Abstract
During vertebrate neural development, positional information is largely specified by extracellular morphogens. Their distribution, however, is very dynamic due to the multiple roles played by the same signals in the developing and adult neural tissue. This suggests that neural progenitors are able to modify their competence to respond to morphogen signalling and autonomously maintain positional identities after their initial specification. In this work, we take advantage of in vitro culture systems of mouse neural stem/progenitor cells (NSPCs) to show that NSPCs isolated from rostral or caudal regions of the mouse neural tube are differentially responsive to retinoic acid (RA), a pivotal morphogen for the specification of posterior neural fates. Hoxb genes are among the best known RA direct targets in the neural tissue, yet we found that RA could promote their transcription only in caudal but not in rostral NSPCs. Correlating with these effects, key RA-responsive regulatory regions in the Hoxb cluster displayed opposite enrichment of activating or repressing histone marks in rostral and caudal NSPCs. Finally, RA was able to strengthen Hoxb chromatin activation in caudal NSPCs, but was ineffective on the repressed Hoxb chromatin of rostral NSPCs. These results suggest that the response of NSPCs to morphogen signalling across the rostrocaudal axis of the neural tube may be gated by the epigenetic configuration of target patterning genes, allowing long-term maintenance of intrinsic positional values in spite of continuously changing extrinsic signals.
Keywords: neural stem/progenitor cells, retinoic acid, Hoxb genes, epigenetic regulation, telencephalon, spinal cord
1. Introduction
A remarkable feature of vertebrate neural development is the early subdivision of the neuroectoderm along its rostrocaudal and dorsoventral axes into an orderly array of sharply delimited domains expressing region-specific combinations of transcription factor-encoding genes [1,2]. For example, expression of Otx1/2 genes identifies presumptive anterior (forebrain and midbrain) territories [3], whereas Hox genes are expressed within posterior (hindbrain and spinal cord) neuroectoderm [4,5]. Other gene families define positional identities along the neural tube dorsoventral axis [2]. Eventually, various combinations of transcription factors become expressed in different areas of the neural tube, leading to specification of cellular compartments with distinct transcriptional programmes and developmental fates [6].
Transcription factors involved in the specification of positional identities acquire their spatially limited expression domains in response to the action of a number of diffusible morphogens that are distributed with concentration gradients across the neural tissue [2,7,8]. Among them, retinoic acid (RA) is a key morphogen controlling rostrocaudal neural patterning [7,9,10]. RA is a product of vitamin A metabolism that is present with a posterior-high to anterior-low gradient in early vertebrate embryos due to the presence of RA-synthesizing enzymes (such as Aldh1a2) in caudal regions and RA-degrading enzymes (such as Cyp26a1) in anterior regions [9–11]. The posterior neural tube develops abnormally in embryos with reduced levels of RA [12–15]. Furthermore, embryos exposed to exogenous RA during early developmental stages undergo dramatic losses of forebrain structures and expansion of posterior neural structures [16–18]. RA regulates gene transcription by binding retinoic acid receptors (RARs), which interact with retinoic acid response elements (RAREs) within regulatory regions of RA-responsive genes to control their expression [11]. In particular, well-characterized RAREs are present within Hox gene clusters [19,20] and transcription of Hox genes is readily upregulated following RA treatments of early vertebrate embryos or in vitro cellular models of early embryonic cells [17,21–25]. Hence, RA can promote posterior neural fates by directly activating Hox gene expression [26], while repressing, directly or indirectly, transcription of Otx2 and other rostral specification genes [17,27].
Although the presumptive anterior neuroectoderm is very sensitive to the caudalizing activity of RA during gastrulation, vertebrate embryos exposed to exogenous RA at later developmental stages become progressively resistant to suppression of anterior development [17,18,28]. Furthermore, at these later stages, endogenous retinoid signalling is essential for proper forebrain formation. For example, RAR-dependent signalling has been implicated in patterning and morphogenesis of the optic cup [29–32], in the differentiation of GABAergic neurons in the basal ganglia of the telencephalon [33], and in the control of neuronal differentiation and migration in the developing cerebral cortex [34–37]. Retinoid signalling also appears to control neural progenitor differentiation in the subventricular zone (SVZ) and dentate gyrus neurogenic niches of the postnatal mammalian forebrain [38,39]. These observations suggest that anterior neural progenitors drastically modify their responsiveness to retinoid signalling during development, losing competence in retinoid-dependent caudalization while becoming able to provide alternative, cell type-specific responses following exposure to locally produced retinoids. Nonetheless, direct evidence that neural progenitors in the embryonic and postnatal forebrain are effectively refractory to posteriorization by retinoid signalling is still lacking. Moreover, it remains unclear how the intrinsic competence of forebrain progenitors to acquire posterior positional identities may be restricted at the molecular level.
Previous studies have shown that neural stem/progenitor cells (NSPCs) derived from the embryonic and the adult murine central nervous system (CNS) can be extensively propagated in vitro using appropriate adherent culture conditions that allow long-term self-renewal [40,41]. Furthermore, NSPCs obtained from different rostrocaudal regions of the mouse neural tube at embryonic day 13.5–14.5 (E13.5–14.5) or from the adult mouse forebrain maintain distinct expression profiles of transcription factor-encoding genes that are consistent with the respective areas of derivation [42,43]. These transcriptional profiles persist even after several passages in culture, suggesting that NSPCs can retain, at least in part, their positional identities in vitro [42,43]. In this work, we took advantage of this experimental system to demonstrate that NSPCs derived from both rostral and caudal regions of E13.5 mouse neural tube can activate retinoid signalling when exposed to RA, but they respond differently to it. In particular, exogenous RA can promote expression of genes of the Hoxb cluster in posterior, but not in anterior NSPCs, which remain resistant to RA-dependent Hoxb transcription also during adulthood and ageing. We further show that Hoxb chromatin, including key RAREs, is differentially enriched for activating and repressing histone modifications in rostral and caudal NSPCs. In agreement with transcriptional effects, RA treatments are unable to reverse Hoxb chromatin repression in rostral NSPCs, but can enhance Hoxb chromatin activation in caudal NSPCs.
Altogether, these results suggest that intrinsic epigenetic differences at the level of key regionally expressed genes may constrain transcriptional responses to morphogen signalling and safeguard maintenance of rostrocaudal positional identities in NSPCs.
2. Material and methods
2.1. Mouse neural stem/progenitor cell culture
NSPCs derived from mouse E13.5 cerebral cortex or spinal cord and protocols for their in vitro culture were previously described [43]. Cortex NSPCs were obtained from the dorsolateral wall of the telencephalon, corresponding to the developing cerebral cortex. Spinal cord NSPCs were derived from spinal cord tissue dissected between the hindbrain/spinal cord boundary and hindlimb buds. Adult or aged SVZ NSPCs were derived from 3-month- or 18-month-old mice, respectively, and cultured in vitro as previously described [44]. NSPCs were routinely expanded in T25 flasks (Corning) that were coated with 10 µg ml−1 poly-ornithine (Sigma-Aldrich) and 5 µg ml−1 laminin (Sigma-Aldrich), using previously described chemically defined media [43,44] supplemented with 20 ng ml−1 human recombinant epidermal growth factor (R&D Systems) and 10 ng ml−1 human recombinant fibroblast growth factor-basic (Peprotech). Media for embryonic cortex and spinal cord NSPC culture also contained 1 : 100 N2 supplement (Invitrogen), while 1 : 50 B27 supplement minus vitamin A (Invitrogen) was used for adult SVZ NSPC cultures. NSPCs were passaged every 3–5 days using Accutase (Sigma-Aldrich) and usually seeded at a density of 10–20 000 cells cm−2. NSPCs expanded for up to 20 passages in vitro since their initial derivations were used for this work.
2.2. Luciferase assays
To detect activation of retinoid signalling in NSPC cultures treated with RA, 2 × 106 embryonic cortex or spinal cord NSPCs were transfected with 2 µg of tk-(βRARE)2-luc plasmid, in which expression of firefly luciferase is under control of RAREs [12], and 100 ng of pRL-TK plasmid, which drives constitutive expression of Renilla luciferase as a control for transfection efficiency [45]. NSPC transfection was carried out using an Amaxa mouse neural stem cell Nucleofector kit (Lonza) on an Amaxa Nucleofector device (Lonza). Transfected cells were seeded in one six-well plate (Corning). The day after transfection, half of the wells received fresh media supplemented with all-trans RA (Sigma-Aldrich) diluted from a 25 mM stock in DMSO. Media added to the other half contained equal volumes of DMSO. After 24 h of treatment, cells were harvested and reporter expression levels were measured using the dual-luciferase reporter assay system (Promega) on a GloMax multi+ detection device (Promega) as previously described [45].
2.3. Real-time RT-PCR
For gene expression analysis by real-time RT-PCR in RA-treated cultures, embryonic cortex or spinal cord NSPCs were seeded in six-well plates (Corning) at a density of 10 000 cells cm−2. Adult SVZ NSPCs were instead seeded in T25 flasks at the same density. After 24 h, cultures received fresh media containing appropriate concentrations of RA or DMSO solvent. Media supplemented with RA or DMSO were again replaced the next day and cultures were harvested for molecular analysis after 48 h of treatment.
Total RNA was extracted from frozen cell pellets using the Qiagen RNeasy mini kit and quantified with a NanoDrop 2000 (Thermo Scientific). For real-time RT-PCR, RNA was reverse-transcribed using the Qiagen QuantiTect reverse transcription kit and amplified on a Rotor-Gene Q (Qiagen), using the Qiagen QuantiFast SYBR Green PCR kit. Primers for real-time RT-PCR were either purchased from Qiagen or designed using Primer3 (http://bioinfo.ut.ee/primer3/). Primer sequences selected with Primer3 are listed in the electronic supplementary material, table S1. Relative gene expression levels in different samples were determined with the built-in comparative quantitation method [46], using Eef1a1 or Rpl19 as reference genes, with similar results. Statistical analysis of experimental data was carried out using the Microsoft Excel software.
2.4. Chromatin immunoprecipitation
For detection of histone H3 modifications by chromatin immunoprecipitation (ChIP), NSPCs were seeded in 100 mm plates at a density of 12–18 000 cells cm−2, until cultures reached approximately 80% confluence. Usually, 10–12 × 106 cells for each experimental sample were used in the following steps. At the time of harvesting, cultures were rinsed with PBS, followed by cross-linking with 1% formaldehyde in PBS for 10 min and quenching with 125 mM glycine for 5 min. Cells were then washed with PBS, harvested with a cell scraper in PBS, centrifuged and lysed in lysis buffer (5 mM PIPES, 85 mM KCl, 0.5% NP-40) for 20 min on ice. This was followed by centrifugation, pellet resuspension in shearing buffer (50 mM Tris (pH 8.1), 10 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate), sonication to an approximate size of 250–500 bps and pre-clearing using Dynabeads-protein G (Invitrogen). The protein content of pre-cleared chromatin was measured using the Bradford assay and 25 µg aliquots of each sample were used for immunoprecipitation with the following antibodies (Cell Signaling): anti-trimethyl-lysine 27 of histone H3 (C36B11), anti-trimethyl-lysine 4 of histone H3 (C42D8), anti-acetyl-lysine 9/14 of histone H3. Samples were incubated overnight at 4°C with approximately 1–5 µg ml−1 of each antibody in modified RIPA buffer (140 mM NaCl, 10 mM Tris (pH 7.5), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.01% SDS, 0.1% sodium deoxycholate). As a negative control, an equivalent concentration of normal rabbit IgG (Cell Signaling) was used. Immunocomplexes were recovered by incubating each sample with 20 µl of Dynabeads-protein G for 90 min at 4°C, followed by a series of washes and elution with 0.1 M NaHCO3, 1% SDS buffer. All steps between cross-linking and elution included protease inhibitor cocktail (Sigma-Aldrich). Eluted chromatin was treated with RNase A and proteinase K, and immunoprecipitated DNA was purified using the Nucleospin gel and PCR clean-up kit (Macherey Nagel), along with DNA from 20 µl of pre-cleared, non-immunoprecipitated chromatin for each sample (input sample). Purified DNA from immunoprecipitated and input samples was used for real-time PCR analysis as described above. Primers for real-time ChIP-PCR were designed using Primer3 and are listed in the electronic supplementary material, table S2. Amplification levels in immunoprecipated samples relative to the corresponding input samples were measured and normalized to a reference amplicon located approximately 1 kb 5' of Eef1a1.
3. Results
3.1. Retinoid signalling can upregulate Hoxb gene expression in spinal cord, but not in telencephalic NSPCs
Previous work showed that NSPCs derived from different districts of mouse E13.5–14.5 neural tube retain position-specific gene expression profiles during in vitro culture that are consistent with the region of derivation [42,43]. In this study, we investigated whether retinoid signalling, a key pathway in the specification of posterior neural fates [7,9,10], could challenge the positional identity of NSPCs derived from the rostral (telencephalon) neural tube region, in comparison with caudal (spinal cord-derived) NSPCs.
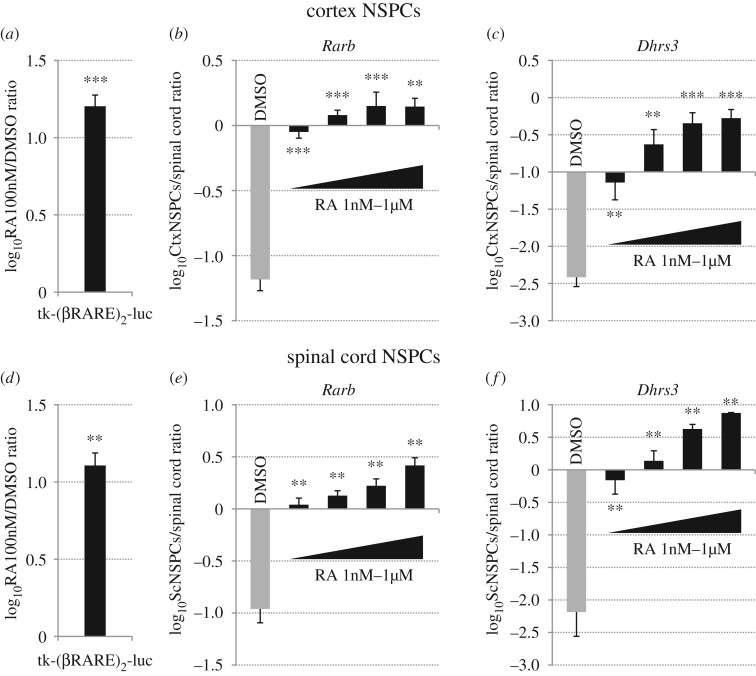
To this aim, we first verified whether retinoid signalling is functional and can be activated by exposure to exogenous RA both in rostral and in caudal NSPCs. Previously described murine NSPC cultures derived from E13.5 cerebral cortex or spinal cord [43] were transfected with RA-responsive tk-(βRARE)2-luc luciferase reporter plasmid [12], as described in the Material and methods section. The day after transfection, NSPCs were treated with DMSO or 100 nM RA and harvested 24 h later for luciferase detection. As shown in figure 1a,d, RA treatments upregulated reporter expression at similar levels in cortex and spinal cord NSPCs. In addition, we analysed RA effects on transcription of Rarb, Dhrs3 and Cyp26a1, which are well-described retinoid-responsive genes in various tissues [47–50], by real-time RT-PCR. Treatments with different concentrations of RA ranging from 1 nM to 1 µM for 48 h caused robust, dose-dependent upregulation of all these genes both in cortex and in spinal cord NSPCs (figure 1b,c,e,f; electronic supplementary material, figure S1a and S1d).
Figure 1.
Retinoid signalling is functional in cortex and spinal cord NSPCs and it can be efficiently activated by exogenous RA. (a,d) Luciferase reporter assays in cortex (a) or spinal cord (d) NSPCs that were electroporated with an RA-responsive luciferase-expressing plasmid (tk-(βRARE)2-luc) and treated with DMSO or 100 nM RA for 24 h. Reporter activity is significantly increased in both cell types at comparable levels by RA treatments. Results are shown as the mean of the log10-transformed ratio between RA and DMSO conditions in three to four biological replicates. (b,c,e,f) Real-time RT-PCR quantification of gene expression in cortex NSPCs (CtxNSPCs) (b,c) or spinal cord NSPCs (ScNSPCs) (e,f) that were treated with either DMSO or 1, 10, 100 or 1000 nM RA for 48 h, showing that RA treatments efficiently upregulate the RA-responsive genes Rarb and Dhrs3 in both cell types. Results are shown as the mean of the log10-transformed ratio between DMSO-treated or RA-treated NSPCs and E13.5 spinal cord tissue in four to five biological replicates. In (a–f), error bars show s.e.m. **p < 0.01; ***p < 0.001 according to a two-tailed Student's t-test performed between DMSO and RA conditions.
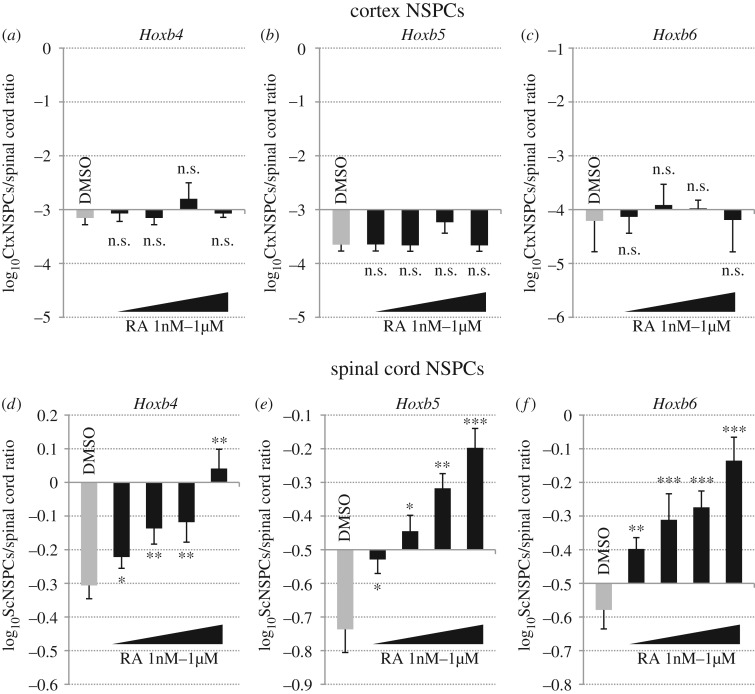
In agreement with previous reports [42,43] and with their expression in the developing mouse spinal cord [20,26], transcription of Hoxb4–9 genes was maintained during in vitro culture of spinal cord NSPCs, while it was barely detectable in cortex NSPCs (figure 2a–f; electronic supplementary material, figure S1b and S1e). We then investigated whether exogenous RA could alter these transcriptional profiles. In spinal cord NSPCs cultured in control conditions, mRNA levels of Hoxb4, Hoxb5, Hoxb6 and Hoxb9 were approximately 10–50% of those detectable in E13.5 spinal cord tissue (figure 2d–f; electronic supplementary material, figure S1e). Treatments with increasing doses of RA (from 1 nM to 1 µM) for 48 h led to significant upregulation of these genes by all RA doses, with 1 µM RA eliciting expression levels comparable with those detectable in embryonic spinal cord tissue (figure 2d–f; electronic supplementary material, figure S1e). By contrast, transcript levels of Hoxb4–9 genes were near the limit of real-time RT-PCR detection in mock-treated cortex NSPCs (approx. 10−3–10−4 relative to their expression in spinal cord tissue) and they were not significantly changed following exposure to exogenous RA (figure 2a–c; electronic supplementary material, figure S1b). To confirm that cortex NSPCs retained a rostral identity after RA treatments, we analysed expression of the anterior neural tube marker Emx2, which is specifically expressed in the developing mouse cerebral cortex [3]. As previously reported [42], in vitro cultures of cortex NSPCs showed comparable Emx2 expression levels to those detected in E13.5 telencephalic tissue (electronic supplementary material, figure S1c). Consistently with the inability of RA to upregulate Hoxb gene transcription in cortex NSPCs, we found no significant effects of RA treatments on Emx2 expression in these cultures (electronic supplementary material, figure S1c).
Figure 2.
Hoxb4–6 genes are differentially expressed in cortex and spinal cord NSPCs and are responsive to retinoid signalling only in spinal cord NSPCs. (a–f) Real-time RT-PCR quantification of Hoxb4, (a,d), Hoxb5 (b,e) and Hoxb6 (c,f) gene expression in cortex (a–c) or spinal cord (d–f) NSPCs that were treated with either DMSO or 1, 10, 100 or 1000 nM RA for 48 h. Transcript levels of Hoxb4–6 genes in DMSO-treated spinal cord NSPCs are approximately 20–50% of those detected in E13.5 spinal cord tissue, but they are more than 1000 times lower in DMSO-treated cortex NSPCs. Furthermore, RA treatments cause significant, dose-dependent upregulation of Hoxb4–6 genes in spinal cord, but not in cortex NSPCs. Results are shown as the mean of the log10-transformed ratio between DMSO-treated or RA-treated NSPCs and E13.5 spinal cord tissue in four to five biological replicates. Error bars show s.e.m. *p ≤ 0.05; **p < 0.01; ***p < 0.001; n.s., non-significant (p > 0.05) according to a two-tailed Student's t-test performed between DMSO and RA conditions.
During postnatal development, NSPCs of the cerebral cortex mostly undergo terminal differentiation, but a limited population of embryonic cortical progenitors contribute to NSPCs persisting within the SVZ neurogenic niche of the adult and the ageing telencephalon [51]. To address whether rostral NSPCs remain resistant to retinoid-dependent caudalization beyond embryonic development, we analysed the effects of RA treatments on Hoxb gene expression in NSPC cultures derived from the adult (3 months old) and the ageing (18 months old) mouse SVZ, as previously described [44]. Similar to cortex NSPCs, treatments of adult and aged SVZ NSPCs with high RA doses (1–2 µM) for 48 h robustly upregulated expression of Rarb, Cyp26a1 and Dhrs3 (electronic supplementary material, figure S2a–f), whereas Hoxb4–6 genes continued to be transcribed at the very low levels detectable in mock-treated cultures without significant changes (electronic supplementary material, figure S3a–f).
Altogether, these results indicate that RA-dependent signal tranduction is functional and can elicit a transcriptional response in both rostral and caudal NSPCs. By contrast, Hoxb genes remain responsive to activation of retinoid signalling in spinal cord NSPCs, whereas in telencephalic NSPCs they are locked in a stably silenced, RA-unresponsive transcriptional state.
3.2. Histone H3 methylation and acetylation are differentially enriched at the level of Hoxb RAREs in telencephalic and spinal cord NSPCs
We set out to investigate whether the different sensitivity of Hoxb genes to retinoid signalling in rostral and caudal NSPCs is epigenetically regulated. To this aim, we focused on Hoxb4, Hoxb5 and Hoxb6 genes, since their expression in the hindbrain/spinal cord region is controlled by retinoid signalling acting through previously characterized RAREs located upstream of and downstream from Hoxb4 [19,20,26]. In particular, one RARE (named ENE-RARE) was identified approximately 3 kb downstream from Hoxb4, and two more (named B4U-RARE and DE-RARE) roughly 4 and 8 kb upstream of Hoxb4, respectively (electronic supplementary material, figure S4; [19,20,26]). We then analysed the levels of key epigenetic modifications around these RAREs.
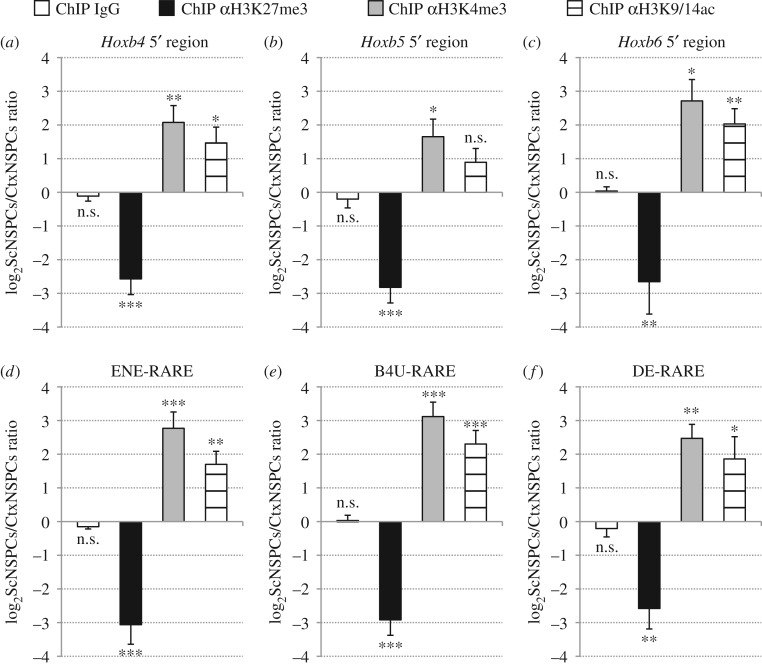
Previous studies have shown that, in embryonic stem cells (ESCs), Hox genes are transcriptionally silent and their chromatin is marked by extensive trimethylation of lysine 27 in histone H3 (H3K27me3), together with limited trimethylation of lysine 4 (H3K4me3) and acetylation of lysines 9 and 14 (H3K9/14ac) [23–25]. Upon ESC differentiation in the presence of exogenous RA, Hox genes are upregulated, along with progressive erasure of H3K27me3 and increased deposition of H3K4me3 and H3K9/14ac [23–25]. These observations prompted us to verify whether these epigenetic marks may also be involved in the long-term maintenance of Hox-off and Hox-on transcriptional states in rostral and caudal NSPCs, respectively, and in the selective ability of caudal NSPCs to upregulate Hoxb gene expression in response to RA. To address this question, we carried out ChIP assays with cortex and spinal cord NSPC cultures, using anti-H3K27me3, anti-H3K4me3 and anti-H3K9/14ac antibodies, as described in the Material and methods section. Immunoprecipitated DNA was then assayed by real-time PCR using primer pairs surrounding ENE-RARE, DE-RARE and B4U-RARE. This analysis was extended to the genomic regions upstream of Hoxb4, Hoxb5 and Hoxb6, by also testing amplicons located approximately 1–2 kb 5' of these genes.
With these assays, we found a significant enrichment of the repressive H3K27me3 mark in cortex NSPCs in comparison with spinal cord NSPCs at the level of both Hoxb4–6 5' regions (figure 3a–c) and ENE-RARE, B4U-RARE and DE-RARE (figure 3d–f). By contrast, the activating H3K4me3 and H3K9/14ac marks were significantly enriched in the same genomic sequences of spinal cord NSPCs (figure 3a–f). No significant differences between cortex and spinal cord NSPCs were detected when samples were immunoprecipitated with a control IgG (figure 3a–f). Since Hoxb genes remain transcriptionally inactive and unresponsive to retinoid signalling in adult and aged SVZ NSPCs, we examined whether they retained an epigenetic signature similar to that of embryonic cortical NSPCs. We carried out ChIP assays with anti-K27me3 and anti-K4me3 antibodies on adult and aged SVZ NSPC cultures and found that the overall enrichment of these modifications was comparable with that detected in cortex NSPCs, both at the level of the Hoxb4 5' region (electronic supplementary material, figure S5a–c) and ENE-RARE (electronic supplementary material, figure S5d–f).
Figure 3.
Cortex and spinal cord NSPCs feature differential deposition of H3K27me3, H3K4me3 and H3K9/14ac at the level of both Hoxb4–6 5' regions and RAREs. (a–f) Real-time PCR quantification of ChIP assays in cortex and spinal cord NSPCs using control (IgG), anti-H3K27me3 (αH3K27me3), anti-H3K4me3 (αH3K4me3) or anti-H3K9/14ac (αH3K9/14ac) antibodies. Primer pairs used for real-time PCR target either the genomic regions 5' of Hoxb4 (a), Hoxb5 (b), Hoxb6 (c), or ENE-RARE (d), B4U-RARE (e), DE-RARE (f), as indicated. A significant increase in H3K4me3 and H3K9/14ac levels and significantly lower H3K27me3 levels are detectable in spinal cord NSPCs relative to cortex NSPCs. No changes between spinal cord and cortex NSPCs are detectable following control ChIP (ChIP IgG). Results are shown as the mean of the log2-transformed ratio between spinal cord and cortex NSPCs in three to six biological replicates, following normalization to a reference amplicon as described in the Material and methods section. Error bars show s.e.m. *p ≤ 0.05; **p < 0.01; ***p < 0.001; n.s., non-significant (p > 0.05) according to a two-tailed Student's t-test performed between spinal cord and cortex NSPC samples.
These results suggest that the levels of H3K27me3, H3K4me3 and H3K9/14ac deposition in Hoxb chromatin, including key RAREs, may underlie the ability of NSPCs to maintain gene expression profiles and transcriptional responses to retinoid signalling matching their rostrocaudal location.
3.3. Retinoid signalling can enhance H3K4me3 and H3K9/14ac deposition in Hoxb chromatin of spinal cord NSPCs, but cannot alter its epigenetic signature in telencephalic NSPCs
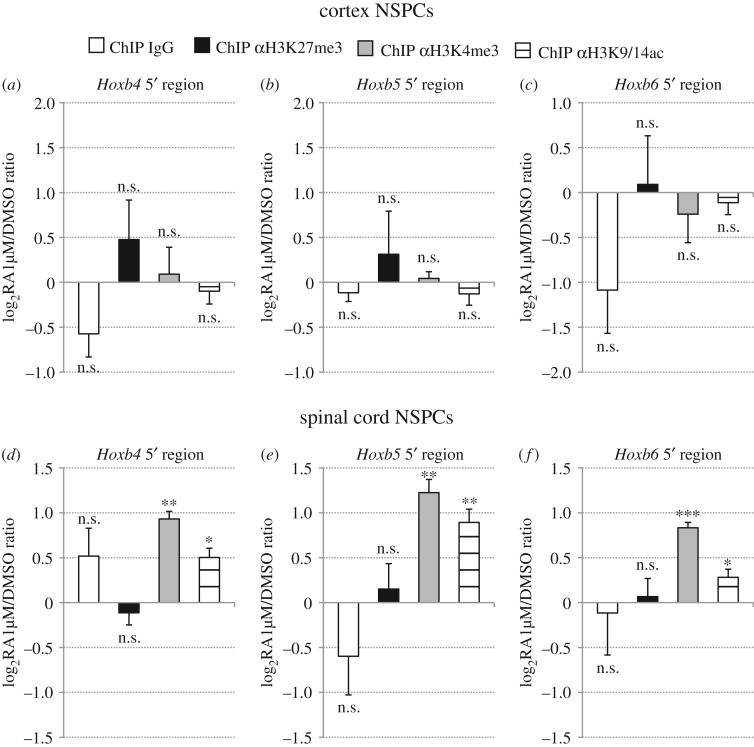
Previous studies have shown that RA treatments of ESCs cause profound epigenetic changes in the Hoxb cluster, by promoting deposition of H3K4me3 and H3K9/14ac at the expense of H3K27me3 [23–25]. We then investigated whether exposure of rostral and caudal NSPCs to exogenous RA could alter the levels of these epigenetic marks in Hoxb4–6 chromatin.
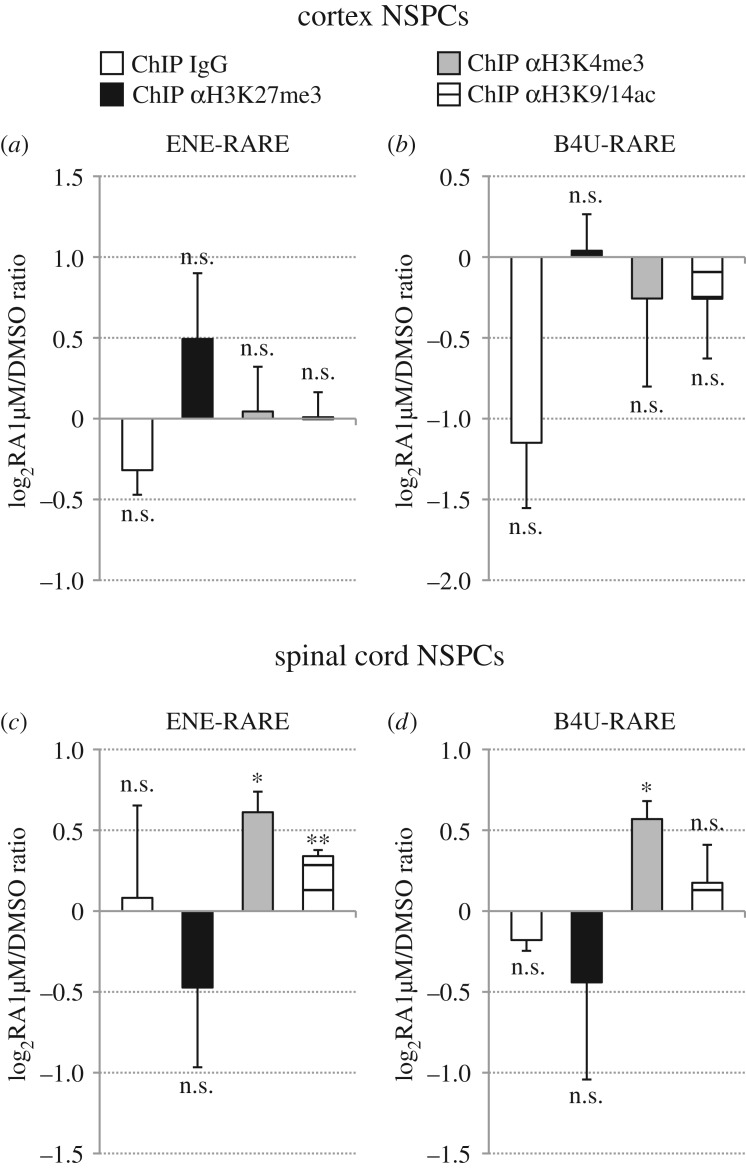
In these assays, cortex and spinal cord NSPC cultures were treated with 1 µM RA for 48 h, followed by ChIP with anti-H3K27me3, anti-H3K4me3 or anti-H3K9/14ac antibodies and real-time PCR analysis. In agreement with RA-dependent upregulation of Hoxb genes in spinal cord NSPCs (figure 2d–f; electronic supplementary material, figure S1e), RA treatments caused in these cells a moderate, but significant increase of H3K4me3 and H3K9/14ac in comparison with mock-treated cultures, at the level of both Hoxb4–6 5' regions (figure 4d–f) and RAREs (figure 5c,d). No significant changes between DMSO-treated and RA-treated spinal cord NSPC cultures were detectable following ChIP with anti-H3K27me3 antibody or control IgG (figures 4d–f and 5c,d). For none of the analysed epigenetic marks, however, did cortex NSPCs show any significant differences between RA-treated cultures and mock-treated cultures (figures 5a–c and figure 6a,b).
Figure 4.
Retinoid signalling promotes H3K4me3 and H3K9/14ac deposition in Hoxb4–6 5' regions of spinal cord NSPCs, but does not alter H3K4me3, H3K27me3 and H3K9/14ac levels in Hoxb4–6 5' regions of cortex NSPCs. (a–f) Real-time PCR quantification of ChIP assays in cortex (a–c) and spinal cord (d–f) NSPCs that were treated with either DMSO or 1 µM RA for 48 h, using control (IgG), anti-H3K27me3 (αH3K27me3), anti-H3K4me3 (αH3K4me3) or anti-H3K9/14ac (αH3K9/14ac) antibodies. Primer pairs used for real-time PCR target the genomic regions 5' of Hoxb4 (a,d), Hoxb5 (b,e) or Hoxb6 (c,f). A significant increase in H3K4me3 and H3K9/K14ac levels is detectable in RA-treated spinal cord NSPCs relative to DMSO-treated cultures, while H3K27me3 levels are not affected. H3K4me3, H3K27me3 and H3K9/K14ac levels do not show significant differences following RA treatments of cortex NSPCs. No changes between RA-treated and DMSO-treated samples are detectable following control ChIP (ChIP IgG). Results are shown as the mean of the log2-transformed ratio between RA-treated and DMSO-treated NSPCs in three to four biological replicates, following normalization to the reference amplicon. Error bars show s.e.m. *p ≤ 0.05; **p < 0.01; ***p < 0.001; n.s., non-significant (p > 0.05) according to a two-tailed Student's t-test performed between RA-treated and DMSO-treated samples.
Figure 5.
Retinoid signalling promotes H3K4me3 and H3K9/14ac deposition in Hoxb RAREs of spinal cord NSPCs, but does not alter H3K4me3, H3K27me3 and H3K9/14ac levels in Hoxb RAREs of cortex NSPCs. (a–f) Real-time PCR quantification of ChIP assays in cortex (a,b) and spinal cord (c,d) NSPCs that were treated with either DMSO or 1 µM RA for 48 h, using control (IgG), anti-H3K27me3 (αH3K27me3), anti-H3K4me3 (αH3K4me3) or anti-H3K9/14ac (αH3K9/14ac) antibodies. Primer pairs used for real-time PCR target ENE-RARE (a,c) or B4U-RARE (b,d). A significant increase in H3K4me3 and H3K9/K14ac levels is detectable in RA-treated spinal cord NSPCs relative to DMSO-treated cultures, while H3K27me3 levels are not significantly affected. H3K4me3, H3K27me3 and H3K9/K14ac levels do not show significant differences following RA treatments of cortex NSPCs. No significant changes between RA-treated and DMSO-treated samples are detectable following control ChIP (ChIP IgG). Results are shown as the mean of the log2-transformed ratio between RA-treated and DMSO-treated NSPCs in three to four biological replicates, following normalization to the reference amplicon. Error bars show s.e.m. *p ≤ 0.05; **p < 0.01; n.s., non-significant (p > 0.05) according to a two-tailed Student's t-test performed between RA-treated and DMSO-treated samples.
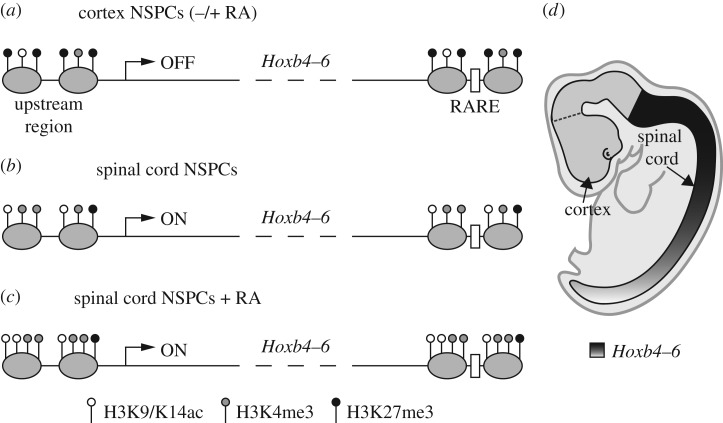
Figure 6.
Proposed model of the epigenetic regulation of Hoxb4–6 expression along the rostrocaudal axis of the neural tube. Rostral (cortex) NSPCs (a) undergo increased deposition of H3K27me3 in comparison with caudal (spinal cord) NSPCs (b), at the level of Hoxb4–6 chromatin (here shown for upstream regions and RAREs flanking these genes). By contrast, H3K4me3 and H3K9/14ac are enriched in spinal cord NSPCs in comparison with cortex NSPCs. This differential deposition of activating and repressive epigenetic marks maintains stable repression or activation of Hoxb4–6 transcription in rostral and caudal NSPCs, respectively. Retinoid signalling cannot alter the epigenetic signature or the repressed transcriptional state of Hoxb4–6 chromatin in rostral NSPCs (a), whereas it can reinforce deposition of activating marks and transcription levels of these genes in caudal NSPCs (c). In (a–c), grey ovals represent nucleosomes, bent arrows represent transcription start sites and white boxes represent RAREs. Panel (d) shows a schematic representation of the Hoxb4–6 expression pattern in the hindbrain/spinal cord region of the developing mouse neural tube. See text for further details.
These results suggest that, at the level of Hoxb chromatin, RA can enhance the active epigenetic configuration of caudal NSPCs, whereas rostral NSPCs are insensitive to retinoid signalling both in terms of transcriptional output and deposition of epigenetic marks.
4. Discussion
Research on both pluripotent stem cells and tissue-specific stem cells has made significant headway over the last two decades, raising increasing expectations over their use in regenerative medicine in the near future. As far as the CNS is concerned, it is hoped that NSPCs derived from the embryonic or the adult CNS and/or by directed differentiation of pluripotent stem cells in vitro may be employed to repair irreversible damage to the neural tissue caused by injury or disease [52,53]. In either case, a key question that needs to be addressed is whether neural progenitors undergoing regional specification due to endogenous patterning cues in vivo or to exogenous inductive signals in vitro are able to stably retain their positional identities when faced with a different extracellular environment. Since it is well established that neural patterning critically depends on extracellular morphogens [1,54], it is crucial to understand how modifications of morphogen signalling impinge on NSPC positional identities and to shed light on the intrinsic mechanisms acting in NSPCs to interpret extrinsic signals at different developmental stages and CNS locations.
Previous studies have suggested that NSPCs derived from different areas of the mouse fetal CNS (E13.5–14.5) along the rostrocaudal axis maintain their positional identities in vitro, as shown by expression profiles of key transcription factors in NSPC cultures that remain consistent with gene expression patterns in vivo [42,43]. In this study, we have challenged the intrinsic ability of region-specific NSPCs to retain their positional identities by exposing them to exogenous RA, a morphogen playing a major role in neuroectoderm rostrocaudal patterning [9,10]. In vertebrate embryos exposed to RA during gastrulation, the anterior expression boundaries of Hox genes and other posterior neural markers are shifted anteriorly at the expense of rostrally expressed genes, and anterior development is suppressed [17,27]. In vitro models of early embryonic progenitors, such as ESCs or embryonic carcinoma cells, respond to exogenous RA applied in the context of neuroectoderm differentiation protocols by efficiently activating Hox gene expression [22–25]. Therefore, pluripotent progenitors and/or early neural progenitors arising from them are highly competent in retinoid-driven posteriorization.
By contrast, we demonstrate that NSPCs derived from the developing neural tube at E13.5, when rostrocaudal patterning has already taken place, display distinct, region-specific responses to retinoid signalling. In particular, NSPCs derived from the anterior neural tube were extremely resistant to the caudalizing activity of retinoids and retained rostral gene expression profiles even when exposed to high doses of exogenous RA. Mean levels of retinoid pathway activation were robustly increased by exogenous RA both in rostral and in caudal NSPCs, indicating that retinoid signalling is functional in both cell types, although we cannot exclude differences in the proportion of RA-responsive cells in rostral and caudal NSPC cultures. Thus, anterior positional identities are very labile during early neural patterning [17,27], but subsequently become intrinsically stable and long-lasting, as shown by continued lack of Hoxb gene response to high RA levels in rostral NSPCs derived from adult or aged mice. These results strengthen previous observations of stage-dependent reduction in the sensitivity of anterior development to exogenous retinoids in vertebrate embryos [17,28]. Notably, analysis of NSPCs derived from posterior regions of the developing neural tube showed that caudal NSPCs retained not only autonomous expression of Hoxb4–9 genes in vitro as previously described [42,43], but also prolonged competence to upregulate their transcription levels in response to RA. The effects of RA were dose-dependent, eliciting peak levels of Hoxb expression at the highest doses used for these assays (1 µM). Significant Hoxb4–9 upregulation, however, was already detectable at the lowest RA concentration tested (1 nM), which is below the estimated physiological range in vivo (2–100 nM [55,56]). This suggests that the continued ability of exogenous RA to promote Hoxb expression in caudal neural progenitors is not due to artificial effects of non-physiological RA levels, but reflects genuine responsiveness of Hoxb loci to retinoid signalling in NSPCs of the posterior CNS well beyond early rostrocaudal patterning. Previous reports have shown that, in developing mouse embryos, retinoid signalling is required until E11.5 to establish the correct anterior boundaries of Hoxb genes in the neural tube [23,26]. Our results indicate that, in caudal NSPCs, Hoxb4–9 genes remain responsive to RA up to at least E13.5. Though not absolutely necessary for Hoxb gene expression, this prolonged window of responsiveness to retinoids and possibly other caudalizing signals may be required to maintain proper Hox transcriptional levels at different rostrocaudal levels.
Over the last two decades, a large number of studies have focussed on Hox genes as an experimental paradigm to investigate the epigenetic regulation of gene expression during embryogenesis. Extensive efforts have been made in understanding how chromatin modifications of Hox clusters can contribute to stage- and position-dependent regulation of Hox gene expression and embryonic patterning. Breakthrough work employing mouse and human ESC cultures has suggested that, in these pluripotent cells, Hox gene chromatin features opposite epigenetic marks. Whereas deposition of the repressive mark H3K27me3 was found throughout Hox clusters, focal deposition of the activating marks H3K4me3 and H3K9/14ac was observed at the level of Hox gene transcription start sites [23,24,57]. Whether this bivalent epigenetic configuration is also a defining feature of Hox loci in pluripotent cells in vivo remains currently unclear [58]. Remarkably, however, RA treatments during ESC differentiation towards the neural lineage were able to dramatically reshape the distribution of these epigenetic marks, causing H3K27me3 erasure across Hox clusters and increased levels and spread of H3K4me3 and H3K9/14ac deposition at Hox regulatory regions [23–25]. Supporting the importance of these epigenetic modifications in Hox gene regulation, landmark studies showed that, in forebrain explants dissected from E10.5 mouse embryos, Hox clusters displayed increased levels of H3K27me3 deposition in comparison to ESCs and were devoid of H3K4me3 marks [57,59]. By contrast, in tissue samples dissected from caudal embryonic regions between E8.5 and E10.5, sequential activation of Hox genes from the 3' end to the 5' end of the cluster was accompanied by progressive erasure of H3K27me3 and deposition of H3K4me3 and H3K9/14ac along the same direction [57,59,60]. Altogether, these observations provide compelling evidence that the switch from H3K27me3-enriched chromatin to H3K4me3 and H3K9/14ac enrichment is a crucial step in the transcriptional activation of Hox genes. They also suggest that these opposite epigenetic signatures may be subsequently employed to maintain stable repression or activation of Hox clusters in anterior and posterior embryonic regions, with the bivalent configuration found in ESCs possibly representing an intermediate, initial state in which Hox genes are silent but poised for activation.
Our analysis of the epigenetic profiles of Hoxb genes in rostral and caudal NSPCs supports the idea that H3K27me3, H3K4me3 and H3K9/14ac may continue to play an important role during the maintenance phase of Hox gene neural expression patterns. As summarized in figure 6, we found that genomic sequences flanking Hoxb4–6 genes, including RAREs controlling their expression in the neural tube, were clearly enriched for H3K27me3 marks in rostral NSPC cultures in comparison with caudal NSPCs, whereas H3K4me3 and H3K9/14ac marks showed opposite profiles. While lacking the power of high-throughput approaches, our results usefully complement previous data obtained with dissected explants from head and trunk regions of developing mice [57,59,61,62]. By taking advantage of culture conditions allowing symmetric NSPC self-renewal in vitro [40], our work provides comparison of nearly homogeneous populations of NSPCs, rather than tissue fragments where neural progenitors coexist with differentiated cells and non-neural cell types. Moreover, our analysis covers later developmental stages than previously described and extends up to adulthood and ageing. We show that region-specific epigenetic profiles of Hoxb genes in NSPCs are very stable, since they were maintained both during embryonic development and adult life in vivo and after long-term passaging in vitro. Finally, our work investigates for the first time the effects of retinoid signalling on the epigenetic signature of Hox genes in region-specific NSPCs, a task previously carried out only in differentiating pluripotent cells. In contrast with the dramatic remodelling of Hox chromatin elicited in ESCs [23–25], RA treatments were utterly unable to modify the levels of H3K27me3, H3K4me3 and H3K9/14ac in rostral NSPCs from E13.5 embryos. Various mechanisms have been proposed to explain temporal and/or positional differences in the competence of progenitor cells to respond to morphogen signalling during neural development [63,64]. We speculate that resistance of rostral neural progenitors to retinoid-driven posteriorization may be linked to disrupted interactions between retinoid signalling and the epigenetic machinery at the level of Hox chromatin. RA treatments, however, could enhance deposition of H3K4me3 and, to a lesser extent, H4K9/14ac in caudal NSPCs, suggesting prolonged cross-talk between retinoid signalling and epigenetic regulators of Hox chromatin during posterior neural development.
A crucial area of investigation that remains relatively unexplored is related to the respective roles played by different histone modifications and chromatin-modifying complexes in controlling the responsiveness of Hox loci to retinoid signalling during neural development. Some initial progress in this direction has been made by analysing the transcriptional response of Hox genes to exogenous RA in ESCs or mouse embryos lacking specific epigenetic activities. For example, abrogation of either the H3K27-demethylase UTX or the H3K9-acetyltransferase MOZ hampered RA-driven activation of Hox gene expression in differentiating ESCs [55,56,65]. Conversely, loss of components of Polycomb repressive complex 1 (PRC1), a transcriptional repressor complex that may be recruited both via H3K27me3 deposition and independently of it [66], caused premature Hox gene response to RA treatments in differentiating ESC cultures and in developing embryos [65,67]. Clearly, much more work is needed in order to understand how the interactions between extracellular signalling pathways and the intracellular epigenetic machinery are modified during development to allow stage- and position-dependent regulation of patterning genes. In recent years, pluripotent stem cell culture systems have allowed steady progress in elucidating the molecular events underlying the early steps of neural patterning [54]. We propose that NSPC in vitro systems, due to their robust, simple, highly homogeneous culture conditions [40] and, as shown in this study, their stable epigenetic memory of rostrocaudal positional identities, can represent an excellent experimental paradigm for dissecting the molecular mechanisms controlling maintenance of gene expression programmes and region-specific cell fates.
Supplementary Material
Acknowledgements
We are grateful to Chiara Soldati for advice in setting up ChIP protocols, to M. Egle De Stefano and Rita Canipari for help with NSPC derivation, and to Bruce Blumberg for the gift of tk-(βRARE)2-luc plasmid. We thank Gabriella Augusti-Tocco, Federico Cremisi and Chiara Mozzetta for their critical reading of this manuscript.
Ethics
Experimental procedures for NSPC derivation from mouse tissues were performed in accordance with EU Directive 2010/63/EU and were approved by the Ethical Committee for Animal Research of the Italian Ministry of Health according to national regulations (Art. 7, D.Lgs. n. 116/1992). This study did not involve human subjects or the use of human samples.
Data accessibility
Datasets used for figure preparation are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dyrad.d22bq [68].
Authors' contributions
N.C. performed experiments and data analysis. E.C. performed experiments and provided NSPC culture protocols. P.S.N. performed experiments. V.L. analysed data and provided ChIP protocols. Y.-L.P. performed experiments and data analysis. S.B., R.N. and P.J.R.-G. contributed to experimental design and data analysis and revised the manuscript. G.L. conceived the study, analysed data and wrote the manuscript. All authors gave their final approval for publication.
Competing interests
We have no competing interests.
Funding
This work was supported by grants from the Italian Ministry of Education, University and Research program ‘Rientro dei Cervelli’, from Istituto Pasteur–Fondazione Cenci Bolognetti and from Sapienza University of Rome (G.L.) and by funding from the Wellcome Trust (WT093736), and Biotechnology and the Biological Sciences Research Council (P.J.R.-G.).
References
- 1.Altmann CR, Brivanlou AH. 2001. Neural patterning in the vertebrate embryo. Int. Rev. Cytol. 203, 447–482. (doi:10.1016/S0074-7696(01)03013-3) [DOI] [PubMed] [Google Scholar]
- 2.Lupo G, Harris WA, Lewis KE. 2006. Mechanisms of ventral patterning in the vertebrate nervous system. Nat. Rev. Neurosci 7, 103–114. (doi:10.1038/nrn1843) [DOI] [PubMed] [Google Scholar]
- 3.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. 1992. Nested expression domains of four homeobox genes in developing rostral brain. Nature 358, 687–690. (doi:10.1038/358687a0) [DOI] [PubMed] [Google Scholar]
- 4.Di Bonito M, Glover JC, Studer M. 2013. Hox genes and region-specific sensorimotor circuit formation in the hindbrain and spinal cord. Dev. Dyn. 242, 1348–1368. (doi:10.1002/dvdy.24055) [DOI] [PubMed] [Google Scholar]
- 5.Tümpel S, Wiedemann LM, Krumlauf R. 2009. Hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 88, 103–137. (doi:10.1016/S0070-2153(09)88004-6) [DOI] [PubMed] [Google Scholar]
- 6.Beccari L, Marco-Ferreres R, Bovolenta P. 2013. The logic of gene regulatory networks in early vertebrate forebrain patterning. Mech. Dev. 130, 95–111. (doi:10.1016/j.mod.2012.10.004) [DOI] [PubMed] [Google Scholar]
- 7.Carron C, Shi D-L. 2016. Specification of anteroposterior axis by combinatorial signaling during Xenopus development. Wiley Interdiscip. Rev. Dev. Biol. 5, 150–168. (doi:10.1002/wdev.217) [DOI] [PubMed] [Google Scholar]
- 8.Pera EM, Acosta H, Gouignard N, Climent M, Arregi I. 2014. Active signals, gradient formation and regional specificity in neural induction. Exp. Cell Res. 321, 25–31. (doi:10.1016/j.yexcr.2013.11.018) [DOI] [PubMed] [Google Scholar]
- 9.Maden M. 2002. Retinoid signalling in the development of the central nervous system. Nat. Rev. Neurosci. 3, 843–853. (doi:10.1038/nrn963) [DOI] [PubMed] [Google Scholar]
- 10.Maden M. 2007. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 8, 755–765. (doi:10.1038/nrn2212) [DOI] [PubMed] [Google Scholar]
- 11.Cunningham TJ, Duester G. 2015. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 16, 110–123. (doi:10.1038/nrm3932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumberg B, Bolado J, Moreno TA, Kintner C, Evans RM, Papalopulu N. 1997. An essential role for retinoid signaling in anteroposterior neural patterning. Development 124, 373–379. [DOI] [PubMed] [Google Scholar]
- 13.Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P. 2002. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development 129, 2851–2865. [DOI] [PubMed] [Google Scholar]
- 14.Maden M, Gale E, Kostetskii I, Zile M. 1996. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr. Biol. 6, 417–426. (doi:10.1016/S0960-9822(02)00509-2) [DOI] [PubMed] [Google Scholar]
- 15.Niederreither K, Subbarayan V, Dollé P, Chambon P. 1999. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 21, 444–448. (doi:10.1038/7788) [DOI] [PubMed] [Google Scholar]
- 16.Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, Heideveld M, Nieuwkoop PD. 1989. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature 340, 140–144. (doi:10.1038/340140a0) [DOI] [PubMed] [Google Scholar]
- 17.Simeone A, Avantaggiato V, Moroni MC, Mavilio F, Arra C, Cotelli F, Nigro V, Acampora D. 1995. Retinoic acid induces stage-specific antero-posterior transformation of rostral central nervous system. Mech. Dev. 51, 83–98. (doi:10.1016/0925-4773(95)96241-M) [DOI] [PubMed] [Google Scholar]
- 18.Sive HL, Draper BW, Harland RM, Weintraub H. 1990. Identification of a retinoic acid-sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 4, 932–942. (doi:10.1101/gad.4.6.932) [DOI] [PubMed] [Google Scholar]
- 19.Mainguy G, In der Rieden PMJ, Berezikov E, Woltering JM, Plasterk RHA, Durston AJ. 2003. A position-dependent organisation of retinoid response elements is conserved in the vertebrate Hox clusters. Trends Genet. 19, 476–479. (doi:10.1016/S0168-9525(03)00202-6) [DOI] [PubMed] [Google Scholar]
- 20.Ahn Y, Mullan HE, Krumlauf R. 2014. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev. Biol. 388, 134–144. (doi:10.1016/j.ydbio.2014.01.027) [DOI] [PubMed] [Google Scholar]
- 21.Papalopulu N, Lovell-Badge R, Krumlauf R. 1991. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a collinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucleic Acids Res. 19, 5497–5506. (doi:10.1093/nar/19.20.5497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simeone A, Acampora D, Arcioni L, Andrews PW, Boncinelli E, Mavilio F. 1990. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature 346, 763–766. (doi:10.1038/346763a0) [DOI] [PubMed] [Google Scholar]
- 23.De Kumar B, et al. 2015. Analysis of dynamic changes in retinoid-induced transcription and epigenetic profiles of murine Hox clusters in ES cells. Genome Res. 25, 1229–1243. (doi:10.1101/gr.184978.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashyap V, Gudas LJ, Brenet F, Funk P, Viale A, Scandura JM. 2011. Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J. Biol. Chem. 286, 3250–3260. (doi:10.1074/jbc.M110.157545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzoni EO, et al. 2013. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 16, 1191–1198. (doi:10.1038/nn.3490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oosterveen T, Niederreither K, Dollé P, Chambon P, Meijlink F, Deschamps J. 2003. Retinoids regulate the anterior expression boundaries of 5′ Hoxb genes in posterior hindbrain. EMBO J. 22, 262–269. (doi:10.1093/emboj/cdg029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudoh T, Wilson SW, Dawid IB. 2002. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development 129, 4335–4346. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham ML, Mac Auley A, Mirkes PE. 1994. From gastrulation to neurulation: transition in retinoic acid sensitivity identifies distinct stages of neural patterning in the rat. Dev. Dyn. 200, 227–241. (doi:10.1002/aja.1002000305) [DOI] [PubMed] [Google Scholar]
- 29.Lupo G, Liu Y, Qiu R, Chandraratna RAS, Barsacchi G, He R-Q, Harris WA. 2005. Dorsoventral patterning of the Xenopus eye: a collaboration of Retinoid, Hedgehog and FGF receptor signaling. Development 132, 1737–1748. (doi:10.1242/dev.01726) [DOI] [PubMed] [Google Scholar]
- 30.Lupo G, Gestri G, O'Brien M, Denton RM, Chandraratna RAS, Ley SV, Harris WA, Wilson SW. 2011. Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc. Natl Acad. Sci. USA 108, 8698–8703. (doi:10.1073/pnas.1103802108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matt N, Dupé V, Garnier J-M, Dennefeld C, Chambon P, Mark M, Ghyselinck NB. 2005. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development 132, 4789–4800. (doi:10.1242/dev.02031) [DOI] [PubMed] [Google Scholar]
- 32.Molotkov A, Molotkova N, Duester G. 2006. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development 133, 1901–1910. (doi:10.1242/dev.02328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatzi C, Brade T, Duester G. 2011. Retinoic acid functions as a key GABAergic differentiation signal in the basal ganglia. PLoS Biol. 9, e1000609 (doi:10.1371/journal.pbio.1000609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi J, Park S, Sockanathan S. 2014. Activated retinoid receptors are required for the migration and fate maintenance of subsets of cortical neurons. Development 141, 1151–1160. (doi:10.1242/dev.104505) [DOI] [PubMed] [Google Scholar]
- 35.Crandall JE, Goodman T, McCarthy DM, Duester G, Bhide PG, Dräger UC, McCaffery P. 2011. Retinoic acid influences neuronal migration from the ganglionic eminence to the cerebral cortex. J. Neurochem. 119, 723–735. (doi:10.1111/j.1471-4159.2011.07471.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajaii F, Bitzer ZT, Xu Q, Sockanathan S. 2008. Expression of the dominant negative retinoid receptor, RAR403, alters telencephalic progenitor proliferation, survival, and cell fate specification. Dev. Biol. 316, 371–382. (doi:10.1016/j.ydbio.2008.01.041) [DOI] [PubMed] [Google Scholar]
- 37.Siegenthaler JA, et al. 2009. Retinoic acid from the meninges regulates cortical neuron generation. Cell 139, 597–609. (doi:10.1016/j.cell.2009.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs S, Lie DC, DeCicco KL, Shi Y, DeLuca LM, Gage FH, Evans RM. 2006. Retinoic acid is required early during adult neurogenesis in the dentate gyrus. Proc. Natl Acad. Sci. USA 103, 3902–3907. (doi:10.1073/pnas.0511294103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T-W, Zhang H, Parent JM. 2005. Retinoic acid regulates postnatal neurogenesis in the murine subventricular zone-olfactory bulb pathway. Development 132, 2721–2732. (doi:10.1242/dev.01867) [DOI] [PubMed] [Google Scholar]
- 40.Conti L, et al. 2005. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283 (doi:10.1371/journal.pbio.0030283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard SM, Conti L, Sun Y, Goffredo D, Smith A. 2006. Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb. Cortex 1, i112–i120. (doi:10.1093/cercor/bhj167) [DOI] [PubMed] [Google Scholar]
- 42.Onorati M, et al. 2011. Preservation of positional identity in fetus-derived neural stem (NS) cells from different mouse central nervous system compartments. Cell. Mol. Life Sci. 68, 1769–1783. (doi:10.1007/s00018-010-0548-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soldati C, Cacci E, Biagioni S, Carucci N, Lupo G, Perrone-Capano C, Saggio I, Augusti-Tocco G. 2012. Restriction of neural precursor ability to respond to Nurr1 by early regional specification. PLoS ONE 7, e51798.. (doi:10.1371/journal.pone.0051798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soldati C, et al. 2015. RE1 silencing transcription factor/neuron-restrictive silencing factor regulates expansion of adult mouse subventricular zone-derived neural stem/progenitor cells in vitro. J. Neurosci. Res. 93, 1203–1214. (doi:10.1002/jnr.23572) [DOI] [PubMed] [Google Scholar]
- 45.Ulbrich L, Favaloro FL, Trobiani L, Marchetti V, Patel V, Pascucci T, Comoletti D, Marciniak SJ, De Jaco A. 2016. Autism-associated R451C mutation in neuroligin3 leads to activation of the unfolded protein response in a PC12 Tet-On inducible system. Biochem. J. 473, 423–434. (doi:10.1042/BJ20150274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warton K, Foster NC, Gold WA, Stanley KK. 2004. A novel gene family induced by acute inflammation in endothelial cells. Gene 342, 85–95. (doi:10.1016/j.gene.2004.07.027) [DOI] [PubMed] [Google Scholar]
- 47.Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. 2010. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev. Biol. 338, 1–14. (doi:10.1016/j.ydbio.2009.10.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashyap V, Gudas LJ. 2010. Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts. J. Biol. Chem. 285, 14 534–14 548. (doi:10.1074/jbc.M110.115345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rydeen A, Voisin N, D'Aniello E, Ravisankar P, Devignes C-S, Waxman JS. 2015. Excessive feedback of Cyp26a1 promotes cell non-autonomous loss of retinoic acid signaling. Dev. Biol. 405, 47–55. (doi:10.1016/j.ydbio.2015.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serpente P, Tümpel S, Ghyselinck NB, Niederreither K, Wiedemann LM, Dollé P, Chambon P, Krumlauf R, Gould AP. 2005. Direct crossregulation between retinoic acid receptor β and Hox genes during hindbrain segmentation. Development 132, 503–513. (doi:10.1242/dev.01593) [DOI] [PubMed] [Google Scholar]
- 51.Fiorelli R, Azim K, Fischer B, Raineteau O. 2015. Adding a spatial dimension to postnatal ventricular-subventricular zone neurogenesis. Development 142, 2109–2120. (doi:10.1242/dev.119966) [DOI] [PubMed] [Google Scholar]
- 52.Casarosa S, Bozzi Y, Conti L. 2014. Neural stem cells: ready for therapeutic applications? Mol. Cell Ther. 2, 31 (doi:10.1186/2052-8426-2-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross CA, Akimov SS. 2014. Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Hum. Mol. Genet. 23, R17–R26. (doi:10.1093/hmg/ddu204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lupo G, Bertacchi M, Carucci N, Augusti-Tocco G, Biagioni S, Cremisi F. 2014. From pluripotency to forebrain patterning: an in vitro journey astride embryonic stem cells. Cell. Mol. Life Sci. 71, 2917–2930. (doi:10.1007/s00018-014-1596-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhar SS, et al. 2016. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 44, 3659–3674. (doi:10.1093/nar/gkv1516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheikh BN, Downer NL, Kueh AJ, Thomas T, Voss AK. 2014. Excessive versus physiologically relevant levels of retinoic acid in embryonic stem cell differentiation. Stem Cells 32, 1451–1458. (doi:10.1002/stem.1604) [DOI] [PubMed] [Google Scholar]
- 57.Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. 2014. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife 3, e02557 (doi:10.7554/eLife.02557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, et al. 2016. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562. (doi:10.1038/nature19362) [DOI] [PubMed] [Google Scholar]
- 59.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334, 222–225. (doi:10.1126/science.1207194) [DOI] [PubMed] [Google Scholar]
- 60.Soshnikova N, Duboule D. 2009. Epigenetic temporal control of mouse Hox genes in vivo. Science 324, 1320–1323. (doi:10.1126/science.1171468) [DOI] [PubMed] [Google Scholar]
- 61.Min H, Lee J-Y, Kim MH. 2012. Structural dynamics and epigenetic modifications of Hoxc loci along the anteroposterior body axis in developing mouse embryos. Int. J. Biol. Sci. 8, 802–810. (doi:10.7150/ijbs.4438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min H, Lee J-Y, Kim MH. 2013. Hoxc gene collinear expression and epigenetic modifications established during embryogenesis are maintained until after birth. Int. J. Biol. Sci. 9, 960–965. (doi:10.7150/ijbs.6739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sasai N, Kutejova E, Briscoe J. 2014. Integration of signals along orthogonal axes of the vertebrate neural tube controls progenitor competence and increases cell diversity. PLoS Biol. 12, e1001907 (doi:10.1371/journal.pbio.1001907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Lupo G, He R, Barsacchi G, Harris WA, Liu Y. 2015. Dorsoventral patterning of the Xenopus eye involves differential temporal changes in the response of optic stalk and retinal progenitors to Hh signalling. Neural Dev. 10, 7 (doi:10.1186/s13064-015-0035-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheikh BN, Downer NL, Phipson B, Vanyai HK, Kueh AJ, McCarthy DJ, Smyth GK, Thomas T, Voss AK. 2015. MOZ and BMI1 play opposing roles during Hox gene activation in ES cells and in body segment identity specification in vivo. Proc. Natl Acad. Sci. USA 112, 5437–5442. (doi:10.1073/pnas.1422872112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tavares L, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678. (doi:10.1016/j.cell.2011.12.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bel-Vialar S, Coré N, Terranova R, Goudot V, Boned A, Djabali M. 2000. Altered retinoic acid sensitivity and temporal expression of Hox genes in polycomb-M33-deficient mice. Dev. Biol. 224, 238–249. (doi:10.1006/dbio.2000.9791) [DOI] [PubMed] [Google Scholar]
- 68.Carucci N, Cacci E, Nisi PS, Licursi V, Paul Y-L, Biagioni S, Negri R, Rugg-Gunn PJ, Lupo G. 2017. Data from: Transcriptional response of Hoxb genes to retinoid signalling is regionally restricted along the neural tube rostrocaudal axis. Dryad Digital Repository. (http://dx.doi.org/10.5061/dyrad.d22bq) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Carucci N, Cacci E, Nisi PS, Licursi V, Paul Y-L, Biagioni S, Negri R, Rugg-Gunn PJ, Lupo G. 2017. Data from: Transcriptional response of Hoxb genes to retinoid signalling is regionally restricted along the neural tube rostrocaudal axis. Dryad Digital Repository. (http://dx.doi.org/10.5061/dyrad.d22bq) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Datasets used for figure preparation are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dyrad.d22bq [68].