Abstract
Helicobacter pylori is the strongest risk factor for gastric adenocarcinoma, yet only a minority of infected persons ever develop this malignancy. One cancer-linked locus is the cag type 4 secretion system (cagT4SS), which translocates an oncoprotein into host cells. A structural component of the cagT4SS is CagY, which become rapidly altered during in vivo adaptation in mice and rhesus monkeys, rendering the cagT4SS nonfunctional; however, these models rarely develop gastric cancer. We previously demonstrated that the H. pylori cag+ strain 7.13 rapidly induces gastric cancer in Mongolian gerbils. We now use this model, in conjunction with samples from patients with premalignant lesions, to define the effects of a carcinogenic host environment on the virulence phenotype of H. pylori to understand how only a subset of infected individuals develop cancer. H. pylori cagY sequence differences and cagT4SS function were directly related to the severity of inflammation in human gastric mucosa in either a synchronous or metachronous manner. Serial infections of Mongolian gerbils with H. pylori strain 7.13 identified an oscillating pattern of cagT4SS function. The development of dysplasia or cancer selected for attenuated virulence phenotypes, but robust cagT4SS function could be restored upon infection of new hosts. Changes in the genetic composition of cagY mirrored cagT4SS function, although the mechanisms of cagY alterations differed in human isolates (mutations) versus gerbil isolates (addition/deletion of motifs). These results indicate that host carcinogenic phenotypes modify cagT4SS function via altering cagY, allowing the bacteria to persist and induce carcinogenic consequences in the gastric niche.
Introduction
Infection with Helicobacter pylori is the strongest known risk factor for gastric cancer, a disease that claims >700,000 lives per year (1), yet the precise mechanisms that regulate cancer development in response to this pathogen are less well defined. In many regions of the world, the rates of H. pylori infection and gastric cancer are concordant; however, this association is not universal. In Colombia, the prevalence of H. pylori is very high throughout the country, but individuals residing in the mountains have high rates of gastric cancer, whereas those on the coast have very low rates (2). Kodaman et al. recently showed that specific interactions between microbial and human genetic ancestries clearly predicted the risk for gastric cancer in Colombia (3). These findings indicate that aberrant co-evolution between H. pylori and its host may affect pathogenesis.
One specific cancer-linked H. pylori locus is the cag pathogenicity island (cagPAI), which encodes a type IV secretion system (T4SS) that translocates the oncoprotein CagA, peptidoglycan, and DNA into host cells (4-6). A structural component of the cagT4SS is CagY, which is required for NF-κB-driven pro-inflammatory cytokine secretion. cagY encodes for an ~1900 amino acid protein, which is susceptible to rearrangements, compromising the length and function of the protein. Previous studies have shown that CagY can become rapidly altered during in vivo adaptation in mice and Rhesus monkeys, raising the hypothesis that CagY can mediate evasion of the host immune response (7,8). However, these models rarely develop gastric cancer. In contrast, infection of Mongolian gerbils with H. pylori leads to gastric adenocarcinoma in approximately 60% of infected animals (9-12).
To address the hypothesis that a carcinogenic environment within the stomach alters H. pylori virulence, we first utilized a unique set of paired H. pylori clinical isolates (13). Archival and recent H. pylori strain J99 are human clinical strains isolated from a single patient that underwent endoscopy six years apart. During this time period, a shift occurred in the pattern of H. pylori-induced inflammation in this patient, from antral-predominant to corpus-predominant gastritis, a crucial step in the cascade to gastric carcinogenesis (14). We now use these unique samples, in conjunction with the Mongolian gerbil model of cancer, to define the effects of a carcinogenic host environment on the virulence phenotype of H. pylori as a means to understand how only a subset of infected individuals develop gastric cancer in response to this pathogen.
Materials and Methods
Bacterial strains
H. pylori strains were maintained on TSA-blood agar plates. Liquid cultures were prepared in Brucella broth supplemented with 10% newborn calf serum. The nomenclature for all in vivo-adapted strains derived from parental H. pylori strain 7.13 is contained in Supplementary Figure S1.
Bacterial-epithelial cell co-cultures
AGS cells (CRL-1739) were obtained from ATCC and maintained in RPMI-1640 supplemented with 10% fetal bovine serum under standard growth conditions. ATCC fully authenticated these cells by Short Tandem Repeat (STR) DNA profiling. After purchase, low passage vials were frozen and maintained in liquid nitrogen for future use. AGS cells were co-cultured with H. pylori at a MOI of 30 for CagA translocation assays or a MOI of 10 for NF-κB assays for all strains included in this study.
Rodent infections and histopathology
Mongolian gerbils were challenged with H. pylori for up to 16 weeks as previously described (5,15). Gastric tissues were harvested and gastric injury and quantitative H. pylori cultures were assessed as described (5).
RFLP
PCR products of cagY, cagA, or components of the cagPAI from H. pylori strains were digested with DdeI or HinfI at 37°C and separated in a 6-9% polyacrylamide gradient gel. Images from stained gels were acquired using a Biorad ChemiDoc analyzer.
Human samples
Archival and recent H. pylori J99 strain were isolated from gastric biopsy specimens from a single patient six years apart (13). Gastric antrum and corpus biopsies from seven additional patients were obtained via endoscopy. Gastric inflammation and quantitative H. pylori cultures were assessed as described (16).
Statistical analysis
The Mann–Whitney test and one-way ANOVA with a Newman–Keuls post-test were used to compare data groups. Data were plotted and analyzed using Prism (GraphPad Inc).
Results
The H. pylori cancer-linked cagY locus harbors a high level of genetic variability
cagY contains 2 repeat domains which are susceptible to in-frame rearrangements compromising the length of the protein (Supplementary Fig S2). Sequence analysis of cagY from H. pylori strain 7.13 revealed the presence of two different motifs (Motif 2A and 2B, Consensus sequences shown in Supplementary Fig S3) within repeat region 2 (Supplementary Fig S2). This particular carcinogenic H. pylori strain has sixteen 2A motifs and six 2B motifs. We therefore used both RFLP and DNA sequence changes to identify cagY diversity among isolates used in this study.
Temporal changes in cagY rearrangements and cagT4SS function are directly related to severity of inflammation within an H. pylori-colonized human pre-malignant gastric environment
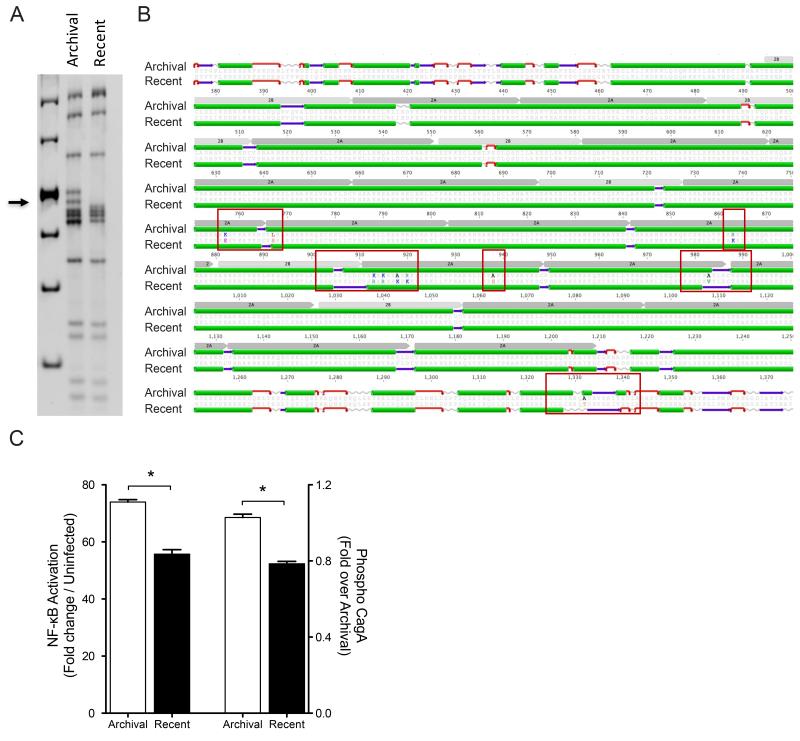
We first utilized paired H. pylori isolates (strain J99), separated in time by 6 years, that were obtained from the gastric antrum of a single patient. The patient did not receive any antibiotic eradication therapy targeting H. pylori during this time interval. In this patient, total inflammation scores in the gastric antrum were three-fold higher compared to scores in the corpus at the initial endoscopy (p<0.05); however, this pattern was reversed six years later as the severity of corpus inflammation was approximately three-fold higher compared to antral inflammation (p<0.05) (14). At the time of the repeat endoscopy, the gastric pH was 5.0, further indicating that acid secretion was impaired. The transition from antral-predominant to corpus-predominant gastritis suggested that this patient was progressing towards a hypochlorhydric phenotype and may be at a higher risk for intestinal-type gastric adenocarcinoma, although he had not developed this malignancy at the time of the second endoscopy. cagY RFLP profiles differed when the archival J99 isolate was compared to the recent isolate (Fig 1A). Concordantly, cagY sequence analysis identified 24 single nucleotide polymorphisms (SNPs) when the archival and recent J99 strains were compared. Among the 24 SNPs, fourteen encoded synonymous changes and ten were non-synonymous (Supplementary Fig S4). Interestingly, 9 of the 10 non-synonymous SNPs were located in the cagY repeat domain 2 (Fig 1B). Comparison of the CagY secondary structures of the archival and recent J99 strains revealed that non-synonymous SNPs induced changes in the predicted secondary structures (Fig 1B).
Fig 1. Relationship between cagY SNPs and cagT4SS function in paired human H. pylori isolates.
(A) cagY RFLP profile of archival and recent H. pylori J99 strains. Arrow indicates area with different bands. (B) Alignment and predicted secondary structure of CagY of archival and recent J99 strains highlighting amino acid substitutions between strains and predicted changes in secondary structure (red boxes). Green: alpha helix; purple: beta-sheets; and red: beta turns. Repeat motifs 2A and 2B are shown in gray. (C) NF-κB activation (Left axis) and CagA translocation (Right axis) in AGS cells following co-culture with archival and recent J99 strains. Values are reported as a fold change over uninfected controls (NF-κB) or archival J99 (CagA translocation). Translocation is shown as a ratio of densitometric values. *: p≤0.05.
To determine if differences in cagY were related to cagT4SS function, we quantified CagA translocation and NF-κB activation in vitro. Archival H. pylori J99, which was isolated from a highly inflamed micro-environment, translocated significantly higher levels of CagA compared to the recent H. pylori J99 isolate. These data mirrored differences in levels of NF-κB activation induced by these strains (Fig 1C). Thus, cagY sequence differences and cagT4SS function were directly related to the severity of inflammation in a patient who had progressed along a histological cascade towards gastric carcinogenesis.
Output strains from Mongolian gerbils infected with a H. pylori cag+ carcinogenic strain contain different cagY rearrangements, which are linked to changes in cagT4SS function
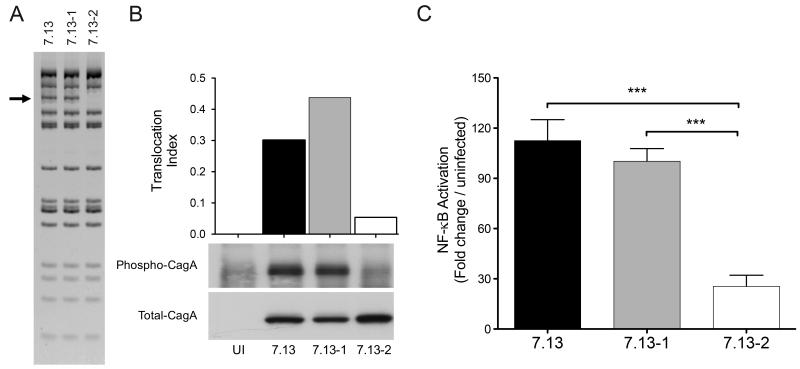
Based on our results in the human stomach, we next determined the mode and stability of cagY and cagT4SS function in an H. pylori cancer model. Gerbils were infected with the prototype clonal H. pylori cag+ carcinogenic strain 7.13 for 12 weeks (Supplementary Fig S1). Two H. pylori strains recovered from independent gerbils with disparate levels of inflammation and injury were then subjected to cagY RFLP analysis. RFLP profiles revealed variation between one output strain (7.13-2) when compared to the parental strain 7.13 or its sibling output strain 7.13-1 (Fig 2A). The ability of H. pylori output strain 7.13-2, which induced low inflammation in vivo, to translocate CagA or activate NF-κB was significantly attenuated compared to the parental input strain 7.13 or the adapted strain 7.13-1 (Fig 2B, 2C). These data indicate that rearrangements in cagY induced by prolonged in vivo adaptation in gerbils could modulate function of the cagT4SS.
Fig 2. cagT4SS function using H. pylori output strains from Mongolian gerbils containing different cagY rearrangements.
(A) cagY RFLP profile of H. pylori strain 7.13, and its derivative strains 7.13-1 and 7.13-2. (B) CagA Western blot of AGS cells co-cultured with H. pylori strains 7.13, 7.13-1, and 7.13-2. Translocation index of CagA is the ratio of phosphorylated CagA to total CagA densitometric value. (C) NF-κB activation induced by H. pylori strains 7.13, 7.13-1, and 7.13-2 after 4 hours of co-culture with AGS cells expressing a Luciferase-based NF-κB reporter. Bars represent mean±SEM of three different assays. Statistical differences were calculated using Mann Whitney test. ***: p≤0.001.
Mongolian gerbils infected with H. pylori derivative strains harboring distinct cagY rearrangements develop different patterns of disease
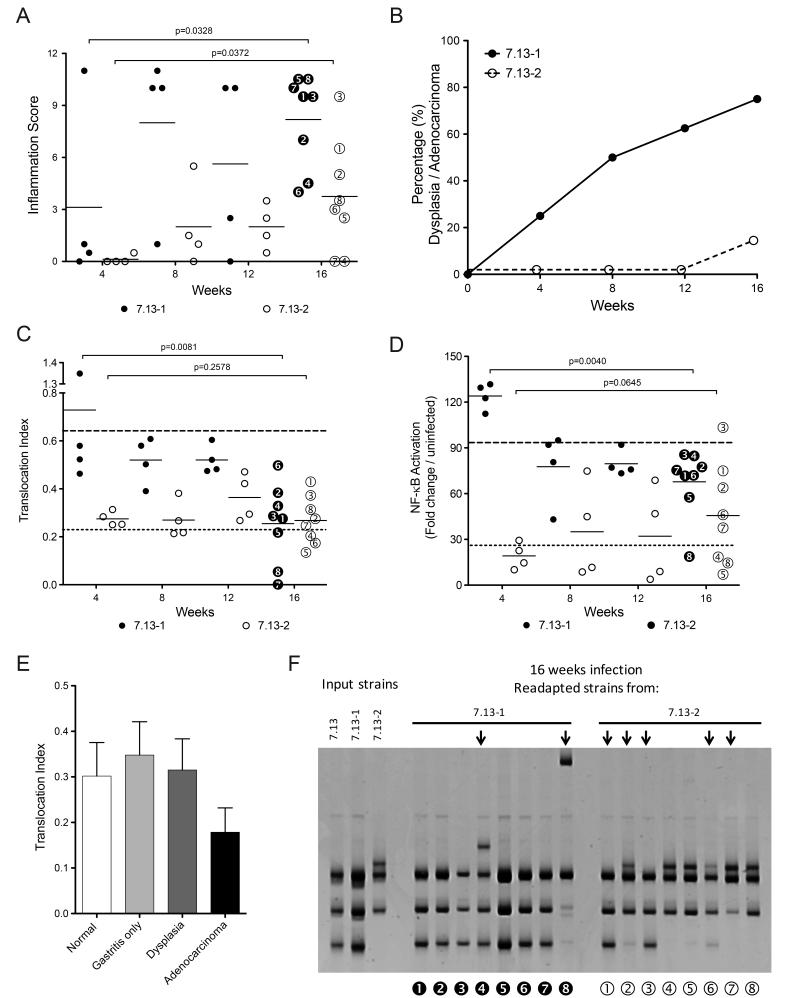
To determine if adapted strains with polar in vitro phenotypes could differentially affect pathogenesis in vivo, new populations of gerbils were infected with the H. pylori derivative strains 7.13-1 or 7.13-2 (Supplementary Fig S1). All challenged animals were successfully colonized and there were no differences in colonization density between the groups. However, strain 7.13-1, which exhibited considerable potency for CagA translocation and NF-κB activation (Fig 2B, 2C), induced high levels of inflammation early (e.g., 8 weeks) post-infection. In contrast, strain 7.13-2 induced a delayed inflammatory response, which began to increase 12-16 weeks post-infection (Fig 3A). Pre-neoplastic lesions were also rapidly induced by strain 7.13-1, and 20% of animals had developed dysplasia 4 weeks post infection, and by 16 weeks post-infection, 80% of the 7.13-1-infected animals developed dysplasia and/or adenocarcinoma. In contrast, only 12% of animals infected with strain 7.13-2 developed dysplasia by 16 weeks post-infection (Fig 3B).
Fig 3. Disease outcome and cagT4SS function using H. pylori gerbil-adapted strains expressing distinct cagY rearrangements.
(A) Inflammation scores and (B) neoplastic lesions from gerbils infected with H. pylori strains 7.13-1 (●) and 7.13-2 (○) for 4, 8, 12, and 16 weeks. (C) CagA translocation index and (D) NF-κB activation induced by H. pylori isolates harvested from gerbils infected with strains 7.13-1 (●) and 7.13-2 (○) for 4, 8, 12, and 16 weeks. Horizontal dotted lines (top line 7.13-1 and bottom line 7.13-2) represent the mean translocation indices of the input strains. (E) Gastric injury and CagA translocation indices of H. pylori derivative strains isolated from gerbils infected with strains 7.13-1 or 7.13-2. Bars represent mean±SEM of CagA translocation. (F) cagY RFLP profiles of H. pylori isolates from gerbils after 16 weeks of infection with strains 7.13-1 or 7.13-2. Arrows denote RFLP profiles that differ compared with the corresponding parental strain profile. Numbered black and white circles represent individual H. pylori isolates which correspond to isolates shown in panels A, C, and D. Statistical differences were calculated using the Mann–Whitney test.
The ability of H. pylori to translocate CagA and activate NF-κB in vitro is directly related to the in vivo inflammatory phenotype, but inversely related to cancer
Having demonstrated different patterns of injury, re-adapted output strains from gerbils infected with the H. pylori derivative strains 7.13-1 and 7.13-2 were then tested in vitro for cagT4SS function. Output isolates from animals infected with the more potent strain 7.13-1, displayed a statistically significant attenuation in the ability to translocate CagA (p=0.008) and activate NF-κB (p=0.004) over time (Fig 3C, 3D). In contrast, output strains isolated from animals infected with strain 7.13-2, maintained a relatively stable ability to translocate CagA and activate NF-κB for the duration of infection (Fig 3C, 3D). Importantly, there was an inverse relationship between the presence of cancer and cagT4SS function, suggesting that host carcinogenic phenotypes may modify the function of specific H. pylori constituents that have been associated with disease (Fig 3E).
Since H. pylori strains 7.13-1 and 7.13-2 exhibited different cagY RFLP patterns, we next determined if the differences observed in CagA translocation and NF-κB activation by the re-adapted derivatives of H. pylori strains 7.13-1 or 7.13-2 were similarly related to new polymorphisms within cagY. Re-adapted strains isolated from animals infected with H. pylori strain 7.13-2 for 16 weeks showed variations in cagY RFLP patterns. Two of the 7.13-2 derived strains (strains 2 and 6) harbored a hybrid RFLP profile containing features present in the profiles of both strains 7.13-1 and 7.13-2; two strains (strains 1 and 3) reverted their RFLP profile to a profile similar to the original parental strain 7.13; and three strains maintained the same profile as input strain 7.13-2 (Fig 3F). This level of variability mirrored the large variability in inflammatory scores in gerbils infected for 16 weeks by H. pylori strain 7.13-2 (Fig 3A). In contrast to this degree of variability, isolates from animals infected with H. pylori strain 7.13-1 showed fewer differences in cagY rearrangements. One 7.13-1 derived strain (strain 4) harbored an intermediate RFLP profile containing elements from strains 7.13-1 and 7.13-2, one (strain 8) had a unique RFLP profile, but six maintained the same profile as the input strain 7.13-1 (Fig 3F). Interestingly, the re-adapted strains that reverted their cagY profiles, also reverted their ability to translocate CagA and activate NF-κB in vitro. In terms of mucosal injury, all H. pylori strains that had cagY RFLP profiles similar to strain 7.13-1 induced high levels of inflammation, while strains that harbored cagY RFLP patterns similar to strain 7.13-2 induced low levels of inflammation. Of interest, strains that possessed unique RFLP patterns (e.g., distinct from either strain 7.13-1 or 7.13-2), induced intermediate levels of inflammation (Fig 3A, 3F).
Rearrangements in cagY play an essential role in modulating function of the H. pylori cagT4SS
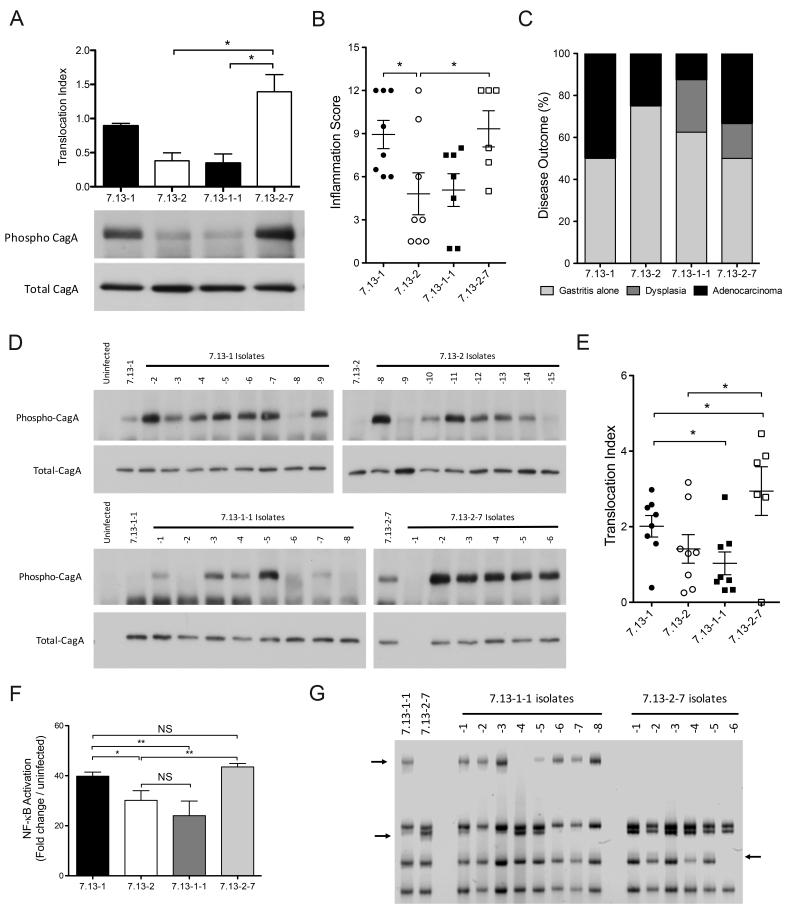
Our previous results indicate that the ability of H. pylori to induce severe inflammation and gastric injury oscillates in this model of cancer, and this parallels changes in cagT4SS function. To determine the stability of in vivo-induced cagY alterations and cagT4SS function, we next selected two output strains from gerbils with the same inflammatory phenotype but which harbored polar cagT4SS function, and infected new populations of gerbils for 16 weeks (Supplementary Fig S1). H. pylori strain 7.13-1-1 is an in vivo-adapted strain obtained from an animal infected for 16 weeks with input strain 7.13-1 and showed an attenuated ability to translocate CagA compared with its parental strain 7.13-1 (Fig 4A). Strain 7.13-2-7 is a derivative strain from an animal infected for 16 weeks with strain 7.13-2 and exhibited substantial potency in CagA translocation compared with its parental strain 7.13-2 (Fig 4A). The data shown in Fig 4B are inflammation scores from new populations of gerbils that were infected with either H. pylori strain 7.13-1 (parent of 7.13-1-1), strain 7.13-2 (parent of 7.13-2-7), strain 7.13-1-1, or strain 7.13-2-7 for 16 weeks. In vivo, the severity of inflammation was directly related to pre-inoculation cagT4SS function, as determined by CagA translocation, in contrast to strain ancestry. Animals infected with strains 7.13-1 or 7.13-2-7 developed higher injury scores compared to animals infected with strains 7.13-2 or 7.13-1-1 (Fig 4B, 4C).
Fig 4. Rearrangements in cagY modify H. pylori cagPAI function.
(A) Translocation index of total and phosphorylated CagA using H. pylori strain 7.13-1 and its derivative strain 7.13-1-1, and strain 7.13-2 and its derivative strain 7.13-2-7. Bars represent mean±SEM of CagA translocation from three independent experiments. (B) Inflammation scores in gastric tissue from gerbils infected with H. pylori strains 7.13-1, 7.13-2, and their derivative strains 7.13-1-1 and 7.13-2-7. (C) Disease outcome of gerbils infected with strains 7.13-1, 7.13-2, 7.13-1-1, and 7.13-2-7 for 16 weeks. (D) Western blot analysis and (E) translocation index for phosphorylated and total CagA in AGS cells co-cultured with H. pylori strains harvested from gerbils infected for 16 weeks with strains 7.13-1, 7.13-2, 7.13-1-1, and 7.13-2-7. (F) NF-κB activation induced by H. pylori derivative strains isolated from gerbils infected with strains 7.13-1, 7.13-2, 7.13-1-1, and 7.13-2-7 for 16 weeks. (G) cagY RFLP profile of H. pylori derivatives from gerbils infected with strains 7.13-1-1, and 7.13-2-7 for 16 weeks. Arrows indicate differential bands between isolates. Statistical differences were calculated by One-way Anova using a Newman-Keuls post-test. *: p≤0.05; **: p≤0.01. NS: Non-significant.
To determine if changes in bacterial phenotypes occurred during these infections, we performed CagA translocation and NF-κB activation assays in vitro. Most of the derivative strains from gerbils infected with H. pylori strains 7.13-2 or 7.13-1-1 recovered the capacity to translocate CagA. Conversely, only a few strains isolated from animals infected with strains 7.13-1 or 7.13-2-7 lost the ability to translocate CagA (Fig 4D, 4E) and these changes in CagA translocation reflected the ability to induce NF-κB (Fig 4F).
There was a direct correlation between levels of CagA translocation and cagY profiles among strains isolated from gerbils infected with strain 7.13-1-1 (Fig 4D, 4G); however, this relationship was not present in strains isolated from gerbils infected with strain 7.13-2-7, as only one isolate (7.13-2-7-6) had a different cagY RFLP profile yet maintained the ability to translocate CagA (Fig 4D, 4G). Conversely, one strain (7.13-2-7-1) did not express CagA (Fig 4D), but harbored the same cagY RFLP profile of other strains than maintained the ability to express CagA, suggesting a different mechanism of regulation.
In vivo-adapted H. pylori strains are panmictic populations that harbor different cagY rearrangements
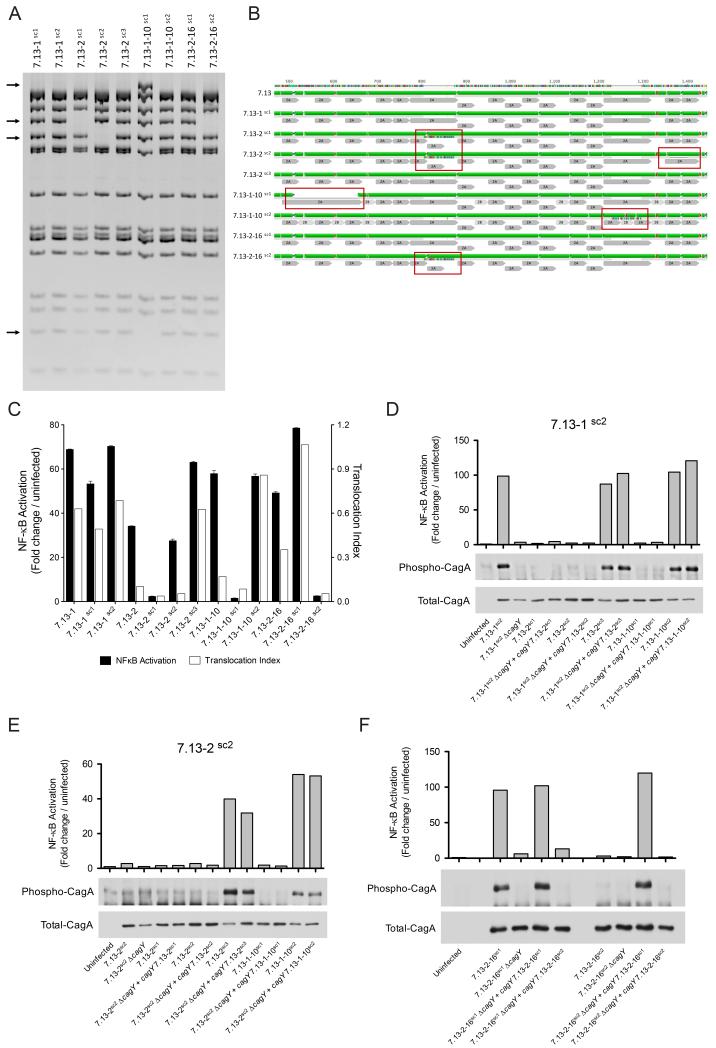
We next determined whether reversions of cagY genotypes are due to: 1) the presence of panmictic populations pre-populated with isolates harboring different rearrangements of cagY or 2) to de novo rearrangements acquired in vivo. To test this hypothesis, we isolated twelve single colonies from H. pylori adapted strains 7.13-1, 7.13-2, 7.13-1-10 and 7.13-2-16 (Supplementary Fig S1) and assessed cagY RFLP profiles. All single colonies from strain 7.13-1 had identical cagY RFLP patterns. Among single colonies from strains 7.13-2, 7.13-1-10 and 7.13-2-16, we identified the presence of clones with different cagY rearrangements (Fig 5A). In total, we found four different RFLP patterns among the collective single colonies. RFLP profiles of strain 7.13-1 single colonies are similar to the original archival strain 7.13; for strain 7.13-2 single colonies, one RFLP profile is similar to 7.13-1 (7.13-2sc3) and two are different (7.13-2sc1 and 7.13-2sc2). For strain 7.13-1-10 single colonies, one RFLP pattern is similar to strain 7.13-1 (7.13-1-10sc2), and one is different (7.13-1-10sc1). For strain 7.13-2-16, one RFLP pattern is similar to that of strain 7.13-1 (7.13-2-16sc1) and one pattern is similar to that of strain 7.13-2sc1 (7.13-2-16sc2) (Fig 5A).
Fig 5. H. pylori gerbil-adapted strains represent panmictic populations harboring diverse cagY rearrangements.
(A) Representative cagY RFLP profiles of H. pylori single colonies generated from strains 7.13-1, 7.13-2, 7.13-1-10, and 7.13-2-16. Arrows denote differential bands between RFLP profiles. (B) Alignment and predicted secondary structure of H. pylori CagY parental strain 7.13 and derivative single colony strains. Red boxes highlight gain or loss of motifs 2A and/or 2B compared with parental strain 7.13. Green: alpha helix; purple: beta-sheets, and red: beta turns. Repeat motifs 2A and 2B are in gray. (C) NF-κB activation (black bars) and CagA translocation (white bars) induced by H. pylori strains and their corresponding single colony isolates. NF-κB activation, and CagA translocation induced by H. pylori strains 7.13-1sc2 (D) and 7.13-2sc2 (E) genetically complemented with different cagY genes. (F) NF-κB activation and CagA translocation induced by H. pylori strains 7.13-2-16sc1 and 7.13-2-16sc2 complemented with either an endogenous cagY motif or with a cagY motif derived from the sibling strain.
We next sequenced cagY from each of these single colony strains. Compared with the parental H. pylori strain 7.13, sequence analysis identified differences in the number and order of 2A and 2B motifs within the cagY repeat region 2 (Fig 5B). Strain 7.13-1-10sc1 has a cluster deletion around amino acid 510, which involves the loss of two 2A motifs and two 2B motifs. Strains 7.13-2sc1, 7.13-2sc2, and 7.13-2-16sc2 have an addition of one 2A motif and one 2B motif around amino acid 800. In addition, strain 7.13-2sc2 also has a deletion that compromises one 2A motif around amino acid 1355. Strain 7.13-1-10sc2 has an addition of one 2A motif and one 2B motif around amino acid 1220, but this insertion was not detectable by RFLP. Sequences from strains 7.13-1sc1, 7.13-2sc3, and 7.13-2-16sc1 did not show any differences when compared with parental strain 7.13 (Fig 5B). When we compared the predicted secondary structure between single colony strains, the addition or deletion of motifs 2A and/or 2B predicted changes in the length of alpha-helices, as well as additions or deletions of beta sheets (Fig 5B). Of interest, the mechanism underpinning changes in cagY sequences was different in isolates adapted to gerbils (deletion and/or addition of entire motifs) when compared to changes seen in the human H. pylori J99 paired strains (SNPs) (Fig 1B).
We also examined cagT4SS function among single colony isolates with varying cagY profiles. H. pylori clones with identical cagY RFLP profiles induced similar levels of NF-κB activation; thus, clones 7.13-1sc1, 7.13-1sc2, 7.13-2sc3, 7.13-1-10sc2, and 7.13-2-16sc1, which have RFLP patterns similar to the parental strain 7.13, induced the highest levels of NF-κB activation. In contrast, clones 7.13-2sc1 and 7.13-2-16sc2, which harbor cagY profiles similar to strain 7.13-2, induced the lowest levels of NF-κB activation. Clones 7.13-2sc2 and 7.13-1-10sc1 had unique cagY rearrangements, with corresponding intermediate and low capacities to activate NF-κB, respectively (Fig 5A, 5C). The ability of each of these clones to translocate CagA into AGS cells mirrored their ability to activate NF-κB (Fig 5C, Supplementary Fig S5).
To exclude the possibility that changes in the cagPAI exogenous to cagY may affect the phenotypes of interest, we performed RFLP analysis of PCR amplicons that spanned the entire cagPAI, excluding cagY. Global cagPAI RFLP profiles from eight different single colony strains were identical (Supplementary Fig S6). To further interrogate the entire cagPAI, sequence analysis of the entire cagPAI from two single colony isolates with different CagA translocation phenotypes (7.13-1sc2 and 7.13-2sc1) was performed (Supplementary Table S1). The only differences identified were within cagY (Supplementary Fig S7).
To further corroborate the role of cagY in modulation of cagPAI function, we genetically exchanged cagY among four different strains of H. pylori (two with high cagPAI phenotypes, 7.13-1sc2 and 7.13-2sc3, and two with low cagPAI phenotypes, 7.13-2sc1 and 7.13-2sc2) using the five different cagY rearrangements found in H. pylori single colonies (Fig 5B, Supplementary Fig S8). We found that independent of the H. pylori recipient background, CagA translocation and NF-κB activation phenotypes were restored or lost based upon the cagY phenotype of the donor (Fig 5D, 5E, Supplementary Fig S9). In addition, to define how more refined rearrangements in cagY motifs may affect cagPAI function, we exchanged cagY between two strains with polar cagPAI phenotypes, 7.13-2-16sc1 and 7.13-2-16sc2, but which only differed in a single cagY domain 2A (Fig 5B, Supplementary Fig S10). We found that cagPAI function could be restored or lost based upon the donor cagY motif, implicating this motif in cagPAI function (Fig 5F).
Mongolian gerbils infected with H. pylori single colonies carrying unique cagY rearrangements develop differences in the severity of inflammation and disease
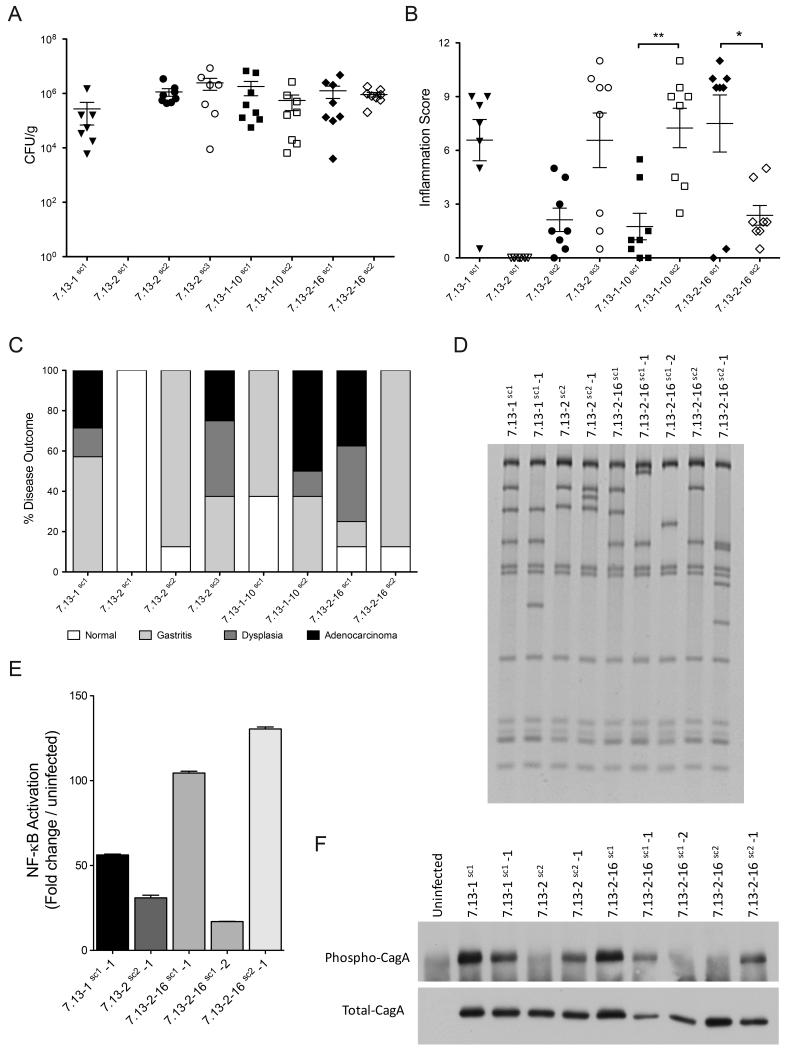
To determine whether H. pylori single colonies induced differences in disease based on cagY RFLP patterns, colonization and inflammation was assessed in gerbils infected with the H. pylori single colony isolates. All animals were successfully colonized with the exception of gerbils challenged with the single colony strain 7.13-2sc1, for which no animals were colonized. Within colonized animals, bacterial density levels were similar (Fig 6A); however, histopathologic analysis revealed significant differences in inflammation. Gerbils infected with H. pylori single colony isolates carrying similar cagY rearrangements developed similar levels of inflammation (Fig 6B). Specifically, gerbils infected with H. pylori isolates 7.13-1sc1, 7.13-2sc3, 7.13-1-10sc2, and 7.13-2-16sc1, which have identical cagY RFLP profiles and induce robust CagA translocation (Fig 5A, 5C), induced high levels of inflammation (Fig 6B). In contrast, animals infected with isolates 7.13-2sc2, 7.13-1-10sc1, and 7.13-2-16sc2, which harbor different cagY profiles and an attenuated ability to translocate CagA (Fig 5A, 5C), induced low levels of inflammation (Fig 6B).
Fig 6. Severity of disease in Mongolian gerbils infected with H. pylori single colony isolates and relationship with cagY rearrangements.
Colonization density (A), inflammation scores (B), and disease outcome (C) of gerbils challenged for 16 weeks with H. pylori single colonies carrying specific cagY rearrangements. (D) cagY RFLP profiles, (E) NF-κB activation, and (F) phosphorylated and total CagA assays of derivative isolates harvested from gerbils infected for 16 weeks with H. pylori single colony isolates carrying new cagY rearrangements. Statistical differences were calculated by One-way Anova using a Newman-Keuls post-test. *: p≤0.05; **: p≤0.01.
Similar to levels of inflammation, only animals infected with strains carrying cagY rearrangements linked to robust CagA translocation developed neoplastic lesions (Fig 6C). In contrast, animals infected with strains carrying cagY rearrangements associated with attenuated levels of CagA translocation only developed gastritis (Fig 6C). These results reinforce the role that cagY exerts as a modulator of cagT4SS function.
We next analyzed the RFLP profiles of isolated strains. As expected, after 16 weeks isolated strains harbored differences in cagY compared to the input clonal H. pylori single colony isolates (Fig 6D); however, all of these rearrangements represented new RFLP patterns with varying capacities to activate NF-κB and translocate CagA (Fig 6E, 6F). Thus, infection with H. pylori single colonies harboring a variety of cagY RFLP profiles can lead to the generation of new cagY rearrangements.
H. pylori clinical isolates can harbor synchronous cagY rearrangements
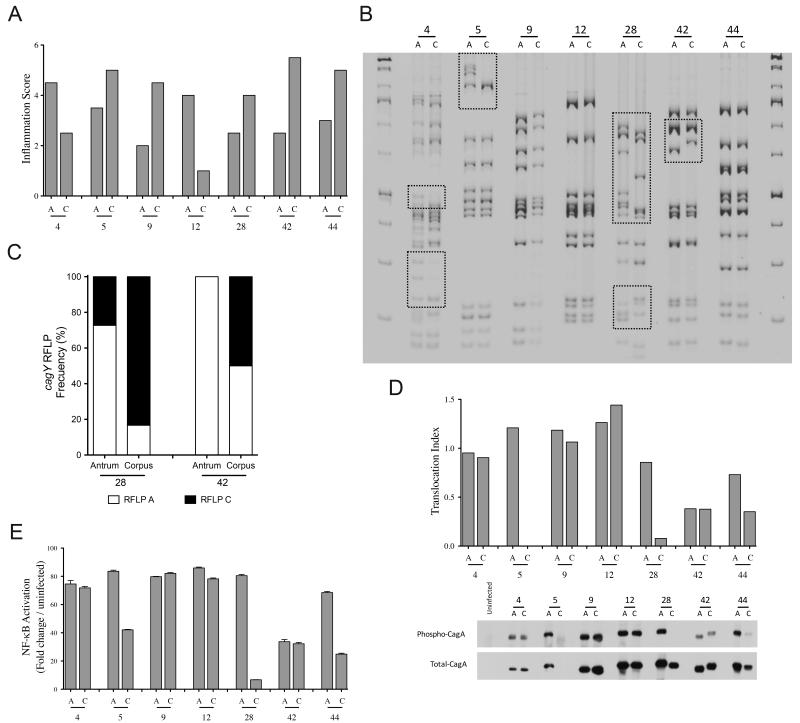
We then validated cagY diversity observed in our gerbil model by using paired H. pylori clinical strains, harvested from 2 different sites (antrum and corpus) within the stomachs of 7 individual patients during a single endoscopy. For these paired sets of samples, two patients (4 and 12) harbored inflammation scores that were higher in the antrum than the corpus, and five patients (5, 9, 28, 42 and 44) harbored the reverse (Fig 7A). In terms of cagY RFLP profiles, strains isolated from patients 4, 5, 28, and 42 harbored different RFLP patterns, while paired isolates from the remaining patients harbored identical profiles (Fig 7B). Five of the seven patients harbored either atrophic gastritis or intestinal metaplasia, but there was no significant relationship between site-specific cagY RFLP patterns and premalignant lesions.
Fig 7. Synchronous cagY rearrangements in H. pylori clinical isolates.
(A) Inflammation scores of human gastric biopsies obtained from the antrum “A” and corpus “C” from the same patient (n=7 patients). (B) cagY RFLP of H. pylori isolates from biopsies obtained from the antrum and corpus of the same patient. Boxes indicate differences in RFLP profiles between sibling strains. (C) Frequency distribution of single colony cagY genotypes within isolates obtained from the antrum and corpus from two patients (28 and 42). CagA translocation (D) and NF-κB activation (E) of H. pylori isolates from the antrum and corpus of seven patients.
We next defined the distribution of cagY rearrangements among single colony isolates (n=12) harvested from patients 9, 12, 28, 42 and 44. Single colony isolates from the antrum and corpus from patients 9, 12, and 44 displayed no differences in cagY RFLP profiles. In contrast, there were 2 different cagY profiles within antral and corpus single colony isolates from patient 28 and 42. Single colony profiles from patient 28 and 42 were unequally distributed between the antrum and corpus, as the predominant clone in the antrum (patient 28: 8/11 colonies (83%); patient 42: 12/12 colonies (100%)) was the minority clone in the corpus (patient 28: 2/12 colonies (17%); patient 42: 6/12 colonies (50%)) (Fig 7C). Finally, we tested the ability of these isolates to translocate CagA and activate NF-κB in vitro. We found no differences between antrum and corpus isolates from patients 4, 9, 12, and 42. In contrast, there were significant differences in cagT4SS function between antrum and corpus H. pylori isolates from patients 5, 28, and 44 (Fig 7D). These results in cagT4SS function mirrored NF-κB activation in vitro (Fig 7E).
Discussion
Our current results indicate that H. pylori harbors the capacity to nimbly modify function of its primary virulence constituent during prolonged colonization in a rodent model of gastric carcinogenesis. Chronic infection in gerbils engenders a portfolio of cagY isoforms, which are selected in conjunction with the intensity of host inflammatory and carcinogenic phenotypes. Importantly, the development of dysplasia and cancer led to the emergence of attenuated cagT4SS phenotypes, but robust function could be restored following adaptation to new hosts. The versatility of cagY isoforms and cagT4SS function was also validated in both synchronous and metachronous paired human samples and different mechanisms of cagY alterations were identified via sequencing. Deletions and additions of entire motifs appeared to predominate in gerbils while the development of single base pair mutations predominated in human samples.
Infection of wild-type mice with H. pylori cagT4SS positive strains frequently leads to deletions within the cag island (17,18), and the mechanism underpinning cag dysfunction appears to be in-frame rearrangements in cagY (7). In contrast to mice, previous studies have shown that H. pylori wild-type cag+ strains colonize gerbils well without loss of cag function (19,20). However, these studies did not examine extensive populations of adapted isolates harvested from a large number of infected gerbils. Our current study has examined changes in cagY and the cagT4SS in this model in much greater depth through the use of serially-passaged strains, investigations of both pools and single colony isolates, and validation in human samples. We now demonstrate that H. pylori strains within gerbil stomachs exist as panmictic populations and that cagT4SS function can be altered based on the severity of disease, which is in contrast to the near complete ablation of cagT4SS function in mice.
Can long-term serial infections modify gain or loss of cagT4SS function and what is the benefit for H. pylori to harbor mechanisms that can up- or down-regulate function of this locus? Our current data indicate that the presence of dysplasia or cancer is associated with reduced cagT4SS function. Therefore, we would predict that serial infections would not enhance CagA translocation and/or the ability to activate NF-κB if there is development of these lesions. However, this needs to be tested formally in the future. In terms of serially infecting derivative strains with minimal cagT4SS function, our data in Figure 4 have shown that strain 7.13-1-1, a derivative strain with low cagT4SS function, when re-infected into gerbils, induces variability in inflammation scores, CagA translocation indices, and NF-κB activation levels (Fig 4B, 4D, 4E, and 4F). Based on our results investigating single colony isolates (Fig 6), we posit that this reflects a panmictic pool of clones with variable cagY function within pre-inoculation strain 7.13-1-1. In terms of strains that never acquire the ability to translocate CagA or activate NF-κB, our data in Figure 5 demonstrate that two derivative strains, 7.13-2sc1 and 7.13-2-16sc2, have little if any ability to translocate CagA or activate NF-κB, and their cagY RFLP profiles are identical (Fig 5A). Therefore, we plan in future experiments to utilize these strains in long-term adaptation studies to more carefully discern whether chronic and repeated serial infections may lead to a gain of cagT4SS function. One downstream ramification of CagA translocation is acquisition of iron sources that fuel H. pylori replication (21). Under conditions of iron deprivation, therefore, it may be beneficial for H. pylori to augment function of the cagT4SS and deliver higher payloads of CagA into host epithelial cells. H. pylori also harbors multiple pathogen-associated molecular patterns (PAMPs) that interact differently with innate immune receptors than the respective counterparts in other mucosal pathogens. H. pylori FlaA is a non-inflammatory molecule in terms of its ability to activate TLR5 (22). H. pylori LPS contains an anergic lipid A core that induces an attenuated TLR4-mediated response (23,24). We and others have shown that deacetylation of peptidoglycan allows H. pylori to evade host clearance (5,25-27). Thus, H. pylori has evolved to express an array of diverse phenotypes to subvert obstacles presented by the host (28), and the ability of H. pylori to alter cagY genotypes is yet another mechanism within the repertoire of this organism to evade host defenses.
In conclusion, we have utilized a robust rodent model of gastric cancer that closely recapitulates human disease to define alterations in H. pylori pathogenesis in vivo. Our findings demonstrate that cagT4SS function is typically maintained during prolonged colonization in this model but that the intensity of cagT4SS function can vary and is related to host disease phenotypes. cagY genotypes are altered concordant with cagT4SS function and can similarly vary in paired human samples obtained from different regions of the stomach or separated in time. The use of this model will facilitate more detailed investigations in the future that can identify new targets and therapeutic strategies to reduce the risk of cancer associated with this pathogen.
Supplementary Material
Acknowledgments
Financial Support: NIH R01-DK58587, R01-CA77955, P01-CA116087, P30-DK058404, P01-CA028842.
Footnotes
Conflicts of Interest: None
References
- 1.Hardbower DM, Peek RM, Jr., Wilson KT. At the Bench: Helicobacter pylori, dysregulated host responses, DNA damage, and gastric cancer. J Leukoc Biol. 2014;96(2):201–12. doi: 10.1189/jlb.4BT0214-099R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Correa P, Cuello C, Duque E, Burbano LC, Garcia FT, Bolanos O, et al. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst. 1976;57(5):1027–35. doi: 10.1093/jnci/57.5.1027. [DOI] [PubMed] [Google Scholar]
- 3.Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci U S A. 2014;111(4):1455–60. doi: 10.1073/pnas.1318093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varga MG, Shaffer CL, Sierra JC, Suarez G, Piazuelo MB, Whitaker ME, et al. Pathogenic Helicobacter pylori strains translocate DNA and activate TLR9 via the cancer-associated cag type IV secretion system. Oncogene. 2016 doi: 10.1038/onc.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suarez G, Romero-Gallo J, Piazuelo MB, Wang G, Maier RJ, Forsberg LS, et al. Modification of Helicobacter pylori peptidoglycan enhances NOD1 activation and promotes cancer of the stomach. Cancer Res. 2015;75(8):1749–59. doi: 10.1158/0008-5472.CAN-14-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5(11):1166–74. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 7.Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, et al. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9(2):e1003189. doi: 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrozo RM, Hansen LM, Lam AM, Skoog EC, Martin ME, Cai LP, et al. CagY is an immune-sensitive regulator of the Helicobacter pylori type IV secretion system. Gastroenterology. 2016 doi: 10.1053/j.gastro.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe T, Tada M, Nagai H, Sasaki S, Nakao M. Helicobacter pylori infection induces gastric cancer in Mongolian gerbils. Gastroenterology. 1998;115(3):642–8. doi: 10.1016/s0016-5085(98)70143-x. [DOI] [PubMed] [Google Scholar]
- 10.Honda S, Fujioka T, Tokieda M, Satoh R, Nishizono A, Nasu M. Development of Helicobacter pylori-induced gastric carcinoma in Mongolian gerbils. Cancer Res. 1998;58(19):4255–9. [PubMed] [Google Scholar]
- 11.Ogura K, Maeda S, Nakao M, Watanabe T, Tada M, Kyutoku T, et al. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J Exp Med. 2000;192(11):1601–10. doi: 10.1084/jem.192.11.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Q, Chen XY, Shi Y, Xiao SD. Development of gastric adenocarcinoma in Mongolian gerbils after long-term infection with Helicobacter pylori. J Gastroenterol Hepatol. 2004;19(10):1192–8. doi: 10.1111/j.1440-1746.2004.03469.x. [DOI] [PubMed] [Google Scholar]
- 13.Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, et al. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci U S A. 2001;98(25):14625–30. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishna U, Romero-Gallo J, Suarez G, Azah A, Krezel AM, Varga MG, et al. Genetic evolution of a Helicobacter pylori acid-sensing histidine kinase and gastric disease. J Infect Dis. 2016;214(4):644–8. doi: 10.1093/infdis/jiw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102(30):10646–51. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J, et al. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010;16(4):369–78. doi: 10.1111/j.1469-0691.2009.02811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philpott DJ, Belaid D, Troubadour P, Thiberge JM, Tankovic J, Labigne A, et al. Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylori isolates. Cell Microbiol. 2002;4(5):285–96. doi: 10.1046/j.1462-5822.2002.00189.x. [DOI] [PubMed] [Google Scholar]
- 18.Sozzi M, Crosatti M, Kim SK, Romero J, Blaser MJ. Heterogeneity of Helicobacter pylori cag genotypes in experimentally infected mice. FEMS Microbiol Lett. 2001;203(1):109–14. doi: 10.1111/j.1574-6968.2001.tb10828.x. [DOI] [PubMed] [Google Scholar]
- 19.Israel DA, Salama N, Arnold CN, Moss SF, Ando T, Wirth HP, et al. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Invest. 2001;107(5):611–20. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peek RM, Jr., Wirth HP, Moss SF, Yang M, Abdalla AM, Tham KT, et al. Helicobacter pylori alters gastric epithelial cell cycle events and gastrin secretion in Mongolian gerbils. Gastroenterology. 2000;118(1):48–59. doi: 10.1016/s0016-5085(00)70413-6. [DOI] [PubMed] [Google Scholar]
- 21.Tan S, Noto JM, Romero-Gallo J, Peek RM, Jr., Amieva MR. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 2011;7(5):e1002050. doi: 10.1371/journal.ppat.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gewirtz AT, Yu Y, Krishna US, Israel DA, Lyons SL, Peek RM., Jr. Helicobacter pylori flagellin evades toll-like receptor 5-mediated innate immunity. J Infect Dis. 2004;189(10):1914–20. doi: 10.1086/386289. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Perez GI, Shepherd VL, Morrow JD, Blaser MJ. Activation of human THP-1 cells and rat bone marrow-derived macrophages by Helicobacter pylori lipopolysaccharide. Infect Immun. 1995;63(4):1183–7. doi: 10.1128/iai.63.4.1183-1187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa T, Suda Y, Kashihara W, Hayashi T, Shimoyama T, Kusumoto S, et al. Immunobiological activities of chemically defined lipid A from Helicobacter pylori LPS in comparison with Porphyromonas gingivalis lipid A and Escherichia coli-type synthetic lipid A (compound 506) Vaccine. 1997;15(15):1598–605. doi: 10.1016/s0264-410x(97)00102-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang G, Olczak A, Forsberg LS, Maier RJ. Oxidative stress-induced peptidoglycan deacetylase in Helicobacter pylori. J Biol Chem. 2009;284(11):6790–800. doi: 10.1074/jbc.M808071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Maier SE, Lo LF, Maier G, Dosi S, Maier RJ. Peptidoglycan deacetylation in Helicobacter pylori contributes to bacterial survival by mitigating host immune responses. Infect Immun. 2010;78(11):4660–6. doi: 10.1128/IAI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci U S A. 2007;104(3):997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lina TT, Alzahrani S, Gonzalez J, Pinchuk IV, Beswick EJ, Reyes VE. Immune evasion strategies used by Helicobacter pylori. World J Gastroenterol. 2014;20(36):12753–66. doi: 10.3748/wjg.v20.i36.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.