Abstract
Triple-negative breast cancers (TNBC) remain clinically challenging with a lack of options for targeted therapy. In this study, we report the development of a second-generation BET bromodomain (BRD) inhibitor, BETd-246, which exhibits superior selectivity, potency and antitumor activity. In human TNBC cells, BETd-246 induced degradation of BET transcription factors at low nanomolar concentrations within 1 hr of exposure, resulting in robust growth inhibition and apoptosis. BETd-246 was more potent and effective in TNBC cells than its parental BET inhibitor compound BETi-211. RNA-seq analysis revealed predominant downregulation of a large number of genes involved in proliferation and apoptosis in cells treated with BETd-246, as compared to BETi-211 treatment which upregulated and downregulated a similar number of genes. Functional investigations identified the MCL1 gene as a critical downstream effector of these BET degraders, which synergized with small molecule inhibitors of BCL-xL in triggering apoptosis. In multiple murine xenograft models of human breast cancer, BETd-246 and a further optimized analogue BETd-260 effectively depleted BET proteins in tumors and exhibited strong antitumor activities at well-tolerated dosing schedules. Overall, our findings show how specific targeting of BET proteins for degradation yields an effective therapeutic strategy for TNBC treatment.
Introduction
TNBC is characterized by the lack of expression of estrogen receptor (ER) α and progesterone receptor, and absence of human epidermal growth factor receptor 2 (HER2) overexpression. Although aggressive chemotherapy can achieve a high response rate in TNBC patients, the risk of recurrence is substantially higher than those with ER+ or HER2+ breast cancers. There is an urgent need to develop effective targeted therapies for TNBC.
Bromodomain and Extra Terminal (BET) proteins, including ubiquitously expressed BRD2, BRD3, BRD4 and testis-specific BRDT, are epigenetic “readers” and play a major role in epigenetic regulation of gene transcription. BET proteins have emerged as new therapeutic targets for human cancer and other diseases. Major breakthroughs in the discovery and development of potent and selective small-molecule BET inhibitors led to several such compounds now in clinical development. Early clinical trials have provided evidence that inhibition of BET proteins is effective against some human cancers, including NUT midline carcinoma, multiple myeloma and acute myeloid leukemia (1-7). Recent preclinical studies further suggested that BET proteins are exciting targets for breast cancer (8-11).
Theoretically, depletion of key oncogenic proteins in tumor cells could achieve much better clinical efficacy than inhibition of the same proteins. Fifteen years ago, Crews proposed the design of PROteolysis TArgeting Chimeric (PROTAC) molecules to recruit targeted proteins for degradation (12,13). This strategy has recently been used to design small-molecule degraders of BET proteins. dBET1 (14) and ARV-825 (15) were designed using JQ-1 for the BET inhibitor portion and thalidomide as the ligand for the Cullin-4A ligase complex. dBET1 efficiently induces degradation of BET proteins in leukemia cells and is more effective than JQ-1 in inhibiting tumor growth in a xenograft model of human acute leukemia cell line in mice (14). ARV-771 uses OTX-015 for the BET inhibitor portion and a ligand for the von Hippel–Landau (VHL) E3 ligase (16). ARV-771 was shown to be a highly effective BET degrader in castration-resistant prostate cancer (CRPC) models and induces partial tumor regression in a CRPC xenograft model (16). These studies suggest that small-molecule BET degraders may be much more effective for cancer treatment than BET inhibitors. To date, the therapeutic potential of BET degraders and their mechanism of action in TNBC have not been reported.
Based upon our optimized, potent small-molecule BET inhibitor BETi-211, we have developed BETd-246 as a highly potent BET degrader and investigated its therapeutic potential and mechanisms of action in TNBC in vitro and in vivo. Our study shows that BET inhibition and degradation are distinctly different in their elicited biological responses, and targeting BET degradation represents a promising therapeutic approach for TNBC.
Materials and Methods
Chemicals
Detailed procedures for the synthesis of BET1-211, BETd-246 and BETd-260 are in SI Scheme I-III. ABT-263 (navitoclax) and ABT-199 (venetoclax) were from Selleck Chemicals. A-1155463 was from ChemieTek.
Cell Lines
The SUM human breast cancer cell lines were developed by Dr. Steve Ethier at the University of Michigan Comprehensive Cancer Center (17) and authenticated at the University of Michigan Comprehensive Cancer Center. All the other breast cancer cell lines were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA) during the course of this study and used within 2 months after initiating from original stocks. The authentication of ATCC breast cancer cell lines was performed by the Promega-ATCC Cell Line Authentication Service by Short Tandem Repeat profiling. All cell lines were cultured as recommended.
Antibodies and Reagents
A detailed list of antibodies and reagents is in SI Materials and Methods
Cell Viability, Apoptosis, Cell Cycle and Immunoblot Analyses
Conventional cell viability, Annexin V-propidium iodide apoptosis, cell cycle and immunoblotting analyses were performed as described previously (18,19).
Lentiviral Constructs and siRNAs
Human MCL1 lentiviral construct was from Applied Biological Materials (Richmond, BC, Canada) and transduced according to the manufacturer's instructions. Lentiviral vectors for human MCL1 shRNA was described previously (19). ON-TARGETplus CRBN and siCONTROL siRNAs were from Dharmacon. Cells were transfected using Lipofectamine RNAiMAX (Thermo Fisher Scientific) following the manufacturer's instructions.
Transcriptomic Profiling
Total RNA was purified using RNeasy Mini Kit (Qiagen) following the manufacturer's instructions. The rRNA depleted total RNA was reversely transcribed into first strand cDNA using random primers. The cDNA libraries were clustered and sequenced with Illumina RNA sequencing flow. RNA-seq data are deposited at GEO as xx. Details of transcriptomic profiling are in SI Materials and Methods.
Quantitative Polymerase Chain Reaction (qPCR)
Real-time PCR was done using a QuantStudio 7 Flex Real-Time PCR System. The relative abundance of gene expression was calculated using comparative CT method which compares the Ct value of target gene to GAPDH (2ΔΔCT). Details of qRT-PCR analysis are in SI Materials and Methods.
Proteomic Profiling
Cell lysis was proteolyzed and labeled with TMT 10-plex (Thermo Fisher Scientific) following the manufacturer's protocol. Details of proteomic profiling are in SI Materials and Methods.
In vivo Pharmacodynamic and Efficacy Studies
WHIM24 PDX model, initially developed at the Washington University School of Medicine, St. Louis, and tumors were passaged in SCID mice. To develop MDA-MB-453 xenografts, five million cells were injected subcutaneously into SCID. For pharmacodynamics studies, when tumors reached 100-200 mm3, mice were treated with vehicle control or a single dose of the drug, sacrificed at the indicated time-point, and tumor tissue was harvested for analyses. For in vivo efficacy experiments, when tumors reached 80-200 mm3, mice were randomized into groups. BETi-211, BETd-246, BETd-260 or vehicle control (10% PEG400: 3% Cremophor: 87% PBS, or 2% TPGS:98% PEG200) was given at the dose and with the duration indicated. Tumor sizes and animal weights were measured 2-3 times per week. Tumor volume (mm3) = (length×width2)/2. Tumor growth inhibition was calculated as TGI (%) = (Vc-Vt)/(Vc-Vo)*100, where Vc, Vt are the median of control and treated groups at the end of the study and Vo at the start. All the in vivo studies were performed under an animal protocol (PRO00005315) approved by the University Committee on Use and Care of Animals of the University of Michigan, in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Statistical Analyses
For the cell viability and apoptosis analyses, data were presented as mean ± SEM. For dose-dependent cell viability assay, data were plotted as mean ± SD and sigmoid fitted (variable slope). Differences in mean values of cell growth inhibition among different groups were analyzed by two-way analysis of variance. For in vivo studies, the significance (P) was calculated by Student's t test. All statistical tests were two-sided. All statistical analyses were performed using GraphPad Prism 6. The P values less than 0.05 are considered statistically significant.
Results
Design of BETd-246 as a potent small-molecule degrader of BET proteins
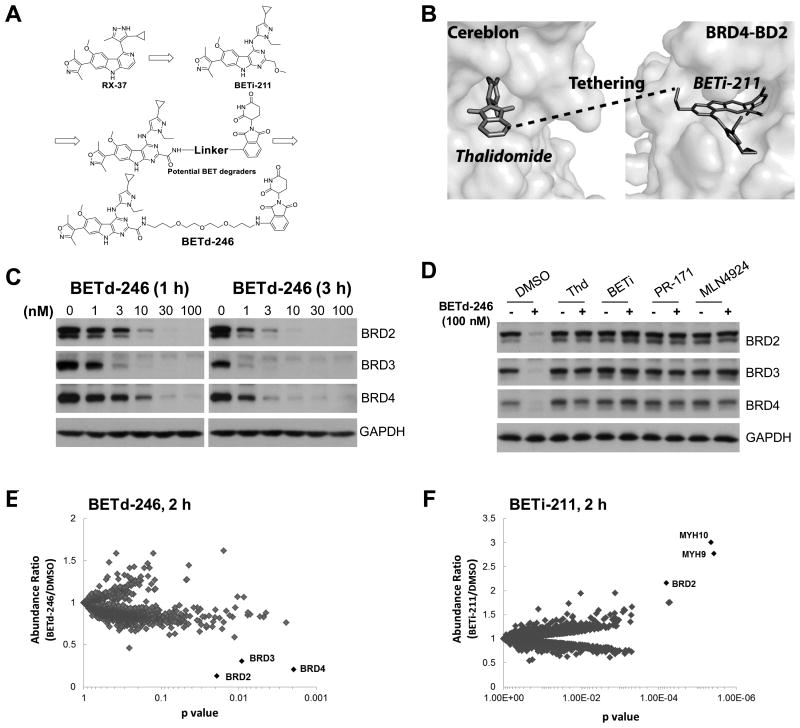
We have optimized our BET inhibitor RX-37 (20) and discovered UM-BETi-211 (BETi-211) (Fig. 1A, SI Scheme I). BETi-211 binds to BET proteins with Ki values of <1 nM and is >10-100 times more potent than RX-37, JQ-1, OTX-015 or IBET-762 (SI Table 1). Based upon the modeled structure of BRD4 in complex with BETi-211 (Fig. 1B), we identified an appropriate site for tethering BETi-211 to thalidomide for the design of BET degraders (Fig. 1B). We designed and synthesized a series of compounds containing linkers with different lengths and physiochemical properties and discovered UM-BETd-246 (BETd-246, Fig. 1A, SI Scheme II) as a potent BET protein degrader.
Fig 1. BETd-246 is a potent and highly selective degrader of BET proteins.
(A). Design and development of BETd-246 as a BET protein degrader based upon the potent and selective BET inhibitor BETi-211 and thalidomide. (B). Identification of an appropriate site in BETi-211 based upon its modeled structure with BRD4 and an appropriate site in thalidomide based upon its crystal structure with Cereblon. (C). MDA-MB-468 cells were treated with BETd-246 at the indicated doses for 1 and 3 h for immunoblotting. GAPDH was included as a loading control. (D) MDA-MB-468 cells were pretreated with thalidomide (2000 nM), BETi-211 (2000 nM), PR-171 (1000 nM) or MNL4924 (500 nM) for 1 h, followed by treatment with BETd-246 (100 nM) for 1 h. (E & F). MDA-MB-468 cells were treated with BETd-246 (E) (100 nM) and (F) BETi-211 (1000 nM) for 2 h. Whole cell lyses were labeled with the Tandem Mass Tag 10-plex reagent for LC-MS/MS analysis.
BETd-246 potently and selectively depletes BET proteins in TNBC cells
We first evaluated BETd-246 for its ability to degrade BET proteins in representative TNBC cell lines. BETd-246 treatment caused a dose-dependent depletion of BRD2, BRD3 and BRD4 in these cell lines (Fig 1C, SI Fig 1A-B). A near-complete depletion of BRD2-4 proteins was observed with 30-100 nM of BETd-246 for 1 h or with 10-30 nM of BETd-246 for 3 h. In contrast, the BET inhibitor BETi-211 did not degrade BET proteins in any cell line.
Hetero-bifunctional BETd-246 was designed to bind concurrently to BET proteins and Cereblon (CRBN) through its BET inhibitor and thalidomide moiety respectively, bringing the CUL4–RBX1–DDB1–CRBN E3 ubiquitin ligase (CRL4CRBN) and BET proteins into close proximity for subsequent ubiquitination and proteasomal degradation of BET proteins. We thus examined the mechanism of BET protein degradation by BETd-246. Thalidomide alone had no significant effect on the levels of BET proteins in all TNBC cell lines evaluated (Fig 1D, SI Fig 1C) and pre-treatment either with excess thalidomide or BETi-211 effectively blocked BETd-246-induced BET protein depletion (Fig 1D, SI Fig 1C). Proteasome inhibitor PR-171 (Carfilzomib) (21) blocked depletion of BET proteins by BETd-246 (Fig 1D, SI Fig 1C). NEDD8 activating E1 enzyme inhibitor MLN4924 (22) also effectively blunted BETd-246-induced depletion of BET proteins (Fig 1D, SI Fig 1C), consistent with the notion that activation of CRL4CRBN requires NEDD8 neddylation (23).
We next performed proteomic analysis to assess the global effect of BETd-246 and BETi-211 on cellular protein levels in MDA-MB-468 cells. Out of ∼5500 proteins quantified, BRD2, BRD3 and BRD4 were the only proteins whose levels were significantly decreased by ≥2-fold (p<0.05) with 100 nM of BETd-246 for 2 h. No protein was increased by ≥2-fold (Fig 1E, SI Table 2). In contrast, BETi-211 increased the BRD2 protein level by 2-fold (Fig 1F, SI Table 3).
Collectively, our data demonstrate that BETd-246 is a highly potent and selective BET protein degrader in TNBC cells.
BETd-246 displays strong growth inhibition and apoptosis induction activity in TNBC cell lines
We next evaluated BETi-211 and BETd-246 for their antiproliferative activity in a panel of 13 TNBC cell lines.
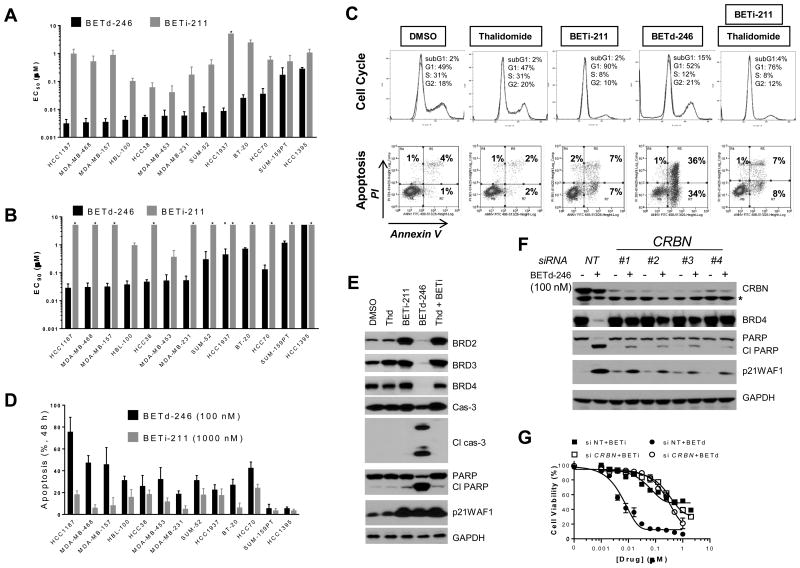
Consistent with a recent study using JQ1 (8), BETi-211 displayed potent growth-inhibitory activity in the TNBC cell lines (Fig 2A, SI Fig 2A), with IC50 < 1 μM in 9 of the 13 cell lines. However, BETi-211 at 2.5 μM only achieved ≥90% growth inhibition in 2 of the 13 cell lines (Fig 2B), suggesting the predominantly cytostatic effect of BETi-211. Flow cytometry analysis confirmed that while BETi-211 at 1000 nM induced a pronounced decrease in S-phase cells after 24 h treatment, it elicited only modest apoptosis induction (3-25%) after 48 h treatment (Fig 2C-D, SI Fig 3A).
Fig 2. BETd-246 displays potent antiproliferative and apoptosis induction activities in TNBC cell lines.
(A & B). Cells were treated with serially diluted BETi-211 or BETd-246 for 4 days for CellTiter Glo cell viability assay. Data are mean ± SEM (n=4). (C). MDA-MB-468 cells were treated with DMSO, thalidomide (1000 nM), BETi-211 (1000 nM), BETd-246 (100 nM) or combination of thalidomide and BETi-211 (1000 nM at 1:1 molar ratio) for 24 h (top panel) or 48 h (lower panel) for flow cytometry analyses. Data are representative of 3 independent experiments. PI: propidium iodide. (D). Cells were treated with BETd-246 (100 nM) or BETi-211 (1000 nM) for 48 h for Annexin V-PI apoptosis analysis. Data are mean ± SEM (n=3). (E). MDA-MB-468 cells were treated with DMSO, thalidomide (1000 nM), BETi-211 (1000 nM), BETd-246 (100 nM) or thalidomide plus BETi-211 (1000 nM at 1:1 molar ratio) for 8 h for immunoblotting. (F & G). MDA-MB-468 cells were transfected with non-targeting or CRBN siRNAs for 40 h, then treated with BETd-246 (100 nM) for 8 h for immunoblotting (F) or serially diluted BETi-211 or BETd-246 for 3 days (G) for cell viability assay. Data are mean ± SD.
BETd-246 on the other hand exhibited strong growth-inhibitory activity with IC50 <10 nM in 9 of the 13 cell lines (Fig 2A). Based upon the IC50 values, BETd-246 is >50-times more potent than BETi-211 in a majority of these cell lines. Significantly, BETd-246 achieved IC90<100 nM in 7 of the 13 cell lines (Fig 2B), suggesting strong cell killing effects in TNBC cells. Flow cytometry and immunoblotting confirmed robust apoptosis induction by BETd-246 in the majority of these cell lines (Fig 2C-E, SI Fig 3B). Treatment with BETd-246 at as low as 10 nM led to profound cleavage of caspase-3 and poly(ADP-ribose) polymerase (PARP) (Fig 2E, SI Fig 3B), and caspase-3/7 activation (SI Fig 4). In comparison, BETi-211 was much less potent in activating caspase-3/7 in these cell lines (SI Fig 4). Profiling of initiator caspases showed that BETd-246 treatment led to the activation of caspase-2, -8 and -9 in MDA-MB-468 cells (SI Fig 5A), suggesting the activation of multiple apoptotic pathways by BETd-246. Immunoblotting confirmed the cleavage of caspase-8, -9 and -3 within 5 h of BETd-246 treatment in MDA-MB-468 cells (SI Fig 5B). Thalidomide alone had no significant effect on apoptosis induction and cell proliferation (Fig 2C, 2E, SI Fig 2A, 3B). BETi-211 in combination with thalidomide had an effect similar to that of BETi-211 alone (Fig 2C, 2E, SI Fig 2A, 3B). These data demonstrate that BET protein degradation by BETd-246, as opposed to BET BRD inhibition by BETi-211, leads to strong apoptosis induction in the majority of the TNBC cell lines.
BET degradation by BETd-246 requires its binding to Cereblon. Consistently, silencing CRBN (encoding Cereblon) by siRNAs blocked BETd-246-induced BET degradation and PARP cleavage in MDA-MB-231 and MDA-MB-468 cells (Fig 2F, SI Fig 6), and reduced the growth-inhibitory activity of BETd-246 to a level comparable to that of BETi-211 (Fig 2G, SI Fig 6). Silencing CRBN had no significant effect on the growth-inhibitory activity of BETi-211 (Fig 2G, SI Fig 6).
BETd-246 and BETi-211 elicit distinct transcriptional responses in TNBC cells
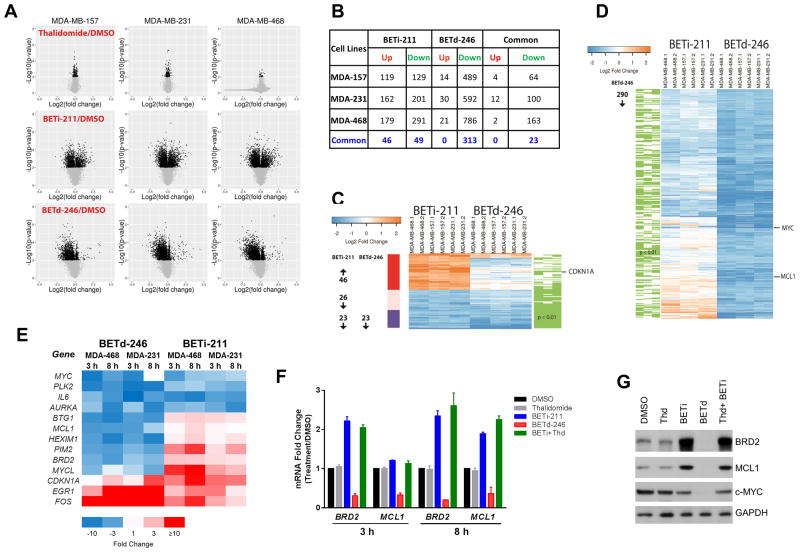
Although genetically and epigenetically heterogeneous, the majority of the 13 TNBC cell lines are highly sensitive to BETd-246, suggesting the presence of certain commonalities in their transcriptomic responses to BET degradation. The greater potency and effectiveness in cell growth inhibition and apoptosis induction by BETd-246 than by BETi-211 also indicate critical differences between these two different BET targeting approaches in their regulation of gene transcription. To understand the common and distinct actions of BETi-211 and BETd-246, we performed RNA-seq analysis on MDA-MB-157, MDA-MB-231 and MDA-MB-468. These cell lines were treated for 3 h to capture the early transcriptional responses to BETi-211 or BETd-246.
Not surprising, thalidomide had a minimal effect on global transcriptome in all three cell lines (Fig 3A). An approximately equal number of genes were up- and down-regulated by BETi-211 in each cell line (Fig 3A. In contrast, BETd-246 caused predominant downregulation of gene expression (Fig 3A).
Fig 3. BET inhibitor and degrader elicit extensive but different transcriptome changes in TNBC cells.
(A). MDA-MB-157, MDA-MB-231 and MDA-MB-468 cells were treated with DMSO, thalidomide (1000 nM), BETi-211 (1000 nM) and BETd-246 (100 nM) for 3 h for RNA-seq analysis. (B). Identification of genes that are up- or down-regulated by BETi-211 or BETd-246 by ≥2-fold (p <0.01) over the DMSO control treatment in each cell line and numbers of overlapped genes that are commonly up or down-regulated by BETi-211 or BETd-246 in all three cell lines. (C). Heatmap of up- or down-regulated genes by BETi-211 (≥2-fold, p <0.01) shared by all three TNBC cell lines by RNA-seq and their alterations by BETd-246 (green lines indicate p < 0.01) in each cell line. (D). Heatmap of up- or down-regulated genes by BETd-246 (≥2-fold, p <0.01) shared by all three TNBC cell lines by RNA-seq and their alterations by BETi-211 (green lines indicate p < 0.01) in each cell line. (E). Heatmap of relative mRNA levels for selected genes in MDA-MB-231 and MDA-MB-468 cells after 3 and 8 h treatment with DMSO, BETi-211 (1000 nM) and BETd-246 (100 nM) by qRT-PCR. Data are average of biological triplicates from one representative experiment. (F). qRT–PCR of relative mRNA levels in MDA-MB-468 cells after 3 and 8 h treatment with DMSO, thalidomide (1000 nM), BETi-211 (1000 nM) and BETd-246 (100 nM), or thalidomide plus BETi-211 (1000 nM at 1:1 molar ratio). Data are mean ± SD. (G) MDA-MB-468 cells were treated with DMSO, thalidomide (1000 nM), BETi-211 (1000 nM), BETd-246 (100 nM), or thalidomide plus BETi-211 (1000 nM at 1:1 molar ratio) for 8h for immunobloting.
Given the pervasive transcriptomic responses to BET protein perturbation by BETi-211 and BETd-246, we focused on the genes that were significantly altered (≥2-fold, p<0.01) by BETi-211 or BETd-246 (Fig 3B-D). This analysis revealed that the expressions of 248-470 genes were significantly affected by BETi-211 with a similar number of genes being up- or down-regulated in each cell line (Fig 3B). In contrast, BETd-246 caused overwhelmingly down-regulation of gene expression in these cell lines (Fig 3B). Despite the distinct transcriptional responses caused by BETi-211 and BETd-246, a set of genes was commonly downregulated by both BETi-211 and BETd-246 in each cell line (Fig 3B). Of note, 23 of the 46 genes which were commonly down-regulated by BETi-211 in all three TNBC cell lines were also down-regulated by BETd-246 (Fig 3B-D).
We then selected 45 representative genes related to gene transcription, cell cycle transition, proliferation and survival for quantitative (q) RT-PCR validation (Fig 3E, SI Fig 7A). High Pearson correlations between the RNA-seq and qRT-PCR data were found in both MDA-MB-231 and MDA-MB-468 (SI Fig 7B-E). The differential regulation of these 45 genes was confirmed by qRT-PCR at 3 and 8 h post treatment in MDA-MB-468 and MDA-MB-231 (Fig 3E, SI Fig 7A).
The antitumor activities of BET inhibitors such as JQ1 in a variety of preclinical tumor models have often been associated with the downregulation of c-MYC and upregulation of cyclin kinase inhibitors such as p21WAF1 (24-28). MYC and CDKN1A (encoding p21WAF1) were indeed down- and up-regulated, respectively, by both BETi-211 and BETd-246, albeit at different levels and with different kinetics. (Fig 3E, SI Fig 7A, 9).
Our transcriptomic analyses also revealed that a set of proliferation and survival-related genes, including BRD2 and MCL1, were oppositely regulated by BETi-211 and BETd-246 (Fig 3E, SI Fig 7A). BRD2 is a direct target for both BETi-211 and BETd-246 but its mRNA level was increased by BETi-211 at 3 h and 8 h after BETi-211 treatment in multiple TNBC cell lines (Fig 3F, SI Fig 8A-C). BRD2 protein increase by BETi-211 was confirmed by immunoblotting (Fig 3G, SI Fig 9), consistent with our proteomic analysis (Fig 1E). The mRNA level of MCL1, an anti-apoptotic BCL-2 family member, was markedly down-regulated by BETd-246 in TNBC cell lines (Fig 3E-F, SI Fig 8A-C) but was significantly up-regulated by BETi-211 at 8 h in MDA-MB-468 (Fig 3F). Opposite regulation of MCL1 protein expression by BETd-246 and BETi-211 was confirmed by immunoblotting in MDA-MB-468 cells (Fig 3G). Downregulation of the mRNA levels of BRD2 and MCL1 by BETd-246 was reversed by excess thalidomide and BETi-211 (SI Fig 8D), confirming its dependence on BET protein degradation.
Hence, BET BRD inhibition by BETi-211 and BET protein degradation by BETd-246 result in distinct transcriptional responses in TNBC cells. Several proliferation and survival-related genes, such as BRD2 and MCL1, are strongly down-regulated by BETd-246, but up-regulated by BETi-211 in TNBC cells.
MCL1 is a key target of BET degrader-induced apoptosis in TNBC
In the majority of the TNBC cell lines, BETd-246 induces much stronger apoptosis than BETi-211 (Fig 2D). Because MCL1 expression is effectively down-regulated by BETd-246 but not BETi-211 and MCL1 is a key apoptosis regulator, we investigated its role in apoptosis induction by BETd-246 and BETi-211 in TNBC cells.
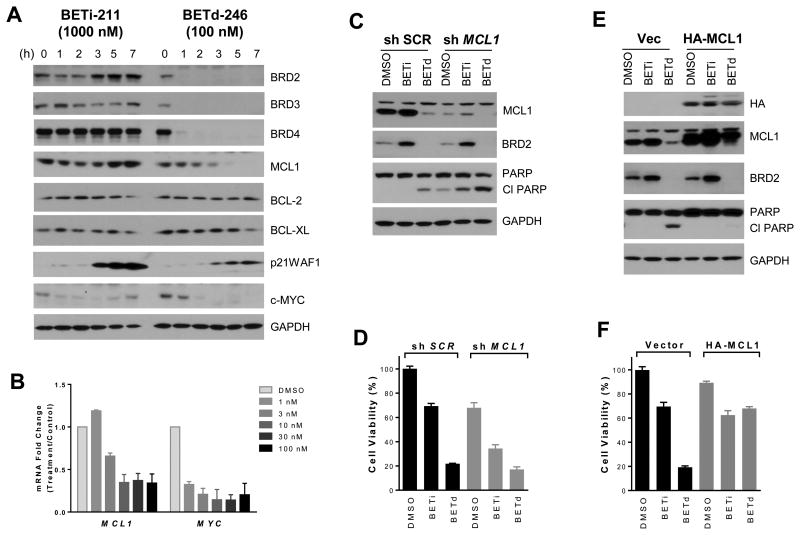
BETd-246 induced a rapid and time-dependent downregulation of MCL1 protein in all the TNBC cell lines evaluated (Fig 4A, SI Fig 9). Significant down-regulation of MCL1 mRNA by BETd-246 was observed at as low as 10 nM, similar to that observed for MYC (Fig 4B). In contrast, MCL1 protein was not down-regulated by BETi-211 in any of the cell lines but instead was up-regulated in MDA-MB-157, MDA-MB-231 and MDA-MB-468 (Fig 4A, SI Fig 9). The expression of anti-apoptotic BCL-2 and BCL-XL in these cell lines was not significantly altered by either BETi-211 or BETd-246 (Fig 4A, SI Fig 9).
Fig 4. Down-regulation of MCL1 by BETd-246 plays a major role in its robust apoptosis induction in TNBC cells.
(A) MDA-MB-468 cells were treated with BETi-211 (1000 nM) and BETd-246 (100 nM) at indicated time points for immunoblotting. (B) qRT–PCR of relative mRNA levels in MDA-MB-468 cells after a 8 h treatment with indicated concentrations of BETd-246 or DMSO. Data are mean ± SD. (C & D) MDA-MB-468 cells were transduced with scrambled (SCR) or MCL1 shRNA lentivirus for 48h followed by 8 h treatment with DMSO, BETi-211 (1000 nM) or BETd-246 (100 nM) for immunoblotting (C) or 2 days for cell viability assay (D). Data are mean ± SD. (E & F). MDA-MB-468 cells were transduced with lentiviral vector or HA-MCL1-expressing lentivirus for 48 h followed by a 8 h treatment with DMSO, BETi-211 (1000 nM) and BETd-246 (100 nM) for immunoblotting (E) or 2 days for cell viability assay (F). Data are mean ± SD.
We next examined the functionality of MCL1 in cell growth inhibition and apoptosis induction by BETd-246 in TNBC cells. Silencing MCL1 promoted apoptosis and inhibited cell proliferation in MDA-MB-468 and MDA-MB-157 (Fig 4C-D, SI Fig 10). Knockdown of MCL1 significantly enhanced the growth-inhibitory activity of BETi-211 but not BETd-246 in these cell lines (Fig. 4D, SI Fig 10). Conversely, ectopic expression of MCL1 attenuated BETd-246-elicited apoptosis induction and cell growth inhibition (Fig 4E-F, SI Fig 10).
Of note, the opposite regulation of MCL1 by BETi-211 and BETd-246 was also observed in non-TNBC cell lines, such as HER2-amplified HCC1954 (SI Fig 11).
Taken together, these data suggest that downregulation of MCL1 by BETd-246 plays a key role in its robust apoptosis induction in TNBC cells.
Small-molecule BCL-XL inhibitors potentiate BET degrader-induced apoptosis in TNBC cells
Though a much more potent and effective apoptosis inducer than BETi-211, BETd-246 was not uniformly effective in apoptosis induction across the cell lines (Fig 2D). Recent studies have demonstrated that MCL1 and BCL-XL are key but independent determinants of cell survival in TNBC (29-31). Therefore, we investigated whether small-molecule BCL-XL/BCL-2 inhibitors could enhance apoptosis induction by BETd-246 in TNBC cells. ABT-263 (32) and BM-1197 (18) were selected as dual BCL-2/BCL-XL inhibitors, A-1153463 as a selective BCL-XL inhibitor (33) and ABT-199 as a selective BCL-2 inhibitor (34).
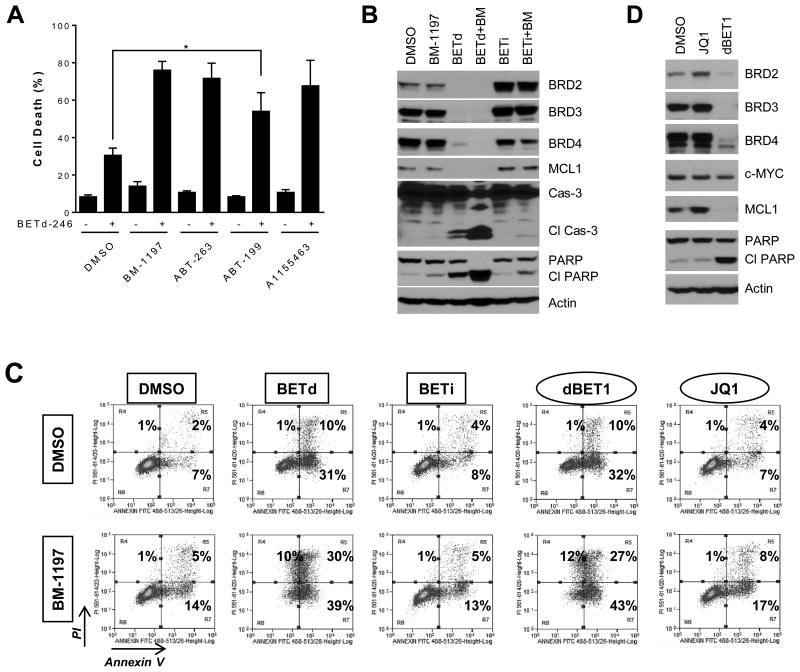
We treated a panel of 7 TNBC cell lines with dose-response matrix and examined the effects of BETd-246 in combination with BCL-2 and/or BCL-XL inhibitors by computing the excess growth inhibition over the Bliss independence model for each combination pairs (35). Modest to strong synergy between BETd-246 and BCL-XL-targeting BM-1197, ABT-263 and A-1155463 was observed in 6 of the 7 cell lines (SI Fig 12). Immunoblotting and/or Annexin-PI staining confirmed that BM-1197, ABT-263 or A-1155463 potentiated BETd-246-induced apoptosis in these cell lines (Fig 5A-B, SI Fig 13-14). In contrast, strong synergistic induction of apoptosis by BETd-246 in combination with ABT-199 was only observed in MDA-MB-468 (Fig 5A, SI Fig 13-14), indicating that BCL-XL, but not BCL2, is a prevalent resistance factor for BETd-246-induced apoptosis in these cell lines. No clear synergism was observed for BETi-211 with either BM-1197 or ABT-263 (Fig 5B-C, SI Fig 14). The synergistic apoptosis induction by the combination of BETd-246 and BM-1197 was also readily detected in additional TNBC cell lines responsive to BETd-246-induced apoptosis (BT-20 and MDA-MB-157) (SI Fig 15), further supporting that BCL-XL inhibitors potentiate BETd-246-induced apoptosis in TNBC.
Fig 5. Small-molecule BCL-XL inhibitors significantly enhance the apoptosis induction by BETd-246.
(A). MDA-MB-468 cells were treated with BETd-246 (50 nM), BM-1197 (250 nM), ABT-263 (250 nM), ABT-199 (250 nM) or A-1155463 (250 nM) as indicated for 24 h for Annexin V-PI apoptosis analysis. Data are the mean ± SEM (n=3). (B). MDA-MB-468 cells were treated with BM-1197 (250 nM), BETd-246 (50 nM) or BETi-211 (1000 nM) as indicated for 6 h for immunoblotting. (C). MDA-MB-468 cells were treated with BM-1197 (250 nM), BETd-246 (50 nM), BETi-211 (1000 nM), JQ1 (2000 nM) or dBET1 (500 nM) as indicated for 24 h for apoptosis analysis. (D). MDA-MB-468 cells were treated with JQ1 (2000 nM) or dBET1 (500 nM) for 12 h for immunoblotting.
We also extended our investigations to dBET1, the first-reported BET degrader designed upon JQ1 (14). dBET at 500 nM effectively depleted BET proteins and downregulated MCL1 expression in MDA-MB-468 cells, concomitant with apoptosis induction (Fig 5D). Apoptosis induction by dBET1 was also markedly enhanced by BM-1197 (Fig 5C). Similar to BETi-211, JQ1 treatment led to increased expression of BRD2 and MCL1 and no obvious synergism between BM-1197 and JQ1 was observed (Fig 5C-D).
Taken together, these data indicate that the mechanisms of action observed for BETd-246 and BETi-211 in TNBC cells are independent of their chemical classes and apply to other classes of BET inhibitors and degraders.
BET degraders decrease BET proteins in xenograft breast tumors and suppress tumor growth
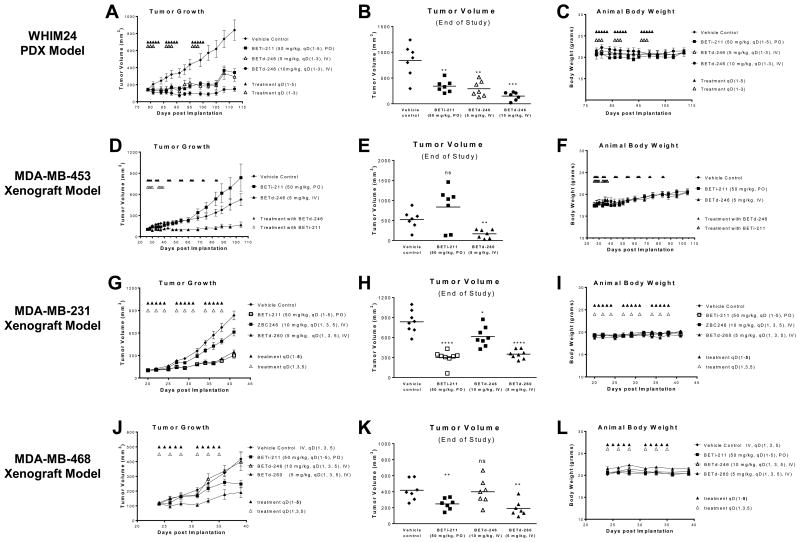
To evaluate the in vivo antitumor activity of BETd-246, we first employed the “Washington Human in Mouse (WHIM)” 24 (WHIM24), a patient derived xenograft (PDX) model developed from a patient with treatment-resistant breast cancer (ESRE380Q, PR- and HER2-) (36). BETd-246 at 5 mg/kg, IV, 3 times per week for 3 weeks effectively inhibited WHIM24 tumor growth, similar to the antitumor activity of BETi-211 at 50 mg/kg, daily, oral dosing, 5 days a week for 3 weeks. BETd-246 at 10 mg/kg, 3 times per week for 3 weeks, induced partial tumor regression during treatment (Fig 6A-B). Neither BETi-211 nor BETd-246 caused significant weight loss (Fig 6C) or apparent toxicity in this model. PD analysis showed that a single IV dose of BETd-246 (10 mg/kg) reduced the levels of BET proteins by >80% at as early as 1 h and this effect lasted for at least 9 h in WHIM24 tumors (SI Fig. 16). Notably, MCL1 protein levels in tumors were markedly reduced at as early as 3 h after BETd-246 treatment (SI Fig. 16). However, the protein levels of BRD2, BRD4 and MCL1 started to rebound at 12 h, indicating that BET degradation by BETd-246 is reversible once the drug is cleared from the tumor tissue. Pharmacokinetics (PK) analysis revealed that, while a single dose of BETd-246 (10 mg/kg) in tumor bearing mice achieved reasonable drug exposure in plasma and WHIM24 tumors at 1 and 3 h, the drug concentrations diminished rapidly in both plasma and tumors (SI Fig 17).
Fig 6. Small-molecule BET degraders are efficacious in multiple xenograft models of breast cancer.
SCID mice bearing xenograft tumors were treated as indicated. BETi-211 was given orally in 2% TPGS: 98% PEG200. BETd-246, BETd-260 and vehicle control was given intravenously in 10% PEG400: 3% Cremophor: 87% PBS. Tumor sizes and body weights were measured every 2-3 days. Data are mean ± SEM (n=7-8). The study in MDA-MB-468 model was terminated early due to profound necrosis in the control tumors. Scatter plots represent the tumor volumes at the end of each study. P values between each treated and the vehicle control group were determined using two-tailed t test. * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001).
We next tested the antitumor activity of BETd-246 in xenograft tumor models of TNBC cell lines. In MDA-MB-453 xenograft model, BETd-246 at 5 mg/kg significantly inhibited tumor growth with TGI (%) of 85% at the end of study (Fig 6D-E). In sharp contrast, BETi-211 at 50 mg/kg, daily, 5 days a week for 2 weeks, failed to achieve tumor growth inhibition. No significant weight loss (Fig 6F) or apparent toxicity was observed with BETi-211 or BETd-246. However, BETd-246 at 10 mg/kg, IV, 3 times per week for 2-3 weeks had very limited or no antitumor activity in MDA-M-231and MDA-MB-468 models, respectively (Fig 6G, J). Our PK analysis revealed that BETd-246 had very limited drug exposure in the xenograft tumor tissue in these two models (SI Fig 18-19), in contrast to the good drug exposure in the WHIM24 xenograft tumor tissue.
To improve its PK, we performed extensive optimization of the linker and the Cereblon-binding moiety of BETd-246 and identified UM-BETd-260 (BETd-260, SI Fig 20A, SI Scheme III). Similar to BETd-246, BETd-260 potently depleted BET proteins at low nanomolar concentrations in TNBC cells. In fact, BETd-260 was 2-7 times more potent than BETd-246 in growth inhibition of TNBC cells (SI Fig 20B-C, 21). Importantly, PK analysis showed that the exposure of BETd-260 in plasma and xenograft tumors was much higher than that of BETd-246 in xenograft models of MDA-MB-231 and MDA-MB-468 (SI Fig 18-19). Accordingly, BETd-260 at 5 mg/kg, IV, 3 times per week for 3 weeks exerted much stronger antitumor activity than BETd-246 at 10 mg/kg with the same dosing-schedule in both MDA-MB-231 and MDA-MB-468 xenograft models (Fig 6G-H, J-K). The antitumor activity of BETd-260 was comparable to or stronger than that of BETi-211 at 50 mg/kg, daily oral dosing, 5 days a week for 3 weeks in these models (Fig 6G-H, J-K). Studies in MDA-MB-468 model were terminated early due to profound necrosis in the control tumors. Neither BETd-246 nor BETd-260 caused significant weight loss (Fig 6I, L) or overt toxicity in these tumor models. PD analysis confirmed that a single dose of BETd-260 at 5 mg/kg effectively reduced BET protein levels in the xenograft tumors of MDA-MB-231 and MDA-MB-468 at 1-3 h post drug exposure (SI Fig 22), as well as upregulation of p21WAF1 and downregulation of MCL1 (SI Fig 22).
We also assessed BETd-246 for its potential toxicity in immune-competent Balb/c mice. Mice were treated with BETd-246 intravenously, daily, 5 days a week for 2 weeks, and then sacrificed at the end of the treatment to examine potential tissue damage. Although a single dose of BETd-246 (5 mg/kg) was able to effectively reduce BRD4 protein in mouse liver tissue, which has a high level of BRD4 protein expression but undetectable levels of BRD2 and BRD3 proteins (SI Fig 23), BETd-246 at 5 mg/kg exhibited no significant toxicity in normal mouse tissues (SI Table 4-6).
Collectively, our data demonstrated that small-molecule BET degraders effectively suppress tumor growth at well-tolerated dosing-schedules in xenograft breast tumor models.
Discussion
In this study, we investigated the therapeutic potential and mechanism of action of small-molecule BET degraders in TNBC. BET degrader BETd-246 efficiently and selectively degrades BET proteins in TNBC cells at low nanomolar concentrations and exhibits exquisite growth-inhibitory activity in the majority of TNBC cell lines evaluated. In comparison, BETd-246 is much more potent than the corresponding BET inhibitor BETi-211 in growth inhibition and apoptosis induction in vitro. In xenograft mouse tumor models, BET degraders effectively suppress breast tumor growth at well-tolerated dosing-schedules.
BET proteins participate in multiprotein complexes comprising of chromatin and transcription regulators to modulate transcription (37-41). In addition to acting in a BRD-dependent manner, BRD4 has BRD-independent functions in breast cancer cells (8,11). Thus, the transcriptional responses to the BET BRD inhibition and BET protein degradation are distinctly different as evidenced by our transcriptome profiling using BETi-211 and BETd-246. Our data further reveal that BET BRD inhibitor and BET degrader disproportionately regulate the expression of a large set of genes, reinforcing a model in which BET proteins act collectively with chromatin and transcription regulators to modulate gene expression output. The distinct transcriptional responses to BET BRD inhibitor and BET protein degrader strongly support the notion that BET proteins modulate gene expression through both BRD-dependent and BRD–independent mechanisms. The detailed mechanisms of their differential effects of BET BRD inhibitor and BET protein degrader on gene expression warrant further investigation.
Previous studies have shown that the effects of BET BRD inhibitors such as JQ-1 are largely cytostatic, especially in solid tumor models, with apoptosis induction limited to a few blood cancer models (25,42). Our data show that a key difference in the actions between BET BRD inhibitor and degrader in TNBC cells is the robust apoptosis induction by BET degrader and minimal to moderate apoptosis induction by BET inhibitor. Conery et al. (43) recently reported that the preclinical antitumor efficacy of BET inhibitors is determined by the magnitude of apoptotic, but not the cytostatic response in melanoma and leukemia models. Earlier studies have shown that BET BRD inhibitors suppress the expression of BCL-2 and/or BCL-XL while sparing MCL1 in multiple cell line models (1,25). We found that MCL1, a key anti-apoptotic BCL-2 member, is significantly down-regulated by BET degraders in TNBC cells in vitro and in vivo. MCL1 silencing by siRNA greatly enhances apoptosis induction by BETi-211 but not by BETd-246, and overexpression of MCL1 significantly reduces apoptosis induction by BETd-246 in TNBC cells. These data suggest that MCL1 is a key target of BET degrader-induced apoptosis in TNBC.
MCL1 expression in breast tumors correlates with high tumor grade and a much lower survival rate in patients (44). Amplification of MCL1 gene loci is prevalent in TNBC (∼20%) (45,46). While ∼30% of TNBC patients receiving neoadjuvant chemotherapy achieve a pathological complete response, the remaining patients with residual tumors exhibit high rates of metastatic disease and one of the most common genetic changes in the chemo-refractory tumors is the amplification of MCL1 loci (54%) (47-49). MCL1 has been identified as both an intrinsic and acquired resistance factor that limits the efficacy of a variety of anticancer agents (45,50) and thus has been intensely pursued as a therapeutic target. Our finding here provides an exciting new strategy to target MCL1 for breast cancer and potentially other types of cancer. Our data also suggest that, with optimal PK, BET degraders are more likely to achieve more favorable clinical responses than BET inhibitors in breast cancer patients.
Although BETd-246 is very effective in suppressing MCL1 expression in TNBC cell lines, it is not uniformly effective in apoptosis induction in these cell lines, suggesting the existence of other apoptotic resistance factors. Using a set of selective BCL-2 and/or BCL-XL inhibitors, we have shown that BCL-XL is a key resistance factor for apoptosis induction by BETd-246 in TNBC. Thus combination of a BCL-XL inhibitor and a BET degrader may provide an effective therapeutic strategy for breast cancer.
In summary, our study provides compelling preclinical data that targeting BET proteins for degradation is a promising therapeutic strategy for TNBC.
Supplementary Material
Acknowledgments
The authors thank the University of Michigan Biomedical Research Core Facilities (DNA Sequencing, Flow Cytometry and Vector Cores), Unit for Laboratory Animal Medicine and the Proteomics Resource Facility of the Department of Pathology for their excellent support.
Grant Support: The Breast Cancer Research Foundation (to S. Wang), the University of Michigan Comprehensive Cancer Center Strategic Fund for Breast Cancer (to S. Wang), and the University of Michigan Comprehensive Cancer Center Core grant (to S. Wang as a program leader) from the National Cancer Institute, NIH (P30CA046592).
Footnotes
Disclosure of Potential Conflicts of Interest: The University of Michigan has filed patent applications on BETd-246 and its analogues, for which S. Wang, B. Zhou, L. Bai, C. Yang, J. Hu, and F. Xu are co-inventors. These patents have been licensed by Medsyn Biopharma, in which Dr. Wang is a co-founder and owns stock.
References
- 1.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odore E, Lokiec F, Cvitkovic E, Bekradda M, Herait P, Bourdel F, et al. Phase I Population Pharmacokinetic Assessment of the Oral Bromodomain Inhibitor OTX015 in Patients with Haematologic Malignancies. Clinical Pharmacokinetics. 2016;55(3):397–405. doi: 10.1007/s40262-015-0327-6. [DOI] [PubMed] [Google Scholar]
- 3.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. The Lancet Haematology. 2016;3(4):e186–e95. doi: 10.1016/S2352-3026(15)00247-1. doi:http://dx.doi.org/10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 4.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord J-P, de La Motte Rouge T, et al. Clinical Response of Carcinomas Harboring the BRD4–NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discovery. 2016;6(5):492–500. doi: 10.1158/2159-8290.cd-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. The Lancet Haematology. 2016;3(4):e196–e204. doi: 10.1016/S2352-3026(16)00021-1. doi:http://dx.doi.org/10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 7.Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, Chaudhry MS, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123(5):697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 8.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529(7586):413–7. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sengupta S, Biarnes MC, Clarke R, Jordan VC. Inhibition of BET proteins impairs estrogen-mediated growth and transcription in breast cancers by pausing RNA polymerase advancement. Breast Cancer Research and Treatment. 2015;150(2):265–78. doi: 10.1007/s10549-015-3319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, et al. Bromodomain Protein BRD4 Is Required for Estrogen Receptor-Dependent Enhancer Activation and Gene Transcription. Cell Reports. 2014;8(2):460–9. doi: 10.1016/j.celrep.2014.06.016. doi:http://dx.doi.org/10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell. 2016;164(1-2):293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences. 2001;98(15):8554–9. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toure M, Crews CM. Small-Molecule PROTACS: New Approaches to Protein Degradation. Angew Chem Int Ed Engl. 2016;55(6):1966–73. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]
- 14.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348(6241):1376–81. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol. 2015;22(6):755–63. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2016;113(26):7124–9. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993;53(3):627–35. [PubMed] [Google Scholar]
- 18.Bai L, Chen J, McEachern D, Liu L, Zhou H, Aguilar A, et al. BM-1197: a novel and specific Bcl-2/Bcl-xL inhibitor inducing complete and long-lasting tumor regression in vivo. PLoS One. 2014;9(6):e99404. doi: 10.1371/journal.pone.0099404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman-Luca CG, Ziazadeh D, McEachern D, Zhao Y, Sun W, Debussche L, et al. Elucidation of Acquired Resistance to Bcl-2 and MDM2 Inhibitors in Acute Leukemia In Vitro and In Vivo. Clin Cancer Res. 2015;21(11):2558–68. doi: 10.1158/1078-0432.CCR-14-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ran X, Zhao Y, Liu L, Bai L, Yang CY, Zhou B, et al. Structure-Based Design of gamma-Carboline Analogues as Potent and Specific BET Bromodomain Inhibitors. J Med Chem. 2015;58(12):4927–39. doi: 10.1021/acs.jmedchem.5b00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, et al. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67(13):6383–91. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- 22.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 23.Chiba T, Tanaka K. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr Protein Pept Sci. 2004;5(3):177–84. doi: 10.2174/1389203043379783. [DOI] [PubMed] [Google Scholar]
- 24.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19(7):1748–59. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacques C, Lamoureux F, Baud'huin M, Calleja LR, Quillard T, Amiaud J, et al. Targeting the epigenetic readers in Ewing Sarcoma inhibits the oncogenic transcription factor EWS/Fli1. Oncotarget. 2016 doi: 10.18632/oncotarget.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodwin CM, Rossanese OW, Olejniczak ET, Fesik SW. Myeloid cell leukemia-1 is an important apoptotic survival factor in triple-negative breast cancer. Cell Death Differ. 2015;22(12):2098–106. doi: 10.1038/cdd.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Y, Nimmer P, Sheppard GS, Bruncko M, Hessler P, Lu X, et al. MCL-1 Is a Key Determinant of Breast Cancer Cell Survival: Validation of MCL-1 Dependency Utilizing a Highly Selective Small Molecule Inhibitor. Mol Cancer Ther. 2015;14(8):1837–47. doi: 10.1158/1535-7163.MCT-14-0928. [DOI] [PubMed] [Google Scholar]
- 31.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24(2):182–96. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68(9):3421–8. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 33.Tao ZF, Hasvold L, Wang L, Wang X, Petros AM, Park CH, et al. Discovery of a Potent and Selective BCL-XL Inhibitor with in Vivo Activity. ACS Med Chem Lett. 2014;5(10):1088–93. doi: 10.1021/ml5001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202–8. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 35.Berenbaum MC. Criteria for analyzing interactions between biologically active agents. Adv Cancer Res. 1981;35:269–335. doi: 10.1016/s0065-230x(08)60912-4. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–30. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12(7):465–77. doi: 10.1038/nrc3256nrc3256. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54(5):728–36. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep. 2016;15(3):519–30. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, et al. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol Cell. 2015;60(6):847–59. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015;58(6):1028–39. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24(6):777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conery AR, Centore RC, Spillane KL, Follmer NE, Bommi-Reddy A, Hatton C, et al. Preclinical Anticancer Efficacy of BET Bromodomain Inhibitors Is Determined by the Apoptotic Response. Cancer Res. 2016;76(6):1313–9. doi: 10.1158/0008-5472.CAN-15-1458. [DOI] [PubMed] [Google Scholar]
- 44.Ding Q, He X, Xia W, Hsu JM, Chen CT, Li LY, et al. Myeloid cell leukemia-1 inversely correlates with glycogen synthase kinase-3beta activity and associates with poor prognosis in human breast cancer. Cancer Res. 2007;67(10):4564–71. doi: 10.1158/0008-5472.CAN-06-1788. [DOI] [PubMed] [Google Scholar]
- 45.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–45. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guarneri V, Broglio K, Kau SW, Cristofanilli M, Buzdar AU, Valero V, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol. 2006;24(7):1037–44. doi: 10.1200/JCO.2005.02.6914. [DOI] [PubMed] [Google Scholar]
- 48.Kuerer HM, Newman LA, Smith TL, Ames FC, Hunt KK, Dhingra K, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17(2):460–9. doi: 10.1200/JCO.1999.17.2.460. [DOI] [PubMed] [Google Scholar]
- 49.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 50.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.