (treated with M-CSF), M1 (LPS + IFN-γ), and M2 (IL-4 + IL-13; as defined in Li et al. [2015]), we found that phagocytic and nonphagocytic macrophages clustered more closely between them for a given tissue than among subtypes defined by phagocytosis, or by the M0, M1, or M2 core signatures (Fig. 4 C). These findings revealed that, whereas the environment dictates the core signature of macrophages for each tissue (Lavin et al., 2014), phagocytic and nonphagocytic macrophages display distinct transcriptomic signatures (Fig. 4 C).
Figure 4.
Distinct transcriptional profile of phagocytic macrophages. (A) Experimental strategy for transcriptomic analysis of phagocytic versus nonphagocytic macrophages. (B) Principal component analysis comparing the transcriptional profile of phagocytic and nonphagocytic macrophages from the bone marrow, spleen, and large intestine. Samples of phagocytic and nonphagocytic macrophages from the same pool of mice are linked by dashed lines. (C) Distance tree illustrating the relative transcriptional proximity of macrophages based on their tissue of residence and phagocytic capacity. For reference, the relative position of in vitro–polarized M0 (M-CSF), M1 (LPS + IFN-γ), and M2 (IL-4 + IL-13) macrophages (obtained from Li et al. [2015]) is shown. (D) K-means clustering (K = 9) of 634 differentially expressed genes in phagocytic (Pha) and nonphagocytic (Non) macrophages; n = 3 samples per group. (E) Validation by quantitative PCR of genes identified in the transcriptomic analyses of phagocytic and nonphagocytic from bone marrow, spleen, and large intestine; n = 5–6 samples per group from two independent experiments. All bars show mean ± SEM. *, P < 0.05; **, P < 0.01, as determined by paired Student’s t test analysis.
Using a criterion of significance (P < 0.05) and expression of the genes in at least one of the six groups under analysis, we identified 634 genes that were differentially expressed between phagocytic and nonphagocytic macrophages. We found gene clusters that were up-regulated in phagocytic macrophages in every tissue, genes whose expression was down-regulated in phagocytic macrophages in all tissues, and genes specifically up- or down-regulated in only one of the analyzed tissues (Fig. 4 D). The differential expression of selected genes was further assessed by quantitative PCR analyses, and confirmed up-regulation of the complement factor C4b in the bone marrow and intestine, down-regulation of the inflammatory gene IL1b in all tissues, and up-regulation of the chemokine Ccl9 and the transcription factor Stat4 in the intestine and spleen, respectively (Fig. 4 E). Expression of other genes associated with phagocytosis and antiinflammatory features of macrophages (C1qb and Nos2) were differentially regulated in phagocytic cells among the different tissues (Fig. S5 G). In contrast, genes predicted a priori to also be regulated by phagocytosis, including Tgfb1 (Fadok et al., 1998) and Abca1 (a transcriptional target of LXR receptors; A-Gonzalez et al., 2009), were not differentially detected in the RNA-seq analyses and did not show differential expression by quantitative PCR (Fig. 4 E and Fig. S5 G). Thus, phagocytosis is associated with distinct transcriptional signatures on resident macrophages across tissues.
CD206 expression allows prospective identification of phagocytic macrophages
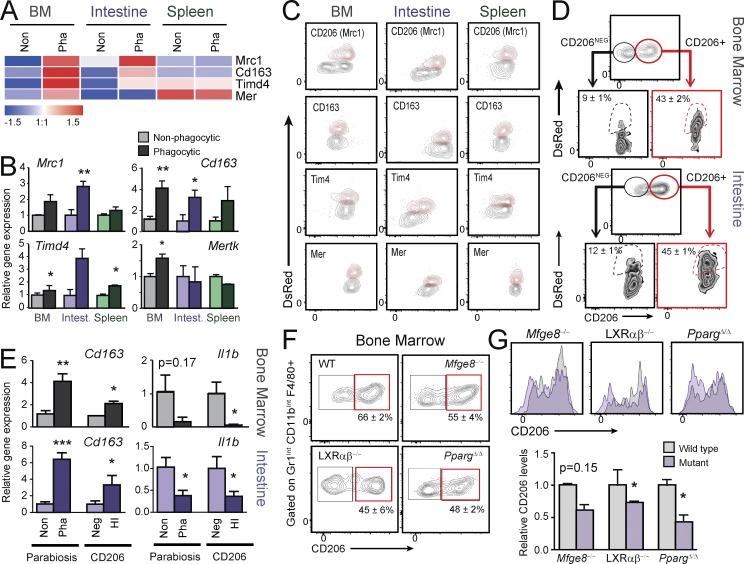
We next aimed at determining if genes from the core signature of phagocytic macrophages could be used to identify phagocytic macrophages in tissues in the steady state, without the need for experimental manipulation (e.g., our parabiosis strategy). Our transcriptomic analyses identified increased expression of Mrc1 (encoding for CD206), CD163, Timd4, and Mertk transcripts in phagocytic cells from the bone marrow and intestine (Fig. 5 A). Because these genes encode for receptors that have been previously linked to phagocytosis of apoptotic cells in vitro or under inflammatory stimuli (Scott et al., 2001; Miyanishi et al., 2007; Jiao et al., 2013; Yoon et al., 2015), we hypothesized that they could also mark bona fide phagocytic macrophages in the steady state and could be useful for their prospective identification. As quantitative PCR analysis of these genes confirmed the expression profile of Mrc1, Cd163, Timd4, and Mer in phagocytic macrophages in the predicted tissues (Fig. 5 B), we evaluated their expression at the protein level by flow cytometry in phagocytic and nonphagocytic macrophages from CD45.1 mice set in parabiosis with DsRedTg partners. We found elevated protein expression of each receptor in the phagocytic population in at least one tissue, with expression of CD206/Mrc1 higher in BM and intestine (Fig. 5 C), as predicted by the transcriptional analyses. To determine whether macrophages prospectively defined by high CD206 expression in the bone marrow and intestinal macrophages were bona fide phagocytic macrophages, we isolated CD206HI and CD206NEG macrophages from CD45.1 mice set in parabiosis with DsRedTg partners, and found that CD206HI cells identified the phagocytic subset that had gained DsRed fluorescence (Fig. 5 D). Transcriptional analyses of CD206HI macrophages showed robust down-regulation of Il1b and increased Cd163 expression compared with CD206NEG cells in the bone marrow and intestine, a finding that closely recapitulated the regulation seen in phagocytic vs. nonphagocytic macrophages (Fig. 5 E). Our findings further predicted that if CD206 was a marker of phagocytic resident macrophages, its expression would be reduced in tissues from mice in which phagocytosis was genetically impaired. Indeed, both the frequency of CD206HI macrophages and the levels of the receptor at the cell surface were reduced in macrophages from the bone marrow of LXRαβ−/−, Mfge8−/−, and PpargΔ/Δ mice relative to control animals (Fig. 5, F and G). These results thus identify CD206 as a bona fide marker of phagocytic macrophages in several tissues, provide further validation of the transcriptomic analyses, and demonstrate that subsets of resident macrophages with differential phagocytic activity coexist within tissues and can be prospectively identified.
Figure 5.
Prospective identification of phagocytic macrophages by Mrc1/CD206 expression. (A) Heat-map showing normalized expression of Mrc1, CD163, Mertk, and Timd4 in phagocytic (Pha) and nonphagocytic (Non) macrophages. (B) Validation of the data shown in A by quantitative PCR analyses of macrophages sorted from bone marrow, spleen, and large intestine (intest.). n = 5–6 samples per group from two independent experiments. (C) Cytometry plots showing the expression levels of Mrc1/CD206, Tim4, Mer, and CD163 in phagocytic (DsRed+; red contours) and nonphagocytic macrophages (DsRedneg; gray contours) from the bone marrow, spleen, and intestine of WT mice in parabiosis with DsRedTg partners. (D) Representative cytometry plots showing expression of CD206 in macrophages from bone marrow and intestine, and the frequency of phagocytic (DsRed+) macrophages within the CD206+ and CD206neg subsets. n = 6 pairs from two independent experiments. (E) Expression of the genes encoding IL1β and CD163 in CD206+ and CD206neg macrophages sorted from the bone marrow and intestine; n = 6 samples per group from two independent experiments. (F) Cytometry plots showing the frequencies of CD206+ macrophages in the bone marrow of wild-type, PpargΔ/Δ, Mfge8−/−, and LXRαβ−/− mice; n = 3–5. (G) Histograms and relative expression levels of CD206 in bone marrow macrophages from the experiments shown in F. All bars show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by paired (B and E) and unpaired (G) Student’s t test analysis.
Phagocytosis controls removal of cellular debris and inflammatory responses in a tissue-specific manner
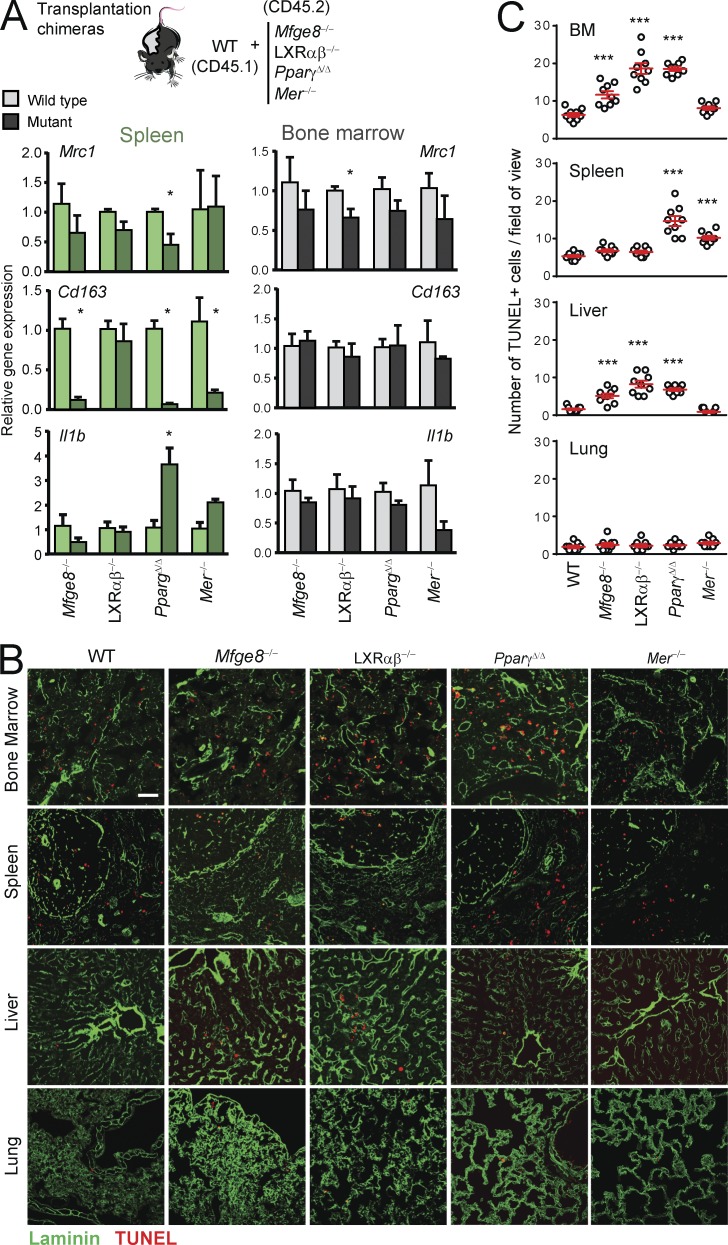
Because impaired phagocytosis by genetic deficiency correlated with increased numbers of inflammatory leukocytes, and this was not caused by local proliferation in those tissues (Fig. S5, E and F), we searched for potential alterations in cytokine production that could drive leukocyte recruitment in those tissues. Using Ingenuity pathway analysis tools, we found that certain genes regulated in phagocytic macrophages of the bone marrow predicted for decreased hematological and tissue morphology–specific functions, including quantity and accumulation of leukocytes (Table S1). To determine whether tissue-specific defects in phagocytosis caused dysregulated expression of factors involved in inflammatory recruitment or removal, we assessed gene expression in mutant and wild-type macrophages from the spleen and bone marrow of transplantation chimeras (Fig. 6 A). Expression of the genes encoding for the scavenging receptors CD206 and CD163 were reduced in splenic macrophages from Mfge8−/−, Mer−/−, and PpargΔ/Δ mice, with minor or no changes in the bone marrow. In contrast, expression of the inflammatory cytokine IL1β was elevated in splenic macrophages from PpargΔ/Δ and, to a lesser extent, Mer−/− mice, but was unaffected in the BM (Fig. 6 A). Together with our previous findings on the expression of other inflammatory chemokines (Cxcl19 and Ccl2; Fig. 3 D), these data suggested that phagocytosis regulates leukocyte recruitment and removal in a tissue-specific manner.
Figure 6.
Phagocytosis limits accumulation of dying cells in tissues. (A) Expression in macrophages sorted from the bone marrow and spleen of chimeric mice of genes encoding for factors required for leukocyte recruitment and disposal; n = 3 pairs per group from one experiment. (B) Representative micrographs of tissues stained for apoptotic cells (TUNEL+; red) in bone marrow, spleen, liver, and lung of wild-type and the indicated mutant mice. Laminin (green) stains for tissue vasculature. Bar, 100 µm. (C) Number of TUNEL+ cells per field of view in the different tissues of mutants shown in (B). Bars show mean ± SEM. *, P < 0.05; ***, P < 0.001, as determined by paired Student’s t test (A) or one-way ANOVA with Dunnett’s multiple comparison test (C). Data are from 15–35 tissue regions from three mice.
To confirm that removal of cellular corpses relies on tissue-specific phagocytosis, we finally quantified the presence of apoptotic cells across various tissues of the different mutant mice. In agreement with the tissue-specific phagocytic pathways identified earlier (Fig. 3 E), we found accumulation of TUNEL+ cells in the bone marrow of PpargΔ/Δ, Mfge8−/−, and LXRαβ−/− mice, in the splenic red pulp of Mer−/− and PpargΔ/Δ mice, and in the livers of Mfge8−/− and LXRαβ−/− mice (Fig. 6, B and C). Apoptotic cells also appeared in the livers of PpargΔ/Δ mice; however, this was likely phagocytosis independent, as this nuclear receptor is known to regulate multiple aspects of hepatic function (Gavrilova et al., 2003). In contrast, we failed to detect accumulation of apoptotic cells in the lung parenchyma, including Mfge8−/− and LXRαβ−/− mice (Fig. 6, B and C), suggesting that alternative, macrophage-independent mechanisms of cell removal may exist in the lung. Thus, tissue-resident macrophages support tissue homeostasis through active phagocytosis, a process that in turn limits the production of cytokines that promote leukocyte recruitment.
Discussion
Although billions of cells die during the normal function of the organism, a more accurate dissection of their in vivo removal by phagocytosis has remained elusive and current knowledge of this process largely derives from in vitro experiments. In this study, we have developed a strategy that allows identification, quantification, and transcriptomic profiling of tissue macrophages from living tissues that engulf leukocytes naturally cleared from the circulation into tissues. We have used a parabiosis-based strategy combined with multiple lines of mice deficient in genes involved in various stages of phagocytosis to characterize the dynamics of the process, and to define tissue specificity in the use of particular mediators of phagocytosis. More importantly, although we find that the tissue of residence is a dominant factor in determining macrophage identity, we provide evidence suggesting that infiltration and engulfment of blood-borne cells and cellular fragments imprint a transcriptional profile that is distinctive of phagocytic macrophages. These findings support previous evidences showing that function, and not only location, classifies tissue-resident macrophages (Haldar et al., 2014).
A limitation of our study is that only fluorescently labeled leukocytes exchanged between parabiotic partners marked phagocytic macrophages, whereas engulfment of other unlabeled cells from the host or extracellular material (e.g., epithelial cells or matrix proteins) remains undetected. In this regard, we find that SpiC, a transcription factor up-regulated by erythrocyte-derived heme in red pulp macrophages (Haldar et al., 2014), is not up-regulated in our phagocytic red pulp macrophages (Fig. 4 D), suggesting that erythrocytes and leukocytes may be engulfed by nonoverlapping subsets of macrophages in the splenic red pulp. Therefore, it will be important to define how phagocytosis of other abundant cell populations, such as epithelial cells or erythrocytes, impact the fate and function of phagocytic cells, and whether different target cells are engulfed by dedicated subsets of phagocytes in living tissues. Consistent with the latter, we find heterogeneity within macrophages in the same tissues, as illustrated by the presence of distinct populations with different levels of expression of CD206 in the bone marrow and intestine. Although the observation that fluorescent labeling of macrophages in our system is relatively stable supports the existence of phagocytic and nonphagocytic subsets within tissues, it is possible that the process of phagocytosis itself (caused, for example, by preferential perivascular localization) also underlies macrophage heterogeneity. In addition, it is possible that degradation of the ingested DsRed material over time prevented accurate separation of phagocytic and nonphagocytic cells, which may have partially blunted the differences in gene expression. One last limitation is the impossibility to unambiguously define whether intact cells or cellular fragments are ultimately being phagocytosed; our data, however, support that both can be taken up and contribute to macrophage heterogeneity. Particularly interesting is the case of the liver macrophages, which show more stable DsRed+ signal over time and no circadian oscillations. This finding correlates with the predominant uptake of microvesicles by this population, and suggests that the kinetics of phagocytosis may rely on the type of preferred targets, whether they are whole cells or cellular fragments.
Cell death in healthy individuals is part of the homeostatic program, as best illustrated by the massive daily elimination of epithelial cells in the intestine (Nakahashi-Oda et al., 2016), photoreceptors in the eye (Law et al., 2015), or autoreactive thymocytes during their selection in the developing thymus (Dzhagalov et al., 2013). Phagocytosis of dead cells in these tissues is accomplished by tissue-resident macrophages, dendritic cells, and even neighboring epithelial cells (Arandjelovic and Ravichandran, 2015). In the case of short-lived circulating cells, such as neutrophils, it is generally accepted that they are eliminated in the spleen, liver, and bone marrow (Suratt et al., 2001). However, we failed to find evidence of apoptosis in neutrophils in the bloodstream or in tissues, thus raising the possibility that viable cells are actively engulfed by tissue macrophages. Nonetheless, we find that Mer and LXRα are preferentially expressed by macrophages in these three tissues, an observation that agrees with the reported role of these receptors in supporting neutrophil homeostasis (Hong et al., 2012). The observation that macrophages within most tissues display a circadian pattern in phagocytic activity, together with the prominence of neutrophils as potential target cells, suggests that phagocytosis may be a timed process that coordinates with the infiltration of tissues by leukocytes (Scheiermann et al., 2012), and may serve to optimize the natural disposal of extravasating cells and their fragments. This finding further parallels the reported circadian fluctuation of macrophage function in vivo (Silver et al., 2012), and aligns with the elimination of neutrophils by bone marrow–resident macrophages at specific times of the day (Casanova-Acebes et al., 2013). In this regard, the demonstration that clearance of neutrophils in the marrow is a functional regulator of hematopoietic niches (Casanova-Acebes et al., 2013) raises the possibility that phagocytosis can regulate other key functions in tissues. Future identification, isolation, and in-depth characterization of phagocytic macrophages across multiple tissues should enable extending the paradigm of bone marrow niches to other tissues.
An important finding was the identification of preferred pathways that tissue resident macrophages use in different tissues to phagocytose blood-borne cells. The process of phagocytosis is finely regulated by multiple molecules that recognize, engage, engulf, and process the cellular material (Arandjelovic and Ravichandran, 2015). We find that the two surface receptors analyzed here, Mer and Tim4, do not appear to play a major role in the recognition and uptake of target cells in the steady state. This may be explained by the large number of receptors that macrophages can use to recognize apoptotic cells, microorganisms, or antigenic structures, including CD36, MARCO, CD169, or CR3/Mac-1 (Gordon, 2016), which suggest extensive redundancy for the recognition of target cells in vivo. This finding, however, is at odds with studies showing the importance of both receptors for the removal of apoptotic cells (Scott et al., 2001; Bosurgi et al., 2013; Mazaheri et al., 2014; Nishi et al., 2014), and suggests that these receptors may act synergistically during the phagocytosis of apoptotic cells, as previously shown (Miyanishi et al., 2012; Nishi et al., 2014). We note, however, that most existing studies have used in vitro or inflammatory conditions and therefore fail to reproduce the physiological context of healthy tissues. In addition, because the individual relevance of Tim4 in peritoneal macrophages (Wong et al., 2010) or the trimming of photoreceptors by Mer (Law et al., 2015) have not been explored here, we cannot rule out their functional predominance in other microanatomical locations. Finally, an additional and intriguing possibility is that recognition of blood-borne cells in tissues proceeds through noncanonical pathways that do not require previous death of target cells (Brown et al., 2015), which would be consistent with our inability to detect apoptosis in circulating or cleared neutrophils. It will be important in future studies to define how macrophages and target cells interact, and to establish the mechanism of death of target cells prior or after their uptake by macrophages, as this may influence the mechanism of phagocytosis.
In stark contrast with the PtdSer receptors, we show that phagocytosis in several tissues is compromised in the absence of the nuclear receptors LXRα and β and PPARγ, suggesting less redundancy in intracellular processing and signal transduction than for the initial recognition and binding of target cells. In agreement with this, nuclear receptors are known to amplify the phagocytic capacity of macrophages and are critical to prevent overt inflammation and autoimmunity (A-Gonzalez et al., 2009; Roszer et al., 2011), functions that have both been speculated to rely on the regulation of the macrophages’ transcriptional activity by metabolites derived from the target cell (A-Gonzalez et al., 2009). We emphasize that, in our studies, we have used young male mice before the onset of autoimmunity, thus ruling out systemic inflammation as the cause of leukocyte accumulation in these mice.
Finally, our transcriptional studies show that macrophages within a tissue share a similar signature regardless of their phagocytic capacity. Despite this dominance of environmental signals, phagocytic cells display a distinct transcriptional profile that varies among tissues but also share commonly regulated genes. Consistent with previous studies (A-Gonzalez et al., 2009; Roszer et al., 2011), factors that promote phagocytosis are elevated in the phagocytic populations across the three tissues analyzed (e.g., C1qb), whereas important inflammatory pathways are suppressed (e.g., Il1b). Together with the observed phenotypic heterogeneity within tissues (Figs. 2, 4, and 5), these findings suggest that macrophage diversity is even broader than anticipated. Hence, we propose that transcriptomic and functional adaptations are imprinted both by the tissue of residence and by the microanatomical localization within a tissue. For example, areas preferentially infiltrated by leukocytes in the steady state are likely enriched in phagocytic macrophages that rid tissues of these cells. This extended complexity in the classification of macrophages within living tissues provides nonetheless a rational context under which to study macrophage biology based not only on it location, but also on their function—in this case, active phagocytosis and processing of unwanted cells.
Material and methods
Mice
All experiments were performed on 8–18-wk-old C57BL/6 male and female mice. Chow and water were available ad libitum. Mice deficient in Mer, Timd4, AnxA1, Mfge8; in both Sele and Selp (PEdKO); and in LXRα and β (Nhr1h3−/−Nhr1h2−/−) have been previously described (Frenette et al., 1996; Lu et al., 1999; Hannon et al., 2003; Hanayama et al., 2004; A-Gonzalez et al., 2009; Rodriguez-Manzanet et al., 2010). Deficiency in PPARγ was induced by injecting 375 µg poli IC intraperitoneally into Mx1CRE Ppargf/f mice (referred to as PpargΔ/Δ) every other day for 1 wk, as previously reported (Hevener et al., 2007). Transgenic mice expressing DsRed under the control of the β-actin promoter were also used. All strains were in the C57BL6 background, except for PpargΔ/Δ mice (129Sv/C57BL6 mixed background) for which littermate controls were used. Mice were maintained on a 12 h light/12 h darkness lighting schedule and housed in a specific pathogen–free facility at Centro Nacional de Investigaciones Cardiovasculares. Experimental procedures were approved by the Animal Care and Ethics Comittee of the Centro Nacional de Investigaciones Cardiovasculares and local authorities.
Parabiosis
To generate parabiotic pairs, we followed previously described procedures. Anesthetized mice were shaved at the corresponding lateral aspects and matching skin incisions were made from the olecranon to the knee joint of each mouse, and the subcutaneous fascia was bluntly dissected to create ∼0.5 cm of free skin. The olecranon and knee joints were attached by a single 5–0 polypropylene suture and tie, and the dorsal and ventral skins were approximated by continuous suture. A single dose of buprenorphine was injected subcutaneously in each partner at the end of the surgical procedure. Mice were analyzed 6–8 wk after surgery.
Generation of mixed bone marrow chimeras
Recipient wild-type C57BL/6 (CD45.2) mice were lethally irradiated (6.5 Gy split doses, 3 h apart), and subsequently received 2 × 106 bone marrow donor cells. Congenic B6.SJL (CD45.1+) and CD45.2 knock-out from the appropriate genotypes (Mer−/−, Mfge8−/−, Nhr1h3−/−Nhr1h2−/−, and PparγΔ/Δ mice) donor BM cells were harvested by flushing both tibiae and femora into RPMI. Equal numbers of wild-type CD45.1 and knock-out CD45.2 BM cells (106 plus 106 cells each) were mixed and intravenously injected into the same recipients.
Tissue processing
Except where indicated, tissues were collected at ZT3 for analysis. Large intestine (colon) was cut into small pieces, first subjected to epithelial segregation by incubation with HBSS with 2mM of EDTA for 20min at 37C and subsequently digested in HBSS with liberase (1U/ml, Roche) and DNase I (10-3U/ml, Sigma) for 40’ at 37°C. Bone marrow, liver, lung and spleen were minced and digested in HBSS with liberase (1U/ml, Roche) and DNase I (10-3U/ml, Sigma) for 30’ at 37°C. After digestion, single-cell suspensions were obtained by pipetting and mechanical dissociation of the remaining pieces through cell strainers (BD Falcon).
Flow cytometry and sorting
For macrophage analysis, CD45.1 (Ly-5.1) and CD45.2 (Ly-5.2; Tonbo Bioscience), CD11b (clone M170; BioLegend), F4/80 (clone CIA31; Serotec), Mertk (R&D Systems), CD64 (clone X54-5/7.1; BioLegend), CD11c (N418; BioLegend), and MHCII (clone M5/114.15.2; BioLegend) antibodies were used. For validation experiments, CD206 (clone C068C2; BioLegend), CD163 (clone M96; Santa Cruz Biotechnology, Inc.), and Tim4 (clone RMT4-54; BioLegend) antibodies were used. Cells were stained with the indicated antibodies for 15 min at 4°C and were analyzed by fluorescence activated cell sorting (FACS) on an LSR Fortessa flow cytometer equipped with DIVA software (BD). FlowJo software was used to analyze data. To measure the degradation of the DsRed fluorescence signal within macrophages, extracted tissues were processed (as explained above) and kept at 37°C or 4°C (control) during flow cytometric analysis of macrophages every hour for a total of 6 h.
Isolation and characterization of exosomes and microvesicles
Extracellular vesicles were purified from blood by sequential ultracentrifugation. In brief, blood circulating cells were removed from plasma by centrifugation at 800 g for 10 min. To obtain microvesicles, plasma was then spun at 12,000 g for 20 min. Finally, exosomes were collected by spinning at 100,000 g for 70 min. Exosomes were washed in 4 ml PBS and pelleted again by ultracentrifugation (50.4Ti rotor; Beckman Coulter). Exosome size and particle number were analyzed using the NS500 or NS300 nanoparticle characterization system (NanoSight; Malvern Instruments) equipped with a blue (405 nm) or green (532 nm) laser, respectively. The final exosome pellet was resuspended in PBS and protein concentration was measured by BCA (Thermo Fisher Scientific). Relative Fluorescence Units (RFU) were measured using Modulus II Microplate Multimode Reader (Turner Biosystems) using green optical kit (Excitation 525 nm, Emission 580–640 nm). For flow cytometry analysis, exosomes and microvesicles (30 µg of protein) were bound to 8 µg aldehyde surface latex beads (Invitrogen) for 1 h at room temperature. The unoccupied sites were saturated with BSA 5% and Glycine 0.1 M overnight at room temperature. After washing with PBS, exosomes were stained with anti–mouse CD9 antibody (eBioscience) then analyze by flow cytometry. For positive fluorescence control, purified exosomes were labeled with PKH26 (Sigma-Aldrich) following the manufacturer's instructions, washed with PBS and pelleted down by centrifugation at 100,000 g for 70 min. These controls demonstrated correct purification of both exosomes and microvesicles.
To determine in vivo uptake of exosomes and microvesicles, 15 µg of exosomes or 30 µg of microvesicles (corresponding to the amount present in the blood of one parabiotic mouse) freshly purified from the blood of DsRed mice were injected intravenously into wild-type mice, and tissues were collected for cytometric analysis 90 min after injection.
In vivo and in vitro phagocytosis of apoptotic thymocytes
To generate apoptotic thymocytes (AT), thymi from 3–4-wk-old DsRedTg mice were harvested and mechanically dissociated, filtered, pelleted, and resuspended in RPMI medium supplemented with 10% FCS. Apoptosis was induced by treatment with 1 µM of dexamethasone (Sigma-Aldrich) for 6 h. Primary peritoneal macrophages were recovered by peritoneal lavage from WT mice. Cells were pelleted and resuspended in RPMI medium supplemented with 10% FCS and 5 × 105 cells were plated. DsRed thymocytes were added to peritoneal macrophages in a 5:1 ratio (AT/macrophages) and cultured at 37°C for 90 min in RPMI supplemented with 10% FBS. Cells were then extensively washed, fresh RPMI with 10% FBS was added and incubated overnight at 37°C before RNA isolation. For in vivo phagocytosis analysis, 107 apoptotic thymocytes were intravenously injected into WT mice. 90 min later, mice were sacrificed and tissues were harvested for cytometric analysis.
Histology
Tissues were collected and fixed in 4% PFA, dehydrated in successive alcohol solutions, embedded in paraffin wax, and sectioned for hematoxylin/eosin staining. Alternatively, after fixation and preservation in a sucrose gradient, tissues were embedded in OCT compound (Tissue-Tek) and snap-frozen in liquid Nitrogen and Isopentane. 6-µm frozen sections were air-dried, blocked with 6% BSA and 2% preimmune serum in PBS. Tissue resident macrophages were stained using anti-F4/80 antibody (clone Cl:A3-1; provided by S. Gordon) diluted in blocking solution and detected by Alexa Fluor secondary antibodies (Molecular Probes). Nuclei were stained with DAPI (Life Technologies) and mounted with Mowiol 4–88 (Sigma-Aldrich).
RNA Isolation, reverse transcription, and real-time PCR
Total RNA from sorted macrophages was prepared with RNA Extraction RNeasy Plus Micro-kit (QIAGEN). For whole tissue RNA extraction, TRIzol (Sigma-Aldrich) extraction method was used. RNA was reverse-transcribed with High-Capacity cDNA Reverse Transcription reagents kit (Applied Biosystems) according to the manufacturer’s protocol. Real-time quantitative PCR (SYBRGreen; Applied Biosystems) assays were performed with an Applied Biosystems 7900HT Fast Real-Time PCR System sequencer detector. Expression was normalized to 36b4 expression. Primer sequences are listed in the Table S2.
RNA-sequencing and data analysis
cDNA amplification from total RNA (200 pg) and index-tagged sequencing libraries generation were performed using the Ovation Single Cell RNA-Seq System (NuGEN Technologies). Libraries were quantified using a Quant-iT dsDNA HS assay with the Q-bit fluorometer (Life Technologies). Average library size and the size distribution were determined using a High Sensitivity DNA assay in an Agilent 2100 Bioanalyzer (Agilent Technologies). Libraries were normalized to 10 nM using Tris-Cl 10 mM, pH 8.5, with 0.1% Tween 20. Libraries were applied to an Illumina flow cell for cluster generation (True Seq SR Cluster kit V2 cBot) and sequence-by-synthesis single reads of 75 base length using the TruSeq SBS kit v5 (Illumina) were generated on the Genome Analyzer IIx following the standard RNA sequencing protocol. Reads were further processed using the CASAVA package (Illumina) to split reads according to adapter indexes and produce fastq files (ref). Read quality was determined by analyzing reads with the application FastQC (Schageman et al., 2013).
For data analysis sequencing adaptor contaminations were removed from reads using cutadapt software and the resulting reads were mapped and quantified on the transcriptome (GRCm38 Ensembl gene-build 70) using RSEM v1.17 (Li and Dewey, 2011). Only genes with at least 1 count per million in at least two samples were considered for statistical analysis. Data were then normalized and differential expression tested using the bioconductor package EdgeR (Robinson et al., 2010). Further clustering, PCA analysis and heat map representations were produced using Genesis (TU Graz; Sturn et al., 2002). Genomic data are available under the Gene Expression Omnibus (GEO) accession no. GSE83222.
For generation of the distance tree, gene expression data series of in vitro activated macrophages (GSE53321) were downloaded from the NCBI GEO. The data were normalized, log transformed and adjusted for batch effects using the comBat function implemented in the R package SVA (Leek and Storey, 2007). Samples were clustered using average linkage and the result was visualized as an unrooted dendrogram using the R package APE (Paradis et al., 2004).
Statistical analysis
Data were expressed as mean ± SEM. Statistical analysis was performed with paired or unpaired Student’s t test when analyzing two groups. For multiple comparisons, data were evaluated by one-way analysis of variance (ANOVA) with Bonferroni correction. P-values <0.05 were considered significant.
Online supplemental material
Fig. S1 shows characterization of target cells and phagocytic and nonphagocytic macrophages. Fig. S2 shows characterization of phagocytosed material, circadian patterns of phagocytosis, and preferential engulfment of neutrophils. Fig. S3 shows apoptosis of target cells in tissues is rare and not sufficient for phagocytosis. Fig. S4 shows phagocytosis by wild-type and mutant macrophages in transplantation chimeras. Fig. S5 shows tissue-specific mutants show alterations in bone marrow and spleen. Table S1 lists diseases and functions predicted for the genes regulated by phagocytosis in bone marrow resident macrophages. Table S2 is a list of primers used for quantitative PCR analyses.
Supplementary Material
Acknowledgments
We thank members of the Hidalgo laboratory for discussion; J.M. Ligos, the Histology unit at CNIC and I. Ortega for animal husbandry, histology, cell sorting and cytometry; Fátima Sánchez and M.J. Gómez for help with transcriptomic analyses and depositing RNA-sequencing data. G. Lemke and O. Soehnlein for the generous gift of mice.
This study was supported by Ministerio de Economia, Competitividad e Industria (MINECO) grants SAF2015-65607 and SAF2013-49662-EXP to A. Hidalgo; SVP-2014-068595 to J.A. Nicolás-Ávila; BES-2013–065550 to J.M. Adrover; JCI-2012-12659 to N. A-Gonzalez; and SAF2014-56819-R and SAF2015-71878-REDT to A. Castrillo. Grant CAM S2010/BMD-2350 RAPHYME to A. Castrillo; SAF2015-64287-R, SAF2015-71878-REDT (MINECO), and CardioNext-ITN-608027 (European Commission FP7) to M. Ricote; and R01 AI089824 (National Institutes of Health) to C.V. Rothlin. H.P. is supported by grants from MINECO (SAF2014-54541-R), ATRES-MEDIA – AXA, Asociación Española Contra el Cáncer, WHRI Academy, and Worldwide Cancer Research. The Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC) is supported by the MINECO and the Pro-CNIC Foundation, and is a Severo Ochoa Center of Excellence (MINECO award SEV-2015-0505).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AT
- apoptotic thymocytes
- EDTA
- ethylenediaminetetraacetic acid
- M-CSF
- macrophage colony-stimulating factor
- OCT
- optimal cutting temperature
- PCR
- polymerase chain reaction
- PFA
- paraformaldehyde
- Poly I/C
- polyinosinic:polycytidylic acid
- PtdSer
- phosphatidylserine
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
- ZT
- zeitgeber time
References
- A-Gonzalez N., Bensinger S.J., Hong C., Beceiro S., Bradley M.N., Zelcer N., Deniz J., Ramirez C., Díaz M., Gallardo G., et al. 2009. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 31:245–258. 10.1016/j.immuni.2009.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- A-Gonzalez N., Guillen J.A., Gallardo G., Diaz M., de la Rosa J.V., Hernandez I.H., Casanova-Acebes M., Lopez F., Tabraue C., Beceiro S., et al. 2013. The nuclear receptor LXRα controls the functional specialization of splenic macrophages. Nat. Immunol. 14:831–839. 10.1038/ni.2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arandjelovic S., and Ravichandran K.S.. 2015. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16:907–917. 10.1038/ni.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosurgi L., Bernink J.H., Delgado Cuevas V., Gagliani N., Joannas L., Schmid E.T., Booth C.J., Ghosh S., and Rothlin C.V.. 2013. Paradoxical role of the proto-oncogene Axl and Mer receptor tyrosine kinases in colon cancer. Proc. Natl. Acad. Sci. USA. 110:13091–13096. 10.1073/pnas.1302507110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.C., Vilalta A., and Fricker M.. 2015. Phagoptosis - cell death by phagocytosis - plays central roles in physiology, host defense and pathology. Curr. Mol. Med. 15:842–851. 10.2174/156652401509151105130628 [DOI] [PubMed] [Google Scholar]
- Casanova-Acebes M., Pitaval C., Weiss L.A., Nombela-Arrieta C., Chèvre R., A-González N., Kunisaki Y., Zhang D., van Rooijen N., Silberstein L.E., et al. 2013. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell. 153:1025–1035. 10.1016/j.cell.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Raposo G., and Théry C.. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30:255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Dalli J., Jones C.P., Cavalcanti D.M., Farsky S.H., Perretti M., and Rankin S.M.. 2012. Annexin A1 regulates neutrophil clearance by macrophages in the mouse bone marrow. FASEB J. 26:387–396. 10.1096/fj.11-182089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damazo A.S., Sampaio A.L., Nakata C.M., Flower R.J., Perretti M., and Oliani S.M.. 2011. Endogenous annexin A1 counter-regulates bleomycin-induced lung fibrosis. BMC Immunol. 12:59 10.1186/1471-2172-12-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L.C., Jenkins S.J., Allen J.E., and Taylor P.R.. 2013. Tissue-resident macrophages. Nat. Immunol. 14:986–995. 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockrell D.H., Lee M., Lynch D.H., and Read R.C.. 2001. Immune-mediated phagocytosis and killing of Streptococcus pneumoniae are associated with direct and bystander macrophage apoptosis. J. Infect. Dis. 184:713–722. 10.1086/323084 [DOI] [PubMed] [Google Scholar]
- Dong Y.S., Hou W.G., Li Y., Liu D.B., Hao G.Z., Zhang H.F., Li J.C., Zhao J., Zhang S., Liang G.B., and Li W.. 2016. Unexpected requirement for a binding partner of the syntaxin family in phagocytosis by murine testicular Sertoli cells. Cell Death Differ. 23:787–800. 10.1038/cdd.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I., St John A., and He Y.W.. 2007. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood. 109:1620–1626. 10.1182/blood-2006-03-013771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhagalov I.L., Chen K.G., Herzmark P., and Robey E.A.. 2013. Elimination of self-reactive T cells in the thymus: a timeline for negative selection. PLoS Biol. 11:e1001566 10.1371/journal.pbio.1001566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok V.A., Bratton D.L., Konowal A., Freed P.W., Westcott J.Y., and Henson P.M.. 1998. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Invest. 101:890–898. 10.1172/JCI1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette P.S., Mayadas T.N., Rayburn H., Hynes R.O., and Wagner D.D.. 1996. Susceptibility to infection and altered hematopoiesis in mice deficient in both P- and E-selectins. Cell. 84:563–574. 10.1016/S0092-8674(00)81032-6 [DOI] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. Immunological Genome Consortium . 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova O., Haluzik M., Matsusue K., Cutson J.J., Johnson L., Dietz K.R., Nicol C.J., Vinson C., Gonzalez F.J., and Reitman M.L.. 2003. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 278:34268–34276. 10.1074/jbc.M300043200 [DOI] [PubMed] [Google Scholar]
- Gordon S. 2016. Phagocytosis: An Immunobiologic Process. Immunity. 44:463–475. 10.1016/j.immuni.2016.02.026 [DOI] [PubMed] [Google Scholar]
- Grabiec A.M., and Hussell T.. 2016. The role of airway macrophages in apoptotic cell clearance following acute and chronic lung inflammation. Semin. Immunopathol. 38:409–423. 10.1007/s00281-016-0555-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M., Kohyama M., So A.Y., Kc W., Wu X., Briseño C.G., Satpathy A.T., Kretzer N.M., Arase H., Rajasekaran N.S., et al. 2014. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 156:1223–1234. 10.1016/j.cell.2014.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R., Tanaka M., Miyasaka K., Aozasa K., Koike M., Uchiyama Y., and Nagata S.. 2004. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 304:1147–1150. 10.1126/science.1094359 [DOI] [PubMed] [Google Scholar]
- Hannon R., Croxtall J.D., Getting S.J., Roviezzo F., Yona S., Paul-Clark M.J., Gavins F.N., Perretti M., Morris J.F., Buckingham J.C., and Flower R.J.. 2003. Aberrant inflammation and resistance to glucocorticoids in annexin 1−/− mouse. FASEB J. 17:253–255. [DOI] [PubMed] [Google Scholar]
- Henson P.M., and Hume D.A.. 2006. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 27:244–250. 10.1016/j.it.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Hevener A.L., Olefsky J.M., Reichart D., Nguyen M.T., Bandyopadyhay G., Leung H.Y., Watt M.J., Benner C., Febbraio M.A., Nguyen A.K., et al. 2007. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Invest. 117:1658–1669. 10.1172/JCI31561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter-Hufford A., and Ravichandran K.S.. 2013. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb. Perspect. Biol. 5:a008748 10.1101/cshperspect.a008748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C., Kidani Y., A-Gonzalez N., Phung T., Ito A., Rong X., Ericson K., Mikkola H., Beaven S.W., Miller L.S., et al. 2012. Coordinate regulation of neutrophil homeostasis by liver X receptors in mice. J. Clin. Invest. 122:337–347. 10.1172/JCI58393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh M.L., Fadok V.A., and Henson P.M.. 2002. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J. Clin. Invest. 109:41–50. 10.1172/JCI0211638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K., Zhang J., Zhang M., Wei Y., Wu Y., Qiu Z.Y., He J., Cao Y., Hu J., Zhu H., et al. 2013. The identification of CD163 expressing phagocytic chondrocytes in joint cartilage and its novel scavenger role in cartilage degradation. PLoS One. 8:e53312 10.1371/journal.pone.0053312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider B., Messier E.M., Janssen W.J., Nahreini P., Wang J., Hartshorn K.L., and Mason R.J.. 2012. Nrf2 protects human alveolar epithelial cells against injury induced by influenza A virus. Respir. Res. 13:43 10.1186/1465-9921-13-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., and Amit I.. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 159:1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law A.L., Parinot C., Chatagnon J., Gravez B., Sahel J.A., Bhattacharya S.S., and Nandrot E.F.. 2015. Cleavage of Mer tyrosine kinase (MerTK) from the cell surface contributes to the regulation of retinal phagocytosis. J. Biol. Chem. 290:4941–4952. 10.1074/jbc.M114.628297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., and Storey J.D.. 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3:1724–1735. 10.1371/journal.pgen.0030161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., and Dewey C.N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ng D.S., Mah W.C., Almeida F.F., Rahmat S.A., Rao V.K., Leow S.C., Laudisi F., Peh M.T., Goh A.M., et al. 2015. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ. 22:1081–1093. 10.1038/cdd.2014.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Gore M., Zhang Q., Camenisch T., Boast S., Casagranda F., Lai C., Skinner M.K., Klein R., Matsushima G.K., et al. 1999. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 398:723–728. 10.1038/19554 [DOI] [PubMed] [Google Scholar]
- Martin C.J., Booty M.G., Rosebrock T.R., Nunes-Alves C., Desjardins D.M., Keren I., Fortune S.M., Remold H.G., and Behar S.M.. 2012. Efferocytosis is an innate antibacterial mechanism. Cell Host Microbe. 12:289–300. 10.1016/j.chom.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaheri F., Breus O., Durdu S., Haas P., Wittbrodt J., Gilmour D., and Peri F.. 2014. Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat. Commun. 5:4046 10.1038/ncomms5046 [DOI] [PubMed] [Google Scholar]
- Medina C.B., and Ravichandran K.S.. 2016. Do not let death do us part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death Differ. 23:979–989. 10.1038/cdd.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., and Nagata S.. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature. 450:435–439. 10.1038/nature06307 [DOI] [PubMed] [Google Scholar]
- Miyanishi M., Segawa K., and Nagata S.. 2012. Synergistic effect of Tim4 and MFG-E8 null mutations on the development of autoimmunity. Int. Immunol. 24:551–559. 10.1093/intimm/dxs064 [DOI] [PubMed] [Google Scholar]
- Mukundan L., Odegaard J.I., Morel C.R., Heredia J.E., Mwangi J.W., Ricardo-Gonzalez R.R., Goh Y.P., Eagle A.R., Dunn S.E., Awakuni J.U., et al. 2009. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 15:1266–1272. 10.1038/nm.2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S., Hanayama R., and Kawane K.. 2010. Autoimmunity and the clearance of dead cells. Cell. 140:619–630. 10.1016/j.cell.2010.02.014 [DOI] [PubMed] [Google Scholar]
- Nakahashi-Oda C., Udayanga K.G., Nakamura Y., Nakazawa Y., Totsuka N., Miki H., Iino S., Tahara-Hanaoka S., Honda S., Shibuya K., and Shibuya A.. 2016. Apoptotic epithelial cells control the abundance of Treg cells at barrier surfaces. Nat. Immunol. 17:441–450. 10.1038/ni.3345 [DOI] [PubMed] [Google Scholar]
- Nguyen K.D., Fentress S.J., Qiu Y., Yun K., Cox J.S., and Chawla A.. 2013. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 341:1483–1488. 10.1126/science.1240636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi C., Toda S., Segawa K., and Nagata S.. 2014. Tim4- and MerTK-mediated engulfment of apoptotic cells by mouse resident peritoneal macrophages. Mol. Cell. Biol. 34:1512–1520. 10.1128/MCB.01394-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley C.A., Brown M.A., McGovern J.L., Jordan C.K., and Ehrenstein M.R.. 2015. Engulfment of activated apoptotic cells abolishes TGF-β-mediated immunoregulation via the induction of IL-6. J. Immunol. 194:1621–1627. 10.4049/jimmunol.1401256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., and Strimmer K.. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 20:289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Pillay J., den Braber I., Vrisekoop N., Kwast L.M., de Boer R.J., Borghans J.A., Tesselaar K., and Koenderman L.. 2010. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 116:625–627. 10.1182/blood-2010-01-259028 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzanet R., Sanjuan M.A., Wu H.Y., Quintana F.J., Xiao S., Anderson A.C., Weiner H.L., Green D.R., and Kuchroo V.K.. 2010. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc. Natl. Acad. Sci. USA. 107:8706–8711. 10.1073/pnas.0910359107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas M., Davies L.C., Giles P.J., Liao C.T., Kharfan B., Stone T.C., O’Donnell V.B., Fraser D.J., Jones S.A., and Taylor P.R.. 2014. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 344:645–648. 10.1126/science.1251414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszer T., Menéndez-Gutiérrez M.P., Lefterova M.I., Alameda D., Núñez V., Lazar M.A., Fischer T., and Ricote M.. 2011. Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator-activated receptor gamma or retinoid X receptor alpha deficiency. J. Immunol. 186:621–631. 10.4049/jimmunol.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schageman J., Zeringer E., Li M., Barta T., Lea K., Gu J., Magdaleno S., Setterquist R., and Vlassov A.V.. 2013. The complete exosome workflow solution: from isolation to characterization of RNA cargo. BioMed Res. Int. 2013:253957 10.1155/2013/253957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann C., Kunisaki Y., Lucas D., Chow A., Jang J.E., Zhang D., Hashimoto D., Merad M., and Frenette P.S.. 2012. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 37:290–301. 10.1016/j.immuni.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiermann C., Frenette P.S., and Hidalgo A.. 2015. Regulation of leucocyte homeostasis in the circulation. Cardiovasc. Res. 107:340–351. 10.1093/cvr/cvv099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., and Kopf M.. 2014. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15:1026–1037. 10.1038/ni.3005 [DOI] [PubMed] [Google Scholar]
- Scott R.S., McMahon E.J., Pop S.M., Reap E.A., Caricchio R., Cohen P.L., Earp H.S., and Matsushima G.K.. 2001. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 411:207–211. 10.1038/35075603 [DOI] [PubMed] [Google Scholar]
- Silver A.C., Arjona A., Walker W.E., and Fikrig E.. 2012. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 36:251–261. 10.1016/j.immuni.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturn A., Quackenbush J., and Trajanoski Z.. 2002. Genesis: cluster analysis of microarray data. Bioinformatics. 18:207–208. 10.1093/bioinformatics/18.1.207 [DOI] [PubMed] [Google Scholar]
- Suratt B.T., Young S.K., Lieber J., Nick J.A., Henson P.M., and Worthen G.S.. 2001. Neutrophil maturation and activation determine anatomic site of clearance from circulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 281:L913–L921. [DOI] [PubMed] [Google Scholar]
- Tak T., Tesselaar K., Pillay J., Borghans J.A., and Koenderman L.. 2013. What’s your age again? Determination of human neutrophil half-lives revisited. J. Leukoc. Biol. 94:595–601. 10.1189/jlb.1112571 [DOI] [PubMed] [Google Scholar]
- Wong K., Valdez P.A., Tan C., Yeh S., Hongo J.A., and Ouyang W.. 2010. Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc. Natl. Acad. Sci. USA. 107:8712–8717. 10.1073/pnas.0910929107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y.S., Kim S.Y., Kim M.J., Lim J.H., Cho M.S., and Kang J.L.. 2015. PPARγ activation following apoptotic cell instillation promotes resolution of lung inflammation and fibrosis via regulation of efferocytosis and proresolving cytokines. Mucosal Immunol. 8:1031–1046. 10.1038/mi.2014.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.