Abstract
Objective
The association of obesity susceptibility variants with change in body mass index (BMI) across the life course is not well understood.
Subjects
In ancestry stratified models of 5,962 European American (EA), 2,080 African American (AA), and 1,582 Hispanic American (HA) individuals from the National Longitudinal Study of Adolescent to Adult Health (Add Health), we examined associations between 34 obesity SNPs with per year change in BMI, measured by the slope from a growth-curve analysis of two or more BMI measurements between adolescence and young adulthood. For SNPs nominally associated with BMI change (p<0.05), we interrogated age differences within data collection Wave and time differences between age categories that overlapped between Waves.
Results
We found SNPs in/near FTO, MC4R, MTCH2, TFAP2B, SEC16B, and TMEM18 were significantly associated (p<0.0015 ≈ 0.05/34) with BMI change in EA and the ancestry-combined meta-analysis. Rs9939609 in FTO met genome-wide significance at p<5e-08 in the EA and ancestry combined analysis, respectively [Beta(se)=0.025(0.004);Beta(se)=0.021(0.003)]. No SNPs were significant after Bonferroni correction in AA or HA, although 5 SNPs in AA and 4 SNPs in HA were nominally significant (p<0.05). In EA and the ancestry-combined meta-analysis, rs3817334 near MTCH2 showed larger effects in younger respondents, while rs987237 near TFAP2B, showed larger effects in older respondents across all Waves. Differences in effect estimates across time for MTCH2 and TFAP2B are suggestive of either era or cohort effects.
Conclusion
The observed association between variants in/near FTO, MC4R, MTCH2, TFAP2B, SEC16B, and TMEM18 with change in BMI from adolescence to young adulthood suggest that the genetic effect of BMI loci varies over time in a complex manner, highlighting the importance of investigating loci influencing obesity risk across the life course.
Keywords: Gene-environment interactions, adolescence, obesity, BMI change, African-American, Hispanic-American
INTRODUCTION
The transition from adolescence to young adulthood is a period of high risk for weight gain and development of obesity [1–3], with high rates of incident obesity (24.1%) and severe obesity (7.9%) [3]. Genome-wide association (GWAS) studies of over 200,000 European descent adults, have identified several independent loci associated with BMI [4, 5]. While recent studies have analyzed BMI loci across the lifecycle [6–14], few longitudinal studies have examined BMI loci associated with change in body mass index (BMI) in the period between adolescence and young adulthood. Several studies have examined the association of FTO and MC4R, two of the earliest identified BMI susceptibility variants, with longitudinal change in BMI from childhood into adulthood [6–8, 15]. Results for BMI change with FTO and MC4R suggest stronger estimated effects encompassing the early adolescent to young adult period compared to later in adulthood [6, 7, 15]. Further, childhood growth trajectories from age one to 16 years have been shown to be associated with established BMI susceptibility loci as a risk score [13], suggesting that genetic determinants associated with BMI at cross-sectional periods are also associated with changes in BMI across the life course. Studies of the influence of genetic variants on BMI change have not yet been interrogated during the late adolescent to early adulthood transition, a developmental period associated with substantial BMI change. More research is warranted to determine the impact of BMI genetic variants across this phase of the life course.
In this study, we examined the association between 34 established BMI variants and change in BMI derived from measured height and weight assessed from adolescence to early adulthood in youth enrolled in an ethnically diverse, nationally representative cohort, the National Longitudinal Study of Adolescent to Adult Health (Add Health), followed for 13 years across three time points, which we call Waves, between 1996 and 2008 when the cohort fell between the ages of 13 years and 34 years. We hypothesized that carriers of obesity risk alleles would have a greater rate of change in increasing BMI across the adolescent and early-adulthood time period. In addition, because of increasing environmental influences that might mask genetic ones as we age, we speculated that some variants might have comparatively stronger estimated effects at earlier versus later ages of the period between adolescence and young adulthood.
METHODS
Add Health
Participants
Add Health is a nationally representative school-based cohort of US adolescents (Wave I: 1994–95, n=20,745, aged 11–20 y, mean age 15.9 y) drawn from a probability sample of 80 high schools and 52 middle schools, representative of US schools in 1994–95 with respect to region, urban setting, school size and type, and race or ethnic background. Wave II (1996, n = 14,738, aged 12–21 y, mean age 16.5 y) included by design Wave I adolescents still of school age, including those currently in high school and high school dropouts. Oversampled subgroups include related and non-related adolescents sharing a Wave I household (n=5,524 Wave I respondents living in 2,639 households) [16] and several race/ethnic subpopulations, including Chinese, Cubans, Puerto Ricans, and Filipinos. Wave III (2001–2002, n= 15,197, aged 18–27 y, mean age 22.3 y) and Wave IV (2008–2009, n=15,701, aged 23–32 y, mean age 28.9 y) followed all Wave I respondents, regardless of Wave II participation. The most recent data collection (Wave IV) included follow-up interviews from 15,701 respondents drawn from 19,962 of the original 20,745 Wave I respondents and included DNA collection and banking for future studies. Survey procedures have been described elsewhere [17–19], and were approved by the Institutional Review Board, University of North Carolina at Chapel Hill.
Literature-based SNPs and Genotyping
We selected 34 SNPs associated with BMI reported by the Genetic Investigation of ANthropometric Traits (GIANT) consortium (Supplementary Table 1) and other studies in European adults [4, 20–24]. Familial relationships were classified according to participant and parental self-report. Twin zygosity was confirmed by 11 molecular genetic markers [25]. Genotyping was performed using TaqMan assays and the ABI Prism 7900® Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Sequences for primers and TaqMan probes are available upon request. Procedures for genotyping (call rate 98%, discordance 0.3%) have been detailed elsewhere [26].
Ancestry
Ancestry was constructed using race and ethnic background and family relationship status (i.e. country of origin, ancestry, and adoption): European American (EA), African American (AA), and Hispanic American (HA), with indicators for subpopulation (e.g., Mexican, Cuban) and immigrant status (e.g., US and non-US born), given differences in BMI by immigrant status [27, 28].
Analysis sample
We included individuals with phenotype data in at least two Waves of data collection, and those that had at least 80% of the SNPs genotyped (n=10,710). In addition, we excluded one from each twin pair (n=142 pairs) with the fewest genotypes SNPs, Asian, Native American or other race/ethnicity (n=719); disabled (n=76); and individuals missing covariate data (n=125) or pregnant at all BMI observations available (n=18). BMI was set to missing for women who were pregnant at a particular observation. The final analytic sample included 9,624 individuals measured at Waves II, III, and/or IV with DNA data (Supplementary Figure 1). Wave I included only self-reported height and weight and was only one year prior to Wave II, thus we did not include these self-reported observations. Within each of three ethnic groups, sample sizes were comprised of 5,962 EA, 2,080 AA, and 1,582 HA. When analyzing AAs, we excluded 12 of 34 SNPs (rs2568958, rs1514175, rs1555543, rs887912, rs2890652, rs13078807, rs2112347, rs4929949, rs4771122, rs11847697, rs571312, rs29941; see notes in all tables) that did not show evidence of association at p<0.20 and consistent direction of effect in AA GWAS [29, 30]. Given the lack of large GWAS in Hispanics, and the observation that 75% of GWAS SNPs for complex traits were replicated in Hispanics [31], all 34 SNPs were considered in analyses of this population.
Statistical analysis
Outcome Measure: Change in Body Mass Index (BMI)
BMI (kg/m2) was calculated from measured height and weight taken at Waves II, III and IV during in-home surveys using standardized procedures. Self-reported height and weight (r=0.95/0.94 with measured weight/height [32]) were substituted for those refusing measurement and/or weighing more than scale (Health-o-meter 844KL digital scale, Jarden Corporation; Rye, NY) capacity (n=163 at Wave II, n=371 at Wave III, and n=82 at Wave IV). The maximum scale at all waves was 200 kg / 440 lbs maximum. We used BMI rather than Z-scores as is common in the literature [33–35], which allows more straightforward interpretation. Individual participant linear slopes were derived using the best linear unbiased prediction method (“BLUP”), regressing BMI on age as both a fixed and random effect. We also adjusted for current smoking at the indicated Wave, and an indicator for whether or not height and weight was self-reported as fixed effects. Models were run by sex to account for sex-specific differences in weight across time and by ancestry to account for ancestry differences in weight gain over time. Of the 9,624 individuals in the analyses, 6,403 had 3 BMI measurements used to create the slope and 3,221 had 2 BMI measurements (1,296 with BMI at Wave II and Wave IV, 1,913 with BMI at Wave III and Wave IV, and 12 with BMI at Wave II and Wave III) to create their slope.
Association analyses
Ancestry-stratified association analyses between change in BMI and SNP genotype were conducted using linear mixed models incorporated in Stata, version 12.1 (Stata Corp, College Station, TX. Each SNP was modeled under an additive model, with SNPs scored for the number of copies of the established (from prior GWAS) risk allele or ‘BMI-increasing’ allele. Covariates included baseline age, sex, geographic region, oversampling of highly educated AAs (n=520), Hispanic ancestry: Cuban (n=284), Puerto Rican (n=275), Central/South American (n=160), Mexican (n=863), other Hispanic (n=102), and if the participant was foreign born (n=432). Study design effects and relatedness were accounted for using random effects for school and family (of the 9,624 total individuals, 1744 (18%) were related or shared a household with another individual in the sample). Effect estimates were combined and meta-analyzed in METAL using the inverse standard-error weighted approach [36]. For each SNP association, we evaluated heterogeneity between race/ethnic groups using Cochran’s Q. We considered evidence for heterogeneity when the chi-square p <0.10 or I2 index >50[37, 38]. While we examined all nominally significant findings (p<0.05), we corrected for multiple testing: α = 0.05/number of SNPs tested (p≤0.0015 in EA, p≤0.0015 in HA, and p≤0.0023 in AA, and p≤0.0015 in the combined meta-analyzed sample). We also ran models without including any SNPs (Supplementary Table 2).
For SNPs with nominally significant effects (p<0.05) on the change in BMI, we interrogated two additional sets of analyses to assess whether the effects on change in BMI were due to different effects across age groups or points in time, the latter for which we use Wave of data collection. Given the narrow age range among participants (approximately 10 year age-span) within each Wave of data collection, cohort and age effects are highly collinear. Thus, to interrogate whether SNP effects vary by age, we performed SNP-by-age interaction analyses on cross-sectional BMI at each Wave. This set of analyses considered that a locus might have a stronger effect in younger compared to older individuals within the full age range studied (e.g., 12–21 years at Wave II) or vice versa. To keep the sample size constant when testing for the SNP-by-age interaction at each Wave, we included a subsample of participants with measured anthropometry at Waves II, III and IV. This reduced our sample size to 6,190 (3,155 females and 3,035 males). To aid in interpretation of the SNP-by-age interaction results, we plotted the results by year of age at each Wave, except for ages 13–14 and ages 18–21, which we combined due to smaller sample sizes. Second, we examined the main genetic effects of each SNP on cross-sectional BMI between individuals at similar ages but at a different Wave of data collection Wave, to verify that the main effect of the SNP on BMI is changing from one Wave to the next (i.e., across time periods). We attempted to age match the groups when testing the same ages between 2 different Waves (i.e. time points). Given the small sample sizes we combined age groups that overlapped including ages 18–20 as one group, both in Wave II and Wave III, and ages 25–26 as a second group, both in Wave III and Wave IV. Then we ran interaction models by testing the SNP×Wave effect separately in each of the two age groupings.
RESULTS
The participants (47% females) were an average of 16.1 years of age (ranging from 13–21 years) in 1996 (Wave II) and 28.5 years (ranging from 25–34 years) in 2008 (Wave IV). Mean BMI in 1996 ranged from 22.9±4.9 kg/m2 in EA to 24.1±5.7 kg/m2 in AA, while in 2008 mean BMI ranged from 28.5±7.1 kg/m2 in EA to 30.5±8.4 kg/m2 in AA (Table 1). The average change per year was largest in AA, 0.54±28 kg/m2 and smallest in EA, 0.45±22 kg/m2. Analyses using a model without SNPs (Supplementary Table 2) showed significant associations with age, sex, and in most cases random effect parameters. Sampling based on education in Africans was not significant. Foreign born and ancestry variables in Hispanics were not significant except that Cuba had significantly lower change in BMI compared to Puerto Ricans (the referent).
Table 1.
Descriptive information for the Add Health analytic sample, by race/ethnicity and in the total sample
| European Americans (N=5,962) | African Americans (N=2,080) | Hispanic Americans (N=1,582) | Total Sample (N=9,624) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Females | Males | Females | Males | Females | Males | Females | Males | |||||||||
| Variable | N | Mean (SD) / % | N | Mean (SD) / % | N | Mean (SD) / % | N | Mean (SD) / % | N | Mean (SD) / % | N | Mean (SD) / % | N | Mean (SD) / % | N | Mean (SD) / % |
| Age in Wave II, years | 2,532 | 15.94 (1.6) | 2,304 | 16.24 (1.63) | 882 | 16.08 (1.63) | 744 | 16.21 (1.67) | 624 | 16.35 (1.66) | 625 | 16.49 (1.60) | 4,038 | 16.03 (1.62) | 3,673 | 16.27 (1.64) |
| Age in Wave III, years | 2,746 | 21.69 (1.77) | 2,445 | 21.95 (1.76) | 991 | 21.74 (1.71) | 797 | 21.96 (1.84) | 670 | 22.15 (1.74) | 679 | 22.27 (1.76) | 4,407 | 21.77 (1.76) | 3,921 | 22.01 (1.78) |
| Age in Wave IV, years | 2,987 | 28.18 (1.76) | 2,831 | 28.43 (1.75) | 1,103 | 28.29 (1.75) | 929 | 28.44 (1.83) | 766 | 28.64 (1.75) | 783 | 28.78 (1.72) | 4,856 | 28.28 (1.76) | 4,543 | 28.49 (1.77) |
| BMI in Wave II, kg/m2 | 2,532 | 22.66 (4.9) | 2,304 | 23.09 (4.76) | 882 | 24.45 (6.03) | 744 | 23.51 (5.07) | 624 | 23.57 (5.32) | 625 | 24.08 (5.10) | 4,038 | 23.19 (5.29) | 3,673 | 23.34 (4.90) |
| BMI in Wave III, kg/m2 | 2,746 | 25.96 (6.43) | 2,445 | 26.28 (5.48) | 991 | 28.4 (7.68) | 797 | 26.61 (6.01) | 670 | 27.14 (6.47) | 679 | 27.42 (5.55) | 4,407 | 26.69 (6.81) | 3,921 | 26.55 (5.62) |
| BMI in Wave IV, kg/m2 | 2,987 | 28.3 (7.74) | 2,831 | 28.66 (6.41) | 1,103 | 31.83 (9.11) | 929 | 29.01 (7.13) | 766 | 29.58 (7.73) | 783 | 30.23 (6.67) | 4,856 | 29.30 (8.20) | 4,543 | 29.00 (6.63) |
|
One year change (slope) in BMI, kg/m2 |
3,131 | 0.44 (0.26) | 2,831 | 0.46 (0.17) | 1,145 | 0.61 (0.28) | 935 | 0.45 (0.24) | 797 | 0.47 (0.24) | 785 | 0.51 (0.22) | 5,073 | 0.48 (0.27) | 4,551 | 0.46 (0.20) |
| Region, N (%) | ||||||||||||||||
| West | 484 | 15.46 | 411 | 14.52 | 158 | 13.8 | 140 | 14.97 | 342 | 42.91 | 305 | 38.85 | 984 | 19.4 | 856 | 18.8 |
| Midwest | 1,142 | 36.47 | 1,032 | 36.45 | 215 | 18.78 | 172 | 18.4 | 59 | 7.4 | 58 | 7.39 | 1416 | 27.9 | 1,262 | 27.7 |
| South | 1,026 | 32.77 | 976 | 34.48 | 700 | 61.14 | 575 | 61.5 | 300 | 37.64 | 310 | 39.49 | 2026 | 39.9 | 1,861 | 40.9 |
| Northeast | 479 | 15.3 | 412 | 14.55 | 72 | 6.29 | 48 | 5.13 | 96 | 12.05 | 112 | 14.27 | 647 | 12.8 | 572 | 12.6 |
Abbreviations: BMI (body mass index), SD (standard deviation)
In EA, FTO SNP rs9939609 was genome-wide significantly associated (p=2.42e-09) with the slope of BMI (e.g. change in BMI over time), suggesting a 0.025 kg/m2 per year of age increase in BMI for each additional copy of the established risk allele compared to non-carriers (Table 2). Similar results were seen for variants (in/near) rs571312 (MC4R), rs3817334 (MTCH2), rs6548238 (TMEM18), rs987237 (TFAP2B), and rs543874 (SEC16B), with estimated effect sizes suggesting increases in BMI ranging from 0.014 to 0.025 kg/m2 per year for each additional copy of their respective established risk alleles as compared to non-risk allele carriers (Table 2). These results were significant after correcting for 34 tests (p<0.0015). Thirty-two of 34 SNPs displayed positive effect estimates on BMI change (consistent with the established risk alleles being associated with higher cross-sectional BMI in prior reports) and 13 of these were at least nominally significant, which is more than expected by chance alone (binomial p=3.9E-09). In AA, no SNPs were significantly associated with BMI slope. However, 17 of 24 tested had positive beta estimates and 5 of these variants in/near SEC16B (rs543874), GNPDA2 (rs10938397), ETV5 (rs7647305), LRRN6C (rs10938397), and MAP2K5 (rs2241423) were also nominally significant, which is more than expected by chance (binomial p=0.005). In HA, no SNPs were significantly associated with BMI slope, but 22 of 34 SNPs had positive beta estimates, and four were nominally significant (rs1514175, rs543874, rs9939609, rs12444979, respectively in/near TNNI3K, SEC16B, FTO and GPRC5B). In the all ancestry meta-analysis, 31 of 34 SNPs had positive effect estimates for the established risk allele on change in BMI and six SNPs in/near MC4R (rs571312), MTCH2 (rs3817334), TMEM18 (rs6548238), TFAP2B (rs987237), FTO (rs9939609), and SEC16B (rs543874) displayed significant associations after correcting for multiple testing (Figure 1). Again, FTO met genome-wide significance. While the statistical significance estimates in the meta-analyses for these 6 loci were dominated by the larger EA results, effect estimates were largely consistent across ethnic groups. Two notable exceptions were for FTO SNP rs9939609 and TFAP2B SNP rs987237, where the effect estimates in AA were noticeably smaller compared to EA and HA (Table 2), possibly because these SNPs are not tagging the relevant signal in Africans. We calculated the variance explained by each SNP based on our meta-analysis results. We calculated the variance explained for a one year change in BMI for each SNP based on our European ancestry analysis results (Table 2). The cumulative variance explained by the six SNPs that met Bonferroni significance or by the 15 SNPs that met nominal significance (p<0.05), is 0.09% 0.37%, respectively.
Table 2.
Association results for the slope of BMIa with each SNP in the Add Health participants by race/ethnicity and in the combined meta-anlayzed sample.b
| SNP information | European ancestry | African ancestryc | Hispanic ancestry | Meta-analyzed sample, All Ancestriesd | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr | BP | Effect/ other allele e |
EAF | Beta(SE) slope of BMI as kg/m2 per year |
Pvalue | N | EAF | Beta(SE) slope of BMI as kg/m2 per year |
Pvalue | N | EAF | Beta(SE) slope of BMI as kg/m2 per year |
Pvalue | N | EAF | Beta(SE) slope of BMI as kg/m2 per year |
Pvalue | N | HetISq | Variance explained f |
| rs2568958 | NEGR1 | 1 | 72765116 | A/G | 0.6283 | 0.009 (0.004) | 0.0363343449806959 | 5899 | 0.6946 | −0.006 (0.009) | 0.537930934892836 | 1571 | 0.6409 | 0.006 (0.004) | 0.1026 | 7470 | 52.3 | 0.0008 | ||||
| rs1514175 | TNNI3K | 1 | 74991644 | A/G | 0.44 | 0.006 (0.004) | 0.120431702081817 | 5928 | 0.5349 | 0.017 (0.008) | 0.0426338583368813 | 1572 | 0.458 | 0.008 (0.004) | 0.02171 | 7500 | 20.2 | 0.00001 | ||||
| rs1555543 | PTBP2 | 1 | 96944797 | C/A | 0.5873 | 0.01 (0.004) | 0.0150685657529825 | 5915 | 0.5688 | 0.007 (0.008) | 0.395956543682014 | 1563 | 0.5861 | 0.009 (0.004) | 0.0106 | 7478 | 0 | 0.0007 | ||||
| rs543874 | SEC16B, LZTR2 | 1 | 177889480 | G/A | 0.1993 | 0.019 (0.005) | 0.000137710423790072 | 5927 | 0.241 | 0.027 (0.01) | 0.00512916970442667 | 2064 | 0.1875 | 0.024 (0.01) | 0.018689836407328 | 1567 | 0.2058 | 0.022 (0.004) | 0.0000001724 | 9558 | 0 | 0.0004 |
| rs6548238 | TMEM18 | 2 | 634905 | C/T | 0.8297 | 0.024 (0.005) | 8.31800216993805E-06 | 5937 | 0.8928 | 0.012 (0.013) | 0.390540350175295 | 2070 | 0.8695 | 0.019 (0.012) | 0.113699082451912 | 1572 | 0.8429 | 0.022 (0.005) | 0.000002315 | 9579 | 0 | 0.00001 |
| rs713586 | POMC | 2 | 25158008 | C/T | 0.4837 | 0.009 (0.004) | 0.0299801260334576 | 5901 | 0.8423 | −0.007 (0.011) | 0.520232429627132 | 2060 | 0.4306 | −0.014 (0.008) | 0.097795157849865 | 1564 | 0.5047 | 0.003 (0.004) | 0.3457 | 9525 | 71.3 | 0.00004 |
| rs887912 | FANCL | 2 | 59302877 | T/C | 0.2803 | 0.003 (0.005) | 0.491160421613799 | 5930 | 0.1879 | 0.004 (0.011) | 0.707969631936108 | 1575 | 0.2663 | 0.003 (0.004) | 0.4354 | 7505 | 0 | 0.0001 | ||||
| rs2890652 | LRP1B | 2 | 142959931 | C/T | 0.1642 | 0.007 (0.006) | 0.217162797065839 | 5920 | 0.1261 | 0.009 (0.012) | 0.448557465796797 | 1569 | 0.1548 | 0.007 (0.005) | 0.1506 | 7489 | 0 | 0.0001 | ||||
| rs13078807 | CADM2 | 3 | 85884150 | G/A | 0.1981 | 0.01 (0.005) | 0.0474965470274247 | 5925 | 0.147 | −0.005 (0.012) | 0.683265705532463 | 1576 | 0.192 | 0.008 (0.005) | 0.09861 | 7501 | 26.8 | 0.0002 | ||||
| rs7647305 | ETV5 | 3 | 185834290 | C/T | 0.7916 | 0.006 (0.005) | 0.234131380762947 | 5916 | 0.597 | 0.021 (0.008) | 0.00933554847364504 | 2062 | 0.8128 | 0.002 (0.01) | 0.864315826922167 | 1568 | 0.749 | 0.009 (0.004) | 0.02417 | 9546 | 35.9 | 0.00001 |
| rs10938397 | GNPDA2 | 4 | 45182527 | G/A | 0.4303 | 0.008 (0.004) | 0.0644008335362849 | 5914 | 0.2411 | 0.031 (0.01) | 0.0013579608823584 | 2067 | 0.3709 | 0.001 (0.009) | 0.8868425665692 | 1571 | 0.3955 | 0.01 (0.004) | 0.005698 | 9552 | 67 | 0.0007 |
| rs13107325 | SLC39A8 | 4 | 103188709 | T/C | 0.08068 | 0.012 (0.007) | 0.105182944382441 | 5938 | 0.0154 | 0.006 (0.034) | 0.851949875091347 | 2073 | 0.04119 | 0.005 (0.021) | 0.822042856372413 | 1576 | 0.0732 | 0.011 (0.007) | 0.1084 | 9587 | 0 | 0.00001 |
| rs2112347 |
FLJ3577, HMGCR |
5 | 75015242 | T/G | 0.6334 | 0.004 (0.004) | 0.386300046195639 | 5909 | 0.6295 | 0.013 (0.009) | 0.123905991663066 | 1572 | 0.63 | 0.006 (0.004) | 0.1455 | 7481 | 0 | 0.0005 | ||||
| rs1077393 |
NCR3,
AIF1, BAT2 |
6 | 31610529 | G/A | 0.4867 | 0.002 (0.004) | 0.655112788718038 | 5883 | 0.345 | −0.001 (0.009) | 0.904136178479392 | 2052 | 0.4852 | −0.006 (0.008) | 0.488099423487185 | 1562 | 0.4687 | 0.0001 (0.003) | 0.9701 | 9497 | 0 | 0.0001 |
| rs206936 | NUDT3 | 6 | 34302869 | G/A | 0.2098 | 0.006 (0.005) | 0.218320267301597 | 5939 | 0.5357 | −0.002 (0.008) | 0.82655403883952 | 2067 | 0.3989 | 0.006 (0.008) | 0.497745024011879 | 1568 | 0.3196 | 0.004 (0.004) | 0.2523 | 9574 | 0 | 0.0001 |
| rs987237 | TFAP2B | 6 | 50803050 | G/A | 0.176 | 0.024 (0.005) | 7.96402874936319E-06 | 5936 | 0.09875 | −0.011 (0.014) | 0.42707797684036 | 2070 | 0.2718 | 0.007 (0.009) | 0.457200851773043 | 1572 | 0.1925 | 0.016 (0.004) | 0.0001747 | 9578 | 71.6 | 0.0004 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.3103 | 0.011 (0.004) | 0.0137325873208902 | 5888 | 0.1722 | 0.026 (0.011) | 0.0129824960281197 | 2058 | 0.2352 | 0.005 (0.01) | 0.605760679610998 | 1558 | 0.2825 | 0.012 (0.004) | 0.00151 | 9504 | 18.3 | 0.0001 |
| rs867559 | LMX1B | 9 | 129465325 | G/A | 0.1912 | 0.007 (0.005) | 0.181945816557514 | 5926 | 0.2988 | 0.006 (0.009) | 0.536527080574311 | 2063 | 0.3307 | 0.008 (0.008) | 0.356649140521792 | 1575 | 0.2409 | 0.007 (0.004) | 0.08423 | 9564 | 0 | 0.0003 |
| rs4929949 | RPL27A, TUB | 11 | 8604593 | C/T | 0.5077 | 0.005 (0.004) | 0.225799407987264 | 5904 | 0.008 (0.008) | 0.312288924912817 | 2065 | 0.4853 | 0.006 (0.008) | 0.455108051652104 | 1562 | 0.506 | 0.005 (0.004) | 0.1565 | 7466 | 0 | 0.0005 | |
| rs10767664 | BDNF | 11 | 27725986 | A/T | 0.7888 | 0.006 (0.005) | 0.266481925639808 | 5921 | 0.93374 | 0.002 (0.017) | 0.895613080276965 | 2064 | 0.8079 | 0.007 (0.01) | 0.510800725004547 | 1566 | 0.8031 | 0.006 (0.004) | 0.2019 | 9551 | 0 | 0.00001 |
| rs3817334 | MTCH2 | 11 | 47650993 | T/C | 0.4026 | 0.014 (0.004) | 0.000597839313208892 | 5901 | 0.2601 | 0.006 (0.009) | 0.531830133787902 | 2067 | 0.3894 | 0.005 (0.008) | 0.537861556694164 | 1568 | 0.3795 | 0.011 (0.003) | 0.0008378 | 9536 | 0 | 0.00001 |
| rs7138803 | FAIM2 | 12 | 50247468 | A/G | 0.3782 | 0.008 (0.004) | 0.0496305504077867 | 5820 | 0.1689 | 0.013 (0.011) | 0.235457647729183 | 2045 | 0.2715 | −0.011 (0.009) | 0.260324258382214 | 1543 | 0.3405 | 0.006 (0.004) | 0.1003 | 9408 | 47.8 | 0.0003 |
| rs4771122 | MTIF3 | 13 | 28020180 | G/A | 0.2199 | 0.006 (0.005) | 0.181961568445123 | 5906 | 0.2016 | 0.009 (0.01) | 0.408092615576273 | 1574 | 0.2164 | 0.007 (0.004) | 0.1189 | 7480 | 0 | 0.0011 | ||||
| rs11847697 | PRKD1 | 14 | 30515112 | T/C | 0.05325 | 0.023 (0.009) | 0.00957949081714604 | 5939 | 0.07183 | 0.024 (0.016) | 0.126117365353668 | 1580 | 0.055 | 0.024 (0.008) | 0.00263 | 7519 | 0 | 0.0001 | ||||
| rs10146997 | NRXN3 | 14 | 79945162 | G/A | 0.7909 | −0.007 (0.005) | 0.165654039718296 | 5943 | 0.6413 | 0.005 (0.008) | 0.520134912588576 | 2070 | 0.7901 | −0.004 (0.01) | 0.659218936596024 | 1580 | 0.7565 | −0.004 (0.004) | 0.336 | 9593 | 0 | 0.00001 |
| rs2241423 | MAP2K5 | 15 | 68086838 | G/A | 0.7672 | 0.01 (0.005) | 0.0416373735766462 | 5930 | 0.6242 | 0.022 (0.008) | 0.00800851227675126 | 2068 | 0.5804 | −0.01 (0.008) | 0.239675502326333 | 1575 | 0.7025 | 0.008 (0.004) | 0.02577 | 9573 | 73.7 | 0.0001 |
| rs2444217 | ADCY9 | 16 | 4038387 | A/G | 0.57 | 0.001 (0.004) | 0.809051948917954 | 5924 | 0.76 | −0.002 (0.01) | 0.870364048427314 | 2062 | 0.43 | −0.007 (0.008) | 0.395081963932239 | 1568 | 0.5694 | −0.001 (0.003) | 0.8358 | 9554 | 0 | 0.00001 |
| rs12444979 | GPRC5B | 16 | 19933600 | C/T | 0.8562 | 0.008 (0.006) | 0.143422025867003 | 5901 | 0.91064 | 0.027 (0.014) | 0.0590406874232221 | 2062 | 0.90834 | 0.039 (0.014) | 0.00471861566537712 | 1570 | 0.8727 | 0.015 (0.005) | 0.003194 | 9533 | 60 | 0.0001 |
| rs4788102 | SH2B1 | 16 | 28873398 | A/G | 0.3923 | 0.011 (0.004) | 0.00550841945096625 | 5928 | 0.2801 | 0.013 (0.009) | 0.153339943320713 | 2061 | 0.4032 | −0.002 (0.008) | 0.812178517765675 | 1573 | 0.3758 | 0.009 (0.003) | 0.006001 | 9562 | 11.1 | 0.0007 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.3929 | 0.025 (0.004) | 2.41962405667095E-09 | 5907 | 0.4731 | 0.004 (0.008) | 0.590061046533582 | 2067 | 0.3283 | 0.025 (0.009) | 0.00475208845717967 | 1570 | 0.3952 | 0.021 (0.003) | 0.0000000005377 | 9544 | 62.5 | 0.0002 |
| rs571312 | MC4R | 18 | 57839769 | A/C | 0.2347 | 0.018 (0.005) | 0.000145767338018077 | 5923 | 0.1638 | 0.017 (0.011) | 0.125813734 | 1571 | 0.2188 | 0.018 (0.004) | 0.0000425 | 7494 | 0 | 0.0001 | ||||
| rs29941 | KCTD15 | 19 | 34309532 | G/A | 0.6803 | 0.004 (0.004) | 0.315950807178992 | 5900 | 0.6435 | 0.015 (0.009) | 0.125813734270379 | 1563 | 0.672 | 0.006 (0.004) | 0.09793 | 7463 | 12.3 | 0.0001 | ||||
| rs2287019 | QPCTL | 19 | 46202172 | C/T | 0.8167 | 0.011 (0.005) | 0.0374216237136888 | 5932 | 0.8833 | 0.003 (0.013) | 0.839650890441376 | 2064 | 0.8702 | −0.005 (0.012) | 0.657435105815855 | 1574 | 0.8343 | 0.008 (0.005) | 0.09055 | 9570 | 0 | 0.0008 |
| rs3810291 | TMEM160 | 19 | 47569003 | A/G | 0.6685 | 0.001 (0.004) | 0.872282995109245 | 5915 | 0.208 | 0.002 (0.01) | 0.843750665430601 | 2060 | 0.5625 | 0.011 (0.008) | 0.203871204936517 | 1568 | 0.5938 | 0.003 (0.004) | 0.4567 | 9543 | 0 | 0.0001 |
Abbreviations: SNP (single nucleotide polymorphism), EAF: Effect Allele Frequency, Chr: Chromosome, BP: base pair position based on NCBI build 37 (hg19), BMI (body mass index), SE (standard error)
Individual participant linear slopes for BMI change were derived using the best linear unbiased prediction method (“BLUP”), regressing BMI on age as both a fixed and random effect. We also adjusted for current smoking at each Wave, and an indicator for whether or not height and weight was self-reported as fixed effects.
Beta and standard error (SE) estimates are presented for the slope of BMI change: Multivariable linear models of slope of BMI regressed baseline age, sex, geographic region. Models in Hispanic Americans also adjusted for an indicator for foreign born and country of origin, and models in African Americans also adjusted for an indicator for selection for high education group (oversampling for education). Random intercepts allowed for individual, family and school. Models were run separately for each SNP.
SNPs that are missing in African American stratum were excluded because it did not generalize based on published African American GWAS (Monda et al. 2012).
Meta-analyzed results were done in METAL software (Willer et al 2012), by combining effect estiamtes from each of the 3 ancestries shown using the inverse-weighted approach.
Effect allele oriented to BMI increasing allele
The variance explained by SNP was calcuated in European ancestry individuals and based on the R-squared after regressing each SNP on the residual of the slope. The residual was calculated by adjusted for fixed effects of baseline age, smoking, gender, self-reported height or weight, and region, and random effects for school and family.
Figure 1.
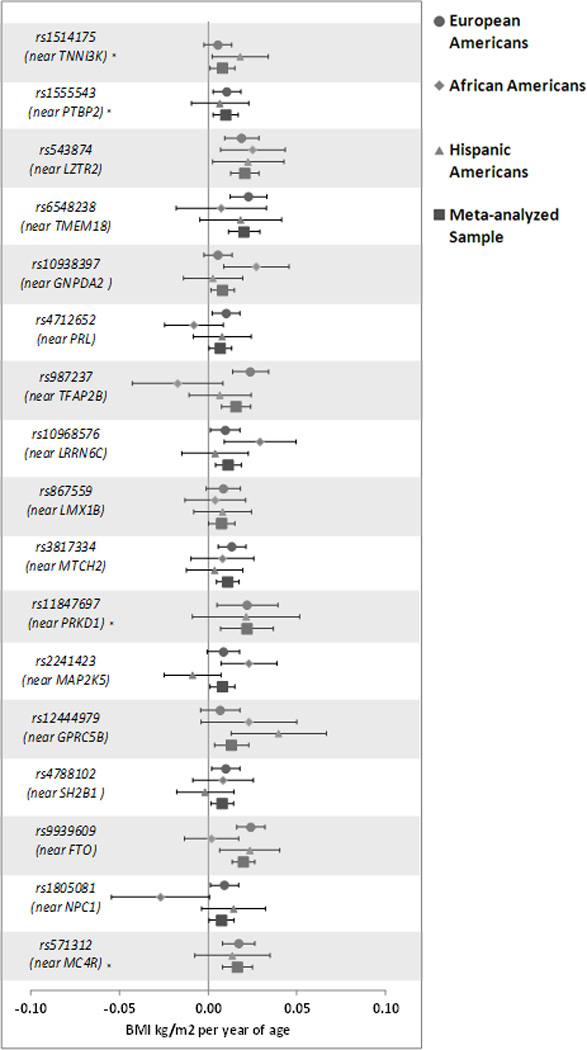
For SNPs that achieved statistical significance at p<0.05 across the meta-anlayzed sample, effect estimates for per allele change in slope of BMI across adolescence to young adulthood for the Add Health cohort by ethnic/race group and combined.
*Denotes SNPs that do not generalize to African Americans and thus were not considered.
We next considered whether the loci significantly associated (p<0.05) with change in BMI within each ancestry might have different magnitudes of effect at younger compared to older ages. Therefore, we tested a SNP-by-age interaction on cross-sectional BMI at each of Waves II, III and IV for nine SNPs in EA and two SNPs each in AA and HA. In EA, we found two interactions that remained statistically significant after correction for multiple testing (p<0.05/9 SNPs=0.0056); a negative estimated SNP-by-age interaction effect on BMI at Wave II for rs3817334 (in MTCH2; βinteraction±[se] = −0.192[0.065]) and a positive estimated interaction effect for rs987237 (in TFAP2B; βinteraction±[se] = 0.308[0.085]) (Table 3 and Supplementary Table 3). Thus, the SNP near TFAP2B had a stronger influence on BMI in EA adolescents who were older, while MTCH2 had a stronger influence on BMI in EA adolescents who were younger. No other SNP-by-age interactions on BMI at any Wave were significant, although SNP-by-age interaction estimates for MTCH2 and TFAP2B on BMI were consistent in direction of effect but smaller in magnitude at Waves III and IV compared to Wave II.
Table 3.
The SNPx Age association results with cross-sectional BMI at each wave for SNPs associated with slope of BMI at Pvalue<0.05 within each race/ethnic group and the meta-analyzed sample.a
| Wave 2 | Wave 3 | Wave 4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr | BP (hg19) | Effect/ other alleleb |
EAF | N | Beta(SE) SNPxAge |
Pvalue SNPxAge |
Beta(SE) SNPxAge |
Pvalue SNPxAge |
Beta(SE) SNPxAge |
Pvalue SNPxAge |
| European Ancestry | ||||||||||||
| rs2568958 | NEGR1 | 1 | 72765116 | A/G | 0.6276 | 3833 | −0.095 (0.067) | 0.15839675 | −0.116 (0.085) | 0.174223 | −0.143 (0.099) | 0.15002094 |
| rs1555543 | PTBP2 | 1 | 96944797 | C/A | 0.588 | 3841 | 0.063 (0.065) | 0.33022502 | 0.045 (0.082) | 0.58172959 | 0.079 (0.095) | 0.40798848 |
| rs543874 | SEC16B /LZTR2 | 1 | 177889480 | G/A | 0.1973 | 3847 | −0.038 (0.079) | 0.63267938 | 0.032 (0.102) | 0.75043498 | −0.012 (0.118) | 0.92137839 |
| rs6548238 | TMEM18 | 2 | 634905 | C/T | 0.8298 | 3853 | −0.071 (0.079) | 0.36787556 | −0.059 (0.102) | 0.56182566 | −0.032 (0.117) | 0.78259877 |
| rs713586 | RBJ/ADCY3/ POMC | 2 | 25158008 | C/T | 0.4827 | 3825 | −0.081 (0.064) | 0.20255856 | −0.053 (0.082) | 0.51419235 | −0.177 (0.095) | 0.06133292 |
| rs987237 | TFAP2B | 6 | 50803050 | G/A | 0.174 | 3858 | 0.308 (0.085) | 0.0002692 | 0.172 (0.109) | 0.11352658 | 0.188 (0.125) | 0.13101385 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.3143 | 3823 | −0.036 (0.068) | 0.59219184 | −0.086 (0.087) | 0.32343075 | 0.007 (0.101) | 0.94512972 |
| rs867559 | LMX1B | 9 | 129465325 | G/A | 0.1917 | 3842 | 0.003 (0.078) | 0.96787912 | 0.065 (0.101) | 0.51830464 | 0.08 (0.117) | 0.4948819 |
| rs3817334 | MTCH2 | 11 | 47650993 | T/C | 0.4056 | 3832 | −0.192 (0.065) | 0.00312145 | −0.157 (0.083) | 0.05902117 | −0.124 (0.095) | 0.19345613 |
| rs11847697 | PRKD1 | 14 | 30515112 | T/C | 0.05271 | 3856 | −0.052 (0.135) | 0.69691654 | −0.1 (0.174) | 0.56412162 | −0.027 (0.203) | 0.8952082 |
| rs4788102 | SH2B1 | 16 | 28873398 | A/G | 0.3924 | 3854 | −0.069 (0.064) | 0.28107862 | −0.129 (0.081) | 0.11123961 | −0.178 (0.094) | 0.05777997 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.3923 | 3834 | 0.066 (0.067) | 0.32476931 | 0.074 (0.084) | 0.37519525 | 0.04 (0.098) | 0.67997773 |
| rs571312 | MC4R | 18 | 57839769 | A/C | 0.2345 | 3846 | 0.03 (0.076) | 0.69118432 | 0.065 (0.096) | 0.50288313 | 0.09 (0.111) | 0.41517632 |
| rs2287019 | QPCTL/GIPR | 19 | 46202172 | C/T | 0.8132 | 3852 | −0.047 (0.083) | 0.57410085 | 0 (0.106) | 0.99808789 | 0.052 (0.122) | 0.66672612 |
| African Ancestry | ||||||||||||
| rs543874 | SEC16B /LZTR2 | 1 | 177889480 | G/A | 0.2411 | 1242 | −0.401 (0.156) | 0.01016935 | −0.28 (0.195) | 0.15141702 | −0.424 (0.225) | 0.05926957 |
| rs7647305 | ETV5/SFRS10/DGKG | 3 | 185834290 | C/T | 0.5983 | 1238 | 0.028 (0.136) | 0.83665072 | 0.173 (0.171) | 0.30973571 | 0.04 (0.202) | 0.84448894 |
| rs10938397 | Gene desert;GNPDA2 | 4 | 45182527 | G/A | 0.2465 | 1238 | −0.004 (0.155) | 0.97974832 | 0.017 (0.197) | 0.93280467 | −0.091 (0.227) | 0.68798342 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.1732 | 1237 | −0.006 (0.171) | 0.97360567 | 0.17 (0.216) | 0.43110424 | 0.256 (0.249) | 0.30517407 |
| rs2241423 | MAP2K5/LBXCOR1 | 15 | 68086838 | G/A | 0.621 | 1244 | 0.033 (0.138) | 0.80981145 | 0.02 (0.174) | 0.90700752 | −0.128 (0.199) | 0.52115135 |
| Hispanic Ancestry | ||||||||||||
| rs1514175 | TNNI3K | 1 | 74991644 | A/G | 0.5425 | 955 | −0.127 (0.14) | 0.36563463 | −0.131 (0.167) | 0.43285491 | −0.058 (0.198) | 0.76942165 |
| rs543874 | SEC16B /LZTR2 | 1 | 177889480 | G/A | 0.1864 | 951 | −0.076 (0.18) | 0.67358686 | 0.011 (0.216) | 0.95944761 | −0.321 (0.258) | 0.21238977 |
| rs12444979 | GPRC5B/IQCK | 16 | 19933600 | C/T | 0.91275 | 955 | 0.089 (0.237) | 0.70626811 | −0.317 (0.282) | 0.26089037 | −0.166 (0.333) | 0.61751673 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.3247 | 955 | −0.115 (0.144) | 0.42517715 | −0.035 (0.172) | 0.84068492 | 0.048 (0.203) | 0.81177089 |
| Meta-analyzed sample, all ancestriesc | ||||||||||||
| rs1514175 | TNNI3K | 1 | 74991644 | A/G | 0.504921987105307 | 6051 | 0.037 (0.054) | 0.4869 | 0.014 (0.067) | 0.8402 | 0.076 (0.079) | 0.3348 |
| rs1555543 | PTBP2 | 1 | 96944797 | C/A | 0.554255430649694 | 6034 | −0.029 (0.054) | 0.594 | −0.072 (0.068) | 0.2858 | −0.005 (0.079) | 0.9536 |
| rs543874 | LZTR2 | 1 | 177889480 | G/A | 0.204572292940982 | 6040 | −0.107 (0.066) | 0.1025 | −0.028 (0.083) | 0.7403 | −0.132 (0.097) | 0.1736 |
| rs6548238 | TMEM18 | 2 | 634905 | C/T | 0.850095966275417 | 6053 | −0.01 (0.07) | 0.8867 | 0.013 (0.088) | 0.8798 | 0.043 (0.102) | 0.6765 |
| rs10938397 | GNPDA2 | 4 | 45182527 | G/A | 0.383783088113738 | 6032 | −0.013 (0.056) | 0.8194 | 0.024 (0.07) | 0.7318 | 0.044 (0.081) | 0.5878 |
| rs987237 | TFAP2B | 6 | 50803050 | G/A | 0.173825852206976 | 6058 | 0.253 (0.07) | 0.0003112 | 0.136 (0.088) | 0.1255 | 0.195 (0.102) | 0.05577 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.273630186807737 | 6005 | −0.027 (0.059) | 0.6502 | −0.032 (0.075) | 0.6723 | 0.061 (0.087) | 0.4863 |
| rs867559 | LMX1B | 9 | 129465325 | G/A | 0.23544250289304 | 6041 | −0.009 (0.062) | 0.8858 | −0.003 (0.079) | 0.9732 | 0.021 (0.091) | 0.8227 |
| rs3817334 | MTCH2 | 11 | 47650993 | T/C | 0.373131542403703 | 6026 | −0.14 (0.054) | 0.009652 | −0.127 (0.069) | 0.0641 | −0.116 (0.079) | 0.1431 |
| rs11847697 | PRKD1 | 14 | 30515112 | T/C | 0.112226528351794 | 6058 | −0.104 (0.092) | 0.2561 | −0.077 (0.118) | 0.5112 | −0.005 (0.136) | 0.9684 |
| rs2241423 | MAP2K5 | 15 | 68086838 | G/A | 0.706667680608365 | 6047 | −0.03 (0.06) | 0.6108 | 0.041 (0.075) | 0.5837 | −0.019 (0.087) | 0.8315 |
| rs12444979 | GPRC5B | 16 | 19933600 | C/T | 0.874909173417094 | 6032 | −0.068 (0.079) | 0.3853 | −0.067 (0.099) | 0.4956 | −0.143 (0.114) | 0.2106 |
| rs4788102 | SH2B1 | 16 | 28873398 | A/G | 0.372053116217557 | 6049 | −0.052 (0.054) | 0.3358 | −0.076 (0.068) | 0.2646 | −0.079 (0.079) | 0.3165 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.397909621425029 | 6031 | 0.06 (0.055) | 0.2711 | 0.078 (0.068) | 0.2519 | 0.083 (0.08) | 0.2973 |
| rs571312 | MC4R | 18 | 57839769 | A/C | 0.246872689700777 | 6043 | 0.012 (0.063) | 0.8455 | 0.012 (0.079) | 0.8812 | 0.006 (0.092) | 0.947 |
Abbreviations: SNP (single nucleotide polymorphism), EAF: Effect Allele Frequency, Chr: Chromosome, BP: base pair position based on NCBI build 37 (hg19), SE (standard error)
Beta and standard error (SE) estimates are presented for the SNP×Age interaction model: Multivariable linear models of cross-sectional BMI regressed on SNP and Age in years, with SNP by Age interaction term, controlling for sex, current smoking (at least one cigarette every day for 30 days), geographic region, and self-reported heights and weights (n=39). Random intercepts allowed for family and school with no sample weighting. Models were run separately for each SNP. Models in Hispanic Americans also adjusted for an indicator for foreign born and country of origin, and models in African Americans also adjusted for an indicator for selection for high education group (oversampling for education).
Effect allele oriented to BMI increasing allele.
Meta-analyzed results were done in METAL software (Willer et al 2012), by combining effect estiamtes from each of the 3 ancestries shown using the inverse-weighted approach.
To aid in interpretation of these effect estimates, we estimated the main effect associations between BMI and rs3817334 (in MTCH2) and rs987237 (in TFAP2B), by age group at each Wave (Figure 2). For rs3817334 (in MTCH2), estimated main effects were comparatively larger in younger respondents across all Waves, with an increasing range in differences by age across time. The magnitude of the estimated effect appears to be driven by the individuals who were younger compared to older (e.g. aged ≤ 15 years versus ≥16 years), with increased variability in effect estimates in 2001 (Wave II) and again in 2008 (Wave III) [Figure 2]. The magnitudes of the estimated main effects were highest across all Waves in the youngest participants that were recruited to Add Health and who were age 13–14 years at Wave II. For rs987237 (in TFAP2B), the estimated main effects were comparatively larger in older respondents across all Waves. We then compared effect sizes in all participants who were similar ages but at different points in time (i.e. those who were aged 18–20 years in Wave II versus Wave III or 25–26 years in Waves III versus Wave IV). While no SNPs were significant for SNP-by-Wave interactions after correcting for multiple testing, both rs3817334 in MTCH2 (stronger effects in Wave III than Wave II) and rs987237 in TFAP2B (weaker effects in Wave III than Wave II) had nominally significant SNP-by-Wave interactions when comparing aged 18–20 year olds between Waves II and III (Table 4 and Supplementary Table 4).
Figure 2.
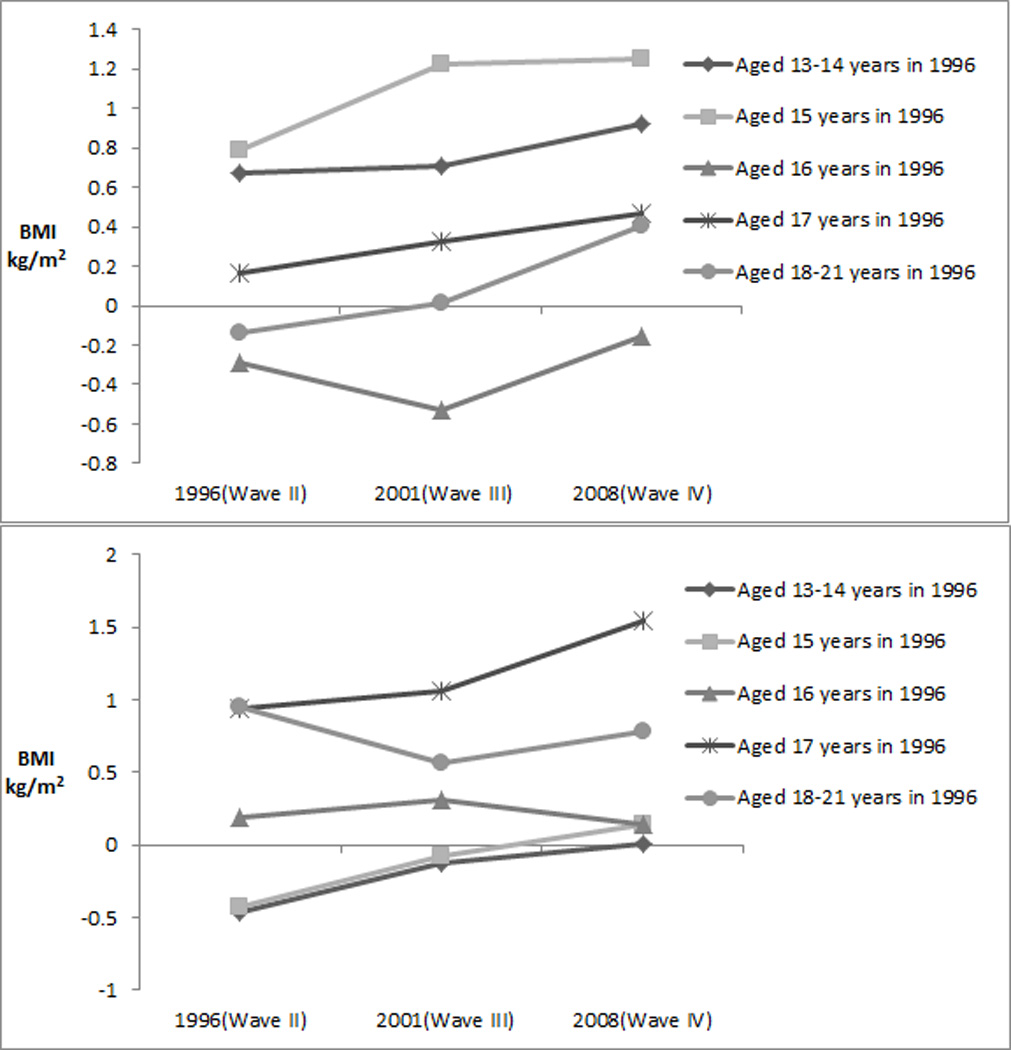
Effect estimates of (a) rs3817334 (near MTCH2) and (b) rs987237 (near TFAP2B) with cross-sectional measures of BMI at each Wave by age group at Wave II.a
a To aid in interpretation, we plotted the results by year of age at each Wave, except for those aged 13–14 and aged 18–21 which we combined due to smaller sample sizes.
Sample sizes for MTCH2:
Aged 13–14 in 1996 (N=961 in Wave II, N=836 in Wave III, N=975 in Wave IV)
Aged 15 in 1996 (N=961 in Wave II, N=836 in Wave III, N=975 in Wave IV)
Aged 16 in 1996 (N=1007 in Wave II, N=840 in Wave III, N=1023 in Wave IV)
Aged 17 in 1996 (N=960 in Wave II, N=782 in Wave III, N=974 in Wave IV)
Aged 18–21 in 1996 (N=1022 in Wave II, N=904 in Wave III, N=1069 in Wave IV)
Sample sizes for TFAP2B:
Aged 13–14 in 1996 (N=966 in Wave II, N=843 in Wave III, N=980 in Wave IV)
Aged 15 in 1996 (N=835 in Wave II, N=715 in Wave III, N=845 in Wave IV)
Aged 16 in 1996 (N=1009 in Wave II, N=842 in Wave III, N=1025 in Wave IV)
Aged 17 in 1996 (N=967 in Wave II, N=789 in Wave III, N=981 in Wave IV)
Aged 18–21 in 1996 (N=1028 in Wave II, N=908 in Wave III, N=1075 in Wave IV)
Table 4.
The SNPx Wave associations with BMI for age-matched individuals across 2 waves for SNPs associated with slope of BMI at p<0.05 within each race/ethnic group and the meta-analyzed sample.a
| Ages 18–20, Waves II and III | Ages 25 and 26, Waves III and IV | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Chr | BP | Effect/ other alleleb |
EAF | N | Beta(SE) SNPxWave |
Pvalue SNPxWave |
N | Beta(SE) SNPxWave |
Pvalue SNPxWave |
| European Ancestry | |||||||||||
| rs2568958 | NEGR1 | 1 | 72765116 | A/G | 0.6276 | 2380 | 0.489 (0.328) | 0.1360018 | 1320 | 0.227 (0.683) | 0.7394976 |
| rs1555543 | PTBP2 | 1 | 96944797 | C/A | 0.588 | 2383 | −0.159 (0.318) | 0.61759384 | 1327 | 0.46 (0.666) | 0.48956544 |
| rs543874 | SEC16B /LZTR2 | 1 | 177889480 | G/A | 0.1973 | 2392 | 0.445 (0.39) | 0.25413935 | 1328 | 0.383 (0.876) | 0.66231479 |
| rs6548238 | TMEM18 | 2 | 634905 | C/T | 0.8298 | 2394 | 0.346 (0.408) | 0.3963025 | 1329 | −0.392 (0.845) | 0.64237252 |
| rs713586 | RBJ/ADCY3/ POMC | 2 | 25158008 | C/T | 0.4827 | ||||||
| rs987237 | TFAP2B | 6 | 50803050 | G/A | 0.174 | 2399 | −0.852 (0.412) | 0.0387382 | 1332 | −0.908 (0.923) | 0.3251806 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.3143 | 2369 | 0.144 (0.343) | 0.67426563 | 1320 | −0.283 (0.713) | 0.69180913 |
| rs867559 | LMX1B | 9 | 129465325 | G/A | 0.1917 | 2389 | −0.378 (0.378) | 0.3181291 | 1327 | −0.028 (0.817) | 0.97297023 |
| rs3817334 | MTCH2 | 11 | 47650993 | T/C | 0.4056 | 2379 | 0.783 (0.317) | 0.01356331 | 1317 | 0.53 (0.673) | 0.4306055 |
| rs11847697 | PRKD1 | 14 | 30515112 | T/C | 0.05271 | 2397 | 0.153 (0.674) | 0.8202924 | 1329 | −0.508 (1.34) | 0.70483292 |
| rs4788102 | SH2B1 | 16 | 28873398 | A/G | 0.3924 | 2392 | 0.246 (0.315) | 0.43535528 | 1330 | 0.464 (0.657) | 0.47997764 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.3923 | 2381 | −0.369 (0.322) | 0.25094021 | 1320 | 0.31 (0.686) | 0.6520413 |
| rs571312 | MC4R | 18 | 57839769 | A/C | 0.2345 | 2390 | 0.056 (0.375) | 0.88096245 | 1330 | 0.464 (0.766) | 0.54458768 |
| rs2287019 | QPCTL/GIPR | 19 | 46202172 | C/T | 0.8132 | 2392 | 0.265 (0.401) | 0.50840104 | 1327 | 1.22 (0.879) | 0.16527409 |
| African Ancestry | |||||||||||
| rs543874 | SEC16B /LZTR2 | 1 | 177889480 | G/A | 0.2411 | 806 | 0.832 (0.724) | 0.25057022 | 429 | 0.223 (1.403) | 0.8734678 |
| rs7647305 | ETV5/SFRS10/DGKG | 3 | 185834290 | C/T | 0.5983 | ||||||
| rs10938397 | Gene desert;GNPDA2 | 4 | 45182527 | G/A | 0.2465 | 810 | 0.755 (0.741) | 0.30796107 | 430 | 0.201 (1.23) | 0.87001824 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.1732 | 804 | 1.114 (0.81) | 0.16906801 | 428 | 1.022 (1.459) | 0.48371119 |
| rs2241423 | MAP2K5/LBXCOR1 | 15 | 68086838 | G/A | 0.621 | 808 | 0.705 (0.64) | 0.27127539 | 427 | 2.152 (1.139) | 0.05887978 |
| Hispanic Ancestry | |||||||||||
| rs1514175 | TNNI3K | 1 | 74991644 | A/G | 0.5425 | 573 | 0.44 (0.672) | 0.51292985 | 288 | 0.276 (1.078) | 0.79756657 |
| rs543874 | SEC16B /LZTR2 | 1 | 177889480 | G/A | 0.1864 | 568 | 0.059 (0.89) | 0.94708825 | 285 | −1.002 (1.488) | 0.50085972 |
| rs12444979 | GPRC5B/IQCK | 16 | 19933600 | C/T | 0.91275 | 571 | 0.433 (1.17) | 0.7117468 | 289 | 1.131 (1.902) | 0.55187899 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.3247 | 571 | 0.637 (0.684) | 0.35131988 | 287 | −1.046 (1.155) | 0.36502999 |
| Meta-analyzed sample, all ancestriesc | |||||||||||
| rs1514175 | TNNI3K | 1 | 74991644 | A/G | 0.504921987105307 | 3778 | −0.202 (0.259) | 0.4347 | 2049 | 0.054 (0.52) | 0.9178 |
| rs1555543 | PTBP2 | 1 | 96944797 | C/A | 0.554255430649694 | 3759 | 0.065 (0.262) | 0.8027 | 2038 | 0.418 (0.521) | 0.423 |
| rs543874 | LZTR2 | 1 | 177889480 | G/A | 0.204572292940982 | 3766 | 0.471 (0.321) | 0.1418 | 2042 | 0.071 (0.665) | 0.9154 |
| rs6548238 | TMEM18 | 2 | 634905 | C/T | 0.850095966275417 | 3777 | 0.03 (0.352) | 0.9316 | 2047 | 0.173 (0.676) | 0.7976 |
| rs10938397 | GNPDA2 | 4 | 45182527 | G/A | 0.383783088113738 | 3767 | 0.096 (0.272) | 0.7234 | 2040 | −0.709 (0.523) | 0.1752 |
| rs987237 | TFAP2B | 6 | 50803050 | G/A | 0.173825852206976 | 3780 | −0.61 (0.335) | 0.06899 | 2050 | −0.397 (0.666) | 0.5509 |
| rs10968576 | LRRN6C | 9 | 28414339 | G/A | 0.273630186807737 | 3739 | 0.079 (0.293) | 0.7881 | 2033 | −0.431 (0.571) | 0.4511 |
| rs867559 | LMX1B | 9 | 129465325 | G/A | 0.23544250289304 | 3774 | 0.023 (0.296) | 0.9389 | 2044 | 0.433 (0.584) | 0.4589 |
| rs3817334 | MTCH2 | 11 | 47650993 | T/C | 0.373131542403703 | 3759 | 0.594 (0.264) | 0.02449 | 2035 | 0.312 (0.523) | 0.5506 |
| rs11847697 | PRKD1 | 14 | 30515112 | T/C | 0.112226528351794 | 3784 | −0.077 (0.445) | 0.8618 | 2051 | 0.466 (0.814) | 0.5669 |
| rs2241423 | MAP2K5 | 15 | 68086838 | G/A | 0.706667680608365 | 3772 | −0.26 (0.283) | 0.3585 | 2048 | 0.794 (0.576) | 0.168 |
| rs12444979 | GPRC5B | 16 | 19933600 | C/T | 0.874909173417094 | 3768 | 0.355 (0.388) | 0.3593 | 2039 | 0.201 (0.801) | 0.8017 |
| rs4788102 | SH2B1 | 16 | 28873398 | A/G | 0.372053116217557 | 3772 | 0.301 (0.263) | 0.2529 | 2045 | 0.869 (0.524) | 0.09725 |
| rs9939609 | FTO | 16 | 53820527 | A/T | 0.397909621425029 | 3761 | −0.156 (0.262) | 0.5513 | 2037 | −0.147 (0.525) | 0.7795 |
| rs571312 | MC4R | 18 | 57839769 | A/C | 0.246872689700777 | 3769 | 0.087 (0.305) | 0.7749 | 2047 | −0.009 (0.594) | 0.9875 |
Abbreviations: SNP (single nucleotide polymorphism), EAF: Effect Allele Frequency, Chr: Chromosome, BP: base pair position, BMI (body mass index), SE (standard error)
Beta and standard error (SE) estimates are presented for the SNP×Wave interaction model: Multivariable linear models of cross-sectional BMI regressed on SNP and wave of data collection in years, with SNP by Wave interaction term, controlling for sex, current smoking (at least one cigarette every day for 30 days), geographic region, and self-reported heights and weights (n=39). Random intercepts allowed for family and school with no sample weighting. Models were run separately for each SNP. Models in Hispanic Americans also adjusted for an indicator for foreign born and country of origin, and models in African Americans also adjusted for an indicator for selection for high education group (oversampling for education).
Effect allele oriented to BMI increasing allele.
Meta-analyzed results were done in METAL software (Willer et al 2012), by combining effect estiamtes from each of the 3 ancestries shown using the inverse-weighted approach.
DISCUSSION
Over the past decade, numerous common genetic loci have been reported to be associated with BMI in primarily European descent adults, as well as in other ancestral groups, and at one or more phases of the life course including adulthood [4, 21, 29, 39–41], childhood [7, 42–44] and adolescence [7, 26, 45]. We extended these findings to interrogate 34 known obesity-related loci for association with change in BMI across the transition from adolescence to adulthood in an ethnically diverse, nationally representative cohort of adolescents followed over 13 years into adulthood.
Among EA, we observed statistically significant positive associations with change in BMI (when oriented on the obesity susceptibility allele) from adolescence into adulthood for 6 of 34 known obesity loci tested, including one FTO variant (rs9939609) that met genome-wide significance. Results were similar in the meta-analyses including EA, HA and AA. Among six Bonferroni-corrected significant SNPs, only FTO, MC4R, TFAP2B, and SEC16B have been shown to be associated with cross-sectional measures of BMI during adolescence in this cohort [26]. The variance explained for a one year change in BMI the by six Bonferroni-corrected significant SNPs or the 15 SNPs that met nominal significance is clinically rather small. How this extends beyond one year is not something we can extrapolate here. However, we have only tested a selection of SNPs and there are likely others that influence change in BMI in addition to those tested here. Our work confirms previous findings in younger children (ages one to 16 years) of European descent of positive associations between BMI-related loci and BMI trajectories of at least nominal significance with FTO, MC4R, SEC16B, TMEM18, TFAP2B and MTCH2 [13]. Other analysis of FTO variant rs9939609 and MC4R variant rs17782313 (R2=0.96 with the MC4R SNP rs571312 we tested) associations with change in BMI from childhood into adulthood in a sample of 2,479 European descent individuals suggested comparatively stronger association with BMI from age two to 20 years and then weakening to age 53 [7]. In our study, FTO variant rs9939609 and MCR4 variant rs571312 were positively associated with change in BMI, however, though consistent in direction with the prior report, the effects of these variants in higher vs. lower ages in Wave-stratified age-by-SNP interaction analyses were not significant. A longitudinal study in over 41,000 European descent adults from three studies, mean ages 60, 45, and 58 years, found no statistically significant association with FTO and change in BMI across a 10-year period of time [46]. Thus, FTO may play a comparatively less prominent role in relation to weight change in later adulthood. A recent study comparing genome-wide genetic effects on BMI in younger adults <=50 years versus older adults (>50 years) showed 11 of 15 loci with greater estimated effects on younger adults [43]. None of the 11 loci were associated with birth weight, yet all but one were nominally associated with increased risk of childhood obesity and BMI in 16-to-25 year-olds, suggesting that some loci exert genetic effects relatively early in life and into young adulthood. Among the 11 loci were variants in FTO, MC4R, SEC16B, and TMEM18 that are in high linkage disequilibrium (R2>0.8) with those identified in the current study.
Change in BMI over time, and the effects of genotype on this change, is likely impacted by many factors including changes in age and environment. Many of the specific environmental factors are either unknown or difficult to adequately measure and/or represent in a social epidemiological study. Over the past couple of decades, there has been a profound increase in obesity in both adolescent and adult populations due to a number of external factors strongly associated with the time period. In our study, like most social and epidemiological longitudinal studies, the impact of change in age and general change in environment captured by year of study (Wave) are highly confounded. In addition to assessing longitudinal change in BMI, we also performed Wave-stratified age-by-SNP interactions and age-matched Wave-by-SNP interactions between adjacent Waves to attempt to tease apart this confounding. Our findings suggest that the etiology for the observed strengthening associations between established BMI variants and BMI during the period of adolescence and young-adulthood is complex and may be a function of both the aging process and exposure to an increasingly obesogenic environment[47]. The interaction between rs987237 (near TFAP2B) and age suggests that this variant has stronger effects on BMI in older adolescents/young adults (in the 13–21 year age range at Wave II, the estimated increase in BMI from each established risk allele is 2.5kg/m2 greater for those at age 21 compared to those at age 13). On the other hand, for MTCH2 variant rs3817334, we observed evidence for stronger estimated effects on BMI at younger ages as suggested by the significant interaction of age and rs3817334 during Wave II when participants ranged from 13 to 21 years old. Given the main and interaction effect sizes, the estimated increase in BMI per T allele of rs3817334 is 1.5kg/m2 greater in those who are 13 years at Wave II versus those who are 21 years at Wave II. Interestingly, we also found supporting evidence for this same variant having stronger effects on BMI in later Waves (e.g. stronger in Wave II vs. Wave III) in age-matched participants. For TFAP2B variant rs987237, the effects appear to be stronger in earlier Waves among age-matched participants. MTCH2 is highly expressed in white adipose tissue and adipocytes, and thought to play a regulatory role in adipocyte differentiation and biology, while TFAP2B mRNA expression has been shown to be correlated negatively with leptin and positively with IL-6 expression in both subcutaneous and omental adipose tissues [48]. Possibly MTCH2 might influence younger individuals more in playing a role during puberty while TFAP2B might have a stronger effect in older individuals in that cytokines (e.g. IL-6) have markedly lower levels in children versus adults [49]. For the other variants associated with change in BMI, we found no significant evidence pointing to differential effects across ages or year of study.
While our study capitalizes on an ancestrally diverse, nationally representative cohort measured during a unique period of the lifecycle, there are limitations. There is a lack of established obesity loci in HA and power in HA for common loci in our study was limited by smaller sample sizes. In addition, the age range of our participants limited our ability to separate differences in effect by age that might be due to cohort, time, and or age. The Add Health sample is largely comprised of post-pubertal adolescents. For example <1% of Add Health females had not achieved menarche by wave I. Nonetheless we conducted a sensitivity analysis with (and without) adjustment for age at menarche in women (we have no such comparable measure for men) (Supplementary Table 5). This sensitivity analysis suggests little difference and therefore would infer that there is little confounding from lack of adjustment for puberty. We were unable to test periods of the life course that might be defined as childhood or middle- to late-adulthood, making comparisons with other studies difficult. However, these questions extend beyond the scope of the current study given our sample. Finally, while it is possible, most common variants are not affected by pop stratification in Europeans which is where we find most results. We are not well powered to detect effects in these samples. Thus, it is not likely that we are reporting any false positives.
In conclusion, we demonstrated that several established BMI variants are positively associated with change in BMI during the period of adolescence and young-adulthood. Through stratified analyses, we demonstrate that MTCH2 variant rs3817334 has stronger effects on BMI in younger participants and in later Wave (i.e. time periods), and that TFAP2B variant rs987237 has stronger effects in older participants and in earlier Waves. Due to the confounding between age and Wave in longitudinal analyses, stratified analyses were necessary to tease apart these directionally conflicted findings for both variants. Our results suggest that the genetic effect of BMI loci varies over time in a complex manner, highlighting the importance of investigating loci influencing obesity risk across the life course.
Supplementary Material
Acknowledgments
This study was funded by National Institutes of Health Grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development R01HD057194. The Carolina Population Center (R24 HD050924) provided general support. This research uses data from Add Health, a program project directed by Kathleen Mullan Harris and designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill and funded by Grant P01- HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations. Special acknowledgment is given to Ronald R. Rindfuss and Barbara Entwisle, of the University of North Carolina at Chapel Hill, for assistance in the original design. Information about how to obtain the Add Health data files is available at http://www.cpc.unc.edu/addhealth. No direct support was received from Grant P01-HD31921 for this analysis.
ABBREVIATIONS
- SNP
Single Nucleotide Polymorphism
- GWA
Genome-Wide Association
- EA
European American
- AA
African American
- HA
Hispanic American
Footnotes
DISCLOSURES: The authors declared no conflicts of interest.
MG, KEN, EML and PG-L designed the study. MG, ASR, and KMY contributed to data analysis. PG-L and KEN are responsible for data acquisition. MIG, EML, KEN, and PG-L drafted the manuscript. All authors contributed to data interpretation and writing of the manuscript. MIG, KEN and PG-L had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors have approved the final version of the manuscript.
REFERENCES
- 1.Dietz W. Critical periods in childhood for the development of obesity. Am J Clin Nutr. 1994;59(5):955–959. doi: 10.1093/ajcn/59.5.955. [DOI] [PubMed] [Google Scholar]
- 2.Gordon-Larsen P, Adair LS, Nelson MC, P BM. Five-year obesity incidence in the transition period between adolescence and adulthood: the National Longitudinal Study of Adolescent Health. American Journal of Clinical Nutrition. 2004;80(3):569–575. doi: 10.1093/ajcn/80.3.569. [DOI] [PubMed] [Google Scholar]
- 3.The, N.S. Suchindran C, North KE, Popkin BM, Gordon-Larsen P. Association of adolescent obesity with risk of severe obesity in adulthood. JAMA. 2010;304(18):2042–2047. doi: 10.1001/jama.2010.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallman DM, Friedel VC, Eissa MA, Boerwinkle E, Huber JC, Jr, Harrist RB, et al. The association of variants in the FTO gene with longitudinal body mass index profiles in non-Hispanic white children and adolescents. Int J Obes (Lond) 2012;36(1):61–68. doi: 10.1038/ijo.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJ, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19(3):545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Zhu H, Dong Y, Podolsky RH, Treiber FA, Snieder H. Influence of common variants in FTO and near INSIG2 and MC4R on growth curves for adiposity in African- and European-American youth. Eur J Epidemiol. 2011;26(6):463–473. doi: 10.1007/s10654-011-9583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44(5):526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wing MR, Ziegler J, Langefeld CD, Ng MC, Haffner SM, Norris JM, et al. Analysis of FTO gene variants with measures of obesity and glucose homeostasis in the IRAS Family Study. Hum Genet. 2009;125(5–6):615–626. doi: 10.1007/s00439-009-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mei H, Chen W, Jiang F, He J, Srinivasan S, Smith EN, et al. Longitudinal replication studies of GWAS risk SNPs influencing body mass index over the course of childhood and adulthood. PLoS One. 2012;7(2):e31470. doi: 10.1371/journal.pone.0031470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warrington NM, Howe LD, Wu YY, Timpson NJ, Tilling K, Pennell CE, et al. Association of a body mass index genetic risk score with growth throughout childhood and adolescence. PLoS One. 2013;8(11):e79547. doi: 10.1371/journal.pone.0079547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croteau-Chonka DC, Marvelle AF, Lange EM, Lee NR, Adair LS, Lange LA, et al. Genome-wide association study of anthropometric traits and evidence of interactions with age and study year in Filipino women. Obesity (Silver Spring) 2011;19(5):1019–1027. doi: 10.1038/oby.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaakinen M, Laara E, Pouta A, Hartikainen AL, Laitinen J, Tammelin TH, et al. Life-course analysis of a fat mass and obesity-associated (FTO) gene variant and body mass index in the Northern Finland Birth Cohort 1966 using structural equation modeling. Am J Epidemiol. 2010;172(6):653–665. doi: 10.1093/aje/kwq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris KM, Halpern CT, Smolen A, Haberstick BC. The National Longitudinal Study of Adolescent Health (Add Health) twin data. Twin Res Hum Genet. 2006;9(6):988–997. doi: 10.1375/183242706779462787. [DOI] [PubMed] [Google Scholar]
- 17.Harris KM. An integrative approach to health. Demography. 2010;47(1):1–22. doi: 10.1353/dem.0.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller WC, Ford CA, Morris M, Handcock MS, Schmitz JL, Hobbs MM, et al. Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA. 2004;291(18):2229–2236. doi: 10.1001/jama.291.18.2229. [DOI] [PubMed] [Google Scholar]
- 19.Resnick MD, Bearman PS, Blum RW, Bauman KE, Harris KM, Jones J, et al. Protecting adolescents from harm. Findings from the National Longitudinal Study on Adolescent Health. JAMA. 1997;278(10):823–832. doi: 10.1001/jama.278.10.823. [DOI] [PubMed] [Google Scholar]
- 20.Thorleifsson G, Walters G, Gudbjartsson D, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nature Genetics. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 21.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heard-Costa NL, Zillikens MC, Monda KL, Johansson A, Harris TB, Fu M, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5(6):e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The Add Health Biomarker Team. Biomarkers in Wave III of the Add Health Study. In: Manhart LE, editor. 1999. p. 57. [Google Scholar]
- 26.Graff M, North KE, Mohlke KL, Lange LA, Luo J, Harris KM, et al. Estimation of genetic effects on BMI during adolescence in an ethnically diverse cohort: The National Longitudinal Study of Adolescent Health. Nutrition and Diabetes. 2012;2:e47. doi: 10.1038/nutd.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bates LM, Acevedo-Garcia D, Alegria M, Krieger N. Immigration and generational trends in body mass index and obesity in the United States: results of the National Latino and Asian American Survey: 2002–2003. Am J Public Health. 2008;98(1):70–77. doi: 10.2105/AJPH.2006.102814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris KM, Perreira KM, Lee D. Obesity in the transition to adulthood: predictions across race/ethnicity, immigrant generation, and sex. Arch Pediatr Adolesc Med. 2009;163(11):1022–1028. doi: 10.1001/archpediatrics.2009.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45(6):690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang SJ, Chiang CW, Palmer CD. Genome-wide association of anthropometric traits in African- and African-derived populations. Human Molecular Genetetics. 2010;19(13):2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11(9):e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodman E, Hinden BR, Khandelwal S. Accuracy of teen and parental reports of obesity and body mass index. Pediatrics. 2000;106(1 Pt 1):52–58. doi: 10.1542/peds.106.1.52. [DOI] [PubMed] [Google Scholar]
- 33.Heo M, Faith MS, Mott JW, Gorman BS, Redden DT, Allison DB. Hierarchical linear models for the development of growth curves: an example with body mass index in overweight/obese adults. Stat Med. 2003;22(11):1911–1942. doi: 10.1002/sim.1218. [DOI] [PubMed] [Google Scholar]
- 34.Kakinami L, Henderson M, Chiolero A, Cole TJ, Paradis G. Identifying the best body mass index metric to assess adiposity change in children. Arch Dis Child. 2014;99(11):1020–1024. doi: 10.1136/archdischild-2013-305163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sovio U, Mook-Kanamori DO, Warrington NM, Lawrence R, Briollais L, Palmer CN, et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: the complex nature of genetic association through growth and development. PLoS Genet. 2011;7(2):e1001307. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 38.Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11(2):193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 39.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasan SK, Fall T, Neville MJ, Antonisamy B, Fall CH, Geethanjali FS, et al. Associations of variants in FTO and near MC4R with obesity traits in South Asian Indians. Obesity (Silver Spring) 2012 doi: 10.1038/oby.2012.64. [DOI] [PubMed] [Google Scholar]
- 41.Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44(3):307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Mei H, Chen W, Jiang Y, Sun W, Li F, et al. Study of eight GWAS-identified common variants for association with obesity-related indices in Chinese children at puberty. Int J Obes (Lond) 2012;36(4):542–547. doi: 10.1038/ijo.2011.218. [DOI] [PubMed] [Google Scholar]
- 43.Zhao J, Bradfield JP, Zhang H, Sleiman PM, Kim CE, Glessner JT, et al. Role of BMI-associated loci identified in GWAS meta-analyses in the context of common childhood obesity in European Americans. Obesity (Silver Spring) 2011;19(12):2436–2439. doi: 10.1038/oby.2011.237. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, Grant SF. Genetics of childhood obesity. J Obes. 2011;2011:845148. doi: 10.1155/2011/845148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.den Hoed M, Ekelund U, Brage S, Grontved A, Zhao JH, Sharp SJ, et al. Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59(11):2980–2988. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CG, et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41,504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;60(5):1637–1644. doi: 10.2337/db10-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravussin E, Bouchard C, et al. Human genomics and obesity: finding appropriate drug targets. Eur J Pharmacol. 2000;410(2–3):131–145. doi: 10.1016/s0014-2999(00)00811-6. [DOI] [PubMed] [Google Scholar]
- 48.Ugi S, Nishio Y, Yamamoto H, Ikeda K, Kobayashi M, Tsukada S, et al. Relation of the expression of transcriptional factor TFAP2B to that of adipokines in subcutaneous and omental adipose tissues. Obesity (Silver Spring) 2010;18(7):1277–1282. doi: 10.1038/oby.2009.442. [DOI] [PubMed] [Google Scholar]
- 49.Lilic D, Cant AJ, Abinun M, Calvert JE, Spickett GP. Cytokine production differs in children and adults. Pediatr Res. 1997;42(2):237–240. doi: 10.1203/00006450-199708000-00018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


