Kräutler et al. show that differentiation of antibody-producing plasma cells from germinal center (GC) B cell precursors is initiated by direct contact with high-affinity antigen within the GC but completed by separate signals delivered by collaborating, GC-resident T follicular helper cells.
Abstract
Plasma cells (PCs) derived from germinal centers (GCs) secrete the high-affinity antibodies required for long-term serological immunity. Nevertheless, the process whereby GC B cells differentiate into PCs is uncharacterized, and the mechanism underlying the selective PC differentiation of only high-affinity GC B cells remains unknown. In this study, we show that differentiation into PCs is induced among a discrete subset of high-affinity B cells residing within the light zone of the GC. Initiation of differentiation required signals delivered upon engagement with intact antigen. Signals delivered by T follicular helper cells were not required to initiate differentiation but were essential to complete the differentiation process and drive migration of maturing PCs through the dark zone and out of the GC. This bipartite or two-signal mechanism has likely evolved to both sustain protective immunity and avoid autoantibody production.
Introduction
Germinal centers (GCs) are transient structures that form around the follicular dendritic cell (FDC) networks located within secondary lymphoid organs 4–7 d after challenge with foreign T cell–dependent antigens (Gatto and Brink, 2010; Victora and Nussenzweig, 2012). Antigen-specific B cells recruited into GCs undergo somatic hypermutation (SHM) of the Ig variable region genes that encode the binding specificity of the clonal B cell receptor (BCR). Clones acquiring increased affinity for antigen via SHM are preferentially retained within the GC in a process known as positive selection (Berek et al., 1991; Jacob et al., 1991). In addition, differentiation of GC B cells into antibody-secreting plasma cells (PCs) is restricted to those with high affinity for antigen (Smith et al., 2000; Phan et al., 2006). Together, these processes ensure that the GC output is made up of the most effective antibodies possible, thus providing the basis for long-term serological immunity after infection and vaccination (Plotkin et al., 2008).
GC B cells consist of spatially and phenotypically distinct light-zone (LZ) and dark-zone (DZ) populations with CXCR4lo CD86hi and CXCR4hi CD86lo cell surface phenotypes, respectively (Victora et al., 2010; Bannard et al., 2013). The signals that sustain GC B cell responses are localized within the LZ in the form of (a) intact antigen displayed on the surface of FDCs and (b) T follicular helper cells (Tfh cells) that bind processed antigenic peptides presented with class II MHC molecules on the B cell surface (Gatto and Brink, 2010; Victora and Nussenzweig, 2012). LZ B cells transit to the DZ where they undergo cell division and SHM before returning to the LZ. Preferential activation of high-affinity GC B cells in the LZ is widely accepted to mediate positive selection. However, PCs appear to exit from the DZ of the GC (Meyer-Hermann et al., 2012), and it remains unclear where and how PC differentiation is initiated within GCs. Conclusions drawn from mathematical modeling (Meyer-Hermann et al., 2006), two-photon microscopy (Allen et al., 2007), and loading of GC B cells with extrinsic peptide (Victora et al., 2010) have led to the suggestion that high-affinity GC B cells receive enhanced Tfh cell help. However, definitive identification of the stimulus that determines selective differentiation of high-affinity GC B cells into PCs awaits detailed characterization of the differentiation process within GCs and the impact of specific abrogation of signals delivered by direct engagement of intact antigen on FDCs versus those provided by Tfh cell help.
Results and discussion
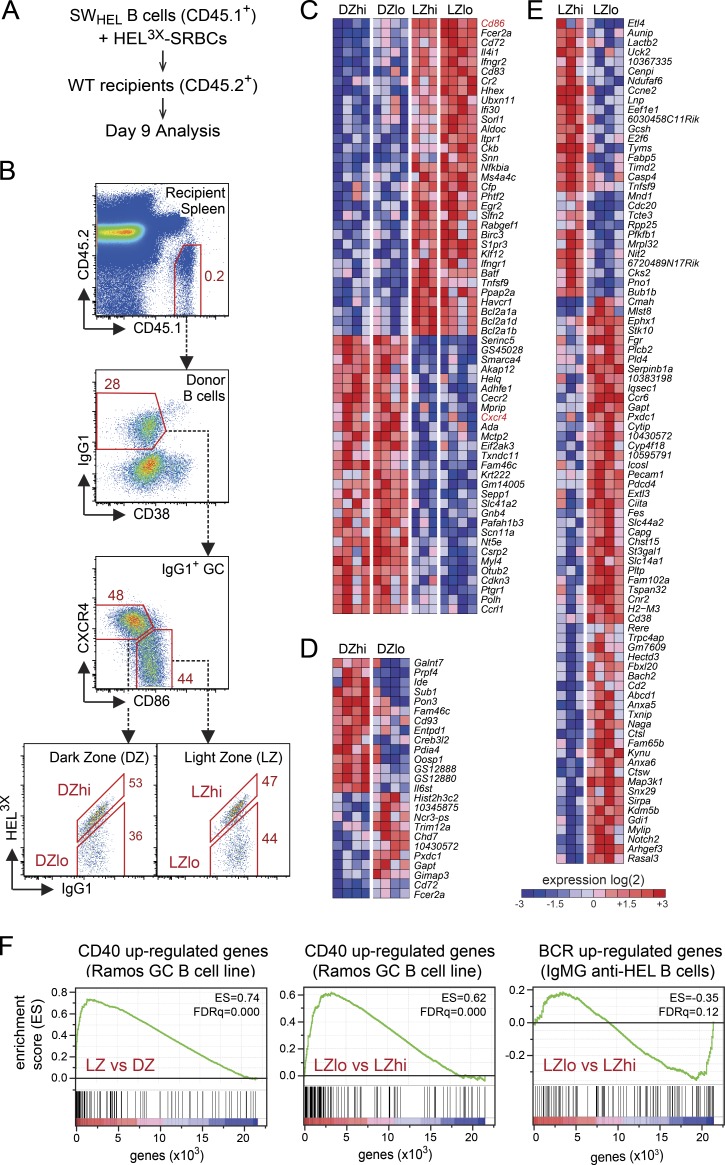
To facilitate such a study, we developed a high-resolution in vivo model in which the phenotype and fate of high- and low-affinity GC B cells are clearly identifiable. CD45.1-marked B cells from SWHEL mice, expressing the anti–hen egg lysozyme (HEL) specificity of the HyHEL10 mAb (Phan et al., 2003), were transferred into wild-type (CD45.2+) recipient mice and challenged with the low-affinity (Κa ∼107 M-1) HEL3X protein coupled to sheep RBCs (SRBCs; HEL3X-SRBCs; Fig. 1 A; Paus et al., 2006; Chan et al., 2012). Donor SWHEL B cells form GCs on days 4–5 of the response (Chan et al., 2009) and undergo affinity-based selection to HEL3X. By day 9, ∼50% of IgG1-switched LZ and DZ B cells possess high affinity for HEL3X (i.e., LZhi/DZhi GC B cells) as defined by flow cytometric staining with limiting HEL3X (Fig. 1 B). High-affinity SWHEL GC B cells carry the Y53D Ig heavy chain substitution (Fig. S1; Phan et al., 2006), which conveys an ∼100-fold increase in HEL3X-binding affinity (Chan et al., 2012).
Figure 1.
Identification of high- and low-affinity LZ and DZ SWHEL GC B cells and their affinity-dependent gene expression signatures. (A) General experimental strategy. (B) Flow cytometric gating used to sort and characterize donor-derived SWHEL GC B cells. IgG1+ GC B cells are resolved into high- and low-affinity DZ and LZ populations (DZhi, DZlo, LZhi, and LZlo). (C) Heat map showing genes differentially expressed between DZ and LZ GC B cells regardless of BCR antigen affinity. Genes encoding markers used to define the DZ and LZ subsets (Cd86 and Cxcr4) are indicated in red. The fold-change cutoff was set to ≥1.5984 (= fold-change up-regulation of Cxcr4 in DZ vs. LZ) with P ≤ 0.0005. (D and E) Heat maps showing genes differentially expressed according to BCR antigen affinity within either the DZ (D) or LZ (E). (F) GSEA of relative gene expression in total LZ versus total DZ GC B cells (left) or LZlo versus LZhi GC B cells (middle) against the gene set identified as up-regulated after CD40L stimulation of the human GC B cell line Ramos (Basso et al., 2004) and LZlo versus LZhi GC B cells against genes up-regulated by antigen (HEL) stimulation of B cells expressing an IgM BCR with an IgG1 cytoplasmic tail (Horikawa et al., 2007). FDRq, false discovery rate. Gene expression data represent four independent experiments of 25 mice per experiment.
Using gene expression microarray analysis, we confirmed that HEL3X-specific LZ and DZ GC B cells differentially express a core set of 62 genes regardless of antigen affinity (Fig. 1 C) and that this gene expression signature corresponds closely to that previously identified in anti-NP GC responses (Fig. S2 A; Victora et al., 2010). However, upon further analysis, we found additional sets of genes displaying affinity-dependent expression, particularly within the LZ compartment (Fig. 1, D and E). Gene set enrichment analysis (GSEA) confirmed previous findings (Victora et al., 2010) that LZ rather than DZ GC B cells possess a gene expression signature that most closely mirrors that of a GC B cell line (Ramos) after CD40 ligand (CD40L) stimulation (Fig. 1 F, left; Basso et al., 2004). Interestingly, this signature was stronger in LZlo compared with LZhi cells (Fig. 1 F, middle), suggesting that LZhi cells may receive signals other than those derived from Tfh cells. A gene expression signature derived from antigen-stimulated B cells expressing a BCR with an IgG1 cytoplasmic tail (Horikawa et al., 2007) showed a slightly stronger similarity to LZhi versus LZlo GC B cells (Fig. 1 F, right). Overall, however, it was not possible to make strong conclusions about the signals delivered to LZhi versus LZlo GC B cells on the basis of gene expression profiling alone.
We next set out to (a) characterize the process of PC differentiation within the GC and (b) identify the impacts on this process of specific abrogation of antigen engagement versus Tfh cell help. We found that DZ B cells displayed a more prominent PC gene signature (Mori et al., 2008) than LZ B cells (Fig. S2 B), with DZhi cells in particular expressing canonical PC genes (e.g., Prdm1/Blimp1, Sdc1, Cd93, Igj, and Xbp1; Fig. 2 A) and exhibiting a strong PC gene expression signature relative to DZlo cells (Fig. 2 B). Nevertheless, a PC gene signature was also evident in LZhi versus LZlo cells (Fig. 2 C), and analysis of responses from SWHEL B cells carrying the Blimp1gfp reporter gene (Kallies et al., 2004) confirmed that PC-lineage cells (Blimp1-GFP+) are present within both the DZhi and LZhi compartments (Fig. 2 D and Fig. S2 C). Based on established PC biology (Kallies et al., 2004), we hypothesized that the GC cells with lowest Blimp1-GFP reporter and highest BCR (IgG1) expression were those most recently committed to PC differentiation. This was supported by the fact that Blimp1-GFPlo IgG1hi cells expressed higher levels of B220 and CD45 than Blimp1-GFPhi IgG1lo PC-lineage cells (Fig. 2 E; Jensen et al., 1989; Lalor et al., 1992). Significantly, we found that Blimp1-GFP+ cells in the LZ almost exclusively possess an early PC-lineage phenotype (Blimp1-GFPlo IgG1hi; Fig. 2 F). Therefore, our data indicated that PC differentiation is initiated by signals delivered to LZhi GC B cells, with subsequent transition to a late PC phenotype (Blimp1-GFPhi IgG1lo) occurring after migration into the DZ. This is consistent with a previous report that presumptive PC-lineage cells expressing high levels of the Blimp1-inducing transcription factor IRF4 (Sciammas et al., 2006) are present in the LZ of human tonsillar GC (Falini et al., 2000) and that the LZhi compartment defined in the present study displays the highest levels of Irf4 mRNA expression in the GC (Fig. 2 A).
Figure 2.
Identification of early and late PC-lineage cells in the GC and enrichment of early PCs in the LZ. (A) Heat map of selected genes known to be up-regulated (top eight) or down-regulated (bottom three) in PCs, indicating a strong PC gene expression signature in the DZhi compartment. (B and C) GSEA of differential gene expression according to antigen affinity within the DZ (B) and LZ (C) compartments, against gene sets either up-regulated (left) or down-regulated (right) in PCs (Mori et al., 2008). FDRq, false discovery rate. (D) Proportion of cells within each of the four GC subcompartments detected as Blimp1-GFP+ on day 9 of the SWHEL.Blimp1gfp/+ response to HEL3X-SRBCs. (E, top) Day 9 GC B cells derived from SWHEL.Blimp1+/+ and SWHEL.Blimp1gfp/+ donor B cells resolving Blimp1-GFPlo, IgG1hi, and Blimp1-GFPhi IgG1lo subpopulations. (Bottom) Histogram overlays showing higher B220 and CD45 staining on the Blimp1-GFPlo IgG1hi population. (F) Flow cytometric analysis of the LZ and DZ phenotypes of Blimp1-GFP+ IgG1+ GC B cells. Data from are representative of five independent experiments of five mice per group. Flow cytometry plots are concatenated data from five recipient mice. P-values were calculated using a paired Student’s t test. *, 0.01 ≤ P < 0.05; **, 0.001 ≤ P < 0.01.
To interrogate the specific roles of Tfh cell help versus antigen engagement in PC differentiation, we again challenged transferred SWHEL.Blimp1gfp/+ B cells with HEL3X-SRBCs. On day 6 of the response (after GC formation), recipients were injected with mAbs that block (anti-CD40L) or deplete (anti-CD4) Tfh cell help, and the effect on GC responses was assessed 3 d later (day 9; Fig. 3, A and B). To block BCR access to FDC-bound antigen (HEL3X), recipients were injected with HyHEL10*, a soluble IgG1 homologue of the SWHEL BCR carrying three heavy chain mutations (S31R, Y53D, and Y58F) that confer very high affinity (Κa >10−10 M-1; Chan et al., 2012) for HEL3X. To exclude possible FcγR-mediated effects in this case, control recipients were injected with HyHEL9, an IgG1 mAb that binds HEL and HEL3X with very high affinity (Κa >10−10 M−1) but does not compete with HyHEL10 (Fig. 3 A; Smith-Gill et al., 1984).
Figure 3.
Blocking antigen access, but not Tfh cell help, prevents initiation of PC differentiation in the GC. (A) Schematic of antigen engagement and reception of cognate CD4+ Tfh cell help by SWHEL B cells in unmanipulated GCs (top), plus the mAb-based approaches used to specifically block Tfh cell help (bottom left) or block engagement of intact antigen by the SWHEL BCR (bottom right). Both HyHEL10* and HyHEL9 bind to HEL3X, but only HyHEL10* blocks access to the antigen by the SWHEL BCR. (B) Experimental design for mAb-blocking experiments. SWHEL.Blimp1gfp/+ B cells were permitted to form GCs in response to HEL3X-SRBCs and recipients and then given a single injection of mAbs 2 or 3 d before spleen harvest and analysis on day 9. (C and D) Impact of 3 d of mAb treatment on IgG1+ PC-lineage cells in GCs derived from SWHEL.Blimp1gfp/+ donor B cells. Representative flow cytometry profiles are shown (C), as well as enumeration of early (Blimp1-GFPlo IgG1hi) and late (Blimp1-GFPhi IgG1lo) PC-lineage populations in individual recipients (D). (E and F) Impact of 2 d of HyHEL10* treatment analyzed as for C and D. Data from each mAb treatment are representative of two to four independent experiments of five mice per group. Flow cytometry plots are concatenated data from five recipient mice. P-values were calculated using an unpaired Student’s t test. **, 0.001 ≤ P < 0.01.
Treatment with either HyHEL10* anti-CD4 or CD40L for 3 d led to similar (40–60%) reductions in the overall size of the GC response, with the DZ compartment most impacted in each case (Fig. S3 A). However, only antigen blockade with HyHEL10* abolished PC differentiation, removing both early (Blimp1-GFPlo IgG1hi) and late (Blimp1-GFPhi IgG1lo) PC-lineage cells (Fig. 3, C and D). The fact that both anti–Tfh cell treatments reduced GC size without preventing PC differentiation indicated that the abrogation of PC differentiation by HyHEL10* cannot be attributed to impaired antigen presentation and reduced Tfh cell help. Instead, HyHEL10* must act by preventing delivery of signals normally imparted to LZhi GC B cells directly upon antigen engagement. The specific impact of antigen blockade on the induction of PC differentiation was also evident after 2 d of HyHEL10* treatment (from days 7 to 9), which selectively depleted early PC-lineage cells (Fig. 3, E and F) including all LZ phenotype cells (not depicted). Analysis of the IgG1− (predominantly IgG2b+) PC-lineage cells in the GC showed similar results (not depicted), indicating that the triggering of the PC differentiation pathway by antigen engagement is a mechanism that is likely to apply generally to IgG-switched GC responses.
Although the overall numbers of PC-lineage cells were largely unchanged by abrogation of Tfh cell help, anti-CD4 (but not anti-CD40L) treatment had a pronounced qualitative impact. Thus, in direct contrast to antigen blockade, Tfh cell depletion resulted in the relative enrichment of early PC-lineage cells (Fig. 3, C and D). Accordingly, anti-CD4 (but not anti-CD40L) treatment also increased the fraction of PC-lineage cells that exhibited an LZ phenotype (Fig. 4, A and B). To determine whether this shift in the phenotype of PC-lineage cells was associated with physical retention in the LZ, immunofluorescence histology was undertaken. PC-lineage cells were identifiable by their strong cytoplasmic expression of IgG1 and localized primarily within the DZ of control GCs before appearing to exit through the base of the DZ (Fig. 4 C and Fig. S3 B; Meyer-Hermann et al., 2012). As has been described previously (Angelin-Duclos et al., 2000; Meyer-Hermann et al., 2012), many PC-lineage cells (30–40% by our enumeration) are found in small clusters in or just outside the GC, suggesting that ongoing cell division may occur during early PC differentiation. LZ PC-lineage cells were in natural proximity to LZ-resident FDCs (Fig. 4 C), raising the possibility that they may access antigen via the immune complexes displayed on the FDC surface. Strikingly, LZ PC-lineage cells were greatly enriched in GCs from mice treated with anti-CD4 (Fig. 4 C). LZ retention of PC-lineage cells was not seen upon treatment with anti-CD40L (Fig. S3 C), confirming that Tfh cell–derived signals other than CD40L are required to drive the LZ egress and phenotypic maturation of LZhi GC B cells that have been previously licensed to initiate PC differentiation upon direct engagement of antigen.
Figure 4.
PC-lineage cells are enriched in the LZ after depletion of CD4+ Tfh cells and colocalize with FDCs. (A and B) GC responses were established from SWHEL.Blimp1gfp/+ B cells and subjected to 3-d treatment with either anti-CD40L (A) or anti-CD4 (B) as outlined in Fig. 3. (Left) Representative flow cytometry profiles indicate the impact of mAb treatments on the LZ and DZ phenotypes of IgG1+ PC-lineage cells (Blimp1-GFP+). (Right) The proportions of IgG1+ PC-lineage cells that fell within the DZhi, DZlo, LZhi, and LZlo compartments in individual recipients were also enumerated. (C) Immunofluorescence histology of spleens from recipient mice 9 d after transfer of SWHEL B cells plus HEL3X-SRBCs and 3 d after injection of isotype control or anti-CD4 mAb. The B cell follicle (Fo) is marked by IgD (white), the LZ by CD35 (FDCs; red), and the DZ by CXCR4 (blue). The unstained T cell zone (TZ) is also indicated. IgG1+ PC-lineage cells are identifiable by bright (cytoplasmic) staining (green). Frequencies of IgG1+ PC-lineage cells identified within the LZ (containing CD35+ FDCs) by immunofluorescence analysis were 7% (5/66) in isotype-treated and 69% (76/110) in anti-CD4–treated mice (enumerated over 17 and 16 individual GCs, respectively). Bars, 50 µm. Data are representative of two to four independent experiments of five mice per group (A and B) or are representative of four independent experiments of five mice per group (C). P-values were calculated using an unpaired Student’s t test. ***, P < 0.001.
The exquisite selectivity of PC differentiation for high-affinity GC cells (Phan et al., 2006) means that its underlying mechanism must be tightly linked to antigen affinity. This condition is clearly satisfied by our finding that signals delivered upon antigen engagement are responsible for the initiation of PC differentiation among LZhi GC B cells. Although it remains unclear whether LZhi cells present more peptide antigen than LZlo cells, our gene microarray data raise the question of whether LZhi cells do preferentially receive Tfh cell help (Victora et al., 2010; Shinnakasu et al., 2016). A determining role for Tfh cells in driving PC differentiation had been proposed based on the finding that GC B cells undergo DZ migration and PC differentiation en masse when they are loaded with exogenous peptide recognized by resident Tfh cells (Victora et al., 2010). Our observations do indeed support the conclusion that Tfh cell help promotes DZ migration of committed PC-lineage cells, interestingly via signals other than CD40L. However, because depletion of Tfh cell help did not impact upon the initiation of PC differentiation in our study, we argue that boosting of Tfh cell help with exogenous peptide provides a supraphysiological stimulus to GC B cells that can drive PC differentiation regardless of BCR affinity, much like B cells stimulated with excess CD40L and cytokines in vitro (Hodgkin et al., 1994). Thus, although Tfh cell help is clearly required to progress PC differentiation and migration within the GC, it is signals delivered directly upon BCR engagement of intact antigen, most likely from the surface of FDCs, that initiate this process under physiological conditions. Therefore, our findings recall the bipartite two-signal paradigm of immune activation originally proposed by Bretscher and Cohn (1970) to prevent activation of self-reactive lymphocytes. Applied to the dynamic BCR repertoire of somatically mutating GC B cells, the need for a discriminating signal 1 (engagement of high-affinity antigen) to be supplemented with signal 2 (cognate Tfh cell help) provides a mechanism not only for selective production of high-affinity antiforeign antibodies, but also of preventing production of high-affinity autoantibodies that can arise because of chance SHM events (Brink, 2014).
The specific nature of the stimulus that initiates PC differentiation after engagement of FDC-bound antigen remains unclear. Although there are conflicting data regarding the ability of the BCRs of GC B cells to deliver intracellular signals (Khalil et al., 2012; Nowosad et al., 2016), many ligands associated with FDCs can engage GC BCRs and may act with or without BCR signaling to initiate PC differentiation. These include complement fragments (e.g., C3d), adhesion molecules (ICAM-1/VCAM-1), IL-6, B cell–activating factor, and C4BP (El Shikh et al., 2010; Goodnow et al., 2010). An intriguing question is whether signals delivered directly upon antigen engagement as opposed to enhanced antigen presentation may also be the primary driver of positive selection in the GC. The iterative nature of positive selection as opposed to the linear progression of PC differentiation makes this a more difficult issue to resolve. However, the clear implication of our findings is that either (a) antigen engagement acts as the master regulator of both positive selection and PC differentiation within the GC or (b) these two key processes rely on distinct selective mechanisms.
Materials and methods
Mice, adoptive transfers, and in vivo antibody treatments
SWHEL mice (Phan et al., 2003) were maintained on a congenic CD45.1 (Ptprca/a) C57BL/6 background. To facilitate tracking of PC differentiation, SWHEL mice were interbred with Blimp1gfp/+ mice (Kallies et al., 2004). For adoptive transfers, 6–10-wk-old wild-type C57BL/6 recipient mice (Australian BioResources) received 3 × 104 HEL-binding SWHEL B cells intravenously in combination with 2 × 108 HEL3X-conjugated SRBCs (Paus et al., 2006). To block interaction of GC B cells with Tfh cells or native antigen, mice were treated with 200 µg anti-CD4 (GK1.5; rat IgG2b; Bio X Cell), 200 µg anti-CD40L (MR-1; hamster IgG; University of California, San Francisco [UCSF] Monoclonal Antibody Core), or 50 µg HyHEL10* (HyHEL10 carrying Y53D/S31R/Y58 mutations; mouse IgG1; produced in house) at the indicated time points. Control groups received equal amounts of the respective isotype control antibodies. Animal studies were approved and conducted in compliance within the guidelines set down by the Garvan/St. Vincent’s Animal Ethics Committee.
HEL proteins and HyHEL antibodies
Recombinant HEL3X was produced in yeast (Pichia pastoris) and purified from culture supernatants as previously described (Paus et al., 2006). HyHEL10* IgG1 mAb was expressed in HEK293 cells and purified as previously described (Butt et al., 2015), and HyHEL9 was supplied by the UCSF Monoclonal Antibody Core.
Flow cytometry
Recipient splenocytes were prepared and stained for cell surface HEL3X and IgG1 as previously described (Chan et al., 2012) and subsequently incubated with the additional directly conjugated antibodies and streptavidin reagents. Donor-derived (SWHEL) GC B cells were identified as CD45.1+, CD45.2−, B220+, and CD38lo (Fig. 1 B). Flow cytometric data were acquired on an LSRII SORP flow cytometer (BD), and data were analyzed by FlowJo software (Tree Star). For sort purification of single (SHM analysis) or bulk GC B cells, samples were isolated using a FACSAria III flow cytometer (BD).
SHM analysis
Single LZhi, LZlo, DZhi, and DZlo IgG1+ SWHEL GC B cells (see gates in Fig. 1 B) were sorted into 96-well plates, and the variable region exon of the SWHEL immunoglobulin heavy chain variable region exon (HyHEL10) was PCR amplified and sequenced as previously described (Paus et al., 2006).
mRNA microarray and computational data analysis
SWHEL B cell subpopulations (minimum 16,800 cells) were gated as outlined in Fig. 1 B and sorted directly into TRIzol (Invitrogen), and RNA extraction was performed according to the manufacturer’s instructions. To improve recovery, RNA was precipitated with GlycoBlue (Thermo Fisher Scientific). RNA quality and quantity was determined using a Bioanalyzer 2100 (Agilent Technologies). RNA samples were reverse transcribed, amplified, labeled, and fragmented using Ovation Pico WTA, WT Ovation Exon Module, and Encore Biotin Module (NuGen) and hybridized with a whole-transcript gene array on a Mouse Gene 1.0 ST array chip by the Ramaciotti Centre for Genomics (University of New South Wales). One of four LZhi samples did not pass the quality control data assessment of the microarray (Console) and was therefore excluded from the analysis. Computational analysis of gene expression was performed on GenePattern (data normalization, determination of differential gene expression by LimmaGP, and GSEA preranked by fold-change; Broad Institute). For generation of heat maps, transcripts were excluded if they failed to reach a minimal mean expression of log2(x) ≥4. Cutoffs (fold-change and p-values) are indicated in the relevant figures. Genes displayed in heat maps were hierarchically clustered using Pearson correlation.
Immunofluorescence histology
Splenic cryosections were prepared and stained as previously described (Chan et al., 2009) using the antibodies and fluorescent reagents. Slides were imaged on an upright microscope (DM5500; Leica Biosystems), and images were analyzed using Photoshop CS5 (version 12.1; Adobe). Strong staining was observed for PC-lineage cells expressing high levels of cytoplasmic IgG1 in addition to weak cell surface staining for IgG1+ GC B cells. For clarity, images of IgG1+ cells presented in this study show only the brightly staining PC-lineage cells.
Statistical analysis
Statistical analyses were undertaken using a paired or unpaired two-tailed Student’s t test in Prism (GraphPad Software). Significant p-values are indicated in figures for the following ranges: *, 0.01 ≤ P < 0.05; **, 0.001 ≤ P < 0.01; ***, P < 0.001. In all summary figures, each data point represents an individual mouse, and bars represent the mean. In bar graphs, error bars represent standard error of the mean.
Accession no.
Gene expression microarray data were deposited to the Gene Expression Omnibus under accession no. GSE94638.
Online supplemental material
Fig. S1 shows the SHM analysis in the four GC compartments on day 9 of the response of SWHEL B cells to HEL3X-SRBCs. Fig. S2 shows GSEA analyses of LZ and DZ gene expression signatures and flow cytometric detection of Blimp1-GFP+ PC-lineage cells in the four GC compartments. Fig. S3 shows the effects of antibody treatments on GC size and composition as well as additional immunofluorescence histology analysis of IgG1+ PC-lineage cells within GC structures.
Supplementary Material
Acknowledgments
We thank the staff of Australian BioResources for animal husbandry, the staff of the Garvan Institute Flow Cytometry Facility for cell sorting, the Ramaciotti Centre for Genomics for microarray analysis, and C. Goodnow and T.G. Phan for discussions and comments on the manuscript.
N.J. Kräutler was supported by the Swiss National Science Foundation (grant PMPDP3_175146), C. Sundling by the Swedish Research Council (grant 2013-7333), D. Butt by an Australian Postgraduate Award, and D. Suan, T.D. Chan, D. Christ, and R. Brink by the National Health and Medical Research Council (NHMRC). This work was funded by NHMRC Program Grant 1016953.
The authors declare no competing financial interests.
Author contributions: N.J. Kräutler and D. Suan designed and performed experiments, interpreted the results, and prepared the manuscript. D. Butt, K. Bourne, and J.R. Hermes performed several experiments. T.D. Chan and C. Sundling helped with experimental design. W. Kaplan helped with microarray analyses. P. Schofield and J. Jackson prepared key reagents. A. Basten and D. Christ helped with experimental design and research supervision. R. Brink designed experiments, supervised research, and wrote the manuscript.
Footnotes
Abbreviations used:
- BCR
- B cell receptor
- DZ
- dark zone
- FDC
- follicular dendritic cell
- GC
- germinal center
- GSEA
- gene set enrichment analysis
- HEL
- hen egg lysozyme
- LZ
- light zone
- PC
- plasma cell
- SHM
- somatic hypermutation
- SRBC
- sheep RBC
- Tfh cell
- T follicular helper cell
References
- Gatto D., and Brink R.. 2010. The germinal center reaction. J. Allergy Clin. Immunol. 126:898–907. 10.1016/j.jaci.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Berek C., Berger A., and Apel M.. 1991. Maturation of the immune response in germinal centers. Cell. 67:1121–1129. 10.1016/0092-8674(91)90289-B [DOI] [PubMed] [Google Scholar]
- Jacob J., Kelsoe G., Rajewsky K., and Weiss U.. 1991. Intraclonal generation of antibody mutants in germinal centres. Nature. 354:389–392. 10.1038/354389a0 [DOI] [PubMed] [Google Scholar]
- Phan T.G., Paus D., Chan T.D., Turner M.L., Nutt S.L., Basten A., and Brink R.. 2006. High affinity germinal center B cells are actively selected into the plasma cell compartment. J. Exp. Med. 203:2419–2424. 10.1084/jem.20061254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.G., Light A., O’Reilly L.A., Ang S.M., Strasser A., and Tarlinton D.. 2000. bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J. Exp. Med. 191:475–484. 10.1084/jem.191.3.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A., Orenstein W.A., and Offit P.A.. 2008. Vaccines. Elsevier, Philadelphia. 1570 pp. [Google Scholar]
- Bannard O., Horton R.M., Allen C.D.C., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924. 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Hermann M., Mohr E., Pelletier N., Zhang Y., Victora G.D., and Toellner K.M.. 2012. A theory of germinal center B cell selection, division, and exit. Cell Reports. 2:162–174. 10.1016/j.celrep.2012.05.010 [DOI] [PubMed] [Google Scholar]
- Meyer-Hermann M.E., Maini P.K., and Iber D.. 2006. An analysis of B cell selection mechanisms in germinal centers. Math. Med. Biol. 23:255–277. 10.1093/imammb/dql012 [DOI] [PubMed] [Google Scholar]
- Allen C.D., Okada T., Tang H.L., and Cyster J.G.. 2007. Imaging of germinal center selection events during affinity maturation. Science. 315:528–531. 10.1126/science.1136736 [DOI] [PubMed] [Google Scholar]
- Phan T.G., Amesbury M., Gardam S., Crosbie J., Hasbold J., Hodgkin P.D., Basten A., and Brink R.. 2003. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J. Exp. Med. 197:845–860. 10.1084/jem.20022144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.D., Wood K., Hermes J.R., Butt D., Jolly C.J., Basten A., and Brink R.. 2012. Elimination of germinal-center-derived self-reactive B cells is governed by the location and concentration of self-antigen. Immunity. 37:893–904. 10.1016/j.immuni.2012.07.017 [DOI] [PubMed] [Google Scholar]
- Paus D., Phan T.G., Chan T.D., Gardam S., Basten A., and Brink R.. 2006. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J. Exp. Med. 203:1081–1091. 10.1084/jem.20060087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T.D., Gatto D., Wood K., Camidge T., Basten A., and Brink R.. 2009. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J. Immunol. 183:3139–3149. 10.4049/jimmunol.0901690 [DOI] [PubMed] [Google Scholar]
- Basso K., Klein U., Niu H., Stolovitzky G.A., Tu Y., Califano A., Cattoretti G., and Dalla-Favera R.. 2004. Tracking CD40 signaling during germinal center development. Blood. 104:4088–4096. 10.1182/blood-2003-12-4291 [DOI] [PubMed] [Google Scholar]
- Horikawa K., Martin S.W., Pogue S.L., Silver K., Peng K., Takatsu K., and Goodnow C.C.. 2007. Enhancement and suppression of signaling by the conserved tail of IgG memory-type B cell antigen receptors. J. Exp. Med. 204:759–769. 10.1084/jem.20061923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Rempel R.E., Chang J.T., Yao G., Lagoo A.S., Potti A., Bild A., and Nevins J.R.. 2008. Utilization of pathway signatures to reveal distinct types of B lymphoma in the Emicro-myc model and human diffuse large B-cell lymphoma. Cancer Res. 68:8525–8534. 10.1158/0008-5472.CAN-08-1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Tarlinton D.M., Dietrich W., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200:967–977. 10.1084/jem.20040973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen G.S., Poppema S., Mant M.J., and Pilarski L.M.. 1989. Transition in CD45 isoform expression during differentiation of normal and abnormal B cells. Int. Immunol. 1:229–236. 10.1093/intimm/1.3.229 [DOI] [PubMed] [Google Scholar]
- Lalor P.A., Nossal G.J., Sanderson R.D., and McHeyzer-Williams M.G.. 1992. Functional and molecular characterization of single, (4-hydroxy-3-nitrophenyl)acetyl (NP)-specific, IgG1+ B cells from antibody-secreting and memory B cell pathways in the C57BL/6 immune response to NP. Eur. J. Immunol. 22:3001–3011. 10.1002/eji.1830221136 [DOI] [PubMed] [Google Scholar]
- Sciammas R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Falini B., Fizzotti M., Pucciarini A., Bigerna B., Marafioti T., Gambacorta M., Pacini R., Alunni C., Natali-Tanci L., Ugolini B., et al. . 2000. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 95:2084–2092. [PubMed] [Google Scholar]
- Smith-Gill S.J., Lavoie T.B., and Mainhart C.R.. 1984. Antigenic regions defined by monoclonal antibodies correspond to structural domains of avian lysozyme. J. Immunol. 133:384–393. [PubMed] [Google Scholar]
- Angelin-Duclos C., Cattoretti G., Lin K.I., and Calame K.. 2000. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J. Immunol. 165:5462–5471. 10.4049/jimmunol.165.10.5462 [DOI] [PubMed] [Google Scholar]
- Shinnakasu R., Inoue T., Kometani K., Moriyama S., Adachi Y., Nakayama M., Takahashi Y., Fukuyama H., Okada T., and Kurosaki T.. 2016. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17:861–869. 10.1038/ni.3460 [DOI] [PubMed] [Google Scholar]
- Hodgkin P.D., Castle B.E., and Kehry M.R.. 1994. B cell differentiation induced by helper T cell membranes: evidence for sequential isotype switching and a requirement for lymphokines during proliferation. Eur. J. Immunol. 24:239–246. 10.1002/eji.1830240138 [DOI] [PubMed] [Google Scholar]
- Bretscher P., and Cohn M.. 1970. A theory of self-nonself discrimination. Science. 169:1042–1049. 10.1126/science.169.3950.1042 [DOI] [PubMed] [Google Scholar]
- Brink R. 2014. The imperfect control of self-reactive germinal center B cells. Curr. Opin. Immunol. 28:97–101. 10.1016/j.coi.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Khalil A.M., Cambier J.C., and Shlomchik M.J.. 2012. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 336:1178–1181. 10.1126/science.1213368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowosad C.R., Spillane K.M., and Tolar P.. 2016. Germinal center B cells recognize antigen through a specialized immune synapse architecture. Nat. Immunol. 17:870–877. 10.1038/ni.3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Shikh M.E., El Sayed R.M., Sukumar S., Szakal A.K., and Tew J.G.. 2010. Activation of B cells by antigens on follicular dendritic cells. Trends Immunol. 31:205–211. 10.1016/j.it.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow C.C., Vinuesa C.G., Randall K.L., Mackay F., and Brink R.. 2010. Control systems and decision making for antibody production. Nat. Immunol. 11:681–688. 10.1038/ni.1900 [DOI] [PubMed] [Google Scholar]
- Butt D., Chan T.D., Bourne K., Hermes J.R., Nguyen A., Statham A., O’Reilly L.A., Strasser A., Price S., Schofield P., et al. . 2015. FAS inactivation releases unconventional germinal center B cells that escape antigen control and drive IgE and autoantibody production. Immunity. 42:890–902. 10.1016/j.immuni.2015.04.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.