Abstract
Background
Vertebrate epithelial appendages constitute a diverse group of organs that includes integumentary structures such as reptilian scales, avian feathers and mammalian hair. Recent studies have provided new evidence for the homology of integumentary organ development throughout amniotes, despite their disparate final morphologies. These structures develop from conserved molecular signalling centres, known as epithelial placodes. It is not yet certain whether this homology extends beyond the integumentary organs of amniotes, as there is a lack of knowledge regarding their development in basal vertebrates. As the ancient sister lineage of bony vertebrates, extant chondrichthyans are well suited to testing the phylogenetic depth of this homology. Elasmobranchs (sharks, skates and rays) possess hard, mineralised epithelial appendages called odontodes, which include teeth and dermal denticles (placoid scales). Odontodes constitute some of the oldest known vertebrate integumentary appendages, predating the origin of gnathostomes. Here, we used an emerging model shark (Scyliorhinus canicula) to test the hypothesis that denticles are homologous to other placode-derived amniote integumentary organs. To examine the conservation of putative gene regulatory network (GRN) member function, we undertook small molecule inhibition of fibroblast growth factor (FGF) signalling during caudal denticle formation.
Results
We show that during early caudal denticle morphogenesis, the shark expresses homologues of conserved developmental gene families, known to comprise a core GRN for early placode morphogenesis in amniotes. This includes conserved expression of FGFs, sonic hedgehog (shh) and bone morphogenetic protein 4 (bmp4). Additionally, we reveal that denticle placodes possess columnar epithelial cells with a reduced rate of proliferation, a conserved characteristic of amniote skin appendage development. Small molecule inhibition of FGF signalling revealed placode development is FGF dependent, and inhibiting FGF activity resulted in downregulation of shh and bmp4 expression, consistent with the expectation from comparison to the amniote integumentary appendage GRN.
Conclusion
Overall, these findings suggest the core GRN for building vertebrate integumentary epithelial appendages has been highly conserved over 450 million years. This provides evidence for the continuous, historical homology of epithelial appendage placodes throughout jawed vertebrates, from sharks to mammals. Epithelial placodes constitute the shared foundation upon which diverse vertebrate integumentary organs have evolved.
Electronic supplementary material
The online version of this article (doi:10.1186/s13227-017-0071-0) contains supplementary material, which is available to authorized users.
Keywords: Homology, Shark, Epithelial appendage, Anatomical placode, Dermal denticle
Background
The diversity of phenotypes among vertebrate epithelial appendages is vast and includes disparate structures of the integument such as feathers, hair, scales and teeth [1, 2]. These organs have evolved to facilitate wide-ranging aspects of survival and reproduction. Despite such diversity, these structures generally develop from patterns of reciprocal interactions between two adjacent tissue layers: the epithelium and underlying mesenchyme [1]. Where scale-like structures also arise from more derived mechanisms, for example the physical cracking of highly keratinised crocodile skin to form randomly spaced, polygonal head scales, placode-derived scales are also present on the body [3].
Recent research has revealed shared ancestry among amniote epithelial appendages, based on the observation that reptilian scales, avian feathers and mammalian hair share a common foundation during early development [4]: the anatomical placode. This structure is characterised by conserved molecular markers and columnar epithelial cells with a reduced rate of proliferation. Placodes constitute a localised thickening of the epithelium together with an underlying dermal condensate (mesenchyme) [5, 6]. Morphogenesis of the placode results in the adult form [7] and is controlled by molecular signals that participate in a complex gene regulatory network (GRN). This placode GRN is thought to be largely conserved throughout amniotes [4, 8]. However, there is a gap in our knowledge regarding the developmental processes guiding placode morphogenesis in non-amniote vertebrates. It is not known whether this GRN is conserved across all jawed vertebrates [9].
Chondrichthyans (sharks, rays and chimaeras) are the sister lineage of osteichthyans and occupy a basal position in jawed vertebrate phylogeny. They possess hard, mineralised epithelial appendages known as odontodes. Odontodes include both teeth and dermal denticles and have been observed in early vertebrates that lived as long as 450 million years ago [10, 11]. Odontodes consist of a central pulp cavity surrounded by a dentine layer, encased within an enameloid (enamel-like) covering [12, 13]. Recent work has provided new genetic evidence for the old hypothesis that teeth and denticles share deep homology and that their development is controlled by a common odontode GRN [14]. Since their likely origin as a form of body armour [15], denticles have evolved to fulfil a plethora of functions: they reduce abrasive damage [16], aid feeding [17], deter parasites [18], enable communication [19] and improve hydrodynamic efficiency [20–22]. Chondrichthyan denticles exhibit broad morphological variation to facilitate these roles [13].
In the S. canicula embryo, this variation can broadly be categorised into 3 classes: (1) the precocious embryonic denticles of the caudal tail, (2) the dorsal trunk and (3) adult type general body denticles (Fig. 1) [14]. Dorsal denticles (Fig. 1d, e) appear in two polarised rows at approximately 60–80 days post-fertilisation (dpf; Stage 31) and may trigger the subsequent emergence of general body denticles [23], as observed during feather tract patterning [24]. They are subsumed into general scalation soon after hatching [14]. General body denticles (Fig. 1f, g) are the most prevalent denticle type, appearing just before hatching at 145–175 dpf (Stage 34) [23], covering the skin in an intricate pattern when space is available and not in discrete rows [25, 26]. Before dorsal and body denticles appear, four rows of caudal denticles emerge at 52–60 dpf (Stage 30) [23]; two rows are present (dorsal and ventral) laterally on either side of the tail fin tip (Fig. 1b, c, j–m) [27]. Caudal denticle number can vary between 9 and 13 units which form on either dorsal row, and between 5 and 10 units which form on either ventral row [23]. The placodes of these denticles consist of a squamous epithelium overlying a basal epithelial layer of columnar cells, with condensing underlying mesenchyme (Fig. 3). They develop sequentially from posterior to anterior, approximately equidistant from one another [23, 27–29]. During morphogenesis, these denticles also mineralise in a posterior to anterior progression [23]. Despite being patterned in rows similarly to dorsal denticles, they display an irregular petaliform shape with variation in cusp number and have a less restricted polarity than other denticle types (Fig. 1l, m). These units have a dentine collar fusing the main cusp to the simple base [27], anchored within the mesenchymal dermis via connective tissues. Caudal denticles are transient epithelial structures that are lost before or during the hatching phase when general body denticles develop to take over their positions. This morphological disparity between caudal denticles and other denticle types from the dorsal trunk and general body extends beyond their macrostructure.
Fig. 1.
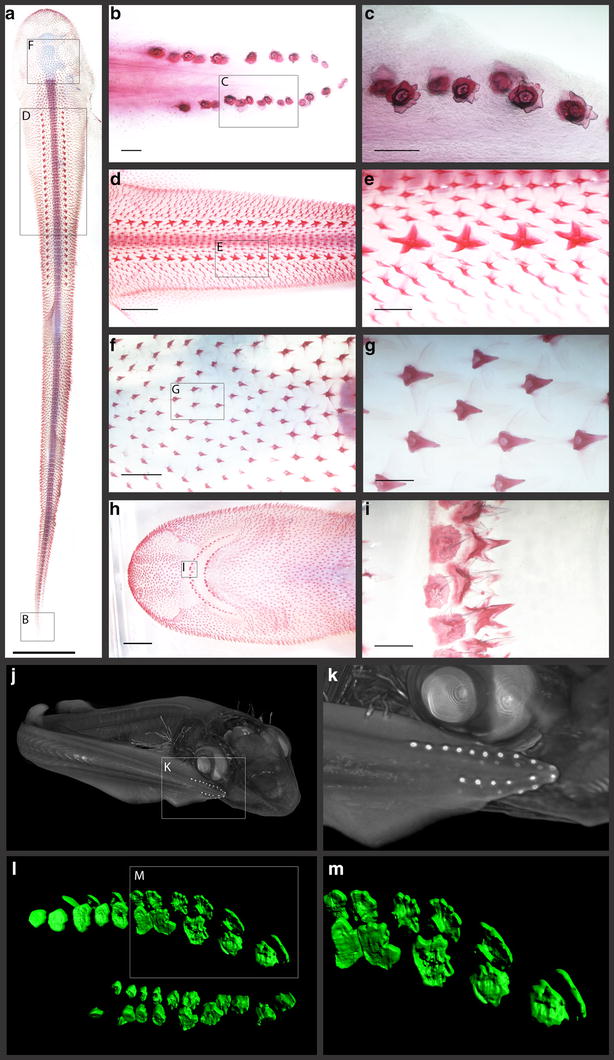
Odontode diversity of the pre-hatchling Catshark (S. canicula). Samples a–i are cleared and stained for calcium-rich tissue using alizarin red dye. Samples j–k are computerised tomography (CT) scans of a Stage 32 whole embryo, and samples l–m are light sheet fluorescence microscopy (LSFM) images of caudal denticles of a Stage 31 embryo, stained with alizarin red. The pre-hatchling (a) possesses three major external denticle types. The caudal denticles are the first to emerge, appearing on either side of the tip of the tail in dorsal and ventral rows (b, c, j–m) [23]. These denticles are not strongly polarised, although cusps generally point towards the posterior [27]. Next, the dorsal denticles emerge along the trunk of the embryo in two polarised rows (d, e). Finally, general body denticles emerge just before hatching, covering the whole body (f, g). These denticles are also highly polarised. Teeth emerge in the jaws at a similar stage to general body denticles (h, i). The scale bar for a = 1000 µm, b, c, g and i = 200 µm, d and h = 2500 µm, e and f = 500 µm
Fig. 3.
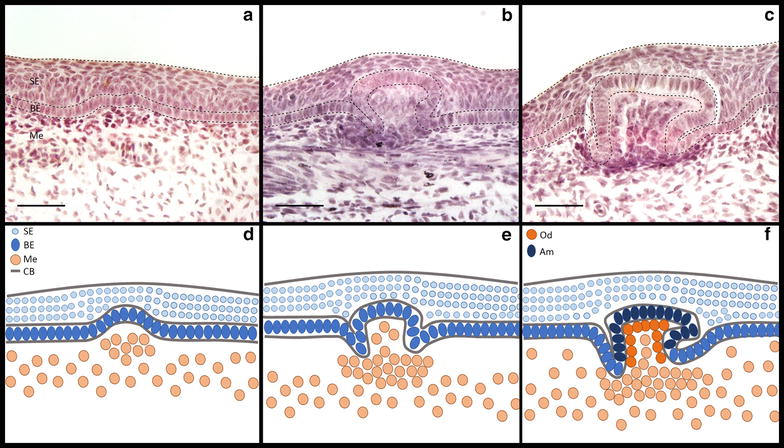
Morphogenesis of a caudal denticle. Caudal denticle placodes consist of a squamous epithelium (SE) overlying columnar cells of the basal epithelium (BE), which overlies the mesenchyme (Me) (a, d). During placode morphogenesis, condensing mesenchymal cells aggregate below columnar cells of the basal epithelium epithelial. The basal epithelium undergoes growth and folding (b, e) to form the posterior facing cusp (c, f). CB is cell layer boundary. Ameloblasts (Am) in the basal epithelial cusp (c, f) and odontodes (Od) in the papilla underlying the basal epithelium produce enameloid and dentine, respectively, to mineralise the unit [40]. Scale bars are 50 µm
Caudal denticles contain a dentine type that shares histological similarity to dentine from odontodes of the Ordovician and Silurian Periods [10, 11, 27]. Unlike the orthodentine observed in the general body denticles of S. canicula, the tubules of this dentine exhibit a branching pattern [27, 30]. The combination of this primitive-type dentine composition and the regulated, iterative patterning mechanism of these denticles, which are found in sharks at least across families Scyliorhindae and Heterodontidae, indicates that caudal denticles may have a deep phylogenetic history and have been retained in extant sharks over 450 million years of evolution [27, 28]. However, little is known about the developmental processes or GRN underlying the formation of caudal denticles or indeed other denticle types in chondrichthyans.
Fibroblast growth factor (FGF) signalling is essential for various aspects of both embryogenesis and adult homeostasis, such as tissue repair and regeneration [31]. FGFs have essential roles throughout vertebrate organogenesis, for example in limb, lung and brain development [32–34]. They mediate their responses by activating cell surface tyrosine kinase FGF receptors (FGFRs) [31]. FGF signalling is also widely involved in the development of taxonomically diverse epithelial appendages of the integumentary system, such as hair, feathers, scutes, scales and teeth [35–39]. Relative to the epithelial appendages of amniotes, little is known about the GRN controlling shark denticle placode formation, although some recent work has documented signalling during shark tooth development [40] and compared it to development of other odontode types [14, 41, 42].
During feather placode development, ligands of the FGF signalling family (such as Fgf4) work together with sonic hedgehog (Shh) in a positive feedback loop, that promotes expression of both Fgf4 and Shh whilst also inducing expression of bone morphogenetic protein 4 (Bmp4) [36, 43]. Bmp4 then has an inhibitory effect upon both Shh and Fgf4, downregulating their expression to control patterning by limiting placode formation exclusively to the site of future organs [36, 44, 45]. This inhibitory action of mesenchymal Bmp4 has also been observed during mouse hair development [46]. The mesenchymal expression of Bmp4 is conserved during morphogenesis throughout amniote epithelial appendage development [4]. It is unknown whether this FGF, Shh and Bmp4 signalling feedback system is conserved throughout all vertebrate epithelial appendage placode GRNs, although conservation of these markers is widely observed during amniote placode formation [4].
This study examines whether the molecular signalling observed during early morphogenesis of amniote integumentary organs is conserved within the development of caudal denticles of the shark (S. canicula). By comparing gene expression to the development of other epithelial appendages and using functional experiments to examine gene interactions, it is possible to infer putative GRN relationships [14]. A combination of anatomical, histological and molecular techniques including whole mount in situ hybridisation and immunohistochemistry was used to examine the development of shark caudal denticles, focusing on the role of the FGF signalling pathway and associated members of the putative core conserved placode GRN, inferred from studies in amniotes. To study the conservation of placode GRN members between different odontode types, gene expression was also examined during development of general body denticles. The fibroblast growth factor receptor (FGFR) inhibitor SU5402 was used to examine the effect of suppressing signalling of this major developmental pathway. By examining the role of FGF signalling during epithelial appendage development in a chondrichthyan model and its effects upon the expression of other putative GRN members, it will be possible to elucidate the degree to which epithelial integumentary organ GRNs are conserved across jawed vertebrates, and evaluate their potential homology.
Results
Caudal denticle placode development reveals conserved morphogenetic mechanisms for integumentary organ formation
To determine the earliest time of caudal denticle morphogenesis in the shark (S. canicula), we charted the sequential development of these units. It has been documented that caudal denticles in S. canicula develop from a posterior to anterior direction in dorsal and ventral rows, on both sides of the caudal-most tip of the tail (Fig. 2) [23, 27]. Previous reports suggest their emergence occurs at 52–60 dpf (Stage 30) [23] (Fig. 2f, g). However, our observations suggest placode development begins earlier, between 42 and 46 dpf (Stage 27) [23] (Fig. 2b, c), although some variation in timing of denticle initiation was noted. One explanation for such variation in development is temperature of the surrounding environment [23]. Caudal denticles arise from distinct placodes (Figs. 2c4, 3a), which form from a thickened condensation of epithelial cells with an underlying mesenchymal condensate (Fig. 3). The first denticle placodes to form (most posterior) are also the first in the sequence to mineralise. This progresses in a posterior–anterior fashion (Fig. 2d4) and can be visualised using alizarin red staining (Fig. 2e4–i5).
Fig. 2.
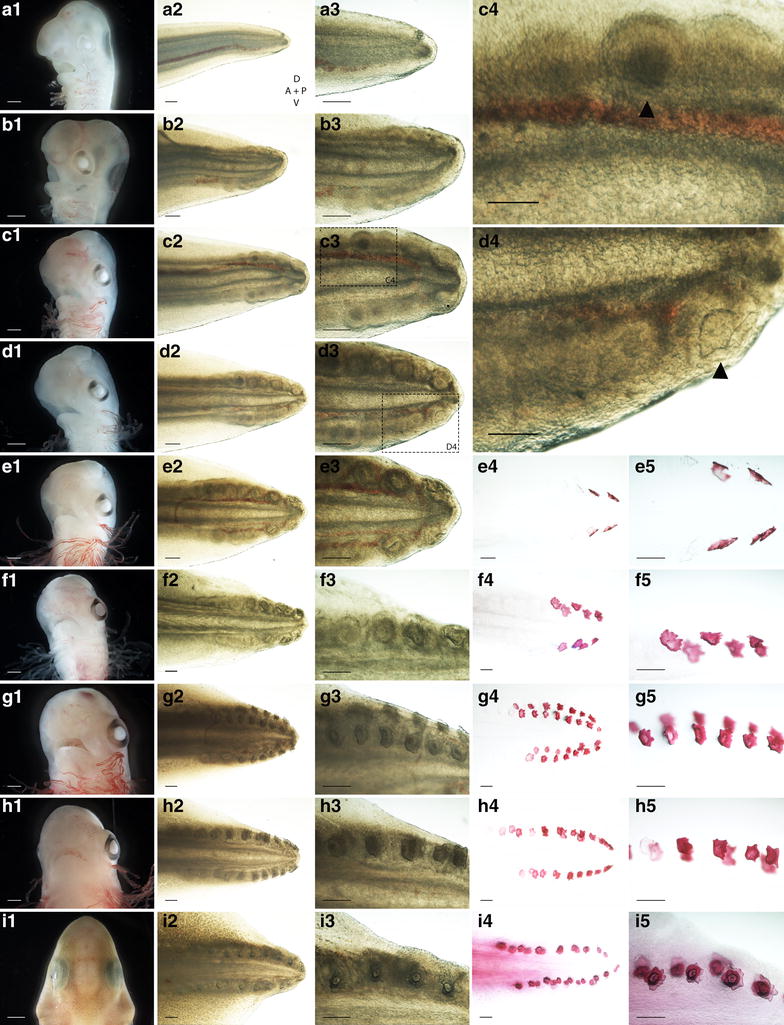
Sequential development of caudal denticles in the Catshark. As the embryo develops from Stage 27 (a) to Stage 33 (i), the gills proliferate, the eyes are encircled with pigment of increasing darkness and the rostrum protrudes anterior to the mouth [23]. During this period, caudal denticles develop from posterior to anterior in dorsal and ventral rows, on either side of the tail tip. At early Stage 27, no placodes can be detected (a2–a2). Epithelial thickenings then form from posterior to anterior (b2–b3, c2–c4). c4 shows a magnified view of c3, highlighting an individual placode (marked with an arrowhead). These placodes then accumulate their first layers of mineralised tissue during morphogenesis (d2–d4). d4 shows a magnified view of d3, highlighting a mineralising placode (marked with an arrowhead). Mineralisation of denticles also occurs sequentially from posterior to anterior (e2–e3, f2–f3, g2–g3, h2–h3 and i2–i3) and can be highlighted with alizarin red staining for calcium-rich tissue (e4–e5, f4–f5, g4–g5, h4–h5 and i4–i5). For the axis, D dorsal, V ventral, P posterior and A anterior. Scale bars are 1000 µm for a1, b1, c1, d1, e1, f1, g1, h1 and i1 and 200 µm for all other images
The sequential development of this morphogenetic placode unit bears remarkable similarity to feather bud development in chicks [4, 36, 47, 48] (Fig. 2). We wished to test whether members of the amniote epithelial appendage placode GRN are conserved in chondrichthyans [4, 8]. Therefore, a selection of well-known GRN components assembled from the literature regarding feather, hair and tooth development were chosen [36, 49, 50], and their expression during early placode morphogenesis of caudal denticles in S. canicula was examined.
Gene expression from integumentary appendage development is conserved in sharks
Recent research has revealed ectodysplasin signalling is conserved throughout development of amniote epithelial appendages [4]. Ectodysplasin-A (Eda) and its receptor (Edar) comprise some of the earliest markers of placode morphogenesis in vertebrates (including zebrafish, chick and mouse) [49, 51–54]. During early morphogenesis of shark caudal denticles, eda and edar expression is detected in the localised epithelial thickening (Fig. 4a–f). eda is also expressed during later denticle morphogenesis in epithelial cells in the signalling centre of the putative enameloid knot (EK) (Fig. 4b, bi). This shares similarity to mammalian tooth development, during which interactions between Eda/Edar and other signalling molecules (e.g. Shh, Fgf4 and Bmp4) regulate morphogenesis of the enamel knot [55]. During hair morphogenesis in mammals, Eda and Edar signalling induces expression of other signalling molecules, such as Shh [56, 57].
Fig. 4.
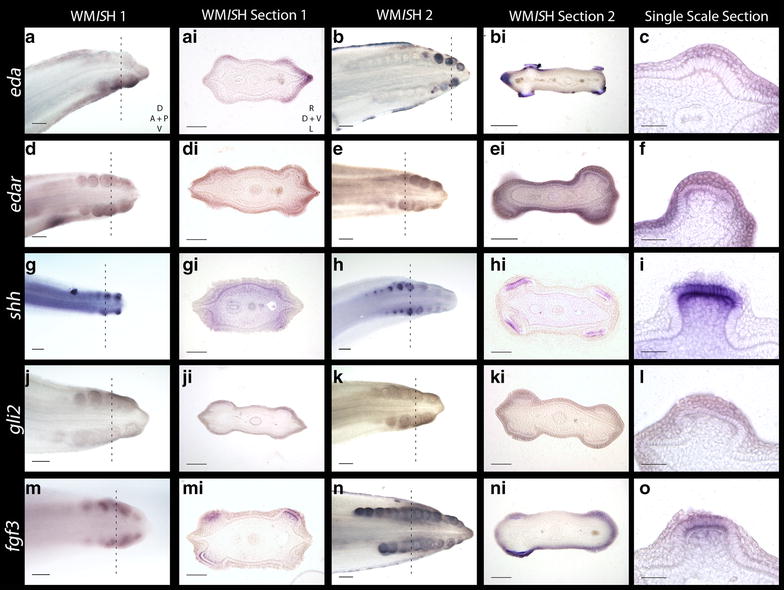
Gene expression analyses of early morphogenesis of caudal denticles. Expression of eda and its receptor edar are observed in the epithelium during early placode morphogenesis (a–f). eda can also be seen in tissue undergoing mineralisation later in morphogenesis (b–bi). shh is first observed in the epithelium during early morphogenesis, before becoming restricted to the basal epithelium later in morphogenesis (g–i). gli2 is also seen in the epithelium early during placode formation (j–l). Expression of fgf3 is first seen in the epithelium, before moving to both the epithelium and mesenchyme later in placode morphogenesis (m–o). The dashed lines show where in the WMISH the section was taken. WMISH Section 1 represents a younger stage specimen than WMISH Section 2. For the WMISH, D dorsal, V ventral, A anterior and P posterior. For WMISH sections, R right, L left, D dorsal and V ventral. For scale bars, a, b, d, e, g, h, j, k, m, n = 200 µm, ai, bi, di, ei, gi, hi, ji, ki, mi, ni = 100 µm, and c, f, i, l, o = 50 µm
Shh is a ligand of the Hedgehog (Hh) signalling pathway that marks early stages of epithelial morphogenesis in a diverse range of integumentary organs [50, 58, 59], including shark teeth and chick feathers [14, 40, 59, 60]. shh is expressed throughout morphogenesis of shark caudal denticles (Fig. 4g–i). During early morphogenesis, shh is first expressed in the superficial squamous epithelium (Fig. 6m), before subsequently becoming restricted to the basal epithelium (Figs. 6n, o, 7). Previous research has shown that Gli2 is expressed both downstream and upstream of Shh signalling [61–63] and is essential in hair follicle development as a promoter of cell proliferation [64]. Here, we found that gli2 is also expressed in the epithelial cells of developing placodes (Fig. 4j–l).
Fig. 6.
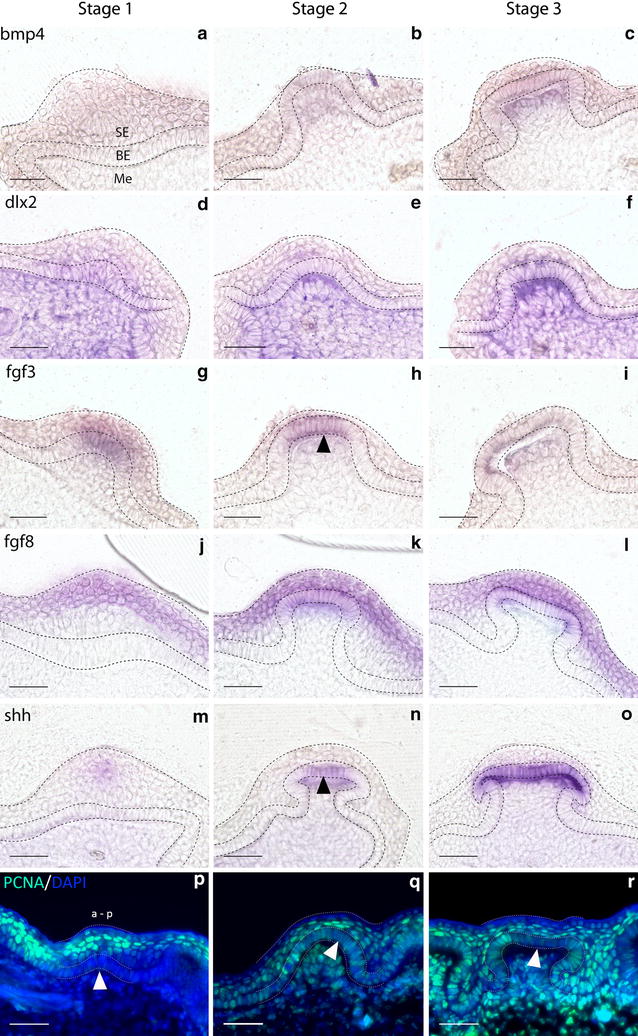
Gene expression/PCNA analysis of early caudal denticle morphogenesis. Gene expression is shown in 30-µm transverse sections of wild-type S. canicula embryo tails post-WMISH, to highlight progressive stages of caudal denticle morphogenesis from the initial epithelial thickening. bmp4 and dlx2 expression is restricted to the mesenchyme throughout morphogenesis (Me) (a–f). shh and fgf8 are first observed in the squamous epithelium (SE) before becoming restricted to the basal epithelium (BE) (m–o, j–l). Expression of fgf3 begins in the squamous and basal epithelium and is subsequently observed throughout the epithelium and mesenchyme (g–i). PCNA immunofluorescence is observed in the epithelium and mesenchyme throughout morphogenesis (p–r). Reduced activity (marked with an arrowhead) was noted in columnar cells of the epithelium during early morphogenesis (p) and in a central region of columnar cells of the basal epithelium during later morphogenesis (q–r). This region (q) overlaps with fgf3 and shh expression in the basal epithelium (k, n) (marked with an arrowhead) and may be indicative of a basic primary enameloid knot. a is anterior, and p is posterior. Dashed lines separate the squamous epithelium (SE), basal epithelium (BE) and mesenchyme (Me). All scale bars are 50 µm in length
Fig. 7.
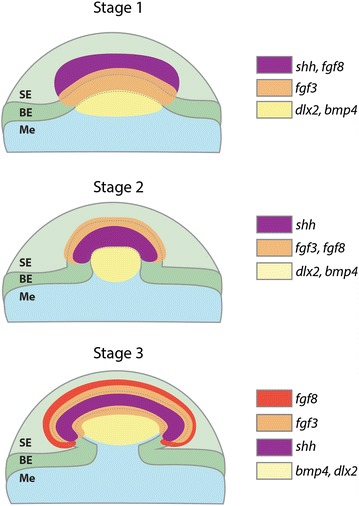
Schematic diagram representing gene expression during early morphogenesis of caudal denticles. This diagram summarises the results from Figs. 4 and 5, representing expression of fgf3, fgf8, shh, bmp4 and dlx2 throughout progressive stages of early morphogenesis. SE is the squamous epithelium, BE is the basal epithelium and Me is the mesenchyme
In various aspects of vertebrate appendage development, Shh and FGFs (Fgf4, Fgf8) exhibit interdependent positive feedback loops that promote the expression of either molecule [65, 66]. Fgf3 expression is mesenchymal during early morphogenesis of both feathers [67] and teeth, although in later tooth morphogenesis it is present in the epithelium of the primary enamel knot [68]. In shark caudal denticles, fgf3 expression is initially epithelial, although it is later seen in both the epithelium and mesenchyme, in a pattern similar to shark tooth and body denticle development [14, 40] (Figs. 4m–o, 6g–i, 7). Fgf8 is an epithelial initiatory signal of mammalian tooth morphogenesis [69]. In the shark, fgf8 expression is observed in the epithelium during early caudal denticle morphogenesis, at a similar stage to shh (Figs. 5a–c, 6j–l, 7), and remains in the epithelium during later morphogenesis of the denticle cusps (Fig. 5l).
Fig. 5.
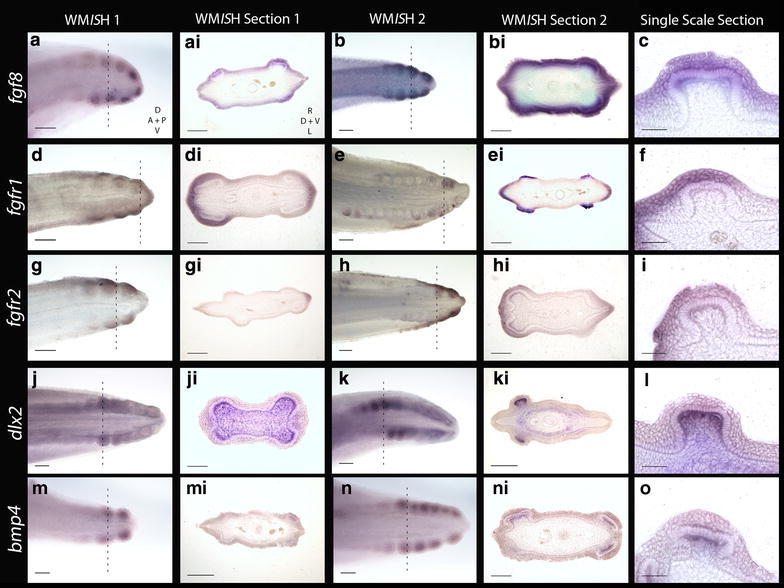
fgf8 signalling is largely retained in the epithelium throughout (a–c). fgfr1 and fgfr2 are both seen in the epithelium during early denticle morphogenesis (d–i). Expression of dlx2 is restricted to the mesenchyme throughout early placode morphogenesis (j–l). Similarly, bmp4 is observed in the mesenchyme during early placode morphogenesis (m–o). The dashed lines show where in the WMISH the section was taken. WMISH Section 1 represents a younger stage specimen than WMISH Section 2. For the WMISH, D dorsal, V ventral, A anterior and P posterior. For WMISH sections, R right, L left, D dorsal and V ventral. For scale bars, a, b, d, e, g, h, j, k, m, n = 200 µm, ai, bi, di, ei, gi, hi, ji, ki, mi, ni = 100 µm, and c, f, i, l and o = 50 µm
Studies from teleosts (medaka) and mammals (mouse) have indicated that Fgf8 is a ligand of Fgfr1 [70–72] that regulates enamel formation during mammalian tooth morphogenesis [73]. During early caudal denticle morphogenesis, we observed expression of fgfr1 in the squamous epithelium of placodes (Fig. 5d–f). fgfr1 is also expressed throughout the epithelium later in morphogenesis during caudal denticle mineralisation (Fig. 5e–ei), which may be indicative of a conserved role regulating enameloid formation. Fgfr2 can transduce Fgf3 during mammalian development [74, 75]. During early morphogenesis of caudal denticles, fgfr2 is expressed in the squamous epithelium of the early developing placodes in S. canicula (Fig. 5g–i). This pattern is similar to epithelial expression of fgf3, suggesting the role of fgfr2 as a fgf3 signal transducer could be conserved.
Dlx2 is a member of the Dlx homeodomain transcription factor family, which is widely important throughout various aspects of vertebrate development, including epithelial appendage formation [76, 77]. Fgf8 regulates Dlx2 expression in the underlying mesenchyme during both mouse tooth and branchial arch development [78, 79]. Previously, dlx gene expression has been documented during caudal denticle morphogenesis in S. canicula [41]. Our results confirm dlx2 is expressed in caudal denticles, and additionally we show that expression is restricted to the mesenchyme throughout early morphogenesis (Figs. 5j–l, 6d–f, 7), as observed during mouse tooth development [78].
Mesenchymal Bmp4 has an inhibitory role during amniote epithelial appendage development [1, 36, 46]. Consistent with expression observed during feather, shark tooth and body denticle development [14, 36, 46], bmp4 is expressed in the mesenchyme during early morphogenesis of caudal denticle placodes (Figs. 5m–o, 6a–c, 7). bmp4 may also be acting as an internal inhibitor here [1, 36], helping to define the size of the placode and therefore the adult caudal denticle.
In addition to examining caudal denticle development, gene expression of these putative core GRN members was also examined in general body denticles to compare signalling between different odontode types (Figs. 1, 8). Expression of shh is restricted to the epithelium throughout early morphogenesis of general body denticles (Fig. 8a–c), whereas fgf3 is first observed most strongly in the epithelium (Fig. 8d) before being expressed in both the epithelium and underlying mesenchyme (Fig. 8e, f). Epithelial fgf3 overlaps with shh expression in the putative enameloid knot, whereas bmp4 is restricted to the mesenchyme throughout morphogenesis (Fig. 8g–i). These results show conservation of gene expression patterns between caudal and general body denticles.
Fig. 8.
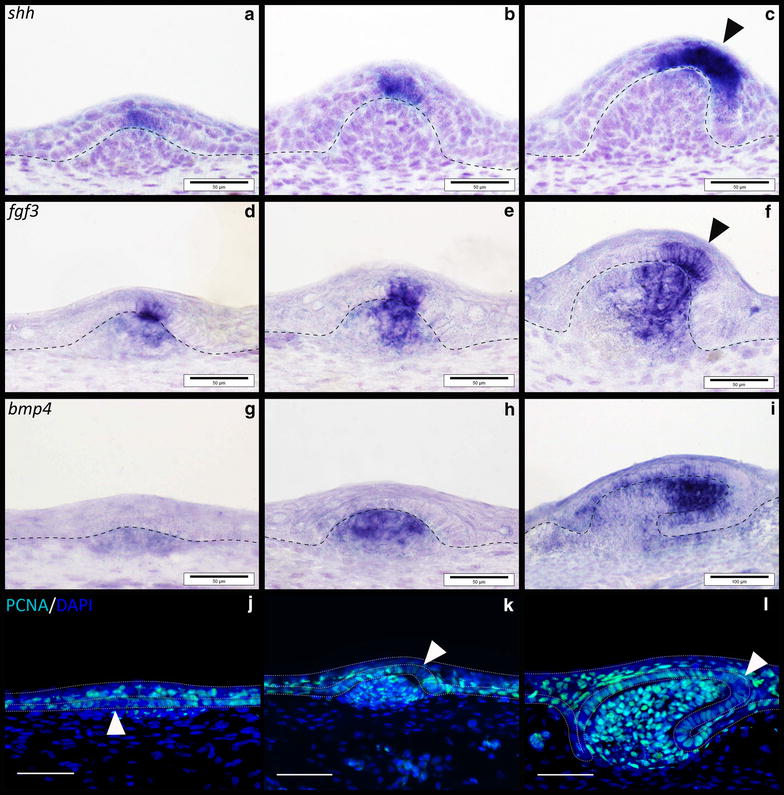
Gene expression analysis of putative placode GRN members, during general body denticle development. Section in situ hybridisation (SISH) was undertaken during early development of body denticles. Expression of shh was epithelial throughout development (a–c), whereas fgf3 was observed in both the epithelium and mesenchyme (d–f). bmp4 was mesenchymal throughout early morphogenesis (g–i). PCNA immunoreactivity was observed in the epithelial cells and condensing mesenchyme of emerging denticles (j–l). Reduced immunoreactivity was noted in columnar cells of the basal epithelium during placode formation (j) (white arrowed). fgf3 and shh expression marks enameloid knot-like cells of the epithelium associated with denticle morphogenesis (c, f), which also show reduced PCNA immunoreactivity (l), characteristic of this signalling centre (black arrowheads). The dashed line separates the epithelium from the underlying mesenchyme (a–i), as well as the basal epithelium and squamous epithelium (j–l). All scale bars are 50 µm in length except for image i for which the scale bar is 100 µm
Proliferating cell nuclear antigen (PCNA) immunoreactivity marks several phases of cell division from late G1 to mitosis [80]. Developing body denticles are highly proliferative units similar to teeth and dental lamina [40, 42] (Fig. 8j–l). During early morphogenesis, the columnar cells of these placodes are characterised by reduced proliferation (Fig. 8j), as also observed throughout amniote skin appendage development [4]. Interestingly, a region of the apical denticle cusp also shows marked reduction in PCNA immunoreactivity (Fig. 8k, l) that corresponds to a putative signalling centre comparable to the enameloid knot in shark teeth [40]. This set of cells appears to overlap with the region of shh and fgf3 expression in the polarised cells that will become the apical cusp (Fig. 8a–f). Caudal denticles display comparable PCNA immunoreactivity during morphogenesis (Fig. 6p–r), including reduced proliferation of columnar cells during early morphogenesis compared to younger anterior epithelial tissue in which placode formation has not begun (Fig. 6p). However, the region of reduced proliferation that occurs in columnar epithelial cells later during morphogenesis appears to be positioned more centrally than observed in body denticles, which have a distinct polarity (Fig. 6q, r). This region overlaps with expression of fgf3 and shh (Fig. 6k–n), and could also be indicative of a putative primary enameloid knot, as observed in general body denticles (Fig. 8) [40]. The positional variation of this enameloid knot could reflect a shift in the morphology of these units, as caudal denticles display a less definitively polarised cusp than general body denticles (Fig. 1).
In the absence of functional data, it is not possible to test for the conserved action of GRN members, which could yield important clues regarding the putative homology of denticles and amniote epithelial appendages. We have therefore initiated a small-molecule-based targeted signalling pathway-knockdown screening assay in S. canicula to test the function of putative epithelial appendage GRN members, based on published results from other vertebrates.
Small molecule inhibition reveals dependency of caudal denticle development on FGF signalling
To elucidate the specific roles of FGF signalling during early caudal denticle placode morphogenesis, in vivo pathway perturbation assays were undertaken using SU5402. This chemical inhibits FGF signalling by blocking FGFR activity [81–83]. Stage 28 S. canicula embryos were treated in their sealed egg cases by injection with SU5402 to a final concentration of ~10 µM for 25 days and then allowed to develop for a further 35 days following the opening of their egg cases and washing with fresh artificial seawater. Treatment with SU5402 resulted in a single denticle knockout in 40% of the treated samples (n = 5) and none of the DMSO-treated control samples (n = 5) (Fig. 9). These units normally form equidistant from each other [23, 28, 29]; however, in the drug treated specimen shown (Fig. 9e–h) the 6th denticle was missing in the left-side dorsal row. This corresponds to the time at which treatment took place, when approximately 5 caudal denticle placodes had developed in sequence on each row (Fig. 2c). Therefore, ~10 µM SU5402 appears to prevent placode formation and subsequent morphogenesis, indicating this process is dependent upon FGF signalling. As only a single denticle was lost from the sequence, it is likely the chemical either diffused out of the egg case or decomposed within it after its initial inhibitory action. We observed a similar result in our preliminary SU5402 treatment trial (see Additional file 1), which revealed a vestige when stained with alcian blue, indicative of denticle abortion. The relatively short window of sensitivity to FGF inhibition by SU5402 treatment coupled with the offset in developmental timing of individual caudal denticle rows is likely to provide an explanation for the unilaterality of this denticle knockout. As subsequent placodes developed, the field of initiatory competence is likely to have already been in place, enabling the sequential, iterative patterning to proceed beyond the disturbance once the effect of SU5402 had subsided.
Fig. 9.
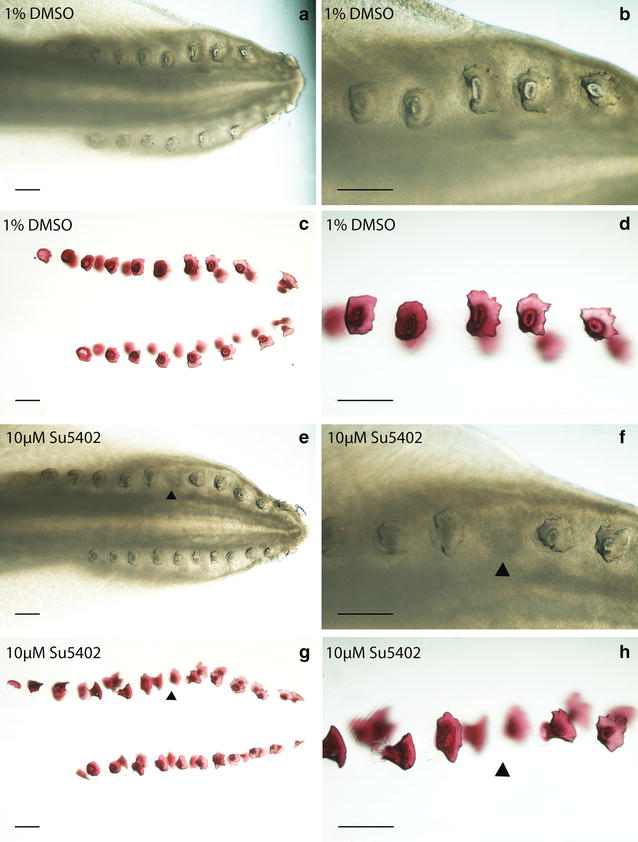
Phenotypic effect of FGF inhibition via SU5402 treatment (10 µM) on caudal denticle development. The DMSO control specimen shown after fixation (a, b) and cleared and stained for calcium-rich tissue using alizarin red (c, d) possesses a full sequence of caudal denticles. However, the specimen treated with the FGF antagonist SU5402 has the 6th denticle missing from the sequence, shown after fixation (e, f) and cleared and stained (g, h). This is marked with a black arrowhead. This denticle knockout corresponds to the stage at which treatment occurred, and was observed in 40% of Su5402-treated specimens (n = 5). Scale bars are 200 µm in length
We sought to confirm a specific effect of SU5402 upon the FGF signalling pathway and the placode-forming GRN by examining expression of participating network members. The prior assay required longer-term development of embryos to observe morphological effects (60 days) and subsequently used a lower drug concentration to avoid mortality, as this was an issue in preliminary trials. Experimental perturbation of FGF signalling was therefore repeated using a higher concentration of SU5402 (1 × 50 µM injection every 24 h, over a 96-h period), in line with previously published assays [37, 84, 85], and embryos were fixed immediately after the 96-h period. Specimens were then processed for in situ hybridisation for a selection of the same putative GRN members examined previously in wild-type embryos (Figs. 4, 5, 6). This allowed us to test whether perturbation of FGF signalling in shark denticles disrupted other members of the placode GRN in a manner consistent with a conserved relationship between network members.
FGF ligands exhibit positive feedback loops with Shh during many aspects of vertebrate embryogenesis, including epithelial appendage, limb bud and gill arch development [36, 65, 66, 86, 87]. We observed a dramatic downregulation of fgf3, fgf8, shh and dlx2 expression in the SU5402-treated individuals compared the DMSO-treated controls, in all but the youngest (most anterior) denticles (Fig. 10c–ji). Expression intensity of bmp4 was also notably reduced compared to the control (Fig. 10a–bi). Two SU5402-treated specimens were used for WMISH for each marker, along with one DMSO control specimen. These results suggest that SU5402 blocked FGF/FGFR signalling [81–83], thereby reducing expression of fgf3 and fgf8 (Fig. 10e–hi). This is likely due to SU5402 blocking earlier FGF signalling required for expression of these ligands [37], and interrupting the FGF–Shh positive feedback loop, which consequently limited expression of shh, bmp4 and dlx2 (Fig. 10a–di, i–ji) [36, 66, 78, 85]. Dlx family members have a role downstream of FGFs during feather bud development [77], indicating that this downregulation of dlx2 is likely a result of FGF inhibition. These results suggest that during caudal denticle formation, the function of FGF signalling in the GRN which guides epithelial appendage morphogenesis is conserved between sharks and other vertebrates.
Fig. 10.
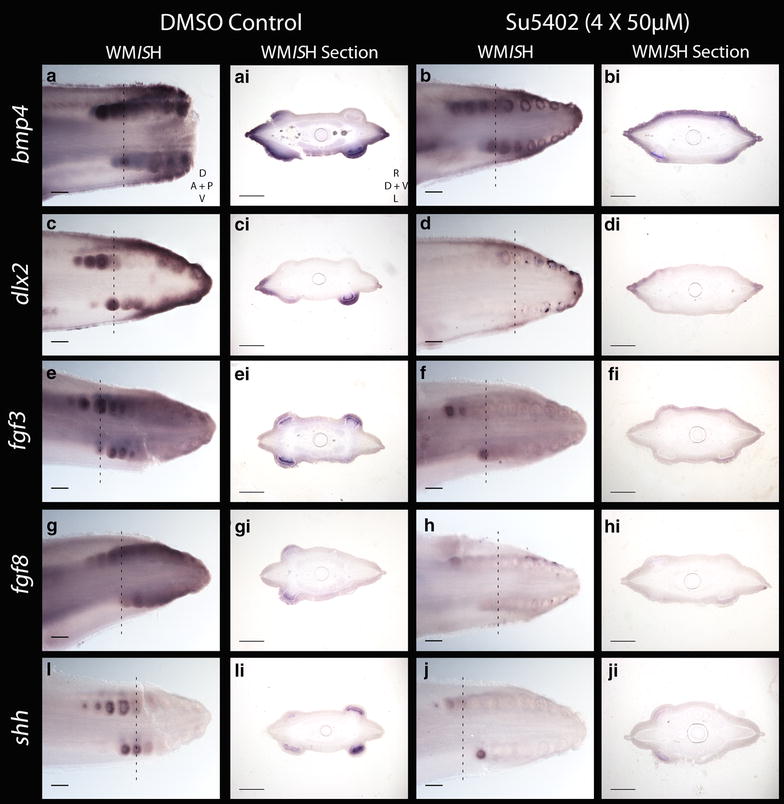
Genetic effect of FGF inhibition via SU5402 treatment (4 × 50 µM) on caudal denticle development. There was a reduction in staining intensity of bmp4 (a–bi), dlx2 (c–di), fgf3 (e–fi), fgf8 (g–hi) and shh (i–ji) in SU5402-treated specimens compared to DMSO-treated controls. We propose this resulted from the interruptions to the following GRN interactions. SU5402 inhibits FGF activity by blocking FGFR activity, thereby reducing expression of fgf3 and fgf8 (e–hi). This reduced shh and dlx2 expression as a FGF—shh positive feedback loops that would normally promote shh and dlx2 expression (as observed during feather development) were interrupted (i–ji, c–di) [78]. The fgf4–shh positive feedback loop that promotes bmp4 was also interrupted by the SU5402 treatment, reducing shh and bmp4 expression (i–ji, a–bi) [36]. SU5402-treated and DMSO control specimens both underwent the colour reaction of the WMISH protocol for the same length of time. The dashed lines show where the section was taken from. Scale bars for WMISH are 200 µm in length, and for the WMISH sections they are 100 µm in length
Discussion
An FGF-dependent GRN constructs the placodes of epithelial appendages throughout jawed vertebrates
Our results suggest that a conserved core GRN, which includes eda/edar, shh, gli2, fgf3, fgf8, bmp4 and dlx2, underlies the development of epithelial integumentary appendage placodes across jawed vertebrates (Figs. 4, 5, 6). These placodes possess columnar epithelial cells with a reduced rate of proliferation (Fig. 6p), which is considered a structural characteristic of amniote skin appendage development [4]. Functional experiments revealed that expression of these GRN members is influenced by the FGF signalling pathway and that normal denticle development is perturbed upon inhibition with SU5402 (Figs. 9, 10). Caudal denticles are considered an ancient epithelial appendage that may have originated in early vertebrates over 450 million years ago and have been retained in some extant chondrichthyans [10, 11, 27]. The historical continuity of the anatomical placode and underlying GRN in both amniotes [4] and chondrichthyans provides evidence for the historical homology of all vertebrate epithelial appendages [88].
Previously, researchers have speculated that epithelial appendages have evolved independently in mammals, reptiles and birds and that therefore molecular similarity of GRNs could be a result of independent genetic co-option or deep homology [9, 89–92]. However, recent evidence has suggested that integumentary epithelial appendages are historically homologous, at least throughout all amniotes on the basis of the anatomical placode with conserved expression and function of GRN members [4]. Our results suggest this historical homology extends even further into vertebrate phylogeny and may encompass the integumentary epithelial appendages of all extant jawed vertebrates.
During both mouse and zebrafish tooth morphogenesis, Fgf8 signalling can promote Dlx2 expression [37, 78, 79]. Inactivation of Fgf8 can result in both misregulation of Fgf4 and Shh [87], which are also known to work together in autocatalytic positive feedback loops during vertebrate development, for example in limb and feather patterning [36, 65]. During early feather placode morphogenesis, this Shh–Fgf4 feedback loop promotes Bmp4 expression, which subsequently acts as an inhibitor to limit their expression in a negative feedback loop [36]. Our results regarding gene expression of FGF-perturbed shark embryos reveal this functional conservation likely extends to the denticles of sharks (Figs. 10, 11).
Fig. 11.
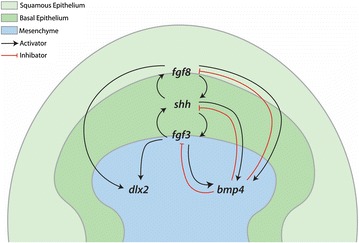
Putative relationship between FGF and associated GRN components during caudal denticle morphogenesis. As observed widely throughout epithelial appendage development, for example during feather placode development, FGF—shh positive feedback loops which promote mesenchymal bmp4 are likely to promote early caudal denticle placode morphogenesis. bmp4 may then act as an internal inhibitor, limiting the size of the final unit. FGF signalling can also promote mesenchymal expression of dlx2. This is a hypothetical GRN based on findings from previous research, gene expression data (Figs. 4, 5, 6, 8) and small molecule inhibition of FGF signalling during early caudal denticle morphogenesis, using SU5402 (Figs. 9, 10)
Despite the broad conservation of this GRN, our observations of gene expression patterns did highlight some taxonomic disparity. We showed that fgf3 is expressed in both the epithelium and later the mesenchyme of denticle placodes, as observed during mouse and shark tooth development [40, 68]. This contrasts with known expression patterns observed during zebrafish pharyngeal tooth and chick feather development [37, 67]. Similarly, Fgf8 is an important inductive signal during mammalian tooth development [69] and is present during caudal denticle development, but is absent from feather or zebrafish tooth morphogenesis [33, 37]. There is potential for evolutionary alterations to gene expression and functionality throughout the FGF signalling family, and paralogs may perform the same developmental role in different taxa in a process known as function shuffling [93], for example mammalian Fgf8 may have a zebrafish specific paralog [37].
Recent research has revealed chondrichthyan general body denticles and teeth are deeply homologous developmental units, despite differences in their regenerative capacities [14, 40]. Our findings suggest this odontode GRN additionally encompasses caudal denticles, as conserved expression patterns were observed throughout early placode morphogenesis between caudal denticles and general body denticles. Caudal denticles are morphologically disparate from general body denticles, dorsal denticles and teeth (Fig. 1), and positional alterations to the putative enameloid knot may contribute to this variation (Figs. 6, 8). As predicted by an hourglass model of development [94], divergence in the GRN later in morphogenesis is also likely to result in alterations to the adult form, constructing different structures upon a homologous foundation: the anatomical placode. Such divergence of networks is known to relate to variation in the adult structure of feathers and teeth [95, 96].
Conclusion
The early morphogenesis of vertebrate epithelial appendages is likely to be a universal and highly conserved process retained over evolutionary time and modified to form the plethora of diverse skin appendages observed throughout all vertebrates, from sharks to mammals. The placodes of vertebrate epithelial appendages constitute the conserved foundations upon which integumentary structures have evolved, via alterations to an otherwise conserved GRN that take effect during later morphogenesis. The shark caudal denticle system provides an ideal set of sequentially developing integumentary epithelial appendages that can be studied further to decipher both complex functional GRNs and patterning mechanisms.
Combining techniques such as small molecule signalling pathway perturbation with gene expression analyses can help us begin to interpret the roles of putative GRN members. The set of genes investigated here were chosen due to their importance in the development of other epithelial appendages; however, there are many important molecules and interactions to further investigate, for example those associated with the Wnt/β-catenin and Notch pathways [97, 98]. A focus on investigating downstream GRN components responsible for later morphogenesis will enable us to elucidate how historically homologous placodes develop into the diverse range of epithelial appendages observed throughout vertebrates.
Methods
Shark husbandry and fixation
The University of Sheffield is a licensed establishment under the Animals (Scientific Procedures) Act 1986. All animals were culled by approved methods cited under Schedule 1 to the Act. Embryos were imported from ‘Station Biologique’ in Roscoff, France, and housed in tanks at The University of Sheffield, Animal and Plant Sciences, at 16 °C. Salinity was adjusted to replicate sea water using ‘Instant Ocean’ salt dissolved in dechlorinated water. Water was oxygenated with a submerged airflow. 50% water changes were undertaken on a weekly basis. Embryos were removed from their egg cases, anesthetised using MS-222 (Tricaine) and fixed overnight at 4 °C in 4% paraformaldehyde (PFA). Samples were dehydrated through a graded series of PBS and MeOH and stored at −20 °C in MeOH.
Alizarin red clear and staining
Fixed specimens were rehydrated through a graded series of MeOH and PBS. Staining took place in darkness overnight in 0.01% alizarin red dissolved in 0.5% KOH. Specimens were treated with trypsin in saturated sodium borate and distilled water. For Additional file 1, the sample was stained with 0.1% alcian blue in EtOH and acetic acid before the alizarin red stain was applied. Samples were then run through a graded series of KOH and glycerol solutions, before imaging took place in glycerol, using a Nikon SMZ1500 stereomicroscope.
Haematoxylin and eosin (H&E) staining
Paraffin-embedded sections were deparaffinised in xylene and rehydrated through a graded series of MeOH and PBS, before staining with haematoxylin. Sections were then rinsed in ddH20, washed with HCl in EtOH and washed with 0.001 M Tris–HCL. Finally, sections were stained with eosin, dehydrated to MeOH and mounted used DePeX mounding medium (VWR). Samples were imaged using an Olympus BX51 microscope and Olympus DP71 Universal digital camera attachment.
Micro-computerised tomography (MicroCT) and light sheet fluorescence microscopy (LSFM)
High-resolution MicroCT scanning was carried out upon a Stage 32 embryo stained with 0.1% PTA (phosphotungstic acid) in 70% EtOH for 3 days, using an Xradia MicroXCT scanner at the Imaging and Analysis Centre of the Natural History Museum (London). Scans were rendered using the 3D volume exploration tool Drishti (www.github.com/nci/drishti) (Fig. 1j, k). LSFM was carried out upon alizarin-stained samples. A Zeiss Z1 light sheet microscope with two sCMOS cameras and an acquisition PC running Zen Black 2014 software was used to scan the tail of a Stage 31 embryo. Rendering was undertaken using the image analysis software Imaris (www.bitplane.com/imaris/imaris) by creating a signal intensity-based isosurface (Fig. 1l, m).
Small molecule gene perturbation experiments
For the first SU5402 treatment trial, Stage 28 S. canicula embryos [23] were treated with the FGF-receptor inhibitor SU5402 (Sigma). At this stage, the egg case is sealed from the external environment, allowing administration of drugs via injection into the vitelline fluid. The egg case acts as a natural treatment chamber. 100 µl of a 500 µM stock solution of SU5402 in 1% DMSO in PBS was injected into 5 egg cases, to achieve a ~10 µM concentration of SU5402 assuming an approximate egg case size of 5 ml. 5 control samples were treated with 100 µl of 1% DMSO in PBS. At Stage 31 of development, the corners of the egg cases naturally open, allowing water to enter the case and replace the vitelline fluid and the chemical gene inhibitor. Once the first egg case had opened, others were artificially opened to ensure that the treatment period remained constant between replicates. Egg cases remained sealed for 25 days before opening and were then allowed to develop for a further 35 days before fixation and morphological examination. After observing inhibition of denticle development (Fig. 9), a second round of drug treatments was conducted to examine the genetic effect of FGF inhibition via SU5402 treatment. WMISH was undertaken to examine caudal denticle morphogenesis for SU5402-treated samples and compared to control samples (treated with DMSO). The concentration of SU5402 was increased, with 10 specimens receiving a 50-µl injection of a 5 mM stock solution of SU5402 in 1% DMSO in PBS, once every 24 h for 96 h, with each individual injection resulting in a ~50 µM concentration. 5 control samples were treated with one 50 µl injection of 1% DMSO in PBS, every 24 h for 96 h. Embryos were immediately fixed after the treatment period, before dissection and WMISH took place. Two SU5402-treated tails and one DMSO control tail were used to investigate expression of each gene. The concentrations used for chemical treatments were gleaned from studies undertaking similar gene perturbation experiments in teleosts and chondrichthyans, and honed using preliminary drug treatment trials in S. canicula [37, 84, 85] (see Additional file 1).
Whole mount in situ hybridisation (WMISH)
Digoxigenin-labelled (DIG) antisense riboprobes were designed using partial skate (Leucoraja erinacea) and catshark (Scyliorhinus canicula) EST assemblies [99] (SkateBase; skatebase.org) and the Vertebrate TimeCapsule (VTcap; transcriptome.cdb.riken.go.jp/vtcap). Riboprobes were cloned from S. canicula cDNA, and DIG-labelled antisense riboprobes were generated using the Riboprobe System Sp6/T7 kit (Promega). WMISH was carried out in accordance with Fraser et al. [84]. Samples were rehydrated through a graded series of MeOH and PBS, and treated with proteinase K (1 µl/mg ProK for 60 min), to facilitate probe penetration. Next, samples were refixed in 4% PFA in PBS and incubated in pre-hybridisation buffer for 1 h at 61 °C. For the hybridisation stage, samples were placed in a shaker incubator overnight at 61 °C in 2 ml tubes (Eppendorf) containing 1 ml aliquots of hybridisation buffer and DIG-labelled antisense RNA probe. Samples were then washed in saline sodium citrate with 0.1% Tween-20 (SSCT), before incubation in blocking reagent (Roche). Antibody labelling occurred overnight at 4 °C in Maleic Acid Buffer with Tween-20 (MABT), using anti-DIG-ALP (0.2 µl/ml) (Roche). This was followed by a series of washes and 48-h incubation in MABT at 4 °C. For the colour reaction, BM purple (Roche) was applied at room temperature, until the staining was sufficiently strong to represent gene expression. For WMISH undertaken upon SU5402 treated specimens, the colour reaction was run for the same length of time for SU5402-treated animals and DMSO controls. Samples were stored and imaged in 10% EtOH in PBS using Nikon SMZ1500 stereomicroscope. After WMISH and imaging, embryos were post-fixed with 4% PFA and embedded in chick albumin cross-fixed with 2.5% glutaraldehyde. A Leica Microsystems VT1000 vibratome was used to cut sections at 30 μm. Vibratome sections were then mounted with Fluoromount (Sigma-Aldrich) and imaged using a BX51 Olympus Microscope.
Section in situ hybridisation (SISH)
Fixed, dehydrated specimens were processed through a graded series of MeOH, chloroform and hot wax before being embedded in paraffin, and sectioned at 14 µm with a microtome (Leica RM2145). Sections were rehydrated from MeOH, and SISH was carried out with solutions as described for WMISH. Sections were incubated in pre-hybridisation buffer, before overnight incubation with a DIG-labelled antisense RNA probe. Sections were then run through post-hybridisation washes. Antibody labelling occurred overnight incubation with anti-DIG-AP (Roche). After post-antibody washes, BM purple (Roche) was used for the colour reaction. Sections were counterstained with haematoxylin and imaged using an Olympus BX51 Microscope and Olympus DP71 Universal digital camera attachment.
Immunofluorescence
Sections were rehydrated from MeOH or EtOH as previously described for SISH. Antigen retrieval occurred in hot 0.01 M sodium citrate (pH 6.0) for 10 min, before blocking and antibody labelling. Primary antibody labelling was undertaken using mouse anti-PCNA antibody (ab29; Abcam), overnight at 4 °C. Secondary antibody incubation was undertaken with goat anti-mouse AlexaFluor-488 (Thermo Fisher), before counterstaining with DAPI (Sigma-Aldrich). Slides were mounted with Fluoromount (Sigma-Aldrich). Imaging was undertaken with an Olympus BX61 upright epifluorescent Microscope and Olympus DP71 Universal digital camera attachment, and visualised with the software Volocity 6.3.
Authors’ contributions
RLC, KJM and GJF designed the research. LJR established ISH protocols and undertook SISH. KJM undertook the additional data collection. RLC undertook all other experiments. RLC, KJM and GJF analysed and interpreted the results. RLC, KJM and GJF wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Dr. Takanori Shono and Alex Thiery for providing laboratory assistance and expertise throughout the course of this project, and Phil Young for animal husbandry. We are grateful to Xavier Bailey and members of Station Biologique de Roscoff for the supply of shark embryos. MicroCT imaging was carried out with assistance from Dr. Zerina Johanson (Department of Earth Sciences), Dr. Farah Ahmed and Dr. Amin Garbout of the Imaging and Analysis Centre (Natural History Museum, London). Other imaging techniques were carried out with assistance from of Dr. Darren Robinson and Dr. Nick van Hateren of the Wolfson Light Microscopy Facility (The University of Sheffield).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article.
Consent for publication
This work does not involve human subjects.
Ethics approval and consent to participate
This work does not involve human subjects, human material, or human data.
Funding
This research was supported by the following grants: Natural Environment Research Council (NERC) Standard Grant NE/K014595/1 (to G.J.F); and Leverhulme Trust Research Grant RPG-211 (to G.J.F.). This work was also funded through ‘Adapting to the Challenges of a Changing Environment’ (ACCE), a NERC funded doctoral training partnership (to R.L.C.) ACCE DTP (NE/L002450/1). LSFM imaging was supported by Biotechnology and Biosciences Research Council (BBSRC) ALERT14 Award BB/M012522/1 for light sheet microscopy.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1. Caudal denticle abortion from preliminary SU5402 treatment trial. A preliminary drug trial involved treating S. canicula embryos with a single injection resulting in a 50 µM concentration of SU5402 in the egg case, which then remained sealed for 30 days. The egg case then opened, and the embryo was allowed an additional 30-day recovery period before fixation. The sample was then stained with alcian blue and alizarin red. There is a clear vestige (B, black arrowhead), indicating a single denticle from the sequence was aborted, as a result of this SU5402 treatment.
Contributor Information
Rory L. Cooper, Email: rcooper2@sheffield.ac.uk
Kyle J. Martin, Email: kyle.martin@sheffield.ac.uk
Liam J. Rasch, Email: liam.j.rasch@gmail.com
Gareth J. Fraser, Email: g.fraser@sheffield.ac.uk
References
- 1.Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/S0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 2.Biggs LC, Mikkola ML. Early inductive events in ectodermal appendage morphogenesis. Semin Cell Dev Biol. 2014;25–26:11–21. doi: 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Milinkovitch MC, Manukyan L, Debry A, Di-Poï N, Martin S, Singh D, et al. Crocodile head scales are not developmental units but emerge from physical cracking. Science. 2013;339:78–81. doi: 10.1126/science.1226265. [DOI] [PubMed] [Google Scholar]
- 4.Di-Poï N, Milinkovitch MC. The anatomical placode in reptile scale morphogenesis indicates shared ancestry among skin appendages in amniotes. Sci Adv. 2016;2:1–8. doi: 10.1126/sciadv.1600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sengel P. Pattern formation in skin development. Int J Dev Biol. 1990;34:33–50. [PubMed] [Google Scholar]
- 6.Thesleff I, Vaahtokari A, Partanen AM. Regulation of organogenesis. Common molecular mechanisms regulating the development of teeth and other organs. Int J Dev Biol. 1995;39:35–50. [PubMed] [Google Scholar]
- 7.Hardy MH. The secret life of the hair follicle. Trends Genet. 1992;8:55–61. doi: 10.1016/0168-9525(92)90350-D. [DOI] [PubMed] [Google Scholar]
- 8.Musser JM, Wagner GP, Prum RO. Nuclear B-catenin localization supports homology of feathers, avian scutate scales, and alligator scales in early development. Evol Dev. 2015;17:185–194. doi: 10.1111/ede.12123. [DOI] [PubMed] [Google Scholar]
- 9.Donoghue P. Evolution of development of the vertebrate dermal and oral skeletons: unravelling concepts, regulatory theories, and homologies. Paleobiology. 2002;28:474–507. doi: 10.1666/0094-8373(2002)028<0474:EODOTV>2.0.CO;2. [DOI] [Google Scholar]
- 10.Sansom IJ, Smith MM, Smith P. Scales of thelodont and shark-like fishes from the Ordovician of Colorado. Nature. 1996;379:628–630. doi: 10.1038/379628a0. [DOI] [Google Scholar]
- 11.Karatajute-Talimaa V. Elegestolepis grossi gen. et sp. nov., ein neuer Typ der Placoidschuppe aus dem oberen Silur der Tuwa. Palaeontogr A. 1973;143:35–50. [Google Scholar]
- 12.Ørvig T. A survey of odontodes (“dermal teeth”) from developmental, structural, functional and phyletic points of view. In: Mahala Andrews S, Miles RS, Walker AD, editors. Problems in vertebrate evolution. New York: Academic Press; 1977. pp. 53–75. [Google Scholar]
- 13.Motta P, Habegger ML, Lang A, Hueter R, Davis J. Scale morphology and flexibility in the shortfin mako Isurus oxyrinchus and the blacktip shark Carcharhinus limbatus. J Morphol. 2012;273:1096–1110. doi: 10.1002/jmor.20047. [DOI] [PubMed] [Google Scholar]
- 14.Martin KJ, Rasch LJ, Cooper RL, Metscher BD, Johanson Z, Fraser GJ. Sox2 + progenitors in sharks link taste development with the evolution of regenerative teeth from denticles. Proc Natl Acad Sci USA. 2016;113:14769–14774. doi: 10.1073/pnas.1612354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser GJ, Cerny R, Soukup V, Bronner-fraser M, Streelman T. The odontode explosion: the origin of tooth-like structures in vertebrates. BioEssays. 2010;32:808–817. doi: 10.1002/bies.200900151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raschi W, Tabit C. Functional aspects of placoid scales: a review and update. Mar Freshw Res. 1992;43:123–147. doi: 10.1071/MF9920123. [DOI] [Google Scholar]
- 17.Southall EJ, Sims DW. Shark skin: a function in feeding. Proc R Soc B Biol Sci. 2003;270:S47–S49. doi: 10.1098/rsbl.2003.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan T, Regan F. The characterization, replication and testing of dermal denticles of Scyliorhinus canicula for physical mechanisms of biofouling prevention. Bioinspir Biomim. 2011;6:46001. doi: 10.1088/1748-3182/6/4/046001. [DOI] [PubMed] [Google Scholar]
- 19.Reif W-E. Functions of scales and photophores in mesopelagic luminescent sharks. Acta Zool. 1985;66:111–118. doi: 10.1111/j.1463-6395.1985.tb00829.x. [DOI] [Google Scholar]
- 20.Reif W-E. Morphology and hydrodynamic effects of the scales of fast swimming sharks. Fortschr Zool. 1985;30:483–485. [Google Scholar]
- 21.Dean B, Bhushan B. Shark-skin surfaces for fluid-drag reduction in turbulent flow: a review. Philos Trans A Math Phys Eng Sci. 2010;368:4775–4806. doi: 10.1098/rsta.2010.0201. [DOI] [PubMed] [Google Scholar]
- 22.Oeffner J, Lauder GV. The hydrodynamic function of shark skin and two biomimetic applications. J Exp Biol. 2012;215:785–795. doi: 10.1242/jeb.063040. [DOI] [PubMed] [Google Scholar]
- 23.Ballard W, Mellinger J, Lechenault H. A series of normal stages for development of Scyliorhinus canicula, the lesser spotted dogfish (Chondrichthyes: Scyliorhinidae) J Exp Zool. 1993;267:318–336. doi: 10.1002/jez.1402670309. [DOI] [Google Scholar]
- 24.Oster GF, Murray JD, Harris AK. Mechanical aspects of mesenchymal morphogenesis. J Embryol Exp Morphol. 1983;78:83–125. [PubMed] [Google Scholar]
- 25.Reif W-E. Evolution of dermal skeleton and dentition in vertebrates—the odontode regulation theory. Evol Biol. 1982;15:287–368. doi: 10.1007/978-1-4615-6968-8_7. [DOI] [Google Scholar]
- 26.Fraser G, Smith M. Evolution of developmental pattern for vertebrate dentitions: an oro-pharyngeal specific mechanism. J Exp Zool B Mol Dev Evol. 2010;316B:99–112. doi: 10.1002/jez.b.21387. [DOI] [PubMed] [Google Scholar]
- 27.Johanson Z, Tanaka M, Chaplin N, Smith M. Early Palaeozoic dentine and patterned scales in the embryonic catshark tail. Biol Lett. 2008;4:87–90. doi: 10.1098/rsbl.2007.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johanson Z, Smith MM, Joss JMP. Early scale development in Heterodontus (Heterodontiformes; Chondrichthyes): a novel chondrichthyan scale pattern. Acta Zool. 2007;88:249–256. doi: 10.1111/j.1463-6395.2007.00276.x. [DOI] [Google Scholar]
- 29.Eames BF, Allen N, Young J, Kaplan A, Helms JA, Schneider RA. Skeletogenesis in the swell shark Cephaloscyllium ventriosum. J Anat. 2007;210:542–554. doi: 10.1111/j.1469-7580.2007.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reif WE. Development of dentition and dermal skeleton in embryonic Scyliorhinus canicula. J Morphol. 1980;166:275–288. doi: 10.1002/jmor.1051660303. [DOI] [PubMed] [Google Scholar]
- 31.Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohuchi H, Kimura S, Watamoto M, Itoh N. Involvement of fibroblast growth factor (FGF)18-FGF8 signaling in specification of left-right asymmetry and brain and limb development of the chick embryo. Mech Dev. 2000;95:55–66. doi: 10.1016/S0925-4773(00)00331-2. [DOI] [PubMed] [Google Scholar]
- 34.Trokovic R, Jukkola T, Saarimäki J, Peltopuro P, Naserke T, Vogt Weisenhorn DM, et al. Fgfr1-dependent boundary cells between developing mid- and hindbrain. Dev Biol. 2005;278:428–439. doi: 10.1016/j.ydbio.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 35.Rosenquist TA, Martin GR. Fibroblast growth factor signalling in the hair growth cycle: expression of the fibroblast growth factor receptor and ligand genes in the murine hair follicle. Dev Dyn. 1996;205:379–386. doi: 10.1002/(SICI)1097-0177(199604)205:4<379::AID-AJA2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 36.Jung H, Francis-west PH, Widelitz RB, Jiang T, Ting-berreth S, Tickle C, et al. Local inhibitory action of BMPs and their relationships with activators in feather formation: implications for periodic patterning. Dev Biol. 1998;196:11–23. doi: 10.1006/dbio.1998.8850. [DOI] [PubMed] [Google Scholar]
- 37.Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Li C-Y, Prochazka J, Goodwin AF, Klein OD. Fibroblast growth factor signaling in mammalian tooth development. Odontology. 2014;102:1–13. doi: 10.1007/s10266-013-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moustakas-Verho JE, Zimm R, Cebra-Thomas J, Lempiainen NK, Kallonen A, Mitchell KL, et al. The origin and loss of periodic patterning in the turtle shell. Development. 2014;141:3033–3039. doi: 10.1242/dev.109041. [DOI] [PubMed] [Google Scholar]
- 40.Rasch LJ, Martin KJ, Cooper RL, Metscher BD, Underwood CJ, Fraser GJ. An ancient dental gene set governs development and continuous regeneration of teeth in sharks. Dev Biol. 2016;415:347–370. doi: 10.1016/j.ydbio.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 41.Debiais-Thibaud M, Oulion S, Bourrat F, Laurenti P, Casane D, Borday-Birraux V. The homology of odontodes in gnathostomes: insights from Dlx gene expression in the dogfish, Scyliorhinus canicula. BMC Evol Biol. 2011;11:307. doi: 10.1186/1471-2148-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Debiais-Thibaud M, Chiori R, Enault S, Oulion S, Germon I, Martinand-Mari C, et al. Tooth and scale morphogenesis in shark: an alternative process to the mammalian enamel knot system. BMC Evol Biol. 2015;15:292. doi: 10.1186/s12862-015-0557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Widelitz RB, Ting-Xin J, Noveen A, Chen C-WJ, Chuong C-M. FGF induces new feather buds from developing avian skin. J Invest Dermatol. 1996;107:798–803. doi: 10.1111/1523-1747.ep12330553. [DOI] [PubMed] [Google Scholar]
- 44.Noramly S, Morgan BA. BMPs mediate lateral inhibition at successive stages in feather tract development. Development. 1998;125:3775–3787. doi: 10.1242/dev.125.19.3775. [DOI] [PubMed] [Google Scholar]
- 45.Ashique AM, Fu K, Richman JM. Signalling via type IA and type IB bone morphogenetic protein receptors (BMPR) regulates intramembranous bone formation, chondrogenesis and feather formation in the chicken embryo. Int J Dev Biol. 2002;46:243–253. doi: 10.1387/ijdb.011535. [DOI] [PubMed] [Google Scholar]
- 46.Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat Cell Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- 47.Hogan BL. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/S0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 48.Stuart ES, Moscona AA. Embryonic morphogenesis: role of fibrous lattice in the development of feathers and feather patterns. Science. 1967;157:947–948. doi: 10.1126/science.157.3791.947-a. [DOI] [PubMed] [Google Scholar]
- 49.Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nüsslein-Volhard C. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 2008;4:e1000206. doi: 10.1371/journal.pgen.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fraser GJ, Graham A, Smith MM. Conserved deployment of genes during odontogenesis across osteichthyans. Proc Biol Sci. 2004;271:2311–2317. doi: 10.1098/rspb.2004.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houghton L, Lindon C, Morgan BA. The ectodysplasin pathway in feather tract development. Development. 2005;132:863–872. doi: 10.1242/dev.01651. [DOI] [PubMed] [Google Scholar]
- 52.Drew CF, Lin CM, Jiang TX, Blunt G, Mou C, Chuong CM, et al. The Edar subfamily in feather placode formation. Dev Biol. 2007;305:232–245. doi: 10.1016/j.ydbio.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mustonen T, Ilmonen M, Pummila M, Kangas AT, Laurikkala J, Jaatinen R, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 54.Fessing MY, Sharova TY, Sharov AA, Atoyan R, Botchkarev VA. Involvement of the Edar signaling in the control of hair follicle involution (catagen) Am J Pathol. 2006;169:2075–2084. doi: 10.2353/ajpath.2006.060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tucker AS, Headon DJ, Schneider P, Ferguson BM, Overbeek P, Tschopp J, et al. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt-Ullrich R, Tobin DJ, Lenhard D, Schneider P, Paus R, Scheidereit C. NF-kappaB transmits Eda A1/EdaR signalling to activate Shh and cyclin D1 expression, and controls post-initiation hair placode down growth. Development. 2006;133:1045–1057. doi: 10.1242/dev.02278. [DOI] [PubMed] [Google Scholar]
- 57.Pummila M, Fliniaux I, Jaatinen R, James MJ, Laurikkala J, Schneider P, et al. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 2007;134:117–125. doi: 10.1242/dev.02708. [DOI] [PubMed] [Google Scholar]
- 58.Buchtová M, Handrigan GR, Tucker AS, Lozanoff S, Town L, Fu K, et al. Initiation and patterning of the snake dentition are dependent on Sonic hedgehog signaling. Dev Biol. 2008;319:132–145. doi: 10.1016/j.ydbio.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Smith MM, Fraser GJ, Chaplin N, Hobbs C, Graham A. Reiterative pattern of sonic hedgehog expression in the catshark dentition reveals a phylogenetic template for jawed vertebrates. Proc Biol Sci. 2009;276:1225–1233. doi: 10.1098/rspb.2008.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ting-berreth SA, Chuong C. Sonic hedgehog in feather morphogenesis: induction of mesenchymal condensation and association with cell death. Dev Dyn. 1996;207:157–170. doi: 10.1002/(SICI)1097-0177(199610)207:2<157::AID-AJA4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 61.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation †. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 63.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, et al. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 64.Mill P, Mo R, Fu H, Grachtchouk M, Kim PCW, Dlugosz AA, et al. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 2003;17:282–294. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 66.Gillis JA, Dahn RD, Shubin NH. Shared developmental mechanisms pattern the vertebrate gill arch and paired fin skeletons. Proc Natl Acad Sci USA. 2009;106:5720–5724. doi: 10.1073/pnas.0810959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mandler M, Neubüser A. FGF signaling is required for initiation of feather placode development. Development. 2004;131:3333–3343. doi: 10.1242/dev.01203. [DOI] [PubMed] [Google Scholar]
- 68.Kettunen P, Laurikkala J, Itäranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 69.Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 70.Yokoi H, Shimada A, Carl M, Takashima S, Kobayashi D, Narita T, et al. Mutant analyses reveal different functions of fgfr1 in medaka and zebrafish despite conserved ligand-receptor relationships. Dev Biol. 2007;304:326–337. doi: 10.1016/j.ydbio.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 71.Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 72.Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takamori K, Hosokawa R, Xu X, Deng X, Bringas P, Chai Y. Epithelial fibroblast growth factor receptor 1 regulates enamel formation. J Dent Res. 2008;87:238–243. doi: 10.1177/154405910808700307. [DOI] [PubMed] [Google Scholar]
- 74.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 75.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family: the complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Panganiban G, Rubenstein JLR. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 77.Rouzankina I, Abate-Shen C, Niswander L. Dlx genes integrate positive and negative signals during feather bud development. Dev Biol. 2004;265:219–233. doi: 10.1016/j.ydbio.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 78.Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- 79.Thomas BL, Liu JK, Rubenstein JL, Sharpe PT. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- 80.Kurki P, Vanderlaan M, Dolbeare F, Gray J, Tan EM. Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp Cell Res. 1986;166:209–219. doi: 10.1016/0014-4827(86)90520-3. [DOI] [PubMed] [Google Scholar]
- 81.Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 82.Poss KD, Shen J, Nechiporuk A, Mcmahon G, Thisse B, Thisse C, et al. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- 83.Paterson JL, Li Z, Wen X-Y, Masih-Khan E, Chang H, Pollett JB, et al. Preclinical studies of fibroblast growth factor receptor 3 as a therapeutic target in multiple myeloma. Br J Haematol. 2004;124:595–603. doi: 10.1111/j.1365-2141.2004.04814.x. [DOI] [PubMed] [Google Scholar]
- 84.Fraser GJ, Bloomquist RF, Streelman JT. Common developmental pathways link tooth shape to regeneration. Dev Biol. 2013;377:399–414. doi: 10.1016/j.ydbio.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Shaughnessy KL, Dahn RD, Cohn MJ. Molecular development of chondrichthyan claspers and the evolution of copulatory organs. Nat Commun. 2015;6:6698. doi: 10.1038/ncomms7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 87.Lewandoski M, Sun X, Martin GR. Fgf8 signalling from the AER is essential for normal limb development. Nat Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- 88.Wagner GP. The developmental genetics of homology. Nat Rev Genet. 2007;8:473–479. doi: 10.1038/nrg2099. [DOI] [PubMed] [Google Scholar]
- 89.Maderson PF. When? Why? and How? Some speculations on the evolution of the vertebrate integument. Am Zool. 1972;12:159–171. doi: 10.1093/icb/12.1.159. [DOI] [Google Scholar]
- 90.Maderson PF. Mammalian skin evolution: a reevaluation. Exp Dermatol. 2003;12:233–236. doi: 10.1034/j.1600-0625.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- 91.Dhouailly D. A new scenario for the evolutionary origin of hair, feather, and avian scales. J Anat. 2009;214:587–606. doi: 10.1111/j.1469-7580.2008.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 93.McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128:2471–2484. doi: 10.1242/dev.128.13.2471. [DOI] [PubMed] [Google Scholar]
- 94.Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, et al. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- 95.Chen C-F, Foley J, Tang P-C, Li A, Jiang TX, Wu P, et al. Development, regeneration, and evolution of feathers. Annu Rev Anim Biosci. 2015;3:169–195. doi: 10.1146/annurev-animal-022513-114127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jernvall J, Thesleff I. Tooth shape formation and tooth renewal: evolving with the same signals. Development. 2012;139:3487–3497. doi: 10.1242/dev.085084. [DOI] [PubMed] [Google Scholar]
- 97.Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010;80:241–248. doi: 10.1016/j.diff.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 98.Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wyffels J, King BL, Vincent J, Chen C, Wu CH, Polson SW. SkateBase, an elasmobranch genome project and collection of molecular resources for chondrichthyan fishes. F1000Research. 2014;3:191. doi: 10.12688/f1000research.4996.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.


