Abstract
The developmental signalling protein Notch can be proteolytically activated following ligand-interaction at the cell surface, or can be activated independently of its ligands, following Deltex (Dx)-induced Notch endocytosis and trafficking to the lysosomal membrane. The means by which different pools of Notch are directed towards these alternative outcomes remains poorly understood. We found that the Drosophila ZO-1 protein Polychaetoid (Pyd) suppresses specifically the Dx-induced form of Notch activation both in vivo and in cell culture assays. In vivo we confirmed the physiological relevance and direction of the Pyd/Dx interaction by showing that the expanded ovary stem cell niche phenotypes of pyd mutants require the presence of functional Dx and other components that are specific to the Dx-induced Notch activation mechanism. In S2 cells we found that Pyd can form a complex with Dx and Notch at the cell surface and reduce Dx-induced Notch endocytosis. Similar to other known activities of ZO-1 family proteins, the action of Pyd on Dx-induced endocytosis and signalling was found to be cell density dependent. Thus, together, our results suggest an alternative means by which external cues can tune Notch signalling through Pyd regulation of Dx-induced Notch trafficking.
Keywords: Notch, endocytosis, Drosophila, signalling, ZO-1
1. Introduction
Notch is a transmembrane, cell–cell signalling receptor molecule associated with the apical epithelial junctions, and mediates an essential and highly conserved signal used in many developmental contexts to determine cell fate [1]. Notch activity is initiated through binding to a membrane bound ligand of the Delta/Serrate/Lag2 (DSL) family, which results in a cascade of proteolytic cleavages that eventually releases the Notch intracellular domain (NICD) [1]. The latter translocates to the nucleus to activate signalling through binding to the transcription factor Suppressor of Hairless. Notch can also be activated independently of its ligands through the activity of the ring finger ubiquitin ligase protein Deltex (Dx), which promotes Notch endocytosis and its activation within the endosomal pathway [2–4]. The latter depends on trafficking of Notch to the late endosome and is associated with the retention of Notch on the outer limiting membrane of this organelle. This maintains cytoplasmic accessibility of the NICD, which is released by a Presenilin-dependent proteolytic activation probably after endo-lysosomal fusion and removal of the Notch extracellular domain (NECD). The Dx-induced route to activation can be distinguished from ligand-induced signalling by the former's requirement for late endosomal trafficking components such as the homotypic fusion and vacuole protein sorting (HOPS) complex. For example, in Drosophila, mutations of the HOPS component carnation (car) prevent Dx-induced Notch activation but have no effect on ligand-induced signalling [2]. Dx also removes Notch from the cell surface, reducing its accessibility to ligands, and therefore Dx can act negatively on the ligand-induced form of signalling [4]. This context-dependency of Dx outcomes on Notch is exemplified by its interplay with the HECT domain ubiquitin ligase Suppressor of deltex (Su(dx)). The latter also binds to the NICD and promotes Notch transfer to a parallel clathrin-independent endocytic route and further promotes Notch entry into the internal luminal vesicles of late endosomes, resulting in Notch degradation [4]. Su(dx) and Dx are in competition with each other to direct Notch through these alternative routes. The phenotypic consequence of removing both Su(dx) and Dx in different cell types is context dependent. When combined, dx and Su(dx) mutations can be mutually suppressive for their respective loss or gain of Notch signalling phenotypes, as observed in the Drosophila wing. Alternatively the double mutant can produce enhanced Notch signalling through increased ligand-dependent activity, as observed in leg development and the embryo nervous system [4]. The latter situation can arise because both Su(dx) and Dx normally act to remove Notch from ligand accessibility by promoting Notch removal from the cell surface. The observed double mutant Notch gain of function can therefore occur through increased ligand-induced signalling.
Polychaetoid (Pyd) is the single Drosophila homologue of the cell junctional scaffolding protein ZO-1 [5] and has previously been implicated in the regulation of Notch signalling [6,7]. In mammalian epithelia ZO-1 is predominantly localized to tight junctions, where it forms complexes with junctional proteins such as Claudins that are responsible for establishing a barrier function. Hence cells deficient in ZO-1 and the close homologue ZO-2 fail to form proper tight junctions [8]. ZO-1 is also found in newly forming fibroblastic spot-like adherens junctions and may accelerate conversion to belt-like adherens junctions during epithelial polarization whereupon ZO-1 is relocalized to tight junctions [9,10]. ZO-1 is further present in adherens junctions of non-epithelial tissue [11]. In insects the tight junction is absent but functionally replaced by the more basally located septate junction. ZO-1 is predominantly localized to the adherens junction in Drosophila epithelia but is also found localized to lateral membranes [12]. Neither Drosophila nor mammalian ZO-1 are essential for overall epithelial cell polarity but both regulate apical domain expansion and morphology [7,13]. Furthermore in the Drosophila eye, pyd mutant cells accumulate the adherens junction proteins Cadherin and Roughest, and in wing disc epithelia Notch accumulation has additionally been observed, [7,14]. While ZO-1 is not required for cell polarity its loss has functional consequences on tissue morphology during development. In Drosophila, pyd mutants cause disruption during embryo morphogenesis through altered cell adhesion and cytoskeletal interactions resulting in defects in tracheal structures and dorsal closure [15,16]; pyd mutants further affect patterning of the eye through altered cell contacts [14]. In Caenorhabditis elegans, ZO-1 contributes to the strength of intercellular adhesion junctions, which are disrupted under stress of morphological movements during embryogenesis of ZO-1 mutants [17].
ZO-1 also interfaces with components of the cellular signalling pathways to regulate proliferation and cell fate. For example mammalian (ZO-1) protein participates in a sensing mechanism that mediates cell density-dependent control of cell proliferation and cell signalling [18–20]. In Drosophila, pyd mutants affect wing growth through an interaction with the Hippo pathway and cell fate through misregulation of Notch signalling [7]. The pyd mutants result in either loss or gain of Notch activity depending on tissue context. In the notum of pyd mutants Notch activity is reduced, resulting in extra sensory macrochaetae. However in the ovary pyd mutants result in increased Notch activity, reflected in an enlarged stem cell niche [7]. Su(dx) mutants prevent the enlargement of the niche which occurs when the fly is heterozygous for a pyd null allele. This antagonistic genetic interaction may reflect a direct regulatory interaction since Su(dx) can form a complex with Pyd. Furthermore, the Su(dx) mutation does not suppress the homozygous null pyd phenotype suggesting that Su(dx) acts negatively on Pyd rather than vice versa [7]. How Pyd then impacts on Notch signalling in niche cells is not understood.
Here we found that pyd mutants upregulate the ectopic ligand-independent Notch signal induced by the overexpression of Dx in the Drosophila wing. We further found that functional Dx is required for full stem cell niche development and that dx mutants suppress the null pyd niche phenotype. The function of dx in the niche is contingent on the presence of Su(dx) since double dx, Su(dx) mutants produced the opposite outcome of an enlarged niche, accompanied by decreased Notch endocytosis. In contrast, the enlargement of the niche of pyd mutants was associated with efficient Notch endocytosis in developing ovaries. We suggest therefore that the niche expansions observed in these different genetic backgrounds are associated with different mechanisms of increased Notch activity, respectively through alternative ligand-dependent or Dx-dependent routes. Using Drosophila S2 cells in which different forms of Notch activity can be studied separately, we found that Pyd downregulated Dx-driven Notch signalling, but had little effect on ligand-activated Notch. We found that Pyd forms a complex with Dx and Notch at the cell surface and acts to suppress Dx-induced Notch endocytosis. As with other reported actions of ZO-1 proteins [18–20], we found that in cell culture Pyd was recruited to the cell surface at high cell density and acted in a density-dependent manner to suppress Dx-induced Notch endocytosis and signalling. Our findings therefore implicate Pyd as an upstream modulator of the late endosomal Dx-induced Notch activation mechanism and establish that, in principle, external cues can modulate Notch signalling levels via Dx regulation.
2. Results
2.1. Pyd suppresses Dx-induced Notch activation
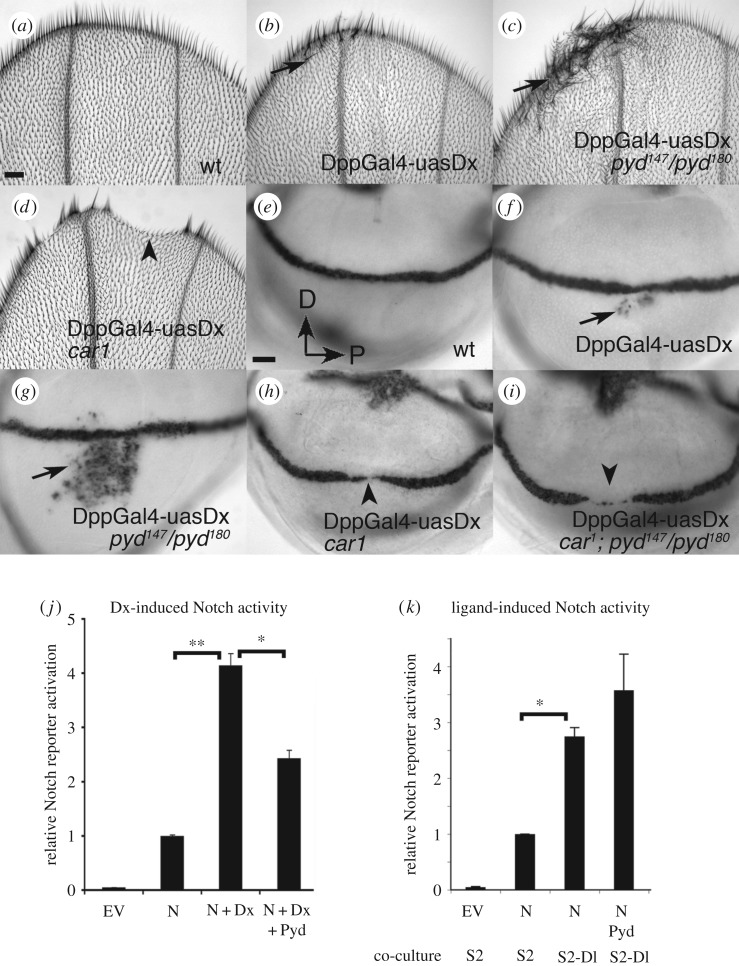
In the wing imaginal disc, Notch signals at the boundary of the dorsal and ventral wing compartments to induce wingless expression and regulate wing margin formation [21]. Notch signalling can be induced independently of its ligands by the overexpression of Dx in the wing imaginal disc [2]. The latter results in ectopic wingless expression in the imaginal discs which is reflected in extra wing margin sensory bristles in the adult wing (figure 1a,b,e,f). We found that pyd mutants result in a strong increase in these Notch activation phenotypes, exhibiting increased levels of ectopic wingless expression and consequently a higher number of ectopic sensory bristles (figure 1c,g). Dx activates an endosomal Notch activation pathway that depends on the late endosomal HOPS component car [2]. When Dx is expressed in a car mutant background the ectopic signalling is suppressed and there is additionally a downregulation of endogenous Notch activity with reduced wingless expression (figure 1d,h). Thus Dx can act negatively on Notch if the latter's trafficking to the late endosome is suppressed. We therefore investigated whether the upregulated Dx-induced signal resulting from pyd mutation was also sensitive to loss of car. Expressing Dx in a car, pyd double mutant background also blocked ectopic Notch signalling, and further exhibited clear gaps in the expression of wingless at the dorsal–ventral boundary of the imaginal discs (figure 1i). Thus loss of Pyd upregulates the late endosome-dependent Notch signal.
Figure 1.
Pyd suppresses Notch signalling induced by Dx expression. Adult distal wing tips showing a wild-type (wt) margin (a), and ectopic margin bristles (arrows) induced by overexpression of Dx with Dpp-Gal4, in a wt background (b), or pyd147/pyd 180 null (c). (d) shows notching associated with overexpression of Dx in a car1 mutant (arrowhead). (e–i) Late third instar wing imaginal discs stained for wingless mRNA expression to report Notch activation. In a wt disc (e) there is a clear line of wingless expression demarcating the dorsal/ventral boundary. (f–i) Dx overexpressed along the A/P axis using the Dpp-Gal4 driver. (f) Mild ectopic wingless expression (arrow) induced in wt background. (g) Enhanced wingless expression in pyd null mutant. Overexpression of Dx in a car1 mutant (h) or in a car1, pyd double mutant (i), leads to a loss of wingless (arrowheads). Scale bar in (a) represents 40 µm in (a–d). Scale bar in (e) represents 20 µm in (e–i). (j) Pyd suppresses Dx-induced signalling in S2 cells. S2 cells were transfected with empty vector (EV) or as indicated. Normalized mean Notch reporter activity measured by luciferase reporter expression is relative to cells expressing Notch only. Error bars are s.e.m., n = 3, * or ** indicates p < 0.05 by Student t-test for comparisons as indicated. (k) Pyd does not suppress ligand-induced signalling. Luciferase assay in S2 cells showing the consequence of Pyd expression on ligand-induced Notch activation. Normalized mean Notch reporter activity is shown relative to cells expressing Notch co-cultured with non-expressing S2 cells. S2 cells were transfected as indicated. Data represent mean of five experimental repeats, each performed in triplicate. Error bars are s.e.m., * indicates p < 0.05, Student t-test for comparison as indicated on graph.
To further investigate the action of Pyd on Dx-induced Notch signalling, we used a previously established cell culture assay that enables us to monitor signalling by using a Notch responsive reporter construct driving luciferase expression [4]. In our cell culture assays we are able to measure Dx-induced signalling separately from any ligand-induced component depending on whether Notch is coexpressed with Dx or if Notch expressing cells are presented with ligand-bearing cells [4]. As previously shown [4], the coexpression of Dx with Notch upregulated Notch signalling observed in S2 cells. The additional expression of Pyd reduced this Dx-induced signal (figure 1j). In contrast Pyd expression did not have any significant effect on Notch activity initiated by exposure to ligand-bearing cells (figure 1k).
2.2. Pyd and Dx regulate cap cell recruitment to the germline stem cell niche
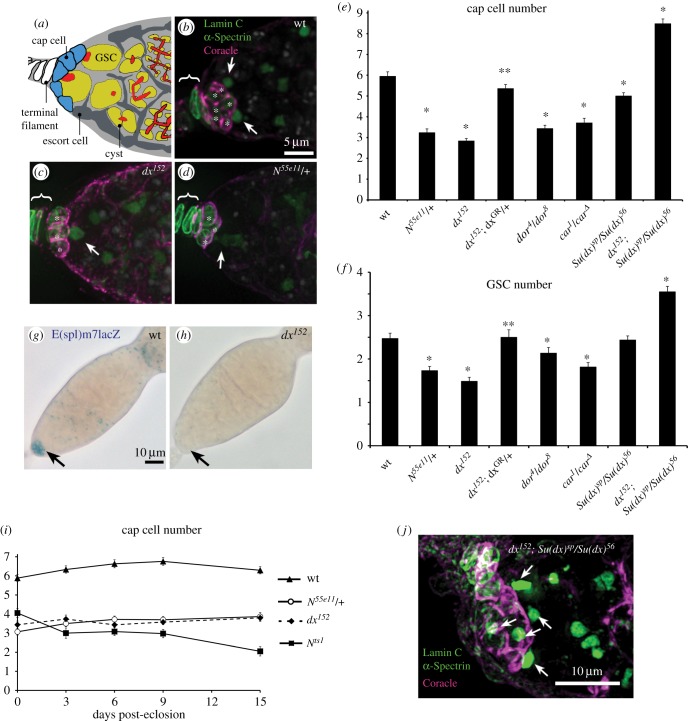
To examine the physiological relevance of the Pyd/Dx functional interaction we examined genetic interactions between these components in vivo. Notch signalling has been shown to control niche size during development, as it is activated in the somatic cell niche precursors by Delta ligand expressed in the terminal filament. The precursor cells differentiate as cap cells that constitute the niche, which is subsequently populated by 2–3 germline stem cells (GSCs) per ovariole (figure 2a) [22–24]. Pyd has previously been identified as a negative regulator of Notch signalling in the cap cells of the Drosophila ovary GSC niche. Pyd mutants display an increased Notch signalling level and increased niche size [7]. In contrast we found that dx mutants have the opposite phenotype showing reduced niche size, populated by fewer GSCs (figure 2a-c,e,f), an outcome similar to the removal of one copy of the Notch gene (figure 2d,e,f). Addition of a Dx genomic rescue construct (DxGR, TX05B) [25] reverted the dx phenotypes (figure 2e,f). We next investigated a Notch reporter, E(spl)M7-lacZ [7], and found reduced Notch signalling in the cap cell niche of dx mutants (figure 2g,h). A key distinguishing feature of the Dx-dependent component of Notch activation is the requirement for HOPS complex components such as Carnation (Car)/VPS33 and Deep orange (Dor)/VPS18 which are involved in late endosomal/lysosomal fusion [2,4,26]. We found that mutations in these components also reduced niche size (figure 2e,f).
Figure 2.
Dx regulates the size of the ovary niche. (a) Diagram of the Drosophila GSC niche showing terminal filament (white), cap cells (blue) and escort cells (dark grey). GSCs (yellow) are attached to cap cells and contain the spectrosome at their anterior membrane (red). Dividing cysts (yellow) show the branching fusome (red) extending through their cytoplasm. (b–d) Germaria labelled for DAPI (grey), anti-Coracle (magenta) strongly marking cap cell membrane and weakly terminal filament and escort cells, anti-α-Spectrin marking the GSC spectrosome (arrows, green) and Lamin C weakly marking cap cells and but more strongly the TF (also green). Images (b–d) represent a merged stack of deconvolved layers. (b) wt niche. (c) dx152 and (d) N55e11/+ niches showing reduced number of cap cells (asterisks) and GSCs compared to wt. (e,f) dx152 and N55e11 have reduced number of cap cells (e) and GSCs (f) compared to wt. dx152 phenotype is rescued by genomic rescue construct (dxGR). HOPS complex mutants, dor and car mutations also reduce niche size and stem cell numbers. * and ** indicate p < 0.05 by two tailed Mann–Whitney U test, respectively compared with wt and dx152. (g,h) E(spl)m7-lacZ expression seen in wt cap cells (arrow) (g) is reduced in dx152 (h). (i) Adult cap cell number for dx152 and N55e11/+ over time since eclosion is stable but cap cell numbers reduce when strong Notch loss of function is induced by shift of temperature-sensitive Nts1 to 29°C as previously reported [22]. Error bars (e,f,i), s.e.m. (j) expanded niche of dx152; Su(dx)sp/Su(dx)56 ovaries, arrows indicate associated spectrosome-containing GSCs. Scoring of cap cell and GSC numbers for this genotype is shown in (e,f).
In the adult niche Notch also plays a role in niche maintenance [22–24]. When Notch activity is switched off in the adult using a temperature-sensitive mutant (Nts1), the size of the niche declines due to loss of cap cells (figure 2i) [22–24]. To determine the contribution of dx to these distinct Notch functions, adults were dissected immediately after eclosion and at various adult ages up to 15 days. In dx null flies, the niche size was reduced in newly eclosed flies but remained stable as adult flies aged (figure 2i). Thus loss of dx specifically reduces cap cell recruitment but does not affect subsequent adult niche maintenance. Flies heterozygous for a null allele of Notch also eclosed with a similarly reduced niche size, which also did not further decline with age (figure 2i). Thus the developmental role of Notch in niche formation is more sensitive to reduction of signalling activity, making the contribution of Dx more critical during this phase. Because of previously reported close functional relationship between Dx and Su(dx) activities, we investigated how the dx phenotype was affected by mutations removing Su(dx). Su(dx)sp/Su(dx)56 mutant ovaries showed only a weakly reduced niche size compared to wild-type controls but maintained a similar GSC number (figure 2e,f). However, we found that in flies double mutant for dx and Su(dx) the phenotype of both mutants was reversed, instead producing an increased niche size (figure 2e,f,j). We have previously reported similar reversals in the direction by which Dx acts on Notch contingent on Su(dx) gene copy number, which likely reflects opposing activities on the ligand-dependent and -independent modes of Notch activation [4].
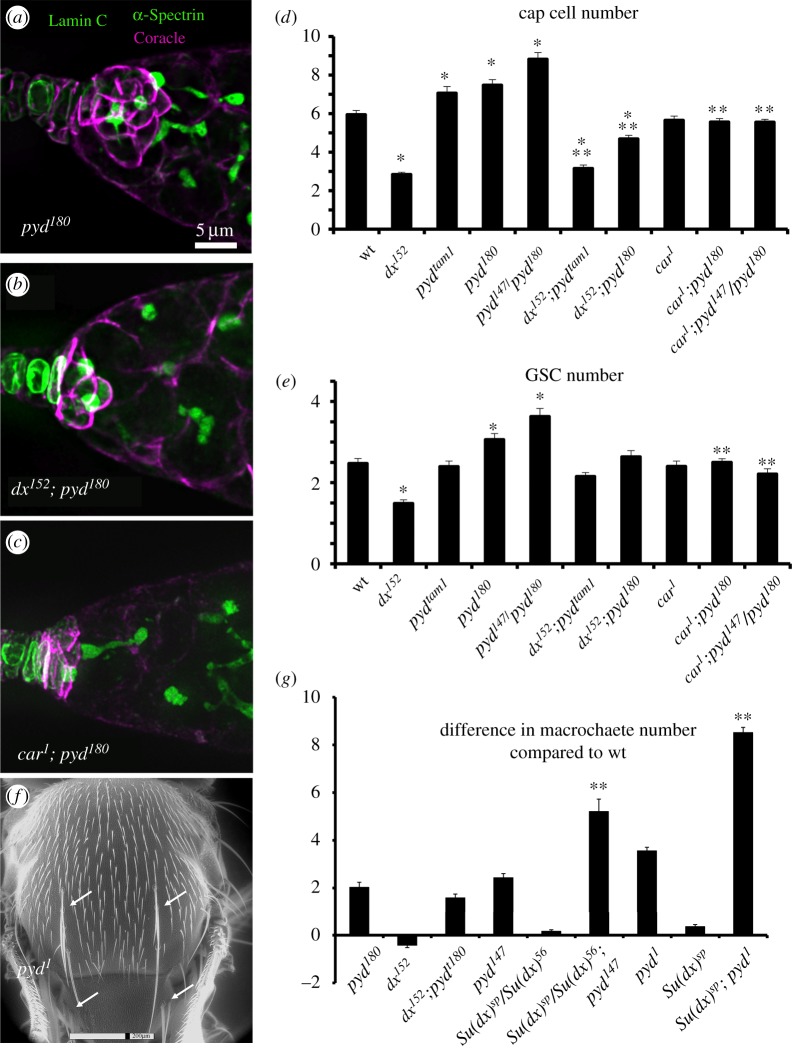
Having established the requirement for Dx-dependent Notch signalling in the full development of the niche, we next investigated genetic interactions with pyd mutants. The pyd mutants displayed an increased number of cap cells and higher GSC numbers for the stronger alleles. We found that introducing a mutation of dx reduced the increased niche size of pyd mutant ovarioles and reduced the number of GSCs although the latter was not statistically significant (figure 3a,b,d,e). However the car1 mutation significantly rescued both the expanded pyd niche and GSC phenotypes (figure 3c,d,e). Thus Pyd normally opposes a Notch signal in the GSC niche that depends on late endosome trafficking, consistent with a model in which ZO-1 acts negatively on the Dx-dependent signalling pathway. In contrast, dx mutants did not have a significant effect on the pyd bristle phenotype (figure 3f,g). Furthermore, while a suppressive interaction of Su(dx) with pyd mutants occurs in the ovary niche [7], the Su(dx) mutation enhanced the bristle gain phenotype of null pyd mutants (figure 3f,g). The differences in the interplay between these mutations suggests an alternative regulatory network involving these components is likely involved in developmental contexts where Pyd acts positively to support Notch signalling levels instead of negatively as it does in the ovary stem cell niche.
Figure 3.
Pyd antagonizes Dx function in the ovary stem cell niche. (a–c) shows the GSC niche region of ovarioles stained for anti-Coracle (magenta), anti-Lamin C (green) and anti-α-Spectrin (also in green, marking the GSC spectrosomes). (a) pyd180 has an enlarged GSC niche; this is reduced to below wild-type levels in flies additionally mutant for dx152 (b). The hypomorphic car1 also reduces the pyd expanded niche phenotype (c). The scale bar in (a) represents 5 µm in (a–c). (d) Genetic interactions between pyd and dx or car mutations. pyd mutants have more cap cells than wild-type. dx152, pyd double mutants have cap cell numbers below the wild-type number. car1 also prevents the pyd niche expansion phenotype. (e) GSC number is reduced by the dx mutation and increased by the stronger pyd mutant alleles. GSC number of pyd mutants is significantly reduced by car1. * and ** indicate p < 0.05 by two tailed Mann–Whitney U test respectively compared to wt and comparative pyd mutations combinations; n = 14 to 48 individuals scored for each genotype. (f) environmental scanning EM of pyd1 mutant fly indicating ectopic macrochaetae phenotype on notum and scutellum (arrows). (g) Scoring of macrochaetae phenotypes as difference compared to wild-type. ** indicates p < 0.05 for comparison with appropriate pyd mutant genotype (Student t-test, n = minimum of 25 flies scored for each genotype. Error bars (d,e,g), s.e.m.
2.3. Pyd alters Dx-regulated Notch trafficking in cells
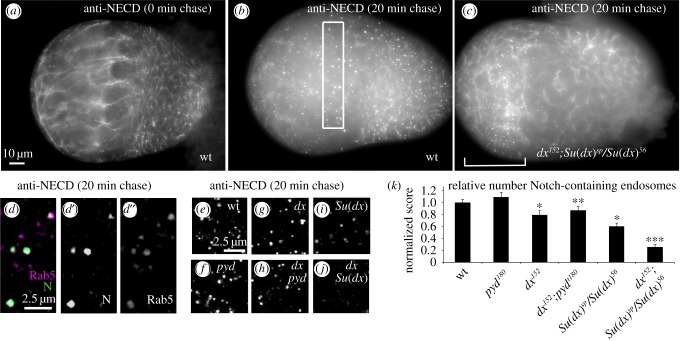
We next investigated how Pyd might regulate Dx-activated Notch signalling. Dx has been shown to act by promoting entry of Notch into an endocytic trafficking pathway [2–4] and we wondered if pyd mutants might increase the amount of Notch endocytosis. This suggestion is somewhat contrary to a previous report of an accumulation of adherens junction Notch in pyd mutants in wing imaginal discs, which might imply that Notch endocytosis is reduced in a pyd mutant [7]. However this apparent accumulation of Notch may more reflect apical domain expansion rather than decreased trafficking [7]. Indeed we found that endocytic uptake of Notch in pyd mutant ovary tissue was not impaired (figure 4), although we were not able to detect a statistically significant increase in the number of Notch containing endosomes compared to wild-type. However, since it was not straightforward to quantitate the total amount of Notch present in the endosomes and because Notch can enter the cell by multiple mechanisms, both Dx-dependent and independent [4], we reasoned that our in vivo assays may not be sufficiently specific to detect pyd mutant outcomes affecting a subset of endocytosed Notch, although at least a proportion of Notch endocytosis occurring in both wild-type and pyd mutant backgrounds is Dx-dependent (figure 4k). This redundancy of Notch entry routes is highlighted by the consequences of removing Dx and Su(dx) function. While dx and Su(dx) mutants each alone resulted in moderate reduction of Notch endocytosis, when the mutants were combined there was an additive effect resulting in loss of most Notch containing endosomes and a corresponding clear surface localization of Notch even after a 20 min endocytic chase period (compare figure 4b and c). Increased ligand-induced signalling through the surface accumulated Notch is the likely explanation for the enlarged niche phenotypes observed in the double mutant case. Notably the loss of endocytosis observed in the Su(dx) and dx double mutant tissue was regionally dependent, as within the terminal filament region (figure 4c) a substantial amount of Notch-containing endosomes was observed, indicating that still further means by which Notch can be internalized into the cell remain to be characterized.
Figure 4.
Notch endocytic uptake assay in developing ovary. (a) 0 h after puparium formation, wt ovary showing Notch staining with anti-NECD with 0 min chase displaying cell surface Notch localization. (b) Wild-type ovary, after 20 min chase. Notch localization is predominantly in endosomal locations. Box represents region just posterior to terminal filaments used for scoring of Notch-containing endosomes in (e–k). (c) A dx152; Su(dx)sp/Su(dx)56 ovary after 20 min chase showing mostly cell surface localization and few Notch containing endosomes, apart from terminal filament region indicated by bracket. (d) Merged image showing localization of Notch (magenta) in Rab5 positive (green) endosomes after 20 min chase. (d',d'') show single channel images of Notch and Rab5 respectively. (e–j) Representative 50 layer merged images to illustrate the differences in endocytosis between the indicated mutant backgrounds. (k) Scoring of mean number of Notch-containing endosomes per standardized unit volume, counted using ImageJ and normalized to wt. * indicates p < 0.05 compared to wt, ** indicates p < 0.05 compared to pyd180, *** indicates p < 0.01 compared to dx152 or compared to Su(dx)sp/Su(dx)56, two tailed Mann–Whitney, n = 8 to 22. Error bars, s.e.m.
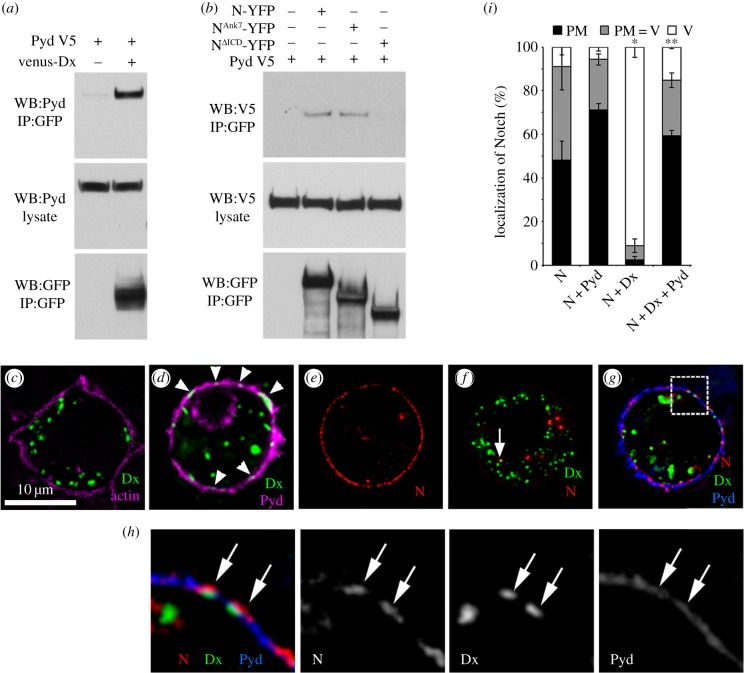
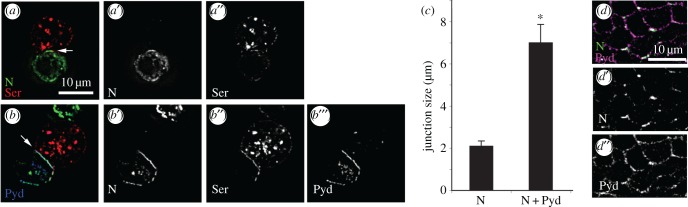
To investigate the consequences of Pyd on Notch in a simpler experimental system, we used a cell-culture model in which the source and route of Notch endocytosis can be controlled by the expression of specific regulatory components [4]. To determine whether Pyd and Dx exist in a complex we used a coimmunoprecipitation assay and found that Pyd coprecipitates with Dx (figure 5a). We similarly found an interaction between Pyd and Notch (figure 5b). When coexpressed in S2 cells, some Dx staining puncta became colocalized in discrete foci at the cell surface together with Pyd, a surface localization not observed when Pyd was not expressed (figure 5c,d). To investigate the mechanism by which Pyd might block Dx-induced Notch signalling, we performed a pulse chase Notch endocytosis assay [4] using antibodies to the Notch extracellular domain. When Notch is expressed alone in S2 cells it is largely localized at the cell surface (figure 5e). Expression of Dx greatly increases Notch internalization into the endocytic pathway (figure 5f). Although Pyd has a wide distribution around the cell periphery, we found that Notch, Pyd and Dx are coincident at discrete foci at the cell membrane (figure 5g,h) and Pyd expression reduces Notch endocytosis (figure 5i). Pyd also colocalized with Notch in the absence of Dx and this was particularly evident when Notch was clustered at cell contacts when forming junctions with ligand-bearing cells in co-culture experiments (figure 6a,b). Indeed Pyd expression increased the junction length of ligand/Notch contact regions (figure 6c), and this may explain the small, although not statistically significant, increase in ligand-induced signalling observed in cell signalling assays (figure 1k). Notch and Pyd also colocalized at discrete, punctate locations at cell–cell contacts in vivo (figure 6d).
Figure 5.
Pyd interacts with Dx to antagonize Dx-induced Notch trafficking. (a) Coimmunoprecipitation of Dx and Pyd. S2 cells were transfected as indicated. Venus-tagged Dx was immunoprecipitated using GFP-TRAP beads and detected using anti-GFP. Pyd protein was detected using anti-Pyd. (b) Notch and Pyd coimmunoprecipitate. YFP-tagged Notch proteins were immunoprecipitated using GFP-TRAP beads and detected using anti-GFP. V5-tagged Pyd was detected using anti-V5. Interaction requires Notch intracellular domain but the region C-terminal to Notch Ankyrin repeats is dispensible for complex formation. (c,d) Recruitment of Dx to plasma membrane by Pyd in S2 cells. (c) S2 cells were transfected with venus-Dx (green) and costained for actin (magenta), (d) venus-Dx (green) cotransfected with Pyd-V5 (magenta). Arrowheads in (d) indicate Dx puncta at the plasma membrane that are seen only with Pyd coexpression. Scale bar, 10 µm. (e-i) Consequence of Pyd expression on Notch endocytic trafficking assayed by uptake of bound anti-NECD antibody. (e) Notch expressed alone in S2 cells is mostly localized to cell surface after 1 h chase. (f) Co-expression of Dx induces endocytic localization of Notch, with Notch only colocalizing with Dx in a small proportion of endosomes after a 1 h chase (arrow). (g) When additionally coexpressed with Pyd, Notch is retained at the plasma membrane. Box in (g) is magnified in (h) to show colocalization of Notch, Dx and Pyd at plasma membrane (arrows in h), separate channels shown in greyscale. (i) Notch ECD antibody localization after 1 h chase in each cell was scored as % cells displaying plasma membrane (PM), vesicular (V), or intermediate (PM = V) Notch localization. Data show mean % localization from three separate experiments. Each experimental repeat quantified Notch localization in a minimum of 80 cells. * indicates p < 0.001 compared to cells transfected with Notch only, ** indicates p < 0.001 compared to cells transfected with N and Dx. Statistical significance determined by two tailed Mann–Whitney U test. Error bars are s.e.m.
Figure 6.
Pyd increases cell–cell contact size of Notch and Serrate (Ser) expressing cells. (a) Notch-Ser co-clustering is observed when Notch-expressing cells (green) and Ser-expressing cells (red) were mixed (arrows). (b) Co-clustering was promoted when Notch cells were expressing GFP-tagged Pyd resulting in longer junctional interfaces. (a,b) Merged pictures of Notch (green), Ser (red) and Pyd (blue). Notch staining (a',b'), Ser staining (a″,b″) and Pyd-V5 staining (b′′′) are shown separately (grey scale). (c) Quantified mean junction length of Notch/Ser interface. n = 27–35, error bars, s.e.m., * indicates p < 0.05, Student t-test). (d) Immunostaining showing colocalization of Notch (green) and Pyd (magenta) at discrete locations around cell boundaries in 0 hours APF ovary. (d',d'') show single channels in grey scale.
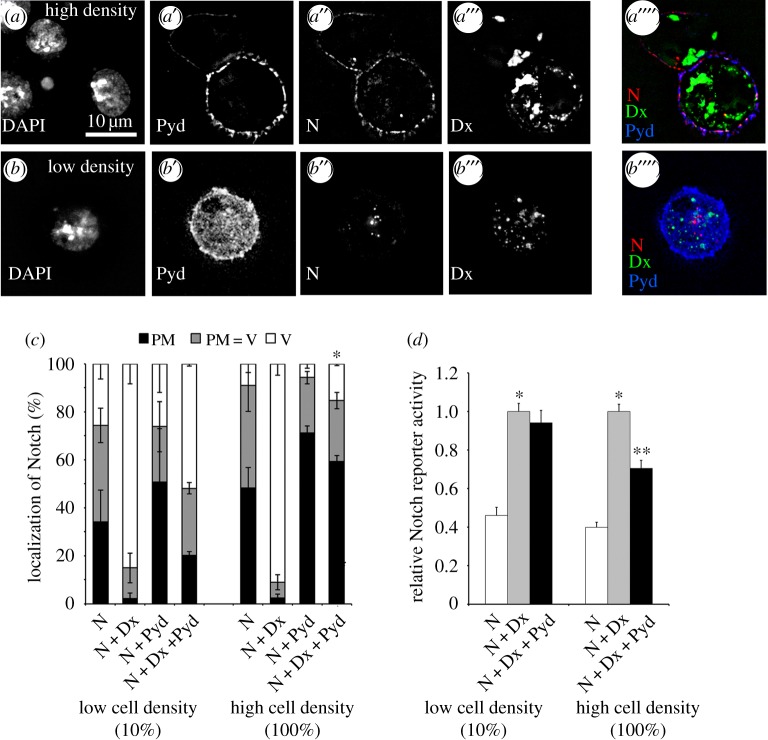
A characteristic feature of mammalian ZO-1 activity is that it has previously been shown to mediate cell density-dependent signal regulation [18,19,27]. We wondered whether Dx-induced Notch signalling would be similarly modulated. Pyd was only strongly localized to the cell membrane at high cell densities, and we found that Pyd expression was less effective at reducing Dx-induced Notch endocytosis at low cell density and also did not significantly suppress Dx-induced Notch signalling (figure 7a–d). Dx mediated signalling, at least in a cell culture context, can therefore be regulated upstream by the cell's external environment through Pyd surface accumulation. Thus in principle Pyd regulation of Notch may act as an alternative means by which external cues can tune the Notch signal output by Dx-regulated Notch trafficking.
Figure 7.
Cell density modulates Pyd regulation of Notch. (a–c) Notch uptake assay at low and high cell density. Transfected cells were confluent (a, high cell density) or diluted 1/10 with medium (b, low cell density) before reseeding, and left 24 h before performing Notch antibody uptake endocytosis assay. (a,b) Representative images of Notch uptake assay at respectively high and low cell density. DAPI, Pyd, Notch and Dx staining are shown in greyscale in (a–a'' and b–b'') respectively. (a"',b"') Merged staining of Notch (red), Dx (green) and Pyd (blue). Pyd is recruited to cell membrane at high cell density. (c) Notch ECD antibody localization after 1 h chase in each cell was scored as plasma membrane (PM), vesicles (V), or intermediate (PM = V). Pyd blocks Dx-induced Notch endocytosis at high cell density but has little effect at low cell density. Data show mean % localization from three separate experiments, error bars are s.e.m.. For each experimental repeat Notch localization was classified in a minimum of 80 cells. Pyd blocks Dx-induced Notch endocytosis at high cell density but is less effective at low cell density. * indicates p < 0.001 for mean % vesicular localization compared to N + Dx + Pyd at low cell density. (d) S2 cells were transfected as indicated and seeded at low or high density. Normalized mean Notch reporter activity is shown relative to cells expressing Notch and Dx. * indicates p < 0.001 compared to N only. ** indicates p < 0.002 compared to N + Dx. Experiments were performed a minimum three times, each with technical triplicates. Error bars, s.e.m. Statistical significance determined by two tailed Mann–Whitney U test.
3. Discussion
Notch signalling can be activated by ligand-dependent or -independent mechanisms [2–4,28]. The latter can be induced by the activity of Dx and, unlike ligand-induced signalling, this mode has a strict requirement for late endosomal Notch trafficking. Although it appears that the Dx-promoted activity of Notch can contribute to full Notch signalling levels during development, it has not been clear whether this alternative form of signalling can itself be modulated in response to upstream inputs. Scaffold adaptor proteins play key roles in enabling cross talk between junctional complexes and signalling pathway components through multiple binding sites [29]. ZO-1 is a conserved adaptor protein that associates with adherens and tight junctions [30]. Pyd, the Drosophila ZO-1 homologue, acts both positively and negatively on Notch signalling depending on developmental context [7]. Here we uncover a mechanism by which Pyd can downregulate Notch activity by suppressing the ligand-independent Dx-driven mechanism of Notch signal activation. Thus Pyd can both regulate Notch signalling levels and select between alternative means of its activation.
We found that Pyd could form a complex with both Dx and Notch and can block signalling by reducing Dx-induced Notch endocytosis. At present the molecular mechanism by which this block on endocytosis occurs is unclear. It is possible that binding prevents an interaction of some unidentified accessory protein, or Notch is sequestered in a membrane protein complex with ZO-1 in a way that prevents Notch assembly into clathrin-coated vesicles. This may represent a similar activity to that previously reported for mammalian ZO-1, whose dissociation from the gap junction complex is linked to the latter's endocytic internalization into the cell [31,32]. In cell culture we found that the ability of full-length Pyd to block Dx-induced endocytosis and signalling also depended on high cell density whereupon Pyd became localized to the cell surface. Thus these cells can sense their local environment to regulate Notch independently of its ligands through Pyd.
To determine if there was a functionally relevant interaction between Pyd and Dx in vivo we investigated genetic interactions between the genes in ovary stem cell niche development. Pyd-dependent suppression of Notch signalling is normally required during the assembly of the GSC niche in the Drosophila ovary to limit niche size [7]. The ovary GSC niche assembles following the larval-pupal transition, after the formation of the terminal filament stacks that prefigure ovariole formation. At this time somatic cells intercalate between the terminal filaments and the germline progenitor cells, and are recruited to differentiate as cap cells which exit mitosis and subsequently provide anchorage for the GSCs. Notch signalling is required in the cap cells for their recruitment by the terminal filament, and up or downregulation of Notch increases or reduces niche size respectively [22–24]. Delta is expressed in the terminal filament and ligand-stimulated Notch signalling contributes to cap cell recruitment and niche formation. Despite this we found Dx is also required for full niche assembly during development. Furthermore, pyd mutants increase Notch signalling and niche size and this is dependent on Dx and also Carnation, a HOPS complex component required for Dx but not ligand-induced Notch activation [2]. Thus we infer that the ligand-independent signalling mode also contributes to the establishment of sufficient cells with cap cell identity to generate the full niche size and normal GSC population level. We further speculate that assembly of cell–cell contacts and the associated junction proteins during assembly of the germline niche ultimately helps determine the population size of the GSCs by dampening this ligand-independent signalling component. We should note however that our in vivo data do not rule out the possibility that there may be additional functions for Pyd in the niche that are not Dx-dependent. The population of the niche by GSCs may depend on a number of factors including the number of cap cells and adhesiveness of GSC/cap cell contacts, as well as the morphology of the niche, which may explain why effects of mutant combinations on GSC number were not as clear in some mutant combinations compared to cap cell number.
Recent work has shown that another apical localized protein, Crumbs, can also act to suppress a Dx-dependent Notch activity in the wing disc through an extracellular domain, which also suppresses endocytosis [33]. This recently published work differs from ours in that they observed a visible increase in Notch endocytosis in crumbs mutant clones in wing disc tissue. The endocytic removal of Notch in crumbs mutants is not prevented by loss of Dx but instead Notch accumulates in the apical region of the cell. They infer that Dx in this case is required for further progression of Notch through the endocytic pathway. This is curious because dx mutants in an otherwise wild-type background cause clear surface accumulation of Notch at the membrane of imaginal wing disc epithelial cells and Dx overexpression causes increased Notch endocytosis. We and others have shown previously that Dx can act in two locations in the cell, to promote Notch endocytosis and to direct Notch to the late endosomal limiting membrane [2–4]. It is possible therefore that Pyd and Crumbs act on separate initial endocytic entry routes into the cell, respectively Dx-dependent and -independent, but downstream the endocytic flux converges on Dx-dependent activation.
In the light of the above discussion it is interesting to contemplate the relationship between Su(dx), Dx and Pyd in the different contexts in which Pyd acts negatively and positively on Notch. In the niche, Su(dx) loss of function appears to increase Pyd activity such that one copy of Pyd is sufficient to produce a normal size niche [7]. The effect of Su(dx) mutation could thus indirectly be antagonistic to Dx-promoted Notch activity since Pyd itself opposes Dx function. However Su(dx) also acts directly on Notch and competes with Dx activity to divert Notch into an alternative endocytic internalization route [4]. Thus in this case Su(dx) mutations could be expected to increase Dx efficacy. This balance of opposing activities may explain why Su(dx) mutants alone have little effect on niche size. Interestingly, when flies are simultaneously homozygous for both dx and Su(dx) mutants there is a phenotypic switch to an increased niche size. Thus both Dx and Su(dx) can each function to either promote or antagonize niche size depending on the genetic background in which they are placed. We have observed similar reversals of phenotype before and attribute this behaviour to the fact that both genes act negatively on ligand-induced Notch signalling [4]. Hence removing both endocytic entry routes increases the availability of Notch for ligand-induced activation, which outweighs any reduction in ligand-independent signalling that results from loss of dx. In this light it is interesting to contemplate that the expanded niche phenotypes arising from the different mutant causes reported here are associated with different Notch sub-cellular distributions and this is likely linked to different cellular locations of Notch activation.
In the peripheral nervous system, where Pyd has the opposite activity to promote Notch activity [7], we found that dx mutants did not significantly suppress the Pyd phenotype and the latter was enhanced by loss of Su(dx) rather than suppressed. Thus in this case Su(dx) and Pyd may act cooperatively through an unknown mechanism to suppress bristle formation. It is possible that in this Notch-promoting context Pyd function acts to stimulate ligand-dependent Notch signalling. Although we did not observe direct evidence for the latter in cell culture Notch reporter luciferase assays, we did find that Pyd promoted Notch/ligand contacts at cell interfaces. It is possible that this in vitro observation may reflect an activity that normally promotes ligand-dependent signalling in an in vivo context where cells are apically polarized and form adherens junctions.
Our results therefore illustrate the complexity by which Notch regulation is integrated into the morphology and functioning of the cell, with endocytic networks interfacing with protein networks controlling cell architecture and cell interactions to tune signalling to physiologically appropriate levels. The complexity of these networks is hence revealed in the often counterintuitive phenotypic outcomes of different combinations of mutants that we observe.
4. Material and methods
4.1. Drosophila stocks
All experiments were at 25°C on standard Drosophila culture medium. car1, N55E11 (Bloomington Stock Center), dx152 [34], dxGR TX05B [25], Su(dx)sp, Su(dx)56 [35], E(spl)m7-lacZ, pyd147, pyd180 [7], pydtamou (tam1) [5]; carΔ [26], y1w67c23 (Bloomington Stock Center) was used as wild-type (wt).
4.2. S2 cell culture
S2 cells were grown at 25°C in Schneider's medium (Invitrogen), with 10% FBS (Hyclone) and antibiotics. S2 cells were transfected using standard Effectene (Qiagen) or CaCl2 methods. Constructs: pUAST-Pyd-GFP as described previously [7]. Pyd constructs were also subcloned into pMT (Invitrogen), removing GFP and inserting a C-terminal V5-tag. pMT-Notch-YFP, pMT-Notch, pMT-NotchΔIntra-YFP, pMT-Notch-Ank7-YFP were derived from pUASTNotch-YFP (gift of K. Matsuno). pMT-NotchΔIntra-YFP and pMT-Notch-Ank7-YFP were generated by removing the intracellular domain, or the region after Ankyrin repeat 7 respectively. pMT-Venus-Dx and pMT-Dx-V5 were derived from pUAST-Venus-Deltex (gift from K. Matsuno). Other vectors were pUAST-EYFP-Rab7 (gift from M.P. Scott), pUAST-Ser and H/N (Heat-Shock inducible Notch) [36,37], and pMT-GAL4 (Drosophila Genomics Resource Center, Indiana).
4.3. Immunocytochemistry and histology
Primary antibodies were mouse anti-Notch extracellular domain (C458.2H, concentrate, 1 : 200, DSHB); mouse anti-Notch intracellular domain, (C17.9C6, concentrate, 1 : 1000, DSHB); rabbit anti-V5 (1 : 1000, Bethyl Laboratories); goat anti-Serrate (1 : 1000, Santa Cruz); guinea pig anti-Coracle (D4.1.3, 1 : 5000, R. Fehon); mouse anti-Lamin C (LC28.26, 1 : 20, DHSB), mouse anti-αSpectrin (3A9, 1 : 10, DSHB), rabbit anti-Pyd (1 : 2000) [7], rabbit anti-Rab5 (ab31261, 1 : 500 Abcam). Actin staining was performed with Alexa Fluor® 647 Phalloidin (1 : 100, Thermo Fisher Scientific).
Ovarioles were immunostained as previously described [23], except primary antibody incubation was at room temperature. S2 cells were grown on poly-lysine (Sigma) coated coverslips. Immunostaining was performed at room temperature. Cells were fixed in 4% formaldehyde (Polysciences) for 30 min, rinsed in PBS, permeabilized in PBS/Triton X-100 (PBS-tx) 0.2%, and blocked for 1 h in 3% skimmed milk/PBS, then incubated with primary antibody in blocking solution for 2 h, and washed in PBS before 1 h incubation with secondary antibody. Tissue and cell preps were washed in PBS-tx 0.1% and mounted in Vectashield with DAPI (Vector Labs). For cell culture Notch endocytic uptake assay, S2 cells grown on coated coverslips were incubated with C458.2H for 15 min on ice, washed with ice cold S2 medium and chased for 60 min at 25°C. Cells were fixed, permeabilized, and stained as above. Aggregation assay with Notch (expressed from H/N vector) and ligand (pUAST-Ser/pMT-Gal4) expressing cells was as described [36]. Images were captured using Volocity (Perkin Elmer) with an Orca-ER digital camera (Hamamatsu) mounted on a M2 fluorescent microscope (Zeiss). Deconvolution of 0.5 µm optical sections was performed with three nearest neighbours using Openlab (Improvision), or by iterative deconvolution (Volocity) and processed in Photoshop (Adobe). Distance measurements were performed with ImageJ.
For endocytic uptake assays in ovary tissue, preps were dissected at 0 h after puparium formation (APF) in ice cold Graces (Sigma) and incubated with anti-NECD (C458.2H, concentrate, DSHB) at 1 : 200 in 200 µl PBS in PCR tubes on ice. The sealed PCR tubes were placed in a 7 ml tube containing ice and gently rotated for 1 h at 10 r.p.m. in a cold room. The contents of the PCR tube were then transferred to a small sieve and rinsed three times with ice cold Graces followed by three 5 min washes in ice cold Graces. The preps were transferred to prewarmed Graces at 25°C for 20 min chase and then fixed for 20 min with 4% formaldehyde, washed three times with PBS-tx 0.1%, and permeabilized for 2 h in PBS-tx 0.3% and 4% normal donkey serum (NDS) (Jackson). The preps were incubated for 2 h in PBS + 4% NDS + anti-Vasa rat (1 : 300, S. Levine), washed in PBS-tx 0.1% three times for 5 min then three times for 15 min and treated with corresponding fluorescent-conjugated secondary antibodies. Preps were mounted between spacers, to avoid crushing, in Vectashield antifade Mounting Medium with DAPI (Vector Labs). Each prep was imaged with the same exposure using Z sections separated by 0.5 µm, and deconvolved with Openlab using the same parameters. Endosomal Notch-containing structures were counted using ImageJ in the region adjacent to the terminal filaments containing the nascent cap cell progenitors. A standardized area of 64 × 11.6 µm was scored for each section and the scores of 10 adjacent Z sections were used to derive a mean score for each 10 layer series. Three to five such sections were processed for each ovariole and a mean score for each ovariole calculated. All data were normalized to y1w67c23.
4.4. Immunoprecipitation and western blotting
For coimmunoprecipitation experiments, S2 cells were grown in 6-well dishes and transfected with pMT plasmids. CuSO4 was added after 24 h to induce expression and after a further 24 h cells were homogenized in lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM CaCl2) and protease inhibitor cocktail (EDTA-free Complete; Roche), and cleared by centrifugation. The lysate was incubated with 10 µl GFP-TRAP agarose (Chromotek) for 1 h at 4°C, and washed four times in lysis buffer. Bound proteins were eluted with sample buffer, run on 3–8% Nupage Gels (Invitrogen) and western blotted with rabbit anti-Pyd (1 : 10 000) [7], mouse anti-V5 (1 : 5000, Invitrogen) or rabbit anti-GFP (1 : 20 000, ImmunoKontact).
4.5. Notch luciferase reporter assay
S2 cells in 12-well dishes were transfected when approximately 50% confluent with combinations of pMT-Notch, pMT-DxV5, pMT-Pyd, NRE:Firefly (gift from S. Bray) and Actin:Renilla (gift from G.Merdes). After 24 h, when cells reached 100% confluence, they were re-suspended and seeded into white 96-well plates (Nunc #136101) and CuSO4 was added after a further 24 h. 24 h after induction, luciferase activity was assayed with Dual-Glo Luciferase (Promega), quantified by a luminometer (Berthold), and Firefly/Renilla ratio was calculated for triplicate samples. For cell-density experiments, cells were re-seeded into white 96-well plates as above (high cell density) or diluted 1/10 with culture medium before reseeding (low cell density). For ligand-induced signalling assays, Notch expressing cells were layered on top of fixed Delta expressing S2 cells (S2-Mt-Dl; DGRC) according to the previously described protocol [4]. Experiments were repeated a minimum of three times.
Acknowledgements
We thank Spyros Artavanis-Tsakonas, Sarah Bray, Rick Fehon, Gunter Merdes, Mark Metzstein, Matthew Scott, Ryu Ueda, Bloomington Drosophila Stock Center (University of Indiana), Developmental Studies Hybridoma Bank (University of Iowa) and Drosophila Genome Resources Center (University of Indiana) for reagents.
Competing interests
We declare we have no competing interests.
Funding
We thank the BBSRC (BB/M020797/1 and BB/P000215/1) for financial support. We also acknowledge technical support from Sanjai Patel and the Manchester Fly Facility.
References
- 1.Kopan R, Ilagan MX. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233. (doi:10.1016/j.cell.2009.03.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkin M, et al. 2008. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of Notch in the endosomal trafficking pathway. Dev. Cell 15, 762–772. (doi:10.1016/j.devcel.2008.09.002) [DOI] [PubMed] [Google Scholar]
- 3.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. 2011. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol. 195, 1005–1015. (doi:10.1083/jcb.201104146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu H, Woodcock SA, Wilkin MB, Trubenová B, Monk NA, Baron M. 2014. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell 157, 1160–1174. (doi:10.1016/j.cell.2014.03.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahisa M, Togashi S, Suzuki T, Kobayashi M, Murayama A, Kondo K, Miyake T, Ueda R. 1996. The Drosophila tamou gene, a component of the activating pathway of extra-macrochaetae expression, encodes a protein homologous to mammalian cell-cell junction-associated protein ZO-1. Genes Dev. 10, 1783–1795. (doi:10.1101/gad.10.14.1783) [DOI] [PubMed] [Google Scholar]
- 6.Chen CM, Freedman JA, Bettler DR Jr, Manning SD, Giep SN, Steiner J, Ellis HM. 1996. Polychaetoid is required to restrict segregation of sensory organ precursors from proneural clusters in Drosophila. Mech. Dev. 57, 215–227. (doi:10.1016/0925-4773(96)00548-5) [DOI] [PubMed] [Google Scholar]
- 7.Djiane A, Shimizu H, Wilkin M, Mazleyrat S, Jennings MD, Avis J, Bray S, Baron M. 2011. Su(dx) E3 ubiquitin ligase-dependent and -independent functions of Polychaetoid, the Drosophila ZO-1 homologue. J. Cell Biol. 192, 189–200. (doi:10.1083/jcb.201007023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umeda K, et al. 2006. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741–754. (doi:10.1016/j.cell.2006.06.043) [DOI] [PubMed] [Google Scholar]
- 9.Ando-Akatsuka Y, Yonemura S, Itoh M, Furuse M, Tsukita S. 1999. Differential behavior of E-cadherin and occludin in their colocalization with ZO-1 during the establishment of epithelial cell polarity. J. Cell. Physiol. 179, 115–125. (doi:10.1002/(SICI)1097-4652(199905)179:2<115::AID-JCP1>3.0.CO;2-T) [DOI] [PubMed] [Google Scholar]
- 10.Ikenouchi J, Umeda K, Tsukita S, Furuse M, Tsukita S. 2007. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J. Cell Biol. 176, 779–786. (doi:10.1083/jcb.200612080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki M, Irie K, Deguchi-Tawarada M, Ikeda W, Ohtsuka T, Takeuchi M, Takai Y. 2003. Nectin-dependent localization of ZO-1 at puncta adhaerentia junctions between the mossy fiber terminals and the dendrites of the pyramidal cells in the CA3 area of adult mouse hippocampus. J. Comp. Neurol. 460, 514–524. (doi:10.1002/cne.10653) [DOI] [PubMed] [Google Scholar]
- 12.Wei X, Ellis HM. 2001. Localization of the Drosophila MAGUK protein Polychaetoid is controlled by alternative splicing. Mech. Dev. 100, 217–231. (doi:10.1016/S0925-4773(00)00550-5) [DOI] [PubMed] [Google Scholar]
- 13.Fanning AS, Van Itallie CM, Anderson JM. 2012. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell 23, 577–590. (doi:10.1091/mbc.E11-09-0791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seppa MJ, Johnson RI, Bao S, Cagan RL. 2008. Polychaetoid controls patterning by modulating adhesion in the Drosophila pupal retina. Dev. Biol. 318, 1–16. (doi:10.1016/j.ydbio.2008.02.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung AC, Ribeiro C, Michaut L, Certa U, Affolter M. 2006. Polychaetoid/ZO-1 is required for cell specification and rearrangement during Drosophila tracheal morphogenesis. Curr. Biol. 16, 1224–1231. (doi:10.1016/j.cub.2006.04.048) [DOI] [PubMed] [Google Scholar]
- 16.Choi W, Jung KC, Nelson KS, Bhat MA, Beitel GJ, Peifer M, Fanning AS. 2011. The single Drosophila ZO-1 protein Polychaetoid regulates embryonic morphogenesis in coordination with Canoe/afadin and Enabled. Mol. Biol. Cell 22, 2010–2030. (doi:10.1091/mbc.E10-12-1014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockwood C, Zaidel-Bar R, Hardin J. 2008. The C. elegans zonula occludens ortholog cooperates with the cadherin complex to recruit actin during morphogenesis. Curr. Biol. 18, 1333–1337. (doi:10.1016/j.cub.2008.07.086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balda MS, Matter K. 2009. Tight junctions and the regulation of gene expression. Biochim. Biophys. Acta 1788, 761–767. (doi:10.1016/j.bbamem.2008.11.024) [DOI] [PubMed] [Google Scholar]
- 19.Balda MS, Matter K. 2000. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 19, 2024–2033. (doi:10.1093/emboj/19.9.2024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kausalya PJ, Phua DC, Hunziker W. 2004. Association of ARVCF with Zonula occludens (ZO)-1 and ZO-2: binding to PDZ-domain proteins and cell–cell adhesion regulate plasma membrane and nuclear localization of ARVCF. Mol. Biol. Cell 15, 5503–5515. (doi:10.1091/mbc.E04-04-0350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Celis JF, Garcia-Bellido A, Bray SJ. 1996. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122, 359–369. [DOI] [PubMed] [Google Scholar]
- 22.Bonfini A, Wilkin MB, Baron M. 2015. Reversible regulation of stem cell niche size associated with dietary control of Notch signalling. BMC Dev. Biol. 15, 8 (doi:10.1186/s12861-015-0059-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song X, Call GB, Kirilly D, Xie T. 2007. Notch signaling controls germline stem cell niche formation in the Drosophila ovary. Development 134, 1071–1080. (doi:10.1242/dev.003392) [DOI] [PubMed] [Google Scholar]
- 24.Hsu HJ, Drummond-Barbosa D. 2011. Insulin signals control the competence of the Drosophila female germline stem cell niche to respond to Notch ligands. Dev. Biol. 350, 290–300. (doi:10.1016/j.ydbio.2010.11.032) [DOI] [PubMed] [Google Scholar]
- 25.Busseau I, Diederich RJ, Xu T, Artavanis-Tsakonas S. 1994. A member of the Notch group of interacting loci, deltex encodes a cytoplasmic basic protein. Genetics 136, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akbar MA, Ray S, Krämer H. 2009. The SM protein Car/Vps33A regulates SNARE-mediated trafficking to lysosomes and lysosome-related organelles. Mol. Biol. Cell 20, 1705–1714. (doi:10.1091/mbc.E08-03-0282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siliciano JD, Goodenough DA. 1988. Localization of the tight junction protein, ZO-1, is modulated by extracellular calcium and cell–cell contact in Madin-Darby canine kidney epithelial cells. J. Cell Biol. 107, 2389–2399. (doi:10.1083/jcb.107.6.2389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baron M. 2012. Endocytic routes to Notch activation. Semin. Cell Dev. Biol. 23, 437–442. (doi:10.1016/j.semcdb.2012.01.008) [DOI] [PubMed] [Google Scholar]
- 29.Tepass U. 2012. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28, 655–685. (doi:10.1146/annurev-cellbio-092910-154033) [DOI] [PubMed] [Google Scholar]
- 30.Fanning AS, Anderson JM. 2009. Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann. N. Y. Acad. Sci. 1165, 113–120. (doi:10.1111/j.1749-6632.2009.04440.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilleron J, Carette D, Fiorini C, Benkdane M, Segretain D, Pointis G. 2009. Connexin 43 gap junction plaque endocytosis implies molecular remodelling of ZO-1 and c-Src partners. Commun. Integr. Biol. 2, 104–106. (doi:10.4161/cib.7626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilleron J, Fiorini C, Carette D, Avondet C, Falk MM, Segretain D, Pointis G. 2008. Molecular reorganization of Cx43, ZO-1 and Src complexes during the endocytosis of gap junction plaques in response to a non-genomic carcinogen. J. Cell Sci. 121, 4069–4078. (doi:10.1242/jcs.033373) [DOI] [PubMed] [Google Scholar]
- 33.Nemetschke L, Knust E. 2016. Drosophila Crumbs prevents ectopic Notch activation in developing wings by inhibiting ligand-independent endocytosis. Development 143, 4543–4553. (doi:10.1242/dev.141762) [DOI] [PubMed] [Google Scholar]
- 34.Fuwa TJ, Hori K, Sasamura T, Higgs J, Baron M, Matsuno K. 2006. The first deltex null mutant indicates tissue-specific Deltex-dependent Notch signaling in Drosophila. Mol. Genet. Genomics 275, 251–263. (doi:10.1007/s00438-005-0087-3) [DOI] [PubMed] [Google Scholar]
- 35.Cornell M, Evans DA, Mann R, Fostier M, Flasza M, Monthatong M, Artavanis-Tsakonas S, Baron M. 1999. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordle J, et al. 2008. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 15, 849–857. (doi:10.1038/nsmb.1457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieber T, Wesley CS, Alcamo E, Hassel B, Krane JF, Campos-Ortega JA, Young MW. 1992. Single amino acid substitutions in EGF-like elements of Notch and Delta modify Drosophila development and affect cell adhesion in vitro. Neuron 9, 847–859. (doi:10.1016/0896-6273(92)90238-9) [DOI] [PubMed] [Google Scholar]