Abstract
Cancer genomic instability contributes to the phenomenon of intratumoral genetic heterogeneity, provides the genetic diversity required for natural selection and enables the extensive phenotypic diversity that is frequently observed among patients. Genomic instability has previously been associated with poor prognosis. However, we have evidence that for solid tumors of epithelial origin, extreme levels of genomic instability, where more than 75% of the genome is subject to somatic copy number alterations, are associated with a potentially better prognosis compared to intermediate levels under this threshold. This has been observed in clonal subpopulations of larger size, especially when genomic instability is shared among a limited number of clones. We hypothesize that cancers with extreme levels of genomic instability may be teetering on the brink of a threshold where so much of their genome is adversely altered that cells rarely replicate successfully. Another possibility is that tumors with high levels of genomic instability are more immunogenic than other cancers with a less extensive burden of genetic aberrations. Regardless of the exact mechanism, but hinging on our ability to quantify how a tumor’s burden of genetic aberrations is distributed among coexisting clones – genomic instability has important therapeutic implications. Herein, we explore the possibility that a high genomic instability could be the basis for a tumor’s sensitivity to DNA damaging therapies. We primarily focus on studies of epithelial-derived solid tumors.
Keywords: genomic instability, clonal evolution, error catastrophe, DNA-damaging therapy, TP53
Introduction
Genomic instability has long been recognized as one of the drivers of carcinogenesis and acquired therapeutic resistance. Genomic instability has been called a “facilitating characteristic” that helps to generate the hallmarks of cancer (1,2). Cancer cells accrue thousands of mutations. They lose and gain entire segments of chromosomes that can involve whole chromosomal arms. Even in cases where cancer cells have the majority of their genomes altered by somatic copy number changes, these cells continue to flourish (3). How cells tolerate such extensive changes in genomic integrity remains a mystery. As long as those changes are confined to gene dosage, they may generally be tolerable at the cell level (4,5). But when genomic instability continues to increase it eventually reaches a level where the consequences of gross genomic changes become lethal to the cell. Evidence that the extent of mutations cannot increase endlessly without adversely affecting cell fitness (6–10) implies the existence of a tolerance limit of genomic instability.
One question is whether the genomic instability limit is related to the rate at which new mutations occur or the cumulative mutation burden that accrues over time. Similarly, what kinds of mutations drive a cell towards the limit; how does the impact of somatic point mutations differ from large-scale somatic copy number alterations (Fig. 1a,b)? In this review we will discuss the possibility of a non-monotonic relation between genomic instability and cancer cell fitness. Hereby, we distinguish between four different perspectives on genomic instability, comparing somatic point mutations to copy number alterations on one hand and mutation rate to cumulative mutation burden on the other hand. We discuss cell-intrinsic and cell-extrinsic mechanisms that could account for this non-monotonic relation (Figure 1c–e). We discuss the challenges of measuring genomic instability in a heterogeneous population and finish with perspectives on potential prognostic and therapeutic implications (Figure 2).
Figure 1. Alternative perspectives on genomic instability and mechanisms that limit genomic instability tolerance.
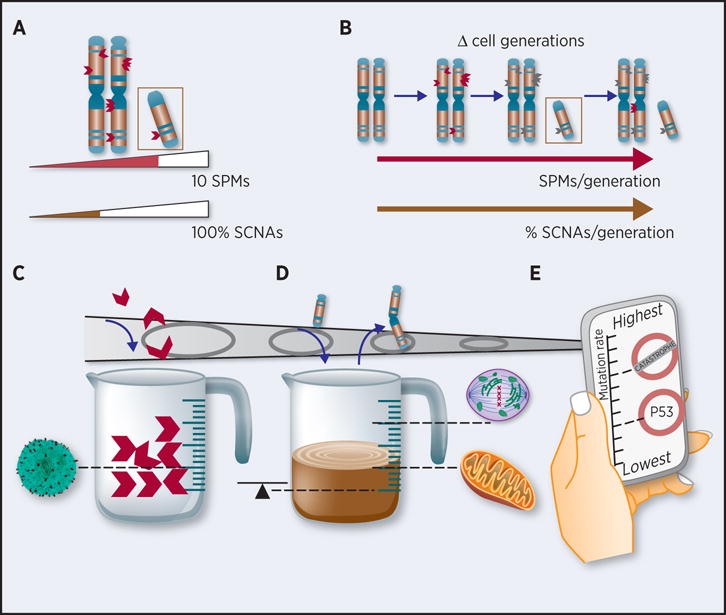
(a) Snapshot of a diploid chromosome’s burden of somatic point mutations (SPMs) and of somatic copy number alterations (SCNAs) at the time of biopsy collection. Mutation burden measured as the total number of SPMs (orange) or as the % chromosome affected by SCNAs (here single-copy gain of one chromosome arm: black). (b) Mutation rate measured as the mutation burden net difference between time points divided by the number of cell generations that took place between those time points. (c–e) Mechanisms that can account for reduced cell fitness due to (c) SPM burden; (d) SCNA burden; (e) SPM- or SCNA rate. Mechanisms include: (c) attack by immune cells, (d) disturbed homeostasis due to gene expression imbalance, impaired mitosis, high energy demands for cell maintenance, (e) mutational meltdown and activation of apoptosis.
Figure 2. Model of DNA damage sensitivity as a function of genomic instability.
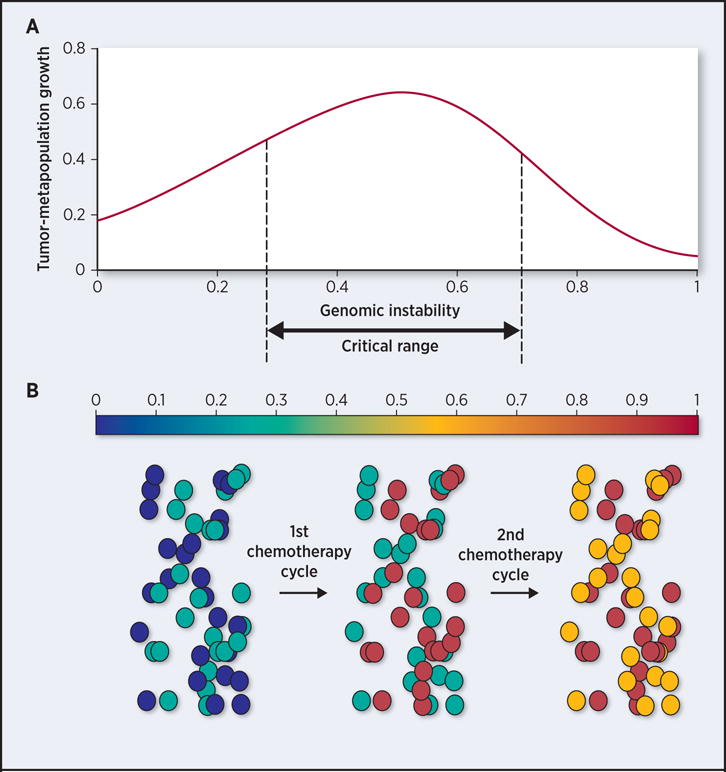
(a) Tumor-metapopulation fitness (y-axis) is maximal when it consists of subpopulations with intermediate levels of genomic instability (critical range), defined as the proportion of the genome that is altered (x-axis). (b) 50 cells (circles) sampled from a heterogeneous tumor-metapopulation consisting of two subclones of variable genomic instability (colorbar). First round of DNA damaging agents causes a shift in the genomic instability of each subclone, such that one subclone (turquoise) is shifted out of the critical range (now orange), while the other one (dark blue) transitions into a critical range of genomic instability (turquoise). A second round of therapy is needed to push both subpopulations out of the critical range of genomic instability and into the range where genomic instability reduces their fitness.
Cumulative somatic point mutation (SPM) burden
A tumor’s SPM burden at a given time point depends on the efficacy of SPM appearance (i.e. SPM rate) and clearance (e.g. TP53 induced apoptosis) and on how many cell generations have occurred after the first tumor cell of origin emerged.
In some cancer types, patients whose tumors have a high SPM burden have a consistently improved prognosis compared to those with a low SPM content. For example, a favorable prognosis is seen in patients with colorectal cancers that have microsatellite instability (MSI) (11). This molecular phenotype results from the loss of DNA mismatch repair, which leads to a dramatically increased mutation rate. The favorable prognosis linked to MSI positive tumors suggests that MSI positive tumor cells have difficulty tolerating either the rate at which SPMs accumulate in these tumors, or the overall SPM burden.
Intolerance to high SPM burden could be accounted for by the immune system (Figure 1c). Tumors with high SPM burden generate new peptides via nonsynomous mutations, frameshifts and other genetic aberrations in coding regions. These mutations lead to new amino acid sequences (i.e. neoantigens) that the immune system has not encountered. The concept of immune surveillance of cancers is based on genomic instability generating non-self epitopes (12). There is evidence that tumors with more point mutations (13,14) respond better to immune checkpoint therapy than more genetically stable tumors, the most dramatic examples being MSI-positive colorectal cancers (15). A similar correlation is observed in metastatic melanomas that are known to have high mutation burden resulting from ultraviolet exposure (16).
Investigating the relationship between genomic instability and immunogenicity is complicated by the fact that most cancers evolve mechanisms to evade the immune system. Thus, in some cases, the number of immunogenic neoantigens may have no effect as tumors have effectively suppressed the immune cellular response (12).
Cumulative burden of somatic copy number alterations (SCNAs)
SCNAs differ from SPMs in that they can encompass vast regions of a cell’s genome. SCNAs have a locus-specific frequency that is two to four orders of magnitude greater than that of point mutations (17). A measure of cumulative SCNA burden is defined as the percentage of a genome affected by SCNAs (18).
SCNAs confer substantial phenotypic plasticity and have been described as the driving force of genetic diversification (19). There is evidence supporting a greater role for SCNAs rather than SPMs in developing and maintaining neoplastic cell population diversity (19–21). Specifically, gene duplication may be the most important evolutionary force on the organism level (22). However, we and others have found evidence for a limit in the cumulative SCNA burden that a tumor with its diverse clonal subpopulations can tolerate (3,23,24).
Using fluorescent in-situ hybridization (FISH) of chromosomes 2 and 15 centromeres, a study of estrogen receptor (ER) negative breast tumors revealed that patients with tumors where greater than 45% of the cells had chromosomal abnormalities had a significantly better prognosis compared to patients with lower number of chromosomal abnormalities (24). Chromosomes 2 and 15 were selected because they are rarely altered in breast cancer, and so copy number changes in those chromosomes are probably indicative of widespread instability across the genome. Along similar lines, another study examined the gene expression of breast cancers that were ER negative and HER2 negative (a.k.a., ERBB2) (23). Those ER-/HER2- tumors with a gene expression indicative of chromosomal instability (highest quartile) were associated with increased time until relapse compared to ER-/HER2- breast cancer with expression levels in the next lowest quartile (23). The same study observed slower progression of squamous non-small cell lung tumors, ovarian and gastric tumors burdened with high genomic instability as compared to intermediate levels (23).
Our study analyzed the exomes of more than 1,000 tumors across 12 cancer types from the Cancer Genome Atlas Project (TCGA) and also found evidence for the existence of a limit in a tumor’s ability to tolerate extensive SCNAs (3). Tumors with 50–75% cumulative SCNA burden (as defined above) represent the highest risk group among individuals with bladder cancer, head and neck cancer, lung adenocarcinoma, stomach adenocarcinoma, cervical cancer and low-grade gliomas (3). Notably, this 75% threshold is not a result of artificially splitting the cohorts into quartiles, but is the fraction of the tumor’s genomes affected by SCNAs. Our results suggest that tumors with greater than 75% SCNA burden have exceeded a threshold of tolerable SCNA burden that limits the tumor’s lethal potential.
We confirmed the same threshold of 75% cumulative SCNA burden when analyzing two additional datasets, including 2,010 tumors across seven distinct cancer types (3). The persistent observation of a tolerance limit when 75% of a cancer’s genome is affected by SCNAs, across several cancer types, is surprising. It suggests that the mechanism that limits the cumulative total of SCNAs may be cell-type independent.
This limit to genomic instability may be explained by several biophysical constraints. For example, changes in chromosome size alter chromosomal alignment to the center of the nucleus and thereby impact metaphase efficiency (25). Chromosome size and shape also influence what force is needed to pull sister chromatids apart during mitosis (26). Large SCNAs may result in altered chromosomal structures that require higher forces than the spindle apparatus can exert. Whether or not the limit on the accumulation of SCNAs is due to such biophysical constraints remains an open question.
An alternative mechanism that could account for the SCNA tolerance limit involves gene dosage. DNA copy number correlates positively with expression levels for 99% of all abundantly expressed human genes (5). Cancers with a significant number of large-scale SCNAs may amplify the expression of neoantigens. Tumors with these amplification characteristics may have a higher degree of immune cellular infiltration compared to tumors that do not have such extensive changes in their genomes.
A special case of large-scale SCNAs occurs with whole-genome doubling, when the ploidy of a given tumor cell increases. Copy number profiling of more than 4,900 primary tumors across 11 cancer types has shown these somatic events to occur in a surprisingly large fraction of 37% of cases (27). For some cancer types, as many as 70% of tumors demonstrate aneuploidy (28). Whole-genome doubling has been attributed as an early event in tumor progression (28,29). There are potential benefits to ploidy increases. For example, a study in flowering plants has shown that masking deleterious mutations can increase the fitness of tetraploids relative to diploids for hundreds to thousands of generations (30). Having four copies of every gene may benefit tetraploid tumors to tolerate more functionally disruptive SPMs, but potentially at the cost of having exceeded the GI limit. Alternatively, even though whole-genome doublings change the copy number status of 100% of the genome, the uniformity of whole-genome doublings may render these events neutral with respect to reaching the GI limit. Distinguishing between these possibilities will require further studies.
Somatic point mutation rate
Among tumors with high levels of SPMs, a proportion has high error rates versus those with low error rates that have steadily accumulated SPMs over extended periods of time. Simulation studies have provided some insights into how natural selection adjusts mutation rate among asexually reproducing populations (7,31). For example, one study found that under fluctuating environmental conditions (e.g. changes in temperature or oxygen availability), the SPM rate increases until it reaches an intolerable level that leads to extinction (32). Importantly, these models suggest that fitness beyond this threshold declines precipitously (7,31).
Another model found that a higher SPM rate acquired as an early event in neoplastic transformation, leads to rapid tumor growth (33). However, beyond an optimum point mutation rate, this model found with higher mutation rates, negative clonal selection occurs that is less favorable for cellular expansion. The location of the optimum in this model was highly variable due to its dependence on the fitness landscape.
Most cancer genome studies provide an individual tumor’s SPMs at a single time point. In general, one needs multiple samples from at least two time points to accurately estimate mutation rate (34,35). The scarcity of longitudinal data makes it difficult to investigate whether a tumor’s mutation rate is bound by an upper limit. Empirical proof for the above-mentioned theoretical models comes from unicellular organisms with small genomes or from viruses such as the human immunodeficiency virus (HIV). For example, even slight increases in the SPM rate of HIV leads to decreased viral fitness, suggesting that the mutation rate in HIV is indeed in close proximity to the error threshold (36). In the case of cancer, measuring SPM rate is difficult because it requires sampling the same tumor at multiple time points, and tracing the evolution of multiple subclones that coexist within that tumor.
Apart from these experimental challenges, the inverse correlation between genome sequence length and SPM tolerance (8,31,37) suggests that the threshold may have limited relevance in cancer because of the vast extent of non-coding regions in the human genome.
The mechanisms that account for a limit of the SPM rate are distinct from those that could explain a limit in SPM burden tolerance. For example, the immune system is likely more sensitive to a cell’s effective overall SPM burden (represented as the net count of neoantigens presented) than to the rate at which these SPMs had accumulated inside the cell. What mechanisms could account for the existence of an SPM rate limit?
The concept of an error rate limit was introduced by Schrodinger in the context of the origin of life (38). For life to evolve and natural selection to work, information must be preserved through inheritance. If the mutation rate exceeds the catastrophe threshold or “error threshold” (7,37) (Figure 1e), then the information in the genome decays and cannot be maintained across generations. This phenomenon has been referred to as a mutational meltdown and is applicable to unicellular organisms, multicellular organisms and neoplastic cells (39,40). Theoretical models and evolutionary experiments support the existence of such an error catastrophe threshold (6–10).
As another alternative mechanism, gatekeeper genes may account for the existence of an SPM rate limit. For example, the TP53 protein, a tumor suppressor, is a regulator of a G1 cell cycle checkpoint and of apoptosis following DNA-damage (41). Oxidative stress is both a potent inducer of TP53 (42) and a primary cause of somatic SPMs (43, 44). The conditions that cause the acquisition of SPMs beyond a certain threshold within a single cell generation may also activate TP53 and other gatekeeper genes, halting the cell cycle and initiating apoptosis (Figure 1e).
Somatic copy number alteration rate
The large size of large-scale SCNAs relative to the human genome renders SCNA rate even more difficult to measure than SPM rate. While the genomic coordinates of two independently acquired SPMs are highly unlikely to overlap, the same is not true for SCNAs. A tumor can loose one copy of a diploid chromosome early in tumor development only to undergo duplication of the remaining copy in later cell generations. While overall SPM burden can be a proxy of SPM rate (when also considering tumor size), the fluctuating presence of SCNAs renders overall SCNA burden less accurate in approximating SCNA rate.
Two studies measured the SCNA rate and clonal dynamics of Barrett’s esophagus (34,45) – a dysplastic condition that has a risk of transforming into invasive carcinoma. In the first study, multiple biopsies from four or more timepoints were subject to analysis with SNP arrays – phylogenetic methods were used to estimate the SCNA rate. The authors found that in general, when patients with Barrett’s esophagus started taking non-steroidal anti-inflammatory drugs, their SCNA rate dropped by an order of magnitude (34). In the second study, multicolor fluorescent in situ hybridization (FISH) was applied to brush cytology specimens from two to three time points. Using this data, the authors measured the frequency and rate of clonal expansions. These studies point to experimental methods that can be used to measure the SCNA rate in other types of tumors.
Apoptosis initiated by DNA damage may account for an SCNA rate limit. The repair of DNA double-strand breaks is a mechanism for SCNAs (17). Moreover, DNA breaks can initiate TP53-dependent signal transduction. In contrast, DNA lesions other than strand breaks, such as thymine dimers, are not directly capable of triggering TP53 induction (46). This observation suggests that gatekeeper genes may limit the tolerable SCNA rate more so than SPM rate, at least until those gatekeeper genes themselves are deleted or inactivated.
Distinction between intra-tumor genetic heterogeneity and genomic instability
Genomic instability is the driving force of intra-tumor genetic heterogeneity. If a cell’s ability to tolerate genomic instability is limited, then population plasticity may be limited as well. We examined over 1,000 treatment-naïve tumors and identified a non-monotonic association between survival and the number of clones detected at more than 10% cell frequency (3). We found that increasing clone number was associated with decreased overall survival. However, this relationship was only observed up to a maximum clonal diversity of four subclones. A clonal count higher than four per a tumor was not associated with further reduction in overall survival. In fact, we conducted a multivariate analysis and discovered that diversity beyond four clonal populations per tumor was associated with an increase in overall SCNA burden and a significant increase in overall survival (3).
Experimentally, it is difficult to distinguish genomic instability events in the context of intratumoral heterogeneity. For example, bulk exome sequencing of a tumor sample represents the aggregate genomes of all of the clones that coexist within the sample. A tumor that demonstrates a SCNA genome percentage of 75% or greater (3) may either be composed of many clones with low levels of SCNA burden per clone or of a few clones, each with high levels of SCNA burden across their genomes. Subsequent analysis suggests that it is important to distinguish between these two scenarios (3). We found that SCNA burden greater than 75% was prognostic of a favorable outcome, but especially so when shared among one or two clones. When high SCNA burden was spread among many clones, the outcome was less favorable, indicating that it is the SCNA burden per clone that limits the viability of the tumor-metapopulation. A limitation of our study is that we only quantified SCNA burden per tumor metapopulation, but did not measure SCNA burden per individual clones. Geographic analysis of tumors will provide a better resolution on the location of a genomic instability limit and on a clone’s proximity to the limit than standard bulk genomic sequences available at TCGA.
In our analysis of the TCGA data, we detected a clonal subpopulation if it accounted for at least 10% of the sample. Rare clones are difficult to detect in most bulk sequencing studies. Yet these are the clones that have evolved most recently, prior to sample collection, giving a more up-to-date picture of the evolutionary dynamics in a tumor than larger clones (47). Resistance to therapy may lie with these small clonal lineages. Deep-sequencing and single cell sequencing methods carry the potential to characterize these small clones (48–50). For example, duplex sequencing enables the detection of a single mutated sequence among tens of millions of wild-type sequences (49).
Therapeutic interventions and the genomic instability limit
DNA damaging agents increase mutation rate within a tumor and the tumor’s cumulative mutation burden (51,52). Comparisons between primary versus relapsed acute myeloid leukemia mutations revealed an increase in transversions – this category of variants is attributable to DNA damage caused by cytotoxic chemotherapy and were not detected in the primary cancer (53).
The mechanisms behind DNA-damage induced increases in mutation rate and burden are well understood. For example, platinum-based compounds, such as cisplatin, bind DNA to generate bulky adducts or intra-strand and inter-strand cross-links between purine bases. These modifications lead to stalled replication forks and the formation of double-strand breaks (54). This type of DNA-damage either leads to cell-death or the double-strand breaks are repaired by homologous recombination (HR) or by non-homologous end-joining. Both repair mechanisms can sometimes generate somatic structural variation that are seen as SCNAs (17).
While there is strong evidence that DNA damaging agents, even at low doses, increase mutation rate and burden (51,52), the magnitude of this effect is variable. It depends in large part on the tumor’s DNA damage response and the functional status of TP53. On the extreme end of the spectrum are elephant cells, which have evolved at least 40 copies of TP53 and undergo apoptosis in response to very low levels of DNA damage (55).
Some studies suggest that increased SCNA burden not only is a consequence of DNA-damaging therapy but also increases the sensitivity to DNA-damage. For example, elevated DNA content, arising from aneuploidy or duplication events in the cancer genome, was associated with higher DNA-damage sensitivity in neuroblastoma (56) and laryngeal carcinoma (57). As a last example, in low-grade gliomas – a tumor type with relatively low SCNA burden – chemotherapy with temozolomide, an alkylating agent, is associated with high rates of progression and frequent temozolomide exposure induces a hypermutator phenotype (58). These studies suggest that tumor cells with higher levels of SCNAs are more sensitive to an initial exposure of DNA damaging agents. If this is true, DNA damaging agents may select genomically stable clones among the various subclones that exist in a tumor. Supporting this concept, a recent study demonstrated that when a transposon–driven, functional genomic mouse model of medulloblastoma was treated with ‘humanized’ in vivo therapy (tumor resection followed by multi-fractionated radiotherapy), after treatment the tumors converged on a single pathway: genomic stability via enrichment in TP53 pathway gene sets (59).
Measuring a tumor’s degree of genomic instability prior to therapy may be informative for assessing chemotherapy. DNA damage therapy dose could be optimized such that it induces additional SCNAs necessary and sufficient to lead the cancer past the limit of tolerable genomic instability. Whether this strategy is feasible will depend on the sharpness of the genomic instability limit, the overall clonal heterogeneity of a tumor population and the genomic instability of each clone (Figure 2). Future studies investigating what mechanisms prevent a cell from tolerating excessive mutations will inform the circumstances under which a sharp threshold is to be expected.
Complementary agents that also increase the mutation burden of a tumor are inhibitors of DNA damage repair. Poly-ADP ribose polymerase (PARP) inhibitors belong to this category. PARP inhibitors lead to synthetic lethality in tumors with BRCA1 and BRCA2 mutations. BRCA-mutated cancers have defective HR. By targeting the base excision repair pathway, PARP inhibitors further impair the cells ability to repair DNA damage in cells lacking BRCA1 and BRCA2 function (60). PARP has a limited role in altering double strand break repair (61). Thus, PARP inhibitors primarily increase the accumulation/rate of SPMs rather than SCNAs.
Conclusions
We describe evidence supporting the existence of a genomic instability limit. The existence of a limit may have significant therapeutic implications. Our studies and others suggest that a subset of tumors lacking genomic instability may not pass this threshold even when treated with maximum tolerable doses of DNA damaging agents, continue to grow and lead to a worse prognosis.
We need to better understand the mechanisms that hinder a cell from tolerating genomic instability beyond a certain threshold and what the cellular consequences of exceeding this threshold are. For example, will passing the genomic instability limit influence a cell’s probability to seed metastasis? If the existence of a genomic instability limit is a manifestation of enhanced immunogenicity, then the ability of the cells to survive in circulation may be compromised due to a higher proportion of immune cells that exist in circulation (62). On the other hand, if the existence of a genomic instability limit is a manifestation of compromised mitosis, then we would expect the bottleneck to metastasis to be growth in the new metastatic niche rather than survival in circulation.
Further research is needed to shed light on the relevance of pre-existing extent of genomic alterations on DNA-damage sensitivity. To influence how clinical decisions are made regarding the use of DNA-damaging agents, such studies must first deconvolute the phenotypic effects of cumulative mutation burden from that of individual mutations. Overcoming this challenge will require analyzing large patient cohorts or genome engineering model systems with techniques such as CRISPRs. These contributions could help guide clinical decisions on how to dose DNA-damaging agents and on whether or not DNA-damaging agents are an appropriate therapy option. This could improve clinical outcome by allowing us to pinpoint those who would respond better and longer to lower doses of DNA-damaging agents, than to higher doses.
Acknowledgments
Financial support
C. Maley was supported by NIH grants P01 CA91955, R01 CA149566, R01 CA170595, R01 CA185138 and R01 CA140657 as well as CDMRP Breast Cancer Research Program Award BC132057. N. Andor and H. Ji were supported by the awards from the Don and Ruth Seiler Fund, the NCI Cancer Target Discovery and Development (CTDD) Consortium (U01CA17629901) and the NCI Integrative Cancer Biology Program (U01CA17629901). Additional support to H. Ji came from the Doris Duke Charitable Foundation Clinical Scientist Development Award, Research Scholar Grant, RSG-13-297-01-TBG from the American Cancer Society and a Howard Hughes Medical Institute Early Career Grant. Additional support to N. Andor came from Stanford University Dean’s Postdoctoral Fellowship Award.
The findings, opinions and recommendations expressed here are those of the authors and not necessarily those of the universities where the research was performed or the National Institutes of Health
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–13. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstein AC, Otto SP. Ploidy and the causes of genomic evolution. J Hered. 2009;100:571–81. doi: 10.1093/jhered/esp057. [DOI] [PubMed] [Google Scholar]
- 5.Fehrmann RSN, Karjalainen JM, Krajewska M, Westra H-J, Maloney D, Simeonov A, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–25. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 6.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. BioEssays News Rev Mol Cell Dev Biol. 2000;22:1057–66. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Nowak M, Schuster P. Error thresholds of replication in finite populations mutation frequencies and the onset of muller’s ratchet. J Theor Biol. 1989;137:375–95. doi: 10.1016/s0022-5193(89)80036-0. [DOI] [PubMed] [Google Scholar]
- 8.Bagnoli F, Bezzi M. Eigen’s Error Threshold and Mutational Meltdown in a Quasispecies Model. Int J Mod Phys C. 1998;09:999–1005. [Google Scholar]
- 9.Herr AJ, Ogawa M, Lawrence NA, Williams LN, Eggington JM, Singh M, et al. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7:e1002282. doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153–62. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vesely MD, Schreiber RD. Cancer immunoediting: antigens, mechanisms, and implications to cancer immunotherapy. Ann N Y Acad Sci. 2013;1284:1–5. doi: 10.1111/nyas.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santarpia M, Karachaliou N. Tumor immune microenvironment characterization and response to anti-PD-1 therapy. Cancer Biol Med. 2015;12:74–8. doi: 10.7497/j.issn.2095-3941.2015.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh PP, Sharma PK, Krishnan G, Lockhart AC. Immune checkpoints and immunotherapy for colorectal cancer. Gastroenterol Rep. 2015;3:289–97. doi: 10.1093/gastro/gov053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hastings P, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cazier J-B, Rao SR, McLean CM, Walker AL, Wright BJ, Jaeger EEM, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun [Internet] 2014 doi: 10.1038/ncomms4756. [cited 2014 Jul 25];5. Available from: http://www.nature.com/ncomms/2014/140429/ncomms4756/full/ncomms4756.html#supplementary-information. [DOI] [PMC free article] [PubMed]
- 19.Mroz EA, Tward AM, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, Mulla WA, Kucharavy A, Tsai H-J, Rubinstein B, Conkright J, et al. Targeting the adaptability of heterogeneous aneuploids. Cell. 2015;160:771–84. doi: 10.1016/j.cell.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–50. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soukup SW. Evolution by gene duplication. In: Ohno S, editor. Teratology. Vol. 9. Springer-Verlag; New York: 1974. pp. 160pp. 250–1. 1970. [Google Scholar]
- 23.Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–52. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2011;20:2183–94. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke K, Cheng J, Hunt AJ. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr Biol CB. 2009;19:807–15. doi: 10.1016/j.cub.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khatibzadeh N, Stilgoe AB, Bui AAM, Rocha Y, Cruz GM, Loke V, et al. Determination of motility forces on isolated chromosomes with laser tweezers. Sci Rep. 2014;4:6866. doi: 10.1038/srep06866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Grönroos E, Endesfelder D, et al. Tolerance of Whole-Genome Doubling Propagates Chromosomal Instability and Accelerates Cancer Genome Evolution. Cancer Discov. 2014;4:175–85. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–37. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 31.Eigen M. Error catastrophe and antiviral strategy. Proc Natl Acad Sci. 2002;99:13374–6. doi: 10.1073/pnas.212514799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang X, Mu B, Huang Z, Zhang M, Wang X, Tao S. Impacts of mutation effects and population size on mutation rate in asexual populations: a simulation study. BMC Evol Biol. 2010;10:298. doi: 10.1186/1471-2148-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckman RA. Mutator mutations enhance tumorigenic efficiency across fitness landscapes. PloS One. 2009;4:e5860. doi: 10.1371/journal.pone.0005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostadinov RL, Kuhner MK, Li X, Sanchez CA, Galipeau PC, Paulson TG, et al. NSAIDs modulate clonal evolution in Barrett’s esophagus. PLoS Genet. 2013;9:e1003553. doi: 10.1371/journal.pgen.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. Measurably evolving populations. Trends Ecol Evol. 2003;18:481–8. [Google Scholar]
- 36.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci U S A. 1999;96:1492–7. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biebricher CK, Eigen M. The error threshold. Virus Res. 2005;107:117–27. doi: 10.1016/j.virusres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Schroedinger E. What is life? The physical aspect of the living cell. London: The Folio Society; 2000. [Google Scholar]
- 39.Zeyl C, Mizesko M, de Visser JA. Mutational meltdown in laboratory yeast populations. Evol Int J Org Evol. 2001;55:909–17. doi: 10.1554/0014-3820(2001)055[0909:mmilyp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Lynch M, Bürger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84:339–44. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 41.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gambino V, De Michele G, Venezia O, Migliaccio P, Dall’Olio V, Bernard L, et al. Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell. 2013;12:435–45. doi: 10.1111/acel.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Degtyareva NP, Heyburn L, Sterling J, Resnick MA, Gordenin DA, Doetsch PW. Oxidative stress-induced mutagenesis in single-strand DNA occurs primarily at cytosines and is DNA polymerase zeta-dependent only for adenines and guanines. Nucleic Acids Res. 2013;41:8995–9005. doi: 10.1093/nar/gkt671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledó JI, Howe DK, et al. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci U S A. 2009;106:16310–4. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez P, Timmer MR, Lau CT, Calpe S, Sancho-Serra MDC, Straub D, et al. Dynamic clonal equilibrium and predetermined cancer risk in Barrett’s oesophagus. Nat Commun. 2016;7:12158. doi: 10.1038/ncomms12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson WG, Kastan MB. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–23. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kostadinov R, Maley CC, Kuhner MK. Bulk Genotyping of Biopsies Can Create Spurious Evidence for Hetereogeneity in Mutation Content. PLOS Comput Biol. 2016;12:e1004413. doi: 10.1371/journal.pcbi.1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang L, Chen H, Pinello L, Yuan G-C. GiniClust: detecting rare cell types from single-cell gene expression data with Gini index. Genome Biol. 2016;17:144. doi: 10.1186/s13059-016-1010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc. 2014;9:2586–606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–60. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gourabi H, Mozdarani H. A cytokinesis-blocked micronucleus study of the radioadaptive response of lymphocytes of individuals occupationally exposed to chronic doses of radiation. Mutagenesis. 1998;13:475–80. doi: 10.1093/mutage/13.5.475. [DOI] [PubMed] [Google Scholar]
- 52.Kumar D, Kumari S, Salian SR, Uppangala S, Kalthur G, Challapalli S, et al. Genetic Instability in Lymphocytes is Associated With Blood Plasma Antioxidant Levels in Health Care Workers Occupationally Exposed to Ionizing Radiation. Int J Toxicol. 2016 doi: 10.1177/1091581815625593. [DOI] [PubMed] [Google Scholar]
- 53.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–52. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA. 2015:1–11. doi: 10.1001/jama.2015.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Look AT, Hayes FA, Nitschke R, McWilliams NB, Green AA. Cellular DNA content as a predictor of response to chemotherapy in infants with unresectable neuroblastoma. N Engl J Med. 1984;311:231–5. doi: 10.1056/NEJM198407263110405. [DOI] [PubMed] [Google Scholar]
- 57.Gregg CM, Beals TE, McClatchy KM, Fisher SG, Wolf GT. DNA content and tumor response to induction chemotherapy in patients with advanced laryngeal squamous cell carcinoma. Otolaryngol–Head Neck Surg Off J Am Acad Otolaryngol-Head Neck Surg. 1993;108:731–7. doi: 10.1177/019459989310800616. [DOI] [PubMed] [Google Scholar]
- 58.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrissy AS, Garzia L, Shih DJH, Zuyderduyn S, Huang X, Skowron P, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature. 2016;529:351–7. doi: 10.1038/nature16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livraghi L, Garber JE. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med. 2015;13:188. doi: 10.1186/s12916-015-0425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rulten SL, Fisher AEO, Robert I, Zuma MC, Rouleau M, Ju L, et al. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Kitamura T, Qian B-Z, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]


