Abstract
Purpose
Using gene-disrupted allogeneic T cells as universal effector cells provides an alternative to and potentially improves current chimeric antigen receptor (CAR) T cell therapy against cancers and infectious diseases.
Experimental Design
The CRISPR/Cas9 system has recently emerged as a simple and efficient way for multiplex genome engineering. By combining the lentiviral delivery of CAR and CRISPR RNA electroporation to co-introduce RNA encoding the Cas9 and gRNAs targeting endogenous TCR, beta-2 microglobulin (B2M) and PD1 simultaneously, to generate gene-disrupted allogeneic CAR T cells deficient of TCR, HLA class I molecule and PD1.
Results
The CRISPR gene-edited CAR T cells showed potent anti-tumor activities, both in vitro and in animal models and were as potent as non-gene edited CAR T cells. In addition the TCR and HLA class I double deficient T cells had reduced alloreactivity and did not cause graft-versus-host disease. Finally, simultaneous triple genome editing by adding the disruption of PD1 led to enhanced in vivo anti-tumor activity of the gene-disrupted CART cells.
Conclusions
Gene-disrupted allogeneic CAR and TCR T cells could provide an alternative as a universal donor to autologous T cells, which carry difficulties and high production costs. Gene-disrupted CAR and TCR T cells with disabled checkpoint molecules may be potent effector cells against cancers and infectious diseases.
Keywords: CRISPR/Cas9, T lymphocytes, PD1, Chimeric antigen receptors, Universal T cells
INTRODUCTION
Engineered T cell receptor (TCR) and CAR T (CART) cell treatments of cancer patients have shown promising results (1–6). The majority of current TCR and CART clinical trials utilize autologous T cells and might therefore be hampered by the poor quality and quantity of T cells as well as the time and expense of manufacturing autologous T cell products. These limitations would be circumvented by the use of allogeneic T cells. However, the endogenous TCR on allogeneic T cells may recognize the alloantigens of the recipient, leading to graft-versus-host disease (GVHD); furthermore, the expression of HLA on the surface of allogeneic T cells causes rapid rejection by the host immune system. Therefore, simple and efficient methods are needed for multiplex genomic editing of T cells.
The CRISPR/Cas9 system has recently emerged as a potentially robust alternative for inducing targeted genetic alterations and as a process for multiplex genome engineering (7–10). In the present study, by using CRISPR/Cas9 system to simultaneously disrupt multiple genomic loci, we have generated CART cells deficient in the expression of endogenous TCR and HLA class I (HLA-I) that can be used as gene-disrupted allogeneic CART and further developed as universal CART cells. We found that TCR and B2M genes could be disrupted with high efficiency through the co-introduction of mRNA encoding the Cas9 with gRNAs targeting these genes by RNA electroporation. We generated gene-disrupted allogeneic CART cells by combining the lentiviral (LV) delivery of CAR and CRISPR RNA electroporation to disrupt endogenous TCR and B2M genes simultaneously. In addition, we demonstrate that disruption of endogenous PD1 enhances the efficacy of gene-disrupted allogeneic CAR therapy in tumor models.
MATERIALS AND METHODS
Primary human lymphocytes
Primary human CD4 and CD8 T cells were isolated from healthy volunteer donors following leukapheresis by negative selection using RosetteSep kits (Stem Cell Technologies, Vancouver BC, Canada). All specimens were collected under a University Institutional Review Board-approved protocol, and written informed consent was obtained from each donor. Primary lymphocytes were stimulated with anti-CD3/CD28 Dynabeads (Life Technologies, Grand Island, NY,). T cells were cryopreserved at day 10 in a solution of 90% fetal calf serum and 10% dimethylsulfoxide (DMSO) at 1×108 cells/vial.
Generation of TCR or CAR constructs for mRNA electroporation and lentiviral transduction
CARs (PSCA or CD19) were synthesized and/or amplified by PCR based on sequencing information provided by the relevant publications (11–13) and subcloned into a pGEM.64A RNA-based vector or pTRPE lentiviral vectors.
Design and construction of CRISPRs
Cas9 and eSpCas9 (1.1) DNA was synthesized by PCR based on Dr. Zhang Feng’s publications (7,14), then inserted to PGEM vector. gRNAs were selected by GN19 with an NGG PAM site, and some were selected from N20 with a NGG PAM site. All gRNAs contained complementary sequences comprised of more than 13 base pairs in order to target sites to exclude potential off-target mRNA events. gRNAs were designed, as shown in Supplementary Figure 1a, and synthesized by overlap PCR. All gRNA PCR products were ligated into the MSGV vector. The in vitro transcribed Cas9 mRNA and gRNA targeted constant regions of TCR α and β chains, beta-2 microglobulin and PD1. gRNAs were designed to target either a sequence within exon 1 of the TCR α constant region, a consensus sequence common to exon 1 of both TCR β constant regions 1 and 2, exon 1 of beta-2 microglobulin or PD1. Sequences encoding the gRNAs were assembled using overlap PCR and cloned into the MSGV vector containing a T7 promoter. These plasmids were linearized with EcoRI before conducting RNA in vitro transcription (IVT). The IVT RNA was stored at −80°C in nuclease-free vials for single use. The following gRNA targeting sequences were used in the study: TRAC-gRNA-1: AGAGTCTCTCAGCTGGTACA. TRAC-gRNA-2: TGTGCTAGACATGAGGTCTA. TRBC-gRNA-1: GCAGTATCTGGAGTCATTGA. TRBC-gRNA-2: GGAGAATGACGAGTGGACCC. B2M-gRNA: CGCGAGCACAGCTAAGGCCA. PD1-gRNA: GGCCAGGATGGTTCTTAGGT
Flow cytometry
The following monoclonal antibodies and reagents were used with the indicated specificity and the appropriate isotype controls. From BD Biosciences (San Jose, CA): APC-conjugated anti-CD3 (555335), FITC-anti-CD8 (555366), PE-anti-CD8 (555635), PE-anti-CD28 (561793), PE-anti-CD107a (555801), and PE-anti-beta-2 microglobulin (551337), FITC-anti-HLA-I (555552), APC-anti-CD137 (550890). From Biolegend (San Diego, CA): APC-anti-PD1 (114102), APC-anti-PDL1 (329702), FITC-anti-CD45RO (304204), APC-anti-CD62L (304814). From Beckman Coulter (Pasadena, CA): PE-anti-Vb13.1 (IM2021U). Data were acquired on a FACS Accuri (BD Biosciences, San Jose, CA) using CellQuest version 3.3 (BD Biosciences, San Jose, CA) and analyzed by FCS Express version 3.00 (De Novo Software, Los Angeles, CA) or FlowJo version 7.6.1 (Tree Star, Inc., Ashland, OR).
Generation of CD3neg T cells
gRNA was in vitro transcribed by a T7 mScript™ Standard mRNA Production System (Cambio, C-MSC100625, Cambridge, England). Cas9, TCR α, TCR β mRNA was in vitro transcribed using mMESSAGE mMACHINE T7 ULTRA kits (Life Technologies, AM1345, Carlsbad, CA). T cells were stimulated by CD3/CD28 Dynabeads for three days prior to RNA electroporation. T cells were electroporated as previously described (15,16). Briefly, T cells were washed three times with OPTI-MEM and re-suspended in OPTI-MEM (Invitrogen) at a final concentration of 1-3×108 cells/ml. Subsequently, 0.1 ml of the cells was mixed with IVT RNA and electroporated in a 2 mm cuvette. 20 μg of Cas9 mRNA and 10 μg of gRNA were electroporated into the cells using a BTX830 (Harvard Apparatus BTX) at 360 V and 1 ms; this process was followed by a second electrotransfer of 5 μg of gRNA 12 to 24 hours later. Following electroporation, the cells were immediately placed in pre-warmed culture media and cultured in the presence of IL-2 (100 IU/ml) at 37°C and 5% CO2.
Enrichment CD3neg T cells
Cells washed with Auto MACS buffer were incubated for 30 min with CD3 microbeads (Miltenyi Biotec, 130-050-101, Auburn, CA) at 4°C. After being washed twice, the cells were passed through an LD column (Miltenyi Biotec, Auburn, CA), and the flow-through fraction was collected for further use.
Proliferation capability of CD3neg T cells test
CD3neg T Cells were electroporated with a total of 15 ug NY-ESO-1 TCR (1G4) α and β chain mRNA (7.5 ug each), using a BTX830 (Harvard Apparatus BTX) at 500 V and 700μs. CD3 expression was measured 24 hours later and then stimulated with CD3/CD28 Dynabeads. Proliferation was monitored every 2 to 3 days.
TCR and B2M double disruption or TCR, B2M and PD1triple disruption
To generate TCR, B2M double and TCR, B2M, PD1 triple knockout T cells, Cas9 mRNA was co-electroporated with different gRNAs targeting TRBC, B2M or TRBC, B2M, PD1. The Cas9 mRNA and gRNAs delivery procedure was the same as generation of CD3neg T cells. The total amount gRNAs of the first electroporation was half of the Cas9 mRNA, the total amount gRNAs of the second electroporation was one fourth of the Cas9 mRNA. The TCR and HLA-I double-negative cell population was sorted on day9 to obtain gene-disrupted T cells. TCR and HLA-I molecule expression was confirmed at each step.
Generation of gene-disrupted and PD1 deficient CART cells
Gene-disrupted CART cells were generated by combing the lentiviral transduction of CD19 or PSCA CAR with the RNA electroporation of CRISPR/gRNAs. 1 day after anti-CD3/CD28 beads stimulation, T cells were transduced with lentiviral-CD19 or PSCA CAR. 2 days later, Cas9 and gRNAs targeting TRBC, B2M or TRBC, B2M, PD1 were transferred into T cells by electroporation. On day 9 after stimulation, T cells negative for CD3, HLA-I were sorted by microbeads depletion.
Measuring allele modification frequencies using T7E1 assay, TIDE, and sequencing of PCR fragments
The level of genomic disruption of TRAC, TRBC1, and TRBC2 in T cells was determined by a T7E1 Surveyor Nuclease assay (NEB). The percent target disruption was quantified by densitometry. PCR products were ligated to TOPO cloning vector (Invitrogen) then transformed in E.coli. Single clone was picked and sequenced to calculate the indels and insertions. PD1 disruption was confirmed by Sanger sequencing. The PCR primers used for the amplification of the target locus were as follows: TRAC forward, 5′-TCATGTCCTAACCCTGATCCTCTT-3′; TRAC reverse, 5′-TTGGACTTTTCCCAGCTGACAGA-3′; TRBC total forward, 5′-TACCAGGACCAGACAGCTCTTAGA-3′; TRBC total reverse, 5′-TCTCACCTAATCTCCTCCAGGCAT-3′; PD1 forward, 5′-GTAATAAAATGCTCAGCACAGAATA-3′; PD1 reverse, 5′-GAGAAAAATATCACCAGCTCATCT-3′. For analyzing allele modification frequencies using TIDE (Tracking of Indels by Decomposition)(17), the purified PCR products were Sanger-sequenced using both PCR primers and each sequence chromatogram was analyzed with the online TIDE software available at http://tide.nki.nl. Analyses were performed using a reference sequence from a Cas9 mock-transfected sample. Parameters were set to the default maximum indel size of 10 nucleotides and the decomposition window to cover the largest possible window with high quality traces. All TIDE analyses below the detection sensitivity of 1.5% were set to 0%. Primers used for TIDE off-target measurement are listed in Supplemental table 1.
ELISA assays
Target cells were washed and suspended at 1×106 cells/ml in R10 medium. Next, 100 μl of each target cell type was added in triplicate to a 96-well round-bottom plate (Corning). Effector T cells were washed and re-suspended at 1×106 cells/ml in R10 medium, and then 100 μl of T cells was combined with the target cells in the indicated wells. The plates were incubated at 37°C for 18 to 24 hours. After the incubation, the supernatant was harvested and subjected to an ELISA (eBioscience).
IFNγ ELISpot
CRISPR edited T cells were plated in EISPOT plates (R&D) at the concentration of 2 × 104 cells per well with irradiated allogenic PBMCs, another experiment were performed by co-culturing of allogenic PBMC with irradiated CRISPR edited T cells. Cells were incubated for 18 h at a stimulator-to-responder ratio of 1:1. Experiments were performed according to the manufacturer’s instructions. The spots were automatically quantified using an Elispot plate reader for scanning and analyzing.
In vivo reactivity of allogeneic T cells against HLA-Ineg T cells
1X107 TCRneg or TCR/HLA-Ineg T cells and 2X106 allogeneic effector T cells were mixed and infused into NSG mice (i.v.). The presence of CD45+CD3+ allogeneic T cells and CD45+ CD3− gene-edited T cells were measured by Trucount assay at day 2, day9 and day16 after T cell infusion.
CD107a staining
Cells were plated at an E:T of 1:1 (1×105 effectors: 1×105 targets) in 160 μl of R10 medium in a 96-well plate. Next, 20 μl of phycoerythrin-labeled anti-CD107a Ab was added, and the plate was incubated at 37°C for 1 hour before the addition of Golgi Stop (2 μl of Golgi Stop in 3 ml of R10 medium, 20 μl/well; BD Biosciences, 51-2092KZ) and incubation for another 2.5 hours. Then, 5 μl of FITC-anti-CD8 and 5 μl of APC-anti-CD3 were added for incubation at 37°C for 30 min. After the incubation, the samples were washed with FACS buffer and analyzed by flow cytometry.
Luciferase-based CTL assay
Nalm6-CBG tumor cells were generated and employed in a modified version of a luciferase-based CTL assay (18). Briefly, click beetle green luciferase (CBG)-T2A-eGFP was lentivirally transduced into Nalm6 tumor cells and sorted for GFP expression. The resulting Nalm6-CBG cells were resuspended at 1 × 105 cells/ml in R10 medium and incubated with different ratios of T cells (e.g., 30:1, 15:1, etc.) overnight at 37°C. Then, 100 μl of the mixture was transferred to a 96-well white luminometer plate. Next, 100 μl of substrate was added, and the luminescence was immediately determined. The results are reported as percent killing based on the luciferase activity in the wells with tumor cells but no T cells (% killing = 100 - ((RLU from well with effector and target cell coculture) / (RLU from well with target cells) × 100)).
Mouse xenograft studies
Studies were performed as previously described with certain modifications (19,20). Briefly, for the Nalm6 tumor model, 6- to 10-week-old NSG mice were injected with 1×106 Nalm6 or Nalm6-PDL1 tumors cells through the tail vein on day 0. The T cell treatment began on day 7 after the tumor inoculation. For the PC3-PDL1 solid tumor model, 6- to 10-week-old NOD/SCID gamma (NSG) mice were injected subcutaneously with 1×106 PC3-PDL1-CBG or PC3-CBG tumors cells in the right flank on day 0. The mice were treated with T cells via the tail vein at day 22 post PC3-PDL1-CBG tumor inoculation, when the tumors were approximately 200 mm3 in volume. T cells were given at 2×106 cells/mouse.
GVHD studies
For the in vivo GVHD studies, we resuspended the cells in FBS and infused them intravenously into the mice after sub-lethal irradiation (175 cGy). We monitored the mice for clinical GVHD 2 to 3 times per week. The following signs were included in the clinical index: weight loss, hunching, activity, fur texture and skin integrity. We euthanized moribund mice for ethical reasons.
RESULTS
Multiple deliveries of gRNAs disrupts genes in human primary T cells with high efficiency without impairing effector function
Efficient multiplex genomic editing is required to generate gene-disrupted T cells that are deficient in TCR, HLA and other genes. We optimized CRISPR/gRNA RNA electroporation to achieve high gene disruption efficiency in T cells. First, Cas9 and gRNAs were generated using an in vitro transcription system (Supplementary Figure 1a), and a “hit-and-run” delivery strategy was developed to transiently deliver the Cas9 mRNA and gRNAs to T cells by electroporation (Supplementary Figure 1b). An initial experiment targeting the TCR α constant region (TRAC) or β constant region (TRBC) with single electroporation resulted in 7.51% and 12% CD3-negative (CD3neg) T cells, respectively (Figure 1a and Supplementary Figure 2a). The optimal molecular ratio of Cas9:gRNA for maximum disruption efficiency was 1:1 to 2:1, and the gene disruption efficiency was correlated with the amount of electro-transferred RNA (Supplementary Figure 2a).
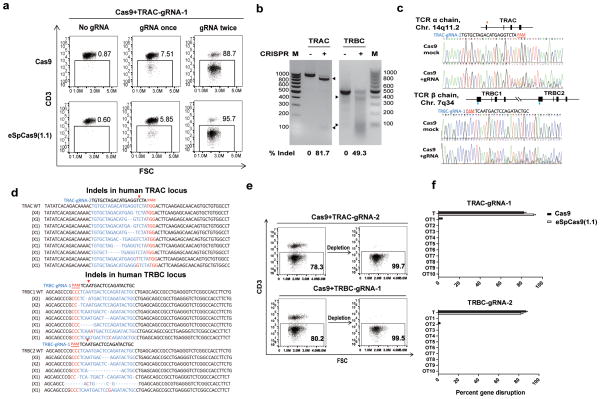
Figure 1. CRISPR/Cas9 mediates efficient TCR disruption in T cells.
(a) CD3 expression of T cells after sequential CRISPR RNA electroporation with Cas9 and eSpCas9 (1.1). (Healthy donors, n=5) (b) Amount of TCR-targeted gene disruption measured by a mismatch-selective T7E1 surveyor nuclease assay on DNA amplified from the cells shown. The calculated amount of targeted gene disruption in TRAC and TRBC is shown at the bottom. Arrows indicate expected bands. (c) A diagram of the human locus encoding the TCR α and β CRISPR gRNA targeting sites within the genomic locus of the TCR α and β constant region. Multiple peaks in the Sanger sequencing results show the CRISPR-mediated events of NHEJ at the TRAC and TRBC genomic loci. Each exon is shown by a block. Red arrow: sense strand gRNA targeting site; blue arrow: anti-sense strand gRNA targeting site. (d) Indels and insertions observed by clonal sequence analysis of PCR amplicons after CRISPR-mediated recombination of the TCR α and β locus. Red arrow indicates putative cleavage site. (e) CD3 expression on purified TCRneg population (n=3). (f) Off-target mutagenesis measurement of TRAC and TRBC. Indel frequencies were measured by TIDE analysis (n=3). All TIDE analyses below the detection sensitivity of 1.5% were set to 0%. Bars, SE, n = 3;T: target; OT: Off-target.
Compared with mRNA, gRNAs are more prone to rapid degradation, which potentially limits the targeting efficiency. Thus, sequential electroporations of gRNA were tested after the initial Cas9/gRNA electroporation. There was a marked increase in disruption frequency at the protein level, as 88.7% (85.7± 6.7, n=5) of cells were CD3neg after the second gRNA electroporation targeting TRAC (Figure 1a). To improve gene targeting specificity, high-fidelity Cas9 mutant eSpCas9 (1.1) was tested. As high as 95.7% (93.6%± 6.2, n=3) CD3 disruption was achieved after a second delivery of gRNA. Clonal sequencing showed that the genomic targeting efficiency reached 89.4% (Supplementary Figure 2b). A surveyor assay confirmed a cleavage rate of 81.7% and 49.3% at the genomic loci of TRAC and TRBC, respectively (Figure 1b). Multiple peaks in the Sanger sequencing data flanking the TRAC and TRBC target sites confirmed that the genomic reading frame shifted downstream of the target sites (Figure 1c). The occurrence of insertions or deletions (indels) caused by NHEJ repair was confirmed by clonal sequencing (Figure 1d). The TCR disrupted TCR/CD3neg population was enriched to over 99% (99.70±0.20%) by a single step of CD3 negative selection (Figure 1e). The CRISPR treated T cells could be expanded 12-folds (12±1.1, n=3), which is 1/3 of the non-genome edited T cells (40±2.6) in a standard 9 days T expansion period due to the toxicity of the second electroporation. Around 8-folds (8.4±0.7, n=3) TCR/CD3 neg T cells can be obtained from the starting T cells (Supplementary Figure 3a). Recent studies showed a low incidence of off-target mutagenesis in T cells using lentivirus and adenovirus-delivered CRISPR/Cas9 to knock out CCR5 (21,22). Another report showed no detectable off-target mutations in the CXCR4 knockout CD4 T cells (23). Consistent with these studies, we observed very rare off-target events when targeting TRAC and TRBC with Cas9 and no detectable of off-target mutagenesis with eSpCas9 (1.1), indicating CRISPR /Cas9 gene editing is more specific in T cells than other cell types (Figure 1f).
To test whether CRISPR/Cas9 gene editing would affect the proliferating capability of the T cells, CD3 expression was restored by electroporation of 1G4 TCR mRNA into the TCR/CD3 neg T cells, and then simulated with CD3/CD28 dynabeads. No Expansion difference was observed between wild type and TCR restored CD3 neg T cells (Supplementary Figure 3).
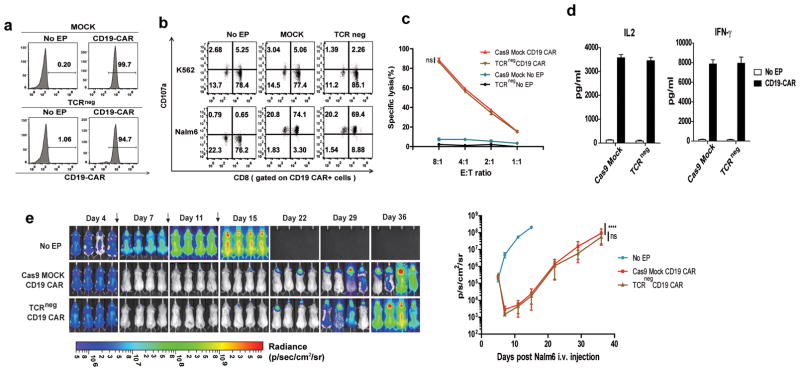
To test whether CRISPR/Cas9 gene editing would affect the effector function of the T cells, the anti-tumor activity was tested after electroporation of CD19 CAR mRNA into TCR/CD3neg T cells. The surface CAR expression of the TCR/CD3neg T cells was equal to that of the control group (Figure 2a). When the TCR/CD3neg CD19-CART cells were stimulated with CD19+ Nalm6 leukemia cells, the CD107a up-regulation, cytokine secretion and killing activity of CD19-CAR TCR/CD3neg T cells was equivalent to those of the wild-type control cells (Figure 2b, c and d). The CD19-CAR TCR/CD3neg T cells were infused into Nalm6-bearing NOD/scid/γc(−/−) mice (NSG) mice to test their in vivo anti-tumor activity. Tumor regression was evident with an efficacy equivalent to that for the CD19-CAR wild-type counterpart cells that were produced using tissue culture conditions used in ongoing clinical trials (Figure 2e). The results indicate that CRISPR/Cas9 editing of the endogenous TCR does not adversely affect the function of primary T cells for adoptive immunotherapy.
Figure 2. CRISPR/Cas9 editing does not impair antitumor efficacy of primary T cells.
(a) Relative CD19-CAR expression after the electrotransfer of CD19-CAR RNA into Cas9 MOCK and TCR/CD3neg cells. No significant functional difference was observed between CD19-CAR redirected Cas9 MOCK and TCR/CD3neg cells as confirmed by (b) CD107 release assay, (c) cytotoxicity assay and (d) IL2 and IFNγ secretion when incubated with the Nalm6 target cell line. Representative data from 3 independent experiments are shown. Bars, SE. (e) NSG mice (n=12) were injected with 1×106 Nalm6 tumor cells (i.v.), and the mice were randomly sorted into three groups. Cas9 MOCK and TCR/CD3neg T cells (10×106) expressing the CD19-CAR after electroporation were injected i.v. every 4 days for a total of three injections (arrows); mice treated with no RNA electroporated T cells from the same donor served as the control. Images were obtained from the surviving animals as indicated. Imaging commenced 1 day before the start of T cell treatment. Bars, SE; E:T, effector to tumor ratio; arrow, time point of T cell infusion; ns, not significant. ****P<0.001, ns, by comparison of the slopes with linear regression.
Reduced alloreactivity of TCR and B2M double-disrupted T cells
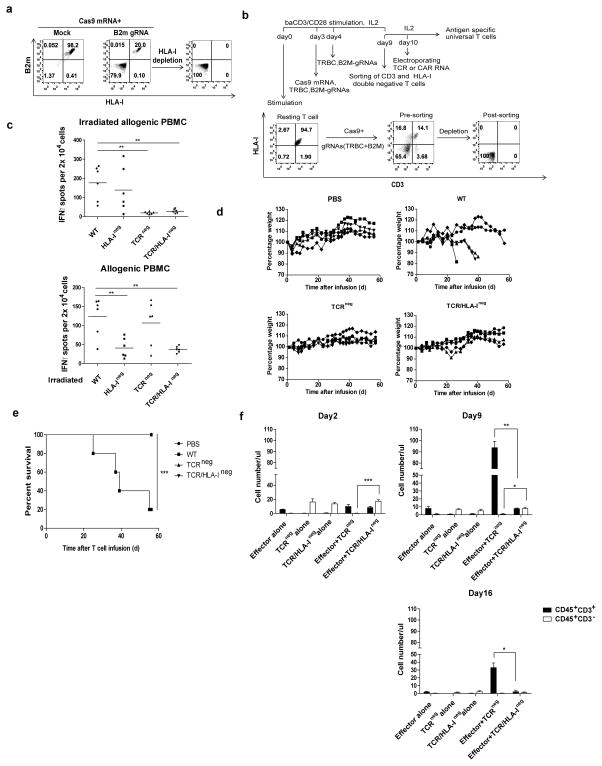
Since disrupting either TCR α or β is sufficient to ablate TCR/CD3 expression and B2M is essential for the assembly and expression of HLA-I complex (24), TCR and B2M double disruption was developed to generate gene-disrupted T cells. First, the ability to eliminate HLA-I expression on the T cells by disrupting B2M was tested. T cells were electroporated with B2M-targeting Cas9/gRNA RNA; this process resulted in a B2M and HLA-I double-negative population of 79.9%. The HLA-Ineg population could be further enriched by negative selection (Figure 3a). To generate double-knockout T cells lacking the TCR and B2M, Cas9 mRNA was co-electroporated with two different gRNAs targeting TRBC and B2M. As a result, the CD3 and HLA-I double-negative cell population was 65.4% (Figure 3b). After enrichment of the double knockout cells, we found that the TCR and B2M double-knockout T cells abrogated the allogeneic killing of HLA unmatched tumor cell lines (Supplementary Figure 4a). We did not observe any response when these cells were challenged by allogeneic whole-blood irradiated PBMCs in an IFNγ Elispot assay (Figure 3c, top panel). The ablation of HLA-I molecules also sharply reduced the alloreactivity, as confirmed by co-culture of allogenic PBMCs with irradiated B2M-disrupted cells (Figure 3c, bottom panel). Although the ability of HLA-I negative T cells to stimulate an allogenic PBMC response caused by T cell was markedly reduced, it was not completely diminished, probably due to the activation of NK cells within the PBMCs, which was supported by the finding that allogeneic T cells activation was completely abrogated as long as the HLA-I of stimulating gene-disrupted T was ablated when the purified CD4 and CD8 T cells, instead of PBMCs, were used as allogeneic effectors (Supplementary Figure 4c). It was further confirmed that B2M-disrupted, HLA-Ineg T cells had reduced target recognition by co-injecting TCRneg or TCR, HLA-I double deficient (TCR/HLA-Ineg) T cells with allogeneic effector T cells into NSG mice. As shown in Figure 3f, significantly reduced number of allogeneic effector T cell (CD45+ CD3+) was found when TCR/HLA-Ineg T cells were co-infused (Effector+TCR /HLA-Ineg), comparing with co-infusing of TCRneg (Effector+ TCRneg ) T cells. However, without disruption of HLA-I, the TCR single gene-disrupted T cells were eliminated when co-infused with allogeneic T cells, while TCR and HLA-I double gene-disrupted T cells remain unchanged, suggesting the rejection of the beta-2m disrupted T cells by the allogeneic effector T cells was reduced. T cells express high levels of HLA class II after being activated, which could potentially lead to the accelerated rejection of infused allogeneic T cells. Although high levels of HLA class II expression on TCR/HLA-Ineg T cells could be by detected after activation (Supplementary Figure 4b), we were unable to detect primed CD4 T cells activation (Supplementary Figure 4c), suggesting this in vitro assay was not sensitive enough to detect CD4 allo-reactivity, even for dendritic cell primed CD4 T cells. To confirm that the TCR and HLA-I editing process abrogates the GVHD reactivity of the gene-modified cells in vivo, we infused TCR ablated, TCR/HLA-I double ablated or non-manipulated cells into 8- to 10-week-old NSG mice that had been conditioned with 175 cGy irradiation. Four of the five mice infused with non-manipulated lymphocytes developed lethal xenogeneic graft-versus-host disease (GVHD) within 2 month after the infusion. GVHD was also evidenced by elevated number of CD8 positive cell infiltrating in different organs, as well as elevated CD45 positive cell counts in the peripheral blood. (Supplementary Figure 5a,b,c), whereas none of the mice receiving TCR single or TCR/HLA-I double ablated cells (n = 5, per group) developed GVHD (Fig. 3d,e).
Figure 3. Multiple gene ablation by CRISPR/Cas9 to generate universal effector cells.
(a) HLA-I disruption with gRNA targeting B2M. (b) Flow chart of the protocol to generate universal effector cells as described in methods. (c) The allo-reactivity of TCR and TCR/HLA disrupted was tested with an IFNγ Elispot assay by challenging the gene-ablated T cells with irradiated allogenic PBMCs (left panel) or co-culturing allogenic PBMCs with irradiated gene-ablated T cells. Specific spots are shown on the y axis as the spots produced in the presence of stimulators minus the spots produced by the effectors alone. **P<0.01 by Mann-Whitney test. (d) Survival without severe GVHD and (e) weight loss in mice after infusion of PBS (n = 5), Cas9 Mock wild type (Cas9 Mock) T cell (n = 5), TCR ablated (TCRneg) cells (n = 5) or TCR/HLA-I double ablated (TCR/HLA-Ineg) (n = 5). ***P<0.005 by the log-rank Mantel-Cox test. (f) Abolishment of target recognition of allogeneic T cells by disrupting MHC-I on target T cells. Allogeneic T cells were primed by dendritic cells of the same donor with gene-disrupted T cells and infused into NSG mice with TCRneg or TCR/HLA-Ineg target T cells. Significant prolonged survival of HLA-I ablated T cells was observed by the presence of CD3neg T cells, which is also confirmed by the failed expansion of allogeneic effector T cells (n=3). ***P<0.001**P<0.01, *P<0.05, by Mann-Whitney test.
Gene-disrupted CART cells retain antitumor efficacy
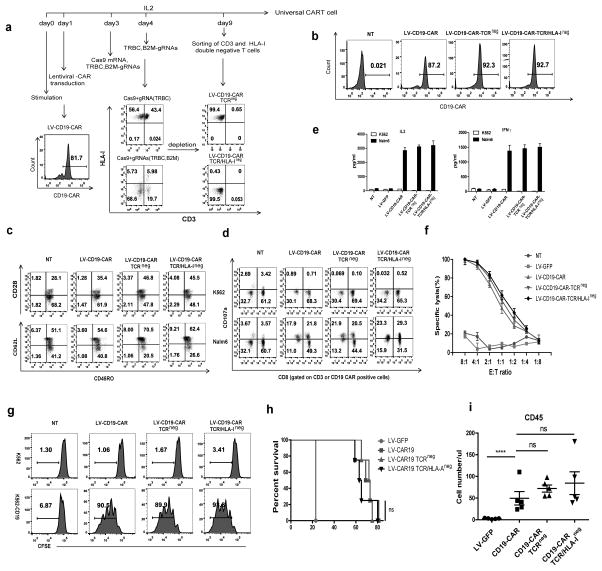
Gene-disrupted CD19 CART cells were generated by combing CD19 CAR lentiviral transduction with the RNA electroporation of Cas9/gRNAs (Figure 4a). The cells were then sorted, and were negative for CD3 with high levels of CD19 CAR expression (Figure 4b). The majority of the gene-disrupted CD19 CART cells expressed high level of CD62L and a medium level of CD28, consistent with central memory status for the major T cell population (Figure 4c). The TCR/HLA-I double-negative CD19 CART showed robust in vitro anti-tumor activities, such as lytic capacity, cytokine secretion and proliferation, that were as potent as those of the wild-type CD19 CART cells (Figure 4d, e, f, g). The T cells were infused into NSG mice bearing disseminated Nalm6 leukemia. Mice treated with CART cells with a disrupted endogenous TCR (LV-CD19 CAR TCRneg) or with a simultaneous disruption of TCR and HLA-I (LV-CD19 CAR TCR/HLA-Ineg) exhibited similar survival rates to that of mice treated with wild-type CD19 CART cells (LV-CD19 CAR) (Figure 4h). The numbers of human T cells in the peripheral blood of the Nalm6-bearing mice treated with TCRneg or TCR/HLA-Ineg were comparable to mice treated with wild type CD19 CART cells (Figure 4i), suggesting that the disruption of TCR alone or together with B2M does not significantly affect CART cell engraftment, in vivo proliferation and anti-tumor activity.
Figure 4. Generation of universal CART cells with a combination of lentiviral gene transfer and CRISPR/Cas9 electroporation.
(a) Flow chart of the generation of universal CD19-CART cells. T cells were transduced with lentiviral CD19-CAR on day 1 after stimulation, and Cas9 mRNA and gRNAs targeting the TCR β chain and B2M were electroporated in the T cells on day 3 followed by a second delivery of gRNAs on day 4. The TCR and HLA-I double-negative cell population was enriched by negative selection using microbeads on day9. (b) CD19-CAR expression of gene-modified lenti-CD19-CAR T cells. (c) Phenotype of universal CD19-CAR T cells. Function of TCR-negative and TCR/HLA-I double-negative CD19-CAR T cells tested by (d) CD107a release, (e) cytokine secretion and (f) tumor lytic capability. Representative data from 3 independent experiments are shown. Bars, SE. (g) CFSE-labeled CD19-CAR and non-transduced T cells were incubated with K562 and target K562-CD19 tumor cells at the indicated E:T ratio for 72 hours. (h) Survival curve of mice receiving gene-edited CD19-CART cells. Tumors were established in NSG mice (n=5 per group) by i.v. injection of 1×106 Nalm6 cells. Beginning on day 7, T cells (5×106) expressing lentiviral (LV) transduced CD19-CAR were infused with a single injection. T cells expressing LV GFP protein were injected as controls. ns, no difference by the log-rank Mantel-Cox test. (i) Peripheral blood from Nalm6-bearing NSG mice treated with CD19 CART cells was obtained on day 21 and quantified for the presence of CD45 T cells by a FACS Trucount assay. Results are expressed as the mean absolute count per μl of peripheral blood±SD with n=5 for all groups. ns, ****P<0.001 by Mann-Whitney test.
Disruption of PD1 in CAR T cells leads to enhanced antitumor efficacy
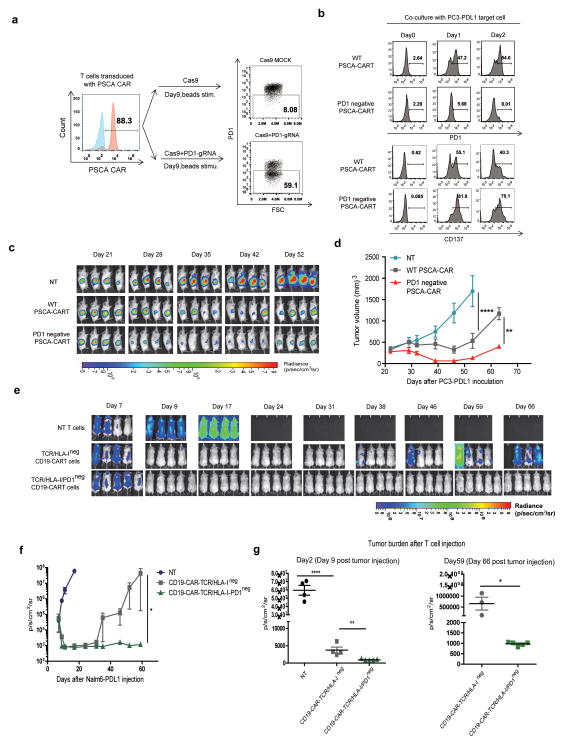
Given the robust antitumor efficacy of PD1 antagonists in multiple clinical trials, and that combination therapy with CAR T cells and PD1 antagonists have enhanced antitumor activity in preclinical models (25), we next tested if disruption of PD1 in CAR T cells would enhance antitumor activity. A CAR specific for prostate stem cell antigen (26) (PSCA) was expressed in T cells using lentiviral vector gene transfer. gRNAs for PD1 were developed, and RNA electroporation of Cas9/gRNAs using the strategy shown in Figure 5a was done to generate a population of PSCA CAR T cells that no longer expressed PD1 upon stimulation. PD1 upregulation were abolished on CRISPR edited PSCA CART cells after co-culture with PC3 tumor cells transfected with PDL1 (PC3-PDL1). Enhanced T cell activation was confirmed by the upregulated expression of CD137 on PD1 ablated CART cells (Figure 5b). The function of PD1 deficient CAR T cells were tested in vivo in NSG mice bearing established large PC3-PDL1 tumors (Figure 5c, d). The PSCA PD1neg CAR T cells showed significantly enhanced antitumor activity compared to the conventional PSCA CAR T cells. Similar results were observed in the setting of adaptive resistance when a native PC3 tumor without forced expression of PDL1was treated with PSCA-CART cells. Over 90% PC3 tumor gained PDL1 expression after encountering PSCA-CART cells in vitro (Supplementary Figure 6c). When tested in vivo, The PSCA PD1neg CAR T cells also showed significantly enhanced antitumor activity compared to wild type PSCA CAR T cells (Supplementary Figure 6d, 5e). To test whether PD1 disruption might improve the function of gene-disrupted CART cells, TCR, B2M and PD1 triple ablated gene-disrupted CD19 CART cells were generated. Enhanced anti-tumor activity of PD1 disrupted gene-disrupted CD19 CART cells were observed in a Nalm6-PDL1 leukemia model, evidenced by more quick and robust anti-tumor response in PD1 ablated gene-disrupted CART cell treatment group, which led to complete elimination of leukemia cells in this aggressive mouse model (Figure 5e, f, g).
Figure 5. PD1 ablation enhances the therapeutic effect of CART cells.
(a) Generation of PD1-negative PSCA-CAR T cells. T cell PD1 ablation was confirmed by flow cytometry after stimulation. PD1 deficient CART cells were sorted. (b) Co-culture of PD1 disrupted CART cells with PC3-PDL1 tumor cells. PD1 and CD137 expression were measured on the CRISPR/Cas9 edited CART cells. (c) PC3-PSCA-PDL1 tumors were established in the flank of NSG mice by inoculating 1×106 tumor cells/mouse (s.c. with Matrigel, n=4). After 3 weeks, the mice were treated with 2×106 PSCA CAR transduced WT (PSCA CAR) or PD1neg (PSCA CAR PD1neg) T cells (i.v.); mice treated with non-transduced T cells (NT) served as the control. BLI conducted before (day 21) and after the mice treated with a single T cell injection. (d) Tumor volume of mice. Results are expressed as the mean tumor volume (mm3±SE) with n=4 for all groups. **P<0.01, ****P<0.001 by comparison of the slopes with linear regression. (e) PD1 ablated universal CD19-CART cells were generated by co-delivery of Cas9 mRNA and gRNAs targeting TRBC, B2M and PD1 after transduction with lenti-CD19-CAR. TCR and HLA-I disruption was confirmed by flow cytometry, PD1 disruption was confirmed Sanger sequencing (Supplementary Figure 6a,b). TCR and HLA-I double negative cells were sorted at day 9. Nalm6-PDL1 tumor were established in NSG mice (n=4 or 5 per group) by i.v. injection of 1×106 cells. Beginning on day 7, T cells (5×106) expressing lentiviral transduced CD19-CAR were infused with a single injection. T cells expressing LV GFP protein were injected as controls. (f) Bioluminescence of mice receiving different treatment. Imaging commenced 1 day before the start of T cell treatment. P<0.05, by comparison of the slopes with liner regression.(g) Tumor burden of different groups were compared at day 2 and day 59 after T cell treatment (n= 4 or 5). *P<0.05, **P<0.01, ****P<0.001 by Mann-Whitney test.
DISCUSSION
Multiplex genome editing is one of most attractive applications of the CRISPR/Cas9 system and holds great promise for advancing T cell-based adoptive immunotherapy. However, the low targeting efficiency of DNA transfection limits the use of multiplex genome engineering in primary T cells (27). A “hit-and-run” delivery strategy was developed to introduce CRISPRs to T cells via the co-electroporation of Cas9 mRNA and gRNA. After two rounds of gRNA electroporation, a targeting efficiency >90% at the protein level was routinely achieved for a single gene disruption. More encouragingly, the triple gene disruption of TRBC, B2M and PD1 yielded double negative CD3 and HLA-I at 65% without any purification or selection. Our results also demonstrate that enrichment to >99% purity of gene-disrupted T cells could be easily achieved using clinically approved paramagnetic beads. The disrupted T cells did not cause GVHD, suggesting that they may be used as allogeneic CAR T cells. Importantly, the gene-edited T cells showed anti-tumor activities both in vitro and in different tumor mouse models that were as potent as non-gene-edited T cells.
In this report, we demonstrate that CRISPR/Cas9 is a powerful multiplex genome editing tool in primary human T cells with high efficiency. This report is the first to describe the simultaneous editing of three different genetic loci, thereby creating checkpoint-resistant T cells that have the potential to be “off the shelf.” Previous reports showed that T cells can be genetically edited by zinc finger nucleases (ZFN) or TAL effector nuclease (TALEN) to eliminate the expression of the endogenous TCR α and β chains to avoid GVHD (28,29) or PD-1 in tumor-infiltrating lymphocytes for the treatment of metastatic melanoma (30). However, due to the complexity in constructing and difficulty in targeting multiple genes with ZFN and TALEN in T cells (31–33), previous studies have not simultaneously targeted 3 gene loci. The advantage of the efficient multiplex genome editing of CRISPR over TALEN is demonstrated by the superior function of PD1-deficient gene-disrupted CART cells that are resistant to an inhibitory pathway other than “off-the-shelf.”
Gene disruption in T cells is not so efficient with lentiviral and Adenoviral CRISPR (21,22). Although Pankaj reported better gene ablation in CD4 T cells with DNA nucleofection of CRISPR reagents, DNA nucleofection associated high toxicity to T cell was a major difficulty for its application (27). By modifying our protocol to clinical settings, we developed a process to generate synthetic cells that could be easily translated into current GMP-compliant manufacturing procedures. We are currently conducting trials to test non-gene-edited CAR T cells that have been manufactured with RNA electroporation (34). Our current protocols generate sufficient cells for a clinical trial to test the safety and feasibility of gene-disrupted CAR T cells. Our data indicates that HLA class I ablation can significantly reduce the surveillance of allogeneic T cells and potentially prevent being rejected rapidly after infusion to the allogeneic recipients. However, testing human T cells in immune deficient mice for the GVHD and host versus graft reaction may not be correlated well to the allogenic setting in humans. Further rigorous testing in non-human primates and further modification of the T cells could establish the safety of these cells as well as their ability to evade the immune system.
CRISPR /Cas9 gene editing has been shown to generate off-target mutations depending upon the experimental setting and cell type (35,36). However, Pankaj et al reported an extremely low incidence of off-target mutagenesis of CRISPR in hematopoietic stem cells (27). Recent studies also showed a low incidence of off-target mutagenesis in T cells using lentivirus and adenovirus-delivered CRISPR/Cas9 to knock out CCR5 (21,22). Another report showed no detectable off-target mutations in the CXCR4 knockout CD4 T cells (23). We observed very rare off-target mutagenesis targeting TRAC or TRBC with Cas9. Although no detectable mutagenesis observed with eSpCas9 (1.1) reduces worries regarding off-target effects in applied and therapeutic settings, deep sequencing of off-target gene disruption should be carefully characterized before conducting human trials. The careful selection of targeting sequences could also be considered to minimize the potential of off-target incidence. Researchers from the Joung lab improved the on-target specificity of the Cas9 nuclease with another independently discovered CRISPR variant SpCas9-HF1 (37) that can be used for more precise genomic edits in mammalian cells. Further comparison of these two Cas9 variants can be carried out for safer clinical use.
The genomic integrity of gene-edited T cells should be considered prior to conducting clinical trials. Unlike the UCAR T cells generated by TALEN (with substantial chromosome translocation (38) from universal CART cells derived from three different donors), no chromosome translocation has been identified (unpublished data).
The therapeutic value of blocking the PD1 inhibitory pathway is confirmed in our system by the ablation PD1 of CART cells in a solid tumor model and by the triple ablation of TCR, B2M and PD1 of CART cells in a leukemia tumor model. One potential limitation of our triple gene-disrupted CAR or TCR T cells is that they may trigger NK cell activation, leading to the eventual rejection of the edited T cells. We showed that although the ability of HLA-Ineg T cells’ allo-stimulation was markedly reduced (Fig 3c), it was not completely diminished due to the activation of NK cells in the absence of HLA-I. Lympho-depletion via chemotherapy, or using NK cell specific antibody (39,40) to deplete most of NK cells could potentially avoid or reduce NK mediated rejection of transferred HLA-Ineg T cells. Furthermore, NK mediated host versus graft reaction could also be minimized via the ablation of stimulatory NK ligands by CRISPR/Cas9 or by the expression of non-classical HLA class I molecules on the T cells, such as HLA-E (32). T cells express high levels of HLA class II after activation, which can potentially lead to the accelerated rejection of infused allogeneic T cells. Therefore, abrogated HLA class II should be considered to incorporate into next generation of gene edited allogeneic CART cells.
In summary, clinical-scale gene-disrupted CART cells with potent anti-tumor activity and reduced alloreactivity can be efficiently generated using multiplex CRISPR technology and potentially could be used as off-the-self universal T cells. This approach can be incorporated into current GMP-compliant manufacturing procedures and has a high potential for translation due to the successful translation of adoptive transfer therapy with ZFNs for HIV/AIDS (41,42). Although they may be compromised due to shorter persistence, gene-disrupted allogeneic CAR and TCR T cells could provide an alternative to autologous T cells, which carry difficulties and high production costs. Gene-disrupted allogeneic CAR and TCR T cells with disabled checkpoint molecules may be potent effector cells against cancers and infectious diseases.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Engineered CAR T cell treatments of cancer patients have shown promising clinical results. The majority of current clinical trials utilizes autologous engineered T cells and might therefore be hampered by the poor quality and quantity of T cells as well as the time and expense of T cell manufacturing. These limitations could be circumvented by the use of allogeneic T cells. The CRISPR/Cas9 system has recently emerged as a unique robust tool for multiplex genome engineering. By combining the lentiviral delivery of CAR and CRISPR RNA electroporation to co-introduce RNA encoding the Cas9 and gRNAs targeting endogenous TCR and beta-2 microglobulin (B2M) simultaneously, we have developed a clinically scalable technology of generating universal CAR T cells that showed potent anti-tumor activities, both in vitro and in animal models and were as potent as non-gene edited CAR T cells, which could be potentially translated into the clinic as an alternative for cancer adoptive immunotherapy.
Acknowledgments
This work was supported by a US National Institutes of Health (NIH) grants to C.H.J and Y.Z (2R01CA120409).
Footnotes
AUTHOR CONTRIBUTIONS
J.R, X. L, C.H.J and Y.Z designed experiments. J.R, X. L, C.F and S.J performed the experiments. J.R, C.H.J and Y.Z wrote the manuscript. All authors discussed and interpreted results.
COMPETING FINANCIALINTERESTS
X.L., C.H.J. and Y.Z. have financial interests due to intellectual property and patents in the field of cell and gene therapy. Conflicts of interest are managed in accordance with University of Pennsylvania policy and oversight. The other authors declare that they have no competing interests.
References
- 1.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Science translational medicine. 2013;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–17. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–33. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(7):917–24. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–18. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kochenderfer JN, Feldman SA, Zhao Y, Xu H, Black MA, Morgan RA, et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. Journal of immunotherapy. 2009;32(7):689–702. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329(5997):1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nature medicine. 2015;21(2):121–31. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Bennett AD, Zheng Z, Wang QJ, Robbins PF, Yu LY, et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. Journal of immunology. 2007;179(9):5845–54. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JL, Stewart-Jones G, Bossi G, Lissin NM, Wooldridge L, Choi EM, et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. The Journal of experimental medicine. 2005;201(8):1243–55. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leyton JV, Olafsen T, Sherman MA, Bauer KB, Aghajanian P, Reiter RE, et al. Engineered humanized diabodies for microPET imaging of prostate stem cell antigen-expressing tumors. Protein engineering, design & selection : PEDS. 2009;22(3):209–16. doi: 10.1093/protein/gzn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Moon E, Carpenito C, Paulos CM, Liu X, Brennan AL, et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer research. 2010;70(22):9053–61. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Tendeloo VF, Ponsaerts P, Lardon F, Nijs G, Lenjou M, Van Broeckhoven C, et al. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98(1):49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- 17.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic acids research. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon EK, Wang LC, Dolfi DV, Wilson CB, Ranganathan R, Sun J, et al. Multifactorial T-cell hypofunction that is reversible can limit the efficacy of chimeric antigen receptor-transduced human T cells in solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(16):4262–73. doi: 10.1158/1078-0432.CCR-13-2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett DM, Zhao Y, Liu X, Jiang S, Carpenito C, Kalos M, et al. Treatment of advanced leukemia in mice with mRNA engineered T cells. Human gene therapy. 2011;22(12):1575–86. doi: 10.1089/hum.2011.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carpenito C, Milone MC, Hassan R, Simonet JC, Lakhal M, Suhoski MM, et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc Natl Acad Sci U S A. 2009;106(9):3360–5. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Guan X, Du T, Jin W, Wu B, Liu Y, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. The Journal of general virology. 2015;96(8):2381–93. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9(12):e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Scientific reports. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serreze DV, Leiter EH, Christianson GJ, Greiner D, Roopenian DC. Major histocompatibility complex class I-deficient NOD-B2mnull mice are diabetes and insulitis resistant. Diabetes. 1994;43(3):505–9. doi: 10.2337/diab.43.3.505. [DOI] [PubMed] [Google Scholar]
- 25.John LB, Devaud C, Duong CM, Yong C, Beavis PA, Haynes NM, et al. Anti-PD-1 antibody therapy potently enhances the eradication of established tumors by gene-modified T cells. Clin Cancer Res. 2013;19(20):5636–46. doi: 10.1158/1078-0432.ccr-13-0458. [DOI] [PubMed] [Google Scholar]
- 26.Abate-Daga D, Lagisetty KH, Tran E, Zheng Z, Gattinoni L, Yu Z, et al. A novel chimeric antigen receptor against PSCA mediates tumor destruction in a humanized mouse model of pancreatic cancer. Hum Gene Ther. 2014 doi: 10.1089/hum.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell stem cell. 2014;15(5):643–52. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Provasi E, Genovese P, Lombardo A, Magnani Z, Liu PQ, Reik A, et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nature medicine. 2012;18(5):807–15. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berdien B, Mock U, Atanackovic D, Fehse B. TALEN-mediated editing of endogenous T-cell receptors facilitates efficient reprogramming of T lymphocytes by lentiviral gene transfer. Gene therapy. 2014;21(6):539–48. doi: 10.1038/gt.2014.26. [DOI] [PubMed] [Google Scholar]
- 30.Beane JD, Lee G, Zheng Z, Mendel M, Abate-Daga D, Bharathan M, et al. Clinical Scale Zinc Finger Nuclease-mediated Gene Editing of PD-1 in Tumor Infiltrating Lymphocytes for the Treatment of Metastatic Melanoma. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23(8):1380–90. doi: 10.1038/mt.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torikai H, Reik A, Liu PQ, Zhou Y, Zhang L, Maiti S, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119(24):5697–705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torikai H, Reik A, Soldner F, Warren EH, Yuen C, Zhou Y, et al. Toward eliminating HLA class I expression to generate universal cells from allogeneic donors. Blood. 2013;122(8):1341–9. doi: 10.1182/blood-2013-03-478255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valton J, Guyot V, Marechal A, Filhol JM, Juillerat A, Duclert A, et al. A Multidrug-resistant Engineered CAR T Cell for Allogeneic Combination Immunotherapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2015;23(9):1507–18. doi: 10.1038/mt.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beatty GL, Haas AR, Maus MV, Torigian DA, Soulen MC, Plesa G, et al. Mesothelin-specific chimeric antigen receptor mRNA-engineered T cells induce anti-tumor activity in solid malignancies. Cancer immunology research. 2014;2(2):112–20. doi: 10.1158/2326-6066.CIR-13-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research. 2014;24(1):132–41. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31(9):822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poirot L, Philip B, Schiffer-Mannioui C, Le Clerre D, Chion-Sotinel I, Derniame S, et al. Multiplex Genome-Edited T-cell Manufacturing Platform for “Off-the-Shelf” Adoptive T-cell Immunotherapies. Cancer research. 2015;75(18):3853–64. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 39.Choi EI, Wang R, Peterson L, Letvin NL, Reimann KA. Use of an anti-CD16 antibody for in vivo depletion of natural killer cells in rhesus macaques. Immunology. 2008;124(2):215–22. doi: 10.1111/j.1365-2567.2007.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi EI, Reimann KA, Letvin NL. In vivo natural killer cell depletion during primary simian immunodeficiency virus infection in rhesus monkeys. J Virol. 2008;82(13):6758–61. doi: 10.1128/JVI.02277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–16. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. The New England journal of medicine. 2014;370(10):901–10. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.