Abstract
Cellular metabolic fluxes are determined by enzyme activities and metabolite abundances. Biochemical approaches reveal the impact of specific substrates or regulators on enzyme kinetics, but do not capture the extent to which metabolite and enzyme concentrations vary across physiological states, and therefore how cellular reactions are regulated. We measured enzyme and metabolite concentrations and metabolic fluxes across 25 steady-state yeast cultures. We then assessed the extent to which flux can be explained by a Michaelis-Menten relationship between enzyme, substrate, product, and potential regulator concentrations. This revealed three new instances of cross-pathway regulation, which we biochemically verified. These included inhibition of pyruvate kinase by citrate, which accumulated and thereby curtailed glycolytic outflow in nitrogen-limited yeast. Overall, substrate concentrations were the strongest driver of the net rates of cellular metabolic reactions, with metabolite concentrations collectively having more than double the physiological impact of enzymes.
Introduction
A crowning achievement of twentieth century biochemistry was determining the enzymatic reactions by which organisms convert diverse nutrients into energy and biomass (1). Despite the extensive knowledge of metabolic reaction networks that resulted, the means by which metabolic reaction rates (fluxes) are controlled remain incompletely understood, even in highly studied model microbes. Most metabolic regulatory mechanisms were derived from studying the kinetics of isolated enzymes in vitro. Although powerful for discovering specific regulatory interactions, this reductionist approach has been less effective at revealing the impact of regulation in the intact cell. Metabolic regulation in vivo depends not only on enzyme kinetics, but also on how much the concentrations of substrates and products change across physiological states, and how enzymes respond in the presence of physiologic concentrations of other metabolites (2-4).
One framework for systematically and quantitatively investigating metabolic flux control in cells is metabolic control analysis. In this approach, the impact of enzyme activities on pathway fluxes is captured by their flux control coefficients ( ), which reflect the fractional change in pathway flux (J) in response to a fractional change in enzyme activity (E): (2). While mathematically elegant, experimental assignment of flux control has proven difficult. The most straightforward approach involves modulating enzyme activities on a one-by-one basis, which is taxing, especially because flux control may also reside in distal cellular reactions, rather than pathway enzymes themselves (3, 5-7). For example, the rate of glycolytic flux may be determined by total cellular ATP demand rather than by glycolytic enzyme expression (8).
To take advantage of growing systems-level data, an alternative approach is differential equation modeling of metabolic dynamics. Through fitting experimental metabolic concentration data, such an approach can identify quantitative kinetic parameters (e.g., kcat, Km, Ki), as well as regulatory interactions (4, 9-12). The difficulty is that parameter and regulator identification requires a global, non-linear search in high-dimensional space and accordingly works poorly when metabolic networks are either large or incomplete. Therefore, there is a need for robust and scalable new approaches (13).
We developed a method that we term Systematic Identification of Meaningful Metabolic Enzyme Regulation (SIMMER). By combining steady-state proteomic, metabolomic, and fluxomic data, SIMMER quantitatively evaluates the physiological mechanisms underlying flux control on a reaction-by-reaction basis. Specifically, given measurements of fluxes, metabolites, and enzymes across multiple steady states, SIMMER tests whether the observed fluxes through an individual reaction can be explained by a Michaelis-Menten relationship between substrate, product, and enzyme concentrations (Fig. 1A). When a misfit is observed, it then searches for potential regulators that significantly rectify the discrepancy (Fig. 1B). Because parameters and regulators are identified one reaction at a time, the approach is scalable. The resulting regulatory insights are local: they describe the factors determining individual reaction fluxes (net reaction rate), not overall control of pathway flux. To apply SIMMER, we analyzed the metabolome, proteome, and fluxome of yeast growing in 25 different steady-state conditions. SIMMER recapitulated much of known yeast metabolic regulation and revealed previously unrecognized regulation. It also revealed the quantitative contribution of substrate, product, enzyme, and allosteric effector concentrations to control of cellular metabolic reaction rates, with small molecule metabolites collectively playing a predominant role in determining flux.
Figure 1.
Integrative analysis of fluxes and metabolite and enzyme concentrations via SIMMER. (A) Measured flux (determined by flux balance analysis constrained with uptake, excretion, and biomass composition data) is related, on a reaction-by-reaction basis, to enzyme and metabolite concentrations via a Michaelis-Menten equation. The extent to which variation in flux across experimental conditions can be explained by enzyme and metabolite abundances is assessed. Heatmaps reflect the measured fluxes, enzyme abundances, and metabolite abundances. Each heatmap contains 5 columns grouped together for each limiting nutrient, arranged from slower to faster growth. The groups of 5 columns are arranged in the order of phosphorus, carbon, nitrogen, leucine, and uracil limitation. For expanded heatmaps, see Fig. S2, S5, and S9. Scenario 1 shows the glycolytic reaction catalyzed by triose phosphate isomerase (see also Fig. 2) and Scenario 2 pyruvate kinase (see also Fig. 5). Each panel compares measured flux (red dots) to the Michaelis-Menten predictions (blue dots) across the 25 nutrient conditions. (B) Identification of allosteric regulators. When a simple Michaelis-Menten expression did not account for the observed fluxes, the impact of adding allosteric regulation by measured metabolites was tested. Physiologically important regulation significantly enhances the fit to the fluxes. Scenario 1 shows regulation of pyruvate kinase by fructose 1,6-bisphosphate and citrate and Scenario 2 shows the lack of regulation of pyruvate kinase by fructose 6-phosphate.
Results
Metabolite and enzyme concentrations and metabolic fluxes in nutrient-limited yeast
Yeast were grown at five different specific growth rates in chemostats in which amounts of carbon (glucose), nitrogen (ammonia), or phosphorus (phosphate), or (in appropriate auxotrophs) leucine or uracil were limited. Replicate experimental measurements were made from single chemostats (14). To determine fluxes, we used flux balance analysis constrained by experimental measurements of nutrient uptake, waste excretion, and biomass generation, including detailed analysis of biomass composition, which varied significantly across conditions. For example, nucleic acid content (mainly in the form of ribosomal RNA) was higher at fast specific growth rates (15, 16), whereas fat and polyphosphate accumulated when nitrogen was limited (Fig. S1). With some important exceptions, such as more amino acid biosynthesis when leucine was limited and less glycolytic flux when glucose was limited, flux correlated strongly with specific growth rate. The range of fluxes that were equally compatible with the experimental observations was determined by flux variability analysis (17, 18), providing fluxes with error estimates for 233 metabolic reactions (Fig. S2). We refer to the fluxes determined by experimentally-constrained flux balance and variability analysis as the “measured fluxes,” to differentiate them from subsequent flux predictions based on metabolite and enzyme concentrations. These measured fluxes agree well with recent literature using 13C-tracers to determine fluxes in carbon limited yeast (Fig. S3 and S4) (19, 20).
The relative concentrations of 106 metabolites were previously measured across these chemostat conditions by LC-MS/MS (21). We augmented these observations by determining absolute metabolite concentrations by an isotope ratio-based approach (22). Overall, metabolite abundances depended strongly on the limiting nutrient (Fig. S5). Products derived from the limiting nutrient, such as nucleotide triphosphates in phosphorous limitation, were depleted at slow specific growth rates, whereas related metabolites lacking the limiting element, such as nucleosides in phosphorous limitation, accumulated.
We also used an isotope ratio-based LC-MS/MS approach to analyze the proteome. To determine relative protein abundances across conditions, we compared each experimental sample to a common 15N-labeled internal reference sample (23, 24). Quantitative data with good reproducibility was obtained for over 20,000 peptides representing 1,187 proteins (Fig. S6). Unlike metabolites, the abundances of many proteins, especially that of ribosomal proteins, depended primarily on specific growth rate irrespective of the limiting nutrient (Fig. S7). Quantitatively, the fraction of concentration variation explained by specific growth rate alone for the proteome was greater than that for the metabolome, but less than that for the transcriptome (Fig. S8) (25). The measured proteins included 370 metabolic enzymes representing over 90% of those covered using selected reaction monitoring (SRM) based approaches tailored for enzyme quantitation (Table S1) (26, 27). Much like metabolite abundances, amounts of metabolic enzymes varied strongly depending on the limiting nutrient. Compared to proteins overall, metabolic enzymes depended less on the specific growth rate and more upon which nutrient was limiting growth (Fig. S8). Notable nutrient-specific trends included down-regulation of glycolytic enzymes and up-regulation of TCA ones in carbon limitation, and increased levels of amino acid biosynthetic enzymes in leucine limitation (Fig. S9). Thus, relative to transcripts or proteins overall, the abundances of metabolites and metabolic enzymes are tied more strongly to the particular nutrient environment. In the case of metabolites, this may directly reflect mass action, whereas for metabolic enzymes it may reflect regulation designed to maintain appropriate flux as amounts of nutrients and thus metabolites change.
Integration of concentration and flux data through SIMMER
In total, we obtained the requisite data for SIMMER analysis (flux, enzyme concentration, substrate concentrations, and optionally product concentrations) for 56 reactions. For all reactions, we applied a reversible Michaelis-Menten rate law that assumes random-order enzyme mechanism and competitive binding of substrates and products (4, 28, 29). Kinetic parameters were identified by non-linear optimization to maximize the consistency of the Michaelis-Menten output (based on the measured enzyme and metabolite concentrations) and measured flux. For 50% of the reactions, Michaelis-Menten kinetics explained much of the physiological flux variation (R2 > 0.35). As an illustrative example, consider the triose-phosphate isomerase (Tpi1) reaction in glycolysis which converts dihydroxyacetone phosphate (DHAP) into glyceraldehyde 3-phosophate (GAP). Triose-phosphate isomerase reaction flux was lowest when carbon was limited but otherwise correlated with specific growth rate, and this was explained by lower enzyme amounts when carbon was limited and higher substrate concentration in fast-growing cells (Fig. 2, A and B).
Figure 2.
Analysis of flux control through triose-phosphate isomerase and PRPP amidotransferase. (A) Substrate, product, and enzyme concentrations for the triose-phosphate isomerase reaction in glycolysis. The 25 experimental conditions are laid out across the X-axis as per Fig. 1. (B) Michaelis-Menten equation relating concentrations to flux and extent of agreement between measured fluxes (red) and the best Michaelis-Menten fit (blue). (C) Substrate, product, and enzyme concentrations for the PRPP aminotransferase reaction, the committed step of purine biosynthesis. (D) Michaelis-Menten equation relating concentrations to flux and extent of agreement between measured fluxes and the best Michaelis-Menten fit. (E) Concentrations of the reaction inhibitor AMP and extent of agreement between measured fluxes and the best Michaelis-Menten fit after including AMP as a regulator. DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; PRPP, phosphoribosyl pyrophosphate.
In the case of triose-phosphate isomerase, although the fit was not perfect, it was not improved significantly, based on the likelihood ratio test with q-value based false discovery rate (FDR) correction (30), by including biochemically annotated inhibitors of the enzyme, such as ATP or phosphoenolpyruvate, as reaction regulators (31). For other reactions, however, inclusion of regulators enhanced the fit. For example, for the first committed step of purine biosynthesis, amidophosphoribosyltransferase (Ade4), our metabolite measurements included one putative regulator from yeast and eight from other organisms. Among these, inhibition by adenosine monophosphate (AMP), which generally accumulates during slow growth, significantly improved the fit (p < 0.00003; q < 0.02) (Fig. 2, C through E); all other putative regulators were not statistically supported (p > 0.05 for each). Inhibition of the first committed step of purine biosynthesis by AMP is a prototypical feedback circuit in yeast (32, 33). Thus, from a set of candidates based on prior knowledge of metabolism, SIMMER identified physiologically relevant regulation of yeast purine biosynthesis.
We were interested in see if this approach could also identify previously unrecognized regulation. In principle, every measured metabolite can be tested as a potential activator or inhibitor of every reaction. The major potential difficulty of this approach, however, is generating false positives due to correlation between metabolites. To explore this issue, we reviewed the literature to identify known physiological metabolic regulatory events. We identified 20 such “gold standard” regulators, occurring across 16 different reactions (33-35). Comprehensive testing of all metabolites as both activators and inhibitors of these reactions identified an average of 49 regulators per reaction that significantly improved the flux fit (q < 0.1 by likelihood ratio test). The identified regulation included 10 of the 20 gold standard regulatory events (50%), which were substantially enriched compared to other previously reported biochemical regulation of these enzymes (24%) and all other metabolites tested as activators and inhibitors (24%) (p < 0.02; Fishers exact test). Nevertheless, the specificity of the predictions was low.
Because the quantitative influence of many metabolites cannot be distinguished, we focused on putative reaction regulators, irrespective of organism, drawn from the BRENDA database (31). This allowed us to capitalize on prior knowledge while identifying regulation that was previously unrecognized in S. cerevisiae. To integrate prior knowledge with our new data, we used a Bayesian approach, trained by the 20 instances of gold standard regulation (33-35). Specifically, we used Bayes’ theorem to determine the probability of each regulatory interaction, in light of the experimental data, based on (i) its ability to account for the measured fluxes, Pr(Data ∣ Regulation) and (ii) its prior probability, Pr(Regulation). A penalty for the additional parameters introduced by regulation was also included based on Akaike information criteria (AICc) (Fig. 3) (36). Due to the small number of gold standard regulatory events relative to all regulation listed in BRENDA, Pr(Regulation) is low (median ~0.03), reflecting that, among putative regulators in BRENDA, physiologically meaningful regulation is a priori rare.
Figure 3.
Integration of experimental data and literature knowledge to predict physiological regulators of yeast metabolism. Candidate reaction equations with and without regulation follow a standard Michaelis-Menten form, with substrate, product, and enzyme taken from standard metabolic reconstructions and regulators selected based on reported regulators in the BRENDA database of the reaction in any organism (in the Michaelis-Menten equation, R = 1 implies no regulation). A list of 20 gold standard instances of physiological yeast metabolic regulation was assembled based on prior literature, and used to determine the prior probability of a candidate regulator i being a physiological regulator, Pr(Regulationi). On average, Pr(Regulationi) is low, consistent with physiological regulation being rare. The extent of fit between the measured fluxes and those predicted by candidate reaction equation determines Pr(Data ∣ Regulationi). By Bayes’ theorem, the product of these two probabilities is Pr(Regulationi ∣ Data), the probability that regulatory event i is physiologically meaningful. A penalty for the additional parameters introduced by regulation was also included, to yield a final determination of whether the regulatory event is supported. Best supported refers to the lowest AICc. Other supported refers to any alternative regulators that improve upon the unregulated model (AICc < AICcnoreg).
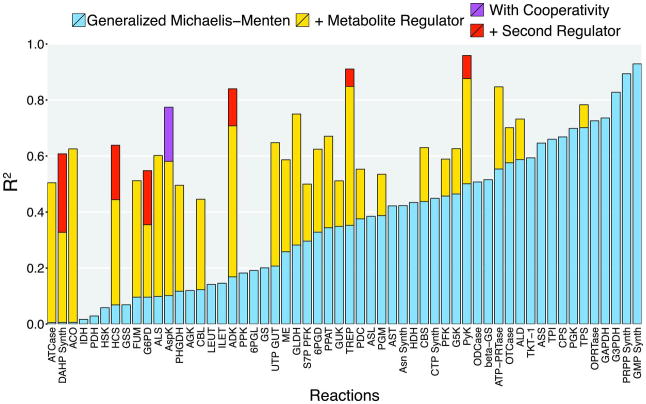
We used the above strategy to assess each of 729 candidate literature regulators of the 56 reactions for which we had sufficient data. For reactions in which one or more regulators were individually supported, we tested also cooperative binding of regulators and the inclusion of a secondary regulator. For 17 reactions, generalized Michaelis-Menten kinetics fit the data reasonably well (R2 > 0.35), and was supported over any regulation. We additionally identified 29 reactions for which physiologically-relevant regulation was supported, including six reactions best described by two physiological regulators and one reaction with cooperative regulation, for a total of 35 regulatory interactions (Fig. 4 and Table S2). Additionally, we identified 22 alternative regulators that were also supported (lower AICc than unregulated; note that each of these individually significantly improves fit with p < 0.01). The distinction between top and secondary predictions was generally modest, but unlikely to be due to noisy measurement of regulators (Fig. S10). Both the best-supported regulators and the alternative regulators were highly enriched for gold standard regulatory interactions, each containing 5 of the 20 total (p < 0.002 by Fishers exact test), validating the overall SIMMER approach (Tables S3, S4, and S5). For each substrate and each instance of regulation, the Michaelis-Menten fit determined metabolite affinity (Km/i/a) based solely on the cellular data. These cellular affinity constants correlated with corresponding values from biochemical literature (Fig. S11), providing further validation (31). In addition to five instances of gold standard yeast regulation (33-35), the 35 best-supported regulatory interactions encompassed four regulators with strong in vitro support in yeast, but no previous evidence for their physiological importance (37-40). A majority of the predicted regulation had not been previously proposed in S. cerevisiae.
Figure 4.
Consistency between measured flux and Michaelis-Menten fits for 56 cellular metabolic reactions. For 29 reactions, inclusion of regulation by a measured metabolite was statistically supported. R2 was determined by Pearson correlation of measured flux with the output of the Michaelis-Menten equation across the 25 experimental conditions. Reaction abbreviations can be found in Table S4.
Biochemical confirmation of three previously unrecognized instances of yeast allosteric regulation
To evaluate the power of SIMMER for finding new yeast regulatory interactions, we tested biochemically 10 of the best-supported predictions, verifying three: inhibition of ornithine transcarbamylase (Arg3) by alanine, pyruvate decarboxylase (Pdc1) by phenylpyruvate, and pyruvate kinase (Cdc19) by citrate. This relatively low validation rate is due in part to the fact that true regulatory interactions are rare (the false positive rate of our top predictions was only 3%) (Tables S6 and S7). It further reflects the difficulty of selecting the correct true regulator from correlated metabolites. For example, we incorrectly predict phenylpyruvate as a regulator of DAHP synthase; the true regulator, phenylalanine, correlates with phenylpyruvate and was predicted as an alternative (Table S4).
Ornithine transcarbamylase sits at the branchpoint between carbamoyl phosphate assimilation into arginine versus pyrimidines. Although Arg3 activity is thought to be regulated through expression (33), SIMMER predicted that alanine is a physiological inhibitor, and we biochemically confirmed inhibition with a Ki of 15 mM, which is below the typical cellular concentration of alanine in yeast (Fig. S12). Alanine concentrations are increased under nutrient conditions where amino acids are more abundant than nucleotides (such as stringent limitation for phosphorus or uracil). Thus, alanine inhibition of Arg3 serves to dampen arginine synthesis when pyrimidines are needed instead.
Pyruvate decarboxylase (Pdc; Pdc1,5,6) shunts pyruvate towards ethanol during yeast fermentation. Although pyruvate sits at one of the most critical branch points in central metabolism, little is known about physiologic regulation of pyruvate of decarboxylase. In Z. bisporus, a spoilage yeast in the Saccharomycetaceae family, phenylpyruvate is an alternative substrate for pyruvate decarboxylase, whose presence inhibits pyruvate decarboxylation (Ki ~5 mM) (41). We confirmed that similarly to Z. bisporus Pdc, S. cerevisiae Pdc1 can make phenylacetaldehyde from phenylpyruvate, which also inhibits the canonical reaction with pyruvate with a Ki of between 1 and 4 mM (Fig. S13). Such inhibition was not shared by the structurally related metabolites phenylalanine or α-ketoglutarate. The downstream phenylacetaldehyde product, phenylethanol, is excreted as a quorum-sensing molecule that promotes foraging behavior in nitrogen-limited yeast (42). Phenylpyruvate concentrations are high under conditions in which nitrogen or leucine are limited and its inhibition of Pdc1 apparently serves to limit carbon wasting. Thus, SIMMER revealed a new means of coordinating carbon and nitrogen metabolism in yeast.
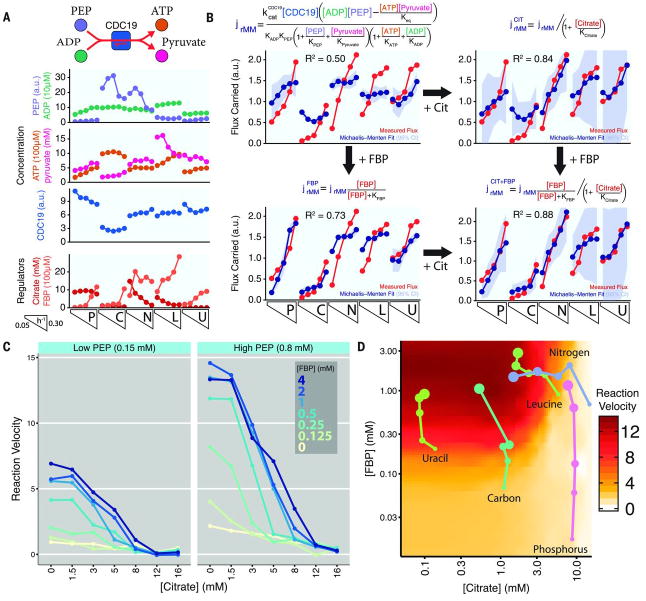
SIMMER also identified citrate as a candidate inhibitor of pyruvate production through the terminal glycolytic enzyme, pyruvate kinase (Cdc19; Pyk2 is a minor isozyme in glucose-fed cells). Due to its role in glycolytic regulation and the switch to a fetal isoform, Pkm2, in human cancer, pyruvate kinase is among the most heavily studied metabolic enzymes. Both human Pkm2 and yeast Cdc19 are strongly activated by fructose 1,6-bisphosphate (FBP) (43, 44). In yeast, upon glucose removal, FBP concentrations drop dramatically, shutting off pyruvate kinase and thereby preparing cells for gluconeogenesis. At the higher FBP concentrations found in glucose-fed cells, however, we observed only moderate sensitivity of Cdc19 to FBP concentrations (Fig. 5, A and B). This suggested that an alternative mode of regulation might control pyruvate kinase flux, with SIMMER pointing to citrate (or equivalently, isocitrate, which co-eluted in our metabolomics method). Citrate is more abundant than isocitrate in cells, and also proved to be the more potent Cdc19 inhibitor with a Ki of ~5 mM (Fig. 5C). Citrate concentrations are high when amounts of phosphate, nitrogen, or leucine are limited. When phosphate was limited, across specific growth rates, abundance of pyruvate kinase was high and counterbalanced by high concentrations of citrate, with flux increasing with specific growth rate primarily because of increased amounts of FBP (Fig. 5D). In contrast, when nitrogen or leucine were limited, flux increased with specific growth rate primarily due to decreasing amounts of citrate (Fig. 5D). Thus inhibition of pyruvate kinase by citrate works in concert with control of abundance of pyruvate kinase and FBP to coordinate glucose catabolism with the availability of other essential elemental nutrients.
Figure 5.
Pyruvate kinase regulation by fructose 1,6-bisphosphate (FBP) and citrate. (A) Substrate, product, enzyme, and regulator concentrations for the pyruvate kinase reaction that controls exit from glycolysis. (B) Extent of agreement between measured fluxes and the best Michaelis-Menten fit, with and without including FBP, citrate, or both as regulators. The improvement in fit due to both regulators was significant (p < 10-4). FBP is a classical activator of pyruvate kinase, whereas citrate had not been described as an inhibitor. (C) Biochemical confirmation that physiological citrate concentrations inhibit the dominant yeast pyruvate kinase isozyme (Cdc19) across the physiologically relevant range of phosphoenolpyruvate (PEP) and FBP concentrations. (D) Impact of cellular FBP and citrate concentrations across the 25 experimental conditions on Cdc19 activity. Pyruvate kinase activity (per enzyme and assuming fixed substrate concentrations of 0.8 mM PEP and 0.2 mM ADP) was determined by interpolation of the biochemical data shown in Panel C. Each data point reflects a single experimental condition, with the size of the point corresponding to the specific growth rate. Increasing flux with faster specific growth rate is accomplished in phosphorous, carbon, and uracil limitation through increasing concentrations of FBP, whereas in nitrogen and leucine limitation it is achieved through decreasing concentration of citrate.
A common feature of the above three new physiological regulatory interactions is that each spans pathways. This contrasts with the nine instances of known yeast regulation that were reconfirmed by SIMMER, each of which was “local” (involving interactions of metabolites and enzymes within the same pathway, including five instances of standard feedback inhibition). Thus, beyond their individual significance, collectively these new discoveries raise the possibility that cross-pathway regulation may be more common and important than currently appreciated.
Quantitative analysis of physiological flux control
In addition to revealing reaction regulators, SIMMER yields quantitative rate laws that can be analyzed to evaluate the mechanisms controlling the rates of cellular metabolic reactions. To assess the impact of physiological variation in enzyme and metabolite concentrations on reaction flux, we can partition the total variability in flux across conditions into the contributions of individual biochemical players. Specifically, the impact of any given biochemical species (substrate, product, enzyme, allosteric regulator) depends on the variance in its concentration across physiological conditions and the sensitivity of net reaction rate to its concentration ( ). Normalization to the sum across all players gives the fractional contribution of the species to control of net reaction rate. We term this normalized partitioning of physiological reaction-rate control, “metabolic leverage:”
For each of the 29 metabolic reactions best described by reaction equations lacking regulation or involving regulation which was biochemically verified in S. cerevisiae, the relative metabolic leverage of substrate and product concentrations, allosteric effector levels, and enzyme abundances is shown in Fig. 6A (for similar analysis involving all of the best-supported SIMMER predictions, see Fig. S14). The observed distributions of metabolic leverage are robust across all well-fitting parameter sets (Fig. S15 and Table S8). For reactions that can change net direction as a function of environmental conditions (“reversible reactions”), metabolic leverage was divided between substrates, products, and enzymes with no contribution from allosteric regulation and a majority of leverage residing in substrates alone (Fig. 6, B and C). For reactions that do not change net direction but are somewhat reversible, allostery made a small contribution, which increased for strongly forward-driven reactions (ΔrG°’ < -5 kJ/mol) (45), with enzymes, substrates and allosteric regulators each playing a major role (Fig. 6, B and C). Overall, the combined impact of metabolites (substrates, products, allosteric effectors) was more than double that of enzymes.
Figure 6.
The primary determinant of net cellular metabolic reaction rates is metabolite concentrations. To capture the impact of metabolite and enzyme concentrations on flux through individual reactions, we determined the metabolic leverage of each species: the fraction of flux variability across experimental conditions that is explained by variation in each species’ concentration, as determined by the sensitivity of the reaction rate to the species and the variance of the species’ concentration across the physiological conditions. (A) Ternary plot displaying breakdown of metabolic leverage into substrates and products (sum of leverage of all such species), enzyme, and regulators. Metabolic reactions containing either no regulation or validated yeast-specific regulation are included (N = 29). (B) The reactions in panel A are grouped according to reversibility. Reactions were classified as reversible or not, based on whether the direction of net flux changes across physiological conditions (18); they were subsequently divided into strongly forward driven versus net forward based on the standard free energy (cutoff -5 kJ/mol) (45). (C) Pie charts displaying the average metabolic leverage of substrate, product, regulator, and enzyme concentrations. Less reversible reactions are subject to greater allosteric regulation.
Discussion
As systems-level measurement technologies steadily improve, there is an increasing need for methods that yield concrete regulatory insights from large-scale data. Previous attempts to infer metabolic regulation from metabolomics data have typically fit dynamic metabolite concentration changes with systems of differential equations. Although this approach works well for small networks, it scales poorly, as inaccurate description of a single reaction leads to global errors. By measuring flux, rather than inferring it based on changing metabolite concentrations, regulation can be evaluated on a reaction-by-reaction basis independent of the scale of the total system. Independent evaluation of each reaction equation enables rapid parameter identification and facilitates discovery of regulatory interactions. Because reaction rates are measured in cells, the approach is well-positioned to identify regulation that depends on the presence of physiological concentrations of other metabolites. We used this approach to identify physiologically relevant regulation, finding three previously unrecognized instances of S. cerevisiae cross-pathway metabolic regulation.
These included new ways of regulating pyruvate flux: both pyruvate production and consumption were inhibited by metabolites that rise during nitrogen limitation, with the main pyruvate kinase isozyme Cdc19 inhibited by citrate and pyruvate decarboxylase inhibited by the phenylpyruvate, the α-ketoacid analogue of phenylalanine. In E. coli, α-ketoacids including α-ketoglutarate inhibit carbon catabolism under low nitrogen conditions, in part by direct inhibition of Enzyme I (46,47). Such inhibition simultaneously cuts off flow into upper glycolysis and out of lower glycolysis, enabling adjustment of glycolytic flux with only modest changes in intermediate concentrations (46). We find that related regulatory mechanisms operate in yeast to help balance upper and lower glycolysis (43, 48), with citrate inhibiting both FBP production (49) and PEP consumption (Fig. 5). Citrate, but not phenylpyruvate, is also abundant when amounts of phosphate are limited, suggesting a common bottlenecking of glycolysis when either nutrient is limiting, but differential regulation of the fate of the pyruvate that is formed. Beyond their specific importance to understand metabolic control, these new regulatory interactions also suggest some general principles: the importance of long-range feedback interactions (involving metabolites outside of the immediate pathway being regulated) and of context-dependent shifts in physiological regulators, such as a switch from FBP as the predominant regulator of pyruvate kinase when carbon is limited (50) to citrate when nitrogen is limited.
In addition to identifying new regulatory interactions, we evaluated the overall mechanisms controlling the physiological rates of individual metabolic reactions in yeast, revealing a predominant role for metabolite concentrations, especially substrate concentrations. This observation builds on previous work in yeast and other microbes which has found that changes in the transcriptome and proteome are insufficient to account for flux changes (51-54). It is in agreement with recent findings that changes in substrate concentrations are a major contributor to overall changes in flux (53, 55).
Pathway-level flux control depends both on how individual reaction rates are determined, as is assessed by metabolic leverage, and on how reactions interact, as is assessed by flux control coefficients (2,3,56). For highly reversible reactions, net flux is sensitive to small shifts in substrate to product ratios. Such reactions respond to other reactions, rather than themselves exerting control over pathway flux (3), with substrate and product concentrations dominating metabolic leverage. In contrast, strongly thermodynamically forward-driven reactions have greater pathway flux control and are major targets of allosteric regulators. We find that substrate concentration also plays an important role in determining flux through these reactions, with the role of metabolites (substrates, products and allosteric effectors) collectively double that of enzymes.
An important question regards the generality of our observation that metabolite concentrations exert more metabolic leverage than enzymes. This observation was made across steady-state yeast cultures differing in terms of their nutrient environment. By working at steady state, we should in principle favor slow regulatory events, like modulation of enzyme concentrations, and thus it is particularly striking that metabolite concentrations play a larger role. This indicates substantial inefficiency in enzyme utilization: If enzymes were running near Vmax in all conditions, the only way to control flux would be through enzyme concentrations. Instead of prioritizing enzyme efficiency (57), it appears that nutrient-limited yeast overexpress enzymes (51, 52), perhaps so that they can grow quickly when nutrient conditions improve (58). This excess enzyme capacity imparts a plasticity to metabolism (7, 57, 58), shifting metabolic leverage onto substrates, products, and allosteric effectors. It is likely that enzyme efficiency is more important in rich nutrient conditions, and that the metabolic leverage of enzymes is greater in such environments. In cases where metabolic variation occurs without variation in environmental nutrient conditions (such as across organs in mammals, which are all fed by the same nutrients carried in the blood stream), enzymes concentrations are likely to play a yet larger role. Nevertheless, even in such settings, many reactions rates may be determined largely by metabolite concentrations, as this allows for proper fluxes to be achieved without fine-grained control of enzyme concentrations. Indeed, mechanisms regulating yeast enzyme concentrations often work at the relatively coarse levels of whole pathways (such as Arg80, Arg81 and Arg82) and even multiple pathways (such as Gcn4) (33, 55, 59, 60). This may reflect an evolutionary prioritization of robustness and flexibility over enzyme efficiency. Alternatively, it could reflect that making some excess enzyme is “cheaper” than building the machinery to more precisely control individual enzyme concentrations.
While we provide a relatively comprehensive metabolic dataset and leverage this data to both confirm canonical regulation and predict regulation that was previously unrecognized in yeast, the majority of SIMMER predictions were invalidated. Predicting regulation is difficult because most metabolites are not physiological regulators of a given enzyme and erroneous predictions can readily occur due to either unmeasured or incorrectly measured variables or an inability to discriminate between potential regulators. More accurate flux measurement through stable isotope labeling experiments (19, 20, 61), deeper metabolome coverage, and evaluation of enzyme post-translational regulation and sub-cellular localization (62-64) should enable more complete understanding of flux regulatory mechanisms. Moreover, extending the conditions studied to additional nutrients (e.g., other carbon sources) and genetic perturbations (e.g. gene knockouts) holds the potential for breaking down correlations between metabolites, and thereby allowing regulators to be more reliably discriminated. With sufficient additional data, rather than relying on candidates from BRENDA, it should be possible to treat all metabolites as potential regulators.
The SIMMER approach can be applied to any pseudo-steady-state metabolic system where metabolite and enzyme concentrations and fluxes can be measured under many conditions. This may allow identification of metabolic regulatory mechanisms in less well-studied microbes. Due to the lack of requirements for organism-specific prior knowledge, SIMMER should be applicable to most microbes that can be cultivated in chemostats. In the meantime, due to the importance of S. cerevisiae as an industrial organism, our quantitative insights into its metabolic regulatory mechanisms may be of immediate utility in metabolic engineering.
Materials and Methods
Overview of SIMMER
Systematic Identification of Meaningful Metabolic Enzyme Regulation (SIMMER) uses ‘omic-scale data to investigate the kinetics of individual metabolic reactions. The four primary steps constituting the SIMMER method are:
Measure the relative concentrations of metabolites (M), enzymes (E), and fluxes (jF) across diverse conditions. Metabolites and enzymes can be directly measured by mass spectrometry; fluxes cannot be directly measured and accordingly are inferred from other experimental data and the known metabolic network stoichiometry.
For each reaction of interest, generate one or more reaction equations that relate measured metabolites (M: substrates, products and putative regulators), enzymes (E) and kinetic parameters (Ω) to reaction flux (jP = g(M,E,Ω)). In this study, reaction equations (g) are based on Michaelis-Menten kinetics and differ in terms of inclusion of regulators.
Fit each reaction equation to determine how well each reaction equation’s prediction (ĵp) agrees with measured flux (jF).
Compare alternative reaction equations to identify which model is best supported. Prior literature is used to assess the plausibility of each model such that models that are less likely a priori will require stronger quantitative evidence to achieve the same overall support.
Strains and culture conditions
FY derivative strains of Saccharomyces cerevisiae were grown at 25 distinct steady states in chemostats by simultaneously modulating both limiting nutrient and dilution rate, as per Boer et al. 2010 (21). Based on previous work showing that technical variability substantially exceeds biological variability for ‘omic analyses of yeast chemostats (14), each ‘omic analysis was based on technical replicates derived from a single biological culture, with proteomics and fluxomics from one culture and metabolomics from a prior culture. For each nutrient condition, the five dilution rates were ~ 0.05, 0.11, 0.16, 0.22, and 0.3 h-1. The strains were prototrophic with the exception of uracil limitation (MATa ura3-52) and leucine limitation (MATa leu241). Each culture was maintained at pH 5 throughout. Culture density was routinely measured using a Klett Colorimeter. Cultures reached steady-state, as determined by stable density over 24 h, after ~5 to 7 days. Intracellular volume per culture volume was determined by directly measuring the volume of packed cells in 5-10 mL of culture (PCV Packed Cell Volume Tube 87005, TPP, Trasadingen, Switzerland).
Strategy for inferring metabolism-wide fluxes in chemostat cultures
Our general approach for estimating metabolism-wide fluxes was to measure the major inputs into metabolism and the major outputs from metabolism (boundary fluxes) in each condition and to use these fluxes as constraints for flux balance analysis. Isotopic labeling was not used in this study to probe internal fluxes due to limitations related to cost and time. Nevertheless, our inferred fluxes agree well with those that have previously been found using 13C-based metabolic flux analysis (MFA) of carbon-limited yeast cultures (19, 20). The generally good agreement can be understood in terms of the sufficiency of boundary fluxes for constraining many fluxes of interest. For example, glucose uptake rate and ethanol excretion rate are generally sufficient to determine glycolytic flux. In contrast, certain branched internal pathways, such as the pentose phosphate pathway, are less well constrained in the absence of isotope tracer measurements. Accordingly, perhaps unsurprisingly, our successful regulation discoveries occur in glycolysis and in anabolic pathways whose fluxes are reliably constrained based boundary fluxes, including the rate of biomass synthesis which constrains the anabolic fluxes.
Determining metabolite uptake and excretion rates through 1H-NMR
Media samples for 1H-NMR analysis were prepared from each of the 25 chemostats by filtering 10 mL of culture through a 0.45 μm HNWP filter (HNWP02500, Millipore, Billerica, MA). The concentration of metabolites in the flow-through (spent media) was analyzed by 1H-NMR, with 2 independent 10 mL samples analyzed per chemostat. D2O, sodium azide, and 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) were added to final concentrations of 10%, 500 μM, and 500 μM, respectively. Using a 500 MHz Advance III (Bruker), 1H-NMR spectra were collected using the following acquisition parameters: TD = 65536, NS = 32, D1 = 10s, SW = 16, O1P = 4.68, P1 = 7.2, P12 = 2400, SPW1 = 0.0009, SPNAM1 = Gaus1 180r.1000. NMR peaks were quantitated using rNMR (65), by choosing the cleanest peak of each metabolite and normalizing this peak’s height relative to the 500 μM DSS internal standard. Absolute quantification was possible through comparison to external standards that were similarly normalized with respect to an internal DSS standard. From the steady state concentration of metabolites in spent media and the culture dilution rate, the rate of excretion (of metabolites not present in the original media) is given by:
| (2) |
The rate of uptake of supplied nutrients can be found analogously by comparing the final concentration of nutrients to their concentration in formulated media:
| (3) |
Limitation-dependence of yeast biomass composition
Because the composition of yeast cells strongly influences the steady-state metabolic fluxes required to support biomass synthesis, the abundances of major biomass constituents (16) were measured for each of the 25 chemostat conditions. For each chemostat, ~200 mL of culture was collected on ice. Yeast were separated from the culture media by centrifugation at 2,600g at 4°C for 30 minutes, the pellet was resuspended in water to remove all extracellular salts, and yeast were again pelleted (1600g at 4°C for 5 min). The resulting pellets were stored at -80°C.
For dry weight, carbohydrate, protein, and phosphate measurement, homogenized cells were prepared as follows: Frozen cell pellets were dehydrated for 24 hours at 60°C using a vacuum concentrator and weighted. Approximately 30 mg of dry yeast was resuspended in 1 mL of homogenization buffer (0.01 M KH2PO4, 1 mM EDTA, pH 7.4) with 0.5% Triton-X, mechanically homogenized (Mini-Beadbeater-16; Biospec Products, Bartlesville, OK), and aliquoted into 96-well assay plates (20 μL homogenized yeast per well) for spectrophotometric measurements. Sample preparation for determination of other biomass components is described in the relevant subsections.
Total carbohydrate
Phenol-sulfuric assay for total carbohydrates was used to assess the total amount of trehalose, glycogen, mannan and glucans found in 96-well plates of homogenized yeast (66). Trehalose concentration, as determined by mass spectrometry (see below), was subtracted from total carbohydrate, leaving a total polysaccharide fraction of dry weight.
Total protein
BCA assay (Pierce BCA Protein Assay Kit; Thermo Fisher Scientific, Waltham, MA) was used to determine the relative amounts of protein across the chemostat conditions, with previous measurements of yeast composition in identical carbon or nitrogen-limited chemostats used as an absolute reference (15, 16).
Total inorganic phosphate
A colorimetric assay (K410-500; BioVision, Milpitas, CA) was used to measure inorganic phosphate. Analysis of polyphosphate standards confirmed that sample preparation steps had already hydrolyzed polyphosphates to monomers; thus, measured phosphate was a combination of inorganic monophosphate and polyphosphates.
RNA
Frozen pellets as above were thawed on ice, and RNA was extracted using phenol-chloroform. The relative abundance of RNA in each sample was measured using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA), with previous measurements of yeast composition in identical carbon or nitrogen-limited chemostats used as an absolute reference (15, 16).
Fatty acids
The concentration of fatty acids was assessed through absolute quantification of the most abundant fatty acid tails (67). Frozen cells were resuspensed in 0.1 M HCl/MeOH (50/50) and extracted with 0.5 mL cold (-20°C) chloroform. After removing the solvent under N2 flow, 1 mL of 90% methanol, 0.3 M KOH was added and samples were heated for one hour at 80°C to saponify lipids. Uniformly 13C-labelled C16-0, C16-1, C18-0, C18-1 were added as internal standards. After acidification with 100 μL glacial formic acid, saponified fatty acids were extracted twice with 1 mL of hexane. Samples were dried under N2 gas and resuspended in chloroform/methanol/H2O (1/1/0.3) to a final concentration of 2 μL cell volume per mL. Fatty acids were quantified by LC-MS as previously described (68), except using a Q-TOF 6550 (Agilent Technologies, Santa Clara, CA) instead of an orbitrap mass analyzer, with absolute concentrations determined by comparing the endogenous unlabeled peaks to the isotope-labeled standard peaks.
DNA
The average DNA content of each cell was inferred from a previously determined relationship between chemostat dilution rate and the fraction of budded cells (fraction budded = 0.936 - 1.971DR) (25). As budded cells are diploid and unbudded cells are haploid, deoxyribonucleotide content could be calculated from genome size, GC content and the number of cells per culture volume.
Soluble metabolites
Intracellular pools of the 20 metabolites with a median concentration of greater than 1 mM were considered in defining biomass composition. For analytical methods for soluble metabolites, see below.
Effluxes to biomass
In a chemostat culture, continual yeast growth (and associated biomass synthesis) is balanced by the loss of yeast cells through dilution:
| (4) |
To relate jsynthesis to metabolic fluxes, we assumed that the relative proportions of monomers (in protein, RNA, etc) were invariant across the chemostats conditions with the following proportions: Amino acids % by weight: A: 8.8%; C: 0.17%; D: 8.42%; E: 9.45%; F: 4.74%; G: 4.67%; H: 2.2%; I: 5.42%; K: 9.03%; L: 8.33%; M: 1.62%; N: 2.88%; P: 4.06%; Q: 3.3%; R: 6.03%; S: 4.18%; T: 4.89%; V: 6.64%; W: 1.24%; Y: 3.96% (18); Ribonucleotides: % by weight: AMP: 24%; CMP: 22%, GMP: 25%; UMP: 29% (18); Polysaccharides: % by weight: β-glucan: 46%; glycogen: 21%; mannan: 33% (18).
Synthesis of each macromolecule pool was represented as a single reaction, consuming monomers in the proportions above, along with polymerization costs (69, 70). For example, the RNA consumption reaction removes ATP, CTP, GTP, and UTP and liberates two phosphates per nucleotide assimilated. This formalism allows a single reaction to represent each biomass component, including the measured uncertainty in its abundance and thereby in the associated biosynthetic flux.
Inferring metabolism-wide fluxes using a constraints-based approach
The stoichiometry and directionality of yeast reactions were taken from a modified version of the YEASTNET 7 Saccharomyces cerevisiae genome-scale model (18). Reactions were added to allow for polyphosphate synthesis (polyphosphate kinase: (ATP + (PO3)n → ADP + (PO3)n+1) and orotate excretion, which we observed experimentally. ATP consumption occurred via three routes: i) a minimal ATP maintenance flux proportional to cell mass (1 mmole per gDW/h) (71), ii) growth-related reactions, iii) an unconstrained and unproductive ATP hydrolysis reaction which was manually added to the model. Reactions that never carried flux under our experimental conditions were removed. This resulted in 2,787 reactions involving 1,843 metabolites that could carry non-zero flux. This count includes equivalent reactions and identical metabolites duplicated across different compartments. The stoichiometry of the 2,787 reactions is defined by the stoichiometric matrix (S).
To determine metabolism-wide fluxes for each experimental chemostat condition, we determined the balanced flux distribution (j such that Sj = 0) that conforms to our experimentally determined boundary fluxes (ĵ) as closely as possible (i.e. min ) (72). Because individual boundary fluxes were measured with variable accuracy, analogously to weighted least squares regression, we weighted squared residuals, (ĵ–j)2 by the experimental precision (σ-2) of that measurement. To minimize unproductive metabolic cycles, absolute fluxes |j| were penalized by a scalar value c(1) or c(2) (i.e. for each reaction k). c(1) was used for high flux reactions (glycolysis, TCA cycle, boundary fluxes and associated transport reactions and the unproductive ATP hydrolysis reaction) and c(2) (which was larger than c(1)) was used for all other reactions. was small relative to the primary penalty for misfitting of the experimental measurements. The optimal vector j can be found using quadratic programming:
| (5) |
Because there may be multiple optimal solutions {j}, we used flux variability analysis (17) to determine the range of fluxes through each reaction (i), which give an identical minimal cost (θ) in Equation 5. This range was taken as the experimentally observed flux range. This approach involves separately minimizing and maximizing the flux through each reaction, treating θ as a constraint:
| (6) |
Both optimization problems were implemented using the Gurobi Optimizer (73). Code to reproduce flux estimation is available on GitHub.
A total of 233 reactions both carried non-zero flux under at least some conditions and were well constrained based on flux variability analysis with median across conditions of (FVA range) / (midpoint of FVA range) < 0.3. These 233 reactions were treated as candidates for further kinetic analysis.
Proteomics
To quantify the relative levels of proteins across 25 chemostats, each experimental sample was paired to an internal 15N-labelled reference sample and analyzed in triplicate.
Protein extraction
Yeast from 300 mL of culture volume were collected by rapid filtration onto a 0.8 μm cellulose acetate filter (CA089025, Sterlitech, Kent, WA). The filter containing the yeast cake was folded and placed into a 50 mL conical vial, which was immediately frozen by immersion in liquid nitrogen, followed by storage at -80°C. Complete yeast particle disruption was accomplished by placing each cryofrozen yeast-laden filter into a 50 mL canister of a Retsch CryoMill (Haan, Germany) precooled on dry ice and pulverizing the total material into a fine powder by cryogenic ball milling at -196°C. Yeast protein was extracted from the powder by resuspension in an 80°C solution of 4% SDS, 100 mM Tris pH 8, 1 mM DTT, and Halt Protease and Phosphatase Inhibitors (ThermoFisher Scientific, MA), followed by boiling for 20 min with periodic agitation. Insoluble material, including filter material, was removed from the samples by centrifugation at 24,000g for 20 minutes. The concentration of extracted protein was determined using the Pierce BCA Protein Assay Kit (ThermoFisher, MA). Experimental samples were mixed 1:1 with an equal protein content of 15N-labeled reference protein solution. The protein reference was yeast cultured in phosphate-limited chemostats (N = 2, mixed together) growing at a dilution rate of 0.05 h-1 in which (15NH4)2SO4 was the sole nitrogen-source.
LC-MS data acquisition
Each experimental sample was subject to buffer-exchange into a urea-based solution, thiol reduction and alkylation, and trypsin digestion using the FASP procedure (74). Peptides were diluted from the digestion solution directly into isoelectric focusing (IEF) buffer, placed above pH 3-10 IPG strips, and subject to IEF using the OffGel apparatus and its reagent kits (Agilent Technologies, Santa Clara, CA). Twenty-four fractions were collected for each sample and each fraction was directly analyzed by nano-LC-MS/MS three times: once using an Agilent 6538 Q-TOF platform and twice using an Agilent 6550 Q-TOF platform. Both Q-TOF platforms were equipped with Agilent 1260 nano-flow HPLC systems and an Agilent ChipCube LC/ion source interface. Large capacity Chip (II) units (150 mm 300 Å Zorbax C18 chip with a 160 nL trap column) were utilized on the 6538 platform, while Polaris-HR-Chip 3C18 units (150 mm 180 Å 3 μm Polaris C18 chip with a 360 nL trap column) were employed on the 6550 system. MS was collected using the high resolution setting (40k resolving power) at 500 ms acquisition time; MS/MS was collected on the top 3 most abundant multiply charged species, at 250 ms acquisition time per spectrum. Raw data was converted from the Agilent .dat format into mzXML format using the software Trapper (Seattle Proteome Center, Trans-proteomic Pipeline) for downstream data analysis.
Peptide identification and quantification
Each mzXML file was analyzed using Mascot (version 2.2.07, Matrix Science, London, UK) with 0.01 Da parent and fragment mass tolerance, methionine oxidation as a variable modification, cysteine carbamidomethylation as a fixed modification, and 15N Metabolic Quantitation. The Mascot results files were processed through Scaffold (version 3.6.0, Proteome Software Inc., Portland, OR) with XTandem (version 2007.01.01.1, The GPM, Alberta, Canada) to confirm peptide identities and report peptides with at least 95% identification confidence in proteins with at least 99% identification confidence and at least 2 peptides identified. The ion counts for the observed peptide mass spectra, which represent the sum of the experimental (light) and reference (heavy) forms of each identified peptide, were fit to theoretical isotopologue distribution (based on the natural abundance of isotopes and the isotopic purity of (15NH4)2SO4) for the experimental and reference peptides to determine their relative abundances and associated error estimates. Using the co-elution probability of every pair of identified peptides, the elution time was predicted for peptides identified in only a subset of the samples. The ion counts for the experimental and reference forms of these peptides were also added together over the elution periods of the peptide in samples without explicit MS2 identifications. When a peptide was identified in multiple fractions, the ion currents from each fraction were summed to achieve a sample-level summary of experimental and reference ion current. 13,135 peptides, where both the experimental and reference ion was quantifiable in at least 15 samples, were chosen for further analysis. For each peptide, i, and condition, c, the log2-ratio of the experimental to reference peptide ion current integrated areas was treated as the peptide relative abundance, .
Protein quantification
To infer the relative abundance of a protein from its corresponding peptides, we need to aggregate multiple noisy estimates into a single consensus. The relative abundance of each peptide was taken as the geometric mean of technical replicates. To downweight noisier peptides, each peptide’s variance was estimated as the mean squared error over technical replicates. The relative abundance (and variance) of a protein in each condition was found by weighting the peptide relative abundances (x) according to their inverse variance:
| (7) |
The relative abundance of proteins with no measured peptides in a condition (4.8% of all measurements and only 0.3% of enzymes catalyzing the 56 reactions of interest) was determined using knn-imputation (75).
Metabolomics
The relative concentrations of 106 metabolites were previously measured across our 25 experimental conditions (21). We further determined: i) absolute concentrations of 56 metabolites, so that growth-associated dilution of large metabolite pools could be accounted for and so that metabolite affinities could be biochemically interpreted, ii) uncertainty of metabolite concentrations (see below).
Metabolite absolute concentrations
Absolute concentrations of 56 of the metabolites measured in Boer et al. 2010 were determined by comparison to commercial standards. The approach depended on whether the metabolite contained nitrogen. For nitrogen-containing metabolites, including amino acids, unlabelled standards of known concentration were added into 15N-labeled phosphate-limited chemostat samples. These samples were CBZ-derivatized and analyzed by LC-MS in negative ionization mode, as per Boer et al. 2010 (21), with absolute concentrations of the metabolites in the phosphate-limited chemostat samples determined based on ratios of the unlabeled and labeled peaks (22). For metabolites that do not contain nitrogen, their relative concentrations in a fast-specific growth rate (DR = 0.30) carbon-limited chemostat were compared to a contemporaneous batch culture containing glucose as a carbon source, for which absolute concentrations have been previously determined by comparison to commercial standards (76). Samples were analyzed by LC-MS in negative mode, as per Xu et al. 2012 (43).
Variance of metabolite relative abundances
The variance of metabolite measurements (in log2 space) was determined from four technical replicates per chemostat. Residuals across chemically similar metabolites were often highly correlated (for example, within amino acids or within nucleotides). Such correlations impact the error estimates for reaction fluxes derived by plugging the concentrations into a Michaelis-Menten reaction equation. For example, if errors in [ATP] and [ADP] measurement are positively correlated, the error in the ratio ([ATP]/[ADP]) is less than that based on standard propagation of error, whereas the error in [ATP]*[ADP] would be greater than that based on standard propagation of error. To enable more accurate propagation of error from metabolite concentrations into the Michaelis-Menten fluxes, the residual covariance across all metabolites was estimated (77).
Since all ‘omic measurements rely only on technical (and not biological) replicates, variances are underestimated, but only modestly as biological variability is less than technical variability (14). Because subsequent analyses are based on finding trends across a continuum of conditions (rather than looking for differences between conditions), underestimation of random biological error does not create a systematic bias in favor of false discoveries.
Summary of integrative ‘omics data
Overall, we determined the relative abundances of 106 metabolites and 304 enzymes and measured flux through 233 reactions. For 56 reactions, fluxes were determined, at least one isoenzyme was measured through proteomics, and major substrates were quantified using metabolomics (Table S9). For 46 of these reactions, ‘omic measurements from all conditions (N = 25) were used to assess kinetic parameters and identify regulation. For the remaining 10 reactions, data in one auxotrophic limitation differed greatly (> 5-fold) from all of the native limitations and from the other auxotrophic limitation. For example, orotate phosphoribosyltransferase (OPRTase) flux is zero under uracil limitation due to the ura3 auxotrophy, and inclusion of uracil limitation data would inappropriately skew the SIMMER results for the OPRTase reaction. Based on a 5-fold difference criterion (relative to the most similar native limitation data), leucine limitation data were excluded for 6 reactions (ALS, AGK, ASL, ASS, LEUT, OTCase) and uracil limitation data for 4 reactions (ATCase, CPS, OPRTase, ODCase). In these 10 cases, SIMMER analysis was conducted using data from the other four limiting nutrients (total of 20 rather than 25 experimental conditions).
Reaction equations
For each reaction, an equation with no small molecule regulation (generalized Michaelis-Menten kinetics) was generated, as were alternative equations involving one or more previously reported metabolite activators or inhibitors. Possible regulators of each reaction were queried based on E.C. number. Using the BRENDA database SOAP API, we drew on both S. cerevisiae specific regulation and non-yeast regulation (31). All putative regulators were matched to ChEBI compounds (78) and their synonyms to determine if they were experimentally measured. Each model of reaction kinetics was translated into a Michaelis-Menten reaction form using an extended form of the convenience kinetics rate law (28, 29), which assumes a random-order mechanism with explicit grouping of metabolites based on binding sites:
| (8) |
E1, E2, … are the concentrations of isoenzymes that catalyze the reaction. Substrates (A) and products (B), together denoted as reactants (R), were assigned to binding sites (Γ) based on structural atom-pair similarity using ChEBI structures (78, 79) and assignments were manually verified. Stoichiometric coefficients of substrates, products and reactants are given by α, β and γ and respectively. Previous work has shown that this reaction form can accurately reproduce bi-bi ping-pong and sequential mechanisms (29). To generate reaction forms containing a potential allosteric regulator D, a pre-multiplier was applied to the Michaelis-Menten equation: for allosteric activation, D/(D + Ka), and for inhibition: 1/(1 + D/Ki). All affinity constants (Ki, Ka, Kd) are treated equivalently when fitting reaction equations and therefore we use Kd to represent any affinity constant in subsequent sections.
Fitting reaction equations to experimental data
Each reaction equation g (Equation 8) is an algebraic relationship that translates enzyme (E) and metabolite (M) abundances and kinetic parameters (Ω = (Kd, kcat, Keq); |Ω| = # metabolites + # enzymes + 1) into predicted flux, jP = g(M, E, Ω). To determine the support for g, we must find a set of kinetic parameters that best approximates FBA-determined flux (jF) with jP. In doing this, we want both an optimal parameter set (e.g. the maximum-likelihood estimator (MLE) or maximum a posteriori probability (MAP) estimator of Ω,Ω̂) and a measure of parameter uncertainty. All parameters (Kd; kcat; Keq) were inferred rather than taken from literature. This approach was chosen due to lack of absolute quantitation for one or more substrates or products of most reactions thereby precluding comparison to literature Keq values, the lack of reliable literature values for many kinetic parameters, and potential for systematic differences between biochemical and in vivo parameters. If we constrained the kinetic parameters based on biochemical literature, such systematic differences could result in over-fitting of regulation to correct for erroneous parameter values.
We assume that deviations between jP and jF are independent and identically distributed (iid) and follow a Normal distribution with common variance, given by the mean squared error. The most popular method of fitting relationships of the form y = g(X, Ω) + ε, is non-linear least squares (NLS) regression which aims to estimate a set of unknown parameters that minimizes the least squares departures between y and g(X, Ω). While widely used, this method is poorly suited to enzyme kinetics for three reasons: i) NLS may become trapped in local minima (80); we found this to be a particularly severe problem for the reaction equations used in this study, ii) NLS may underestimate parameter standard errors because of the non-linear relationships between parameters (80), iii) NLS does not readily allow for uncertainty in the response variable, which in our case is flux. This latter issue is particularly critical for SIMMER, as the measured fluxes span the interval determined by flux variability analysis.
To avoid the limitations of non-linear regression, we employed a Bayesian approach to estimate the kinetic parameters Ω:
| (9) |
The posterior distribution of Ω was estimated using Markov Chain Monte-Carlo. Each round of the algorithm involves: i) proposing a set of kinetic parameters (Ω) drawn from the proposal distribution, ii) evaluating jP = g(M, E, Ωproposed), iii) determining how well jP agrees with jF (i.e. the likelihood of Ωproposed), iv) evaluating the posterior probability of Ωproposed (Equation 9) to determine whether Ωproposed should be accepted or rejected.
Prior and proposal distribution on Ω
The prior and proposal distributions for each kinetic coefficient were equivalent and constructed to appropriately reflect reaction chemistry.
Since each disassociation constant (Kd) reflects the affinity of an enzyme for a metabolite (M), the value of Kd is only meaningful when compared to this metabolite’s concentration: . To symmetrically span the extremes of near unsaturation and near saturation, the prior on each metabolite’s Kd was a Log-Uniform distribution centered about the metabolite’s median log concentration:
| (10) |
The range of this distribution was chosen such that near unsaturation or saturation could be achieved across all conditions even when metabolite concentrations varied greatly across conditions.
Similarly to Kd, the value of Keq is meaningful only when compared to the reaction quotient Qr. The disequilibrium ratio (Qr/Keq) equals one when the reaction is at equilibrium and approaches zero when the reaction is far from equilibrium (in the forward direction). To symmetrically span meaningful values of free energy (about thermodynamic equilibrium), the prior on Keq was a Log-Uniform distribution centered about the log of the median reaction quotient:
| (11) |
The range of this distribution was chosen such that near irreversibility could be achieved across all conditions even when the reaction quotient varied greatly across conditions.
For numerical reasons, kcat parameters were not drawn from a proposal distribution, but were instead optimized based on the current values of Kds and Keq. Their prior probability can be considered as an improper Uniform prior over the non-negative real numbers: Pr(kcat)~ Unif[0,∞).
Evaluating g(M, E, Ωproposed): Every reaction equation, g(M, E, Ω), can be represented as the product of a non-linear equation, f(M, Kd, Keq), and a linear equation, h(E, kcat), where g = fh. For the simplest case of Michaelis-Menten kinetics: f = [S]/([S] + KM) and h =kcat[E] (i.e. Vmax).
Once values of Kds and Keq have been drawn from the proposal distribution, the fraction of maximum activity of each condition (o = f(M, Kd, Keq)) can be found. This fraction of maximum activity, scaled by the measured enzyme concentration and kcat, approximates measured flux (i.e. jF = kcat[E]o + ε). To find the value of kcat (or multiple kcats if isozymes are measured) that minimizes the residual squared error, we can use linear regression to solve for kcat, treating [E]o as predictors. To enforce that kcat values are strictly nonnegative, kcat parameters are found using non-negative least squares (NNLS). Once all coefficients in Ω have either been drawn from the proposal distribution, or directly estimated in the case of kcats, jP = g(M, E, Ωproposed) can be evaluated. Metabolites that were not experimentally measured were treated as having invariant concentrations.
Determining the likelihood of Ω
As is the case for NLS, we assume that deviations between jP and jF follow a Normal distribution with variance given by the mean squared error. If measured fluxes ( ) are point estimates, the log-likelihood of a proposed parameter set, Ω, l(Ω∣ M, E, jF) would be:
| (12) |
Here, ϕ is the Normal probability density function (pdf). Since we are accounting for experimental uncertainty in fluxes through flux variability analysis, Equation 12 must be modified to represent as a uniform density between some lower ( ) and upper bound ( ). This log-likelihood function reflects the equivalent likelihood of jP falling anywhere within the interval determined by flux variability analysis:
| (13) |
Evaluating the posterior probability of proposed
Due to the component-wise Uniform/Log-Uniform priors on Ω (Equations 10 and 11), the posterior probability of Ω, Pr(Ω∣ M, E, jF), is proportional to the likelihood (natural exponential of Equation 13):
| (14) |
Algorithm for calculating posterior distribution of Ω
For a single reaction with K non-linear kinetic parameters ( ) tracked over the course of I sets of metropolis updates, the joint values of these kinetic parameters can be optimized using Algorithm 1.
Algorithm 1. MCMC-NNLS inference of kinetic parameters.
Input: Metabolite concentrations (M), enzyme concentrations (E), measured flux (jF) and a reaction equation (jP = g(M, E, Ω)).
Output: Posterior distribution of kinetic parameters (Ω).
Initialization:
begin
- for k ← 1 to K do
- ▪ drawn from Log-Uniform prior
Calculate ocurrent = f(M, Ωcurrent) using reaction form
Find kcat parameters using NNLS
Update lcurrent = l(Ωcurrent∣ M, E, jF)
Iteration:
- for i ← 1 to I do
- ▪ for k ← 1 to K do
- ○ drawn from Log-Uniform prior
- ○ Calculate oproposed and kcat values
- ○ lproposed = l(Ωproposed∣ M, E, jF)
- ○ draw d from Unif(0, 1)
- ○ if lproposed < lcurrent or : then
- Ωcurrent = Ωproposed
- lcurrent = lproposed
- ▪
return Ωtrack
To obtain distributions of parameter values that appropriately reflect the true distribution, Pr(Ω∣ M, E, jF), the Markov algorithm was implemented as follows: i) the first 8,000 samples from the Markov chain were discarded (as the initial values are determined by the starting assumptions regarding the parameter distribution, not the experimental data fit), ii) thereafter, only every 300th parameter set of the Markov chain was recorded as an element of Ω (as values from nearby iterations of the chain are inappropriately highly correlated) until a total of 200 Markov samples had been generated, iii) the algorithm was run 10 times from different initial conditions. By comparing the 10 Markov chains from each model using the multivariate potential scale reduction factor (MPSRF) (81), we verified that in all cases the distinct initial conditions had converged to equivalent posterior distributions.
From the 10 runs of Algorithm 1, for each reaction equation, we generated 2,000 samples from the posterior distribution of Ω. For purposes of model comparison, we calculated the MAP of each model and the corresponding MAP estimate of parameters:
| (15) |
| (16) |
When estimates of Ω using non-linear least squares were possible (for most reaction equations NLS failed to converge from over 100 initial conditions), the MAP likelihood was either indistinguishable from the NLS fit or substantially higher (due to NLS not reaching a global least squares minima).
Allowing for cooperativity of regulator binding
Cooperative binding of regulators was assessed for each literature-informed allosteric activator or inhibitor by adding an appropriate Hill coefficient (n) to the pre-multiplier; for activation and for inhibition 1/(1+(D/ki)n). Parameters for models with cooperativity were inferred as above, with the inclusion of an additional proposal distribution for the Hill coefficient. The prior and proposal distributions on Hill coefficients were distributed as Log-Uniform: Pr(log2n) ~ Unif(-3, 3). This distribution provides symmetry of negative cooperativity (n < 1) and positive cooperativity (n > 1) about non-cooperativity (n = 1).
Code to apply the MCMC-NNLS algorithm to reaction-level data for the 56 studied reactions is available on GitHub.
Evaluating how well reaction forms fit given experimental uncertainty
To determine whether discrepancies between measured fluxes and Michaelis-Menten predictions can be accounted for by inaccurate measurements of reaction species, the uncertainty of the Michaelis-Menten prediction based on the collective error of all reaction species was calculated using the multivariate delta method (82):
| (17) |
Here, s is the set of all measured enzymes and metabolites involved in a reaction. j refer to pathway fluxes, while v are reaction-level fluxes. Σ is a square residual covariance matrix (if 4 measured metabolites and 2 proteins = 6 × 6 matrix). Diagonal entries are the variance of species in condition c, and off-diagonal entries are covariances between species (i.e. how correlated is the measurement error of two species weighted by their residual uncertainty). Covariances between proteins were unknown and assumed to be zero, while covariances between metabolites were calculated as above.
Assessing regulator significance
Our general strategy was to first identify all putative regulators that significantly (after FDR correction) improved the fit. We then used a Bayesian approach to assess the plausibility of each regulator. This incorporated how well-fit a model was in terms of its maximum a posteriori probability (Equation 16), the model’s a priori plausibility based on literature evidence, and the model’s complexity. Bayesian integration of these factors allowed us to identify the most likely model.
Determining regulators that significantly improve fit
Each reaction equation containing regulation was compared to the unregulated Michaelis-Menten fit for the same reaction to determine if the improvement in fit due to regulation was significant given the increased number of fitted parameters. Because we are comparing nested models with differing numbers of degrees of freedom, a likelihood ratio test was used (because of Uniform priors, the posterior probability of each model is proportional to the likelihood: Equation 14). An increased likelihood (Equation 16) indicates better fit. The expected increase in fit (2 Δ l) due to an irrelevant parameter will follow a distribution with p equaling the number of additional parameters. By this approach, a p-value was determined for each regulator. Regulators that were not significant at a FDR of 0.1 (30) were removed from further consideration to yield a set of candidate regulators. Models involving multiple regulators were built in a step-wise fashion from individual significant regulators, with a more complex model accepted only when it was significantly superior to all simpler models.
Integrating quantitative support and prior knowledge to compare alternative models of reaction kinetics
The above approach facilitates the quantitative evaluation of many reaction mechanisms without regards to their plausibility.
To integrate our data and prior literature knowledge, we took a Bayesian approach, aiming to maximize Pr(Model ∣ Data), where “Model” is a Michaelis-Menten equation including regulation and “Model” is “True” if the regulation is physiologically meaningful. Using Bayes theorem:
| (18) |
Pr(Data ∣ Model) accounts for how well concentrations predict fluxes and is directly assessed by Equation 16. Pr(Data) is independent of the choice of the model, thus:
| (19) |
After empirically estimating Pr(Model) and accounting for differences in model complexity (see below), alternative models of kinetics for each reaction can be simultaneously compared balancing each model’s plausibility and quantitative support.
Literature support for regulation, Pr(Model): to carryout the above strategy, we needed to estimate the a priori probability that each tested regulator is physiologically meaningful in yeast: Pr(Model). From literature reviews and recent primary literature (33-35), 20 reported physiological regulators of 16 reactions were assembled. This gold standard list was initially treated as a priori true regulation. We also constructed a list of 186 BRENDA regulators of the 16 gold standard reactions (31); we assume that the vast majority of regulation in this list is non-physiological. Accordingly, all members (except for the 20 members of the gold standard list) were initially treated as a priori false regulation.
Using BRENDA, for each regulator, i, we determined how many times it has been reported in S. cerevisiae ( ), and in other species ( ). We also determined the total number of regulators that are tested for the reaction corresponding to i (Ni). The log (base 2) of this number was treated as a third covariate ( ). Treating gold standard regulation as true regulation (yi = 1) and other BRENDA regulators as false regulation (yi = 0), the predictive power of xsce, xother, and xn were determined using logistic regression:
| (20) |
The effect of yeast annotations (β̂sce = 0:84) and non-yeast annotations (β̂other= 0:08) were both significantly positive (p < 0.001), while the effect of a higher number of total regulators (β̂n= -0.91) was significantly negative (p < 0.005). The output of the logistic regression (i.e.p̂) was applied across all regulators to determine the a priori Pr(Model). The prior probabilities for models with multiple regulators were the product of individual regulator prior probabilities. For models without any regulation, we used a prior probability of 1. This equals the maximum prior probability that could exist for any model including regulation.
Predicting the most likely reaction form for each reaction based on Pr(Model ∣ Data)
For each reaction, Michaelis-Menten kinetics and all regulatory reaction forms that significantly improve fit of simpler models (q < 0.1) were simultaneously considered. The quantitative support for each model, Pr(Data ∣ Model) (Equation 16), and literature support for each regulatory mechanism, Pr(Model) (Equation 20), were combined to estimate each model’s total support, Pr(Model ∣ Data) (Equation 19). The relative support for each model was corrected for overparameterization by using the Akaike Information Criterion with correction (AICc) (36):
| (21) |
Here, n is the number of conditions and k is the number of fitted parameters. The reaction form with the lowest AICc was considered most plausible. Alternative significant regulators are reported in Table S4.
Experimental verification of predicted regulation
Ten regulators of seven reactions (ATP-PRTase: His1, DAHP Synthase: Aro3,4, G6PD: Zwf1, 6PGD: Gnd1, OTCase: Arg3, PYK: Cdc19 (Pyk1),Pyk2, PDC: Pdc1,5,6), predicted by SIMMER, were tested using in vitro biochemistry (Table S2). For the PYK and PDC reactions, while minor isozymes exist (Pyk2, Pdc5,6), Pyk1 and Pdc1 are the predominant isozymes in glucose, where all of our experimental data were collected.
Purified S. cerevisiae enzymes were commercially available for G6PD, 6PGD and PDC (10127655001, P4553, P9474; Sigma-Aldrich, MO). Using proteomics, we confirmed that commercial PDC was primarily composed of Pdc1 (total spectral counts from Pdc1 were more than three times higher than those from Pdc5 or Pdc6). A strain containing an N-terminal 6-His affinity tagged version of S. cerevisiae Cdc19 was obtained from Xu et al. 2012 (43). For Arg3, Aro3, Aro4, and His1, a C-terminal 6-His affinity tag was attached to the native gene using PCR (amplified from BY4742 genomic DNA) and incorporated into a p426GAL plasmid (high-copy plasmid, URA3 selectable, gene products are galactose-inducible). This plasmid was transformed into DBY12045 (MATa, HAP1+ ura3DΔ0) to generate strains expressing Arg3-His6, Aro3-His6, Aro4-His6 and His1-His6. Protein expression of all pGal-His6-tagged strains was induced by first growing overnight cultures until an OD600 of 0.6 on SC - URA media + 2% Raffinose, followed by 12 hours of induction on SC - URA media + 2% Galactose. Total protein was extracted through mechanical homogenization in the presence of Y-PER reagent (Thermo Scientific, MA) containing 10 μL/mL EDTA-free HALT protease inhibitors (Thermo Scientific, MA) and 2 mM PMSF. Tagged enzymes were purified using HisPur Cobalt Spin Columns (Thermo Scientific, MA), and successful purification was confirmed by SDS-PAGE.
Activities of G6PD (ZWF1), 6PGD (Gnd1), ATP-PRTase (His1), and DAHP synthase (Aro3,4) were assessed using previously published kinetic spectrophotometric assays (83-85). Assays used to identify or confirm previously unrecognized regulation are detailed below. Putative regulators of each enzyme were adjusted to the pH of the reaction buffer.
Spectrophotometric (NADH-coupled) analysis of pyruvate kinase (PYK: Cdc19) activity
Production of pyruvate by Cdc19 was coupled to NADH oxidation via lactate dehydrogenase (LDH) (86). Initial concentrations were 0.15 or 0.8 mM PEP (as indicated), 0.2 mM ADP, 0.2 mM NADH, and 4.3 unit/mL LDH (L2500, Sigma-Aldrich) in 50 mM Tris, 4 mM MgCl2, 75 mM KCl, pH 7.5 buffer. Reaction progress was tracked in real time at 340 nm and 30°C for one hour. Initial reaction velocities were fitted and blank (Cdc19-free) activity was subtracted.
Spectrophotometric (direct) confirmation of pyruvate kinase regulation
The stoichiometric conversion of PEP (ɛ232 = 2.84 mM-1cm-1) to pyruvate (ɛ232 = 0.6 mM-1cm-1) as the reaction progressed was tracked by capitalizing on the difference in molar absorptivity at 232 nm between these two species (86). Initial concentrations were 0.15 and 0.4 mM PEP, 0.2 mM ADP in 50 mM Tris, 4 mM MgCl2, 75 mM KCl, pH 7.5 buffer. Reaction progress was tracked in real time at 232 nm and 30°C for one hour. Initial reaction velocities were fitted and blank (Cdc19-free) activity was subtracted.
Spectrophotometric (NADH-coupled) analysis of pyruvate decarboxylase (PDC: Pdc1) activity
Production of acetaldehyde by Pdc1 was coupled to NADH oxidation via alcohol dehydrogenase (ADH). Initial concentrations were 0.1-30 mM pyruvate, 0.1 mM NADH, 3 unit/mL ADH (A7011, Sigma-Aldrich) in 200 mM citric acid, pH 6.0 buffer. Reaction progress was tracked in real time at 340 nm and 30°C for one hour. Initial reaction velocities were fitted and blank (Pdc1-free) activity was subtracted.
NMR-based confirmation of pyruvate decarboxylase (PDC: Pdc1) regulation
Acetaldehyde accumulation by PDC was directly detected using 1H-NMR. PDC was incubated at 30°C for one hour in 10 mM pyruvate, 200 mM citric acid, pH 6.0 buffer. At the end of incubation, the reaction was stopped by a five-minute incubation at 98°C. Acetaldehyde was quantified by 1H-NMR as above (see “Determining metabolite uptake and excretion rates through 1H-NMR”).
Mass spectrometry based detection of acetaldehyde and phenylacetaldehyde produced by pyruvate decarboxylase (PDC: Pdc1)
PDC was incubated at 30°C for one hour with either 10 mM pyruvate or 10 mM phenylpyruvate in 200 mM citric acid, pH 6.0 buffer. An equal volume of acetonitrile was added to quench the reactions. Acetaldehyde and phenylacetaldehyde were TSH (p-Toluenesulfonyl hydrazide) derivatized and quantified by LC-MS in positive ionization mode, as per Boer et al. 2010 (21).
Spectrophotometric (NADP-coupled) analysis of ornithine transcarbamylase (OTCase: Arg3) activity
OTCase activity was tracked by coupling the loss of phosphate from carbamoyl phosphate to glycogen phosphorylase and ultimately NADP reduction (87). Initial concentrations were 5 mM ornithine, 100 μM carbamoyl phosphate, 0.8 mg/mL glycogen, 0.6 mM NADP+, 0.5 unit/mL glycogen phosphorylase a (P1261, Sigma-Aldrich), 0.24 unit/mL phosphoglucomutase (P3397, Sigma-Aldrich), 0.85 unit/mL glucose 6-phosphate dehydrogenase (G7877, Sigma-Aldrich), 2 μM glucose 1,6-bisphosphate, 20 μM AMP in 10 mM Tris, 10 mM Bis-Tris, 10 mM CAPS, 4 mM DTT, 0.4 mM MgCl2, pH 7.0 buffer. Because this approach tracks phosphate liberated from carbamoyl phosphate, it was necessary to minimize both the amount of contaminating phosphorous and the amount of non-enzymatic phosphate released through the spontaneous breakdown of carbamoyl phosphate. Before quantification, all reagents except carbamoyl phosphate were combined and incubated for one hour at room temperature to consume contaminating phosphate. Freshly prepared carbamoyl phosphate was added to initiate the assay. Reaction progress was tracked in real time at 340 nm and 30°C for one hour. The signal from non-enzymatic breakdown of carbamoyl phosphate was accounted for using Arg3p-free blanks whose predictable baseline could be reliably subtracted from assays including Arg3p.
Mass spectrometry based confirmation of ornithine transcarbamylase regulation
Citrulline accumulation by OTCase was directly detected using LC-MS. Arg3p was incubated at 30°C for two hours in 5 mM ornithine, 100 μM carbamoyl phosphate, 10 mM Tris, 10 mM Bis-Tris, 10 mM CAPS, 4 mM DTT, 0.4 mM MgCl2, pH 7.0.The reaction was stopped by the addition of -20°C extraction solvent (acetonitrile:methanol:water, 40:40:20). Citrulline was quantified by LC-MS in positive ionization mode, as per Boer et al. 2010 (21).
Metabolic leverage
The concept of metabolic leverage is to partition the cause of flux changes across conditions (at the individual reaction level) into underlying changes in the concentrations of enzymes, substrates, products, and allosteric effectors. This concept can be formulated as partitioning the environmental-driven variance in flux across conditions into the contributions from the concentrations of the relevant molecular players. Mathematically, this is the inverse of propagation of uncertainty. For example, to determine the uncertainty in a flux calculation via a Michaelis-Menten expression, one would classically propagate uncertainty from the technical variation in individual species measurements (see section “Evaluating how well reaction forms fit given experimental uncertainty”). To determine metabolic leverage, instead, we decompose the overall environmental variance in flux into the variance contributed by individual reaction species:
| (22) |
Here, Vare(vF) is the variance in measured flux across chemostats. To focus on physiologically relevant variation in flux, only the 15 prototrophic chemostats limited by an elemental nutrient were considered. are sensitivities that describe how the reaction equations output (vP) responds to changes in the mean concentration (across chemostats) of each reaction species. Sensitivities were evaluated at the mean concentration of all other reaction species using the best-supported kinetic parameters. Σe is the environmental covariance matrix describing how all reaction species covary across the chemostat conditions. Diagonal entries are the environmental variance of each reaction species . In order to determine the individual contributions of each species i, we make the simplifying assumption that environmental covariances between species are negligible and therefore Σe is diagonal:
| (23) |
The fractional contribution of each reaction species k to overall flux changes is its metabolic leverage (ψk), which is calculated by normalizing each term in Equation 23 relative to their sum such that these metabolic leverages sum to one across all reaction species:
| (24) |
Supplementary Material
Acknowledgments
Data described in the paper are presented in Table S9. This research was supported by grants from the U.S. DOE (DE-SC0012461), NIH (R01CA16359-01A1), and Agilent Technologies (Thought Leader Award) to J.D.R., NIH (R01HG002913) to J.D.S., NIH (GM046406, GM071508), and Agilent Technologies (Thought Leader Award) to D.B., and a DOE SCGF fellowship (DE-AC05-06OR23100) to S.R.H.
Footnotes
Supplementary Materials
Table S1-S9
Figure S1-S15
References
- 1.Caspi R, et al. Nucleic acids research. 2014;42:D459. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kacser H, Burns JA. Symposia of the Society for Experimental Biology. 1973;27:65. [PubMed] [Google Scholar]
- 3.Fell D. Understanding the control of metabolism. Portland Press Ltd; 1997. [Google Scholar]
- 4.Tummler K, Lubitz T, Schelker M, Klipp E. The FEBS journal. 2014;281:549. doi: 10.1111/febs.12525. [DOI] [PubMed] [Google Scholar]
- 5.Hauf J, Zimmermann FK, Muller S. Enzyme and microbial technology. 2000;26:688. doi: 10.1016/s0141-0229(00)00160-5. [DOI] [PubMed] [Google Scholar]
- 6.Kochanowski K, Sauer U, Chubukov V. Current Opinion in Biotechnology. 2013;24:987. doi: 10.1016/j.copbio.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Cornish-Bowden A, Hofmeyr JHS, Cardenas ML. Bioorganic Chemistry. 1995;23:439. [Google Scholar]
- 8.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. Journal of bacteriology. 2002;184:3909. doi: 10.1128/JB.184.14.3909-3916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzi M, Baltes M, Theobald U, Reuss M. Biotechnology and bioengineering. 1997;55:592. doi: 10.1002/(SICI)1097-0290(19970820)55:4<592::AID-BIT2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 10.Teusink B, et al. European Journal of Biochemistry. 2000;267:5313. doi: 10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed] [Google Scholar]
- 11.Chassagnole C, Noisommit-Rizzi N, Schmid JW, Mauch K, Reuss M. Biotechnology and bioengineering. 2002;79:53. doi: 10.1002/bit.10288. [DOI] [PubMed] [Google Scholar]
- 12.Link H, Kochanowski K, Sauer U. Nature Biotechnology. 2013;31:357. doi: 10.1038/nbt.2489. [DOI] [PubMed] [Google Scholar]
- 13.Gerosa L, Sauer U. Current Opinion in Biotechnology. 2011;22:566. doi: 10.1016/j.copbio.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Robinson DG, Wang JY, Storey JD. Nucleic acids research. 2015;43:e131. doi: 10.1093/nar/gkv636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulze U. Ph D thesis. Department of Biotechnology, Techinical University of Denmark; 1995. Anaerobic Physiology of Saccharomyces cerevisae. [Google Scholar]
- 16.Lange HC, Heijnen JJ. Biotechnology and bioengineering. 2001;75:334. doi: 10.1002/bit.10054. [DOI] [PubMed] [Google Scholar]
- 17.Mahadevan R, Schilling CH. Metabolic engineering. 2003;5:264. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Aung HW, Henry SA, Walker LP. Industrial Biochemistry. 2013;9:215. doi: 10.1089/ind.2013.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frick O, Wittmann C. Microbial cell factories. 2005;4:30. doi: 10.1186/1475-2859-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouhten P, et al. BMC Systems Biology. 2008;2:60. doi: 10.1186/1752-0509-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. Molecular biology of the cell. 2010;21:198. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Nature Protocols. 2008;3:1299. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6591. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong S-E, et al. Molecular & cellular proteomics : MCP. 2002;1:376. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Brauer MJ, et al. Molecular biology of the cell. 2008;19:352. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costenoble R, et al. Molecular Systems Biology. 2011;7:464. doi: 10.1038/msb.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zampar GG, et al. Molecular Systems Biology. 2013;9:651. doi: 10.1038/msb.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebermeister W, Klipp E. Theoretical biology & medical modelling. 2006;3:41. doi: 10.1186/1742-4682-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohwer J, Hanekom AJ, Hofmeyr JHS. Proc 2nd Int Symp on Experimental Standard Conditions of Enzyme Characterizations (ESEC 2006); 2007. [Google Scholar]
- 30.Storey JD, Tibshirani R. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheer M, et al. Nucleic acids research. 2011;39:D670. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyngaarden JB, Ashton DM. Nature. 1959;183:747. doi: 10.1038/183747a0. [DOI] [PubMed] [Google Scholar]
- 33.Jones EW, Fink GR. Cold Spring Harbor Monograph Archive. 1982;11B:181. [Google Scholar]
- 34.Sekine T, Kawaguchi A, Hamano Y, Takagi H. Applied and environmental microbiology. 2007;73:4011. doi: 10.1128/AEM.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraenkel DG. Yeast intermediary metabolism. Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 36.Hurvich CM, Tsai C-l. Biometrika. 1989;76:297. [Google Scholar]
- 37.Li Y, Zhang Y, Yan H. Journal of Biological Chemistry. 1996;271:28038. doi: 10.1074/jbc.271.45.28038. [DOI] [PubMed] [Google Scholar]
- 38.Khoo JC, Russell PJ. Journal of Biological Chemistry. 1970;245:4163. [PubMed] [Google Scholar]
- 39.Majtan T, et al. PloS one. 2014;9:e105290. doi: 10.1371/journal.pone.0105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandercammen A, Francois J, Hers HG. European Journal of Biochemistry. 1989;182:613. doi: 10.1111/j.1432-1033.1989.tb14870.x. [DOI] [PubMed] [Google Scholar]
- 41.Neuser F, Zorn H, Richter U, Berger RG. Biological Chemistry. 2000;381:349. doi: 10.1515/BC.2000.046. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Fink GR. Genes & development. 2006;20:1150. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, et al. Molecular cell. 2012;48:52. doi: 10.1016/j.molcel.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Nature. 2008;452:181. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 45.Noor E, Haraldsd′ottir HS, Milo R, Fleming RMT. PLoS Computational Biology. 2013;9:e1003098. doi: 10.1371/journal.pcbi.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doucette CD, Schwab DJ, Wingreen NS, Rabinowitz JD. Nature Chemical Biology. 2011;7:894. doi: 10.1038/nchembio.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You C, et al. Nature. 2013;500:7462. [Google Scholar]
- 48.van Heerden JH, et al. Science. 2014;343:1245114. doi: 10.1126/science.1245114. [DOI] [PubMed] [Google Scholar]
- 49.Mlakar T, Legisa M. Applied and environmental microbiology. 2006;72:4515. doi: 10.1128/AEM.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huberts DHEW, Niebel B, Heinemann M. FEMS yeast research. 2012;12:118. doi: 10.1111/j.1567-1364.2011.00767.x. [DOI] [PubMed] [Google Scholar]
- 51.Daran-Lapujade P, et al. Journal of Biological Chemistry. 2004;279:9125. doi: 10.1074/jbc.M309578200. [DOI] [PubMed] [Google Scholar]
- 52.Daran-Lapujade P, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15753. doi: 10.1073/pnas.0707476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chubukov V, et al. Molecular Systems Biology. 2013;9:709. doi: 10.1038/msb.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valgepea K, Adamberg K, Seiman A, Vilu R. Molecular bioSystems. 2013;9:2344. doi: 10.1039/c3mb70119k. [DOI] [PubMed] [Google Scholar]
- 55.Gerosa L, et al. Cell systems. 2015;1:270. doi: 10.1016/j.cels.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 56.Liao JC, Delgado J. Biotechnology Progress. 1993;9:221. [Google Scholar]
- 57.Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10039. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fendt S-M, et al. Molecular Systems Biology. 2010;6:356. doi: 10.1038/msb.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Natarajan K, et al. Molecular and cellular biology. 2001;21:4347. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu J, et al. PLoS Biology. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan J, Bennett BD, Rabinowitz JD. Nature Protocols. 2008;3:1328. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kitamoto K, Yoshizawa K, Ohsumi Y, Anraku Y. Journal of bacteriology. 1988;170:2683. doi: 10.1128/jb.170.6.2683-2686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiedler D, et al. Cell. 2009;136:952. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulz JC, Zampieri M, Wanka S, von Mering C, Sauer U. Science signaling. 2014;7:rs6. doi: 10.1126/scisignal.2005602. [DOI] [PubMed] [Google Scholar]
- 65.Lewis IA, Schommer SC, Markley JL. Magnetic resonance in chemistry : MRC. 2009;47(Suppl 1):S123. doi: 10.1002/mrc.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masuko T, et al. Analytical Biochemistry. 2005;339:69. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 67.Pramanik J, Keasling JD. Biotechnology and bioengineering. 1997;56:398. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 68.Kamphorst JJ, Fan J, Lu W, White E, Rabinowitz JD. Analytical Chemistry. 2011;83:9114. doi: 10.1021/ac202220b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6 W H Freeman; 2006. [Google Scholar]
- 70.Ramanagoudr-Bhojappa R, et al. Journal of Biological Chemistry. 2013;288:16185. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Famili I, Forster J, Nielsen J, Palsson BØ. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13134. doi: 10.1073/pnas.2235812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yizhak K, Benyamini T, Liebermeister W, Ruppin E, Shlomi T. Bioinformatics. 2010;26:i255. doi: 10.1093/bioinformatics/btq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurobi Optimization, Inc. Gurobi Optimizer Reference Manual. 2015 [Google Scholar]
- 74.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Nature methods. 2009;6:359. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 75.Troyanskaya O, et al. Bioinformatics. 2001;17:520. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 76.Park JO, et al. Nature Chemical Biology. 2016 [Google Scholar]
- 77.Schaefer J, Opgen-Rhein R, Strimmer K. R package version. 2010:15. [Google Scholar]
- 78.Degtyarenko K, et al. Nucleic acids research. 2008;36:D344. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao Y, Charisi A, Cheng LC, Jiang T, Girke T. Bioinformatics. 2008;24:1733. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson KA. FEBS letters. 2013;587:2753. doi: 10.1016/j.febslet.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks SP, Gelman A. Journal of computational and graphical statistics. 1997;7:434. [Google Scholar]
- 82.Lynch M, Walsh B. Genetics and analysis of quantitative traits. first edn Sinauer Associates; 1998. [Google Scholar]
- 83.Furdui C, Zhou L, Woodard RW, Anderson KS. Journal of Biological Chemistry. 2004;279:45618. doi: 10.1074/jbc.M404753200. [DOI] [PubMed] [Google Scholar]
- 84.Pedreño S, Pisco JP, Larrouy-Maumus G, Kelly G, de Carvalho LPS. Biochemistry. 2012;51:8027. doi: 10.1021/bi300808b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greenberg AJ, Hackett SR, Harshman LG, Clark AG. Molecular Systems Biology. 2011;7:563. doi: 10.1038/msb.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pon NG, Bondar RJL. Analytical Biochemistry. 1967;19:272. doi: 10.1016/0003-2697(67)90163-7. [DOI] [PubMed] [Google Scholar]
- 87.Foote J, Lipscomb WN. Journal of Biological Chemistry. 1981;256:11428. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.