Abstract
The CCAAT-binding factor CBF/NF-Y is needed for cell proliferation and early embryonic development. NF-Y can regulate the expression of different cell type-specific genes that are activated by various physiological signaling pathways. Dysregulation of NF-Y was observed in pathogenic conditions in humans such as scleroderma, neurodegenerative disease, and cancer. Conditional inactivation of the NF-YA gene in mice demonstrated that NF-Y activity is essential for normal tissue homeostasis, survival, and metabolic function. Altogether, NF-Y is an essential transcription factor that plays a critical role in mammalian development, from the early stages to adulthood, and in human pathogenesis.
1. Introduction
The CCAAT-binding factor CBF/NF-Y is a unique DNA-binding protein consisting of three different subunits: NF-YA (CBF-B), NF-YB (CBF-A), and NFY-C (CBF-C). Part of the amino acid sequences of each subunit is evolutionarily conserved with segments of yeast HAP2, HAP3, and HAP5, respectively. The HAP genes in yeast specifically regulate the expression of nuclear genes that function in mitochondrial respiration. The conserved segments of NF-YB and NF-YC that are needed for DNA binding contain histone-fold motifs that are similar to histones H2B and H2A, respectively. NF-YB and NF-YC together form a stable heterodimer that interacts with the conserved segment of NF-YA, forming a heterotrimeric complex, which then binds to the CCAAT motif DNA [1, 2].
For the last three decades, the role of CCAAT-binding factor (CBF/NF-Y) has been studied in the context of many mammalian promoters that are regulated by different signaling events [3]. Although initial studies described the CCAAT motif as a basal proximal promoter element that is located upstream to the transcription start site, studies of different mammalian genes have defined the role of the CCAAT motif and its binding partner NF-Y in transcription in specific cell types, which can be regulated by diverse cellular signaling or pathogenic conditions, such as mechanical stress [4], endoplasmic reticulum (ER) stress [5, 6], cholesterol and fatty acid metabolism [7–9], interferon gamma [10, 11], cell cycle progression [12, 13], DNA damage response [14–18], neurodegenerative disease [19, 20], and cancer [21–23] (Table 1).
Table 1.
Regulation of NF-Y dependent gene expression by various specific cell types or signaling linked to normal development or pathogenic conditions
| Genes | Cell type | Signaling | Normal/Pathophysiology | References |
|---|---|---|---|---|
| Type I collagen (COL1A1, COL1A2) |
Fibroblasts | TGF-beta, Mechanical strain |
Systemic sclerosis, Fibrosis, Hypertension |
4, 36, 37 |
| Dentin Sialophospho protein (DSPP) |
Odontoblasts | BMP2 | Tooth development | 39 |
| SOX9 | Chondrocytes | BMP2 | Cartilage development | 40 |
| HOXB4 | Hematopoietic stem cells (HSC) |
HSC self- renewal |
Hematopoiesis | 46 |
| Major histocompatibility class II |
B cells | Interferon- gamma |
Immune response | 10, 11 |
| Tet 1/2 | T regulatory cells |
Inflammatory response |
Immune homeostasis | 47 |
| OCT4/Nanog/Prdm14 | Embryonic stem cells (ESC) |
ESC identity | Early development | 50 |
| TopoIIalpha, CCNB1, CCNB2, CDC25C, and other cell cycle regulated genes |
Cancer cells | Cell cycling, Oncogenic signaling by wild-type and mutant p53 |
Cell proliferation, DNA damage response, Cancer progression |
12– 18, 21–23 |
| GRP78, GRP94, PDI, and other endoplasmic reticulum (ER) chaperones |
Hepatocytes, Islet beta-cells, Neuronal cells, Various secretory cells |
Unfolded protein response or ER stress mediated by ATF6 activation |
Genetic disorders associated with protein misfolding, Metabolic disease, Neurodegenerative disease, Diabetes |
5, 6, 19, 20, 24–26, 44, 45 |
| HMG-CoA synthase, SQLE, DHCR24, and other Cholesterol biosynthesis genes, and FASN, SCD, and lipid biosynthesis genes |
Hepatocytes, Adipocytes, Various lipogenic cells |
Sterol balance, nutritional response mediated by SREBP1 activation |
Metabolic syndrome, Hepatosteatosis, Lipotoxicity associated disease |
7– 9, 27, 28 |
The context-specific function of NF-Y is often mediated by its ability to interact with or recruit different pathway-specific promoter-binding factors. For example, during ER stress, NF-Y recruits ATF6, which is activated by proteolysis; this is followed by nuclear localization to stimulate the expression of various chaperone genes that are associated with pathogenic conditions in protein misfolding diseases, such as neurodegenerative disease, and metabolic diseases, such as diabetes and non-alcoholic hepatic steatosis [24–26]. In contrast, during sterol depletion, NF-Y collaborates with SREBP1, which is also activated by proteolysis, to increase the expression of cholesterol or fatty acid metabolism genes that are associated with metabolic syndrome [27, 28].
A mutational analysis of NF-YA identified three domains: DNA binding, subunit interaction, and transactivation in the NF-YA protein. The mutants in the DNA-binding domain of NF-YA that interacted with the NF-YB/NF-YC heterodimer but did not bind DNA were used as a dominant-negative NF-YA mutant to inhibit NF-Y’s function in different cell lines. Another NF-YA mutant that lacks the transactivation domain that binds to DNA was also used to inhibit NF-Y-mediated transactivation in several cell lines. These mutants, as well as RNAi-mediated knockdown of NF-Y subunits, were used to define the role of NF-Y in cultured cells [13, 29–31].
To understand the role of NF-Y in vivo in mouse models, we generated a conditional NF-YA (CBF-B) mutant by introducing loxP sites in the murine NF-YA gene that can be deleted by regulated expression of Cre recombinase (Cre) in the embryo or in various tissues after birth. The study of the NF-YA mutant mouse strain in the context of various tissues provided critical information regarding the role of NF-Y in tissue homeostasis and pathogenesis [32–36].
2. Function in different cell type contexts
Fibroblast proliferation, type I collagen, and TGF-beta signaling
A dominant-negative NF-Y mutant was used to determine the role of NF-Y in cultured mouse fibroblast cells [13]. Regulated expression of the dominant-negative mutant resulted in retardation of fibroblast growth and reduced expression of type I collagen and cell cycle-regulated genes. The results of this study indicated that NF-Y is needed for the proliferation of fibroblasts in culture.
NF-Y binds and activates the transcription of the promoters of both subunits of the type I collagen genes COL1A1 and COL1A2 [37]. Type I collagen is abundantly expressed as extracellular matrix protein in fibroblasts and can be regulated under various physiological and pathological conditions. The CCAAT motif of the COL1A1 promoter can be responsible for high-level expression of the gene in fibroblasts derived from human scleroderma or systemic sclerosis, causing excessive fibrosis in the tissues and fibroproliferative lesions in the small blood vessels [38]. The promoter analysis showed that a higher level of NF-Y-binding activity regulated promoter activity of COL1A1 in scleroderma patient fibroblasts than that of normal human fibroblasts.
The role of the CCAAT motif in the COL1A1 promoter was studied in the context of the cardiopulmonary system, in which interstitial fibroblasts play a critical role during tissue remodeling and repair, in response to mechanical strain, under normal physiological conditions as well as in systemic hypertension [4]. The mechanical strain also induces the expression of transforming growth factor-beta (TGF-beta), which stimulates the proliferation of the interstitial fibroblasts and the expression of type I collagen. The study of COL1A1 promoter demonstrated that TGF-beta enhanced the binding of NF-Y to the COL1A1 promoter in rat cardiac fibroblasts, resulting in stimulation of promoter activity. The role of TGF-beta in NF-Y activity is further supported by another study that showed that TGF-beta increases nuclear localization of the NF-YA subunit that involves mitogenactivated protein kinase cascades [39]. The results of this study suggest that TGF-beta regulation of NF-Y activity is cell type dependent due to the differences in protein kinase activity.
The results of these studies together suggest that NF-Y plays an important role in fibroblast proliferation and expression of type I collagen gene regulated under various physiological and pathological conditions that are regulated by TGF-beta signaling.
Odontoblasts, chondrocytes, and BMP2 signaling
NF-Y regulates the expression of specific genes in odontoblasts and chondrocytes in response to bone morphogenetic protein 2 (BMP2), a member of the TGF-beta super family.
Dentin sialophosphoprotein (DSPP) is specifically expressed in odontoblasts and is necessary for tooth development and mineralization. Expression of the DSPP gene is activated by BMP2, which is mediated by the NF-Y binding site in the DSPP promoter [40]. The results of this study showed that BMP2 treatment induces NF-Y accumulation in the nucleus and increases DSPP gene expression in mouse preodontoblasts.
Sox9 is a transcription factor that plays a crucial role in chondrogenesis during cartilage development and is activated by BMP2 during chondrocyte differentiation. The NF-Y binding site in the Sox9 promoter is needed for BMP2-mediated activation of the Sox9 promoter during the differentiation of mouse embryonic fibroblasts into chondrocytes [41]. The study showed that BMP2 increases NF-Y occupancy in the Sox9 promoter during chondrocyte differentiation that involves the p38 kinase pathway.
Muscle cell differentiation
The expression of NF-YA, but not NF-YB and NF-YC, was not detected in adult mouse skeletal and cardiac muscle cells. Similarly, down-regulation of NF-YA was observed in terminally differentiated muscle cells but not in proliferating muscle cells in culture [42, 43]. The loss of NF-Y activity during muscle cell differentiation coincided with the down-regulation of several cell cycle genes, and conversely, forced expression of NF-YA in differentiated muscle cells resulted in a delay of the early muscle-specific differentiation program. Interestingly, an NF-YA splice variant, NF-YAI, which contains a longer transactivation domain but has intact DNA binding activity, plays a specific role in the expression of muscle-specific genes [44]. Inactivation of NF-YA expression also resulted in the inhibition of both the proliferation and differentiation of muscle cells, indicating that NF-Y activity is needed for the muscle cell differentiation program.
Neuronal cells and neurodegeneration
NF-Y is expressed in mature neuronal cells and is deregulated in a group of hereditary neurodegenerative disorders that result from polyglutamine expansion in various proteins. The polyglutamine proteins are generated by the abnormal expansion of the trinucleotide CAG repeat that encodes the polyglutamine tract in specific proteins that cause disease. The polyglutamine proteins selectively cause neurodegeneration in the brain, although the polyglutamine proteins are expressed in many different tissues. This is possibly due to misfolding of the proteins and abnormal interaction with other key cellular proteins in neurons of the brain.
Spinal and bulbar muscular atrophy (SMBA) is caused by polyglutamine expansion in the androgen receptor (AR) gene. SMBA is a gradually progressive neuromuscular disease with degeneration of lower motor neurons and occurs primarily in males. The mouse model of SMBA that expresses the polyglutamine AR shows decreased expression of the NF-Y-regulated transforming growth factor-beta (TGF-beta) receptor type II gene and disruption of TGF-beta signaling in the spinal motor neurons of male mice [45]. The polyglutamine mutant AR pathogenic protein associates with NF-Y and p300/CBP-associated factor, resulting in sequestration of NF-Y in molecular aggregates and inhibition of NF-Y-mediated transcription.
Spinocerebellar ataxia type 17 (SCA17) is caused by polyglutamine expansion in the general transcription factor TATA box-binding protein (TBP). SCA17 disorder has a broad spectrum of phenotypes that includes seizures, cognitive dysfunction, psychiatric symptoms, and Parkinsonism. In the mouse model of SCA17, expressing the polyglutamine TBP caused age-dependent neurological symptoms and cerebellar Purkinje cell degeneration and inhibited the expression of NF-Y-regulated chaperone genes, such as HSP70 and HSPA5 (GRP78) [19, 46]. The polyglutamine TBP mutant interacted with NF-YA and inhibited NF-Y’s binding to the cellular HSPA5 promoter.
Huntington’s disease is caused by polyglutamine expansion in the Huntington (Htt) protein, which forms nuclear aggregates in neurons of the striatum and cortex. The mutant Htt protein interacts with multiple transcription factors, including NF-Y. Importantly, the mutant Htt protein inhibited NF-Y recruitment to the HSP70 promoter and inhibited its expression [20].
The results of these studies indicate that one possible cause of SCA17 and Huntington’s disease is the inactivation of NF-Y, resulting in the inhibition of chaperone proteins that might lead to protein misfolding or aggregation and neuronal apoptosis. In this regard, the promoters of many chaperone genes contain multiple NF-Y sites. Importantly, the expression of HSPA5 and several other chaperone genes involved in protein folding in the ER can be regulated by the unfolded protein response (UPR) or ER stress. Many ER stress-regulated promoters contain a composite promoter element, ER stress element that binds to NF-Y constitutively and allows recruitment of the ER-stress-regulated transcription factor ATF6 during UPR activation [6]. The results of these studies suggest that NF-Y regulates both basal- and UPR-regulated activation of the chaperone genes expression that plays a critical role in normal neuron function, which might be disrupted by the polyglutamine mutant proteins that cause neuronal disease.
Hematopoietic cell lineages
Hematopoietic stem cells (HSCs) produce myeloid and lymphoid precursor cells and balance both differentiation and self-renewal throughout the animal’s lifetime. NF-Y is expressed in bone marrow HSCs and regulates the expression of the homeobox gene family factor HOXB4, which plays a key role in the expansion of HSCs [47]. The result of this study showed that overexpression of NF-YA in primitive hematopoietic cells activates the expression of several key genes regulating HSC self-renewal. The role of NF-Y in HSCs’ function is also further established by conditional knockout of the NF-YA gene in the bone marrow in mice, which is discussed later in this review.
The major histocompatibility complex (MHC) class II genes play a central role in the immune response [10, 11]. A study of MHC class II promoters identified a highly conserved promoter element with spatial-helical arrangements that bind to NF-Y, RFX, and CREB transcription factors, which together recruit a non-DNA-binding class II transactivator, CIITA, that mediates gamma-interferon-activated transcription in B cells. A detailed analysis of NF-Y’s function in MHC class II transcription provided a mechanistic paradigm by which NF-Y could regulate the expression of a specific gene under specific cellular conditions.
NF-Y is part of the transcriptional circuitry for regulatory T (Treg) cells’ lineage specification [48]. Treg cells are needed to maintain immune homeostasis and to suppress inflammatory responses. NF-Y controls the expression of Tet1 and Tet2, which control epigenetic changes to induce the expression of FOXP3, a master transcription factor for Treg cell development. Interestingly, the activity of NF-Y in T cells is regulated by hydrogen sulfide-mediated sulfhydrating of the NF-YB subunit, which allows increased binding to promoters of both Tet1 and Tet2 genes.
The results of these studies indicate that NF-Y plays a critical role in hematopoietic cell development and differentiation and immune response homeostasis.
Embryonic stem cell differentiation
NF-Y is expressed in embryonic stem cells (ESCs) and is needed for ESC proliferation. The expression of NF-Y, particularly the NF-YAs isoform and NF-YB subunits, is down-regulated during ESC differentiation [49, 50]. NF-Y regulates the expression of several pluripotency-associated genes, including Oct4, Nanog, and Prdm14 [51]. NF-Y co-localizes with Oct4 and Sox2 in many ESC-specific distal enhancers of genes that regulate ESC identity. A comparison of NF-Y occupancy in ESCs and ESC-derived neuronal progenitor cells revealed cell type-specific binding of NF-Y specifically in distal enhancers, in which ESC-specific transcription factors and NF-Y are co-localized in the ESC-specific enhancers; neuronal regulators and NF-Y are co-localized in the enhancers of neuronal-specific genes. These studies provided mechanistic insight by which NF-Y could regulate cell type-specific functions in collaboration with specific master transcription factors that drive ESC identity or neuronal cell differentiation.
3. Conditional deletion of NF-YA gene in mice
Essential role in early development
The physiological function of NF-Y in intact animals was studied by generating a conditional NF-YA (CBF-B) mutant allele, in which loxP sites were introduced in the mouse NF-YA gene, allowing deletion of the gene by controlled expression of the Cre enzyme. The mouse NF-YA gene consists of nine exons, of which exons 7 and 8 encode DNA binding and subunit interaction of the NF-YA subunit. The loxP sites were introduced flanking exon 3 and exon 8 so that Cre could delete most of the functional domains of the NF-YA gene. Deletion of both floxed NF-YA alleles resulted in early embryonic lethality, with no embryo found older than 8.5 days postcoitum (d.p.c.) [32]. However, heterozygous NF-YA mutant mice were developed similarly as the wild-type NF-YA mice, indicating that NF-Y activity is needed for early mouse embryo development.
Mouse embryonic fibroblasts (MEFs) isolated from 13.5 d.p.c. embryos of floxed NF-YA mice were used to delete the NF-YA gene after Cre had been expressed in culture. This showed that deletion of the NF-YA gene resulted in inhibition of proliferation and induction of apoptosis, indicating that NF-Y activity is essential for the proliferation and viability of MEFs.
Conditional deletion of the NF-YA gene postnatally in different tissues
The floxed NF-YA allele mice were used to delete the NF-YA gene after birth in the liver, forebrain neuron, adipose tissue, and hematopoietic system in bone marrow.
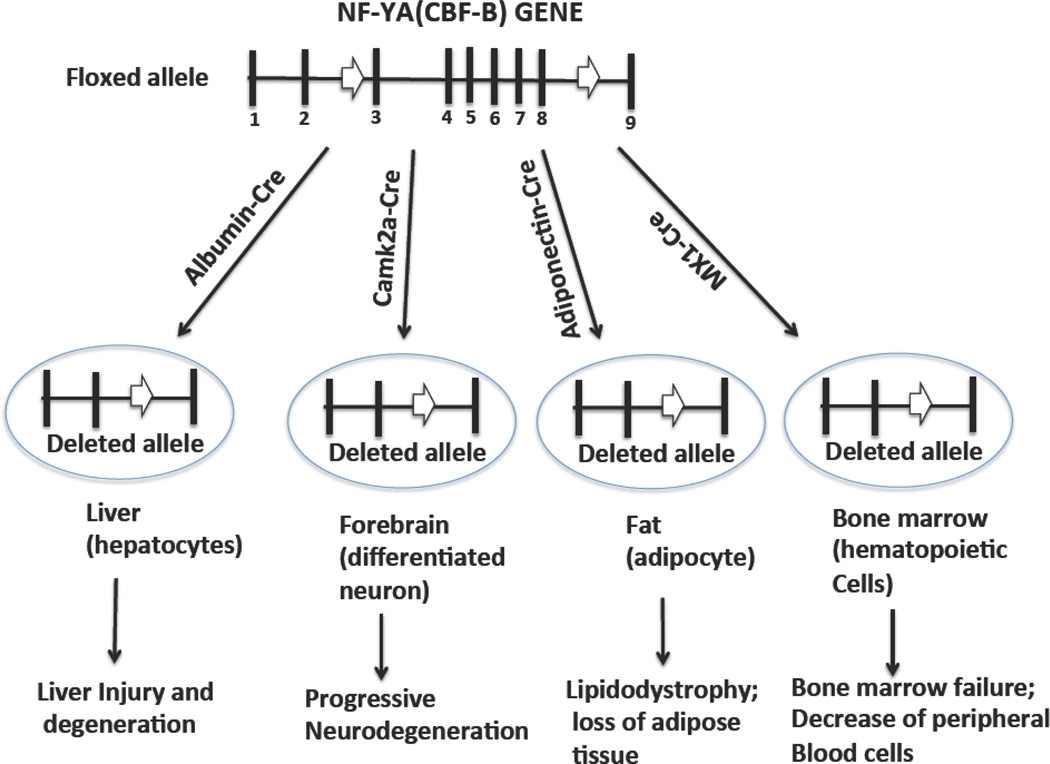
This was performed using mice that expressed tissue-specific Cre, which is expressed after birth and thus results in the conditional deletion of floxed NF-YA allele in specific tissues or cell types in developed mice (Figure 1).
Figure 1.
Conditional knockout of the NF-YA gene in four different tissues in mice after birth. The murine NF-YA consists of 346-amino acids encoded by nine exons (black bars), of which the DNA binding region and the subunit interaction domains are encoded in exons 7 and 8. The conditional murine NF-YA allele that deletes all of the functional NF-YA domains was generated by the insertion of loxP sites (thick white arrows) flanking exon 3 and exon 8 in the murine NF-YA gene. Mice with a floxed NF-YA allele were mated with transgenic mice expressing tissue-specific Cre enzyme, which deleted exons 3 to 8 of the NF-YA gene in different tissues at the postnatal stage. The deletion of the NF-YA gene resulted in inactivation of NF-Y activity in vivo in mice.
The albumin-Cre (Alb-Cre) transgenic mice were mated with the NF-YA allele mice to generate mice for the postnatal deletion of the NF-YA gene, exclusively in the liver. The Alb-Cre transgene expresses Cre specifically in the liver [52], and the level of Cre expression progressively increases with age, resulting in almost 60 percent deletion of the floxed NF-Y allele in the liver at 4 weeks after birth. NF-Y inactivation in the liver resulted in progressive liver injury and severe hepatocellular degeneration, and most mice with NF-YA-deleted livers died between 4 and 6 weeks of age [35]. The hepatocytes of the liver underwent pleotropic changes, with disorganized ER and abnormal mitochondria, indicating that NF-Y regulates organelle functions in hepatocytes. The results of the gene expression study indicated that the activation of ER stress pathway genes and loss of C/EBP alpha-controlling liver metabolic genes might be responsible for the liver injury in NF-Y-inactivated mice. However, the inactivation of NF-Y did not alter the expression of the cell cycle genes. The results of these analyses indicate that NF-Y regulates genes are associated with metabolic and ER functions in mature differentiated hepatocytes.
The role of the ER stress pathway in liver function was also revealed in the study of ATF6 knockout mice. As noted earlier, NF-Y recruits ATF6 to activate transcription during ER stress. ATF6 consists of two isoforms, ATF6-alpha and ATF6-beta. NF-Y mostly recruits ATF6-alpha to activate transcription during ER stress. Neither ATF6-alpha-null nor ATF6-beta-null mice have any obvious phenotype. However, burdening mice with an ER stress-inducing agent resulted in liver dysfunction and steatosis in ATF6-alpha-null but not ATF6-beta-null mice [26]. The results of this study suggest that the NF-Y-ATF6-alpha pathway plays a critical role in ER stress response in mature hepatocytes.
NF-Y is expressed in mature neurons of the adult brain and is suppressed by various neurodegenerative diseases, as described earlier. Alpha-calcium-calmodulin-dependent kinase-2-Cre (CAMK2a-Cre) transgenic mice expressed Cre selectively in postmitotic neurons of the forebrain starting at 3 weeks of age [53]. The mating of CAMK2a-Cre transgenic mice with the floxed NF-YA allele mice generated mice with deletion of the NF-YA gene specifically in the cortex and hippocampus of the forebrain region. The inactivation of NF-Y in the forebrain resulted in progressive neurodegeneration, accompanied by gliosis, a reduction of body weight, and a shortened lifespan [36]. Distinctive neuropathological characteristics are associated with neurodegeneration, including the accumulation of ubiquitin and a scaffold protein, together with insoluble membrane proteins, on the ER. The inactivation of NF-Y also resulted in an aberrant increase in smooth ER and selective down-regulation of GRP94, an ER chaperone with no activation of the ER stress pathway, suggesting that NF-Y is involved in ER organization function by a unique mechanism in mature neurons.
Recently, the function of NF-Y was also analyzed in different sets of neurons of central nervous system (CNS) in mice. Vesicular acetylcholine transporter-Cre (VAChT-Cre) transgenic mice expressed Cre selectively in a subset of motor neurons after birth starting at one week of age [54]. The mating of VAChT-Cre transgenic mice with the floxed NF-YA allele generated mice with deletion of the NF-YA gene specifically in the motor neurons of spinal cord and brainstem [55]. The inactivation of NF-Y in the motor neurons resulted progressive weight loss with abnormal posture and showed tremor-like movement that became more prominent with aging; histopathology analysis showed progressive neurodegeneration and loss of motor neurons but not any obvious abnormal protein accumulation. The inactivation of NF-Y induces downregulation of ER chaperones including GRP78 and GRP94 in the motor neurons. Interestingly, RNAi mediated knockdown of GRP78 and GRP94 in the motor neurons recapitulated the pathology as observed after the NF-Y inactivation, indicating loss of GRP78/GRP94 expression by the NF-Y inactivation caused the motor neurons degeneration. The results of these studies indicated that NF-Y is needed for neuronal maintenance in the CNS by differential regulation of ER chaperones expression in various types of neurons.
The role of NF-Y in adipose tissue was studied in the context of leptin, an adipocyte hormone that regulates adipose tissue mass and homeostasis. Leptin plays a critical role in metabolism and body weight. NF-Y binds to a distal enhancer of the leptin gene that drives adipose tissue-specific expression of leptin in vivo in mice [34]. Knockdown of NF-YA by RNAi inhibited adipocyte differentiation in culture and resulted in decreased expression of several adipocyte-specific genes, including leptin. To test NF-Y’s function in vivo, the floxed NF-YA allele mice were mated with the adiponectin-Cre transgenic mice that express Cre exclusively in both white and brown adipose tissue at 6 weeks after birth [56]. The inactivation of NF-Y in the adipose tissue resulted in progressive loss of body fat and lipodystrophy phenotype characteristics, including metabolic abnormalities in mice at 28 weeks of age [34]. A gene expression analysis showed that NF-Y inactivation resulted in decreased expression of several adipocyte-specific markers and adipokines, including leptin, which was reduced 25-fold at 28 weeks of age. When NF-YA knockout mice were treated with leptin for 2 weeks, the metabolic syndrome was partly reversed to a normal state, indicating that the phenotype is partly due to loss of leptin expression. The results of this study demonstrated that NF-Y is needed for adipocyte differentiation and the expression of leptin and other adipokines in vivo. The role of NF-Y in the expression of leptin and other adipokines suggests that NF-Y plays a key role in maintaining metabolic homeostasis in adult animals.
To define NF-Y’s function in the hematopoietic system, the floxed NF-YA allele mice were mated with the MX-Cre transgenic mice, in which Cre expression was regulated by the interferon-inducible promoter that can be activated in bone marrow by treatment with polyinosinic-polycytidylic acid (pIpC) [57]. The bone marrow of floxed-NF-YA/MX-Cre mice was transplanted into the wild-type host animal, and 8 weeks after transplantation, the mice were treated with pIpC, followed by an analysis of the bone marrow and peripheral blood cells. NF-Y inactivation resulted in rapid loss of peripheral blood cell counts and the death of mice, indicating that bone marrow failure caused mice death [33]. A cellular analysis demonstrated that NF-Y is needed for cycling, but not for quiescent HSCs. The NF-YA deletion in HSCs resulted in cell cycle arrest at G2/M phase and induced apoptosis, causing HSC depletion and inhibition in hematopoiesis. The dysregulation of cyclin B1, p21, and Bcl-2, but not p53, might be responsible for cell cycle arrest and apoptosis in HSCs due to the loss of NF-Y activity. In addition, NF-Y inactivation caused a reduction in the expression of several HSC-specific self-renewal genes, such as HOXB4, Notch1, and Bmi-1, indicating that NF-Y functions in the regulation of HSC self-renewal and survival.
4. Perspective
NF-Y regulates the expression of many mammalian cell cycle and house-keeping genes and is needed for cell proliferation and early mouse development. However, it also plays a key role in regulating various cell type-specific genes under different developmental signaling or pathogenic conditions. Our understanding is that NF-Y’s function in vivo is enriched by conditional knockout of the NF-YA gene in different tissues in developed mice after birth. These studies demonstrated that NF-Y is needed for normal tissue homeostasis, metabolism, and survival in adult animals. Inactivation of NF-Y resulted in dysregulation of specific genes in each tissue type, indicating that the mechanism of NF-Y’s function is context specific in different tissues in adult animals. Part of the tissue-specific response can be mediated by NF-Y binding to the tissue-specific distal enhancers that also bind the various cell type-specific master transcription factors that control differentiation and tissue function. In addition, NF-Y’s function in ER stress and cholesterol and fatty acid metabolism can have differential effects in different tissues. Thus, NF-Y can be at the crossroads of different tissue-specific signals, regulating normal homeostasis in adult animals. This finding suggests that NF-Y could be an important biomarker that needs to be evaluated in various human pathogenic conditions, such as in tissue degeneration, metabolic syndrome, and cancer. Such future studies could unravel the role of NF-Y in the context of different tissues during human pathogenesis.
Acknowledgments
The author thanks Ann Sutton for editing the manuscript. The author’s work in this review was supported previously by the National Institutes of Health and is currently supported by the Prostate Cancer Research Program of MD Anderson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 3.Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29–49. doi: 10.3109/10409238.2011.628970. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl GE, Chambers RC, Papakrivopoulou J, Dawson SJ, Jacobsen MC, Bishop JE, Laurent GJ. Activation of fibroblast procollagen alpha 1(I) transcription by mechanical strain is transforming growth factor-beta-dependent and involves increased binding of CCAAT-binding factor (CBF/NF-Y) at the proximal promoter. J Biol Chem. 2002;277:6153–6161. doi: 10.1074/jbc.M108966200. [DOI] [PubMed] [Google Scholar]
- 5.Luo R, Lu JF, Hu Q, Maity SN. CBF/NF-Y controls endoplasmic reticulum stress induced transcription through recruitment of both ATF6(N) and TBP. J Cell Biochem. 2008;104:1708–1723. doi: 10.1002/jcb.21736. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol Cell Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong S, Chirala SS, Wakil SJ. Sterol regulation of human fatty acid synthase promoter I requires nuclear factor-Y- and Sp-1-binding sites. Proc Natl Acad Sci U S A. 2000;97:3948–3953. doi: 10.1073/pnas.040574197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dooley KA, Millinder S, Osborne TF. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 10.Villard J, Peretti M, Masternak K, Barras E, Caretti G, Mantovani R, Reith W. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol Cell Biol. 2000;20:3364–3376. doi: 10.1128/mcb.20.10.3364-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu XS, Linhoff MW, Li G, Chin KC, Maity SN, Ting JP. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013;23:1195–1209. doi: 10.1101/gr.148080.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Q, Maity SN. Stable expression of a dominant negative mutant of CCAAT binding factor/NF-Y in mouse fibroblast cells resulting in retardation of cell growth and inhibition of transcription of various cellular genes. J Biol Chem. 2000;275:4435–4444. doi: 10.1074/jbc.275.6.4435. [DOI] [PubMed] [Google Scholar]
- 14.Basile V, Mantovani R, Imbriano C. DNA damage promotes histone deacetylase 4 nuclear localization and repression of G2/M promoters, via p53 C-terminal lysines. J Biol Chem. 2006;281:2347–2357. doi: 10.1074/jbc.M507712200. [DOI] [PubMed] [Google Scholar]
- 15.Imbriano C, Gnesutta N, Mantovani R. The NF-Y/p53 liaison: well beyond repression. Biochim Biophys Acta. 2012;1825:131–139. doi: 10.1016/j.bbcan.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manni I, Mazzaro G, Gurtner A, Mantovani R, Haugwitz U, Krause K, Engeland K, Sacchi A, Soddu S, Piaggio G. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and cdc25C promoters upon induced G2 arrest. J Biol Chem. 2001;276:5570–5576. doi: 10.1074/jbc.M006052200. [DOI] [PubMed] [Google Scholar]
- 18.Yun J, Chae HD, Choi TS, Kim EH, Bang YJ, Chung J, Choi KS, Mantovani R, Shin DY. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J Biol Chem. 2003;278:36966–36972. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- 19.Lee LC, Chen CM, Wang HC, Hsieh HH, Chiu IS, Su MT, Hsieh-Li HM, Wu CH, Lee GC, Lee-Chen GJ, Lin JY. Role of the CCAAT-binding protein NFY in SCA17 pathogenesis. PLoS One. 2012;7:e35302. doi: 10.1371/journal.pone.0035302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamanaka T, Miyazaki H, Oyama F, Kurosawa M, Washizu C, Doi H, Nukina N. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–839. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Peart MJ, Prives C. Mutant p53 gain of function: the NF-Y connection. Cancer Cell. 2006;10:173–174. doi: 10.1016/j.ccr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Shi Z, Chiang CI, Labhart P, Zhao Y, Yang J, Mistretta TA, Henning SJ, Maity SN, Mori-Akiyama Y. Context-specific role of SOX9 in NF-Y mediated gene regulation in colorectal cancer cells. Nucleic Acids Res. 2015;43:6257–6269. doi: 10.1093/nar/gkv568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimano H. SREBPs: physiology and pathophysiology of the SREBP family. FEBS J. 2009;276:616–621. doi: 10.1111/j.1742-4658.2008.06806.x. [DOI] [PubMed] [Google Scholar]
- 28.Soyal SM, Nofziger C, Dossena S, Paulmichl M, Patsch W. Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol Sci. 2015;36:406–416. doi: 10.1016/j.tips.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Hu Q, Lu JF, Luo R, Sen S, Maity SN. Inhibition of CBF/NF-Y mediated transcription activation arrests cells at G2/M phase and suppresses expression of genes activated at G2/M phase of the cell cycle. Nucleic Acids Res. 2006;34:6272–6285. doi: 10.1093/nar/gkl801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 31.Benatti P, Dolfini D, Vigano A, Ravo M, Weisz A, Imbriano C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011;39:5356–5368. doi: 10.1093/nar/gkr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharya A, Deng JM, Zhang Z, Behringer R, de Crombrugghe B, Maity SN. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- 33.Bungartz G, Land H, Scadden DT, Emerson SG. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood. 2012;119:1380–1389. doi: 10.1182/blood-2011-06-359406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu YH, Dallner OS, Birsoy K, Fayzikhodjaeva G, Friedman JM. Nuclear Factor-Y is an adipogenic factor that regulates leptin gene expression. Mol Metab. 2015;4:392–405. doi: 10.1016/j.molmet.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo R, Klumpp SA, Finegold MJ, Maity SN. Inactivation of CBF/NF-Y in postnatal liver causes hepatocellular degeneration, lipid deposition, and endoplasmic reticulum stress. Sci Rep. 2011;1:136. doi: 10.1038/srep00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanaka T, Tosaki A, Kurosawa M, Matsumoto G, Koike M, Uchiyama Y, Maity SN, Shimogori T, Hattori N, Nukina N. NF-Y inactivation causes atypical neurodegeneration characterized by ubiquitin and p62 accumulation and endoplasmic reticulum disorganization. Nat Commun. 2014;5:3354. doi: 10.1038/ncomms4354. [DOI] [PubMed] [Google Scholar]
- 37.Maity SN, Golumbek PT, Karsenty G, de Crombrugghe B. Selective activation of transcription by a novel CCAAT binding factor. Science. 1988;241:582–585. doi: 10.1126/science.3399893. [DOI] [PubMed] [Google Scholar]
- 38.Saitta B, Gaidarova S, Cicchillitti L, Jimenez SA. CCAAT binding transcription factor binds and regulates human COL1A1 promoter activity in human dermal fibroblasts: demonstration of increased binding in systemic sclerosis fibroblasts. Arthritis Rheum. 2000;43:2219–2229. doi: 10.1002/1529-0131(200010)43:10<2219::AID-ANR9>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Alabert C, Rogers L, Kahn L, Niellez S, Fafet P, Cerulis S, Blanchard JM, Hipskind RA, Vignais ML. Cell type-dependent control of NF-Y activity by TGF-beta. Oncogene. 2006;25:3387–3396. doi: 10.1038/sj.onc.1209385. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang HH, Chen L, Dong J, Gay I, MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Q, Yu Y, Chen Q, Li C, Wu H, Wan Y, Ma J, Sun F. Sox9, a key transcription factor of bone morphogenetic protein-2-induced chondrogenesis, is activated through BMP pathway and a CCAAT box in the proximal promoter. J Cell Physiol. 2008;217:228–241. doi: 10.1002/jcp.21496. [DOI] [PubMed] [Google Scholar]
- 42.Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, Mantovani R, Sacchi A, Piaggio G. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18:2818–2827. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- 43.Gurtner A, Manni I, Fuschi P, Mantovani R, Guadagni F, Sacchi A, Piaggio G. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol Biol Cell. 2003;14:2706–2715. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basile V, Baruffaldi F, Dolfini D, Belluti S, Benatti P, Ricci L, Artusi V, Tagliafico E, Mantovani R, Molinari S, Imbriano C. NF-YA splice variants have different roles on muscle differentiation. Biochim Biophys Acta. 2016;1859:627–638. doi: 10.1016/j.bbagrm.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 45.Katsuno M, Adachi H, Minamiyama M, Waza M, Doi H, Kondo N, Mizoguchi H, Nitta A, Yamada K, Banno H, Suzuki K, Tanaka F, Sobue G. Disrupted transforming growth factor-beta signaling in spinal and bulbar muscular atrophy. J Neurosci. 2010;30:5702–5712. doi: 10.1523/JNEUROSCI.0388-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang S, Ling JJ, Yang S, Li XJ, Li S. Neuronal expression of TATA box-binding protein containing expanded polyglutamine in knock-in mice reduces chaperone protein response by impairing the function of nuclear factor-Y transcription factor. Brain. 2011;134:1943–1958. doi: 10.1093/brain/awr146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci U S A. 2005;102:11728–11733. doi: 10.1073/pnas.0503405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y, Chen C, Liu S, Liu D, Chen Y, Zandi E, Chen W, Zhou Y, Shi S. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity. 2015;43:251–263. doi: 10.1016/j.immuni.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolfini D, Minuzzo M, Pavesi G, Mantovani R. The short isoform of NF-YA belongs to the embryonic stem cell transcription factor circuitry. Stem Cells. 2012;30:2450–2459. doi: 10.1002/stem.1232. [DOI] [PubMed] [Google Scholar]
- 50.Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, Jothi R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell. 2014;55:708–722. doi: 10.1016/j.molcel.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 53.Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 54.Misawa H, Nakata K, Toda K, Matsuura J, Oda Y, Inoue H, Tateno M, Takahashi R. VAChT-Cre. Fast and VAChT-Cre.Slow: postnatal expression of Cre recombinase in somatomotor neurons with different onset. Genesis. 2003;37:44–50. doi: 10.1002/gene.10224. [DOI] [PubMed] [Google Scholar]
- 55.Yamanaka T, Tosaki A, Miyazaki H, Kurosawa M, Koike M, Uchiyama Y, Maity SN, Misawa H, Takahashi R, Shimogori T, Hattori N, Nukina N. Differential roles of NF-Y transcription factor in ER chaperone expression and neuronal maintenance in the CNS. Sci Rep. 2016;6:34575. doi: 10.1038/srep34575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, Kahn BB, Kahn CR. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gudmundsson KO, Oakley K, Han Y, Du Y. Analyzing gene function in adult long-term hematopoietic stem cells using the interferon inducible Mx1-Cre mouse system. Methods Mol Biol. 2014;1194:313–325. doi: 10.1007/978-1-4939-1215-5_17. [DOI] [PubMed] [Google Scholar]