Abstract
Local transmission of chikungunya virus (CHIKV) was first detected in the Americas in December 2013, after which it spread rapidly throughout the Caribbean islands and American mainland, causing a major chikungunya fever epidemic. Previous phylogenetic analysis of CHIKV from a limited number of countries in the Americas suggests that an Asian genotype strain was responsible, except in Brazil where both Asian and East/Central/South African (ECSA) lineage strains were detected. In this study, we sequenced thirty-three complete CHIKV genomes from viruses isolated in 2014 from fourteen Caribbean islands, the Bahamas and two mainland countries in the Americas. Phylogenetic analyses confirmed that they all belonged to the Asian genotype and clustered together with other Caribbean and mainland sequences isolated during the American outbreak, forming an ‘Asian/American’ lineage defined by two amino acid substitutions, E2 V368A and 6K L20M, and divided into two well-supported clades. This lineage is estimated to be evolving at a mean rate of 5 × 10−4 substitutions per site per year (95% higher probability density, 2.9–7.9 × 10−4) and to have arisen from an ancestor introduced to the Caribbean (most likely from Oceania) in about March 2013, 9 months prior to the first report of CHIKV in the Americas. Estimation of evolutionary rates for individual gene regions and selection analyses indicate that (in contrast to the Indian Ocean Lineage that emerged from the ECSA genotype followed by adaptive evolution and with a significantly higher substitution rate) the evolutionary dynamics of the Asian/American lineage are very similar to the rest of the Asian genotype and natural selection does not appear to have played a major role in its emergence. However, several codon sites with evidence of positive selection were identified within the non-structural regions of Asian genotype sequences outside of the Asian/American lineage.
Keywords: Caribbean, chikungunya virus, complete genome, evolution, Americas, selection analysis
1. Introduction
Chikungunya virus (CHIKV; family Togaviridae, genus Alphavirus) is a mosquito-borne virus that causes chikungunya fever (CHIKF), an acute febrile illness characterised by severe, debilitating joint pain often associated with a rash, muscle pain, headache, nausea and fatigue. Patients often make a full recovery and infection is thought to result in life-long immunity. However chronic arthritis in individuals who have otherwise recovered is also common (Brighton et al. 1983; Gerardin et al. 2011), with an estimated 60 per cent experiencing recurrent and severe joint pain months and even years after infection (Schwartz and Albert 2010).
CHIKV is thought to exist as a single serotype and was first isolated from a febrile individual in 1952 in Tanganyika, part of what is now Tanzania (Ross 1956). Until the early 2000s it was primarily restricted to regions of Africa and Asia where it caused numerous sporadic outbreaks (Weaver 2014). Three major lineages have been defined, including the West African (WAf), East/Central/South African (ECSA) and Asian (Powers et al. 2000; Volk et al. 2010) based on their previous geographic distributions. In 2005–2006, a strain of the ECSA lineage [Indian Ocean Lineage (IOL)] was associated with a massive epidemic in Indian Ocean islands and the Indian subcontinent, the first after 32 years of quiescence (Arankalle et al. 2007; Volk et al. 2010). Small outbreaks initiated by travellers returning from this region also occurred in Italy in 2007 (Rezza et al. 2007) and France in 2010 (Grandadam et al. 2011).
It has been proposed that CHIKV emerged in the Western hemisphere as early as the 19th century but that cases were misdiagnosed, for example as dengue fever (Carey 1971). This is plausible given the similarities in signs and symptoms between CHIKF and dengue; however, there is no well-documented evidence of local transmission in the Americas prior to December 2013 when a CHIKF outbreak was reported in the French Caribbean island of Saint Martin (Leparc-Goffart et al. 2014). The immunologically naive human population in the Americas, widespread occurrence of Aedes aegypti, and frequent travel among islands facilitated rapid spread of the virus throughout the Caribbean and eventually to the American mainland. By 28 February 2014, autochthonous transmission had been confirmed in ten territories in the Americas with 2,582 confirmed CHIKF cases (Pan American Health Organization 2014b) and by 29 December 2014, thirty-three territories had confirmed transmission with 1,071,696 and 22,796 suspected and confirmed cases, respectively (Pan American Health Organization 2014a).
Some strains of the IOL are characterized by sequential adaptive mutations in the E1 and E2 envelope glycoproteins that increase the infectivity for Ae. albopictus (Tsetsarkin et al. 2007; Vazeille et al. 2007), providing an opportunity for CHIKV transmission in regions where this species predominates, including beyond the tropical and sub-tropical boundaries where Ae. aegypti thrives. Combined with the thousands of infected travellers documented between 2006 and 2008, it was expected that the IOL would have been the first to emerge in the Western hemisphere (Weaver 2014). However, sequence analysis confirmed that CHIKV sequences isolated from Saint Martin at the beginning of the outbreak in the Americas belonged to the Asian lineage (Leparc-Goffart et al. 2014). Subsequent sequencing of CHIKV from other Caribbean islands [e.g. British Virgin Islands (Lanciotti and Valadere 2014), Guadeloupe, Martinique (Stapleford et al. 2016) and Trinidad (Sahadeo et al. 2015)] and from the American mainland [i.e. Colombia (Mattar et al. 2015), Panama (Diaz et al. 2015), Mexico (Diaz-Quinonez et al. 2015) and Brazil (Nunes et al. 2015)] also confirmed circulation of the Asian lineage. However, in Brazil, concurrent with the Asian genotype CHIKV circulation, there was an outbreak in the east-central region of Feira de Santana that was caused by a newly introduced ECSA strain (Nunes et al. 2015). Since sequence data were only available for a small portion of the CHIKV-affected countries in the Americas, it was not clear whether the ECSA genotype was in fact more widespread, whether other genotypes were in circulation, or if (with the exception of Feira de Santana) the outbreak in the Americas was the result of a single introduction. Analysis of complete genomic sequences from the wider region would also facilitate more accurate estimation of evolutionary rates and selection pressures driving evolution.
To thoroughly address these questions, and to identify mutations and potential selection pressures on CHIKV circulation in the Americas that may have implications for virulence and transmission, we sequenced CHIKV isolates from seventeen countries (fourteen Caribbean islands, the Bahamas and two mainland countries) and performed phylogenetic analyses together with previously published sequences.
2. Methods
2.1 Virus Isolation and Illumina sequencing
Forty-six serum samples that were positive for CHIKV by real-time reverse transcription-polymerase chain reaction (cycle threshold median, 21; range, 14–31) were obtained from the existing collection of the Caribbean Public Health Agency. The samples originated from adult individuals presenting with acute onset of fever (>38.5 °C) and severe undiagnosed arthralgia/arthritis within 5 days of fever onset in fourteen Caribbean islands (Trinidad, Anguilla, Antigua, Dominica, Jamaica, St. Vincent, St. Lucia, Turks and Caicos, St. Kitts, Grenada, Montserrat, Haiti, Cayman Islands and Barbados), the Bahamas and two mainland countries (Suriname and Guyana) and were selected to maximize the temporal and spatial range of samples. Serum samples were passaged once in Vero cells and viral RNA subsequently extracted and subjected to Illumina sequencing using methods as described previously (Auguste et al. 2015; Sahadeo et al. 2015). Genbank accession numbers are shown in Supplementary Table S1.
2.2 Variant analysis
Reads were aligned to the reference sequences with bowtie2 (Langmead and Salzberg 2012), version 2.2.5 with the local setting. The samtools mpileup command was used to create an mpileup file (Samtools version 0.1.19). VarScan2 (Koboldt et al. 2012) (version 2.3.9) mpileup2snp was used to call variants with P value set at 0.1 and minimum variant frequency at 0.009.
2.3 Data sets
Sequences derived in this study (n = 33) were combined with 198 complete CHIKV genomic sequences including all available Caribbean sequences and representatives for all three genotypes available on GenBank and the European Virus Archive up to March 2016 (Supplementary Table S1). Sequences (n = 231) were aligned using the ClustalW alignment tool within Geneious version 7.1.9 (Biomatters Limited) and then trimmed to the boundaries of the open reading frames (ORFs) to eliminate ambiguous alignment of the 5' and 3' untranslated regions (UTR) (final length = 11,198 bp). This data set was designated CHIKV231. Two other data sets (both subsets of CHIKV231) were similarly prepared, i.e. (1) Asian149 that included all the Asian genotype sequences from the Caribbean, Central and South America as well as closely related Asian sequences (n=149) and (2) Asian50, a subset of Asian149 (n =50), that excluded sequences related to the American CHIKV epidemic (Supplementary Table S1). Nucleotide and amino acid position numberings used are based on NCBI reference sequences KJ451622 (Micronesia, 2013), AB860301 (Philippines, 2013) and KF318729 (China, 2012). All sequences were tagged with the date of sampling and location from which they originated.
2.4 Phylogenetic analyses
Sequences were confirmed as non-recombinant using SBP and GARD (Kosakovsky Pond et al. 2006) from the HyPhy online package. Data sets were tested for temporal evolutionary signal prior to molecular clock analysis using Path-O-Gen version 1.4 (Rambaut et al. 2016) to calculate regressions of root-to-tip genetic distance against sampling time. Phylogenetic trees were inferred using the maximum-likelihood method in the PAUP* program version 4.0b10 and the maximum clade credibility (MCC) from BEAST version 1.8.3 under the best fit model as determined by jModeltest 2 (Darriba et al. 2012), i.e. general time reversible substitution model with a discrete gamma distribution with four-rate categories (GTR + G4).
Using the Asian149 data set, substitution rates and times to the most recent common ancestor (TMRCA) for the complete phylogeny and sub-lineages were jointly estimated using BEAST, with the GTR + G4 model of nucleotide substitution, along with a relaxed lognormal molecular clock model and the Skygrid tree prior (Gill et al. 2013). Analyses were run in duplicate for ninety million generations with 10 per cent removed as burn-in, and convergence of parameters was assessed by calculating the effective sample size (ESS >200) using TRACER version 1.6. Substitution rates were estimated with and without partitioning the complete ORFs into codon positions [i.e. positions 1 and 2 combined (CP1 + 2) and position 3 only (CP3) with substitution rate and base frequencies unlinked] and for individual genes. Substitution rates were similarly estimated for the Asian50 data set.
For phylogeographic reconstruction, Asian149 data set sequences were tagged according to the geographic region from which they originated, i.e. Greater Antilles (Cayman Islands, Dominican Republic, Haiti, Jamaica and Puerto Rico), Windward Islands (Barbados, Dominica, Grenada, Martinique, St. Lucia, St. Vincent and Trinidad), Leeward Islands (Anguilla, Antigua, British Virgin Islands, Guadeloupe, Montserrat, Saint Barthélemy, St. Kitts and Saint Martin), Lucayan Archipelago (Bahamas and Turks & Caicos), Asia (China, Indonesia, Malaysia, Philippines and Thailand), Central America (Mexico and Panama), South America (Brazil, Colombia, Guyana and Suriname) and Oceania (American Samoa, French Polynesia, Micronesia and New Caledonia).
2.5 Selection analyses
Selection analyses were performed on the complete Asian149 data set and on a subset of ninety-nine sequences (i.e. ninety-eight American sequences and a 2015 sequence from Polynesia) belonging to an ‘Asian/American’ lineage defined by the aforementioned phylogenetic analyses. Data sets were trimmed to the boundaries of the non-structural (7,485 bp) and structural (3,744 bp) ORFs (excluding the non-coding region between the two), and each region investigated separately. Individual sites with evidence of positive or negative selection were identified using SLAC and FEL analyses from the online Datamonkey webserver (Pond and Frost 2005; Delport et al. 2010) (http://datamonkey.org). Also using the Datamonkey webserver, sites with evidence of pervasive diversifying selection and episodic diversifying selection were identified using FUBAR and MEME analyses respectively. A significance level of P < 0.05 was used for analyses except for FUBAR for which a posterior P >0.95 was used. The ratio of non-synonymous (dN) to synonymous (dS) substitutions per site for each region was estimated using SLAC.
2.6 Ethics statement
Serum samples used in this study were kindly provided de-identified by CARPHA from excess samples submitted from member states for diagnostic testing. None of the samples were collected specifically for this study. Study protocols were approved by the Ethics Committees of CARPHA and the University of the West Indies, St. Augustine.
3. Results
3.1 Virus isolation and nucleotide sequencing
CHIKV was isolated on Vero cells (passage 1) from thirty-four of the forty-six serum samples screened and whole-genome sequences were recovered from thirty-three of these (Supplementary Table S1). The genomic sequences recovered ranged from 11,970 to 12,053 nucleotides in length, and sequence identities across the ORFs (11,238 nucleotides) ranged from 99.9 to 100 per cent at both the nucleotide and amino acid levels. Sequence read depths suggested the presence of a 177-bp duplication within the 3'UTR of all thirty-three sequences as described previously (Stapleford et al. 2016).
3.2 Phylogenetic analysis of American outbreak CHIKV complete ORF sequences
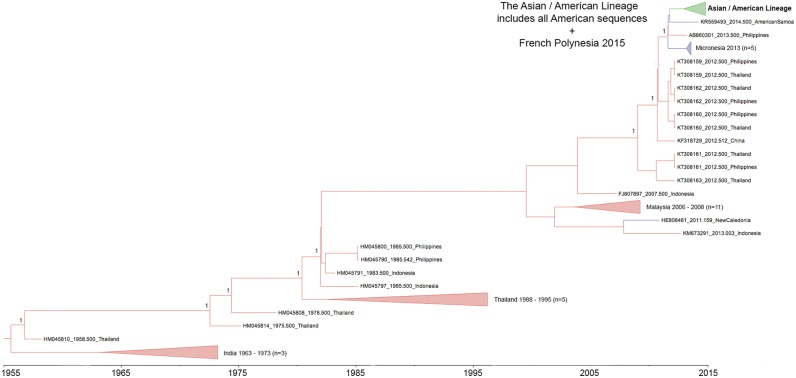
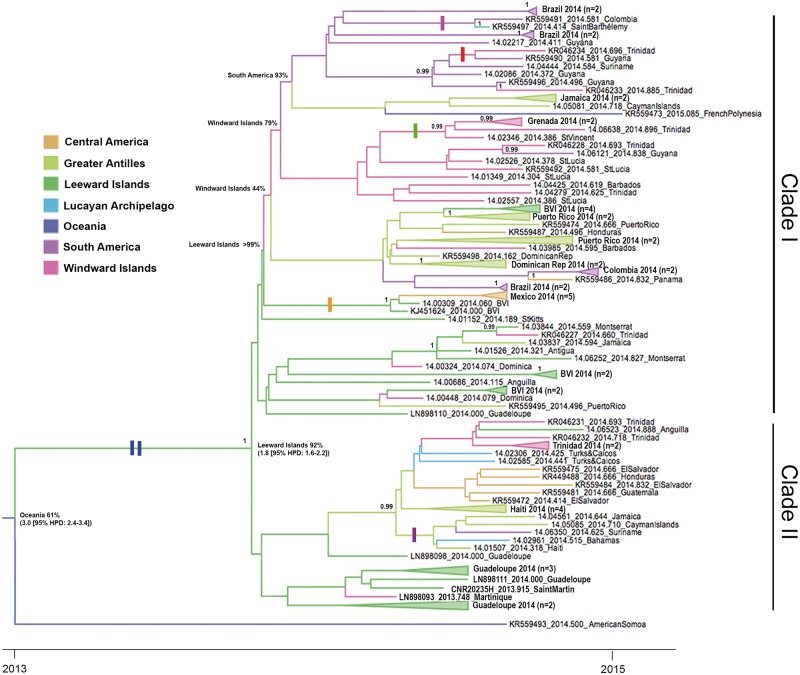
Phylogenetic analysis of 231 CHIKV complete ORF sequences representing the three CHIKV lineages confirmed that the thirty-three complete genomic sequences derived in this study all belonged to the Asian lineage (Supplementary Fig. S1) and clustered together in a well-supported clade (clade credibility ≥0.99; Fig. 1) that includes previously published sequences from the Americas, a 2014 sequence from American Samoa, a 2013 sequence from the Philippines, a group of five 2013 sequences from Micronesia and a single 2015 sequence from French Polynesia. The latter fell within the sub-clade containing all of the American sequences designated the Asian/American lineage (Fig. 1). The support for the node separating the Asian/American lineage from the 2014 American Samoan sequence was weak (clade credibility = 20%), but the Asian/American lineage (shown in detail in Fig. 2) was distinguished from this and all other Asian lineage sequences by two non-synonymous nucleotide changes (T9053C and C9870), which result in a V → A substitution at amino acid position 368 of E2 and an L → M substitution at amino acid position 20 of 6K, respectively. Both of these changes (indicated by blue bars in Fig. 2) were conservative, with a small, non-polar, aliphatic residue replaced by a small, non-polar residue in one case, and a sulphur containing reside replacing a non-polar aliphatic residue in the other. In addition to the latter, there were twenty-four other non-synonymous nucleotide substitutions distributed across both the non-structural and structural ORFs of one or more Asian/American lineage sequences that were not found in the other closely related Asian sequences (Table 1), five of these, i.e. nsP4 R99Q, nsP4 H179Y, E2 V113A, E1 T101M and nsP3 Stop521R (indicated by coloured bars in Fig. 2) defined clades within the Asian/American lineage. Of these, three were non-conservative amino acid changes, i.e. nsP4 R99Q [polar, charged (+ve), basic → polar, uncharged, acidic], nsP4 H179Y [aromatic, polar, charged (+ve), basic → aromatic, polar, uncharged] and nsP3 Stop521R [stop → polar, charged (+ve), basic].
Figure 1.
MCC phylogeny based on the complete ORFs of 149 Asian lineage sequences (Asian149), including all Caribbean sequences. Taxon labels include year of isolation, accession number/sequence ID and country of isolation, except in collapsed clades, which are comprised of two or more sequences from the same country and year. The Asian/American lineage comprises all Caribbean sequences, including the thirty-three newly generated sequences, as well as Asian sequences from Central and South America and a 2015 sequence from French Polynesia (Genbank Accession No. KR559473). The most closely related sequences to the Asian/American lineage were sequences from American Samoa (2014), the Philippines (2013) and Micronesia (2013). Posterior probabilities ≥ 99 per cent are indicated at relevant nodes.
Figure 2.
The Asian/American lineage and its most closely related sequence from American Samoa (2014). Taxon labels include year of isolation, accession number/sequence ID and country of isolation except in collapsed clades, which are comprised of two or more sequences from the same country and year. The number of sequences in each collapsed clade is shown in parentheses, (the accession numbers for sequences in these collapsed clades are shown in Supplementary Figure S2). Amino acid changes defining clades within the phylogeny are indicated by coloured bars on the relevant branches (blue = 6K L20M and E2 V368A; red= nsP4 R99Q; green = nsP4 H179Y; orange = E2 V113A, purple = nsP3 Stop521R; pink = E1 T101M). Terminal branches of the tree are colored according to the region to which the country from which the sequence at the tip was derived, i.e. Central America (Mexico); Greater Antilles (Cayman Islands, Dominican Republic, Haiti, Jamaica, Puerto Rico); Leeward Islands (Anguilla, Antigua, British Virgin Islands, Guadeloupe, Montserrat, Saint Barthélemy, St. Kitts and Saint Martin); Lucayan Archipelago (Bahamas and Turks & Caicos); Oceania (American Samoa, French Polynesia, Micronesia and New Caledonia); South America (Brazil, Colombia, Guyana and Suriname); Windward Islands (Barbados, Dominica, Grenada, Martinique, St. Lucia, St. Vincent and Trinidad). Internal branches are colored according to the most probable (model) location of their parental nodes and the location state probability is indicated as a percentage at selected nodes. Also at two selected nodes, in square brackets, are node ages. and corresponding 95% HPDs. Nodes with posterior probabilities (clade credibilities) ≥0.99 are labelled.
Table 1.
Non-synonymous amino acid changes that distinguish CHIKV sequences from the Americas from other closely related Asian genotype sequences (American Samoa 2014, Philippines 2013, Micronesia 2013).
| Gene | Nt change | AA change | Sequence ID | Country (year) |
|---|---|---|---|---|
| Capsid | T7735 C | 56 V → A | 14.03985 | Barbados (2014) |
| C8158T | 197 S → L | 14.01349 | St. Lucia (2014) | |
| E2 | C8824T | 94 T → I | KR046233.1 | Trinidad (2014) |
| T8881C | 113 V → A | 14.00309 | BVI (2014) | |
| KJ451624.1 | BVI (2014) | |||
| CH0008 | Mexico (2014) | |||
| CH0045 | Mexico (2014) | |||
| CH0072 | Mexico (2014) | |||
| LI0036 | Mexico (2014) | |||
| TA0006 | Mexico (2014) | |||
| T9646C | 368 V → A | All | All | |
| 6K | C9870A | 20 L → M | All | All |
| T9909A | 33 C → S | 14.03985 | Barbados (2014) | |
| E1 | C10219T | 101 T → M | KR559491 | Colombia (2014) |
| G10452A | 153 A → T | KR559497 | St. Barts 2014) | |
| 14.04425 | Barbados (2014) | |||
| C10498T | 168 S → L | 14.06638 | Trinidad (2014) | |
| T10867C | 291 V → A | 14.01152 | St. Kitts (2014) | |
| A10890T | 299 M → L | 14.03985 | Barbados (2014) | |
| nsP1 | G590A | 171 R → Q | 14.02961 | Bahamas (2014) |
| 14.02086 | Guyana (2014) | |||
| G603A | 176 V → I | 14.0444 | Suriname (2014) | |
| G765A | 230 G → R | 14.0444 | Suriname (2014) | |
| T775A | 233 L → Q | KR046230.1 | Trinidad (2014) | |
| nsP2 | C3389T | 569 R → C | KR046232.1 | Trinidad (2014) |
| C2416T | 245 P → L | 14.0444 | Suriname (2014) | |
| G2481T | 267 A → S | 14.02086 | Guyana (2014) | |
| C1932A | H644Q | KP164571 | Brazil (2014) | |
| nsP3 | G4430A | 118 G → R | 14.01526 | Antigua (2014) |
| T5646C | 521 stop → R | 14.05081 | Cayman Is (2014) | |
| 14.02961 | Bahamas (2014) | |||
| 14.05085 | Cayman Is (2014) | |||
| 14.01507 | Haiti (2014) | |||
| 14.04561 | Jamaica (2014) | |||
| 14.06350 | Suriname (2014) | |||
| CNR20235H | Saint Martin (2013) | |||
| nsP4 | G5962A | 99 R → Q | 14.0444 | Suriname (2014) |
| KR046233.1 | Trinidad (2014) | |||
| 14.02086 | Guyana (2014) | |||
| KR559490.1 | Guyana (2014) | |||
| KR559496.1 | Guyana (2014) | |||
| C6201T | 179 H → Y | 14.06638 | Trinidad (2014) | |
| 14.02346 | St. Vincent (2014) | |||
| 14.03562 | Grenada (2014) | |||
| 14.0256 | Grenada (2014) | |||
| G6762A | 366 A → T | 14.05085 | Cayman Is (2014) | |
| C7276T | 537 A → V | 14.02961 | Bahamas (2014) |
Nucleotide positions are numbered beginning at the start codon for the complete coding sequence; amino acid positions are numbered from the first amino acid of each gene product.
As shown in Fig. 2, the Asian/American lineage was comprised of two major clades: designated I and II (clade credibility ≥0.99). Four of the aforementioned clade-defining, non-synonymous substitutions were in clade I. The fifth, an opal stop codon replaced by an arginine codon at position 521 within nsP3, defined a 2014 lineage within clade II [Haiti (14.01507), Jamaica (14.04561), Suriname (14.06350), Cayman Islands (14.05081, 14.05085), Bahamas (14.02961)] and is also present in the St. Martin sequence in clade II that was isolated at the beginning of the outbreak (CNR20235H). Alphavirus genomes are known to vary between sense and stop codons at this position (Rupp et al. 2015).
The Asian/American lineage was estimated to be evolving at a rate of 5.0 × 10−4 substitutions per site per year [95% higher probability density (HPD): 2.9–7.9 × 10−4 subs/site/yr] (Table 2) with the mean estimate for when the MRCA existed being March 2013 [i.e. mean estimate for time to MRCA (tMRCA) = 1.8 yrs (95% HPD: 1.6–2.2 yrs)]. The MRCA for the Asian/American lineage and the 2014 sequence from American Samoa is estimated to have existed around February 2012 [tMRCA = 3 yrs (95% HPD 2.4–3.4 yrs)] most likely in Oceania, although the location state probability (61%) was relatively low (Fig. 2). The phylogeographic reconstruction also suggested that the introduction from Oceania to the Western Hemisphere was most likely via the Leeward Islands (location state probability for MRCA of Asian/American lineage = 92%). The inferred location states along the backbone of the phylogeny suggested that after initial spread of the virus from the Leeward Islands, the Windward Islands and later the South American mainland served as the major source population for the region.
Table 2.
Substitution rates estimated for the complete coding sequence and for individual genes of CHIKV Asian lineages.
| Substitution rate (95% HPD)/ × 10−4 subs/site/yr |
|||
|---|---|---|---|
| Data set | Asian149 |
Asian50 | |
| Region | Asian genotype | Asian/ American lineage | Asian lineages paraphyletic to Asian/ American lineage |
| Complete cds | 4.6 (4.1–5.2) | 5.0 (2.9–7.9) | 4.4 (3.8–5.0) |
| CP1 + 2 of complete cds | 0.6 (0.6–0.7) | 0.8 (0.6–0.9) | 0.6 (0.5–0.6) |
| CP3 of complete cds | 5.5 (5.2–5.7) | 4.9 (4.4–5.5) | 5.8 (5.5–6.0) |
| C | 4.2 (3.0–6.0) | 5.0 (2.0–9.0) | 3.7 (2.3–5.3) |
| 6k | 10 .0 (6.0–20.0) | 11.0 (3.0–20.0) | 2.6 (0.3–8.1) |
| E1 | 6.0 (4.0–8.0) | 8.0 (1.0–19.0) | 5.3 (3.5–7.2) |
| E2 | 7.0 (5.0–8.0) | 9.0 (3.0–13.0) | 5.7 (4.2–7.3) |
| E3 | 6.0 (3.0–9.9) | −a | 6.6 (2.3–12.2) |
| nsP1 | 4.0 (3.0– 5.0) | 5.0 (2.0–8.0) | 3.7 (2.6–4.8) |
| nsP2 | 4.0 (3.0– 5.0) | 4.0 (3.0–6.0) | 4.2 (3.2–5.0) |
| nsP3 | 7.0 (6.0–9.0) | −a | 6.0 (4.5–7.5) |
| nsP4 | 4.0 (3.0– 5.0) | −a | 4.5 (2.5 – 17.1) |
aThe Asian/American lineage was not apparent as a distinct clade in phylogenies based on these gene regions.
As shown in Table 2, substitution rates estimated for the Asian/American lineage (complete coding sequence and at the level of individual genes) were not significantly different from those estimated for the paraphyletic set of fifty Asian lineage strains. As expected, the substitution rate at the third codon position (CP3) was higher than at the first and second codon position partition (CP1 + 2), but there were no significant differences between CP1 + 2 or CP3 substitution rates for the Asian/American lineage and the paraphyletic Asian lineages.
3.3 Variant analysis
For thirty of the thirty-three virus isolates made in this study sequencing was of sufficient depth to allow reliable reporting of variation at individual nucleotide positions within both the non-structural and structural polypeptide genes. A total of 101 individual variants were observed, with seventy-nine (78.2%) in the non-structural ORF and twenty-two (21.8%) in the structural ORF. The sequences and individual variants observed within each strain are listed in Supplementary Table S2. Within the ORFs, the frequency of individual variants observed within individual isolates ranged from 0.95 to 13.78 per cent with the following two exceptions: (1) A → C at nucleotide position 8,385 found in 33.1 per cent of reads from Haitian isolate 14.01507 and (2) a T → C at nucleotide position 8,091 found in 45.57 per cent of the reads from isolate 14.06252 from Montserrat. The most commonly observed variant within the structural ORF was a G → A transition at nucleotide position 9,365, occurring in nine of the thirty virus isolates at low frequencies (1.35–2.21%). This change resulted in a non-synonymous substitution (G279Q) of the E2 protein. Within the non-structural ORF, however, the two most commonly observed variants occurred at nucleotide positions 4,424 and 5,633, in sixteen and eighteen sequences, respectively. At position 4,424, a G → A transition was observed resulting in a non-synonymous substitution (G117R) within the nsP3 protein. At nucleotide position 5,633, a T → C transition was observed, which resulted in the replacement of an opal stop codon with an arginine residue at position 521 in the nsP3 peptide. This variant corresponded to the amino acid substitution observed in seven Caribbean sequences (see Table 1).
3.4 Selection analyses
Ratios of non-synonymous (dN) to synonymous (dS) substitutions per site for the structural and non-structural regions of the complete Asian149 data set were 0.17 (95% CI 0.14–0.21) and 0.17 (95% CI 0.14–0.19), respectively. There were no significant differences between the ratios for the Asian/American lineage (structural = 0.20 [95% CI, 0.13–0.29]; non-structural = 0.22 [95% CI, 0.16–0.30]) and the paraphyletic Asian lineages (structural = 0.13 [95% CI = 0.11–0.16]; non-structural = 0.14 (95% CI = 0.11–0.18) and they were all <1, indicating similar evolutionary dynamics and that purifying selection was the predominant evolutionary force. However, tests for individual sites with evidence of negative and positive selection (SLAC and FEL), and specifically for episodic diversifying selection (MEME) and pervasive diversifying selection (FUBAR) did provide evidence of positive selection at specific sites (Table 3). Negatively selected sites were also identified by the SLAC and FEL methods and are listed in Supplementary Table S3.
Table 3.
Positively selected sites within CHIKV Asian lineages.
| Method | Data set | No. of sites | Codon position | P value | AA position | Mutation | Sequences with derived amino acid state |
|---|---|---|---|---|---|---|---|
| Structural region | |||||||
| SLAC | Asian149 | 0 | – | – | – | – | – |
| Asian/American | 0 | – | – | – | – | – | |
| FEL | Asian149 | 0 | – | – | – | – | – |
| Asian/American | 0 | – | – | – | – | – | |
| MEME | Asian149 | 1 | 546 | 0.01 | 221 E2 | K → G | Thailand 1958 (HM045810) |
| Asian/American | – | – | – | ||||
| FUBAR | Asian149 | 2 | 323 | 116.04 | 62 E3 | Q → R | Malaysia 2006 (FN295483-4) |
| 546 | 59.83 | 221 E2 | K → G | Malaysia 2006 (EU703759-63) | |||
| Malaysia 2009 (KX168429) | |||||||
| Thailand 1958 (HM045810) | |||||||
| Asian/American | 0 | – | – | – | – | ||
| Non-structural region | |||||||
| SLAC | Asian149 | 1 | 1,728 | 0.02 | 395 nsP3 | V → P | Malaysia 2006 (FN295483, FN295484) |
| V → S | New Caledonia 2011 (HE806461) | ||||||
| Malaysia 2006 (EU703759-62) | |||||||
| Malaysia 2007 (KM923917-20) | |||||||
| Philippines 1985 (HM045790, HM045800) | |||||||
| Indonesia 1983 (HM045791) | |||||||
| Indonesia 1985 (HM045797) | |||||||
| India 1963 (HN045803) | |||||||
| India 1963 (HM045813) | |||||||
| India 1973 (HM045788) | |||||||
| Asian/American | 0 | – | – | – | – | – | |
| FEL | Asian149 | 0 | – | – | – | – | – |
| Asian/American | 0 | – | – | – | – | – | |
| MEME | Asian149 | 2 | 1,727 | 0.00 | 394 nsP3 | P → M | Malaysia 2007 (KM923917-20) |
| 1,728 | 0.00 | 395 nsP3 | V → P | Malaysia 2006 (EU70359-62) | |||
| V → S | Thailand 1995 (HM045787, HM045796) | ||||||
| Thailand 1996 (KX262987) | |||||||
| Philippines 1985 (HM045790, HM045800) | |||||||
| Indonesia 1983 (HM045791) | |||||||
| Indonesia 1985 (HM)45797) | |||||||
| Thailand 1978 (HM045808) | |||||||
| Thailand 1975 (HM045814) | |||||||
| Thailand 1958 (HM045810) | |||||||
| India 1963 (HM045803, HM045813) | |||||||
| India 1973 (HM045788) | |||||||
| Malaysia 2006 (FN295483, FN295484) | |||||||
| New Caledonia 2011 (HE806461) | |||||||
| Malaysia 2006 (EU703759-62) | |||||||
| Malaysia 2007 (KM923917-20) | |||||||
| Philippines 1985 (HM045790, HM045800) | |||||||
| Indonesia 1983 (HM045791) | |||||||
| Indonesia 1985 (HM045797) | |||||||
| India 1963 (HN045803) | |||||||
| India 1963 (HM045813) | |||||||
| India 1973 (HM045788) | |||||||
| Asian/American | 1 | 1,510 | 0.01 | 177 nsP3 | R → Q | Guadeloupe (2014) LN898098 | |
| FUBAR | Asian149 | 1 | 1728 | 153.20 | 395 nsP3 | V → P | Malaysia 2006 (FN295483, FN295484) |
| V → S | New Caledonia 2011 (HE806461) | ||||||
| Malaysia 2006 (EU703759-62) | |||||||
| Malaysia 2007 (KM923917-20) | |||||||
| Philippines 1985 (HM045790, HM045800) | |||||||
| Indonesia 1983 (HM045791) | |||||||
| Indonesia 1985 (HM045797) | |||||||
| India 1963 (HN045803) | |||||||
| India 1963 (HM045813) | |||||||
| India 1973 (HM045788) | |||||||
| Asian/American | 0 | – | – | – | – | – | |
For the Asian/American lineage, the only site with evidence of positive selection was codon 1,510 corresponding to amino acid position 177 of nsP3, where there was an R → Q amino acid change in the 2014 Guadeloupe sequence (LN898098). When all Asian lineage sequences were considered, one positively selected site within the structural region corresponding to amino acid position 221 within the E2 protein was identified by both the MEME and FUBAR algorithms. At this position all sequences possessed a lysine residue except for a 1958 sequence from Thailand (HM045810), which possessed a glycine residue. Within the structural region, FUBAR also identified a Q → R substitution at position 62 of the E3 protein (present in three Malaysian sequences from 2006 and 2009) as having been positively selected. Analysis of the Asian genotype non-structural genes identified two sites with evidence of positive selection, i.e. amino acid positions 39 (P → M) and 395 (V → P/S) within the nsP3 protein. The former was identified by the MEME method and the latter by SLAC, MEME and FUBAR.
4. Discussion
At the time of writing, complete CHIKV genomic sequences from the 2013/2014 American epidemic were available for twenty countries in the Western Hemisphere including ten Caribbean islands (British Virgin Islands, Martinique, Haiti, Dominican Republic, Puerto Rico, Trinidad, Saint Martin, Saint Lucia, Saint Barthélemy and Guadeloupe) (Lanciotti and Valadere 2014; Sahadeo et al. 2015; Stapleford et al. 2016). The thirty-three complete genomic CHIKV sequences generated in this study increase the number of countries sampled to twenty-nine (nineteen Caribbean islands) facilitating more robust analyses and reliable conclusions about the initial evolution and spread of CHIKV in the Americas.
Phylogenetic analyses and phylogeographic reconstruction indicated that the sampled lineages all belong to a monophyletic Asian/American lineage within the Asian lineage, which arose from an MRCA estimated to have existed around March 2013 (95% HPD December 2012–May 2013) in the Leeward Islands. This suggests that CHIKV was present in the Caribbean 7 to 12 months before being first detected in December of 2013 in St. Martin (one of the Leeward Islands). Nunes et al. (2015) estimated previously the MRCA for the Asian/American lineage to have existed as early as 23 August 2012, but this was based on a data set with far fewer Caribbean sequences and the 95 per cent Bayesian confidence intervals on their estimate (i.e. 3 October 2011 to 2 June 2013) were considerably wider. Our data provide no evidence for circulation of lineages other than the Asian in the countries represented (with the exception of Brazil, where an ECSA lineage strain has also been detected). The ECSA strain is estimated to have been introduced to Feira de Santana in Brazil around June 2014, six months after CHIKV was first reported in the Caribbean islands, and almost a year after our estimated date of existence for the MRCA of the Asian/American lineage. It is therefore likely that herd immunity was already established in the Caribbean, restricting the spread of the ECSA strain to the islands.
Although there is overwhelming support for the Leeward Islands as the location of origin for the MRCA of the Asian/American lineage, our phylogeographic inference cannot rule out the possibility that the MRCA existed in a location that is not represented in our data set, or in a location such as Oceania that is under-represented. CHIKV was active in this region between 2011 and 2013 (Roth et al. 2014) and the most closely-related sequence to the Asian/American lineage is a 2014 sequence from American Samoa, separated by a node with posterior support of only 20 per cent. The Asian/American lineage is estimated to have diverged from the American Samoan lineage in early 2012. The location state probability for this divergence favors Oceania (61%), but it is also possible that the American Samoan sequence is the descendant of a precursor that existed in Asia (location state probability 35%).
The Zika virus (ZIKV) epidemic in the Americas (Hennessey et al. 2016) that followed soon after the CHIKV epidemic waned in the Caribbean is also thought to have originated in Oceania (Lanciotti et al. 2008; Duffy et al. 2009; Lanciotti et al. 2016). In contrast to CHIKV, the evidence suggest that ZIKV was introduced first to the mainland (possibly northeastern Brazil) (Faria et al. 2016) but its rate of spread throughout the Americas was similarly rapid (Fischer et al. 2014). Previous authors have noted the similarities between the phylogeny and global movement of ZIKV and CHIKV (from East Africa to Asia to Pacific then New World) and suggested that similar ecologic and/or human social factors might be responsible for the nearly simultaneous spread of CHIKV and ZIKV to the New World (Musso et al. 2015).
While there is evidence of geographic structure within the CHIKV Asian/American lineage phylogeny, the presence of several mixed clades indicates considerable gene flow among countries and regions within the Americas. The overall pattern and rate of CHIKV spread described by our tree [i.e. CHIKV rapidly spreading throughout the Leeward Islands, then onwards to the wider Caribbean (Windward Islands and the Greater Antilles), and then to the American mainland] mirrors data from epidemiological reports based on virus isolation, and it is possible that our inclusion of sequences associated with such reports could account for the agreement between the phylogenetic and epidemiological data. However, to avoid this bias as far as possible, our study included sequences representing both the beginning and end of outbreaks in each of the Caribbean countries.
Previously published estimates of CHIKV rates of evolution vary among the three main lineages (Supplementary Table S4). We estimated the Asian/American lineage to be evolving at a rate of 5.0 × 10−4 subs/site/yr (95% HPD: 2.9 to 7.9 × 10−4 subs/site/yr) which is not significantly different from our current and previous estimates for the Asian lineage (Volk et al. 2010; Nunes et al. 2015). This is in contrast to the IOL, which has a substitution rate at least three times that estimated for the ancestral, enzootic ECSA lineage from which it evolved. However, the substitution rates for the older ECSA and WAf lineages are also significantly lower than for the Asian lineage, and the estimated rate for the IOL (while slightly higher than the Asian lineage) is not significantly different from the rate estimated for the Asian/American lineage.
Twenty-four non-synonymous substitutions were observed within consensus sequences for the Asian/American lineage ORFs, but none corresponded to the adaptive mutations observed in the IOL (Schuffenecker et al. 2006; Tsetsarkin et al. 2014). Furthermore, the amino acid changes that defined the Asian/American lineage i.e. V → A at position 368 in the E2 protein and L → M at position 20 in the 6K peptide (previously reported by Sahadeo et al. 2015) were both conservative, suggesting that adaptive evolution (at least within the ORFs) was not responsible for the emergence of the Asian/American lineage. The latter was supported by our selection analyses, which provide no evidence that these two mutations were under positive selection. Also none of the sites we identified as being positively selected corresponded to the adaptive mutations in E1 and E2 that gave rise to the IOL. Furthermore, with the exception of a non-conservative R → Q change found at amino acid position 177 in nsP3 of a single 2014 sequence from Guadeloupe, none of the mutations that defined the clades within the Asian/American lineage showed evidence of positive selection.
Of those sites with evidence of positive selection, the K → G change at amino acid position 221 within the E2 protein is of interest because Tsetsarkin et al. (2014) noted that all second-step Ae. albopictus-adaptive mutations in the E2 protein share a spatial arrangement along the acid sensitive axis formed by E2 residues 210–252, and hypothesized that substitution of additional amino acids along this axis may further increase fitness for Ae. albopictus infection. This mutation, however, is restricted to only one sequence, i.e. Thailand 1958 (HM045810).
Of the four substitutions that define clades within the Asian/American lineage, three could reasonably be expected to impact the properties of the peptides they encode as they involve changes in charge and/or acid-base properties in two cases (both in nsP4) and replacement of an opal stop codon with an arginine residue in nsP3 of the other. These three non-conservative lineage-defining substitutions occurred late in the evolution of the Asian/American lineage. It would be useful to compare the in vitro replication characteristics of strains with and without these lineage-defining substitutions and to track their progress in subsequent outbreaks to assess whether they represent mutations that are moving towards fixation.
The aforementioned replacement of the opal stop codon by arginine was noted in a single strain within clade II and in the St. Martin sequence isolated at the beginning of the American outbreak, which not only belongs to a different sub-lineage but also falls into clade 2. A corresponding stop codon, located seven amino acids before the C-terminal of nsP3, was reported in CHIKV sequences isolated during the 2005–2006 Indian Ocean outbreak by Schuffenecker (Schuffenecker et al. 2006), and differentiates sequences isolated from the Indian Ocean outbreak from the S27 strain, which has an arginine codon (Schuffenecker et al. 2006). The presence of this opal stop codon before the C-terminal of nsP3 has been shown to enhance in vitro replication of CHIKV within the ECSA lineage (Chen et al. 2013). Whether the effect would be the same on isolates within the Asian/American lineage is unclear but this mutation has also been observed in other alphaviruses (Rupp et al. 2015) and there is evidence to suggest that its presence increases nsP3 stability and enhances infectivity, relative to viruses encoding an arginine at the corresponding position (Myles et al. 2006). However, for both eastern equine encephalitis and o’nyong-nyong viruses, replacement of the opal stop codon by an arginine codon has been demonstrated, under experimental conditions, after multiple passages in mosquito cell cultures (Levinson et al. 1990; Lanciotti et al. 1998; Weaver et al. 1999). Of the sequences in the current study with this mutation, one was derived directly from serum and is therefore not a consequence of in-vitro adaptation to mosquito cell culture. The others were passaged only once [and in mammalian (Vero) as opposed to mosquito cells] but we cannot rule out the possibility that the mutation was acquired in vitro.
The nucleotide substitutions described above are based on comparison of consensus sequences derived from millions of reads from individual CHIKV isolates derived from human serum after one passage in Vero cells. Analysis of reads from the coding region of individual isolates revealed 101 individual synonymous and non-synonymous nucleotide changes, 78.2 per cent (n = 79) of which were in non-structural genes. The majority of these SNPs occurred at low frequencies (<14%) within any given isolate, and occurred in reads from less than one third of the virus isolates. The most commonly observed variant within the structural region was E2 G279Q which occurred at low frequency in about one third of the isolates. The latter resembles the replacement of other E2 residues by glutamine, which has been shown to enhance infection of Ae. albopictus by IOL CHIKV strains (Tsetsarkin et al. 2014). This substitution therefore deserves further study in light of the potential implications for vector transmission. In the non-structural region, the most common variant was a T → C change at nucleotide position 5,633 that was observed in eighteen sequences, at frequencies ranging from 1.5 to 37.8 per cent. This variant would lead to the replacement of the aforementioned opal stop codon in nsP3 with an arginine residue. Four of these eighteen sequences also contained an A → T transversion at nucleotide position 5,635 leading to a replacement of the opal stop codon with a cysteine residue. The frequencies of this variant ranged from 2.9 to 35.5 per cent.
The sequences derived in this study include both 3' and 5' UTRs and although Sanger sequencing would be required to confirm, read depths suggest that the 177 nucleotide duplication 3'UTR reported by Stapleford et al. (2016) in CHIKV sequences isolated from Mexico, the Dominican Republic and Trinidad during the 2013/2014 epidemic is present in all of our new sequences (data not shown). Stapleford et al. (2016) also showed that this duplication led to increased viral titers in mosquito cells compared to similar strains lacking the duplication. The presence of the duplication in all thirty-three newly generated sequences supports the suggestion that it is fixed in the circulating Caribbean (Asian/American) CHIKV strains.
In summary, our data suggest that the 2013/2014 outbreak in the Caribbean, which spread to the American mainland, arose from a single introduction to the region between mid-2012 and late 2013 (most likely from Oceania to the Leeward Islands). Our data provide no evidence for the circulation of the ECSA or other lineages outside of Brazil in 2014. Our data also suggest that, in contrast to the relationship between the IOL and the ECSA, the evolutionary dynamics of the Asian/American lineage are very similar to the Asian lineage from which it arose and its emergence is not likely to have been a consequence of adaptive evolution. There were, however, several sites within the non-structural regions of non-Asian/American lineage sequences that appear to have been subject to positive selection. Also, analysis of reads from the coding region of individual isolates identified a common E2 substitution similar to mutations that enhance infection of Ae. albopictus by IOL CHIKV strains (Tsetsarkin et al. 2014). Further investigation to determine if this mutation affects vector transmission is warranted.
Supplementary data
Supplementary data are available at Virus Evolution online.
Funding
This study was supported by the UWI-Trinidad and Tobago Research and Development Impact Fund [to C.V.F.C], National Institutes of Health [AI120942 and AI121452 to S.C.W.]. A.J.A. was supported by the James W. McLaughlin endowment fund.
Data availability
All sequences derived in this study are available on Genbank accession numbers KY435454 - KY435486.
Conflict of interest: None declared.
Supplementary Material
References
- Arankalle V. A. et al. (2007) ‘Genetic divergence of Chikungunya viruses in India (1963-2006) with special reference to the 2005-2006 explosive epidemic’, Journal of General Virology, 88/Pt 7: 1967–76. [DOI] [PubMed] [Google Scholar]
- Auguste A. J. et al. (2015) ‘A newly isolated reovirus has the simplest genomic and structural organization of any reovirus’, Journal of Virology, 89/1: 676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighton S. W., Prozesky O. W., de la Harpe A. L. (1983) ‘Chikungunya virus infection. A retrospective study of 107 cases’, South African Medical Journal, 63/9: 313–5. [PubMed] [Google Scholar]
- Carey D. E. (1971) ‘Chikungunya and dengue: a case of mistaken identity?’, Journal of the History of Medicine and Allied Sciences, 26/3: 243–62. [DOI] [PubMed] [Google Scholar]
- Chen K. C. et al. (2013) ‘Comparative analysis of the genome sequences and replication profiles of chikungunya virus isolates within the East, Central and South African (ECSA) lineage’, Virology Journal, 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D. et al. (2012) ‘jModelTest 2: more models, new heuristics and parallel computing’, Nature Methods, 9/8: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W. et al. (2010) ‘Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology’, Bioinformatics, 26/19: 2455–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Y. et al. (2015) ‘Chikungunya virus infection: first detection of imported and autochthonous cases in Panama’, The American Journal of Tropical Medicine and Hygiene, 92/3: 482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Quinonez J. A. et al. (2015) ‘Complete genome sequences of chikungunya virus strains isolated in Mexico: first detection of imported and autochthonous cases’, Genome Announcements, 3/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M. R. et al. (2009) ‘Zika virus outbreak on Yap Island, Federated States of Micronesia’, The New England Journal of Medicine, 360/24: 2536–43. [DOI] [PubMed] [Google Scholar]
- Faria N. R. et al. (2016) ‘Zika virus in the Americas: Early epidemiological and genetic findings’, Science, 352/6283: 345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. et al. (2014) ‘Notes from the field: chikungunya virus spreads in the Americas - Caribbean and South America, 2013-2014’, Morbidity and Mortality Weekly Report, 63/22: 500–1. [PMC free article] [PubMed] [Google Scholar]
- Gerardin P. et al. (2011) ‘Perceived morbidity and community burden after a Chikungunya outbreak: the TELECHIK survey, a population-based cohort study’, BMC Medicine, 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M. S. et al. (2013) ‘Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci’, Molecular Biology and Evolution, 30/3: 713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandadam M. et al. (2011) ‘Chikungunya virus, southeastern France’, Emerging Infectious Diseases journal, 17/5: 910–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessey M., Fischer M., Staples J. E. (2016) ‘Zika Virus spreads to new areas - region of the Americas, May 2015-January 2016’, Morbidity and Mortality Weekly Report, 65/3: 55–8. [DOI] [PubMed] [Google Scholar]
- Koboldt D. C. et al. (2012) ‘VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing’, Genome Research, 22/3: 568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L. et al. (2006) ‘GARD: a genetic algorithm for recombination detection’, Bioinformatics, 22/24: 3096–8. [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S., Valadere A. M. (2014) ‘Transcontinental movement of Asian genotype chikungunya virus’, Emerging Infectious Diseases journal, 20/8: 1400–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S. et al. (1998) ‘Emergence of epidemic O'nyong-nyong fever in Uganda after a 35-year absence: genetic characterization of the virus’, Virology, 252/1: 258–68. [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S. et al. (2008) ‘Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007’, Emerging Infectious Diseases journal, 14/8: 1232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S. et al. (2016) ‘Phylogeny of Zika Virus in Western Hemisphere, 2015’, Emerging Infectious Diseases journal, 22/5: 933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast gapped-read alignment with Bowtie 2’, Nature Methods, 9/4: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leparc-Goffart I. et al. (2014) ‘Chikungunya in the Americas’, Lancet, 383/9916: 514. [DOI] [PubMed] [Google Scholar]
- Levinson R. S., Strauss J. H., Strauss E. G. (1990) ‘Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees’, Virology, 175/1: 110–23. [DOI] [PubMed] [Google Scholar]
- Mattar S. et al. (2015) ‘Outbreak of Chikungunya virus in the north Caribbean area of Colombia: clinical presentation and phylogenetic analysis’, The Journal of Infection in Developing Countries, 9/10: 1126–32. [DOI] [PubMed] [Google Scholar]
- Musso D., Cao-Lormeau V. M., Gubler D. J. (2015) ‘Zika virus: following the path of dengue and chikungunya?’, Lancet, 386/9990: 243–4. [DOI] [PubMed] [Google Scholar]
- Myles K. M. et al. (2006) ‘Effects of an opal termination codon preceding the nsP4 gene sequence in the O'Nyong-Nyong virus genome on Anopheles gambiae infectivity’, Journal of Virology, 80/10: 4992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M. R. et al. (2015) ‘Emergence and potential for spread of Chikungunya virus in Brazil', BMC Medicine, 13: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan American Health Organization (2014a), Number of reported cases of Chikungunya fever in the Americas—EW 52 (December 29, 2014) http://www2.paho.org/hq/index.php? option=com_topics&view=readall&cid= 5927&Itemid=40931 &lang=en.
- Pan American Health Organization (2014b), Number of reported cases of Chikungunya fever in the Americas—EW 09 (February 28, 2014) http://www2.paho.org/hq/index.php?option=com_topics&view=readall&cid= 5927&Itemid=40931&lang=en.
- Pond S. L., Frost S. D. (2005) ‘Datamonkey: rapid detection of selective pressure on individual sites of codon alignments’, Bioinformatics, 21/10: 2531–3. [DOI] [PubMed] [Google Scholar]
- Powers A. M. et al. (2000) ‘Re-emergence of Chikungunya and O'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships’, Journal of General Virology , 81/Pt 2: 471–9. [DOI] [PubMed] [Google Scholar]
- Rambaut A. et al. (2016) ‘Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen)’, Virus Evolution, 2/1: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezza G. et al. (2007) ‘Infection with chikungunya virus in Italy: an outbreak in a temperate region’, Lancet, 370/9602: 1840–6. [DOI] [PubMed] [Google Scholar]
- Ross R. W. (1956) ‘The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic’, The Journal of Hygiene (London), 54/2: 177–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. et al. (2014) ‘Preparedness for threat of chikungunya in the pacific’, Emerging Infectious Diseases journal, 20/8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp J. C. et al. (2015) ‘Alphavirus RNA synthesis and non-structural protein functions’, Journal of General Virology, 96/9: 2483–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahadeo N. et al. (2015) ‘Correction: molecular characterisation of chikungunya virus infections in trinidad and comparison of clinical and laboratory features with dengue and other acute febrile cases’, PLOS Neglected Tropical Diseases, 9/12: e0004305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffenecker I. et al. (2006) ‘Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak’, PLoS Med, 3/7: e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O., Albert M. L. (2010) ‘Biology and pathogenesis of chikungunya virus’, Nature Review Microbiology, 8/7: 491–500. [DOI] [PubMed] [Google Scholar]
- Stapleford K. A. et al. (2016) ‘Whole-genome sequencing analysis from the chikungunya virus caribbean outbreak reveals novel evolutionary genomic elements’, PLOS Neglected Tropical Diseases, 10/1: e0004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K. A. et al. (2007) ‘A single mutation in chikungunya virus affects vector specificity and epidemic potential’, PLoS Pathog, 3/12: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin K. A. et al. (2014) ‘Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes’, Nature Communications, 5: 4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazeille M. et al. (2007) ‘Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus’, PLoS One, 2/11: e1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk S. M. et al. (2010) ‘Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates’, Journal of Virology, 84/13: 6497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C. (2014) ‘Arrival of chikungunya virus in the new world: prospects for spread and impact on public health’, PLOS Neglected Tropical Diseases, 8/6: e2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S. C. et al. (1999) ‘Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells’, Journal of Virology, 73/5: 4316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences derived in this study are available on Genbank accession numbers KY435454 - KY435486.
Conflict of interest: None declared.