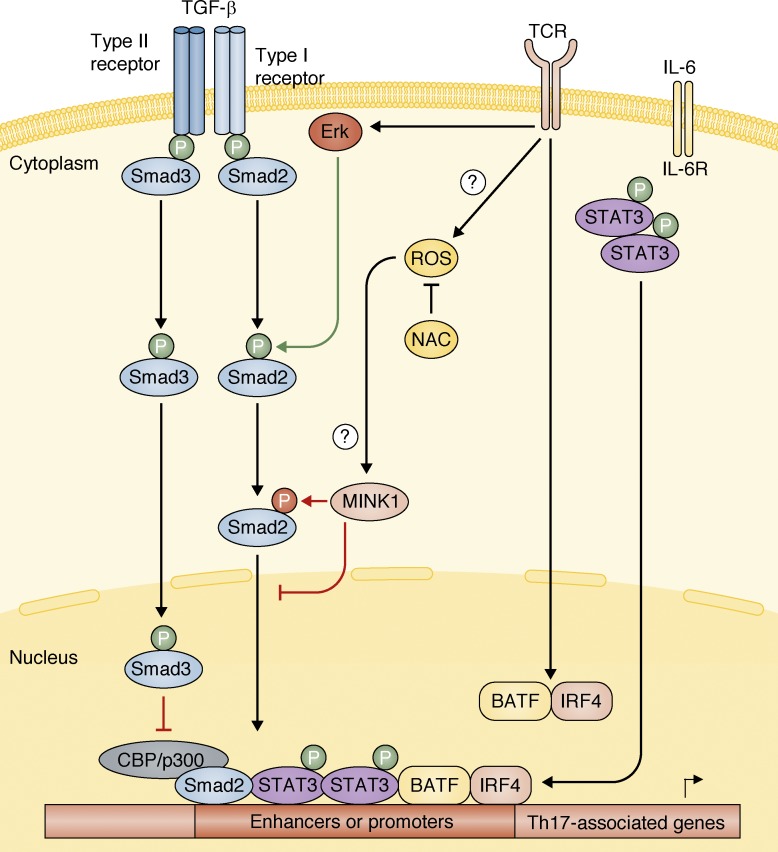
In this issue of JEM, Fu et al. identified the kinase Mink1 as a novel negative regulator of Th17 cell generation. Mink1, activated by reactive oxygen species (ROS), prevents TGF-β activation of Smad2, therefore limiting Th17 cell differentiation.
Abstract
In this issue of JEM, Fu et al. (https://doi.org/10.1084/jem.20161120) identified the kinase Mink1 as a novel negative regulator of Th17 cell generation. Mink1, activated by reactive oxygen species (ROS), prevents TGF-β activation of Smad2, therefore limiting Th17 cell differentiation.

Insight from Gustavo J. Martinez
Naive CD4+ T helper cells can differentiate into distinct lineages depending on the cytokine signals received during T cell activation. Upon TCR and costimulatory molecules signaling during Th17 cell differentiation, BATF and IRF4 transcription factors cooperate and function as pioneer transcription factors promoting chromatin remodeling and nucleosome repositioning, allowing the binding of lineage-specific transcription factors to DNA (Ciofani et al., 2012). In cooperation with STAT3, activated by IL-6 signaling, these transcription factors lead to induction of the lineage-specifying transcription factors RORα and RORγt (Ivanov et al., 2006; Yang et al., 2008; Ciofani et al., 2012; see figure). TGF-β signaling is also required for Th17 cell generation, through activation of Smad2 (Malhotra et al., 2010; Martinez et al., 2010), which then leads to up-regulation of Th17-associated genes (Yoon et al., 2015; see figure).
Mink1 inhibits Smad2-mediated Th17 cell induction. Upon TCR and co-stimulatory molecules signals, ROS are induced, which in turn can lead to the activation of the kinase Mink1. Also, the pioneering transcription factors BATF and IRF4 are activated upon TCR/co-stimulation signals, which induce chromatin remodeling at Th17-associated genes, allowing chromatin to be accessible to other transcription factors. TGF-β signaling induces activation of Smad2 and Smad3. Smad2 is required for proper Th17 cell generation (Malhotra et al., 2010; Martinez et al., 2010; Yoon et al., 2015), whereas Smad3 inhibits the generation of Th17 cells (Martinez et al., 2009). Previous studies have shown that Smad2, phosphorylated by Erk upon TCR and TGF-β stimulation, cooperates with Stat3 in the induction of Th17 cells (Yoon et al., 2015). In this issue of JEM, Fu et al. (2017) now demonstrate that the activation of Mink1 by ROS species, which can be blocked by NAC treatment, leads to an inhibitory phosphorylation of Smad2 in T324 residue, preventing its nuclear localization and therefore induction of Th17-associted genes.
In this issue of JEM, Fu et al. described a novel mechanism in which reactive oxygen species (ROS)–activated Mink1 directly binds to and phosphorylates Smad2 at the inhibitory residue T324, preventing its nuclear localization (see figure). Consistent with its role in inhibiting Smad2 activation, cells lacking Mink1 displayed an opposite phenotype to Smad2-deficient cells (Malhotra et al., 2010; Martinez et al., 2010; Yoon et al., 2015). Not only TGF-β1 but also TGF-β3 stimulation in the presence of IL-6 results in enhanced Th17 cell generation in Mink1-deficient cells. Thus, it is plausible that Smad1, activated by TGF-β3 (Lee et al., 2012), could be directly phosphorylated and inhibited by Mink1 because it also contains this same inhibitory residue. Moreover, although both Smad1 and Smad2 possess the inhibitory T324 residue, Smad3 does not and is not phosphorylated by Mink1 (see figure). It is therefore possible that the inhibitory role of Mink1 in Th17 cell differentiation is not only through reduced Smad2 nuclear translocation, which is required to induce expression of RORγt and IL-17 in a Stat3-dependent manner (Yoon et al., 2015), but also results in unaffected or increased Smad3 levels in the nucleus. Smad3 has been shown to inhibit the commitment toward Th17 cells; the mechanism behind this inhibitory role remains controversial because it has been shown to interact with and prevent RORγt-mediated transcription (Martinez et al., 2009) as well as directly prevent RORγt expression itself (Yoon et al., 2015). Further understanding on how Smads, in cooperation with distinct transcription factors, regulate Th17 cell generation is needed for the potential development of therapeutic strategies targeting this pathway.
Mink1 deficiency results not only in a T cell–intrinsic enhanced Th17 cell commitment, but also in an increase in activated/memory phenotype. By using mixed bone marrow chimeras in Rag1−/− mice, Fu et al. (2017) showed that compared with WT counterparts in the same recipient mice, Mink1-deficient cells displayed enhanced IL-17 production and expression of the activation marker CD44 and reduced expression of the lymph node homing receptor CD62L, which is highly expressed in naive cells. The exact mechanism leading to this increased activated status by T cells lacking Mink1 expression was not addressed by Fu et al. (2017). Further research to delineate how Mink1 prevents activation in steady-state conditions is required to fully comprehend its role in T cell biology. Given that Smad signaling is enhanced in Mink1-deficient cells, it seems unlikely that these cells are less susceptible to T reg cell–suppressive activity, although this remains to be determined.
The impact ROS have on Th17 cell differentiation has been controversial. Zhi et al. (2012) have suggested that deficiency in IEX-1 in T cells results in enhanced mitochondrial ROS, which in turn induces higher Th17 cell generation. On the contrary, Gerriets et al. (2015) have shown that inhibition of pyruvate dehydrogenase kinase with dichloroacetate (DCA) leads to increased ROS that negatively affect Th17 cell generation. Moreover, the DCA inhibitory effect on Th17 cell generation can be blocked by the ROS scavenger N-acetyl cysteine (NAC) treatment. Similarly, Fu et al. (2017) also demonstrate that inhibition of ROS by NAC treatment leads to a decrease in Mink1 activity, resulting in enhanced Smad2 activation, which in turn leads to enhanced Th17 cell generation in WT cells. This study demonstrated that Mink1 is the molecular mechanism that links ROS with reduced Th17 cell differentiation because NAC treatment has no effect in Mink1-deficient cells. Interestingly, the authors showed that although administration of the ROS scavenger NAC in drinking water did not influence the disease severity of EAE, probably because of the pleiotropic effect of ROS in the body, T cells isolated from mice treated with NAC during EAE induction and adoptively transfered into naive mice showed enhanced disease score with higher IL-17–producing cells infiltrating the CNS. These novel results highlight the importance and precautions that need to be taken when administering antioxidants in patients. Antioxidant treatments have been extremely popular in recent years, especially in inflammatory-mediated diseases such as cancer, aging, and metabolic disorders. The results presented in this issue of JEM by Fu et al. (2017), together with growing evidence on the role of ROS in Th17 cell activation, suggest that caution should be taken as inhibition of ROS could be a detrimental approach by further increasing Th17-mediated pathogenesis. The exact mechanism that leads to Mink1 activation by ROS remains to be elucidated, as well as the pathways that converge in ROS induction upon T cell activation. Research to fill these gaps in our current knowledge would be crucial to evaluate the therapeutic potential of ROS inhibitors in T cell–mediated diseases.
References
- Ciofani M., et al. Cell. 2012 doi: 10.1016/j.cell.2012.09.016. [DOI] [Google Scholar]
- Ivanov I.I., et al. Cell. 2006 doi: 10.1016/j.cell.2006.07.035. [DOI] [Google Scholar]
- Yang X.O., et al. Immunity. 2008 doi: 10.1016/j.immuni.2007.11.016. [DOI] [Google Scholar]
- Malhotra N., et al. J. Biol. Chem. 2010 doi: 10.1074/jbc.C110.156745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G.J., et al. J. Biol. Chem. 2010 doi: 10.1074/jbc.C110.155820. [DOI] [Google Scholar]
- Yoon J.H., et al. Nat. Commun. 2015 doi: 10.1038/ncomms8600. [DOI] [Google Scholar]
- Martinez G.J., et al. J. Biol. Chem. 2009 doi: 10.1074/jbc.C109.078238. [DOI] [Google Scholar]
- Fu G., et al. J. Exp. Med. 2017 doi: 10.1084/jem.20161120. [DOI] [Google Scholar]
- Lee Y., et al. Nat. Immunol. 2012 doi: 10.1038/ni.2416. [DOI] [Google Scholar]
- Zhi L., et al. J. Immunol. 2012 doi: 10.4049/jimmunol.1200528. [DOI] [Google Scholar]
- Gerriets, et al. J. Clin. Invest. 2015 doi: 10.1172/JCI76012. [DOI] [Google Scholar]