Abstract
Reprogramming cell fates towards pluripotent stem cells and other cell types has revolutionized our understanding of cellular plasticity. During the last decade, transcription factors and microRNAs have become powerful reprogramming factors for modulating cell fates. Recently, many efforts are focused on reprogramming cell fates by non-viral and non-integrating chemical approaches. Small molecules not only are useful in generating desired cell types in vitro for various applications, such as disease modeling and cell-based transplantation, but also hold great promise to be further developed as drugs to stimulate patients’ endogenous cells to repair and regenerate in vivo. Here we will focus on chemical approaches for generating induced pluripotent stem cells, neurons, cardiomyocytes, hepatocytes and pancreatic β cells. Significantly, the rapid and exciting advances in cellular reprogramming by small molecules will help us to achieve the long-term goal of curing devastating diseases, injuries, cancers and aging.
Keywords: reprogramming, small molecules, stem cells, cell fates
INTRODUCTION
Studies of developmental biology have demonstrated that the zygote is totipotent and can gradually differentiate into all tissues and organs of the human body, that is composed by more than 200 types of specialized cells. However, the demonstration of interchanging cell fates and converting one cell type into another by different strategies, such as nuclear transfer or cell fusion, has challenged the traditional cognition of development and regeneration (Gurdon, 1962; Tada et al., 1997). In 2006, Yamanaka and Takahashi reported the development of the induced pluripotent stem cell (iPSC) technology, which can convert somatic cells to iPSCs using four transcription factors (TFs) (Takahashi and Yamanaka, 2006). During the last 10 years, iPSC technology has provided many fundamental insights into the molecular mechanisms of cell fate transition, and already demonstrated promising results in various applications, including disease modeling, drug screening, and cell-based therapy (Takahashi and Yamanaka, 2016). Recently, different combinations of lineage-specific TFs have been screened and applied to generate various cell types, including neurons, cardiomyocytes, hepatocytes, and pancreatic β cells (Huang et al., 2011; Ieda et al., 2010; Vierbuchen et al., 2010; Zhou et al., 2008). However, TF-based reprogramming approaches face many challenges in efficiency, safety, and in vivo delivery. As a novel and promising solution, small molecules are easy to apply and remove, more efficient and amenable to scale up. Small molecules not only are useful in generating desired cell types in vitro for various applications, but also can be further developed as drugs to stimulate patients’ endogenous cells to repair and regenerate in vivo.
In this review, we will focus on making iPSCs, neurons, cardiomyocytes, hepatocytes and pancreatic β cells by small molecules (Fig. 1). Many interesting small molecules have been identified that can significantly promote cellular reprogramming (Table 1). The rapid and exciting advances in cellular reprogramming by small molecules will undoubtedly advance biomedical studies and clinical translation.
Figure 1.
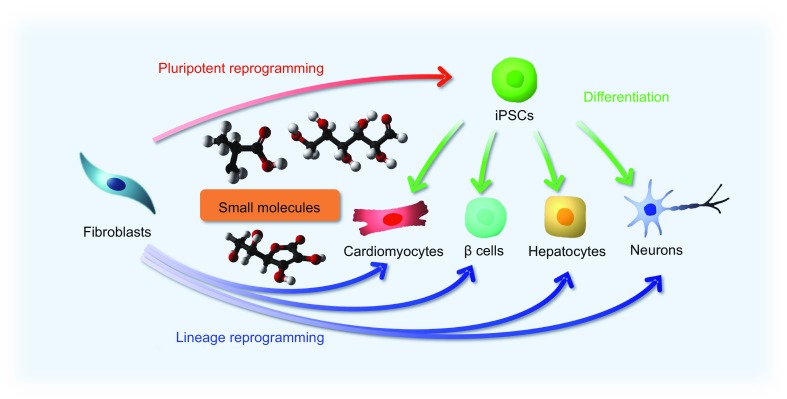
Reprogramming cell fates by small molecules. Chemical approaches can be widely applied to manipulate cell fates and states, including pluripotent reprogramming, directed differentiation, and lineage reprogramming. Small molecules not only are useful in generating functional cell types, such as cardiomyocytes, hepatocytes, pancreatic β cells, and neurons, but also can provide a better understanding of the detailed mechanisms underlying specific reprogramming processes
Table 1.
Representative small molecules for reprogramming cell fates
| Name | Chemical formula | Function | Application |
|---|---|---|---|
| 5-Azacytidine (5-aza) |
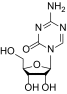
|
DNA methyltransferase inhibitor | iPSC reprogramming |
| 5-Aza-2’-deoxycitidine (5-aza-dc) |
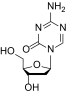
|
DNA methyltransferase inhibitor | iPSC reprogramming |
| RepSox (E616452) |
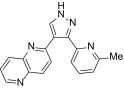
|
ALK5 inhibitor | iPSC reprogramming |
| Kenpaullone |
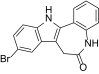
|
GSK3β inhibitor, CDK inhibitor | iPSC reprogramming |
| BIX01294 |
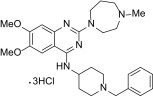
|
G9a inhibitor | iPSC, iCM, pancreatic β-like cell reprogramming |
| Bayk8644 |
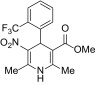
|
L-type Ca2+ channel agonist | iPSC and pancreatic β-like cell reprogramming |
| RG108 |
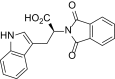
|
DNA methyltransferase inhibitor | iPSC, iNSC, pancreatic β-like cell reprogramming |
| CHIR99021 |
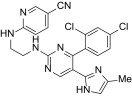
|
GSK3β inhibitor | iPSC, iN, iNSC, iCM, iCPC, iHep reprogramming |
| Tranylcypromine (Parnate) |
|
Lysine-specific demethylase 1 inhibitor | iPSC,iNSC, iCM reprogramming |
| Valproic acid (VPA) |
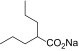
|
Histone deacetylase inhibitor | iPSC, iNSC, iCM reprogramming |
| AMI-5 |
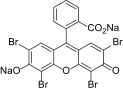
|
Protein arginine methyltransferase inhibitor | iPSC reprogramming |
| A83-01 |
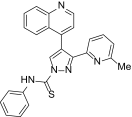
|
TGFβ signaling pathway inhibitor | iPSC, iNSC, iCM, iHep reprogramming |
| Forskolin |
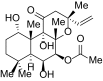
|
cAMP signaling activator | iPSC, iN, iCM reprogramming |
| 2-Methyl-5-hydroxytryptamine (2-Me-5-HT) |
|
5-HT receptor agonist | iPSC reprogramming |
| D4476 |
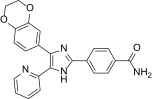
|
Casein kinase inhibitor | iPSC reprogramming |
| 3-Deazaneplanocin (DZNep) |
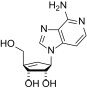
|
DNA methyltransferase inhibitor | iPSC reprogramming |
| AM580 |
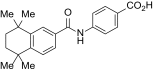
|
RARα agonist | iPSC reprogramming |
| EPZ004777 |
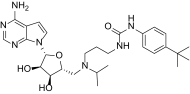
|
DOT1L inhibitor | iPSC reprogramming |
| SGC0946 |
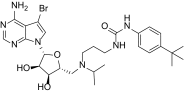
|
DOT1L inhibitor | iPSC reprogramming |
| Sodium butyrate (NaB) |
|
Histone deacetylase inhibitor | iPSC and iNSC reprogramming |
| PS48 |
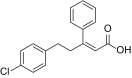
|
PDK1 activator | iPSC reprogramming |
| PD0325901 |
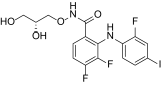
|
MAPK/ERK signaling pathway inhibitor | iPSC reprogramming |
| ISX9 |
|
Activates the expression of endogenous neurogenic transcription factors | iN reprogramming |
| I-BET151 |
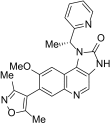
|
BET family protein inhibitor | iN reprogramming |
| SP600125 |
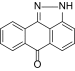
|
JNK inhibitor | iN and iNSC reprogramming |
| GO6983 |
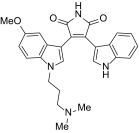
|
Pan-PKC inhibitor | iN reprogramming |
| Dorsomorphin |
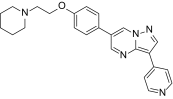
|
BMP signaling inhibitor | iN reprogramming |
| LDN193189 |
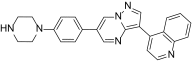
|
BMP signaling inhibitor | iN, iNSC reprogramming |
| SB431542 |
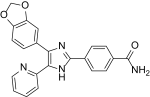
|
TGFβ signaling pathway inhibitor | iN, iCM, pancreatic β-like cell reprogramming |
| TTNPB |
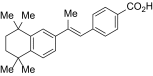
|
Synthetic retinoic acid receptor ligand | iN reprogramming |
| Thiazovivin (Tzv) |
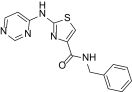
|
ROCK inhibitor | iPSC, iN reprogramming |
| DAPT |
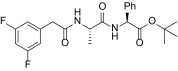
|
γ-secretase inhibitor | iN reprogramming |
| Smoothened agonist (SAG) |
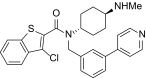
|
Hedgehog signaling activator | iN reprogramming |
| Purmorphamine (Purmo) |
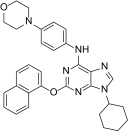
|
Hedgehog signaling activator | iN reprogramming |
| Lysophosphatidic acid (LPA) |
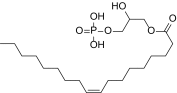
|
GPCR activator | iNSC reprogramming |
| Rolipram |
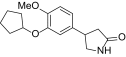
|
Phosphodiesterase-4 inhibitor | iNSC, iCM reprogramming |
| Retinoic acid (RA) |
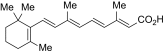
|
RAR receptor | iNSC reprogramming |
| Hh-Ag1.5 |
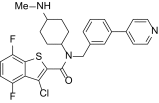
|
Hedgehog signaling activator | iNSC reprogramming |
| SMER28 |
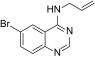
|
Autophagy enhancer | iNSC reprogramming |
| Y-27632 |
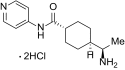
|
ROCK1 inhibitor | iN, iCM reprogramming |
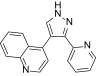
| JAK inhibitor I |
|
JAK inhibitor | iCM reprogramming |
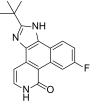
| LY-364947 |
|
TGFβR-I inhibitor | iCM reprogramming |
| SD-208 |
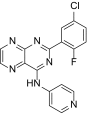
|
TGFβR-I inhibitor | iCM reprogramming |
| GW788388 |
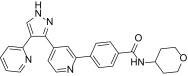
|
ALK5 inhibitor | iCM reprogramming |
| ICARIIN |
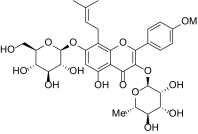
|
Exhibits a variety of pharmacological actions | iCM reprogramming |
| PD169316 |
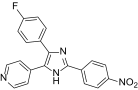
|
p38 inhibitor | iCM reprogramming |
| AS8351 |
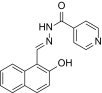
|
KDM5B inhibitor | iCM reprogramming |
| SC1 |
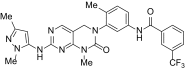
|
ERK1, RasGAP inhibitor | iCM reprogramming |
| OAC2 |
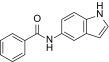
|
Oct4 activator | iCM reprogramming |
| SU16F |
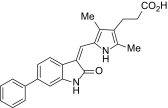
|
PDGFR inhibitor | iCM reprogramming |
| JNJ10198409 |
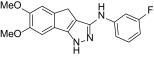
|
PDGFR inhibitor | iCM reprogramming |
| SU5402 |
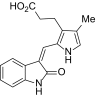
|
FGFR, VEGFR, and PDGFR inhibitor | iCPC reprogramming |
| Compound E |
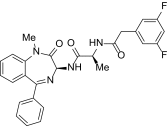
|
Notch signaling inhibitor | iHep, pancreatic β-like cell reprogramming |
| Nicotinamide |
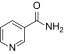
|
Coenzyme, cofactor | Pancreatic β-cell reprogramming |
| 5’-N-ethylcarboxamidoadeno-sine (NECA) |
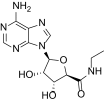
|
Adenosine receptor agonist | Pancreatic β-like cell reprogramming |
| BRD7552 |
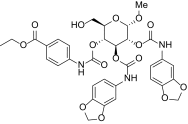
|
PDX1 expression inducer | Increases insulin expression |
| Vitamin C |
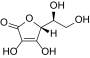
|
Cofactor of epigenetic modulators | Increases reprogramming efficiency |
PLURIPOTENT REPROGRAMMING
In 2006, the first generation of iPSCs were derived by using four transcription factors (Oct4, Sox2, Klf4, and c-Myc, termed OSKM) (Takahashi and Yamanaka, 2006). During the last 10 years, substantial efforts have been made to improve the efficiency and safety of iPSC reprogramming by non-integrating viral vectors, synthetic RNAs, recombinant proteins and small molecules (Gonzalez et al., 2011). Particularly, screenings have been applied to identify chemical compounds that can replace individual or multiple Yamanaka factors. Ichida et al. carried out a high-content chemical screening and identified RepSox (also named as E616452, an ALK5 inhibitor) as a chemical replacer of Sox2 (Ichida et al., 2009). The authors suggested that RepSox promoted the completion of reprogramming through induction of Nanog. In addition, RepSox possibly promoted mesenchymal-to-epithelial transition (MET) during the early phase of reprogramming (Li et al., 2010). Using a Nanog-Luciferase reporter as readout, Kenpaullone was found to functionally replace Klf4 in the presence of OSM (Lyssiotis et al., 2009). Shi et al. found that the combination of BIX01294 and Bayk8644 or BIX01294 and RG108 enabled OK-induced reprogramming of mouse fibroblasts, and Li et al. further reported successful OK-reprogramming of human somatic cells by using CHIR99021 and Tranylcypromine (also named Parnate) (Li et al., 2009; Shi et al., 2008). Huangfu et al. reported that Valproic acid (VPA) could promote OS-induced reprogramming of human fibroblasts (Huangfu et al., 2008). Taken together, these studies demonstrated that epigenetic regulators play important roles in iPSC reprogramming. Subsequently, several groups have focused on Oct4-only reprogramming by employing different chemical cocktails. Chen et al. found that bone morphogenetic proteins (BMPs) could support efficient mouse Oct4-alone reprogramming partly through the promotion of MET (Chen et al., 2011). Interestingly, Yuan et al. reported that AMI-5 and A83-01 enabled Oct4-induced reprogramming of mouse fibroblasts. Remarkably, these Oct4-only iPSCs were truly pluripotent and could give rise to live-born pups by tetraploid complementation assay (Yuan et al., 2011). Furthermore, Li et al. identified a specific chemical combination, including VPA, CHIR99021, E616452 and Tranylcypromine, which was sufficient to reprogramming mouse fibroblasts to iPSCs with Oct4 alone (Li et al., 2011). Subsequently, some interesting chemical substitutes of Oct4 in mouse iPS-reprogramming were identified, including Forskolin, D4476, and 2-Methyl-5-hydroxytryptamine (2-Me-5-HT) (Hou et al., 2013).
All chemical induced pluripotent reprogramming can provide a promising paradigm for cell fate transitions. In 2013, Hou et al. demonstrated successful reprogramming of mouse cells into iPS cells for the first time by using a novel cocktail of seven small molecules, including VPA, CHIR99021, E616452, Tranylcypromine, Forskolin, 3-deazaneplanocin A (DZNep), and TTNPB (Hou et al., 2013). Further refinement of this all-chemical reprogramming led a 1000-fold greater efficiency with additional small molecules, including AM580, EPZ004777, SGC0946, and 5-aza-2-deoxycitidine (5-aza-dC) (Zhao et al., 2015b). Interestingly, chemical induced pluripotent reprogramming process required the early formation of extra-embryonic endoderm (XEN)-like cells and a late transition from XEN-like cells to iPSCs, which fundamentally distinct from the pathway of TF-induced pluripotent reprogramming. However, it is still largely unknown how small molecules activate the endogenous gene regulation network, and gradually enable cell fate transitions. The remaining question is obvious: how to achieve chemical induced pluripotent reprogramming of human cells? We previously made some progress and reported that a chemical cocktail of Sodium butyrate (NaB), Parnate, PS48, CHIR99021, A83-01 and PD0325901 enabled OCT4-induced reprogramming of human primary somatic cells (Zhu et al., 2010). These human OCT4-only iPSCs were pluripotent and could give rise to cells of all three germ layers both in vitro and in vivo. Further mechanistic studies demonstrated that a metabolic switch from oxidative phosphorylation to glycolysis is an important step in iPSC reprogramming. More efforts are required to identify chemical substitutes for OCT4 in human cell reprogramming. The demonstration of human pluripotent reprogramming by an all-chemical approach will greatly benefit the study of stem cell biology and regenerative medicine.
NEURAL REPROGRAMMING
An increasing number of people suffer from neurodegenerative disorders, such as Alzheimer’s disease and Parkinson’s disease, as life expectancy further extends. It is not feasible to obtain sufficient amounts of patient specific neural cells for disease modeling and drug development. In the last few years, great progress has been made using TF-based direct reprogramming towards induced neurons (iNs) or induced neural stem cells (iNSCs).
In 2010, Vierbuchen et al. firstly reported that mesodermal mouse fibroblasts could be rapidly and efficiently reprogrammed into ectodermal iNs by using three TFs: Brn2, Ascl1, and Myt1 l (BAM) (Vierbuchen et al., 2010). Subsequently, BAM plus NEUROD1 was applied for neuronal reprogramming of human cells (Pang et al., 2011). So far, a number of groups have generated different subtypes of neurons by combining BAM with specific TFs that play an important role in the development of specific neuron subtypes (Caiazzo et al., 2011; Kim, 2011; Pang et al., 2011; Son et al., 2011). Besides TFs, microRNAs, such as miR-9 and miR-124, can also facilitate neuronal reprogramming (Ambasudhan et al., 2011; Yang et al., 2016; Yoo et al., 2011). Obviously, it is desirable to develop non-viral and non-integrative iN reprogramming approaches by small molecules. In 2015, two simultaneous articles reported the successful generation of iN from mouse and human fibroblasts via chemical-only approaches (Hu et al., 2015; Li et al., 2015). Li et al. showed that mouse fibroblasts could be converted into neurons by using an optimal cocktail of four small molecules (Forskolin, ISX9, CHIR99021, and I-BET151, termed FICB) (Li et al., 2015). The authors suggested that I-BET151, a BET family protein inhibitor, suppressed the fibroblast-specific program, and ISX9 activated the expression of endogenous neurogenic transcription factors, which synergistically promoted neuronal conversion (Li et al., 2015). Hu et al. demonstrated the generation of neurons from human fibroblasts using a small molecule cocktail (VPA, CHIR99021, RepSox, Forskolin, SP600125, GO6983, Y-27631, and Dorsomorphin) (Hu et al., 2015). Furthermore, via this chemical cocktail, fibroblasts from familial Alzheimer’s disease patients could also be reprogrammed to iNs. Such cells can be used for in vitro disease modeling and drug screenings (Hu et al., 2015). Subsequently, Zhang et al. reported that sequential addition of a cocktail of small molecules (LDN193189, SB431542, TTNPB, Thiazovivin (Tzv), CHIR99021, VPA, DAPT, Smoothened agonist (SAG), and Purmorphamine) can reprogram human astrocytes into functional neurons (Zhang et al., 2015). Mechanistically, these small molecules inhibited glial but activated neuronal signaling pathways through epigenetic and transcriptional modulation. Remarkably, these human iNs were functional and could survive more than 5 months under cell culture conditions.
Compared with neurons, expandable and multipotent iNSCs are desirable for downstream applications, like disease modeling and drug screening. In the past few years, many groups reported the generation of iNSCs using neural lineage-specific TFs. These iNSCs are multipotent and can differentiate into functional neurons, astrocytes, and oligodendrocytes both in vitro and in vivo (Ring et al., 2012; Zhou and Tripathi, 2012). Recently, we achieved both mouse and human iNSC reprogramming by a cell-activation signaling-directed (CASD) strategy (Kim et al., 2011; Zhu et al., 2015). The CASD strategy uses transient exposure of somatic cells to reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) in conjunction with soluble lineage-specific signals to reprogram cells into other cell types, such as iNSCs. Several interesting small molecules could promote OCT4-mediated iNSC reprogramming process, including A83-01, CHIR99021, NaB, Lysophosphatidic acid (LPA), Rolipram and SP600125 (Zhu et al., 2014a). Furthermore, similar to iN reprogramming, there are also great advances in iNSC reprogramming by using small molecules alone. In 2014, Cheng and colleagues used three small molecules VPA, CHIR99021, and RepSox to derive iNPCs from somatic cells (Cheng et al., 2014). More recently, Zhang et al. achieved more efficient mouse iNSC reprogramming by using a cocktail of nine components (CHIR99021, LDN193189, A83-01, Retinoic acid (RA), Hh-Ag1.5, RG108, Parnate, SMER28, and bFGF) (Zhang et al., 2016a). They provided definitive evidence that these iNSCs could be reprogrammed from fibroblasts using a genetic lineage-tracing system. Interestingly, further mechanistic studies uncovered that these small molecules could gradually and specifically activate key neurogenic regulators, such as Sox2, and then facilitated the neural cell fate transition.
Direct in vivo reprogramming will provide a perspective for cell-based clinical regenerative therapy (Chen et al., 2015; Li and Chen, 2016). Glial cells are the most abundant cells in adult brains and several groups have reported the successful TF-based reprogramming of glial cells to neurons or iNPCs. Niu et al. found that delivery of Sox2 could reprogram endogenous astrocytes to proliferating neuroblasts and these neuroblasts further differentiated to functional neurons that integrated into neural networks in the brain (Niu et al., 2013). Guo et al. demonstrated that cortical glial cells activated by injury or disease could be reprogrammed by NeuroD1 in vivo (Guo et al., 2014). The further application of knowledge learned from in vitro chemical screening and ambitious in vivo chemical screening will undoubtedly advance this field.
CARDIAC REPROGRAMMING
The adult mammalian heart possesses little regenerative capacity following injury. Cardiac fibroblasts account for a majority of cells in the heart, and cardiac reprogramming holds great potentials. In 2010, Ieda et al. reported that postnatal cardiac fibroblasts could be directly reprogrammed into induced cardiomyocyte-like cells (iCMs) by transfection with a combination of three TFs (Gata4, Mef2c, Tbx5, termed GMT) (Ieda et al., 2010). Lineage-tracing experiments showed that the cardiac reprogramming with GMT was a direct conversion process. Subsequently, other groups showed that addition of TFs such as Hand2 and Nkx2.5 to GMT promoted the reprogramming efficiency or maturation of iCMs (Addis and Epstein, 2013; Ifkovits et al., 2014; Song et al., 2012). Additionally,miRNAs, such as miR-1 and miR-133, also play important roles in cardiac reprogramming (Ieda, 2016; Jayawardena et al., 2012; Muraoka et al., 2014; Nam et al., 2013; Zhao et al., 2015a). Although the efficiency of cardiac reprogramming has been improved in recent years, the molecular mechanisms of this process are largely unknown. More recently, Zhou et al. carried out a small-scale functional screening and identified that loss of Bmi1 significantly promoted mouse cardiac reprogramming. Mechanistically, Bmi1 blocked cardiac reprogramming through direct interactions with the regulatory regions of many cardiogenic genes (Zhou et al., 2016). Compared with mouse cardiac reprogramming, human cardiac reprogramming is much more challenging to be achieved. Fu et al. showed that GMT plus ESRRG, MESP1, Myocardin, and ZFPM2 could reprogram human fibroblasts to iCMs (Fu et al., 2013). Nam et al. demonstrated that human fibroblasts could be converted to iCMs by introduction of GATA4, HAND2, TBX5, Myocardin, miR-1, and miR-133 under long-term culture (Nam et al., 2013). Wada et al. found that GMT plus MESP1 and Myocardin could reprogram human fibroblasts to iCMs (Rie et al., 2013). Such TF-reprogrammed iCMs exhibited sarcomere formation, calcium transients, and action potentials, but could not contract spontaneously and robustly. These results indicated that further improvements are still required for complete human cardiac reprogramming.
Small molecules can potentially replace TFs and provide a novel approach for cardiac reprogramming. Interestingly, the addition of TGF-β inhibitor SB431542 or A83-01 improved the efficiency of iCM generation (Fu et al., 2013; Zhao et al., 2015a). Other small molecules, including Y-27632, JAK inhibitor I, LY-364947, SD-208 and GW788388, did also enhance cardiac reprogramming (Lalit et al., 2016; Zhao et al., 2015a, b). Tamakawa et al. showed that adding fibroblast growth factor (FGF) 2, FGF10, and vascular endothelial growth factor (VEGF) to GMT enhanced cardiac reprogramming about 100-fold compared with GMT only (Yamakawa et al., 2015). Wang et al. identified a small molecule cocktail, including CHIR99021, SB431542, Parnate, and Forskolin, that enabled efficient conversion of mouse fibroblasts into iCMs with Oct4 alone employing the CASD paradigm. These iCMs spontaneously contracted and exhibited a ventricular phenotype (Wang et al., 2014). Recently, there have been significant achievements in cardiac reprogramming by using small molecules alone. Fu et al. showed that a chemical cocktail (CHIR99021, RepSox, Forskolin, and VPA) could induce beating clusters of cardiac cells from mouse fibroblasts. Other small molecules, including ICARIIN, PD169316 and Rolipram also increased the efficiency of cardiac reprogramming (Fu et al., 2015). Nan et al. showed that human iCMs could be generated from fibroblasts by a specific set of small molecules (Cao et al., 2016). This combination of nine compounds was termed 9C, including CHIR99021, A83-01, BIX01294, AS8351, SC1, Y-27632, OAC2, SU16F, and JNJ10198409. Remarkably, at both transcriptome and epigenetic levels, these human iCMs resembled human cardiomyocytes. The authors observed enrichment of H3K4me3 as well as H3K27ac, and decrease of H3K27me3 on a cohort of heart developmental genes during 9C-induced cardiac reprogramming. Mechanistically, 9C treatment epigenetically and transcriptionally activated key cardiac developmental genes and facilitated the cardiac fate transition. Interestingly, unlike human iCMs from TF-based reprogramming, these human 9C-induced iCMs uniformly contracted. More efforts are required to apply this chemical cardiac reprogramming approach to patient cells.
Expandable and multipotent induced cardiac progenitor cells (iCPCs) are more suitable for basic studies and translational applications. Islas et al. found that human dermal fibroblasts could be converted into cardiac progenitors by ETS2 and MESP1 overexpression (Islas et al., 2012). After a screening of multiple cardiac TFs, Lalit et al. reported that a combination of Mesp1, Tbx5, Gata4, Nkx2.5, and Baf60c could reprogram mouse fibroblasts into iCPCs (Lalit et al., 2016). Meanwhile, Zhang et al. demonstrated that fibroblasts could be induced into expandable iCPCs by the CASD lineage conversion strategy and these iCPCs could be further long-term expanded using a combination of growth factors and small molecules (BMP4, Activin A, CHIR99021 and SU5402) (Zhang et al., 2016b). The activation of Wnt signaling by CHIR99021 not only promoted iCPC proliferation, but also inhibited further differentiation of iCPCs into cardiomyocytes. In addition, treating iCPC with Wnt inhibitor IWP2 facilitated even more efficient cardiac differentiation. Reprogramming mouse and human fibroblasts into expandable cardiac progenitors by small molecules alone could likely be achieved with further screening and modifications in the future.
Cardiac fibroblasts account for a majority of cells in the heart and represent a potential cell source for cardiac reprogramming. Direct in vivo cardiac reprogramming has already demonstrated some promising results. Qian et al. converted mouse resident non-myocytes into iCMs in vivo by local delivery of GMT after coronary ligation (Qian et al., 2012). Song et al. showed that GMT plus Hand2 could cooperatively reprogram dividing non-cardiomyocytes into functional iCMs in situ (Song et al., 2012). These cardiac reprogramming TF-delivered hearts had decreased infarct size and better cardiac function. Therefore, reprogramming cardiac fibroblasts to iCMs in their native environment by chemical compounds will hold great potentials in cardiac regenerative therapies.
HEPATIC REPROGRAMMING
The liver is a fascinating organ that can regenerate itself; but unfortunately, this remarkable capacity is lost in chronic liver diseases. Currently, there are over 600 million people with liver diseases and 1 million deaths per year worldwide. Because the limitation of organ donors, cell-based therapy may represent an attractive approach for treating life-threatening liver failure, if a sufficient amount of functional hepatocytes can be generated from easily accessible cell sources. Many research groups focus their efforts on the generation of induced hepatocyte-like cells (iHeps) via reprogramming approaches. In 2011, Huang et al. showed that functional iHep cells could be converted from p19 null mouse fibroblasts via transduction of Gata4, Hnf1α, and Foxa3 (Huang et al., 2011). Moreover, these iHep cells restored liver functions in vivo and rescued the recipients from death, which provided a promising proof of principle example for the use in regenerative medicine. Meanwhile, Sekiya et al. converted mouse embryonic and adult fibroblasts into iHep cells using a combination of Hnf4α plus Foxa1, Foxa2 or Foxa3 (Sekiya and Suzuki, 2011). Remarkably, after overexpressing Hnf1β and Foxa3, mouse embryonic fibroblasts were converted to induced hepatic stem cells, which had the capability of long-term expansion and bipotential differentiation to hepatocytes and cholangiocytes (Yu et al., 2013). Subsequently, human fibroblasts were successfully reprogrammed into iHep cells by different combinations of TFs (Du et al., 2014; Huang et al., 2014). Huang et al. generated human iHeps from fibroblasts by introduction of FOXA3, HNF1α, and HNF4α (Huang et al., 2014). Meanwhile, Du et al. showed that human fibroblasts could be reprogrammed to functional iHeps by overexpressing HNF1α, HNF4α, HNF6, ATF5, PROX1, and CEBPα (Du et al., 2014). These human iHeps expressed liver-specific markers, and could integrate into mouse liver and demonstrated many metabolic functions of the liver.
Recently, the non-viral and non-integrating transduction systems, including synthetic RNAs, recombinant proteins, and episomal plasmids, become more attractive (Fusaki et al., 2009; Kim et al., 2009, 2015; Warren et al., 2010; Zhou and Freed, 2009). For example, Simeonov et al. used a method of repeating transfection with synthetic modified mRNAs encoding hepatic reprogramming TFs and successfully induced iHep cells from human fibroblasts (Simeonov and Uppal, 2014). Kim et al. applied an oriP/EBNA1-based episomal system to deliver a set of transcription factors, including Gata4, Hnf1α, and Foxa3, to convert fibroblasts into iHep cells (Kim et al., 2015). Small molecules also played a critical role in iHep reprogramming process (Lim et al., 2016; Pournasr et al., 2015). We previously generated induced endodermal progenitor cells by the CASD lineage conversion strategy, and greatly expanded these endodermal progenitor cells by a chemically defined condition, consisting of EGF, bFGF, A83-01, and CHIR99021. Further treatment with soluble growth factors and small molecules, such as A83-01 and Compound E, these induced endodermal progenitor cells efficiently differentiated into functional hepatocytes. After transplantation, human iHeps integrated into immunodeficient mouse livers, expanded extensively, and acquired mature hepatocyte functions (Zhu et al., 2014b). Moreover, Lim et al. reported that direct conversion into iHeps is a stepwise transition involving the sequential erasure of somatic memory, MET transition, and induction of hepatic cell fate (Lim et al., 2016). Through chemical screening, they found that CHIR99021 and A83-01 facilitated one factor Hnf1α-induced hepatic reprogramming. Mechanistically, these small molecules facilitated the robust iHep generation through promoting MET process. In the future, utilizing all-chemical method to induce iHep reprogramming will be a promising way to promote the development of this field.
More recently, in vivo hepatic reprogramming was fulfilled by injection of hepatic reprogramming factors in situ (Rezvani et al., 2016; Song et al., 2016). Rezvani et al. developed in vivo reprogramming of myofibroblasts into hepatocytes using adeno-associated virus (AAV) vectors expressing hepatic transcription factors: Foxa1, Foxa2, Foxa3, Gata4, Hnf1a, and Hnf4a. These iHeps converted from myofibroblsts were functional and reduced liver fibrosis (Rezvani et al., 2016). Song et al. showed that overexpression of FOXA3, GATA4, HNF1A, and HNF4A could convert mouse myofibroblasts into iHeps cells in fibrotic mouse livers and also reduced liver fibrosis (Song et al., 2016). More efforts are required to develop better delivery methods and improve the conversion efficiency and cell maturity. Obviously, in vivo hepatic reprogramming by small molecules will facilitate the translation of the iHep technology into clinical treatment of chronic liver diseases.
PANCREATIC REPROGRAMMING
Diabetes mellitus represents a global health epidemic and affects more than 300 million people worldwide according to the International Diabetes Federation. Diabetes is caused and developed as a deficiency and/or dysfunction of pancreatic β cells. A key method to study diabetes and treat patients is to obtain unlimited numbers of functional pancreatic β cells. Reprogramming other types of somatic cells to pancreatic β cells is under active investigation. In 2005, Minammi et al. reported that adult mouse pancreatic exocrine cells could be converted into insulin-producing cells in vitro by suspension culture with EGF and nicotinamide (Minami et al., 2005). In 2008, Zhou et al. reprogrammed adult pancreatic exocrine cells to β-like cells in vivo by introducing three TFs: Pdx1, Ngn3, and Mafa (Zhou et al., 2008). Then, Li et al. developed an improved method for in vivo pancreatic conversion (Li et al., 2014b). These induced pancreatic β-like cells formed the islet-like structures and could persist in vivo for more than one year (Li et al., 2014a). Additionally, Chen et al. reported that transient intestinal expression of Pdx1, Ngn3, and Mafa promoted rapid conversion of intestinal crypt cells into endocrine cells (Chen et al., 2014). Besides mouse cells, human pancreatic ductal cells could also be converted into pancreatic β-like cells by overexpression of PDX1, NGN3, MAFA, and PAX6 (Lee et al., 2013). These converted pancreatic β-like cells could secrete insulin in response to high concentration of glucose. More interestingly, Ariyachet et al. reported that cells of the antral stomach were ideal cell source for pancreatic reprogramming (Ariyachet et al., 2016). These reprogrammed insulin+ β-like cells performed molecularly and functionally similar to bona fide pancreatic β cells. These results suggested that the initial cell type can affect the reprogramming efficiency and induced cell phenotype and its functional properties.
Introducing exogenous genetic material and altering the genome raise safety concerns (Barrilleaux and Knoepfler, 2011). Therefore, using small molecules to active endogenous TFs is an attractive method for cellular reprogramming. By high-throughput screening, BRD7552 was shown to be an inducer of PDX1 expression (Yuan et al., 2013). In another report, DNA methyltransferase inhibitor 5-aza-dC was capable of increasing the expression of Ngn3 in PANC-1 cells (Lefebvre et al., 2010). In our previous study, we found that several specific small molecules could enhance the endodermal conversion process, including CHIR99021, NaB, Parnate, RG108, and 5’-N-ethylcarboxamidoadenosine, and promote pancreatic β-like cell differentiation and maturation, including Compound E, Vitamin C, and Bayk8644 (Zhu et al., 2016). More recently, Wang et al. described the derivation of human induced endodermal progenitor cells from gastrointestinal epithelial cells using a cocktail of defined small molecules, including Bayk8644, BIX01294, RG108, and SB431542, along with support from tissue-specific mesenchymal feeders (Wang et al., 2016). Induced endodermal progenitor cells could subsequently differentiate into more specified cell types, including pancreatic β-like cells. However, the therapeutic potentials of these pancreatic β-like cells have not been investigated so far. Obviously, using small molecules to replace master TFs as well as promote reprogramming to mature pancreatic β cells is a promising therapy for treating diabetes. However, transferring these findings for efficient in vivo pancreatic reprogramming, preferably be an all-chemical approach is waiting to be achieved in the future.
PERSPECTIVE
Cell fate transition and regeneration have fascinated biologists for centuries. After the discovery of iPSC technology, a lot of progress has been made in TF-mediated cellular reprogramming into many different defined cell types. Compared with the introduction of TFs by viral methods, small molecules have several advantages, can enhance reprogramming efficiency and improve the quality of the reprogrammed cells. Currently, identification of novel and effective small molecules for cellular reprogramming remains very labor intense. New technologies, such as high-throughput screening (HTS2) platform (Li et al., 2012) and reporter systems generated by CRISPR tools (Hockemeyer and Jaenisch, 2016), can be applied to this field and will accelerate the discovery process. In addition, the molecular mechanisms of most reprogramming processes are largely unknown, and how TFs or small molecules mediate cell fate transitions is still unclear. New technologies, such as single-cell analysis (Wen and Tang, 2016) and CRISPR-based genome-wide screening (Shalem et al., 2014), can provide new insights and help us to better understand the underlying molecular mechanisms. A better understanding of the detailed mechanisms during reprogramming processes can also help to improve the reprogramming efficiency and acquire mature cells with complete functionality for disease modeling, drug development, and cell-based transplantation (Fig. 2). Optimistically, future studies in cellular reprogramming by small molecules will overcome current hurdles, generate new discoveries, and benefit human health.
Figure 2.
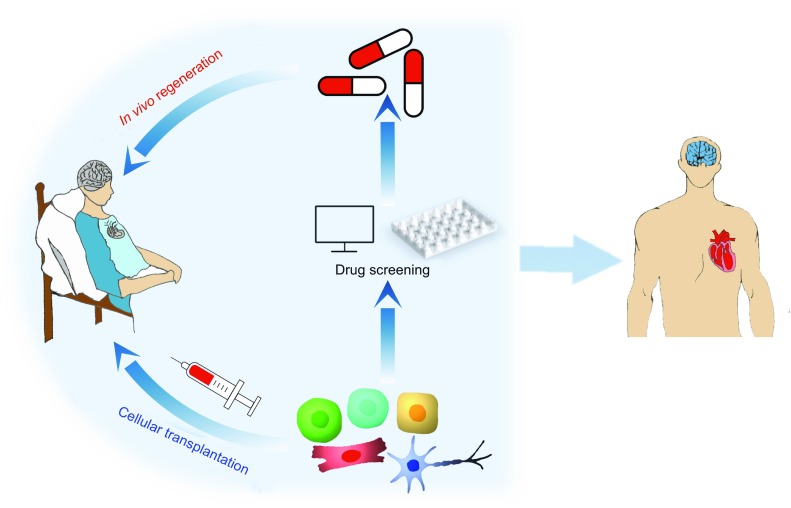
Potential applications of cellular reprogramming. Cellular reprogramming can provide a large number of functional cells, which can be used for cell-based transplantation and high-throughput chemical screenings. This technology will help develop drugs to stimulate patients’ endogenous cells to repair and regenerate in vivo in the near future. Cellular reprogramming by only small molecules will significantly advance biomedical studies and clinical applications, and realize the long-term goal of curing degenerative diseases, injuries, and aging
ACKNOWLEDGEMENTS
We would like to thank Dr. Wanguo Wei at Shanghai Advanced Research Institute, Chinese Academy of Sciences and Dr. Holger A. Russ at the Barbara Davis Center for Childhood Diabetes at the University of Colorado for their generous help. This work was funded by the national key research and development programme of China (2016YFC1305300) and the outstanding youth fund of Zhejiang Province (R17C120002). S.Z. is a scholar of the National 100 Young Talents Program. We apologize to those whose findings are relevant but not cited in this review due to space limitation.
ABBREVIATIONS
2-Me-5-HT, 2-Methyl-5-hydroxytryptamine; AAV, adeno-associated virus; BAM, Brn2, Ascl1, and Myt1 l; BMP, bone morphogenetic protein; CASD, cell-activation signaling-directed; FGF, fibroblast growth factor; iCM, induced cardiomyocyte-like cell; iCPC, induced cardiac progenitor cell; iN, induced neuron; iNSC, induced neural stem cell; iPSC, induced pluripotent stem cell; LPA, Lysophosphatidic acid; NaB, Sodium butyrate; RA, Retinoic acid; SAG, Smoothened agonist; TF, transcription factor; Tzv, Thiazovivin; VEGF, vascular endothelial growth factor; XEN, extra-embryonic endoderm.
COMPLIANCE WITH ETHICS GUIDELINES
Xiaojie Ma, Linghao Kong, and Saiyong Zhu declare that they have no conflict of interest.
REFERENCES
- Addis RC, Epstein JA. Induced regeneration–the progress and promise of direct reprogramming for heart repair. Nat Med. 2013;19:829–836. doi: 10.1038/nm.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyachet C, Tovaglieri A, Xiang G, Lu J, Shah MS, Richmond CA, Zhou Q. Reprogrammed stomach tissue as a renewable source of functional beta cells for blood glucose regulation. Cell Stem Cell. 2016;18:410–421. doi: 10.1016/j.stem.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrilleaux B, Knoepfler P. Transduction of human cells with polymer-complexed ecotropic lentivirus for enhanced biosafety. J Vis Exp. 2011;53:1–7. doi: 10.3791/2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu J, Nie B, Xie M, Zhang M, Wang H, Ma T, Xu T, Shi G, Srivastava D, Ding S. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu J, Yang J, Chen Y, Chen J, Ni S, Song H, Zeng L, Ding K, Pei D. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 2011;21:205–212. doi: 10.1038/cr.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Finkbeiner SR, Weinblatt D, Emmett MJ, Tameire F, Yousefi M, Yang C, Maehr R, Zhou Q, Shemer R, Dor Y, Li C, Spence JR, Stanger BZ. De novo formation of insulin-producing “neo-beta cell islets” from intestinal crypts. Cell Rep. 2014;6:1046–1058. doi: 10.1016/j.celrep.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wernig M, Berninger B, Nakafuku M, Parmar M, Zhang CL. In vivo reprogramming for brain and spinal cord repair. eNeuro. 2015;2:e0106-15. doi: 10.1523/ENEURO.0106-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan W, Wang M, Yang W, Pei G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;24:665–679. doi: 10.1038/cr.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, Hou Z, Su X, Liu B, Jiang T, Zhao D, Sun Y, Shu J, Guo Q, Yin M, Sun D, Lu S, Shi Y, Deng H. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Fu J, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Huang C, Xu X, Gu H, Ye Y, Jiang C, Qiu Z, Xie X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015;25:1013–1024. doi: 10.1038/cr.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proce Jpn Acad Ser B. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Boue S, Izpisua Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nat Rev Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Hu W, Qiu B, Guan W, Wang Q, Wang M, Li W, Gao L, Shen L, Huang Y, Xie G, Zhao H, Jin Y, Tang B, Yu Y, Zhao J, Pei G. Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell. 2015;17:204–212. doi: 10.1016/j.stem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, Lai D, Hu Z, Chen L, Zhang Y, Cheng X, Ma X, Pan G, Wang X, Hui L. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M. Heart development, diseases, and regeneration- new approaches from innervation, fibroblasts, and reprogramming. Circ J. 2016;80:2081–2088. doi: 10.1253/circj.CJ-16-0815. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu J, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifkovits JL, Addis RC, Epstein JA, Gearhart JD. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One. 2014;9:e89678. doi: 10.1371/journal.pone.0089678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng K-C, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, Mercola M, Oshima RG, Willerson JT, Potaman VN, Schwartz RJ. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS. Converting human skin cells to neurons: a new tool to study and treat brain disorders? Cell Stem Cell. 2011;9:179–181. doi: 10.1016/j.stem.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim KP, Lim KT, Lee SC, Yoon J, Song G, Hwang SI, Schöler HR, Cantz T, Han DW. Generation of integration-free induced hepatocyte-like cells from mouse fibroblasts. Sci Rep. 2015;5:15706. doi: 10.1038/srep15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalit PA, Salick MR, Nelson DO, Squirrell JM, Shafer CM, Patel NG, Saeed I, Schmuck EG, Markandeya YS, Wong R, Lea MR, Eliceiri KW, Hacker TA, Crone WC, Kyba M, Garry DJ, Stewart R, Thomson JA, Downs KM, Lyons GE, Kamp TJ. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell. 2016;18:354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sugiyama T, Liu Y, Wang J, Gu X, Lei J, Markmann JF, Miyazaki S, Miyazaki J, Szot GL, Bottino R, Kim SK. Expansion and conversion of human pancreatic ductal cells into insulin-secreting endocrine cells. Elife. 2013;2:e00940. doi: 10.7554/eLife.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Belaich S, Longue J, Vandewalle B, Oberholzer J, Gmyr V, Pattou F, Kerr-Conte J. 5’-AZA induces Ngn3 expression and endocrine differentiation in the PANC-1 human ductal cell line. Biochem Biophys Res Commun. 2010;391:305–309. doi: 10.1016/j.bbrc.2009.11.054. [DOI] [PubMed] [Google Scholar]
- Li H, Chen G. In vivo reprogramming for CNS repair: regenerating neurons from endogenous glial cells. Neuron. 2016;91:728–738. doi: 10.1016/j.neuron.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Schöler HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, Wu Y, Li H, Liu K, Wu C, Song Z, Zhao Y, Shi Y, Deng H. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou H, Wang D, Qiu J, Zhou Y, Li X, Rosenfeld MG, Ding S, Fu X. Versatile pathway-centric approach based on high- throughput sequencing to anticancer drug discovery. Proc Natl Acad Sci USA. 2012;109:4609–4614. doi: 10.1073/pnas.1200305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cavelti-Weder C, Zhang Y, Clement K, Donovan S, Gonzalez G, Zhu J, Stemann M, Xu K, Hashimoto T, Yamada T, Nakanishi M, Zhang Y, Zeng S, Gifford D, Meissner A, Weir G, Zhou Q, Nakanishi M, Zhang Y, Zeng S, Gifford D, Meissner A, Weir G, Zhou Q. Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells. Nat Biotechnol. 2014;32:1223–1230. doi: 10.1038/nbt.3082. [DOI] [PubMed] [Google Scholar]
- Li W, Nakanishi M, Zumsteg A, Shear M, Wright C, Melton DA, Zhou Q. In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. Elife. 2014;3:e01846. doi: 10.7554/eLife.01846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zuo X, Jing J, Ma Y, Wang J, Liu D, Zhu J, Du X, Xiong L, Du Y, Xu J, Xiao X, Wang J, Chai Z, Zhao Y, Deng H. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell. 2015;17:195–203. doi: 10.1016/j.stem.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Lim KT, Lee SC, Gao Y, Kim KP, Song G, An SY, Adachi K, Jang YJ, Kim J, Oh KJ, Kwak TH, Hwang SI, You JS, Ko K, Koo SH, Sharma AD, Kim JH, Hui L, Cantz T, Schöler HR, Han DW. Small molecules facilitate single factor-mediated hepatic reprogramming. Cell Rep. 2016;15:814–829. doi: 10.1016/j.celrep.2016.03.071. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, Bollong M, Kunick C, Brinker A, Cho CY, Schultz PG, Jaenisch R. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci USA. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K, Okuno M, Miyawaki K, Okumachi A, Ishizaki K, Oyama K, Kawaguchi M, Ishizuka N, Iwanaga T, Seino S. Lineage tracing and characterization of insulin-secreting cells generated from adult pancreatic acinar cells. Proc Natl Acad Sci USA. 2005;102:15116–15121. doi: 10.1073/pnas.0507567102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, Nishiyama T, Kaneda R, Fukuda T, Takeda S, Tohyama S, Hashimoto H, Kawamura Y, Goshima N, Aeba R, Yamagishi H, Fukuda K, Ieda M. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–1581. doi: 10.15252/embj.201387605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, Zhang CL. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang T, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournasr B, Asghari-Vostikolaee MH, Baharvand H. Transcription factor-mediated reprograming of fibroblasts to hepatocyte-like cells. Eur J Cell Biol. 2015;94:603–610. doi: 10.1016/j.ejcb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani M, Espanol-Suner R, Malato Y, Dumont L, Grimm AA, Kienle E, Willenbring H. In vivo hepatic reprogramming of myofibroblasts with aav vectors as a therapeutic strategy for liver fibrosis. Cell Stem Cell. 2016;18:809–816. doi: 10.1016/j.stem.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rie W, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, Tohyama S. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci USA. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Simeonov KP, Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS One. 2014;9:e100134. doi: 10.1371/journal.pone.0100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang G, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Pacher M, Balakrishnan A, Yuan Q, Tsay HC, Yang D, Reetz J, Brandes S, Dai Z, Pützer BM, Araúzo-Bravo MJ, Steinemann D, Luedde T, Schwabe RF, Manns MP, Schöler HR, Schambach A, Cantz T, Ott M, Sharma AD. Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell. 2016;18:797–808. doi: 10.1016/j.stem.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang Z, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D, Ding S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014;6:951–960. doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin J, Wang S, Zhang W, Duan J, Zhang J, Wang X, Yan F, Chang M, Liu X, Feng B, Liu J, Pei X. Conversion of human gastric epithelial cells to multipotent endodermal progenitors using defined small molecules. Cell Stem Cell. 2016;19:449–461. doi: 10.1016/j.stem.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Tang F. Single-cell sequencing in stem cell biology. Genome Biol. 2016;17:71. doi: 10.1186/s13059-016-0941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y, Kurokawa J, Furukawa T, Fukuda K, Ieda M. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports. 2015;5:1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhang L, An J, Zhang Q, Liu C, He B, Hao D. MicroRNA-mediated reprogramming of somatic cells into neural stem cells or neurons. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0115-9. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, He Z, You P, Han Q, Xiang D, Chen F, Wang M, Liu C, Lin X, Borjigin U, Zi X, Li J, Zhu H, Li W, Han C, Wangensteen KJ, Shi Y, Hui L, Wang X, Hu Y. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell. 2013;13:328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Yuan X, Wan H, Zhao X, Zhu S, Zhou Q, Ding S. Brief report: combined chemical treatment enables Oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells. 2011;29:549–553. doi: 10.1002/stem.594. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Hartland K, Boskovic Z, Wang Y, Walpita D, Lysy PA, Zhong C, Young DW, Kim YK, Tolliday NJ, Sokal EM, Schreiber SL, Wagner BK. A small-molecule inducer of PDX1 expression identified by high-throughput screening. Chem Biol. 2013;20:1513–1522. doi: 10.1016/j.chembiol.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yin J, Yeh H, Ma N, Lee G, Chen X, Wang Y, Lin L, Chen L, Jin P, Wu G, Chen G. Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell. 2015;17:735–747. doi: 10.1016/j.stem.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lin Y, Sun Y, Zhu S, Zheng J, Liu K, Cao N, Li K, Huang Y, Ding S. Pharmacological reprogramming of fibroblasts into neural stem cells by signaling-directed transcriptional activation. Cell Stem Cell. 2016;18:653–667. doi: 10.1016/j.stem.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao N, Huang Y, Spencer CI, Fu J, Yu C, Liu K, Nie B, Xu T, Li K, Xu S, Bruneau BG, Srivastava D, Ding S. Expandable cardiovascular progenitor cells reprogrammed from fibroblasts. Cell Stem Cell. 2016;18:368–381. doi: 10.1016/j.stem.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O’Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM, McKinsey TA, Song K. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhao T, Guan J, Zhang X, Fu Y, Ye J, Zhu J, Meng G, Ge J, Yang S, Cheng L, Du Y, Zhao C, Wang T, Su L, Yang W, Deng H. A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell. 2015;163:1678–1691. doi: 10.1016/j.cell.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Tripathi P. How to remake a fibroblast into a neural stem cell. Cell Stem Cell. 2012;10:347–348. doi: 10.1016/j.stem.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin C, Fu J, Wang G, Liu J, Qian L. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Ambasudhan R, Sun W, Kim HJ, Talantova M, Wang X, Zhang M, Zhang Y, Laurent T, Parker J, Kim HS, Zaremba JD, Saleem S, Sanz-Blasco S, Masliah E, McKercher SR, Cho YS, Lipton SA, Kim J, Ding S. Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 2014;24:126–129. doi: 10.1038/cr.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, Willenbring H, Ding S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Wang H, Ding S. Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nat Protoc. 2015;10:959–973. doi: 10.1038/nprot.2015.059. [DOI] [PubMed] [Google Scholar]
- Zhu S, Russ HA, Wang X, Zhang M, Ma T, Xu T, Tang S, Hebrok M, Ding S. Human pancreatic beta-like cells converted from fibroblasts. Nat Commun. 2016;7:10080. doi: 10.1038/ncomms10080. [DOI] [PMC free article] [PubMed] [Google Scholar]


