Dear Editor,
Histone deacetylase 6 (Hdac6) was discovered as a deacetylase of α-tubulin and functions in cell migration, immunity and resistance to virus infection in vitro (Hubbert et al., 2002; Valenzuela-Fernandez et al., 2008). Overexpression of Hdac6 enhances resistance to virus infection in embryonic stem (ES) cells and in mice (Wang et al., 2015). Hdac6 also can function to deacetylate protein and is involved in protein ubiquitination and degradation (Seigneurin-Berny et al., 2001; Zhang et al., 2014), and self-clearance of misfolded proteins, promoting autophagy and preventing neurodegeneration (Lee et al., 2010; Pandey et al., 2007). HDAC6 also is implicated in DNA damage response and depletion or inhibition of HDAC6 induces DNA damage and apoptosis (Namdar et al., 2010; Zhang et al., 2014), suggesting that HDAC6 could be important for DNA repair and integrity maintenance.
DNA damage accumulates with somatic aging and reproductive aging (Titus et al., 2013), and enhanced DNA damage repair can reverse aging (Maynard et al., 2015). Reproductive aging in females mainly results from ovarian senescence, as shown by reduced number and quality of follicles and germ cells. Interestingly, Hdac6 is highly expressed in germ cell tissues, testis and spermatogenic cells, relative to other somatic tissues, such as liver, heart, muscle, spleen and kidney (Seigneurin-Berny et al., 2001; Zhang et al., 2008), and yet Hdac6-deficient mice are viable and fertile, and show normal development and function of testis (Zhang et al., 2008). Initially, we tested whether DNA damage is reduced in Hdac6 overexpression embryonic stem (ES) cells, which expressed higher Hdac6 protein levels and lower levels of α-acetylated tubulin than did WT ES cells (Wang et al., 2015). Treatment of mouse embryonic fibroblasts (MEFs) with mitomycin C can generate mitotically inactive feeder cells and these cells showed high levels of DNA damage as shown by many γH2AX foci served as positive control (Fig. S1A and 1B). Fewer γH2AX foci were seen in WT ES cells, and comparatively, even fewer γH2AX foci in Hdac6 overexpression ES cells than in WT ES cells (Fig. S1A and 1B). Also, Hdac6 overexpression ES cells expressed lower γH2AX protein levels than did WT ES cells (Fig. S1C). These data show that high levels of Hdac6 can reduce DNA damage or induce more robust DNA repair.
Further, we compared telomere lengths of Hdac6 overexpression ES cells with those of WT ES cells. Telomeres were longer in Hdac6 overexpression ES cells than in WT ES cells shown as T/S ratio by qPCR method (Fig. S1D), telomere restriction fragment (TRF) analysis using conventional Southern blot (Fig. S1E) and quantitative fluorescence in situ hybridization (Q-FISH) (Fig. S1F). These data suggest that high expression levels of Hdac6 can lead to telomere elongation in ES cells.
To look at down-stream genes regulated by overexpression of Hdac6, we performed the genome-wide expression analysis using Agilent Mouse Gene Expression Microarray, 8X60K chips (heatmap showing differential gene expression as 1.5 fold changes as cut-off, Fig. S1G). The microarray data was confirmed by QPCR analysis (Table S1). Genes for telomere elongation, e.g. Tbx3 and Zscan4 were up-regulated in Hdac6 overexpression ES cells. Other prominently up-regulated genes included Trp73, Gadd45a, Sox17, Gata4 and Foxa2 (Fig. S1H and S1I). Sox17, Gata4 and Foxa2 are important in gonad and germ cell development, fertility and suppression of cell senescence. Genes involved in apoptosis, ubiquitin mediated proteolysis and DNA damage response also showed expression changes in Hdac6 overexpression ES cells.
We tested whether Hdac6 influences epigenetic modification. H3K9me3 levels negatively regulate telomere lengths, but the histone acetylation levels positively regulate telomere lengths (Liu, 2017). H3ac and H3K9ac protein levels did not differ between Hdac6 overexpression and WT ES cells, whereas H3K9me3 levels were reduced in Hdac6 overexpression ES cells compared with WT ES cells (Fig. S1J). By ChIP-qPCR analysis, levels of H3K9ac at subtelomeres of selected chromosomes 7 and 13 were generally low, as expected, and did not differ between Hdac6 overexpression and WT ES cells (Fig. S1K). However, H3K9me3 was enriched at subtelomeres in WT ES cells and the enrichment reduced in Hdac6 overexpression ES cells (Fig. S1K). Moreover, H3K9me3 levels at Zscan4 loci also were declined in Hdac6 overexpression ES cells. These data show that Hdac6 can repress H3K9me3 and its enrichment at subtelomeres, associated with telomere elongation in ES cells.
Hdac6 overexpression mice were generated from Hdac6 overexpression ES cells (Wang et al., 2015). During breeding of Hdac6 overexpression transgenic mice, we observed that Hdac6 overexpression females in B6C3F1 background could still reproduce good number of pups by middle age, and even at the old age, in contrast to none or minimal number of pups produced from age-matched WT mice. Higher expression levels of Hdac6 in the ovary of Hdac6 transgenic mice than in WT mice were confirmed by immunohistochemistry (Fig. S2A), qPCR analysis (Fig. S2B), Western blot (Fig. S2C) and immunofluorescence microscopy (Fig. S2D). Hdac6 was expressed mostly in the cytoplasm and also in nuclei of cells in the cortex, oocytes, granulosa cells and corpus luteum (Fig. S2A and S2D). Hdac6 overexpression female mice in hybrid background at the age of 12–16 months produced significantly larger litter size compared with age-matched WT mice (Fig. 1A). By backcross breeding of the hybrid F1 mice, we achieved a few litter of Hdac6 overexpression inbred mice in C57BL/6 genetic background. Hdac6 overexpression C57BL/6 females at the age of 10–12 month old still reproduced an average of four pups compared to none from age-matched WT mice (Fig. 1B). Higher levels of Hdac6 are associated with extended reproductive age. Histological analysis of sections of ovaries collected from old mice revealed increased folliculogenesis and particularly secondary and antral follicles in Hdac6 overexpression mice, compared with WT mice (Fig. 1C and 1D).
Figure 1.
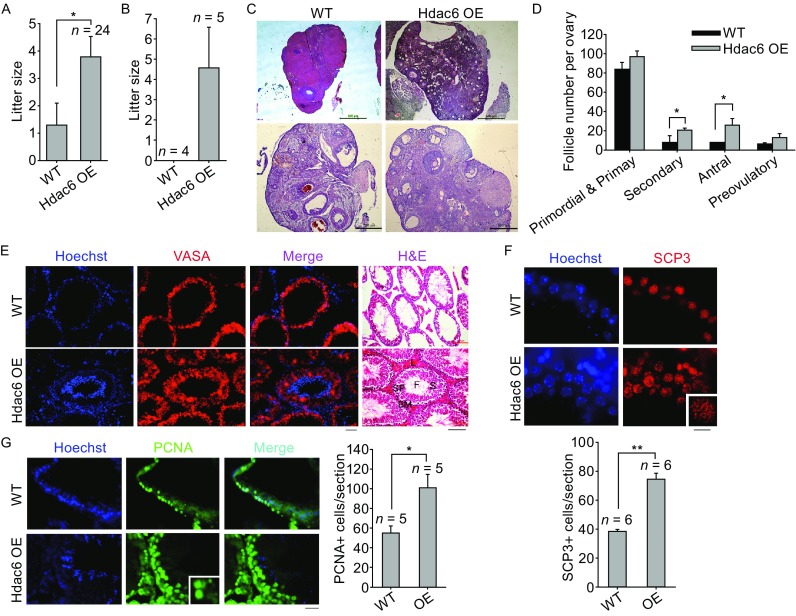
Overexpression of Hdac6 extends reproductive performance. (A–D) Litter size and folliculogenesis in Hdac6 overexpression and WT females. (A) Average litter size based on the number of old (age of 12 to 16 months) B6C3F1 female mice that were successfully mated, showing mating plugs. P < 0.05 by t test. (B) Average litter size produced from successfully mated old (about 10–12 months) C57BL/6 inbred females. n = number of successfully mated females. More old C57BL/6 mice were set to mate but failed from plugging. (C) Representative morphology of ovaries by hematoxylin and eosin (H&E) histology. Shown are ovaries from WT at 16-month-old and Hdac6 OE mice at 19-month-old in B6C3F1 hybrid background. Scale bar = 500 μm. (D) Follicle development at the primordial and primary, secondary, antral, preovulatory stages. *P < 0.05. (E–G) Spermatogenesis and meiosis in old male mice. (E) Robust spermatogenesis in Hdac6 OE mice shown by hematoxylin and eosin (H&E) histology of testis sections and immunofluorescence (IF) of VASA, indicative of germ cells, compared with much reduced spermatogenesis in WT mice at B6C3F1 hybrid background. Scale bars for IF = 20 μm; Scale bars for H&E = 100 μm. Testis section includes basement membrane (BM), Leydig cells (L), Sertoli cells (S), spermatogonia (SP) and filament (F). (F) Early meiocytes showing distinct synaptonemal complex protein-3 (SCP3) lateral filament structure at pachytene stage and quantification of average SCP3 positive cells per seminiferous tubule section. Inset, SCP3 lateral filaments at higher magnification. (G) Immunofluorescence of PCNA in testis from WT and Hdac6 OE males and relative quantification of average PCNA positive cells per seminiferous tubule section. Scale bar = 20 μm (SCP3) or 50 μm (PCNA). *P < 0.05, **P < 0.01. n = number of tubule sections counted
Overexpression of Hdac6 in the testis of Hdac6 transgenic mice was shown by immunohistochemistry, immunofluorescence microscopy and qPCR analysis (Fig. S2E, S2F and S2G). Hdac6 expression was distributed in both nuclei and cytoplasm of various germ cells. Seminiferous tubules of old Hdac6 overexpression males (19–21 month-old) were enriched with various germ cell types, while the age-matched WT controls showed much reduced number of germ cells in some seminiferous tubules (Fig. 1E). Abundant VASA positive cells were found in sections of seminiferous tubules in Hdac6 overexpression testis, in contrast to some testis of WT mice (Fig. 1E), indicating that testis of old Hdac6 overexpression males still actively undergo spermatogenesis and meiosis. Early meiocytes/spermatocytes are characterized by distinct synaptonemal complex protein-3 (SCP3) filament structure. Abundant PCNA+ proliferative germ cells and spermatocytes with homologous pairing marked by SCP3 filaments were found in testis of Hdac6 overexpression mice, unlike age-matched WT mice (Fig. 1F and 1G). By TUNEL assay, fewer apoptotic cells were found in testis of Hdac6 overexpression mice than in WT mice at the old age (Fig. S2H). These data indicate that germ cell attrition occurs in old males, but higher expression levels of Hdac6 can reduce germ cell attrition and preserve spermatogenesis.
Follicle atresia and germ cell attrition could be related to autophagy. We did not find evident changes in the expression levels by qPCR of key genes for autophagy between Hdac6 overexpression and WT mouse gonads (Fig. 2A). Autophagy-LC3B protein expression levels and distribution also did not differ between Hdac6 overexpression and WT ES cells (Data not shown). We analyzed telomere lengths of somatic tails, and ovaries and testis from Hdac6 overexpression mice in comparison with age-matched WT mice. Relative telomere lengths shown as T/S ratio were longer in tails and testis, and slightly longer in ovaries of Hdac6 overexpression mice than those of age-matched WT mice (Fig. 2C). Interestingly, telomeres were longer in female tails than those of male tails in general. In males, telomeres were longer in testis than in somatic tails. Analysis of expression of genes important for telomere regulation showed that expression levels of telomerase genes Tert and Terc, telomerase activating gene Sirt1, and genes for meiosis recombination, Spo11 or Dmc1, or cell senescence genes p53, p16 or mTOR basically did not differ between Hdac6 overexpression and WT gonads (Fig. 2B).
Figure 2.
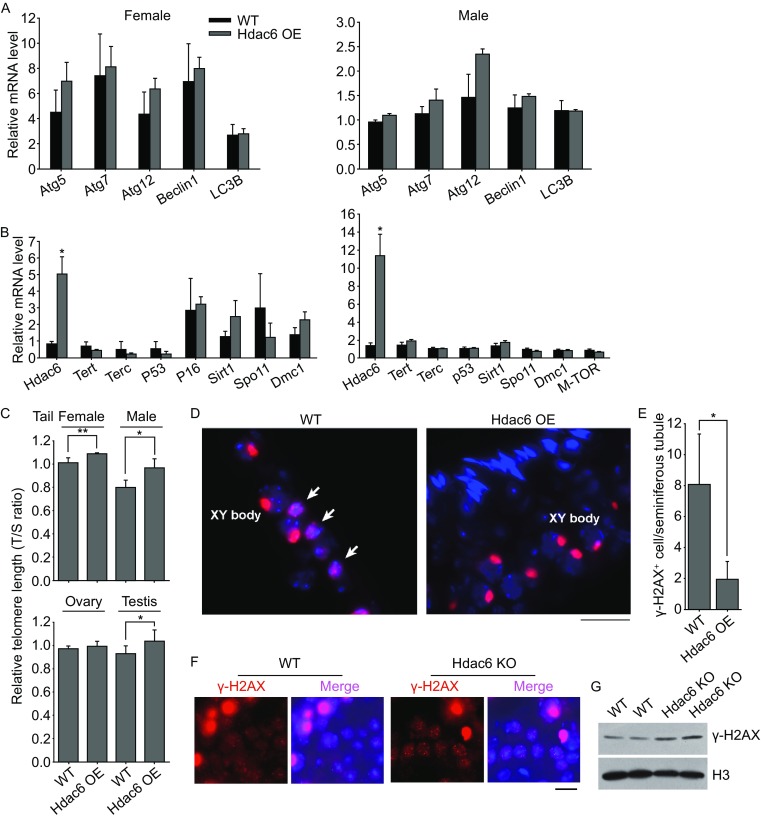
Influences of Hdac6 overexpression on telomere length maintenance and DNA damage response in vivo. (A) Relative mRNA levels by qPCR analysis of genes associated with autophagy of ovaries or testis of old WT and Hdac6 OE females and males in B6C3F1 hybrid background. (B) Relative mRNA levels of genes related to telomere, meiosis and aging in ovaries or testis of old mice. WT mice were 15–16 months old and Hdac6 OE mice 19 months old. (C) Relative telomere length shown as T/S ratio by qPCR analysis. For telomere analysis of tails, females were aged at 14–15 months, and males at 12–14 months (n = 4). For telomere analysis of gonads, female mice were 17–18 months old, and WT males at 15 months old, and Hdac6 OE males at 19 months old. The data are expressed as mean ± SD (n = 4). (D and E) DNA damage shown by γH2AX foci is reduced in the seminiferous of Hdac6 OE testis than in WT mice (19 month old). (D) Immunofluorescence of γH2AX indicated DNA damage with fragmented DNA (white arrows) and paired XY chromosomes at pachytene (indicated as XY body) in the seminiferous tubule. (E) Quantification of average number of γH2AX positive cells with fragmented DNA per seminiferous tubule of WT and Hdac6 OE testis sections. (F and G) DNA damage is increased in Hdac6 knockout (KO) mice compared with WT mice. (F) Immunofluorescence of γH2AX showing noticeable DNA damage foci in spermatocytes of Hdac6 KO testis but fewer foci in WT mice. Large spots in red are XY bodies, shown as in (D). (G) Western blot analysis of γH2AX protein levels. H3 served as a loading control. Scale bar = 20 μm. *P < 0.05, **P < 0.01
Furthermore, γH2AX positive cells with fragmented DNA foci, indicative of DNA damage response, were reduced in the testis of Hdac6 overexpression mice compared with WT mice (Fig. 2D and 2E). γH2AX staining can detect DNA damage as well as the XY sex body for pairing at pachytene in spermatocytes that form unique large spot but not foci (Vasileva et al., 2013). On the contrary, Hdac6 knockout mice exhibited increased DNA damage as evidenced by increased γH2AX foci in spermatocytes as well γH2AX protein levels in the testis, compared with WT mice at the same age (Fig. 2F and 2G).
Together, high levels of Hdac6 can prolong reproductive life span in both males and females using transgenic mouse model. Furthermore, telomere maintenance, reduced DNA damage or increased DNA repair and less apoptosis together may contribute to the extended reproductive age by overexpression of Hdac6. The impact of Hdac6 on DNA damage response and telomere regulation could be indirect and the underlying regulatory mechanisms still warrant further investigation. Excitingly, ES cells with hyper-long telomeres generate healthier chimera mice displaying longer telomeres, reduced cell senescence and DNA damage with age, and better skin wound healing (Varela et al., 2016), further supporting the notion that longer telomeres from ES cells can be passed to their offspring that can achieve healthy aging.
Concerns may exist regarding potential risks of Hdac6 overexpression in tumorigenesis. Inactivation or selective inhibition of Hdac6 by inhibitors can greatly decrease cancer cell growth and proliferation (Lee et al., 2008), and the inhibition causes DNA damage (Namdar et al., 2010), in line with our observation here that overexpression of Hdac6 reduces DNA damage, elongates telomeres, and increases cell proliferation. Presumably, longer telomeres and reduced DNA damage may reduce tumorigenesis in Hdac6 overexpression mice. Further studies are required to investigate whether Hdac6 overexpression mice may produce tumor at much older age or are more or less susceptible to tumor induction with age.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
FOOTNOTES
We thank Yushan Zhu and Shiyu Liu for providing materials and valuable discussion. This work was supported by the National Natural Science Foundation of China (Grant No. 31430052), the National Basic Research Program (973 Program) (2010CB94500), and PCSIRT (No. IRT13023).
X. Z., J. Y., H. W., R. G., H. W., D. Z. and Q. Z. performed experiments; X. Z., J. Y. and Y. Y. analyzed the data; J. Z. and L. C. provided the reagents or materials, and revised the manuscript. L. L. conceived and supervised the study, wrote and edited the manuscript.
Xiaoxi Zhang, Jiao Yang, Haiying Wang, Renpeng Guo, Yu Yin, Dongdong Zhang, Qian Zhang, Hua Wang, Zhongcheng Zhou, Lingyi Chen, Jun Zhou and Lin Liu declare that they have no conflict of interest. All institutional and national guidelines for the care and use of laboratory animals were followed.
Footnotes
Xiaoxi Zhang and Jiao Yang have contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s13238-017-0375-9) contains supplementary material, which is available to authorized users.
Contributor Information
Jun Zhou, Email: junzhou@nankai.edu.cn.
Lin Liu, Email: liulin@nankai.edu.cn.
References
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Lee YS, Lim KH, Guo X, Kawaguchi Y, Gao Y, Barrientos T, Ordentlich P, Wang XF, Counter CM, Yao TP. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res. 2008;68:7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Linking telomere regulation to stem cell pluripotency. Trends Genet. 2017;33:16–33. doi: 10.1016/j.tig.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Maynard S, Fang EF, Scheibye-Knudsen M, Croteau DL, Bohr VA. DNA damage, DNA repair, aging, and neurodegeneration. Cold Spring Harb Perspect Med. 2015 doi: 10.1101/cshperspect.a025130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namdar M, Perez G, Ngo L, Marks PA. Selective inhibition of histone deacetylase 6 (HDAC6) induces DNA damage and sensitizes transformed cells to anticancer agents. Proc Natl Acad Sci USA. 2010;107:20003–20008. doi: 10.1073/pnas.1013754107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol. 2001;21:8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus S, Li F, Stobezki R, Akula K, Unsal E, Jeong K, Robson M, Robson F, Moy F, Goswami S, et al. Impairment of BRCA1-related DNA double-strand break repair leads to ovarian aging in mice and humans. Sci Transl Med. 2013;5:172ra121. doi: 10.1126/scitranslmed.3004925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Varela E, Munoz-Lorente MA, Tejera AM, Ortega S, Blasco MA. Generation of mice with longer and better preserved telomeres in the absence of genetic manipulations. Nat Commun. 2016;7:11739. doi: 10.1038/ncomms11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A, Hopkins KM, Wang X, Weisbach MM, Friedman RA, Wolgemuth DJ, Lieberman HB. The DNA damage checkpoint protein RAD9A is essential for male meiosis in the mouse. J Cell Sci. 2013;126:3927–3938. doi: 10.1242/jcs.126763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Meng Q, Huo L, Yang M, Wang L, Chen X, Wang J, Li Z, Ye X, Liu N, et al. Overexpression of Hdac6 enhances resistance to virus infection in embryonic stem cells and in mice. Protein Cell. 2015;6:152–156. doi: 10.1007/s13238-014-0120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol Cell Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xiang S, Joo HY, Wang L, Williams KA, Liu W, Hu C, Tong D, Haakenson J, Wang C, et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol Cell. 2014;55:31–46. doi: 10.1016/j.molcel.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


