Abstract
Host-intrinsic factors as well as environmental changes are known to be strong evolutionary drivers defining the genetic foundation of immunity. Using a novel set of teleost genomes and a time-calibrated phylogeny, we here investigate the family of Toll-like receptor (TLR) genes and address the underlying evolutionary processes shaping the diversity of the first-line defence. Our findings reveal remarkable flexibility within the evolutionary design of teleost innate immunity characterized by prominent TLR gene losses and expansions. In the order of Gadiformes, expansions correlate with the loss of major histocompatibility complex class II (MHCII) and diversifying selection analyses support that this has fostered new immunological innovations in TLRs within this lineage. In teleosts overall, TLRs expansions correlate with species latitudinal distributions and maximum depth. By contrast, lineage-specific gene losses overlap with well-described changes in palaeoclimate (global ocean anoxia) and past Atlantic Ocean geography. In conclusion, we suggest that the evolvability of the teleost immune system has most likely played a prominent role in the survival and successful radiation of this lineage.
Keywords: adaptive evolution, innate immunity, Toll-like receptors, gene loss, gene expansion, past climatic change
1. Background
The evolutionary success of ray-finned fishes (class Actinopterygii) is characterized by large species radiations [1]. Actinopterygii comprises an exceptionally diverse group of fishes with species inhabiting numerous aquatic habitats spanning from Arctic to Antarctic oceans, deep-sea benthos to the shore, along coastlines and rivers as well as freshwater systems. Moreover, the high degree of diversity is mirrored in the array of life-history strategies, morphological varieties, distinct migratory behaviour and reproductive strategies displayed ([2,3] and references therein). The teleost lineage is the largest within the class of ray-finned fishes [4]. Genome sequencing efforts of non-model organisms have provided new insight into the extreme diversity of the teleost lineage including evidence for several alternate immunological strategies. The discoveries of the genetic loss of the major histocompatibility complex (MHC) class II pathway in Atlantic cod (Gadus morhua) as well as the functional loss in the more distant broadnosed pipefish (Syngnathus typhle) [5,6] show that MHCII is not crucial for the defence against pathogens and survival in some fish species. These findings are further supported in a recent study by Malmstrom et al. [7], which demonstrated that the loss of MHCII is shared by the entire Gadiformes lineage. Accompanying the loss of MHCII, highly variable MHCI copy number within the Gadiformes was reported, with several species having more than 40 copies including Atlantic cod found to have 80–100 copies [7,8]. Furthermore, it was hypothesized that the expanded repertoire of MHCI had undergone sub- or neofunctionalization as a possible adaptation to the MHCII loss. However, Malmstrom et al. also identified large numbers of MHCI in many Percomorphaceae lineages (all containing MHCII) demonstrating an extreme evolutionary plasticity of teleost immunity, and that it most likely is influenced by species habitat. Additional analyses revealed a correlation between high MHCI copy number and elevated speciation rates, and thus being a key to the success of this group of fishes [7].
The teleost immune system also displays important strategies with respect to the innate immune system such as the alternative set of Toll-like receptor (TLR) genes compared with other vertebrates [9–11]. Again, Atlantic cod is reported to be divergent compared with other investigated teleosts. In a recent study, the TLR repertoire in Atlantic cod was characterized and compared with that of other genome-sequenced fish species, revealing that Atlantic cod displays large gene expansions and several gene losses. These findings were attributed to the loss of MHCII, which may have boosted evolutionary innovation leading to a more complex TLR repertoire [12].
In general, it is the genetic basis of teleost alternative immunological strategies that has been investigated and studies beyond this tend to focus on genes related to the adaptive immune system. However, the underlying selective mechanisms driving the variety of immunological strategies observed and why they arose are poorly understood—especially for the innate immune system. Using genome assemblies from 66 teleost species, our aim was to characterize teleost TLRs with emphasis on the Gadiformes lineage and thereby investigate the possible link between the loss of MHCII, past and present environmental conditions and the genetic architecture of the innate immune system. We show that the teleost TLR repertoire contains an array of lineage-specific losses and expansions, with the Gadiformes lineage as an extreme outlier. Importantly, within the Gadiformes, we discovered expansions of TLR genes to be correlated with the loss of MHCII and to display different patterns of selection. Furthermore, in teleosts overall, we found that TLR copy number variation correlated with species latitudinal distribution in teleosts overall. By contrast, a weak correlation was found with species maximum depth for TLR9 and TLR22. This suggests that there is a strong ongoing selection of the innate immune system linked to specific environmental and host-intrinsic factors. Furthermore, timing of the lineage-specific losses overlaps with well-described changes in palaeoclimate and continental drift, and hence unveils past adaptive signatures driving the genetic change within the teleost immune system. Our study reveals a remarkable evolutionary flexibility of teleost innate immunity, which has played an essential role in the survival and radiation of the teleost lineage.
2. Material and methods
(a). Sequencing and assembly summary
The 66 teleost genomes and species phylogeny were generated by Malmstrom et al. [7,13]. In short, DNA was isolated from 66 teleost species and subjected to Illumina HiSeq sequencing (2 × 150 bp paired-end reads) which after trimming resulted in an overall coverage between 9 and 34×. The genomes were assembled using the Celera Assembler. For the phylogenetic reconstruction, nine reference fish species were added from Ensembl together with Salmo salar. An alignment of 71 418 bp was used as input for phylogenetic reconstruction with the Bayesian software BEAST [14]. The phylogeny was made using BEAST combined with fossil time-calibration. Note: all timings derived from the phylogeny presented in this study include the confidence interval to illustrate the uncertainty underlying the time fossil calibration performed by Malmstrom et al. [7], and thus spans a longer time period than the branches depicted in the phylogeny (figure 1).
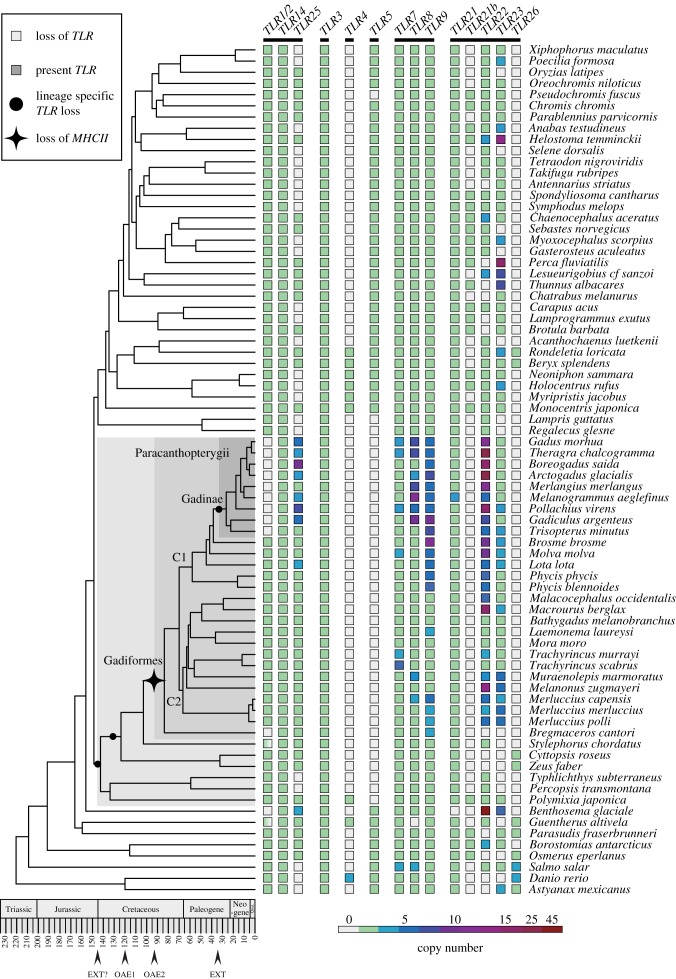
Figure 1.
The TLR repertoires of 76 teleosts mapped onto a time-calibrated species phylogeny. All TLRs characterized in the new 66 teleost genomes as well as in 10 reference teleosts genomes (Ensembl and GenBank) mapped onto a species phylogeny generated by Malmstrom et al. The phylogeny demonstrates the loss of MHCII 110–64 Ma (branch range time, black star) reported by Malmstrom et al. Lineage-specific TLR losses are marked by black circles (Gadinae TLR1/2, Paracanthopterygii TLR5 and TLR21beta). The individual species' repertoires are depicted with boxes where the coloration represents the number of copies of each individual TLR. The six major TLR families: TLR1-family, TLR3-family, TLR4-family, TLR5-family, TLR7-family and TLR11-family are indicated with black bars underneath the TLR names. See the electronic supplementary material, table S1 for copy number details. For TLR1/2, a gradient-filled box indicates the presences of either TLR1 or TLR2. The Paracanthopterygiian lineage, Gadiformes order and Gadinae family display shaded grey backgrounds.
(b). Gene searches
Protein query TIR (Toll/interleukin-1 receptor) domain sequences from Atlantic cod [12], all fish genomes available at Ensembl [15] and channel catfish [16–18], collectively representing all known vertebrate TLR genes to date, were used for TBLASTN searches towards the 66 fish genomes supplied by Malmstrom et al. (see below for parameters). TLR copy numbers for the Ensembl species were taken from [12]. The NCBI BLAST tool was used to search the S. salar genome (ICSASG_v2, GCA_000233375.4) with default settings using the same query sequences. TBLASTN from BLAST + 2.2.26 [19] was used with an e-value cut-off at 1e−10 (and in some cases lower, to capture the largest expansions), otherwise default settings. The number of detected TIR domains was counted for each TLR gene. Owing to the fragmented nature of the genomes, conservative estimates of copy numbers were used and are shown in the electronic supplementary material, table S1. These copy numbers form the foundation for the TLR repertoires depicted in figure 1.
Note on gene annotation: TLR gene annotation varies greatly between species. In this study, the following annotations are used (similar to that of [12]): TLR1, TLR1/6 (in cases where annotation has not been provided and phylogeny cannot determine stronger homology towards TLR1 or TLR6), TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9, TLR10, TLR11, TLR12, TLR13, TLR14, TLR15, TLR16, TLR18 is by phylogeny determined to be TLR14, TLR15, TLR16, TLR19 is by phylogeny determined to be TLR26, TLR20 is by phylogeny determined to be TLR26, TLR21, TLR22, TLR23, TLR25 and TLR26.
(c). TLR, MHC, latitude and depth correlations using stochastic linear Ornstein–Uhlenbeck models for comparative hypotheses
For genes displaying more than four different gene copy numbers (TLR8, TLR9, TLR22, TLR23, TLR25), we ran SLOUCH—stochastic linear Ornstein–Uhlenbeck models for comparative hypotheses. This is a phylogenetic comparative method designed to study adaptive evolution of a trait along a phylogeny implemented in the R program SLOUCH [20–22]. The output of models analysed in SLOUCH can be summarized by a regression, which includes information on whether the analysed traits are evolving towards the estimated optima, how fast (or slow) this evolution is and how much of the trait variation that is explained by evolution towards these optima. We used SLOUCH to test whether TLR copy numbers have evolved towards optima that are influenced by the species' latitudinal distribution (values obtained from Fishbase.org [23]), species maximum depth (values obtained from Fishbase.org [23]) and evolutionary loss of the MHCII complex. We defined six latitudinal categories: 75, 50, 25, 0 (equator), −25 and −50. If a species' latitudinal distribution includes or crosses one of these categories, it was assigned to that respective category (multiple assignments are possible). Some species were not included in any of the categories owing to failure to cross the defined latitudes. Similarly, where data on depth were unavailable, species were excluded from the phylogeny resulting in a reduced tree used as input for SLOUCH.
The model of evolution in SLOUCH is based on an Ornstein–Uhlenbeck process and assumes that a trait (e.g. gene copy number) has a tendency to evolve towards a ‘primary’ optimum Θ. We assume that average copy number in a lineage can take any non-negative real number (i.e. intraspecies variation in copy numbers exist). A primary optimum is defined as the average optimal state that species will reach in a given environment when ancestral constraints have disappeared [20], at a rate proportional to a parameter α. As an example, in some of our analyses, we investigated whether species sharing the same latitudinal distribution have a tendency to evolve similar copy numbers for a given TLR locus. Lag in adaptation towards primary optima is quantified by a half-life parameter, t1/2 = ln(2)/α, which can be interpreted as the average time it takes a species to evolve half the distance from the ancestral (copy number) state towards the predicted optimal (copy number) state. For example, a half-life of zero signifies immediate adaptation of the trait to any change in the optimum for every lineage present in the phylogeny. A half-life above zero indicates adaptation is not immediate, with the amount of constrained evolution increasing with an increasing half-life. The model of evolution used in SLOUCH also includes a stochastic component with standard deviation σ, which can be interpreted as evolutionary changes in the trait (e.g. copy numbers) owing to unmeasured selective forces and genetic drift. This component of the model is reported as vy = σ2/2α, and can be interpreted as the expected residual variance when adaptation and stochastic changes have come to an equilibrium.
Our latitudinal categories, maximum depth and evolutionary losses of MHCII represent ‘niches’ and the model estimates one primary optimum for each niche included in any particular model. The different states of niches (e.g. presence and absence of MCHII) are known for all extant species in our phylogeny, but are unobserved for internal branches in the tree. We therefore mapped a separate state called ancestral to all internal nodes in the phylogeny to avoid having to infer uncertain primary optima. The method uses generalized least squares for estimation of the regression parameters (i.e. the influence of the predictor on the primary optimum) and maximum-likelihood for estimation of α and σ2 in an iterative procedure. For a full description of the model implemented in SLOUCH, see Hansen et al. [21]. All analyses were performed in R v. 3.0 [22].
We used SLOUCH to estimate the phylogenetic effect in the data. A phylogenetic effect indicates that some part of the variation in the trait is explained by shared ancestry (i.e. phylogeny), which means closely related species tend to have more similar trait values compared with more distantly related species. The phylogenetic effect can be estimated in SLOUCH by running a model without any predictor variables (i.e. no latitudinal categorical variables). The half-life parameter in such a model will represent an estimate for how important shared history is in explaining the distribution of trait means (average) on the phylogeny: a half-life of zero indicates that the trait data are not phylogenetically structured, while a half-life greater than 0 indicates that there exists an influence of phylogeny on the data. A phylogenetic effect can be owing to slowness of adaptation, adaptation towards phylogenetically structured optima or a combination of both. To investigate which of these scenarios we find support for, we contrasted the phylogenetic effect model with a model run with predictor variables (e.g. latitudinal distribution or maximum depth) using the bias-corrected Akaike information criterion (AICc), which balances goodness of fit (log-likelihood) with the number of parameters in the model (model complexity). The model with the lowest AICc value is the best supported. A better (lower) AICc value for a model including predictor variables indicate evidence for a scenario where the traits in our models are evolving towards optima that are shared by species across niches (e.g. the same latitudinal section). r2 was not used for assessing model support, but represents the amount of the total variation in the response trait (TLR gene copy number) that is explained by the optimal regression.
(d). Diversifying selection analysis using mixed effects model of evolution and branch-site random effects likelihood
As there were different degrees of TLR gene expansions throughout our dataset, and because expansions were more prominent within the Gadiformes order, we wanted to determine if any individual positions within the coding sequence or certain lineages have been affected by diversifying selection. Owing to the fragmented nature of our dataset, this analysis was not feasible unless we selected a set of species as well as a set of TLRs. We selected nine species from the draft genome dataset: Melanogrammus aeglefinus, Macrourus berglax and Muraenolepsis marmoratus from the Gadiformes, Stylephorus chordatus which is a putative ancestral clade of Gadiformes, Cyttopsis roseus and Zeus faber from the Zeiformes (Gadiformes + S. chordatus sister clade), Polymixia japonica at the base of the Paracanthopterygii superorder, Rondeletia loricata and Beryx splendens as two closely related species outside the Paracanthopterygii. We also included TLR sequences from the second version of the Atlantic cod genome (GadMor2) as an additional Gadiformes representative [24]. Finally, we added the respective TLR sequences from fish species whose genomes are available through Ensembl [15]. Collectively, these species cover the entire range of the teleost phylogenetic tree obtained from Malmstrom et al. [7].
We selected three TLR genes for investigation: TLR3—a single copy gene present in all investigated teleosts, TLR9—expanded in most Gadiformes as well as present in all investigated teleosts and TLR25—mainly expanded in the C1 clade of the Gadiformes but also displaying both presence and absence patterns in our data. Collectively, these genes represent the range of different patterns observed. Query TLR sequences were identical to those used for the overall TLR characterization described above except the full-length protein sequence was used in a TBLASTN with an e-value cut-off equal to 1 × 10−10 and otherwise default parameters towards the draft genomes and GadMor2. The target unitigs (draft genomes) and linkage group (GadMor2) regions were extracted and aligned towards the coding sequences obtained from Ensembl using ClustalW in MEGA5 [25]. The resulting alignment was manually curated to ensure that the reading frame was maintained. We chose to only investigate the ecto-domain of the TLR as the transmembrane and TIR domain are known to be under purifying selection. For all alignments, the coverage of unitig sequence was variable. Therefore, the alignments were divided into sections to obtain alignments with the least amount of missing data. This resulted in one alignment for TLR3, two for TLR9 and four for TLR25. The alignments are available in our GitHub repository.
These alignments were uploaded to www.datamonkey.org [26,27] where we performed model selection analysis to find the best fitting model of nucleotide evolution for each of the alignments (reported in the electronic supplementary material). We then performed Mixed Effects Model of Evolution (MEME) analysis on all alignments as well as Branch-Site Random Effects Likelihood (BSR) analysis on TLR9 and TLR25 alignments allowing for the generation of gene trees based on the alignments. MEME is based on the ratio between non-synonymous to synonymous substitutions where this ratio can vary from site to site as well between lineages. In this way, MEME can detect both pervasive and episodic positive (diversifying) selection. MEME compares its estimates with a null hypothesis for which all sites are evolving neutrally (worst-case scenario) and thus, the results given by MEME are conservative estimates. BSR is also based on the ratio between non-synonymous to synonymous substitutions. MEME implements this analysis for each individual site, but we also ran BSR alone to obtain on overall impression of any likely diversifying selection affecting lineages or individual branches. By contrast, in BSR, there is no need to define any branches a priori as neutral or under negative selection. Thus, detecting episodic diversifying selection in a few sites or in a few lineages becomes more reliable by using BSR [28,29].
3. Results
Mapping all the identified teleost TLRs—extracted from the 66 genome assemblies—onto the phylogeny of Malmstrom et al. [7] demonstrates the presence of comprehensive TLR repertoires in all investigated teleosts (figure 1) similar to that found in other vertebrates [9,11,12]. However, most notable was the observation of three lineage-specific gene losses, several lineage-specific gene expansions and a substantial number of recorded species-specific repertoire variants (figure 1). Specifically, TLR1/2 are lost from the Gadinae (40–16 Ma) in addition to being completely or partially lost in Bregmaceros cantori, Benthosema glaciale, S. chordatus and Guentherus altivela. TLR5 is lost from the entire Paracanthopterygii superorder and the order Lampridiformes (175–130 Ma) in addition to Pseudochromis fuscus. Further, we discovered a new TLR, here annotated as TLR21beta based on sequence homology, which is also absent in all Paracanthopterygiian species with the exception of P. japonica, and Lampridiformes. However, in contrast with TLR5, the presence of TLR21beta does not follow any clear phylogenetic pattern outside Paracanthopterygii/Lampridiformes (figure 1). The Gadinae is the only clade consistent with the recently reported alternative TLR repertoire in Atlantic cod [5,12] owing to the prominent gene losses of TLR1/2.
Three TLRs are found in all species; TLR3, TLR14 and TLR21, the latter with the exception of Be. glaciale. Within the Gadiformes, we find gene expansions for TLR7, TLR8, TLR9, TLR22, TLR23 and TLR25, especially within the C1 clade (the Gadiformes segregate into two distinct clades here named C1 and C2 (figure 1)). Outside the Gadiformes, TLR25 displays no obvious phylogenetic pattern. This is in contrast with TLR7, TLR8 and TLR9 which are present in all species with the exception of a single TLR8 loss in G. altivela. TLR22 and TLR23 are found in all Gadiformes except in Br. cantori and show a substantial degree of gene expansion within the Gadiformes lineage—particularly for TLR22. Outside the Gadiformes, the expansion of TLR22 is less pronounced, whereas, by contrast, TLR23 is frequently expanded. However, TLR22 and TLR23 display phylogenetically non-structured patterns of presence and gene loss outside the Gadiformes order (figure 1; electronic supplementary material, table S1). Finally, there are two rare teleost TLRs, i.e. TLR4 and TLR26. TLR4 is found in the Holocentriformes and in three out of four Beryciformes species in addition to Danio rerio, P. japonica and G. altivela. TLR26 is mainly found in species basal to the Gadiformes and in two Beryciformes: R. loricata and B. splendens (figure 1; electronic supplementary material, table S1).
To identify episodic diversifying selection, MEME and BSR selection analyses were performed on the ecto-domain of three TLR representatives—TLR3, TLR9 and TLR25. MEME reported 19 sites for TLR3, 35 sites for TLR9 and 18 sites for TLR25 likely to have experienced diversifying selection (figure 2). The BSR analysis identified multiple nodes and branches encompassing most TLR9 paralogues in the Gadiformes (mainly Gadinae) subject to diversifying selection. Diversifying selection was also detected in one of the TLR25 alignments at one node and on one branch encompassing some of the Gadinae TLR25 paralogues (electronic supplementary material, figures S1 and S2).
Figure 2.
Overview of sites reported by the MEME analysis performed on TLR3, TLR9 and TLR25 in the selected species. A schematic drawing of the TLR3 (a), TLR9 (b) and TLR25 (c) protein domains with the ecto-domain (dimerization and ligand interaction), transmembrane (TM) domain and TIR domain (signalling domain). Only the ecto-domain was subjected to selection analysis as the TM and TIR domains are known to be under purifying selection. Grey boxes indicate which parts of the ecto-domain were included in the alignment and also show how many alignments were generated per gene. There is a 22 codon overlap between TLR25 section 2 and 3. Arrows indicate sites reported by the MEME analysis. For site details, see the electronic supplementary material. (Online version in colour.)
Associations between specific TLR expansions, species latitudinal distributions, species maximum depth as well as the absence of MHCII—specific for the Gadiformes lineage (figure 1)—were further investigated using SLOUCH [21]. Models using the specified latitudinal categories as predictor variables showed that latitude explained 19–32% of the TLR copy number variation for TLR8, TLR9, TLR22 and TLR25 (table 1), whereas species maximum depth explained 4–10% of the variation seen in TLR9 and TLR22 (electronic supplementary material). Especially northern latitudinal categories were found to be associated with higher copy numbers of TLR8, TLR22 and TLR25, while increased copy numbers of TLR9 were associated with more tropical latitudes—particularly in the equatorial region (table 1; electronic supplementary material, table S1). However, for TLR23, there was no indication that the copy number has evolved as a consequence of changes in latitude or depth (table 1, data not shown for depth correlation). Moreover, within the Gadiformes lineage, we found strong support for scenarios where TLR8, TLR9, TLR22 and TLR25 have evolved additional gene copies with the loss of MHCII explaining between 14% and 27% of the copy number variation (table 2). The explained variation in copy numbers was 3–6% larger (compared with latitude alone) and 3–16% larger (compared with MHCII loss alone) when we ran models where copy numbers of TLR8, TLR22 and TLR25 evolved towards optima jointly defined by latitudinal categories and presence/absence of MHCII. This indicates that both latitude and loss of MHCII have contributed to the expansion of these TLRs. However, we were not able to distinguish the relative contribution of MHCII and latitude, respectively. This is contrary to the striking result obtained for TLR9 where the combination of latitude and loss of MHCII explained 50% of the copy number variation—compared with 20% and 22% for latitude and MHCII loss separately (table 2).
Table 1.
Phylogenetic comparative analyses of the evolution of TLR copy numbers in relation to species latitudinal distributions using SLOUCH. (For each model, we show the phylogenetically corrected r2, and the AICc score. AICc balances goodness of fit (log-likelihood) with the number of parameters in the model (model complexity). The model with the lowest AICc value is the best supported. r2 represents the amount of the total variation that is explained by the model. Detailed output from each model is given in the electronic supplementary material. The model called ‘phylogeny’ does not include any explanatory variables and is given as a reference point for comparison to models with predictor variables. The best AICc scores and corresponding r2 values for each of the investigated TLR expansion are italicized.)
| category |
TLR8 |
TLR9 |
TLR22 |
TLR23 |
TLR25 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| AICc | r2 | AICc | r2 | AICc | r2 | AICc | r2 | AICc | r2 | |
| phylogeny | 266.41 | 0.00 | 243.91 | 0.00 | 430.27 | 0.00 | 307.65 | 0.00 | 241.36 | 0.00 |
| group 75 latitude | 260.29 | 18.32 | 239.07 | 18.91 | 418.86 | 24.63 | 311.72 | 0.96 | 226.61 | 32.26 |
| group 50 latitude | 259.75 | 19.02 | 240.67 | 15.49 | 427.26 | 13.70 | 310.88 | 2.30 | 232.46 | 21.96 |
| group 25 latitude | 259.98 | 18.72 | 240.34 | 17.24 | 429.86 | 8.86 | 307.22 | 7.91 | 233.31 | 20.89 |
| group 0 latitude | 259.90 | 20.13 | 238.24 | 19.99 | 427.05 | 13.99 | 311.27 | 1.69 | 232.56 | 21.84 |
| group −25 latitude | 260.06 | 18.63 | 239.78 | 16.69 | 429.34 | 9.62 | 309.78 | 4.00 | 233.38 | 20.80 |
| group −50 latitude | 260.31 | 16.35 | 240.16 | 16.18 | 429.62 | 9.21 | 311.54 | 1.24 | 233.45 | 20.71 |
Table 2.
Phylogenetic comparative analyses of the evolution of TLR copy numbers in relation to species latitudinal distributions and MHCII status using SLOUCH. (For each model, we show the phylogenetically corrected r2, and the AICc score. AICc balances goodness of fit (log-likelihood) with the number of parameters in the model (model complexity). The model with the lowest AICc value is the best supported. r2 represents the amount of the total variation that is explained by the model. Detailed output from each model is given in the electronic supplementary material. The model called ‘phylogeny’ in table 1 does not include any explanatory variables and is given as a reference point for comparison to models with predictor variables. The best AICc scores and corresponding r2 values for each of the investigated TLR expansion are italicized.)
| category |
TLR8 |
TLR9 |
TLR22 |
TLR23 |
TLR25 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| AICc | r2 | AICc | r2 | AICc | r2 | AICc | r2 | AICc | r2 | |
| group MHCII | 259.43 | 19.44 | 239.01 | 22.41 | 427.99 | 14.53 | 328.13 | 2.65 | 231.30 | 26.94 |
| group MHCII + group 75 lat. | 264.37 | 19.52 | 240.39 | 31.`32 | 420.41 | 30.23 | 315.34 | 3.21 | 228.98 | 35.08 |
| group MHCII + group 50 lat. | 262.31 | 22.16 | 243.11 | 25.72 | 431.07 | 17.15 | 314.69 | 4.23 | 235.60 | 27.76 |
| group MHCII + group 25 lat. | 263.76 | 20.32 | 239.13 | 32.69 | 431.69 | 16.32 | 311.21 | 9.41 | 234.82 | 28.67 |
| group MHCII + group 0 lat. | 261.00 | 27.11 | 228.48 | 53.53 | 430.16 | 18.36 | 314.90 | 3.92 | 233.63 | 30.02 |
| group MHCII + group −25 lat. | 263.37 | 20.82 | 230.06 | 52.33 | 432.07 | 15.80 | 313.49 | 6.01 | 234.84 | 28.64 |
| group MHCII + group −50 lat. | 264.39 | 19.50 | 240.15 | 29.17 | 432.19 | 15.63 | 314.92 | 3.81 | 235.60 | 27.76 |
4. Discussion
Overall, vertebrate and teleost genome duplications may explain some of the teleost TLR repertoire variation demonstrated here with respect to gene expansions. However, the extreme numbers seen for some of the TLR expansions within the Gadiformes indicate that these genes have undergone additional lineage-specific duplication events—a phenomenon also seen for other genes in teleost species [30]. Gene duplicates preserved after a duplication event commonly undergo neo- or subfunctionalization ([31] and references therein). In Atlantic cod, we have previously demonstrated that the TLR expansions and their paralogues show signs of diversifying selection. For some expansions, this was indicative of neofunctionalization owing to high numbers of sites under selection in putative dimerization and ligand-interacting regions. For other expansions, it was more indicative of subfunctionalization owing to fewer sites under selection combined with tissue-specific expression patterns [12]. The selection analyses on the chosen TLR representatives demonstrated that TLR3 and TLR25 display similar amounts of sites subject to diversifying selection, despite their highly different patterns in our dataset (single copy present in all species versus expanded in Gadiformes combined with both presence and absence in the remaining species). By contrast, TLR9 displayed almost double the number of sites reported as under diversifying selection (figure 2). In the human system, and by proxy in teleosts, the TLR3 protein is located to the endosomal membranes and signals for an antiviral response upon interaction with double-stranded RNA [32]. It has recently been demonstrated that mammalian TLR3 also can detect structured RNAs [33]. This could explain the presence of sites under diversifying selection in fish TLR3 adapting the protein towards different structured RNAs or other possible ligands not presently known.
TLR25 is a relatively newly identified fish-specific TLR where ligand and subcellular localization is yet to be determined [17]. We have earlier suggested that this TLR in Atlantic cod is located to the cell surface and interacts with ligands similar to other TLR1-family members (TLR1, TLR2 and TLR6)—such as bacterial or parasitic lipoproteins [12]. In humans, TLR1, TLR2 and TLR6 form both homo- and heterodimers actively increasing their ligand repertoire [32]. Gadinae do not have TLR1, TLR2 or TLR6 (figure 1) and thus, in their case, TLR14 and the expanded TLR25 may be a replacing set of TLR1-family members. In the remaining Gadiformes, both TLR1, TLR2 as well as TLR14 and TLR25 are present (figure 1). The MEME analysis reported a similar number of sites under diversifying selection in TLR25 compared with TLR3, which could suggest that they are subjected to similar selective pressures. However, the BSR analysis indicated that nodes and branches representing only some of the Gadinae TLR25 paralogues are subject to diversifying selection (electronic supplementary material, figure S2). Overall, this demonstrates that TLR25 paralogues may be affected by different selection pressures within expansions, whereas TLR25 generally is adapted towards unknown species-specific factors similar to that of TLR3.
In Atlantic cod, TLR9 paralogues showed clear signs of diversifying selection and differences in expression patterns [12]. The MEME analysis reported a large amount of sites under diversifying selection and the BSR analysis strongly indicates diversifying selection on nodes and branches leading to different clades of Gadinae TLR9 paralogues. In humans, TLR9 interacts with unmethylated single-stranded CpG DNA, both viral and bacterial, within the endosomal track in a highly sequence-dependent manner. However, dependent on the sequences, TLR9-ligand interaction can result in both antagonistic and agonistic signalling [34] suggesting a regulatory role. Diversification of TLR9 paralogues indicates adaptation towards lineage-specific pathogen loads or diversity within Gadiformes. It may also suggest a larger regulatory role for TLR9 in this lineage. Furthermore, in mammals, TLR9 signalling can induce MHCI antigen cross-presentation [35] which overlaps with the hypothesized subfunctionalization of some MHCI copies in Gadiformes [7]. Overall, our findings demonstrate that TLR9 paralogues have experienced a different selection pressure compared with TLR25 paralogues. Collectively, the gene expansions observed in Gadiformes, as well as in teleosts overall, are probably subject to different levels of neo- and subfunctionalization contributing to the further adaptation of the teleost innate immune system. Extreme northern or southern distributions are proxy indicators for temperature as these regions are cooler but also have undergone a larger degree of palaeoclimatic changes compared with the more tropical regions [36]. The observed expansions for TLR7, TLR8, TLR9, TLR22, TLR23 and TLR25, especially within the Gadiformes, indicate selection towards higher copy number optima. This could potentially be explained by different pathogen loads or pathogen community compositions connected to highly variable palaeoclimatic arctic environments. We found correlations between increased copy number of TLR8, TLR22 and TLR25 with more northern species distributions (table 1). By contrast, TLR9 showed higher optimal copy numbers in tropical regions—especially combined with the loss of MHCII (tables 1 and 2), most probably driven by the specific biotic or abiotic factors encountered in the tropics. Collectively, our findings indicate that, for the Gadiformes, both the palaeogeographical distribution (reflecting the environments these species have inhabited through time) and the loss of MHCII have been vital drivers for the expansion of TLR8, TLR22, TLR25 and in particular TLR9.
By using a dated phylogeny, we find that the successive alterations to the teleost immune system occurred in periods with substantial palaeoclimatic fluctuations as well as oceanographic changes owing to continental drift. Such events are often associated with periods of extinction followed by population diversification and subsequent speciation enabling the invasion of new niches [37,38]. Our data suggest that the overall loss of TLR5 (previously reported [39]) and TLR21beta (175–130 Ma) overlap the Jurassic–Cretaceous (J–K) boundary (figure 1). Although this transition between geological periods does not harbour any well-defined events, there is accumulating evidence of both species extinctions and radiations [40–44]. The loss of TLR5 and TLR21beta may have occurred as adaptations to new habitats such as the expanding Central Atlantic Ocean. Even though both TLR5 and TLR21b display lineage-specific loss, their presence/absence pattern outside the Paracanthopterygii (figure 1) indicate that they have experienced different selection pressures before the J–K boundary.
Within the Gadiformes clade, we find that the loss of MHCII coincides with the overall gene expansion patterns of TLR7, TLR8, TLR9, TLR22, TLR23 and TLR25, spanning a total interval 110–64 Ma. This further overlaps with the early–late Cretaceous transition which includes one of the late Cretaceous global anoxia events (95 Ma). This anoxic environment, although probably allowing a small degree of specialized adaptation, generally deprived the deep seas of species [45,46]. Anoxic conditions led to higher extinction rates during this time period [47–50], fitting with the metabolic cost scenario proposed to promote the loss of MHCII [51]. In this scenario, the benefits of maintaining the MHCII system in some environments could not compensate for the metabolic cost of expressing it. Coinciding with the anoxic event is the further northward opening of the Central Atlantic Ocean [52] and the propagation of the South Atlantic Ocean to meet the Central Atlantic Ocean [53–55]. The stress imposed by global ocean anoxia therefore appears simultaneously with the appearance of new habitats. Further, this time period is associated with a decrease in bony fish family richness, indirectly derived from fossil data [56], indicating that these secondary changes to the Gadiformes immune system may have had slightly more adverse effects here compared with the initial ones occurring at the J–K boundary. However, this probably had a positive effect supporting species survival and radiation in the long term. The more recent loss of TLR1/2 from the Gadinae subfamily (40–16 Ma) is probably a temperature-driven adaptation caused by an abrupt cooling of global climate and loss of habitat owing to the drastic decrease in eustatic sea levels approximately 34 Ma [50,57,58] overlapping with the opening of the North Atlantic Ocean between Greenland and Norway [52].
5. Conclusion
Overall, our findings reveal unprecedented variability within the teleost innate immune system, particularly within the Gadiformes, characterized by significant gene expansions and losses. Intriguingly, we find that higher copy numbers of TLRs correlate with species latitudinal distribution and the loss of MHCII. Further evidence of diversifying selection indicates that the paralogues probably experience different selection pressures. The successive nature of these changes to the ancestral teleost immune system, combined with the extensive evolvability of the innate immune system described here, have probably contributed to the overall survival and successful radiation of this lineage.
Supplementary Material
Acknowledgements
This work was supported by The Research Council of Norway. Some of the teleost genomes were assembled using the Abel Cluster, owned by the University of Oslo and the Norwegian metacenter for High Performance Computing (NOTUR), and operated by the Department for Research Computing at USIT, the University of Oslo IT-department http://www.hpc.uio.no/. The authors would like to thank Dr Mark Ravinet and Helle Tessand Baalsrud for comments on the manuscript.
Data accessibility
All novel teleost sequence and genome resources are available at European Nucleotide Archive (ENA) and the Dryad digital repository, submitted by the Malmstrom et al. [7]: all raw data (sequencing reads) are available at ENA with study accession number PRJEB12469 (sample identifiers ERS1199874-ERS1199939). Genome assemblies, available at Dryad, exist in two versions (UTGs and scaffolds) under http://dx.doi.org/10.5061/dryad.326r8. All additional resources needed to generate the findings presented here are available in our GitHub repository including, but not limited to, scripts, BLAST and SLOUCH output files: https://github.com/MonicaSolbakken/TLR [7,13].
Authors' contributions
M.H.S., K.S.J. and S.J. conceived, planned and oversaw the project. M.H.S. generated all TLR-related data based on BLAST searches towards the teleost genome resources as well as extracting information about latitude and depth from online databases. K.L.V. performed all SLOUCH analyses. M.H.S. made all figures/tables and wrote the overall text with significant aid of S.J. and K.S.J. K.L.V. wrote all sections related to SLOUCH. All authors contributed with comments, edits and proofreading of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by The Research Council of Norway (grant numbers 222378/F20 and 199806/S40 to K.S.J./S.J.). The funding agency was not involved with any aspects of the project beyond providing funding.
References
- 1.Eschmeyer WN, Fricke R, van der Laan R (eds). 2015. Catalog of fishes: genera, species, references. See http://researcharchive.calacadamy.org/research/ichthyology/catalog/fishcatmain.asp (accessed 15 January 2016). [DOI] [PubMed]
- 2.Faircloth BC, Sorenson L, Santini F, Alfaro ME. 2013. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of ultraconserved elements (UCEs). PLoS ONE 8, e65923 ( 10.1371/journal.pone.0065923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sallan LC. 2014. Major issues in the origins of ray-finned fish (Actinopterygii) biodiversity. Biol. Rev. Camb. Philos. Soc. 89, 950–971. ( 10.1111/brv.12086) [DOI] [PubMed] [Google Scholar]
- 4.Volff JN. 2005. Genome evolution and biodiversity in teleost fish. Heredity (Edinb) 94, 280–294. ( 10.1038/sj.hdy.6800635) [DOI] [PubMed] [Google Scholar]
- 5.Star B, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477, 207–210. ( 10.1038/nature10342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haase D, Roth O, Kalbe M, Schmiedeskamp G, Scharsack JP, Rosenstiel P, Reusch TB. 2013. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: evidence from deep transcriptome sequencing. Biol. Lett. 9, 20130044 ( 10.1098/rsbl.2013.0044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malmstrom M, et al. 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 48, 1204–1210. ( 10.1038/ng.3645) [DOI] [PubMed] [Google Scholar]
- 8.Malmstrom M, Jentoft S, Gregers TF, Jakobsen KS. 2013. Unraveling the evolution of the Atlantic cod's (Gadus morhua L.) alternative immune strategy. PLoS ONE 8, e74004 ( 10.1371/journal.pone.0074004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palti Y. 2011. Toll-like receptors in bony fish: from genomics to function. Dev. Comp. Immunol. 35, 1263–1272. ( 10.1016/j.dci.2011.03.006) [DOI] [PubMed] [Google Scholar]
- 10.Rebl A, Goldammer T, Seyfert HM. 2010. Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 134, 139–150. ( 10.1016/j.vetimm.2009.09.021) [DOI] [PubMed] [Google Scholar]
- 11.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. 2005. The evolution of vertebrate Toll-like receptors. Proc. Natl Acad. Sci. USA 102, 9577–9582. ( 10.1073/pnas.0502272102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solbakken MH, Tørresen OK, Seppola M, Gregers TF, Jakobsen KS, Jentoft S. 2016. Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions. Sci. Rep. 6, 25211 ( 10.1038/srep25211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malmstrom M, Matschiner M, Tørresen OK, Jakobsen KS, Jentoft S. 2017. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci. Data 4, 160132 ( 10.1038/sdata.2016.132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham F, et al. 2015. Ensembl 2015. Nucleic Acids Res. 43, D662–D669. ( 10.1093/nar/gku1010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res. 41, D36–D42. ( 10.1093/nar/gks1195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quiniou SM, Boudinot P, Bengten E. 2013. Comprehensive survey and genomic characterization of Toll-like receptors (TLRs) in channel catfish, Ictalurus punctatus: identification of novel fish TLRs. Immunogenetics 65, 511–530. ( 10.1007/s00251-013-0694-9) [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, et al. 2016. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat. Commun. 7, 11757 ( 10.1038/ncomms11757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen TF. 1997. Stabilizing selection and the comparative analysis of adaptation. Evolution 51, 1341–1351. ( 10.2307/2411186) [DOI] [PubMed] [Google Scholar]
- 21.Hansen TF, Pienaar J, Orzack SH. 2008. A comparative method for studying adaptation to a randomly evolving environment. Evolution 62, 1965–1977. ( 10.1111/j.1558-5646.2008.00412.x) [DOI] [PubMed] [Google Scholar]
- 22.R Core Team. 2015. R: a language and environment for statistical computing. See http://www.R-project.org. [Google Scholar]
- 23.Froese R, Pauly D (eds). 2015 Fishbase, version (01/2016). See www.fishbase.org.
- 24.Torresen OK, et al. 2017. An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18, 95 ( 10.1186/s12864-016-3448-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. ( 10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457. ( 10.1093/bioinformatics/btq429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533. ( 10.1093/bioinformatics/bti320) [DOI] [PubMed] [Google Scholar]
- 28.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 28, 3033–3043. ( 10.1093/molbev/msr125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murrell B, Wetheim JO, Moola S, Weighill T, Scheffler K, Pond SLK. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 ( 10.1371/journal.pgen.1002764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Zou M, He S. 2013. Genome-wide identification, characterization, and expression analysis of lineage-specific genes within zebrafish. BMC Genomics 14, 65 ( 10.1186/1471-2164-14-65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. 2009. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 19, 1404–1418. ( 10.1101/gr.086827.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietretti D, Wiegertjes GF. 2014. Ligand specificities of Toll-like receptors in fish: indications from infection studies. Dev. Comp. Immunol. 43, 205–222. ( 10.1016/j.dci.2013.08.010) [DOI] [PubMed] [Google Scholar]
- 33.Tatematsu M, Seya T, Matsumoto M. 2014. Beyond dsRNA: Toll-like receptor 3 signalling in RNA-induced immune responses. Biochem. J. 458, 195–201. ( 10.1042/BJ20131492) [DOI] [PubMed] [Google Scholar]
- 34.Vabret N, Bhardwaj N, Greenbaum BD. 2017. Sequence-specific sensing of nucleic acids. Trends Immunol. 38, 53–65. ( 10.1016/j.it.2016.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brutkiewicz RR. 2016. Cell signaling pathways that regulate antigen presentation. J. Immunol. 197, 2971–2979. ( 10.4049/jimmunol.1600460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson AM, Weir JT. 2014. Latitudinal gradients in climatic-niche evolution accelerate trait evolution at high latitudes. Ecol. Lett. 17, 1427–1436. ( 10.1111/ele.12346) [DOI] [PubMed] [Google Scholar]
- 37.Wellborn GA, Langerhans RB. 2015. Ecological opportunity and the adaptive diversification of lineages. Ecol. Evol. 5, 176–195. ( 10.1002/ece3.1347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simões M, Breitkreuz L, Alvarado M, Baca S, Cooper JC, Heins L, Herzog K, Lieberman BS. 2016. The evolving theory of evolutionary radiations. Trends Ecol. Evol. 31, 27–34. ( 10.1016/j.tree.2015.10.007) [DOI] [PubMed] [Google Scholar]
- 39.Solbakken MH, Rise ML, Jakobsen KS, Jentoft S. 2016. Successive losses of central immune genes characterize the Gadiformes’ alternate immunity. Genome Biol. Evol. 8, 3508–3515. ( 10.1093/gbe/evw250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson RB, Butler RJ, Lindgren J, Smith AS. 2010. Mesozoic marine tetrapod diversity: mass extinctions and temporal heterogeneity in geological megabiases affecting vertebrates. Proc. R. Soc. B 277, 829–834. ( 10.1098/rspb.2009.1845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson RB, Druckenmiller PS. 2014. Faunal turnover of marine tetrapods during the Jurassic-Cretaceous transition. Biol. Rev. Camb. Philos. Soc. 89, 1–23. ( 10.1111/brv.12038) [DOI] [PubMed] [Google Scholar]
- 42.Bambach RK. 2006. Phanerozoic biodiversity mass extinctions. Annu. Rev. Earth Planet. Sci. 34, 127–155. ( 10.1146/annurev.earth.33.092203.122654) [DOI] [Google Scholar]
- 43.Alroy J. 2010. The shifting balance of diversity among major marine animal groups. Science 329, 1191–1194. ( 10.1126/science.1189910) [DOI] [PubMed] [Google Scholar]
- 44.Cavin L. 2010. Diversity of Mesozoic semionotiform fishes and the origin of gars (Lepisosteidae). Naturwissenschaften 97, 1035–1040. ( 10.1007/s00114-010-0722-7) [DOI] [PubMed] [Google Scholar]
- 45.Priede IG, Froese R. 2013. Colonization of the deep sea by fishes. J. Fish Biol. 83, 1528–1550. ( 10.1111/jfb.12265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers AD. 2000. The role of the oceanic oxygen minima in generating biodiversity in the deep sea. Deep Sea Res. Part II Top. Stud. Oceanogr. 47, 119–148. ( 10.1016/S0967-0645(99)00107-1) [DOI] [Google Scholar]
- 47.Takashima R, Nishi H, Huber B, Leckie RM. 2006. Greenhouse world and the Mesozoic ocean. Oceanography 19, 82–92. ( 10.5670/oceanog.2006.07) [DOI] [Google Scholar]
- 48.Wilson PA, Norris RD. 2001. Warm tropical ocean surface and global anoxia during the mid-Cretaceous period. Nature 412, 425–429. ( 10.1038/35086553) [DOI] [PubMed] [Google Scholar]
- 49.Sinninghe Damstéa JS, van Bentum EC, Reichart G-J, Pross J, Schouten S. 2010. A CO2 decrease-driven cooling and increased latitudinal temperature gradient during the mid-Cretaceous Oceanic Anoxic Event 2. Earth Planet. Sci. Lett. 293, 97–103. ( 10.1016/j.epsl.2010.02.027) [DOI] [Google Scholar]
- 50.Harnik PG, et al. 2012. Extinctions in ancient and modern seas. Trends Ecol. Evol. 27, 608–617. ( 10.1016/j.tree.2012.07.010) [DOI] [PubMed] [Google Scholar]
- 51.Star B, Jentoft S. 2012. Why does the immune system of Atlantic cod lack MHC II? Bioessays 234, 648–651. ( 10.1002/bies.201200005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melankholina EN, Sushchevskaya NM. 2015. Development of passive volcanic margins of the Central Atlantic and initial opening of ocean. Geotectonics 49, 75–92. ( 10.1134/S0016852115010033) [DOI] [Google Scholar]
- 53.Granot R, Dyment J. 2015. The Cretaceous opening of the South Atlantic Ocean. Earth Planet. Sci. Lett. 414, 156–163. ( 10.1016/j.epsl.2015.01.015) [DOI] [Google Scholar]
- 54.Voigt S, Jung C, Friedrich O, Frank M, Teschner C, Hoffmann J. 2013. Tectonically restricted deep-ocean circulation at the end of the Cretaceous greenhouse. Earth Planet. Sci. Lett. 369, 169–177. ( 10.1016/j.epsl.2013.03.019) [DOI] [Google Scholar]
- 55.Murphy DP, Thomas DJ. 2013. The evolution of Late Cretaceous deep-ocean circulation in the Atlantic basins: neodymium isotope evidence from South Atlantic drill sites for tectonic controls. Geochem. Geophys. Geosyst. 14, 5323–5340. ( 10.1002/2013GC004889) [DOI] [Google Scholar]
- 56.Guinot G, Cavin L. 2016. ‘Fish’ (Actinopterygii and Elasmobranchii) diversification patterns through deep time. Biol. Rev. Camb. Philos. Soc. 91, 950–981. ( 10.1111/brv.12203) [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Pagani M, Zinniker D, DeConto R, Huber M, Brinkhuis H, Shah SR, Leckie RM, Pearson A. 2009. Global cooling during the Eocene-Oligocene climate transition. Science 323, 1187–1190. ( 10.1126/science.1166368) [DOI] [PubMed] [Google Scholar]
- 58.Goldner A, Herold N, Huber M. 2014. Antarctic glaciation caused ocean circulation changes at the Eocene-Oligocene transition. Nature 511, 574–577. ( 10.1038/nature13597) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All novel teleost sequence and genome resources are available at European Nucleotide Archive (ENA) and the Dryad digital repository, submitted by the Malmstrom et al. [7]: all raw data (sequencing reads) are available at ENA with study accession number PRJEB12469 (sample identifiers ERS1199874-ERS1199939). Genome assemblies, available at Dryad, exist in two versions (UTGs and scaffolds) under http://dx.doi.org/10.5061/dryad.326r8. All additional resources needed to generate the findings presented here are available in our GitHub repository including, but not limited to, scripts, BLAST and SLOUCH output files: https://github.com/MonicaSolbakken/TLR [7,13].