Abstract
Shallow‐water coral reef ecosystems, particularly those already impaired by anthropogenic pressures, may be highly sensitive to disturbances from natural catastrophic events, such as volcanic eruptions. Explosive volcanic eruptions expel large quantities of silicate ash particles into the atmosphere, which can disperse across millions of square kilometres and deposit into coral reef ecosystems. Following heavy ash deposition, mass mortality of reef biota is expected, but little is known about the recovery of post‐burial reef ecosystems. Reef regeneration depends partly upon the capacity of the ash deposit to be colonised by waterborne bacterial communities and may be influenced to an unknown extent by the physiochemical properties of the ash substrate itself. To determine the potential for volcanic ash to support pioneer bacterial colonisation, we exposed five well‐characterised volcanic and coral reef substrates to a marine aquarium under low light conditions for 3 months: volcanic ash, synthetic volcanic glass, carbonate reef sand, calcite sand and quartz sand. Multivariate statistical analysis of Automated Ribosomal Intergenic Spacer Analysis (ARISA) fingerprinting data demonstrates clear segregation of volcanic substrates from the quartz and coral reef substrates over 3 months of bacterial colonisation. Overall bacterial diversity showed shared and substrate‐specific bacterial communities; however, the volcanic ash substrate supported the most diverse bacterial community. These data suggest a significant influence of substrate properties (composition, granulometry and colour) on bacterial settlement. Our findings provide first insights into physicochemical controls on pioneer bacterial colonisation of volcanic ash and highlight the potential for volcanic ash deposits to support bacterial diversity in the aftermath of reef burial, on timescales that could permit cascading effects on larval settlement.
1. Introduction
Coral reefs are unique, biodiverse ecosystems of high socio‐economic importance on both global and local scales (Nicholls et al., 2007). Anthropogenic disturbances, such as sedimentation and eutrophication, increasingly pressure fragile coral reef ecosystems worldwide (Wilkinson 1999). The deterioration of water quality in coastal regions consequently favours macro‐algal dominance (Fabricius, 2005; Schaffelke, Mellors, & Duke, 2005) and increases the risk of disease for coral reef‐building species, including sponges and corals (Haapkyla et al., 2011; Webster, Xavier, Freckelton, Motti, & Cobb, 2008), which further exacerbates coral reef vulnerability to catastrophic natural disturbances, such as volcanic ash deposition (Vroom & Zgliczynski, 2011). After an explosive volcanic eruption, widespread dispersal and deposition of volcanic ash over areas up to millions of square kilometres, in thickness of up to several centimetres, may be damaging to ash‐affected coral reef ecosystems; both Maniwavie, Rewald, Aitsi, Wagner, and Munday (2001) and Vroom and Zgliczynski (2011) have reported the destruction and mass mortality of reef biota following heavy ash deposition. However, the capacity of reef ecosystems to recover after burial by ash remains uncertain. Maniwavie et al. (2001) reported that 2 years after burial by volcanic ash corals had only re‐colonised the surfaces of protruding or unburied objects (e.g., boulders, tree stumps), while the ash substrate itself remained barren; in contrast, Schils (2012) noted that a period of frequent ash deposition into a tropical reef ecosystem promoted a change in benthic microbial and macrofloral communities on a similar timescale. These varying responses indicate a clear need to better understand the factors that may dictate the recovery of vulnerable and valuable coral reef ecosystems after ash deposition.
In the aftermath of large‐scale burial, recovery of the reef ecosystem may depend on pioneer colonisation of the new substrate by free‐living bacteria from the water column. After attachment to the surface, these bacteria produce an extracellular polymeric matrix that embeds further microbial organisms, forming so‐called biofilms (Costerton, Lewandowski, Caldwell, Korber, & Lappin‐Scott, 1995). Biofilm communities are highly abundant in coral reefs and are crucial in biogeochemical nutrient cycling and the degradation of anthropogenic pollutants (reviewed in Davey & O'Toole, 2004). Further, they provide an essential settlement surface for larvae of important reef‐building invertebrates (e.g., corals and sponges) and influence larval settlement cues and metamorphosis (Webster et al., 2004; Wieczorek & Todd, 1998). Accordingly, any changes in bacterial biofilm communities may influence invertebrate larval settlement, coral reef establishment and further development. Therefore, the capacity of a volcanic ash substrate to support bacterial settlement, particularly compared to the marine substrates it overlies, may play a crucial role in shaping the recovery of ash‐affected reef ecosystems.
Previous studies on aquatic biofilm formation using an array of natural and artificial substrates, including basaltic glasses and borosilicate (Thorseth, Furnes, & Tumyr, 1995), biotite (Ward, 2013), granite (Chung et al., 2010), coral skeletons and clay tiles (Witt, Wild, & Uthicke, 2011), highlight the importance of physicochemical properties (e.g., granulometry, surface morphology, mineralogy and chemistry, colour) in promoting initial substrate colonisation. Crucially, volcanic ash materials are subject to a wide variation in all of these properties, which are the product of magma composition and eruption history (Dingwell, Lavallée, & Kueppers, 2012). Ash particles range in size from the millimetre to submicron scale and vary in morphology from smooth, blocky particles, to rough‐textured vesicular clasts (Heiken, 1974). They commonly contain crystalline and amorphous silicates of various compositions, and range in colour from light to dark (Ayris & Delmelle, 2012). Ash surfaces can be a source of variably extractable elements, some of which (e.g., Al, Ca, Co, Cu, Fe, K, Mg, Mn, Ni, Mo, P, S, Zn; Jones & Gislason, 2008) may be important macro‐ or micronutrients for bacteria and phytoplankton (Duggen, Croot, Schacht, & Hoffmann, 2007; Munn, 2003), while others (e.g., Al, Cu) may be toxic (Duggen et al., 2007b).
Investigating the propensity for volcanic ash to promote pioneer bacterial colonisation in situ is hampered by the difficulties associated with substrate accessibility, geographic location and the dangers associated with sampling near active volcanoes. Laboratory experiments conducted in simulated tropical coral reef aquaria, therefore, offer a viable method to approximate the properties of volcanic ash that may dictate its capacity to act as a colonisable substrate. In this study, we incubate a selection of volcanogenic, terrigenic and biogenic substrates in a coral reef‐like marine aquarium system for 3 months and correlate differences in the bacterial colonising consortia with physicochemical properties (i.e., chemical composition, mineralogy, granulometry, morphology, colour) of particulate substrates.
2. Materials and methods
2.1. Substrate selection, preparation and characterisation
The effect of physicochemical parameters on the composition of bacterial community colonisation was explored using five different substrates in a marine aquaria under controlled conditions for bacterial colonisation: two volcanogenic materials (volcanic ash and synthetic volcanic glass), two biogenic materials of reef origin (carbonate coral reef sand and calcite sand) and a terrigenic quartz sand (Table 1). Fresh, crystal‐bearing volcanic ash was obtained from Sakurajima volcano, Japan, on 18 July 2013. Sakurajima was selected because it prevalently erupts andesitic magma, which is of intermediate volcanic composition and represents the dominant composition at island arc volcanoes, and because the physicochemical properties of its eruptive products are well characterised (Hillman et al., 2012). The synthetic volcanic glass substrate was employed to provide a crystal‐free system of comparable chemical composition to the volcanic ash substrate and was prepared by subjecting the Sakurajima ash to five cycles of high temperature melting in a platinum crucible at 1500°C to produce a homogeneous melt. This melt was quenched rapidly to produce a glass and then mechanically crushed. The terrigenic quartz sand was selected to provide a silicate reference material for comparison with the volcanogenic substrates and was obtained commercially (Sigma‐Aldrich ID no. 274739). To draw comparisons between the silicate substrates and biogenic reef sands, we utilised two different carbonate materials, each with different origins. The coral reef sand was obtained from a shallow fringing reef in the north‐eastern Gulf of Aqaba, Red Sea, located within a marine reserve close to the Marine Science Station in Aqaba, Jordan (29°27′N, 34°58′E), and was mechanically crushed before use. The calcite sand was sourced from Longcliffe Quarries Ltd. (Brassington, UK).
Table 1.
Physicochemical data of the five substrates
| Substrate | Volcanic Glass | Volcanic Asha | Carbonate reef sandb | Calciteb | Quartzc | |
|---|---|---|---|---|---|---|
| Chemical composition (wt. %) | SiO2 | 58.2 | 58.5 | 0.2 | 0.9 | >99.5 |
| Al2O3 | 16.8 | 16 | 0.1 | 0.3 | n.s. | |
| Fe2O3 | 7.9 | 7.9 | 0 | 0 | n.s. | |
| MnO | 0.2 | 0.2 | 0 | 0 | n.s. | |
| MgO | 3.4 | 3.6 | 1.3 | 0.4 | n.s. | |
| CaO | 7.4 | 7.2 | 52.2 | 54.4 | n.s. | |
| Na2O | 3.4 | 3 | 0.3 | 0.1 | n.s. | |
| K2O | 1.4 | 1.4 | 0 | 0 | n.s. | |
| TiO2 | 0.8 | 0.8 | 0 | 0 | n.s. | |
| P2O5 | 0.2 | 0.2 | 0.1 | 0 | n.s. | |
| Total | 99.6 | 100.1 | 99.0 | 100.0 | >99.5 | |
| L.O.I | −0.17 | 0.02 | 44.49 | 44.43 | <0.5 | |
| Physical properties | SSA (mb/g) | 0.02 | 0.1 | 0.49 | 0.22 | 0.11 |
| Median diameter (μm) | 234 | 221 | 224 | 230 | 263 | |
| Interquartile diameter (μm) | 180–260 | 190–260 | 190–270 | 190–260 | 220–310 | |
| Colour | Dark | Dark | White | White | White | |
| Morphology | Angular | Subangular | Aggregate | Aggregate | Subangular | |
| Surface appearance | Smooth | Rough | Rough | Rough | Rough | |
Comprised of glass, plagioclase, pyroxenes, Fe–Ti oxides (Miwa et al., 2013).
Comprised of CaCO3.
Compositional data of pure quartz from Sigma–Aldrich product specifications (n.s. not stated).
The two volcanic substrates were subjected to a preliminary leaching period of four hours in deionised water at an ash:water mass ratio of 1:5 and dried at 105°C for six hours. The remaining three substrates were washed in deionised water to remove very fine particulate adhering to the surface of bigger clasts, and dried at 105°C for six hours. All substrates were then leached for 12 hr in sea water taken from the aquarium system at a 1:5 ash:water mass ratio and dried at 105°C for six hours, spread out in large Petri dishes, UV‐sterilised by 2 × 30‐min cycles and stirred in between the cycles. These treatments were intended to isolate the effect of the substrate by eliminating any potential contribution from mechanical crushing of the glass, calcite and quartz substrates, any adsorbed chemical species (Ayris et al., 2014) that may be toxic to the other organisms in the aquarium water system, and any pre‐existing contamination by bacteria prior to or between sample collection/synthesis. The absence of bacteria on the samples was confirmed at the onset of the experiment using the DNA fingerprinting tool of Automated Ribosomal Intergenic Spacer Analysis (ARISA) (see details in Methods section below). As this study intentionally targets the physicochemical properties of substrates, the preliminary leaching of volcanic ash was intended to remove any soluble salts emplaced on ash surfaces during transport through the volcanic eruption plume. While the relevance of surface salts as a readily available nutrient source in ocean surface waters has been previously considered (e.g., Duggen et al., 2007b), their capacity for rapid dissolution on first contact with sea water likely limits their relevance to any subsequent influence once deposited as a substrate on the ocean floor.
Bulk chemical compositions of the substrates were determined by X‐ray fluorescence spectroscopy (Philips, MagiX Pro). The major and minor elements were measured using glass beads prepared by fusion with a lithium borate flux in a Panalytical Eagon 2 furnace fusion system. Specific surface area was determined by application of the Brunauer–Emmett–Teller (BET) theory to argon adsorption measurements conducted at −196°C using a Micrometrics Gemini surface area analyser. Samples were dried overnight at 105°C overnight prior to analyses. Data are the mean of three repeated measurements. Particle size distributions were determined by laser particle analysis using a Beckman‐Coulter LS230. The particle morphology of the leached substrates was examined by scanning electron microscopy (SEM; Leo 1430 VP) with a maximum operating voltage of 15 kV. Samples were prepared for imaging on SEM stubs and sputter coated with gold. Representative images of single particles were taken for each sample to qualitatively document differing particle morphologies amongst the substrates.
2.2. Design of the marine colonisation experiment
The aquarium system consists of a main tank (330 L) containing a 7‐year established coral reef community, including corals, algae, diverse marine invertebrates and fish. The main tank supplies sea water to a series of 80‐L subtanks. Those subtanks were used for experiments and did not contain coral reef organisms. The subtank utilised in this study was configured for a 12:12‐hr light:dark cycle and was covered by shade cloth to ensure low light availability of <0.2 klux. Light availability and temperature were recorded at 30‐min intervals over the course of the experiment using loggers (Onset HOBO TidBit). Tank sea water was maintained at constant flow rate, as to not perturb the substrates in the dishes (see below), constant average temperature (25.4°C ± 0.05), pH (8.13 ± 0.05) and conductivity (51.8 ± 1.14 mS), and was tested weekly to ensure that nutrient concentrations remained low (<8 nmol/L NO3, <0.2 nmol/L PO4; see Table S1).
The duration of the experiment was 3 months, with sampling of all substrates after one (T1), two (T2) and three (T3) months. For each sampling time‐point, each substrate was represented by three separate Petri dishes. Sterile glass Petri dishes of 50 mm diameter and 10 mm depth were filled with 1 g of substrate (a uniform depth of ~5 mm) and positioned approximately 50 mm apart at the bottom of one of the 80‐L subtanks, immersed to a depth of 250 mm in sea water. The granulometry of the substrate was selected to minimise the possibility of any disturbance by the water flow. To prevent loss of material during the initial sample placement, the dishes were carefully filled with sea water via manual pipetting and loaded into the tank with their lids in place. After placement, the lids were removed. In total, 45 dishes were utilised in the experiment (5 substrates × 3 sampling points × 3 replicates). At each sampling time‐point, 2 × 0.3 g of substrate was recovered from each replicate dish using a sterilised spatula. DNA from the substrate samples was either extracted immediately, or the sampled substrates were directly snap‐frozen in liquid nitrogen and stored at −80°C and then extracted within 2 weeks after sampling. To ensure that observed bacterial differences amongst substrates and time‐points were not due to changes in water quality parameters (i.e., temperature, pH, conductivity, nutrient concentrations), 2 × 500 mL seawater samples were collected at the time of substrate immersion (T0 beginning of the experiment) and at each sampling time‐point (n = 2), filtered through 0.2 μm polycarbonate filters using a vacuum pump, and filters were stored and processed following the same protocols as the substrate samples.
2.3. Determination of differences in bacterial community structures
Differences in bacterial community structure amongst substrates and time‐points were determined by the DNA fingerprinting method of ARISA using capillary electrophoresis. The technique targets the ITS region (Intergenic Transcribed Spacer) between the 16S and the 23S rRNA gene regions, which is highly variable in nucleotide sequence and length (from 50 bp to 1500 bp). Different bacterial species exhibit specific nucleotide lengths relative to the ITS region and, hence, the bacterial community structure (community composition and relative abundance of dominant bacterial species) can be determined (Brown & Fuhrman, 2005). While next‐generation sequencing (NGS) enables more in‐depth studies, Gobet, Boetius, and Ramette (2014) emphasise that the DNA fingerprinting of ARISA retains value in determining differences in community structure. It is particularly beneficial for the current study, as it permits the identification of fundamental differences in bacterial community composition amongst many different samples. DNA fingerprinting is a cost‐effective and rapid approach to identify taxonomic affiliations or diversity estimates and to separate samples on local spatio‐temporal scales, and correlate environmental variables with bacterial community structure (van Dorst et al., 2014), prior to embarking on more cost‐intensive NGS campaigns. Hence, this tool was chosen to gain an initial overview of community structure amongst different substrates. DNA was extracted from 0.25 g of each of the collected subsamples (three replicates and two separate extractions from each) and water samples in separate tubes. Extraction utilised a PowerLyzer DNA Isolation Kit according to the manufacturer's instructions (MoBio, Carlsbad, CA, USA) with the following alterations: bead beating cycles 2 × 30 s for higher yields of bacterial DNA and elution in 2 × 50 μL 1 × TE buffer to lower the possibility of disturbing polymerase chain reaction (PCR) amplifications.
DNA from all samples was stored in aliquots at −20°C until further processing. Extracted DNA from all replicates (three per substrate and time‐point) was amplified by PCR using the ITSF‐FAM and ITSReub primer pair (Cardinale et al., 2004) in three separate reactions per replicate sample. The ARISA‐PCR mixture (25 μL) contained 5 × PCR buffer (Promega, Madison, WI, USA); 2.5 mm MgCl2 (Promega); 0.25 mm deoxynucleoside triphosphate mix (Promega); 1.5 μg/μL bovine serum albumin (BSA); 400 nm universal primer ITSF‐FAM (5′‐GTCGTAACAAGGTAGCCGTA‐3′), 5′‐labelled with the phosphoramidite dye FAM and eubacterial ITSReub (5′‐GCCAAGGCATCCACC‐3′); and 0.025 U of GoTaq polymerase (Promega). To each reaction, approximately 20 ng of extracted DNA (quantified by a ND‐1000 Nanodrop; Peqlab Biotechnology, Erlangen, Germany) was added and the volume was adjusted to 25 μL using PCR water. Thermal cycling was carried out in an Eppendorf MasterCycler (Eppendorf, Hamburg, Germany) with an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 55°C for 45 s, 72°C for 90 s, with a final extension at 72°C for 5 min and then cooled to 15°C. PCR products were purified utilising the Nucleospin PCR clean‐up kit (Macherey‐Nagel, Düren, Germany).
Diluted PCR products (300–1200 bp) were prepared for analysis by ARISA using capillary electrophoresis as follows. A standardised amount of DNA (100 ng) was added to a separation cocktail containing 0.5 μL of internal GeneScan 1200LIZ size standard (50–1000 bp) (Applied Biosystems, Foster City, CA, USA) and 14 μL of deionised Hi‐Di formamide. The preparation was denatured for 3 min at 95°C and kept on ice for at least 5 min before being further processed by the sequencer. Separation of the PCR‐amplified fragments via capillary electrophoresis was carried out on an 80 cm‐capillary ABI PRISM 3730xl genetic analyzer (Applied Biosystems) with the following run parameters: 14.6 kV run voltage, 2.4 kV injection voltage, 20‐s injection time, and 60°C oven temperature. Raw profiles were checked for stable baselines and voltage, and peak sizes and absolute areas were then determined using the GeneMapper software v4.0 (Applied Biosystems) with minimum peak heights of 50 fluorescence units for all dyes. The best‐fit size‐calling curves were built using a second‐order least‐squares method (compensating for anomalously running fragments in the standard) and the Local Southern method as the size‐calling algorithm. A perfect fit for the calibration curves in the range of 100–1000 bp was always checked before further processing the samples. The fragments/peaks matrix generated with GeneMapper was transferred into the T‐align software (Smith et al., 2005). A difference of 1.5 bp between peaks was defined as a single operational taxonomic unit (OTU) (Danovaro, Luna, Dell'Anno, & Pietrangeli, 2006). The values of the relative fluorescence intensity of the peak area of the ARISA fingerprinting data were normalised, square‐root transformed and standardised prior to statistical analysis.
2.4. Statistical analysis
To explore the variation in the community structure and to determine whether bacterial assemblages grouped by substrate, multivariate statistics were applied. Pairwise distance matrices were calculated from the relative abundance data (ARISA) using the Bray–Curtis dissimilarity index. Non‐metric multidimensional scaling (nMDS) was applied to the distance matrices to explore the variation in community structure. Differences in the bacterial community structure according to substrate were assessed for significance by applying the ANalysis Of SIMilarity (ANOSIM) test based on permutation (9999 permutations) procedures using the Bray–Curtis distance measure. The contribution of each single taxon to the overall dissimilarities of factors was determined using the SIMilarity PERcentage (SIMPER) routine.
To visualise the community structure and relationships between substrate‐specific and shared operational taxonomic units (OTUs) amongst the different substrates, a Venn diagram was constructed. To determine differences in the number of OTUs between substrate groups and the water column, one‐way Analysis of Variance (ANOVA) was applied. Homogeneity of variance was tested using Levene's test, and the Tukey–Kramer test was used as a post hoc test. To determine biodiversity differences from the fingerprinting data, the Shannon–Wiener (H′) and Simpson's (1‐D) indices for bacterial OTUs in each substrate (average value for 6 replicates) were calculated. To test for significant differences between the bacterial communities on the different substrates, the Shannon index values (n = 6 per substrate) for bacterial communities on the five substrates after 3 months of incubation were analysed using the t‐test. All statistical analyses were performed using the statistical program PAST (Hammer, Harper, & Paul, 2001). The level of significance was <0.05 for all statistical tests.
3. Results
3.1. Particle substrate characterisation
Bulk chemical analysis by XRF confirms the volcanic ash, glass and quartz as silicates and the reef sand and calcite substrate as calcium carbonates (Table 1). The volcanic ash and glass are both dark in colour, whereas the quartz, reef sand and calcite are all white. Particle size data showed mono‐modal Gaussian distributions for all tested substrates. Ash, glass, calcite and reef sand had median particle diameters of 220–234 μm, while the quartz sand had a median diameter of 263 μm (Table 1). Particle morphology of the substrates was investigated by SEM analyses (Figure 1). Ash comprised subangular blocky particles with rough surfaces, often with micron‐sized vesicles. Glass generated from ash melting and crushing shows smooth surfaces with sharp edges (angular clasts) and conchoidal fractures imparted during preparation. Quartz particles display similar morphological characteristics to the volcanic ash substrates, being blocky, subangular particles with rough surfaces. Reef sand particles are blocky aggregates, composed of micron‐sized fragments of carbonate biogenic material (mainly shell and exoskeleton fragments of marine organisms) cemented by crystalline calcite. Calcite clasts are similarly blocky and equant and appear to consist of aggregates of micron‐sized calcite minerals. The specific surface area varies between 0.02 and 0.5 m2/g, with the highest surface area materials being the biogenic substrates (carbonate reef sand = 0.5 m2/g, calcite = 0.2 m2/g; Table 1).
Figure 1.
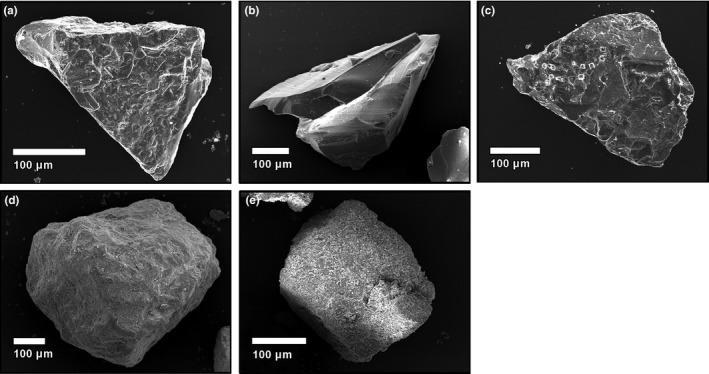
Scanning electron microscopy (SEM) images of substrates: (a) volcanic ash, (b) synthetic volcanic glass, (c) quartz, (d) carbonate reef sand and (e) calcite. Images were collected at 15 kV
3.2. Marine colonisation experiment
Multivariate statistical analysis of the ARISA fingerprinting data of the ITS region of the biofilm communities established upon the five substrates repeatedly showed overall significantly different bacterial community compositions over the 3‐month period and was consistently distinct from the community within the water column, represented by replicates from the beginning (T0) and additional data points as time progressed. These differences are reflected by the variations in the total number of OTUs and in the number of unique OTUs for each substrate at each time‐point. We summarise these trends in the following paragraphs but, for brevity, we further interpret the data for month three only, as this represents the most well‐established bacterial community.
Total OTUs successively increased by over 2.5‐fold from the first to the third month of the experiment (from 108 OTUs at T1 to 273 OTUs at T3). At each time‐point, most OTUs were shared amongst the substrate communities, with few substrate‐specific ones. Both shared and substrate‐specific OTUs increased with time from 99 shared and 10 substrate‐specific OTUs at T1 to 259 and 13 substrate‐specific OTUs at T3. Amongst the five‐substrates bacterial communities, volcanic ash showed the most substrate‐specific OTUs, followed by glass, carbonate and calcite, with quartz exhibiting no substrate‐specific OTUs at all.
The grouping patterns visualised in nMDS (Figure 2) showed significant differences amongst the bacterial communities on the five substrates at T1 (ANOSIM R = .272, P = .0001), T2 (ANOSIM R = .557, P = .0002) and T3 (ANOSIM R = .507, P = .0001), apart from quartz, which was different to the volcanic substrates but very similar to the biogenic substrates carbonate and calcite at T2 (ANOSIM R = .191, P = .273) and T3 (ANOSIM R = .033, P = .065 and R = .209, P = .303, respectively, Table S2). At T1 and T2, Quartz did not exhibit any specific bacterial community. The overall community for all five substrates showed distinct groupings of bacterial communities associated to the substrates of volcanic origin and coral reef origin, while quartz and the carbonate substrates both share bacterial OTUs. SIMPER analysis at T1 revealed that the OTUs contributing the most to the community differences were 545 bp (5%), 890 bp (4%) and 620 bp (4%). Of these, the 545‐ and 890‐bp OTUs were exclusive to, and dominant within, the volcanogenic substrate bacterial community, whereas 620‐bp OTU was dominant in the volcanogenic substrates, occurred occasionally in the biogenic substrates and was absent in the quartz substrate. In addition to the groupings according to substrate, the bacterial assemblages on each substrate differed significantly from those detected in the water column (ANOSIM R = .469, P = .0002). In the water column, 112 OTUs were detected, of which 2 were specific to the water community. SIMPER analysis at T2 identified the fragments 530 bp (6%), 890 bp (4%) and 721 bp (3%) contributing most to the overall community differences. We determined that the 530‐bp fragment represents a dominant OTU present in all substrates. Substrate‐associated bacterial communities differed significantly from those in the water column (ANOSIM R = .294, P = .0002). In the bacterial community in the water column, 119 OTUs were detected, of which 3 OTUs were water‐specific.
Figure 2.
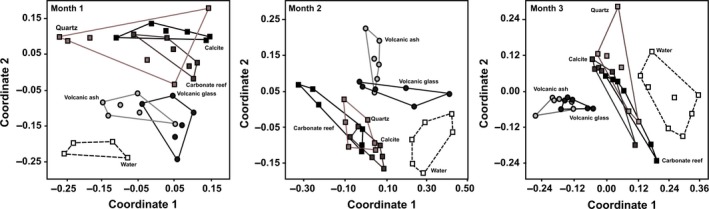
Non‐metric multidimensional scaling (nMDS) ordination (Bray–Curtis distance) of ARISA‐derived bacterial community profiles at months 1, 2 and 3, with six replicates per substrate per time‐point. Groupings are specified according to substrate. Data from an initial analysis (T0) of the bacterial community in the water column (n = 2) are plotted alongside the monthly water data. The water groupings are of cumulative data at each time‐point, inclusive of T0, for reference. Points plotting close to each other show a more similar community than distant ones
After 3 months (T3), a total of 273 OTUs could be detected in the overall bacterial community from the ARISA data. Of these, 272 OTUs were detected on the substrates, with 13 substrate‐specific OTUs and 259 shared ones (Figure 3). Of the shared OTUs, 144 were shared amongst all five substrates. The volcanic ash substrate showed the largest bacterial community with the most substrate‐specific OTUs (7), followed by volcanic glass (3), while a further 30 shared OTUs were specific to the volcanogenic substrates only. The biogenic substrates showed very low substrate specificity with <2 substrate‐specific OTUs each (reef sand 1 OTU, calcite 2 OTUs). The bacterial community in the water column had a total of 127 OTUs, of which solely 1 OTU was water‐specific.
Figure 3.
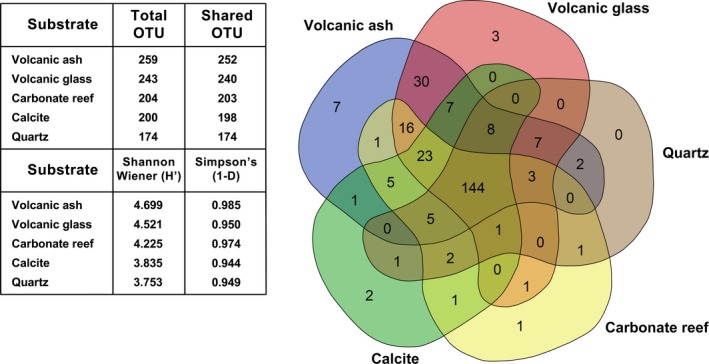
Relationship between the substrate‐specific and shared operational taxonomic units (OTUs) amongst all five substrates after 3 months. The diagram was constructed using the program VENN (http://bioinformatics.psb.ugent.be/webtools/Venn). Table (left) shows the total number of OTUs per substrate after 3 months (T3), the Shannon–Wiener (H′) and Simpson's (1‐D) diversity indices. The values were calculated by taking the average of OTUs of the ARISA data of the replicate samples of each substrate (n = 6)
Multivariate statistical analysis of the ARISA fingerprinting data of the ITS region of the biofilm communities established upon the five substrates after 3 months showed distinct groupings of bacterial communities associated with the substrates of volcanic origin and coral reef origin (Figure 2), and of water. The two biogenic substrates had a higher variability amongst replicates than those of volcanic origin, but the greatest variability was detected amongst quartz substrate replicate samples. Bacterial communities on all substrates were significantly different to each other (ANOSIM R = .507, P = .0001), apart from quartz, which was different to the volcanic substrates but the same as the biogenic substrates (ANOSIM R = .209, P = .303 and R = .033, P = .065, respectively, Table S2). Substrate specificity was further confirmed as the bacterial assemblages on each substrate differed significantly from those detected in the water column (ANOSIM R = .241, P = .049; Table S2). SIMPER analysis revealed an overall dissimilarity pooled over all groups of 52.37% with the OTUs contributing most to community structure differences being the 332‐bp (4.6%), 422‐bp (1.7%) and 481‐bp (1.5%) fragments, but with unspecified taxonomic affiliations. These OTUs were detected amongst all substrates, but were all higher in the volcanogenic than in the biogenic and terrigenic substrates; 481‐bp OTU was a dominant OTU throughout all substrates.
All five substrates and the water column varied significantly from each other with respect to the number of associated OTUs at the 3‐month time‐point (one‐way ANOVA, degrees of freedom = 5, mean square = 13 844, F = 1044, P = <.0001, Tukey–Kramer post hoc test P = .0001; Table S3). The volcanic substrates supported the highest number of total OTUs and, according to Shannon–Wiener and Simpson's diversity indices, also harboured the most diverse bacterial community compared to the quartz and biogenic substrates (Table in Figure 2). Further, statistical t‐tests confirmed significant differences in diversity of bacterial OTUs between the volcanogenic substrates and the biogenic and quartz substrates, while calcite, carbonate reef sand and quartz were not significantly different from each other (Table S4).
4. Discussion
No previous in situ studies have investigated bacterial colonisation of volcanic ash as a substrate within a reef environment, and the only in situ studies documenting the aftermath of ash deposition are both focused on macroflora and fauna and are contrasting in their results: Maniwavie et al. (2001) implied that recolonisation of ash substrates by reef flora did not occur, whereas Schils (2012) reported a sudden change in benthic microbial and macrofloral communities of an ash‐affected reef. Therefore, while comparisons between the aquarium experiment of the current study and in situ reef settings should be treated with caution, our study makes an important contribution to a highly uncertain subject by demonstrating that bacteria can colonise ash substrates and can establish a significantly different community structure than those on co‐existing substrates commonly found in reef habitats. Importantly, larval settlement is driven by bacterial community composition and settlement success rates increase with the age of the biofilm, and the timescale over which significant differences in bacterial community structure were observed in the current study are similar, and relevant, to the timescales required for the settlement of invertebrate larva (days to weeks; Bao, Satuito, Yang, & Kitamura, 2007; Campbell et al., 2011). Consequently, the significant differences in pioneer bacterial colonisation between volcanogenic substrates, and the terrigenic and biogenic substrates in the present study, even after 3 months, could impart further differences in invertebrate larval settlement, and so have cascading effects on the subsequent reef formation and succession. This fast response time echoes recent studies of fresh basaltic lava flows in terrestrial environments, where it was suggested that bacterial communities were rapidly established within days or months of lava flow emplacement (Kelly, Cockell, Thorsteinsson, Marteinsson, & Stevenson, 2014).
4.1. Differences in bacterial community structure
Evaluation of the bacterial communities colonising volcanogenic, biogenic and terrigenic substrates in a coral reef environment mesocosm, as determined by DNA fingerprinting of the ITS region using Automated Ribosomal Intergenic Spacer Analysis (ARISA), showed both substrate‐specific and shared bacterial OTUs. Such differences in the establishment of biofilms on different substrates are compatible with previous investigations on biofilm formation on different silicate and carbonate substrates incubated in tropical marine waters, which also noted the importance of the substrate for biofilm formation (Chung et al., 2010; Dobretsov, Abed, & Voolstra, 2013; Witt et al., 2011). Further, DNA fingerprinting revealed distinct differences between bacterial biofilm communities on the substrates and the communities found in the water column over the 3‐month experiment. Differences in the structure and diversity between the attached and free‐living bacterial communities are in line with in situ observations in different marine coastal systems (Mohit, Archambault, Toupoint, & Lovejoy, 2014; Zhang, Liu, Lau, Ki, & Qian, 2007) and have also previously been detected in coral reefs in situ (Santavy & Colwell, 1990; Schöttner et al., 2009).
Of the five substrates, we observed that the volcanogenic group (ash, glass) carried a more diverse, and significantly different, bacterial community compared to the communities associated with biogenic (carbonate and calcite sand) and terrigenic (quartz) substrates. The SIMPER analysis revealed substrate‐specific bacterial OTUs in the volcanogenic substrates contributing the most to the observed differences. These were either OTUs found in all substrate communities, which were more prevalent in the volcanogenic substrates, or were unique to the volcanogenic substrates, highlighting the substrate specificity of the colonising bacterial community. Further, statistical tests of the Shannon–Wiener diversity index confirmed significantly higher diversity within the volcanogenic substrates when compared to the other three substrates, while, amongst the biogenic and terrigenic substrates, no significant differences in diversity were detected. However, within the volcanogenic and biogenic substrate groups, the number of total OTUs and diversity indices indicate that the ash and carbonate reef sands carried a higher bacterial diversity than the calcite and glass. The observed differences in bacterial abundance and community differences amongst substrates indicate a substantive control of substrate physicochemical properties on bacterial community settlement and structure.
4.2. Physicochemical controls on bacterial communities
Differences in substrate physical properties (surface morphology and granulometry) may account for the higher bacterial diversity on the ash and carbonate reef sand compared to the glass and calcite sands. The former materials exhibit higher specific surface areas, reflecting higher surface roughness, porosity or the presence of smaller particles adhering to larger particle surfaces; these properties are known to affect growth in other bacterial systems (Gerasimenko, Orleanskii, Karpov, & Ushantinskaya, 2013; Yamamoto & Lopez, 1985). We detected significantly different bacterial communities on white carbonate and calcite substrates, while quartz was the same as both of these biogenic substrates. In contrast, in a comparable study of silicate and carbonate substrates in situ (Red Sea), Schöttner et al. (2011) found that both sands showed similar bacterial density and diversity, although with differing bacterial community structure over seasonal changes. However, the carbonate sand investigated by Schöttner et al. (2011) was poorly sorted and significantly coarser (median particle size = 553 μm) than the quartz sand (median particle size = 326 μm). Yet, no evident influence of substrate chemistry between quartz and carbonate materials in Schöttner et al. (2011) and the current study implies that differing substrate granulometry may have caused bacterial niche‐partitioning.
An effect of substrate colour could explain our observation that the two dark volcanogenic substrates carried more diverse bacterial communities than the three light substrates. The colour of a substrate has recently been observed to affect attraction of different microbial communities, whereby black substrates carry a higher bacterial density than white substrates (Dobretsov et al., 2013). Notably, diverse invertebrate settlement assays have shown that some larvae and algal spores prefer dark substrates over light‐coloured ones, likely due to diminished light reflection and greater heat retention of dark substrates, conditions that are preferred by negatively photo‐tactic organisms (Svane & Dolmer, 1995). Further, invertebrates often prefer dark substrates for colonisation to benefit from better protection from grazers (Swain, Herpe, Ralston, & Tribou, 2006). These factors are likely to apply to bacterial colonisation as well. Therefore, Dobretsov et al. (2013) and the current study emphasise the effects of colour on bacterial settlement, which should be tested further in future settlement assays.
The chemistry and/or mineralogy of the substrates could equally govern the observed differences in community structure, as substrate colour is strongly influenced by composition and the coordination of atoms within a material (Nassau, 1978). These properties will, in turn, influence the distribution and availability of nutrients at the substrate surfaces. Nutrients may be extracted from the substrate surface by leaching and dissolution by organic compounds and by water within the established biofilm (Brehm, Gorbushina, & Mottershead, 2005). Pre‐leaching of the materials in the present study was essential to ensuring that nutrients were substrate‐derived. However, the response to available nutrients is likely to be species‐dependent (Gerasimenko et al., 2013; Nies & Silver, 1989); thus, differences in substrate chemistry and/or mineralogy may promote or inhibit growth of different bacterial species on the different substrates.
Dependences of bacterial community structure on substrate mineralogy have been previously invoked by Kelly et al. (2011, 2009), Gleeson et al. (2006) and Hutchens, Gleeson, McDermott, Miranda‐CasoLuengo, and Clipson (2010). The first two studies listed above note correlations between bacterial communities on weathered glasses and crystalline rocks, depending on their composition (basaltic to rhyolitic). The last two studies document significant differences in bacterial community structuring in biofilms growing on adjacent (on length scales of cm to m) silicate minerals (quartz, albite, K‐feldspar, muscovite) at the surface of a terrestrial granite. Accordingly, there may therefore be significant differences in bacterial community structure both spatially across a single ash deposit, and in deposits of differing composition produced by different volcanoes.
Direct comparison of the fresh volcanic ash substrates used here and the weathered terrestrial rocks should be made carefully as weathering alters nutrient availability and introduces mineral phases (e.g., palagonite; Kelly et al., 2010) absent in the fresh material. It may even be difficult to compare fresh ash surfaces and those of fresh lava flows (e.g., Kelly et al., 2014), as the surface of the latter is altered by crystallisation (Burkhard, 2002) and volatile degassing processes (White & Hochella, 1992) during and immediately after emplacement. However, if similar differences in bacterial communities depending on the silicate mineral substrate are found in our substrates as in terrestrial studies, our data would imply a shared mineralogy between the ash and glass substrate. As ash from Sakurajima volcano is predominantly glassy (70%–90%, Miwa, Geshi, & Shinohara, 2013), otherwise comprising plagioclase and mafic minerals, this may suggest that the glass component is driving the bacterial community structure of both volcanogenic substrates. As neither of the volcanic materials contain a significant quartz component, their different community structures relative to that of the terrigenic quartz would be consistent with the mineralogical dependence invoked by Gleeson et al. (2006) and Hutchens et al. (2010).
Volcanic ash is characterised by a wide range of physical (morphology, particle size, surface area, colour) and chemical properties (mineralogy, composition), and volcanic ash deposits can contain a diverse array of particles, including entirely glassy or crystalline particles from fresh magma, remobilised older ejecta, and fragments of weathered lithic rocks. Furthermore, ash can deposit variably according to eruption dynamics as well as ash dispersal and sedimentation patterns with increasing distance from the volcano (Bonadonna, Costa, Folch, & Koyaguchi, 2015). Accordingly, there is the potential for significant variation amongst bacterial communities that establish on ash‐buried reefs at different positions relative to a source volcano as well as amongst different volcanoes. Hence, additional studies should be undertaken to disentangle the contribution of variable ash properties to bacterial colonisation as only one ash sample was considered here.
5. Conclusion
This study is the first to investigate volcanic ash as a substrate for pioneer bacterial colonisation using a simulated coral reef environment. We show that volcanogenic substrates support a notably diverse bacterial community, exhibiting higher numbers of OTUs over the course of 1–3 months than both terrigenic and biogenic substrates. The observed diversity amongst substrates indicates that the initial community structure is likely dictated by differences in substrate physicochemical properties. We identify greater diversity in substrates with higher specific surface areas compared to those with lower surface areas but are compositionally similar, which, coupled with comparisons to in situ studies, suggests possible controls associated with particle physical properties (e.g., granulometry, surface morphology). Our findings also suggest a significant control of substrate composition (bulk chemistry and mineralogy), which could result from a direct influence of nutrient availability or an indirect influence through substrate colour, whereby compositionally diverse dark‐coloured volcanogenic substrates favoured development of a larger community structure relative to light‐coloured quartz and biogenic substrates. Identification of the bacterial community diversity using next‐generation sequencing and an “omics” approach would provide additional information about the pioneer bacterial communities on the different substrates and help to understand their function. Critically, our findings indicate the potential for volcanic ash to promote bacterial diversity in an immediate post‐burial scenario in ash‐affected coral reefs, on timescales that could permit cascading effects on larval settlement and ultimately reef recovery. Further investigation of coral reef recovery and resilience following large‐scale natural disturbances, such as volcanic ash deposition, may contribute to predictions on ecosystem recovery and hazard management strategies.
Supporting information
Acknowledgments
VW, PMA, CC and DED thank the AXA Research Grant “Risk from volcanic ash in the Earth system” for supporting the research and DBD acknowledges the ERC Advanced Investigator Grant No. 247076 (EVOKES). We acknowledge R. Melzer for providing access to the scanning electron microscope at the Zoologische Staatssammlung München. Thanks to Larry Miller for providing an internal USGS review.
Witt V, Ayris PM, Damby DE, et al. Volcanic ash supports a diverse bacterial community in a marine mesocosm. Geobiology 2017;15:453–463. https://doi.org/10.1111/gbi.12231
Contributor Information
V. Witt, Email: wittverena@gmx.de
C. Cimarelli, Email: cimarelli@min.uni-muenchen.de
References
- Ayris, P. M. , & Delmelle, P. (2012). The immediate environmental effects of tephra emission. Bulletin of Volcanology, 74(9), 1905–1936. [Google Scholar]
- Ayris, P. M. , Delmelle, P. , Cimarelli, C. , Maters, E. C. , Suzuki, Y. J. , & Dingwell, D. B. (2014). HCl uptake by volcanic ash in the high temperature eruption plume: mechanistic insights. Geochimica and Cosmochimica Acta, 144, 188–201. [Google Scholar]
- Bao, W.‐Y. , Satuito, C. G. , Yang, J.‐L. , & Kitamura, H. (2007). Larval settlement and metamorphosis of the mussel Mytilus galloprovincialis in response to biofilms. Marine Biology, 150(4), 565–574. [Google Scholar]
- Bonadonna, C. , Costa, A. , Folch, A. , & Koyaguchi, T. (2015). Tephra dispersal and sedimentation In Sigurdsson H. (Ed.), Encyclopedia of Volcanoes (2nd edn, pp. 587–597). Amsterdam: Academic Press, (Ch. 33). [Google Scholar]
- Brehm, U. , Gorbushina, A. , & Mottershead, D. (2005). The role of microorganisms and biofilms in the breakdown and dissolution of quartz and glass: Palaeogeography. Palaeoclimatology, Palaeoecology, 219(1), 117–129. [Google Scholar]
- Brown, M. V. , & Fuhrman, J. A. (2005). Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquatic Microbial Ecology, 41(11), 15–23. [Google Scholar]
- Burkhard, D. J. (2002). Kinetics of crystallization: Example of micro‐crystallization in basalt lava. Contributions to Mineralogy and Petrology, 142(6), 724–737. [Google Scholar]
- Campbell, A. H. , Meritt, D. W. , Franklin, R. B. , Boone, E. L. , Nicely, C. T. , & Brown, B. L. (2011). Effects of age and composition of field‐produced biofilms on oyster larval setting. Biofouling, 27(3), 255–265. [DOI] [PubMed] [Google Scholar]
- Cardinale, M. , Brusetti, L. , Quatrini, P. , Borin, S. , Puglia, A. M. , Rizzi, A. , … Daffonchio, D. (2004). Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Applied and Environmental Microbiology, 70(10), 6147–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H. , Lee, O. , Huang, Y.‐L. , Mok, S. , Kolter, R. , & Qian, P.‐Y. (2010). Bacterial community succession and chemical profiles of sub tidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. The ISME Journal, 4, 817–828. [DOI] [PubMed] [Google Scholar]
- Costerton, J. W. , Lewandowski, Z. , Caldwell, D. E. , Korber, D. R. , & Lappin‐Scott, H. M. (1995). Microbial biofilms. Annual Revision Microbiology, 49, 711–745. [DOI] [PubMed] [Google Scholar]
- Danovaro, R. , Luna, G.M. , Dell'Anno, A. , & Pietrangeli, B. (2006). Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments. Applied and Environmental Microbiology, 72(9), 5982–5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, M. , & O'Toole, G. A. (2004). Microbial biofilms: From ecology to molecular genetics. Microbial Molecular Biology Reviews, 70(10), 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwell, D. B. , Lavallée, Y. , & Kueppers, U. (2012). Volcanic ash: A primary agent in the Earth system. Physics and Chemistry of the Earth, Parts A/B/C(45), 2–4. [Google Scholar]
- Dobretsov, S. , Abed, R. M. , & Voolstra, C. R. (2013). The effect of surface colour on the formation of marine micro and macrofouling communities. Biofouling, 29(6), 617–627. [DOI] [PubMed] [Google Scholar]
- van Dorst, J. , Bissett, A. , Palmer, A. S. , Brown, M. , Snape, I. , Stark, J. S. , ··· Ferrari, B. C. (2014). Community fingerprinting in a sequencing world. FEMS Microbiology Ecology, 89(2), 1574–6941. [DOI] [PubMed] [Google Scholar]
- Duggen, S. , Croot, P. , Schacht, U. , & Hoffmann, L. (2007). Subduction zone volcanic ash can fertilize the surface ocean and stimulate phytoplankton growth: Evidence from biogeochemical experiments and satellite data. Geophysical Research Letters, 34(1), 1–5. [Google Scholar]
- Fabricius, K. E. (2005). Effects of terrestrial runoff on the ecology of corals and coral reefs: Review and synthesis. Marine Pollution Bulletin, 50(2), 125–146. [DOI] [PubMed] [Google Scholar]
- Gerasimenko, L. M. , Orleanskii, V. K. , Karpov, G. A. , & Ushantinskaya, G. T. (2013). Interaction of cyanobacteria with volcanic ashes. Microbiology, 82(1), 111–118. [DOI] [PubMed] [Google Scholar]
- Gleeson, B. G. , Kennedy, N. M. , Clipson, N. , Melville, K. , Gadd, G. M. , & McDermott, P. F. (2006). Characterization of bacterial community structure on a weathered pegmatitic granite. Microbial Ecology, 51(4), 526–534. [DOI] [PubMed] [Google Scholar]
- Gobet, A. , Boetius, A. , & Ramette, A. (2014). Ecological coherence of diversity patterns derived from classical fingerprinting and Next Generation Sequencing techniques. Environmental Microbiology, 16(9), 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapkyla, J. , Unsworth, R. K. F. , Flavell, M. , Bourne, D. G. , Schaffelke, B. , & Willis, B. L. (2011). Seasonal rainfall and runoff promote coral disease on an inshore reef. PLoS ONE, 6(2), e16893. doi: 10.1371/annotation/365162ee‐363718‐365144ce‐b365162e365169‐388302d365165e360801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, O. , Harper, D.A.T. , & Paul, D.R. (2001). AST: Paleontological statistics software package for education and data analysis. Paleontologia Electronica, 4, 1–9. [Google Scholar]
- Heiken, G. (1974). An Atlas of volcanic ash. Washington: Smithsonian Institute Press, 108 p. [Google Scholar]
- Hillman, S. E. , Horwell, C. J. , Densmore, A. L. , Damby, D. E. , Fubini, B. , Ishimine, Y. , & Tomatis, M. (2012). Sakurajima volcano: A physico‐chemical study of the health consequences of long‐term exposure to volcanic ash. Bulletin of Volcanology, 74(4), 913–930. [Google Scholar]
- Hutchens, E. , Gleeson, D. , McDermott, F. , Miranda‐CasoLuengo, R. , & Clipson, N. (2010). Meter‐scale diversity of microbial communities on a weathered pegmatite granite outcrop in the Wicklow Mountains, Ireland; evidence for mineral induced selection? Geomicrobiology Journal, 27(1), 1–14. [Google Scholar]
- Jones, M. T. , & Gislason, S. R. (2008). Rapid releases of metal salts and nutrients following the deposition of volcanic ash into aqueous environments. Geochimica et Cosmochimica Acta, 72(15), 3661–3680. [Google Scholar]
- Kelly, L. C. , Cockell, C. S. , Herrera‐Belaroussi, A. , Piceno, Y. , Andersen, G. , DeSantis, T. , ··· LeRoux, X. (2011). Bacterial diversity of terrestrial crystalline volcanic rocks, Iceland. Microbial Ecology, 62(1), 69–79. [DOI] [PubMed] [Google Scholar]
- Kelly, L. C. , Cockell, C. S. , Piceno, Y. M. , Andersen, G. L. , Thorsteinsson, T. , & Marteinsson, V. (2010). Bacterial diversity of weathered terrestrial Icelandic volcanic glasses. Microbial Ecology, 60(4), 740–752. [DOI] [PubMed] [Google Scholar]
- Kelly, L. C. , Cockell, C. S. , Thorsteinsson, T. , Marteinsson, V. , & Stevenson, J. (2014). Pioneer microbial communities of the Fimmvörðuháls lava flow, Eyjafjallajökull, Iceland. Microbial Ecology, 68(3), 504–518. [DOI] [PubMed] [Google Scholar]
- Kelly, L. C. , Herrera, A. , Olsson‐Francis, K. , Andersen, G. , Piceno, Y. , & Cockell, C. (2009). A geobiological comparison of high‐and low‐Silica containing weathered volcanic glass. Geochimica et Cosmochimica Acta Supplement, 73, 637. [Google Scholar]
- Maniwavie, T. , Rewald, J. , Aitsi, J. , Wagner, T. P. , & Munday, P. (2001). Recovery of corals after volcanic eruptions in Papua New Guinea. Coral Reefs, 20, 24. [Google Scholar]
- Miwa, T. , Geshi, N. , & Shinohara, H. (2013). Temporal variation in volcanic ash texture during a vulcanian eruption at the Sakurajima volcano, Japan. Journal of Volcanology and Geothermal Research, 260, 80–89. [Google Scholar]
- Mohit, V. , Archambault, P. , Toupoint, N. , & Lovejoy, C. (2014). Phylogenetic differences in attached and free‐living bacterial communities in a temperate coastal lagoon during summer, revealed via high‐throughput 16sRNA gene sequencing. Applied and Environment Microbiology, 80(7), 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn, C. (2003). Marine microbiology: Ecology and applications (p. 312). Oxon: Garland Science, BIOS Scientific Publishers. [Google Scholar]
- Nassau, K. (1978). The origins of color in minerals. American Mineralogist, 63(3‐4), 219–229. [Google Scholar]
- Nicholls, R. J. , Wong, P. P. , Burkett, V. R. , Codignotto, J. O. , Hay, J. E. , McLean, R. F. , … Woodroffe, C.D. (2007). Coastal systems and low‐lying areas In Parry M. L., Canziani O. F., Palutikof J. P., van der Linden P. J. & Hanson C. E. (Eds.), Climate Change 2007: Impacts, Adaptation and Vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (pp. 315–356). Cambridge: Cambridge University Press. [Google Scholar]
- Nies, D. H. , & Silver, S. (1989). Metal ion uptake by a plasmid‐free metal‐sensitive Alcaligenes eutrophus strain. Journal of Bacteriology, 171(7), 4073–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santavy, D. L. , & Colwell, R. R. (1990). Comparison of bacterial communities associated with the Caribbean sclerosponge Ceratoporella nicholsoni and ambient seawater. Marine Ecology Progress Series, 67, 73–82. [Google Scholar]
- Schaffelke, B. , Mellors, J. , & Duke, N. C. (2005). Water quality in the Great Barrier Reef region: Responses of mangrove, seagrass and macroalgal communities. Marine Pollution Bulletin, 51(1–4), 279–296. [DOI] [PubMed] [Google Scholar]
- Schils, T. (2012). Episodic eruptions of volcanic ash trigger a reversible cascade of nuisance species outbreaks in pristine coral habitats. PLoS ONE, 7(10), e46639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttner, S. , Hoffmann, F. , Wild, C. , Rapp, H. T. , Boetius, A. , & Ramette, A. (2009). Inter‐ and intra‐habitat bacterial diversity associated with cold‐water corals. The ISME Journal, 3(6), 756–759. [DOI] [PubMed] [Google Scholar]
- Schöttner, S. , Pfitzner, B. , Grünke, S. , Rasheed, M. , Wild, C. , & Ramette, A. (2011). Drivers of bacterial diversity dynamics in permeable carbonate and silicate coral reef sands from the Red Sea. Environmental Microbiology, 13(7), 1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. J. , Danilowicz, B. S. , Clear, A. K. , Costello, F. J. , Wilson, B. , & Meijer, W. G. (2005). T‐Align, a web‐based tool for comparison of multiple terminal restriction fragment length polymorphism profiles. Fems Microbiology Ecology, 54(3), 375–380. [DOI] [PubMed] [Google Scholar]
- Svane, I. , & Dolmer, P. (1995). Perception of light at settlement: A comparative study of two invertebrate larvae, a scyphozoan planula and a simple ascidian tadpole. Journal of Experimental Marine Biology and Ecology, 187, 561–591. [Google Scholar]
- Swain, G. W. , Herpe, S. , Ralston, E. , & Tribou, M. (2006). Short‐term testing of antifouling surfaces: The importance of colour. Biofouling, 22, 425–429. [DOI] [PubMed] [Google Scholar]
- Thorseth, I. H. , Furnes, H. , & Tumyr, O. (1995). Textural and chemical effects of bacterial activity on basaltic glass: An experimental approach. Chemical Geology, 119, 139–160. [Google Scholar]
- Vroom, P. , & Zgliczynski, B. J. (2011). Effects of volcanic ash deposits on four functional groups of a coral reef. Coral Reefs, 30, 1025–1032. [Google Scholar]
- Ward, S. (2013). Investigating the role of microbes in mineral weathering: Nanometre‐scale characterisation of the cell‐mineral interface using FIB and TEM. Micron, 47, 10–17. [DOI] [PubMed] [Google Scholar]
- Webster, N. S. , Smith, L. D. , Heyward, A. J. , Watts, J. E. M. , Webb, R. I. , Blackall, L. L. , & Negri, A. P. (2004). Metamorphosis of a scleractinian coral in response to microbial biofilms. Applied and Environmental Microbiology, 70(2), 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, N. S. , Xavier, J. R. , Freckelton, M. , Motti, C. A. , & Cobb, R. (2008). Shifts in microbial and chemical patterns within the marine sponge Aplysina aerophoba during a disease outbreak. Environmental Microbiology, 10(12), 3366–3376. [DOI] [PubMed] [Google Scholar]
- White, A. F. , & Hochella, M. F. (1992). Surface chemistry associated with the cooling and subaerial weathering of recent basalt flows. Geochimica et Cosmochimica Acta, 56(10), 3711–3721. [Google Scholar]
- Wieczorek, S. K. , & Todd, C. D. (1998). Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling, 12(1–3), 81–118. [Google Scholar]
- Wilkinson, C. R. (1999). Global and local threats to coral reef functioning and existence: Review and predictions. Marine and Freshwater Research, 50, 867–878. [Google Scholar]
- Witt, V. , Wild, C. , & Uthicke, S. (2011). Effect of substrate type on bacterial community composition in biofilms from the Great Barrier Reef. FEMS Microbiology Letters, 323(2), 188–195. [DOI] [PubMed] [Google Scholar]
- Yamamoto, N. , & Lopez, G. (1985). Bacterial abundance in relation to surface area and organic content of marine sediments. Journal of Experimental Marine Biology and Ecology, 90(3), 209–220. [Google Scholar]
- Zhang, R. , Liu, B. , Lau, S. C. , Ki, J. S. , & Qian, P. Y. (2007). Particle‐attached and free‐living bacterial communities in a contrasting marine environment: Victoria Harbor, Hong Kong. FEMS Microbiol Ecology, 61(3), 496–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


