Rodero et al. report the direct quantification of IFNα protein in monogenic interferonopathies, autoimmunity, and infectious disease states, made possible by the combination of digital ELISA and high-affinity autoantibodies isolated from APECED patients, revealing differential levels and cellular sources dependent on underlying pathology.
Abstract
Type I interferons (IFNs) are essential mediators of antiviral responses. These cytokines have been implicated in the pathogenesis of autoimmunity, most notably systemic lupus erythematosus (SLE), diabetes mellitus, and dermatomyositis, as well as monogenic type I interferonopathies. Despite a fundamental role in health and disease, the direct quantification of type I IFNs has been challenging. Using single-molecule array (Simoa) digital ELISA technology, we recorded attomolar concentrations of IFNα in healthy donors, viral infection, and complex and monogenic interferonopathies. IFNα protein correlated well with functional activity and IFN-stimulated gene expression. High circulating IFNα levels were associated with increased clinical severity in SLE patients, and a study of the cellular source of IFNα protein indicated disease-specific mechanisms. Measurement of IFNα attomolar concentrations by digital ELISA will enhance our understanding of IFN biology and potentially improve the diagnosis and stratification of pathologies associated with IFN dysregulation.
Introduction
The identification of a soluble factor that protects cells from viral infection was first made by Isaacs and Lindenmann in 1957 (Isaacs and Lindenmann, 1957; Isaacs et al., 1957). We now know that multiple species of type I IFN exist, with this heterogeneity arising from the presence of 13 functional α genes and one β gene situated syntenically on human chromosome 9p (Manry et al., 2011). However, despite almost 60 years of active research in this field, the direct measurement of type I IFN protein in biological samples has remained elusive. Type I IFN mRNA is usually present at only trace levels in PBMCs from healthy individuals, and current ELISAs have proven either insensitive or unreliable, leading to the development of proxy assays based on type I IFN signaling (Hua et al., 2006; Niewold et al., 2009; Seo et al., 2009; Li et al., 2010; Berger Rentsch and Zimmer, 2011). Such presumed low levels of circulating IFN protein likely reflect the high biological potency of these cytokines, with most cell types expressing a type I IFN receptor.
Balanced against its beneficial role in antiviral protection, nonphysiological exposure to IFN can have major detrimental effects (Hunt et al., 2014). This point is well illustrated by the complex disorders systemic lupus erythematosus (SLE) and dermatomyositis (DM) and the recently defined group of monogenic autoinflammatory diseases referred to as the type I interferonopathies, where persistent type I IFN-induced signaling is considered causal to pathology (Hooks et al., 1979; Greenberg et al., 2005; Crow, 2011; Rodero and Crow, 2016). However, the mechanistic dissection of these pathologies has been hampered by the inability to directly quantify the disease-causing protein. This also represents a major unmet clinical need because such a test could improve diagnosis and therapeutic monitoring in the context of infection, autoimmunity and type I interferonopathies.
To overcome this limitation, we took advantage of a new digital ELISA technology based on counting individual enzyme-labeled immunocomplexes of proteins captured on paramagnetic beads in single-molecule arrays (Simoa; Rissin et al., 2010; Wilson et al., 2016). Combining this technology with unique high-affinity antibodies isolated from APS1/APECED mutation patients (Meyer et al., 2016) enabled the direct quantification of IFNα at attomolar (femtograms per milliliter) concentrations. With this 5,000-fold increase in sensitivity over commercial ELISAs, we could directly measure elevated IFNα protein in patients with adult and juvenile-onset SLE (JSLE) and juvenile-onset dermatomyositis (JDM), a group of molecularly distinct type I interferonopathies and acute viral meningitis. IFN protein quantification by Simoa was highly correlated with IFN antiviral activity and interferon-stimulated gene (ISG) expression measured in serum and in the cerebrospinal fluid (CSF) of all patients examined. Finally, differences within disease groups were observed based on serum IFNα protein levels and between diseases as indicated by the results of our studies on the cellular source of IFNα protein.
These data show that the measurement of IFNα protein by digital ELISA is a sensitive, reliable, and biologically relevant method that can be used for the diagnosis, stratification, and therapeutic monitoring of pathological states associated with an up-regulation of type I IFN signaling. The ability to directly measure IFNα protein levels will facilitate a better understanding of the nature, regulation, and impact of the human IFN-induced response.
Results
Direct quantification of IFNα in plasma, serum, and CSF
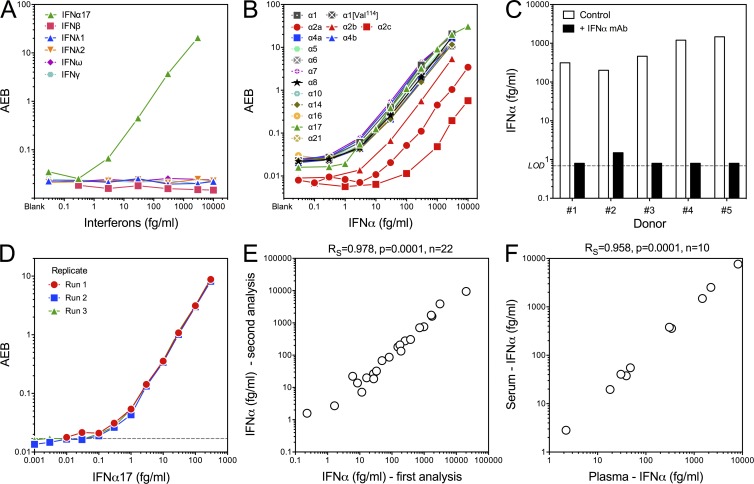
To confirm the specificity of our Simoa assay for human IFNα, and non–cross-reactivity for other IFNs, we tested these antibodies against IFNβ, IFNλ1, IFNλ2, IFNω, and IFNγ recombinant proteins (Fig. 1 A) and 16 subtypes of IFNα that included three commercially available IFNα2 types; namely, IFNα2a, IFNα2b, and IFNα2c (Fig. 1 B). Our assay did not cross-react with other IFNs and was able to detect all IFNα subtypes, although a lower affinity for IFNα2 was observed (Fig. S1 A). Because its standard curve was representative of all other subtypes, IFNα17 (IFNαI) was chosen as our reference protein. To further define the specificity of these reagents, we performed a competition assay in plasma samples from five SLE patients. Addition of anti-IFNα antibody–depleted the signal, showing a specific detection of IFNα in the tested samples (Fig. 1 C). Reproducibility was assessed across three assays (Fig. 1 D), and the mean of three blanks +3 SD was used to calculate the limit of detection (LOD). The mean of all LODs was calculated at 0.23 fg/ml, and this value was multiplied by our standard dilution factor (×3) to give a measure of 0.69 fg/ml, which was used to replace all undetectable values for presentation purposes. Assay reproducibility was also assessed by the measurement of IFNα protein in 22 plasma samples from 8 SLE, 8 JDM, 3 Aicardi–Goutières syndrome (AGS), and 3 TMEM173-mutated/STING patients, using two independently prepared lots of beads, performed by different users at different times (Rs = 0.978, P = 0.0001; Fig. 1 E). Analyzing 10 matched plasma and serum samples from AGS patients revealed a strong correlation (Rs = 0.958, P = 0.0001; Fig. 1 F), indicating a negligible influence of blood processing on IFNα concentration and the ability to use either sample for retrospective patient screening.
Figure 1.
Specificity, sensitivity, and reproducibility of the Simoa IFNα assay. (A) Simoa IFNα assay reactivity with IFNα17, IFNβ, IFNλ1, IFNλ2, IFNω, and IFNγ recombinant proteins. Lowest concentration is the blank. (B) Simoa IFNα assay cross-reactivity with IFNα1, IFNα1 (Val114), IFNα2a, IFNα2b, IFNα2c, IFNα4a, IFNα4b, IFNα5, IFNα6, IFNα7, IFNα8, IFNα10, IFNα14, IFNα16, IFNα17, and IFNα21. (C) Simoa IFNα competition assay; measurement of IFNα in five SLE patient plasma samples after preincubation with the human anti-IFNα capture antibody for 30 min before analysis. (D) Reproducibility testing for each concentration, acquired as duplicates across three independent runs. Dashed line represents the LOD, defined by mean blank average enzyme per bead (AEB) + 3 SD of all runs. (E) 22 plasma samples (8 SLE, 8 JDM, 3 AGS, and 3 STING) were analyzed with two independently prepared lots of beads by different users and at different times. Spearman correlation is reported. (F) Correlation of IFNα protein measured by Simoa in paired plasma and serum samples from 10 AGS patients. Spearman correlation is reported.
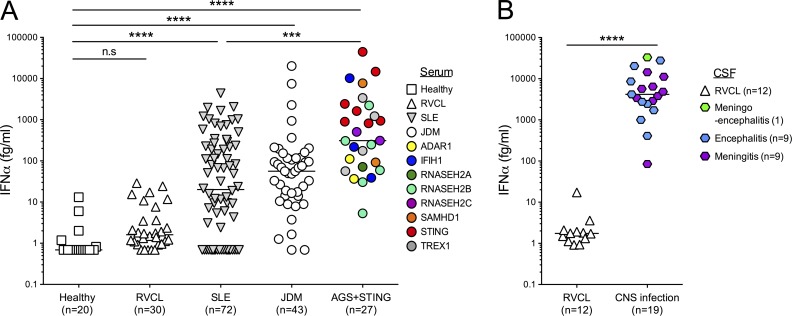
To determine the potential diagnostic capability of our assay, we examined collections of plasma and serum established during the study of Mendelian type I interferonopathies (27 patients in total), attributable to mutations in TREX1 (n = 4), RNASEH2A (n = 1), RNASEH2B (n = 6), RNASEH2C (n = 2), SAMHD1 (n = 2), ADAR1 (n = 2), IFIH1 (n = 3), and TMEM173 gain of function (n = 7; STING). We also analyzed samples from patients with JDM (n = 43) and SLE (n = 72; both juvenile and adult forms; n = 6 and 66, respectively). We compared these data with samples from 20 healthy control subjects and patients with specific autosomal-dominant mutations in the 3′ end of TREX1 causing retinal vasculopathy and cerebral leukodystrophy (RVCL) which has not previously been associated with increased type I IFN signaling (n = 30). Clinical characteristics of these patient and control groups are described in Tables S1, S2, S3, S4, S5, and S6. A one-way ANOVA test (Kruskal–Wallis) and Dunn’s post test controlling for multiple comparison testing revealed significantly elevated IFNα in the monogenic interferonopathy (P < 0.0001), JDM (P < 0.0001), and SLE (P < 0.0001) groups as compared with healthy controls, confirming the previously recognized associations with these clinical phenotypes (Fig. 2 A). In contrast, RVCL patients showed no difference in comparison to healthy controls (median 1.6 fg/ml; interquartile range [IQR] 0.95–4.6 fg/ml; Fig. 2 A). Multiple group testing also demonstrated that patients with a molecularly determined interferonopathy had significantly higher IFNα (median 310 fg/ml; IQR 71–2,223 fg/ml) in comparison to SLE (median 20 fg/ml; IQR 0.69–234 fg/ml; P < 0.001), but not to JDM (median 56 fg/ml; IQR 14–120 fg/ml) patients (Fig. 2 A).
Figure 2.
Quantification of plasma, serum, and CSF IFNα in patient cohorts. (A) Plasma from healthy controls (n = 20) and patients with RVCL (n = 30), SLE (n = 72), JDM (n = 43), and molecularly defined interferonopathies (n = 27) were assayed by Simoa for IFNα protein. Values were assessed by one-way ANOVA test (Kruskal–Wallis) and Dunn’s multiple comparison testing between groups. (B) CSF samples from acute meningitis (n = 9), acute encephalitis (n = 9), acute meningoencephalitis (n = 1), and RVCL (n = 12) were assayed by Simoa for IFNα protein. Values were assessed by Mann–Whitney T test. ***, P < 0.001; ****, P < 0.0001; n.s., not significant; horizontal lines indicate the median.
As expected, given the fundamental role of IFN in antiviral protection, testing of CSF from patients with central nervous system (CNS) infection (acute meningitis, n = 9; acute encephalitis, n = 9; acute meningoencephalitis, n = 1) revealed very high levels of IFNα protein (Fig. 2 B: median 4,174.2 fg/ml; IQR 2,437.4–11,173 fg/ml). In contrast, all but one patient with RVCL (n = 12) demonstrated an IFNα protein level of less than 10 fg/ml (P < 0.001, Mann–Whitney test as compared with infection samples).
IFNα concentration correlates with functional antiviral activity
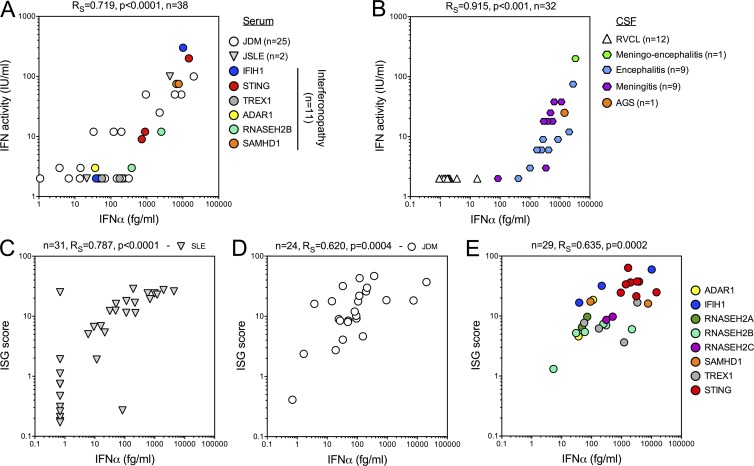
To investigate the functional relevance of such low protein levels, we compared IFNα protein measurements with IFN activity as assessed by a cytopathic protection assay (Lebon et al., 1979, 1988; Palmer et al., 2007). Specifically, IFN activity is determined by dilution of patient material incubated with Madin–Darby bovine kidney cells. Cells are challenged the next day with vesicular stomatitis virus and examined 18 h postinfection to measure the viral cytopathic effect. Cell protection against infection reveals the biological activity of type I IFN, which is compared with serial dilutions of an IFN standard. Comparing these assays on serum samples (Fig. 3 A: 25 JDM, 2 JSLE, and 11 monogenic interferonopathy patients, either AGS or STING; Table S7) demonstrated a positive correlation (Rs = 0.83; P < 0.0001). Considering the potential contribution of type I IFNs to CNS disorders, we performed a similar comparison on the CSF samples presented in Fig. 2 B. This analysis revealed an even stronger positive correlation between IFN activity and concentration (Rs = 0.924, P < 0.001). To note, statistical analysis was performed on samples with either detectable IFN activity (≥2 IU/ml) or IFN protein (>0.69 fg/ml; Fig. 3 B). Interestingly, 10 serum samples and 1 CSF sample had IFNα concentration >10 fg/ml with no detectable antiviral activity, reflecting either the increased sensitivity of the Simoa assay or the presence of posttranslational (or other) modifications that may alter functional potency.
Figure 3.
Comparison of IFNα concentration with antiviral activity and ISG expression. (A) Correlation of Simoa IFNα protein measurement with IFN activity measured by a cytopathic assay for interferonopathy (n = 10), JDM (n = 26), and JSLE (n = 2) patients. (B) Correlation of Simoa IFNα protein measurement with IFN activity measured by a cytopathic assay for CSF samples from acute viral meningitis (n = 9), acute viral encephalitis (n = 9), acute viral meningoencephalitis (n = 1), AGS (n = 1), and RVCL (n = 12). (C–E) Correlation of Simoa IFNα concentration with the ISG score in SLE (C; n = 21), JDM (D; n = 23), and molecularly defined interferonopathy patients (E; n = 29). Spearman correlations were calculated for each patient group, excluding samples where both the ISG score and the IFNα concentration were negative (SLE n = 10, JDM n = 1).
Comparison of IFNα concentration with ISG expression
We have previously described a screening tool for the identification of monogenic type I interferonopathies based on quantitative PCR assessment of six ISGs expressed by leukocytes (Rice et al., 2013, 2017). In this test, the median fold change of the ISGs when compared with the median of healthy controls is used to create an IFN score. Scores two SDs above the median of 29 controls are designated as positive (score of >2.466). We compared Simoa IFNα measurements with the ISG score of samples from 31 SLE (Fig. 3 C), 24 JDM (Fig. 3 D), and 29 monogenic interferonopathy (Fig. 3 E and Table S8) patients, excluding patients who had negative results for both tests (10 SLE and 1 JDM patients). This analysis revealed a positive correlation between the two parameters in all three groups (SLE, Rs = 0.787, P < 0.0001 [n = 31]; JDM, Rs = 0.665, P = 0.0004 [n = 24]; interferonopathies, Rs = 0.635, P = 0.0002 [n = 29]). As type I interferonopathies are associated with a broad range of phenotypes at least partially determined by genotype, we grouped patients according to identified mutations (color coded in Fig. 3 E), but this did not obviously affect the correlation. To note, two outliers in the SLE cohort showed either high ISG expression with no detectable IFNα or vice versa, although the explanation for such phenotypes is not clear.
Association of disease phenotypes with serum IFNα in lupus patients
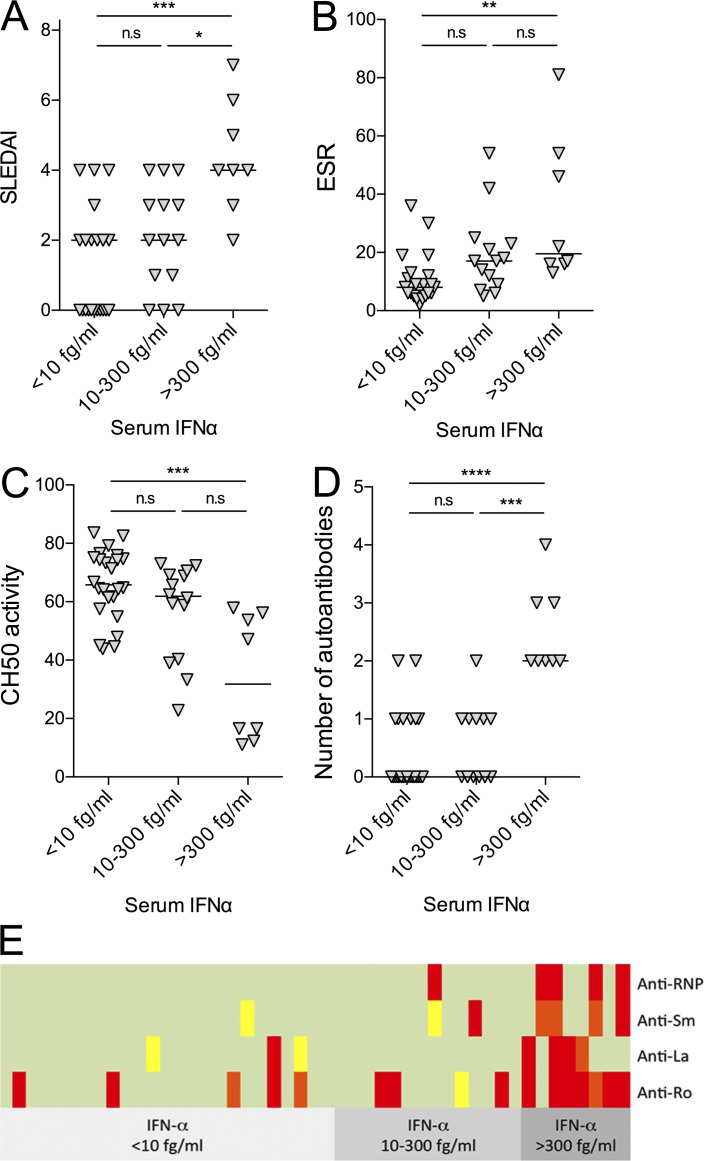
To examine whether serum IFNα concentrations were relevant to disease status, we focused on the SLE cohort. SLE is a complex and heterogeneous disease, and as such, a number of different biomarkers are used to monitor disease severity and progression. For this analysis, we divided the SLE patients into three groups based on serum IFNα concentrations: <10 fg/ml (n = 25), 10–300 fg/ml (n = 14), and >300 fg/ml (n = 8; Fig. 4). In these subgroups, we examined (a) SLE disease activity index (SLEDAI), (b) erythrocyte sedimentation rate (ESR), (c) CH50 activity, (d) number of autoantibodies, and (e) profile of autoantibodies directed against ribonucleoproteins. An ANOVA with Dunn’s post testing revealed significantly higher α-RNP antibodies (P < 0.0001), ESR (P < 0.01), and SLEDAI (P < 0.01) and lower CH50 activity (P < 0.001) in patients who had the highest levels of serum IFNα. In contrast, anti–double-stranded DNA antibodies and C-reactive protein (CRP) activity showed no differences between the subgroups (Table S9).
Figure 4.
Disease associations of serum IFNα in SLE patients. (A–D) Higher serum IFNα levels associate with higher SLEDAI (P < 0.001; A), ESR (P < 0.01; B), lower CH50 activity (P < 0.001; C), and number of specific autoantibodies against ribonucleoproteins (anti-Ro, La, Sm, RNP; P < 0.0001; D). IFNα <10 fg/ml: n = 25; IFNα = 10–300 fg/ml: n = 14; IFNα >300 fg/ml: n = 8; one-way ANOVA (Kruskal–Wallis) p-values are reported. (E) Profile of autoantibodies directed against ribonucleoproteins in patients with low, intermediate, and high IFN levels. Green, <25 U/ml; yellow, 25–50 U/ml; orange, 50–100 U/ml; red, >100 U/ml. A positive result is >25 U/ml. The total number of autoantibodies against ribonucleoproteins (anti-Ro, La, Sm, RNP) is significantly increased in patients with the highest levels of serum IFNα (two-way ANOVA, P < 0.0001). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant; horizontal lines indicate median.
Identification of cellular sources of IFNα
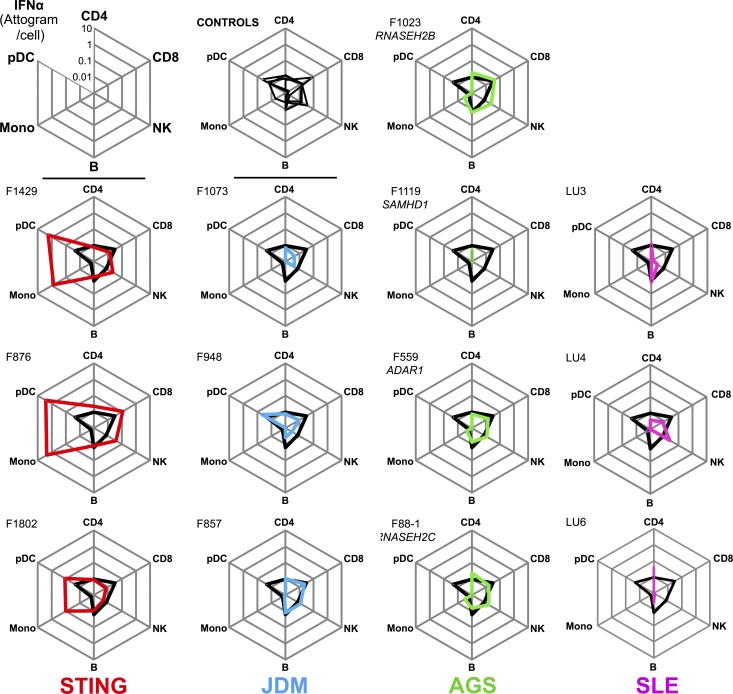
To explore cellular mechanisms driving disease pathogenesis, we next considered the source of IFNα detected in patients. We sampled blood from three STING, four AGS, three JDM, three SLE patients, and four healthy donors, and we isolated CD4+ and CD8+ T cells, B cells, NK cells, CD14+ monocytes, and plasmacytoid DCs (pDCs). A representative flow cytometry panel with the gating strategy is shown in Fig. S2. The percentage of each cell population is presented in Table S10, the purity of sorted cell numbers are detailed in the materials and Table S11, and clinical characteristics of these patients are detailed in Table S12. Cell subsets were isolated, lysed, and assessed with our IFNα Simoa assay. To calculate the level of protein present per cell, we divided the concentration of IFNα by the number of cells sorted and normalized for the volume in which the cells were lysed. This revealed a striking presence of IFNα in the monocytes and pDCs of STING patients, with a mean of 1.03 attograms/cell in pDCs and 1.53 attograms/cell in monocytes (Fig. 5). Notably, no other cell types from the STING patients, or any cell type analyzed from the other patient groups tested, demonstrated levels of IFNα above those in healthy controls. This was despite the high levels of IFNα observed in plasma, suggesting a noncirculating cellular source of IFNα protein in these diseases. This discrepancy in the cellular source of IFNα between monogenic interferonopathy patients highlights a potential cell type–specific mechanism dependent on genes involved in nucleic acid metabolism or sensing.
Figure 5.
Identification of circulating IFNα-producing cells in STING patients. IFNα protein levels presented as median attograms per cell in sorted CD4 and CD8 T cells, NK cells, B cells, monocytes, and pDCs from STING mutation (red, n = 3), JDM (blue, n = 3), AGS (green, n = 4), and SLE (purple n = 3) patients. The black line on each plot represents the median of four control healthy donors.
Discussion
In conclusion, we report here that the ultrasensitive detection of IFNα protein in human material can provide novel insights into disease-causing pathways. The transformational increase in sensitivity over conventional methods that we present derives from the combination of the Simoa digital ELISA and the extremely high affinity of the human mAbs isolated from APS1/APECED mutation patients (Meyer et al., 2016). In this way, we were able to identify circulating pDCs and monocytes as a constitutive source of IFNα protein in STING mutation patients. Although pDCs are recognized as the major type I IFN-producing leukocyte (Siegal et al., 1999), they were not apparently implicated in AGS, JDM, or SLE patients in our study, possibly suggesting tissue sequestration of these cell types or an additional cellular source of IFNα in these conditions. However, the number of pDCs isolated was low, in particular from SLE patients, so that these results will require additional confirmatory experiments. The current lack of a test to measure type I IFN protein in routine medical practice represents a major unmet clinical need. The potential of direct measurement of IFN protein per se as a disease biomarker is obvious and will be immediately relevant in SLE, where anti-IFNα and anti-IFNAR therapies are currently being tested (Lauwerys et al., 2014). Indeed, our data identified lupus patients with low or high IFNα concentrations, which differentiated patients based on a number of disease-relevant phenotypes, and as such could be used for future patient stratification, as well as for on-treatment monitoring. Such changes might also be of clinical relevance in other situations, as evidenced by the risk of developing IFN-driven pathology in the context of TNF blockade (Conrad et al., 2015). Furthermore, our ability to detect and quantify IFNα in different sample types from virally infected patients suggests possible clinical utility in the assessment of disease severity. For these reasons, we consider that digital ELISA technology can play a central role in future management of many different disease states.
Materials and methods
Subjects and samples
Historical serum, plasma, and CSF samples were collected from patients demonstrating the presence of biallelic mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR1, as well as recognized dominant disease-causing mutations in TREX1, and from patients with dominant mutations in IFIH1 and TMEM173 (STING). Samples from patients with SLE and DM demonstrated clinical criteria conforming to American College of Rheumatology and Bohan and Peter criteria, respectively. Historical samples for IFN activity studies were collected across the period 1985–2015. All samples were collected with informed consent. The study was approved by the Leeds (East) Research Ethics Committee (reference number 10/H1307/132), by the Comité de Protection des Personnes (ID-RCB/EUDRACT: 2014-A01017-40), and South-East Scotland Research Ethics Committee (0114/SS/0003). In Fig. 2, only the first sample collected from each individual patient was plotted (Tables S1, S2, S3, S4, and S5). For the correlation analysis in Fig. 3, serial samples from the same patient were used (Tables S7 and S8). SLE disease activity was assessed by an experienced rheumatology nurse specializing in SLE, who interviewed each patient and had access to all medical data and blood results using the SLEDAI 2000. SLE patients also had blood tests at the relevant clinic visits including CH50, C3, C4, autoantibody testing, CRP, and erythrocyte sedimentation rate (ESR), which were performed in a fully accredited NHS laboratory (Wiseman et al., 2016).
Simoa assay development
The Simoa IFNα assay was developed using a Quanterix Homebrew Simoa assay according to the manufacturer’s instructions and using two autoantibodies specific for IFNα isolated and cloned from 2 APS1/APECED patients recently described (Meyer et al., 2016 and patent application WO2013/098419). The 8H1 antibody clone was used as a capture antibody after coating on paramagnetic beads (0.3mg/ml), and the 12H5 was biotinylated (biotin/antibody ratio = 30/1) and used as the detector. EC50 binding of the mAbs was determined by ELISA as previously described (Meyer et al., 2016) and observed to be 4.02 ng/ml and 4.4 ng/ml for 8H1 and 12H5, respectively, for IFNα5. The IC50 was determined using an ISRE-Luciferase reporter assay (described in Meyer et al., 2016), with results presented for both antibodies against all IFNα subtypes in Fig. S1 A. For the Simoa assay, recombinant IFNα17/αI (PBL Assay Science) was used as a standard curve after cross-reactivity testing (Fig. 1 B). The LOD was calculated by the mean value of all blank runs + 3 SDs and was 0.23 fg/ml. Nonspecificity was demonstrated against IFNβ, IFNλ1, IFNλ2, IFNω, and IFNγ (PeproTech and PBL Assay Science), and cross-reactivity was tested against IFNα1, IFNα1(Val114), IFNα2b, IFNα4a, IFNα4b, IFNα5, IFNα6, IFNα7, IFNα8, IFNα10, IFNα14, IFNα16, IFNα17, and IFNα21 (all PBL Assay Science), IFNα2a (PeproTech), and IFNα2c (eBioscience). Additional specificity of the assay was demonstrated in a competition assay where SLE patient plasma samples (n = 5) were 1/3 diluted with PBS and preincubated with 50 µg/ml of the IFNα capture antibody for 30 min at room temperature before Simoa analysis. For Simoa measurements, biological samples were diluted from 1/3 to 1/30 depending on the amount of material available and to avoid saturation. Samples with signal below the LOD were normalized to 0.69 fg/ml (LOD × 3, the minimal dilution factor) for presentation and analysis purposes.
IFN activity assay
Type I IFN activity was measured by determining the cytopathic reduction (i.e., protection of Madin–Darby bovine kidney cells against cell death after infection with vesicular stomatitis virus) afforded by patient CSF/serum. A reference of human IFNα, standardized against the National Institutes of Health reference Ga 023–902-530, was included with each titration. IFNα activity in normal healthy serum is <2 IU/ml.
Assessment of ISG expression in blood cells
Blood was collected into PAXgene tubes (PreAnalytix), and total RNA was extracted using a PAXgene (PreAnalytix) RNA isolation kit. RNA concentration was assessed using a spectrophotometer (FLUOstar Omega; Labtech). Quantitative reverse transcription polymerase chain reaction (qPCR) analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems) and cDNA derived from 40ng total RNA. Using TaqMan probes for IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs00356631_g1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1), and SIGLEC1 (Hs00988063_m1), the relative abundance of each target transcript was normalized to the expression level of HPRT1 (Hs03929096_g1) and 18S (Hs999999001_s1) and assessed with the Applied Biosystems StepOne Software v2.1 and DataAssist Software v.3.01. For each of the six probes, individual (patient and control) data were expressed relative to a single calibrator (control C25). The median fold change of the six ISGs, when compared with the median of the combined 29 healthy controls, was used to create an IFN score for each patient. RQ is equal to 2−ΔCt (i.e., the normalized fold change relative to a control). When a patient was assayed on more than one occasion, the data for repeat measurements were combined to calculate a mean value (using DataAssist software v.3.01; Applied Biosystems).
Cell sorting and lysis for Simoa analysis
PBMCs were isolated from blood using lymphocyte separation medium. Just after isolation, PBMCs were labeled with CD3 Krome Orange, CD8 pCp-Cy5, CD11c PE, CD19 PE-Cy7, CD56 FITC, CD14 APC-Alexa Fluor 750 and HLA-DR Pacific Blue. PBMC subsets were isolated using a BD FACS Aria II according to the gating strategy presented in Fig. S1. Purity of the cell sorting was verified for nine individual donors and was high for all populations (mean ± SD for CD4: 97% ± 3.2, CD8: 97.2% ± 1.4, B cells: 97.5% ± 2.3, natural killer cells: 97.9% ± 1.6, monocytes: 97.7% ± 2.6, pDCs: 96.9% ± 2.3). After sorting, cells were pelleted and lysed in 50 µl RIPA buffer containing 1x of Halt protease inhibitor cocktail. Details of cell numbers and IFNα concentration are provided in Table S11. Clinical characteristics of these patients are described in Table S12. Cell populations were not sorted from virally infected patients because of ethical restrictions on obtaining sufficient blood volumes.
Statistical analysis
GraphPad Prism was used for statistical analysis. ANOVA tests (Kruskal–Wallis) with Dunn’s post testing for multiple comparisons were used to test for differences between patient groups, with median and IQRs reported. Correlations between the different assays were calculated using Spearman test.
Online supplemental material
Fig. S1 shows affinity measurements of the antibodies used in the Simoa assay. Fig. S2 shows the gating strategy for cell sorting. Tables S1–S7 list clinical characteristics of the patient cohorts. Table S8 provides information on data used for Fig. 3 (C–E). Table S9 provides demographic and clinical associations of raised serum IFNα levels in SLE patients. Table S10 shows circulating leukocyte frequencies in STING, AGS, JDM, SLE, and controls. Table S11 shows sorted cell numbers and IFNα concentrations. Table S12 shows clinical information of patients studied for cell subset IFNα content.
Supplementary Material
Acknowledgments
We acknowledge technical help from the flow cytometry platform at SFR Necker (INSERM US24-CNRS UMS 3633) and the contribution of Dr. Neeraj Dhaun, University of Edinburgh, for helping to collect the SLE cellular samples, and thanks are given to Professor John Isaacs, Newcastle University, for additional SLE samples. Thanks are given to the Clinical Research Unit, Institut Imagine for protocol assistance.
Y.J. Crow acknowledges support from the European Research Council (fellowship GA 309449; fellowship), the European Leukodystrophy Association (ELA 2012-008I1), and a state subsidy managed by the Agence Nationale de la Recherche (ANR; France) under the “Investments for the Future” program bearing the reference ANR-10-IAHU-01. Y.J. Crow and D. Duffy acknowledge support from the ANR (CE17001002). Y.L. Crow and A.M.J.M. van den Maagdenberg acknowledge the EU FP7 project NIMBL (241779). D. Duffy acknowledges funding from a PasteurInnov grant and the EU FP7 project PoC-HCV (261365) for development of Simoa assays. S. Wiseman and J. Wardlaw acknowledge funding from Lupus UK. A. Hayday acknowledges support from the Wellcome Trust (grant 106292/Z/14/Z). F. Rieux-Laucat and Y.J. Crow acknowledge the ANR (ANR-14-CE14-0026-01 “Lumugene”). D. Hunt is supported by the Wellcome Trust.
A. Hayday is cofounder and shareholder and C. Hertel is an employee of ImmunoQure AG. The remaining authors declare no competing financial interests.
Author contributions: M.P. Rodero, J. Decalf, G.I. Rice, and F. Rozenberg performed experiments, analyzed data, and wrote the paper. D. Hunt led the studies and analysis of the SLE patients. S. Werneke, S.L. McGlasson, and V. Bondet performed experiments and analyzed data. M.-A. Alyanakian, B. Bader-Meunier, C. Barnerias, N. Bellon, A. Belot, C. Bodemer, T.A. Briggs, I. Desguerre, M.-L. Frémond, M. Hully, A.M.J.M. van den Maagdenberg, I. Melki, I. Meyts, L. Musset, N. Pelzer, P. Quartier, G.M. Terwindt, J. Wardlaw, S. Wiseman, F. Rieux-Laucat, Y. Rose, and B. Neven recruited patients. C. Hertel and A. Hayday provided key reagents and critical review of the manuscript. M.L. Albert provided critical review and intellectual contribution. Y.J. Crow and D. Duffy designed the study, analyzed data, and wrote the paper.
Footnotes
Abbreviations used:
- AGS
- Aicardi–Goutières syndrome
- CNS
- central nervous system
- CSF
- cerebrospinal fluid
- DM
- dermatomyositis
- ESR
- erythrocyte sedimentation rate
- JDM
- juvenile-onset DM
- JSLE
- juvenile-onset SLE
- IQR
- interquartile range
- ISG
- interferon-stimulated gene
- LOD
- limit of detection
- pDC
- plasmacytoid DC
- RVCL
- retinal vasculopathy with cerebral leukodystrophy
- SLE
- systemic lupus erythematosus
- SLEDAI
- SLE disease activity index
- CRP
- C-reactive protein
- Simoa
- single-molecule array
References
- Berger Rentsch M., and Zimmer G.. 2011. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 6:e25858 10.1371/journal.pone.0025858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C.D., Domizio J.D., Mylonas A., Belkhodja C., Demaria O., Navarini A., Lapointe A.-K., French L., Vernez M., and Gilliet M.. 2015. ID: 2: Paradoxical psoriasis—Unabated type I IFN production induced by TNF blockade. Cytokine. 76:66–112. 10.1016/j.cyto.2015.08.045 [DOI] [Google Scholar]
- Crow Y.J. 2011. Type I interferonopathies: A novel set of inborn errors of immunity. Ann. N. Y. Acad. Sci. 1238:91–98. 10.1111/j.1749-6632.2011.06220.x [DOI] [PubMed] [Google Scholar]
- Greenberg S.A., Pinkus J.L., Pinkus G.S., Burleson T., Sanoudou D., Tawil R., Barohn R.J., Saperstein D.S., Briemberg H.R., Ericsson M., et al. . 2005. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann. Neurol. 57:664–678. 10.1002/ana.20464 [DOI] [PubMed] [Google Scholar]
- Hooks J.J., Moutsopoulos H.M., Geis S.A., Stahl N.I., Decker J.L., and Notkins A.L.. 1979. Immune interferon in the circulation of patients with autoimmune disease. N. Engl. J. Med. 301:5–8. 10.1056/NEJM197907053010102 [DOI] [PubMed] [Google Scholar]
- Hua J., Kirou K., Lee C., and Crow M.K.. 2006. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 54:1906–1916. 10.1002/art.21890 [DOI] [PubMed] [Google Scholar]
- Hunt D., Kavanagh D., Drummond I., Weller B., Bellamy C., Overell J., Evans S., Jackson A., and Chandran S.. 2014. Thrombotic microangiopathy associated with interferon beta. N. Engl. J. Med. 370:1270–1271. 10.1056/NEJMc1316118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A., and Lindenmann J.. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258–267. 10.1098/rspb.1957.0048 [DOI] [PubMed] [Google Scholar]
- Isaacs A., Lindenmann J., and Valentine R.C.. 1957. Virus interference. II. Some properties of interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:268–273. 10.1098/rspb.1957.0049 [DOI] [PubMed] [Google Scholar]
- Lauwerys B.R., Ducreux J., and Houssiau F.A.. 2014. Type I interferon blockade in systemic lupus erythematosus: where do we stand? Rheumatology (Oxford). 53:1369–1376. 10.1093/rheumatology/ket403 [DOI] [PubMed] [Google Scholar]
- Lebon P., Ponsot G., Aicardi J., Goutières F., and Arthuis M.. 1979. Early intrathecal synthesis of interferon in herpes encephalitis. Biomedicine. 31:267–271. [PubMed] [Google Scholar]
- Lebon P., Badoual J., Ponsot G., Goutières F., Hémeury-Cukier F., and Aicardi J.. 1988. Intrathecal synthesis of interferon-alpha in infants with progressive familial encephalopathy. J. Neurol. Sci. 84:201–208. 10.1016/0022-510X(88)90125-6 [DOI] [PubMed] [Google Scholar]
- Li Y., Lee P.Y., Kellner E.S., Paulus M., Switanek J., Xu Y., Zhuang H., Sobel E.S., Segal M.S., Satoh M., and Reeves W.H.. 2010. Monocyte surface expression of Fcgamma receptor RI (CD64), a biomarker reflecting type-I interferon levels in systemic lupus erythematosus. Arthritis Res. Ther. 12:R90 10.1186/ar3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manry J., Laval G., Patin E., Fornarino S., Itan Y., Fumagalli M., Sironi M., Tichit M., Bouchier C., Casanova J.L., et al. . 2011. Evolutionary genetic dissection of human interferons. J. Exp. Med. 208:2747–2759. 10.1084/jem.20111680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Woodward M., Hertel C., Vlaicu P., Haque Y., Kärner J., Macagno A., Onuoha S.C., Fishman D., Peterson H., et al. APECED patient collaborative . 2016. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 166:582–595. 10.1016/j.cell.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewold T.B., Kariuki S.N., Morgan G.A., Shrestha S., and Pachman L.M.. 2009. Elevated serum interferon-alpha activity in juvenile dermatomyositis: associations with disease activity at diagnosis and after thirty-six months of therapy. Arthritis Rheum. 60:1815–1824. 10.1002/art.24555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer P., Tovey M.G., Raschilas F., Brassart L., Meritet J.F., Porcher R., and Lebon P.. 2007. Type I interferon subtypes produced by human peripheral mononuclear cells from one normal donor stimulated by viral and non-viral inducing factors. Eur. Cytokine Netw. 18:108–114. 10.1684/ecn.2007.0093 [DOI] [PubMed] [Google Scholar]
- Rice G.I., Forte G.M., Szynkiewicz M., Chase D.S., Aeby A., Abdel-Hamid M.S., Ackroyd S., Allcock R., Bailey K.M., Balottin U., et al. . 2013. Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 12:1159–1169. 10.1016/S1474-4422(13)70258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Melki I., Frémond M.-L., Briggs T.A., Rodero M.P., Kitabayashi N., Oojageer A., Bader-Meunier B., Belot A., Bodemer C., et al. . 2017. Assessment of type I interferon signaling in pediatric inflammatory disease. J. Clin. Immunol. 37:123–132. 10.1007/s10875-016-0359-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J., et al. . 2010. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28:595–599. 10.1038/nbt.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., and Crow Y.J.. 2016. Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J. Exp. Med. 213:2527–2538. 10.1084/jem.20161596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y.J., Kim G.H., Kwak H.J., Nam J.S., Lee H.J., Suh S.K., Baek K.M., Sohn Y.W., and Hong S.H.. 2009. Validation of a HeLa Mx2/Luc reporter cell line for the quantification of human type I interferons. Pharmacology. 84:135–144. 10.1159/000235158 [DOI] [PubMed] [Google Scholar]
- Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., and Liu Y.J.. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science. 284:1835–1837. 10.1126/science.284.5421.1835 [DOI] [PubMed] [Google Scholar]
- Wilson D.H., Rissin D.M., Kan C.W., Fournier D.R., Piech T., Campbell T.G., Meyer R.E., Fishburn M.W., Cabrera C., Patel P.P., et al. . 2016. The Simoa HD-1 analyzer: A novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J. Lab. Autom. 21:533–547. 10.1177/2211068215589580 [DOI] [PubMed] [Google Scholar]
- Wiseman S.J., Bastin M.E., Jardine C.L., Barclay G., Hamilton I.F., Sandeman E., Hunt D., Amft E.N., Thomson S., Belch J.F., et al. . 2016. Cerebral Small Vessel Disease Burden Is Increased in Systemic Lupus Erythematosus. Stroke. 47:2722–2728. 10.1161/STROKEAHA.116.014330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.