Abstract
Tumour antigens have attracted much attention because of their importance to cancer diagnosis, prognosis and targeted therapy. With the development of cancer genomics, the identification of tumour-specific neoantigens became possible, which is a crucial step for cancer immunotherapy. In this study, we developed software called the tumour-specific neoantigen detector for detecting cancer somatic mutations following the best practices of the genome analysis toolkit and predicting potential tumour-specific neoantigens, which could be either extracellular mutations of membrane proteins or mutated peptides presented by class I major histocompatibility complex molecules. This pipeline was beneficial to the biologist with little programmatic background. We also applied the software to the somatic mutations from the International Cancer Genome Consortium database to predict numerous potential tumour-specific neoantigens. This software is freely available from https://github.com/jiujiezz/tsnad.
Keywords: cancer somatic mutation, tumour antigen, neoantigen, major histocompatibility complex, membrane protein
1. Introduction
Tumour antigens have attracted much attention for their importance in cancer diagnosis, prognosis and targeted therapy, as they are crucial tumour biomarkers for identifying tumour cells and are potential targets for cancer therapy [1–3]. Tumour antigens can be broadly classified into two categories based on their specificity: tumour-specific antigens, which are only present in tumour cells; and tumour-associated antigens, which are overexpressed or aberrantly expressed in tumour cells and are also expressed in some normal cells [1]. In addition to abnormal expression patterns, tumour cells also contain a range of cancer somatic mutations and mutations in protein-coding regions might produce tumour-specific mutant proteins [4,5]. Tumour antigens derived from these tumour-specific mutant proteins are unparalleled tumour biomarkers, as they are only produced by tumour cells and are potential tumour-specific mutant antigens or neoantigens [3].
Tumour antigens recognized by T cells or antibodies should present on the surface of tumour cells [6,7]. A major part of tumour antigens used as drug targets are membrane proteins, such as HER2 and CD19, which are targets of the antibody trastuzumab [8] and chimaeric antigen receptor T-cell immunotherapy (CAR-T) for B-cell cancer [9,10], respectively. Additionally, tumour antigens presented by class I major histocompatibility complex (MHC) molecules for recognition by T cells (i.e. tumour-specific neoantigens) could also be used as drug targets [2,11,12]. On the other hand, in the immune checkpoint blockade therapy, the neoantigen load is associated with the therapy efficacy (i.e. PD-1, CTLA-4 blockade), which indicates that the neoantigen load is a great biomarker in cancer immunotherapy [13]. Because of their potential application to be targets and biomarkers in cancer immunotherapy [1,12,14,15], tumour-specific neoantigens have attracted much attention in biomedical research. Several prediction tools have been developed to predict tumour-specific neoantigens from cancer somatic mutations, such as pVAC-seq [16] and INTEGRATE-neo [17], which can predict neoantigens produced by non-synonymous somatic mutations and gene fusions, respectively. However, these tools only predict neoantigens presented by class I MHC molecules that can be recognized by T cells, they do not consider the mutations in the extracellular regions of membrane proteins that can be recognized by mutation-specific antibodies [18,19].
In this study, we developed integrated software with a graphical user interface (GUI), called the tumour-specific neoantigen detector (TSNAD), which can identify cancer somatic mutations following the best practices of the genome analysis toolkit (GATK v. 3.5) [20] from the genome/exome sequencing data of tumour-normal pairs. We also provided a filter for calling tumour-specific mutant proteins. Then, we conducted two strategies to predict neoantigens. First, we extracted the extracellular mutations of membrane proteins according to the protein topology. Second, we invoked NetMHCpan (v. 2.8) [21] to predict the binding information of mutant peptides to class I MHC molecules. Finally, we applied TSNAD on the cancer somatic mutations collected in the International Cancer Genome Consortium (ICGC) database to predict potential neoantigens.
2. Material and methods
2.1. Tools
Standard sequencing data processing consists of preprocessing, alignment, variants calling, annotation and further analysis. Given that the existing software or tools are designed for specific functions, it was necessary to develop an automated and user-friendly framework that calls a series of software. This section summarizes the required software and its main features.
2.1.1. Data filtering software
Trimmomatic (v. 0.35) [22]. Original raw sequences have random lengths and contain adaptors that will be harmful to the subsequent data processing. This software can trim and crop raw reads and remove artefacts.
2.1.2. Genome mapping software
Burrows-Wheeler Aligner (BWA, v. 0.7.12) [23,24]. This alignment toolkit is used for mapping short sequences to a reference genome. This software is based on the Burrows-Wheeler transformation and is highly efficient at finding locations of low-divergent sequences on a large genome.
2.1.3. Alignment manipulating tool
Samtools (v. 1.3) [25]. Its view and sort functions transform sequencing data format from SAM (sequence alignment/map) to BAM (binary alignment/map), which will save an enormous amount of storage space. Moreover, it can manage duplicate reads and index alignments.
2.1.4. Data processing tool
Picard tools (v. 1.140) [26]. This program consists of a set of Java command lines to handle with different sequencing data format (such as SAM, BAM and VCF). Given redundancy data may influence further processing, Picard MarkDuplicates tool can thus be applied to remove repeat sequences.
2.1.5. Variant calling software
Genome Analysis Toolkit (GATK v. 3.5) [20], Mutect2 [27]. The main function of GATK is variant discovery in high-throughput sequencing data. Mutect2 is a package in GATK to identify somatic SNVs and INDELs.
2.1.6. Mutation annotation software
Annovar (14 December 2015) [28,29]. We use it to functional annotate somatic mutations, including position, change of nucleotide, change of amino acid for protein-coding region, and other functions. We can then extract tumour-specific mutant proteins.
2.1.7. Human leucocyte antigen typing software
SOAP-HLA (v. 2.2) [30]. This software detects the human leucocyte antigen (HLA—the MHC in humans) types for each sample. The program takes sorted aligned sequencing data (BAM format) as the input and outputs HLA types. The HLA types are critical for the MHC-binding predictions.
2.1.8. Protein topology indicating software
TMHMM (v. 2.0) [31]. This tool is used to predict the topology of membrane proteins based on a hidden Markov model (HMM). The prediction of transmembrane helices and membrane proteins is highly accurate [32].
2.1.9. Major histocompatibility complex-binding predicting software
NetMHCpan (v. 2.8) [21]. This software can forecast peptides that can bind to MHC class I molecules using artificial neural networks.
2.2. Datasets
The somatic mutations were collected from the whole-genome/exome sequencing data of 9155 tumour-normal pairs in the ICGC database (Release 20, http://icgc.org). This dataset has compiled over 1.5 million sample somatic mutations in coding regions, among which 828 129 missense variants have caused amino acid changes with a frequency range from 1 to 476 out of 9155 tumour samples.
The HLA types were extracted from the 1000 Genome Project. We choose 16 HLA alleles with frequencies of more than 5% in the population collected in the 1000 Genome Project [33], which includes five HLA-A (HLA-A*01:01, HLA-A*02:01, HLA-A*03:01, HLA-A*11:01 and HLA-A*24:02), four HLA-B (HLA-B*07:02, HLA-B*35:01, HLA-B*40:01 and HLA-B*51:01) and seven HLA-C (HLA-C*01:02, HLA-C*03:03, HLA-C*03:04, HLA-C*04:01, HLA-C*06:02, HLA-C*07:01 and HLA-C*07:02) alleles.
2.3. Identification of extracellular region of membrane proteins
The list of human membrane proteins was extracted from the human protein atlas [34]. The amino acid sequences of these membrane proteins were downloaded from Ensembl (GRCh37 v. 75) [35]. TMHMM (v. 2.0) was used to identify the transmembrane topology and extracellular region of each membrane protein [31].
2.4. Prediction of class I major histocompatibility complex binding
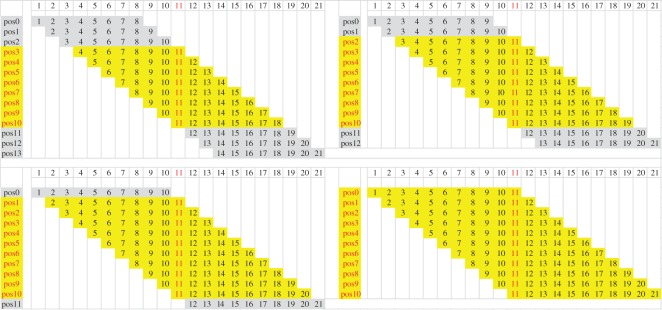
After we obtained the list of the tumour-specific mutant proteins, we extracted the peptide sequences around the mutation sites. As MHC molecules always bind to peptides 8–11 amino acids in length, we extracted peptides 21 amino acids in length, with 10 amino acids upstream and 10 amino acids downstream of mutation sites for NetMHCpan prediction (figure 1). Wild-type peptides with the same length as the mutant peptides were extracted as references. These wild-type and mutant peptides were measured for their binding affinities (50% inhibitory concentration [IC50], nM) to each class I HLA allele. The binding was considered strong if the IC50 value was less than 150 nM, and a weak binding had an IC50 value between 150 and 500 nM. Non-binding occurred if the IC50 value was more than 500 nM [11].
Figure 1.
Mutant peptides with 21 amino acids and corresponding 8–11 mer peptides. MHC molecules always bind to 8–11 mer peptides, so we extracted peptides 21 amino acids in length, with 10 amino acids upstream and 10 amino acids downstream of mutation sites for NetMHCpan prediction. The number 11 in red indicates the mutated site, and the peptides in yellow represent all the possible peptides which may bind to MHC molecules.
2.5. Experimental validation of peptide biding to class I major histocompatibility complex molecular
Peptides were obtained lyophilized (more than 95% purity) from Bankpeptide Biological Technology Co., Ltd (Hefei, China), dissolved in 10% DMSO in sterile water and tested for sterility, purity, endotoxin and residual organics. Peptide binding to HLA-A*02:01 was determined by T2 assay [36]. T2 cells were washed in phosphate buffered saline (PBS) and RPMI-1640 without serum. In total, 5 × 105 cell ml–1 were incubated with 5 µg ml−1 peptide and 10 µg ml−1 human beta-2-microglobulin in serum-free RPMI-1640 for 4 h or overnight at 37°C. The pulsed cells were pelleted and followed by 3 × 1 ml rinses in PBS with centrifugation at 500g for 5 min at 4°C. Cells were resuspended in 200 µl PBS and stained with 1 µl of w6/32 (Thermo Fisher) for 30 min on ice, followed by three rinses with 1 ml PBS at 4°C. Cells were then resuspended in 200 µl PBS and 1 µl of goat anti-mouse antibody-FITC (Beyotime Biotechnology) for 30 min on ice, followed by three rinses at 4°C. Then, cells were resuspended in 500 µl PBS. Stained T2 cells were analysed using a FACSCalibur.
3. Results
3.1. Software overview
We developed integrated software, called TSNAD, under the Linux operation system through a GUI. The platform is completely automated and is mainly designed for users who have little programming experience. There are several neoantigen prediction pipelines such as pVAC-seq, INTEGRATE-neo: pVAC-seq combined the tumour mutation and expression data to predict neoantigens by invoking NetMHC v. 3.4; INTEGRATE-neo was designed to predict neoantigens from fusion genes based on the pipeline INTERGRATE and NetMHC v. 4.0. Similar with these pipelines, TSNAD also used widely approved software NetMHCpan v. 2.8 to predict neoantigens. Compared with other neoantigen prediction pipelines, TSNAD has lists of advantages: first, TSNAD offered a pipeline for mutation calling from sequencing data; second, TSNAD not only considered the neoantigens presented by class I MHC molecules, but also took mutations in membrane proteins into consideration; third, unlike other pipelines that performed through command lines, TSNAD provided a GUI for biologists without programming background to analyse their data easily. The software consists of two toolkits: mutation detection and neoantigen prediction. Each toolkit is a two-step process as follows: configure the parameters and run the corresponding toolkit.
The first step is to configure the software paths and parameters. This step is of great significance, and users are expected to ensure the appropriateness and correctness of the configurations. Users can find the detailed instructions about how to set paths and parameters in the user's manual. For the software paths, the users do not need to change these parameters once they are set because TSNAD will import the existing configuration files by default. Users can also edit partial parameters by GUI or by manually modifying the configuration files. It is worth noting that TSNAD requires its own naming convention for the input files. The users can choose to either manually or use the tool we provided to rename the names of sequencing files to suit the criteria of TSNAD.
After setting the configurations, non-expert users can run the pipeline by just clicking on the appropriate toolbar. In the processing monitoring window, the users can observe the pipeline progression. The pipeline, which was written in Python programming language (v2.7), calls for standard third-party software and applies multiprocessing strategy to speed up the data processing.
When the pipeline is finished, all of the results will be stored in a user-specified folder. The mutation detection pipeline returns the list of somatic mutations with annotations. The neoantigen prediction pipeline returns extracellular mutations of the membrane proteins and the MHC-binding information (all in TXT format).
3.2. Detection of cancer somatic mutations
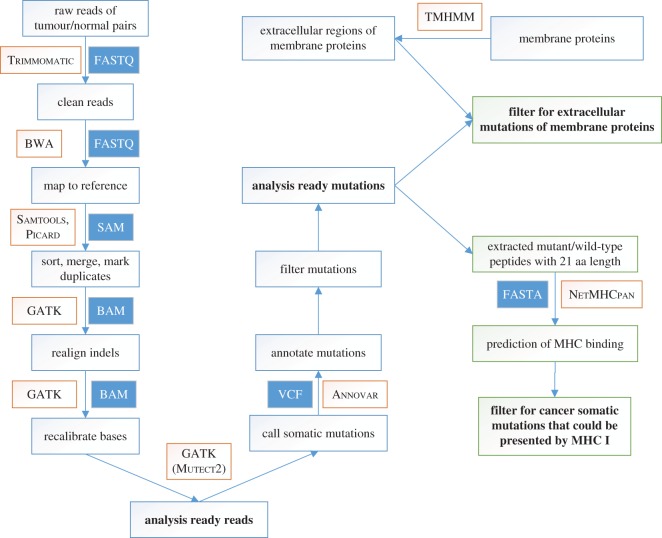
The software can detect single-nucleotide variants (SNVs) and small insertions or deletions (INDELs) according to the pipeline as depicted in figure 2. The raw paired-end sequence data were in FastQ format from the whole-genome sequencing, the whole-exome sequencing or the targeted gene panel sequencing using the Illumina platform. The raw data were cleaned using Trimmomatic [22]. BWA-MEM was used to map the reads to the reference genome sequences [23,24]. Samtools [25] and Picard [26] were used to address files in SAM or BAM formats, including transform, sort, merge and mark duplicates. GATK [20] was used to pre-process the BAM files, such as realigning the INDELs and recalibrating the bases. Mutect2 [27] in GATK was used to call the somatic SNVs and INDELs between tumour and normal samples. Annovar [28,29] was used to annotate the detailed mutation information.
Figure 2.
The software pipeline of TSNAD. The pipeline performs best practices for somatic SNVs and INDELs in whole-genome/exome sequence with GATK. Then, we extracted the extracellular mutations of membrane proteins according to the protein topology, and invoked NetMHCpan to predict the binding information of mutant peptides to class I MHC molecules.
We further provide a filter to detect the somatic mutations in the protein-coding regions and the somatic missense variants which fit the cut-off (tumour reads > 10, normal reads > 6, tumour alteration reads > 5, variant allele frequency (VAF) in tumour DNA > 0.05 and VAF in normal DNA = 0).
3.3. Prediction of neoantigens
When peptides differ by only one amino acid change, specific antibodies can be generated [19,37]. Therefore, missense mutations that are present on the surfaces of tumour cells are important targets for antibody-based immunotherapy. We performed two strategies to predict the neoantigens that would present on the surfaces of tumour cells [1,2]. First, we extracted the somatic mutations in the extracellular regions of the membrane proteins. Second, we predicted the neoantigens that would present on the cell surface by evaluating the binding affinity between the peptides and class I MHC molecules.
According to the Human Protein Atlas, there were 5462 predicted membrane proteins [34]. We identified the transmembrane topologies and the extracellular regions of these proteins using TMHMM [31]. To identify the extracellular mutations of membrane proteins, the filtered cancer somatic missense variants were mapped to the extracellular regions of membrane proteins. We further verified the characteristics of the mutant amino acids. Mutations that change the polarity of the amino acids have gained more attention, as they may be more likely to cause differences in binding features to antibodies between wild-type and mutant proteins.
In addition to the membrane proteins, peptides could be present on the cell surface because of the antigen presenting system, which is mediated by MHC molecules. SOAP-HLA was used for the HLA typing of each sample [30]. NetMHCpan was used to predict the binding affinity between the class I MHC and wild-type/mutant peptides [21]. We further compared the binding information of the HLA molecules to the wild-type and mutant peptides. The mutant peptides that can bind to the HLA-A/B/C molecules were extracted for further analysis; the specific bindings of the HLA proteins to the mutant peptides were preferred for their potential to be drug targets without affecting normal tissues.
3.4. Prediction of neoantigens based on the somatic mutation data from the International Cancer Genome Consortium database
In previous study, we performed oncogene targeted depth sequencing on a malignant peritoneal mesothelioma [38]. Applying the TSNAD to analyse the sequence data of the tumour sample and the paired peripheral blood sample, we detected 2897 somatic SNVs and 218 somatic INDELs. Four SNVs of NOTCH2, PDE4DIP, ATP10B and NSD1 and one frameshift INDEL of BAP1 were validated by Sanger sequencing on tumour RNA. We also predicted the neoantigens on these mutated proteins, and found specific-binding of neo-peptide generated by BAP1 frameshift INDEL to HLA-B*35:42 of the patient. A polyclonal antibody of the neo-peptide of BAP1 were produced in rabbits and showed a good antibody-neoantigen specificity, which indicates that the neo-peptide of BAP1 could be a potential tumour-specific neoantigen [38].
In addition to handling original sequencing data, TSNAD could also analyse exiting mutations data to predict potential neoantigens. We applied TSNAD to the simple somatic mutations of 9155 samples from the ICGC database and predicted numerous neoantigens, including extracellular mutations of membrane proteins and peptides presented by the class I MHC molecules.
3.5. Prediction of neoantigens from membrane proteins
To identify the extracellular mutations of membrane proteins, we mapped all of the missense mutations to the extracellular regions of membrane proteins. A dataset containing 88 354 extracellular mutations was obtained. A majority of these extracellular mutations (89.6%, 79 198 out of 88 354) occurs only once in the 9155 donors (electronic supplementary material, table S1 and figure S1), which illustrates the high heterogeneity in tumour samples. However, membrane proteins with mutations that occur in more samples are also ideal drug targets for antibody-based immunotherapy. The top 20 frequent extracellular mutations are listed in table 1 and MUC4:H4205Q is the most frequent extracellular mutation (44 out of 9155).
Table 1.
Top 20 most frequent extracellular mutations in 9155 donors.
| Chr | Pos | ID | gene | DNA mutation | protein mutation | mutation frequency |
|---|---|---|---|---|---|---|
| 3 | 195505836 | MU10935 | MUC4 | 12615C>G | H4205Q | 44 out of 9155 |
| 1 | 29138975 | MU68226 | OPRD1 | 80G>T | C27F | 25 out of 9155 |
| 1 | 120611960 | MU869951 | NOTCH2 | 61G>A | A21T | 23 out of 9155 |
| 19 | 1065018 | MU68245 | ABCA7 | 6133G>T | A2045S | 18 out of 9155 |
| 15 | 22369378 | MU4380351 | OR4M2 | 803C>T | S268F | 16 out of 9155 |
| 17 | 37868208 | MU85975 | ERBB2 | 929C>T | S310F | 15 out of 9155 |
| 3 | 195509676 | MU586249 | MUC4 | 8775G>C | Q2925H | 15 out of 9155 |
| 7 | 55233043 | MU589341 | EGFR | 1793G>T | G598 V | 15 out of 9155 |
| 3 | 195515449 | MU605883 | MUC4 | 3002T>A | V1001E | 14 out of 9155 |
| 19 | 9072091 | MU4382243 | MUC16 | 15355C>T | P5119S | 13 out of 9155 |
| 20 | 17639816 | MU4585427 | RRBP1 | 1337A>C | Q446P | 12 out of 9155 |
| 5 | 179071958 | MU4110168 | C5orf60 | 64G>C | D22H | 12 out of 9155 |
| 7 | 146829338 | MU4413315 | CNTNAP2 | 1085G>A | G362E | 12 out of 9155 |
| 1 | 158261127 | MU4408485 | CD1C | 265C>T | R89C | 11 out of 9155 |
| 11 | 5345040 | MU4383907 | OR51B2 | 488C>T | S163 L | 11 out of 9155 |
| 17 | 21319519 | MU613603 | KCNJ12 | 865G>C | E289Q | 11 out of 9155 |
| 2 | 46707884 | MU70561 | TMEM247 | 458A>G | Q153R | 11 out of 9155 |
| 2 | 137814319 | MU4440003 | THSD7B | 469G>A | E157 K | 11 out of 9155 |
| 3 | 195511286 | MU4617526 | MUC4 | 7165G>A | D2389N | 11 out of 9155 |
| 7 | 139167934 | MU66261 | KLRG2 | 455A>C | K152T | 11 out of 9155 |
3.6. Prediction of neoantigens through major histocompatibility complex-binding information
Peptides could also present on the cell surface via the antigen presenting system, mediated by MHC class I molecules. In this manner, mutant peptides that are present exclusively in tumour cells are the potential neoantigens, and the MHC–peptide complexes are called neoantigens.
Based on the missense mutations of the 9155 tumour samples from the ICGC, we extracted peptides 21 amino acids length, with 10 amino acids upstream and 10 amino acids downstream of the mutation sites. Both the mutant and reference peptides were extracted. Combined with the 16 HLA alleles whose frequencies were more than 5% in the population collected in the 1000 Genome Project, we used our software, invoking NetMHCpan (v2.8) [21] to predict the binding affinity between HLA and the collected peptides. Then, we compared the binding information of the HLA proteins to wild-type and mutant peptides, and the specific bindings of the HLA proteins to mutant peptides were collected. These mutant peptides are seen as potential neoantigens. Finally, we obtained a dataset containing 1 420 785 records. We also analysed the distribution of the dataset (electronic supplementary material, table S2 and figure S2). The results showed a similar phenomenon with that in membrane proteins.
Mutations with more frequencies in the samples may play important roles in tumorigenesis. There are 65 potential common neoantigens whose corresponding mutations appear in at least 20 out of the 9155 donors from the ICGC database and had an IC50 of less than 500. The 65 neoantigens are related to the 23 somatic mutations of 12 genes (table 2; electronic supplementary material, table S3). KRAS, PIK3CA and TP53 occupy more potential neoantigens than other genes, indicating that these genes play more important roles in tumour immunotherapy, corresponding to former research results that KRAS and PIK3CA are oncogenes and that TP53 is a tumour suppressor gene [39]. Moreover, we also found some genes that have not been identified as tumour-associated genes by Cancer Gene Census also encode potential neoantigens, such as MUC4, FAM194B, OPRD1 and FRG1.
Table 2.
Sixty five potential common neoantigens and their corresponding genes and mutation frequency.
| gene | role in tumour | no. mutation | no. neoantigen |
|---|---|---|---|
| KRAS | oncogene | 5 | 11 |
| PIK3CA | oncogene | 5 | 21 |
| TP53 | tumour suppressor gene | 3 | 8 |
| SF3B1 | tumour-related gene | 1 | 2 |
| MUC4 | — | 1 | 1 |
| CHEK2 | tumour suppressor gene | 1 | 2 |
| PTEN | tumour suppressor gene | 2 | 3 |
| FAM194B | — | 1 | 2 |
| OPRD1 | — | 1 | 5 |
| CTNNB1 | oncogene | 1 | 5 |
| FRG1 | — | 1 | 4 |
| GNAS | tumour-related gene | 1 | 1 |
We found that the most frequent potential neoantigens are encoded by gene KRAS, which has been identified as an oncogene in vivo. There are six potential neoantigens related to the KRAS gene in the top 10 potential neoantigens, with two different mutations: G12D and G12 V. Among the six peptides, three of them (KLVVVGADGV, KLVVVGAVGV and KLVVVGAV) are presented by HLA-A*02:01, one (TEYKLVVVGAV) is presented by HLA-A*40:01, one (GAVGVGKSAL) is presented by HLA-A*03:04 and one (GAVGVGKSAL) is presented by HLA-C*03:03 (table 3).
Table 3.
Top 10 neoantigens with the highest mutation frequency in 9155 donors.
| gene | HLA allele | position | peptide | mutation | affinity (nM) | mutation frequency |
|---|---|---|---|---|---|---|
| KRAS | HLA-A*02:01 | 8 | KLVVVGADGV | G12D | 214 | 322 out of 9155 |
| KRAS | HLA-A*02:01 | 8 | KLVVVGAVGV | G12 V | 112 | 239 out of 9155 |
| KRAS | HLA-A*02:01 | 8 | KLVVVGAV | G12 V | 163 | 239 out of 9155 |
| KRAS | HLA-B*40:01 | 11 | TEYKLVVVGAV | G12 V | 90 | 239 out of 9155 |
| KRAS | HLA-C*03:04 | 3 | GAVGVGKSAL | G12 V | 172 | 239 out of 9155 |
| KRAS | HLA-C*03:03 | 3 | GAVGVGKSAL | G12 V | 172 | 239 out of 9155 |
| PIK3CA | HLA-C*07:02 | 2 | ARHGGWTTKM | H1047R | 218 | 200 out of 9155 |
| PIK3CA | HLA-C*06:02 | 3 | ARHGGWTTKM | H1047R | 457 | 200 out of 9155 |
| PIK3CA | HLA-C*07:01 | 2 | ARHGGWTTKM | H1047R | 249 | 200 out of 9155 |
| PIK3CA | HLA-A*11:01 | 11 | STRDPLSEITK | E545 K | 81 | 182 out of 9155 |
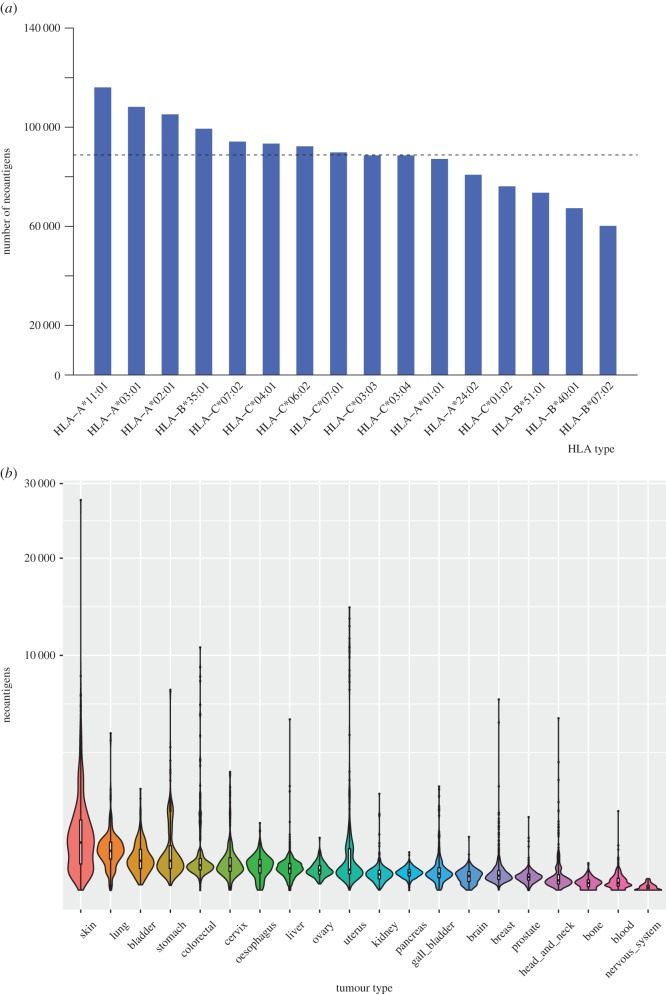
To study the distribution of the neoantigens across different HLA type, we classified the 1 420 785 records into 16 parts according to the HLA type we used (figure 3a). It was found that approximately 10 mutant peptides could bind to each HLA type in each sample, which means that we can find about 60 neoantigens in each tumour sample on average.
Figure 3.
The distribution of tumour-specific neoantigens across 16 HLA types and 20 tumour types. (a) The number of tumour-specific neoantigens with each HLA type is shown in decreasing order. The dashed line indicates the average number of neoantigens. (b) Distribution of tumour-specific neoantigens across 20 tumour types. The width of each violin indicates the proportion of donors sharing a certain number of neoantigens in each tumour type. Upper limit and lower limit of white bar and the black line in it denote upper quartile, lower quartile and median number for each type.
Because of the highly heterogeneity of tumours, we further investigated the distribution of neoantigens in each tumour type (based on the tissue origin; figure 3b). The results showed that the neoantigen load is related to the somatic mutation burden. The cancer types have more mutation load, such as skin and lung cancer, have more neoantigens in average. Interestingly, uterus cancer has the largest number of neoantigens on average (715.98, electronic supplementary material, table S4), but the median number of neoantigens of uterus cancer ranks 10th among the 20 cancer types (figure 3b). The reason may be that the number of neoantigens varies greatly among different patients of uterus cancer, several uterus tumours have large numbers of neoantigens. The nervous system cancer possesses the least neoantigens (2.39) on average. The results indicated that the neoantigen load is not only quite different between different cancer types, but also quite different between different tumours from the same tissue.
3.7. Specific binding of the TP53 mutant peptide to HLA-A*02:01
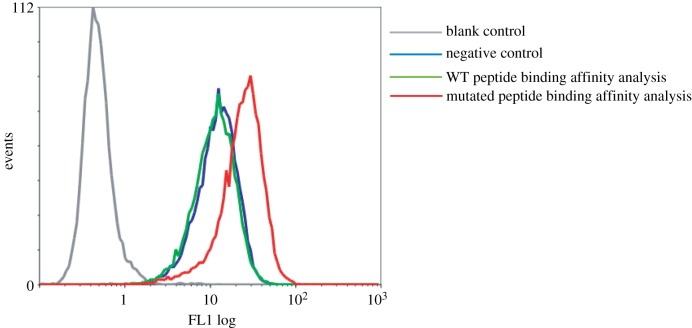
We choose one of the 65 potential common neoantigens, which was generated by the TP53 R248 W mutation, to experimentally confirm the specific-binding of neoantigen to HLA-A*02:01 using T2 assay [36]. We predicted that the wild-type (WT) peptide (GMNRRPILTII) could not bind to HLA-A*02:01, while the mutant peptide (GMNWRPILTII) could weakly bind to HLA-A*02:01 with the IC50 value = 350 nM (electronic supplementary material, table S3). The T2 cell line was widely used to confirm the binding of the peptides to HLA-A*02:01 as its HLA levels can be stabilized by the addition of exogenous HLA-binding peptides but unable to present the endogenous HLA-associated peptides [15,36]. To assess binding strength, we first incubated T2 cells with the WT and mutant peptides, respectively, and then used the W6/32 antibody that targets HLA molecules stabilized by any HLA-binding peptides. The strength of peptide binding between WT and mutant peptides were comparable as suggested by W6/32 staining. Analysis of the pulsed cells by flow cytometry showed that binding of the TP53 (R248 W) mutant peptide to T2 cells was more significant than the background levels of staining to the WT peptide or negative control cells (figure 4), which confirms the specific binding of the TP53 mutant peptide to HLA-A*02:01. Therefore, the R248 W mutation of TP53 can generate a potential tumour-specific neoantigen in the patient with HLA-A*02:01, which can be an ideal target for neoantigen-specific cancer immunotherapy.
Figure 4.
Specific binding of mutant peptide of TP53 to HLA-A*02:01. Blank control: FITC-goat anti-mouse IgG + T2 cells; negative control: human beta-2-microglobulin were incubated with T2 cells overnight at 37°C + W6/32 + FITC-goat anti-mouse IgG; wide-type (WT) peptide binding affinity analysis: WT peptide (GMNRRPILTII) and human beta-2-microglobulin were incubated with T2 cells overnight at 37°C + W6/32 + FITC-goat anti-mouse IgG; mutated peptide binding affinity analysis: mutated peptide (GMNWRPILTII) and human beta-2-microglobulin were incubated with T2 cells overnight at 37°C + W6/32 + FITC-goat anti-mouse IgG.
4. Discussion
TSNAD is a tool for detecting cancer somatic mutations following the best practices of GATK [20]. TSNAD can also provide potential neoantigens [1], which can be either extracellular mutations of membrane proteins or mutant peptides presented by class I MHC molecules. It is critical for biologists without programming background. We applied the antigen-predicting tool of TSNAD to predict neoantigens, including extracellular mutations of membrane proteins and neoantigens presented by MHC class I molecules. And we experimentally verified the specific-binding of the mutated peptide of TP53 we predicted (R248 W, wild-type: GMNRRPILTII, mutant: GMNWRPILTII) to HLA-A*02:01. The predicted neoantigens in our study were important sources for selecting suitable drug targets. In further study, these predicted neoantigens would need more experimental validation for their potential to be employed as drug targets of T cell or antibody-based immunotherapy.
Supplementary Material
Acknowledgements
We would like to thank Dr Binbin Zhou from Zhejiang University for her help with programming. We also gratefully acknowledge the clinical contributors and the data producers from the International Cancer Genome Consortium (ICGC) for referencing the ICGC datasets.
Data accessibility
The software and codes are freely available from https://github.com/jiujiezz/tsnad and the predicted neoantigens are freely available from http://biopharm.zju.edu.cn/lab/database/tsnadb.
Authors' contributions
Z.Z., Z.X.S., X.G. and S.Q.C. designed and directed the research; Z.Z., X.Z.L. wrote the programs; Z.Z., J.C.W., S.S.W. and J.Z. performed data analysis; X.Y.Y. performed experimental validation. Z.Z., X.Z.L., J.C.W. and Z.X.S. wrote the manuscript. All authors reviewed the manuscript.
Competing interests
We declare that we have no competing interests.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31501021 and 81430081), the Zhejiang Provincial Natural Sciences Foundation of China (LY15C060001), the Fundamental Research Funds for the Central Universities and the State Key Laboratory of Genetic Engineering at Fudan University.
References
- 1.Ilyas S, Yang JC. 2015. Landscape of tumor antigens in T cell immunotherapy. J. Immunol.. 195, 5117–5122. (doi:10.4049/jimmunol.1501657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbins PF, et al. 2013. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 19, 747–752. (doi:10.1038/nm.3161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, et al. 2011. Mutant proteins as cancer-specific biomarkers. Proc. Natl Acad. Sci. USA 108, 2444–2449. (doi:10.1073/pnas.1019203108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stratton MR, Campbell PJ, Futreal PA. 2009. The cancer genome. Nature 458, 719–724. (doi:10.1038/nature07943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov LB, et al. 2013. Signatures of mutational processes in human cancer. Nature 500, 415–421. (doi:10.1038/nature12477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin RW, Embleton MJ, Price MR. 1983. Monoclonal antibody-defined antigens on tumor cells. Biomembranes 11, 285–312. [PubMed] [Google Scholar]
- 7.Van den Eynde BJ, van der Bruggen P. 1997. T cell defined tumor antigens. Curr. Opin. Immunol. 9, 684–693. [DOI] [PubMed] [Google Scholar]
- 8.Hudis CA. 2007. Trastuzumab--mechanism of action and use in clinical practice. N. Engl. J. Med. 357, 39–51. (doi:10.1056/NEJMra043186) [DOI] [PubMed] [Google Scholar]
- 9.Lee DW, Barrett DM, Mackall C, Orentas R, Grupp SA. 2012. The future is now: chimeric antigen receptors as new targeted therapies for childhood cancer. Clin. Cancer Res. 18, 2780–2790. (doi:10.1158/1078-0432.CCR-11-1920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kochenderfer JN, Rosenberg SA. 2013. Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat. Rev. Clin. Oncol. 10, 267–276. (doi:10.1038/nrclinonc.2013.46) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajasagi M, et al. 2014. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 124, 453–462. (doi:10.1182/blood-2014-04-567933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher TN, Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science 348, 69–74. (doi:10.1126/science.aaa4971) [DOI] [PubMed] [Google Scholar]
- 13.Sharma P, Allison JP. 2015. The future of immune checkpoint therapy. Science 348, 56–61. (doi:10.1126/science.aaa8172) [DOI] [PubMed] [Google Scholar]
- 14.Desrichard A, Snyder A, Chan TA. 2016. Cancer neoantigens and applications for immunotherapy. Clin. Cancer Res. 22, 807–812. (doi:10.1158/1078-0432.CCR-14-3175) [DOI] [PubMed] [Google Scholar]
- 15.Carreno BM, et al. 2015. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808. (doi:10.1126/science.aaa3828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, Griffith M. 2016. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 8, 11 (doi:10.1186/s13073-016-0264-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Mardis ER, Maher CA. 2016. INTEGRATE-Neo: a pipeline for personalized gene fusion neoantigen discovery. Bioinformatics 32, 511–517. (doi:10.1093/bioinformatics/btw674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker KF, et al. 1999. Analysis of E-cadherin in diffuse-type gastric cancer using a mutation-specific monoclonal antibody. Am. J. Pathol. 155, 1803–1809. (doi:10.1016/S0002-9440(10)65497-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, et al. 2009. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin. Cancer Res. 15, 3023–3028. (doi:10.1158/1078-0432.CCR-08-2739) [DOI] [PubMed] [Google Scholar]
- 20.McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. (doi:10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. 2009. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13. (doi:10.1007/s00251-008-0341-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. (doi:10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. (doi:10.1093/bioinformatics/btp698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. (doi:10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. (doi:10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. (doi:10.1038/ng.806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cibulskis K, et al. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219. (doi:10.1038/nbt.2514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Wang K. 2015. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 10, 1556–1566. (doi:10.1038/nprot.2015.105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (doi:10.1093/nar/gkq603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. (doi:10.1093/bioinformatics/btp336) [DOI] [PubMed] [Google Scholar]
- 31.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182. [PubMed] [Google Scholar]
- 32.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. (doi:10.1006/jmbi.2000.4315) [DOI] [PubMed] [Google Scholar]
- 33.Gourraud PA, Khankhanian P, Cereb N, Yang SY, Feolo M, Maiers M, Rioux JD, Hauser S, Oksenberg J. 2014. HLA diversity in the 1000 genomes dataset. PLoS ONE 9, e97282 (doi:10.1371/journal.pone.0097282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlen M, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (doi:10.1126/science.1260419) [DOI] [PubMed] [Google Scholar]
- 35.Yates A, et al. 2016. Ensembl 2016. Nucleic Acids Res. 44, D710–D716. (doi:10.1093/nar/gkv1157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elvin J, Potter C, Elliott T, Cerundolo V, Townsend A. 1993. A method to quantify binding of unlabeled peptides to class I MHC molecules and detect their allele specificity. J. Immunol. Methods 158 161–171. [DOI] [PubMed] [Google Scholar]
- 37.Skora AD, et al. 2015. Generation of MANAbodies specific to HLA-restricted epitopes encoded by somatically mutated genes. Proc. Natl Acad. Sci. USA 112, 9967–9972. (doi:10.1073/pnas.1511996112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai J, Zhou Z, Tang XJ, Gao ZB, Zhou J, Chen SQ. 2016. A tumor-specific neo-antigen caused by a frameshift mutation in BAP1 is a potential personalized biomarker in malignant peritoneal mesothelioma. Int. J. Mol. Sci. 17, 739 (doi:10.3390/ijms17050739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. 2004. A census of human cancer genes. Nat. Rev. Cancer. 4, 177–183. (doi:10.1038/nrc1299) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The software and codes are freely available from https://github.com/jiujiezz/tsnad and the predicted neoantigens are freely available from http://biopharm.zju.edu.cn/lab/database/tsnadb.