Abstract
We studied non-driver mutations in 62 subjects with myeloproliferative neoplasm (MPN)-associated myelofibrosis upon diagnosis, including 45 subjects with primary myelofibrosis (PMF) and 17 with post-polycythemia vera or post-essential thrombocythemia myelofibrosis (post-PV/ET MF). Fifty-eight subjects had ≥1 non-driver mutation upon diagnosis. Mutations in mRNA splicing genes, especially in U2AF1, were significantly more frequent in PMF than in post-PV/ET MF (33 vs. 6%; P = 0.015). There were also striking differences in clonal architecture. These data indicate different genomic spectrums between PMF and post-PV/ET MF.
Electronic supplementary material
The online version of this article (doi:10.1186/s13045-017-0472-5) contains supplementary material, which is available to authorized users.
Keywords: Myeloproliferative neoplasm-associated myelofibrosis, Non-driver mutation, Targeted gene sequencing
To the Editor
MPN-associated myelofibrosis is a clinical descriptor encompassing primary myelofibrosis (PMF) and post-polycythemia vera and post-essential thrombocythemia (ET) myelofibrosis (post-PV/ET MF) [1]. Most individuals with MPN-associated myelofibrosis have so-called driver mutations in JAK2, CALR, or MPL. Although the distribution of these mutations is similar in PMF and post-PV/ET MF, some data indicate that the impact of the type of driver mutation on prognosis differs [2]. Recently, non-driver mutations were reported to be associated with the initiation, progression, and prognosis of MPNs [3–6]. We analyzed whether the landscape of non-driver mutations differed among cohorts.
Forty-five subjects with PMF and 17 with post-PV/ET MF (13 post-PV MF and 4 post-ET MF) were studied. The subjects provided informed consent in compliance with the Declaration of Helsinki (The Ethics Committee of Institute of Hematology and Blood Diseases Hospital; No. KT2015010-EC-1). Diagnoses were based on the revised 2016 criteria of the World Health Organization (WHO) [7]. The median interval from diagnosis of PV or ET to post-PV/ET MF was 12 years (range, 5–30 years). Prognosis was evaluated with the Dynamic International Prognostic Scoring System (DIPSS) [8]. The cytogenetic findings were classified as favorable or unfavorable using the DIPSS-plus classification [9]. Subject-, disease-, and therapy-related variables are displayed in Additional file 1: Table S1.
Targeted gene sequencing was carried out upon diagnosis of myelofibrosis. Non-driver mutations were compared against expected patterns and categorized into oncogenic mutations, possibly oncogenic variants or unknown significance. Germline control DNA was used to ascertain the somatic mutations. To account for the absence of a matched control for some subjects, a bespoke variant selection pipeline was used. Details of the methods are described in the Additional file 2 and Additional file 1: Table S2–S3.
Forty-two subjects with PMF (93%) had ≥1 non-driver mutation. Two of the 17 subjects with no driver mutation in JAK2 V617F, JAK2 exon 12, CALR, or MPL W515 had mutations in SH2B3 or PIAS3 of the JAK-STAT signaling pathway, 13 had mutations in other genes, and 2 had no mutations. Non-driver mutations were detected in 16 subjects with post-PV/ET MF (94%). There was no difference in the median numbers of non-driver mutations in the subjects with PMFs vs. those with post-PV/ET MF (3; [range, 0–5] vs. 3; [range, 0–7]; P = 0.885). There was also no difference in the median numbers of non-driver mutations (3; [range, 0–5] vs. 3 [range, 0–5]; P = 0.668) in the subjects with PMF with and without driver mutations.
The distribution of non-driver mutations in PMF vs. post-PV/ET MF is shown in Fig. 1a, b. Twelve mutations in non-driver genes were detected in >5% of the subjects with PMF, including ASXL1 (N = 15), U2AF1 (N = 10), TET2 (N = 7), FAT1 (N = 7), SETBP1 (N = 6), SRSF2 (N = 4), CUX1 (N = 4), EP300 (N = 4), FAT2 (N = 3), NOTCH3 (N = 3), EZH2 (N = 3), and GATA3 (N = 3). In subjects with post-PV/ET MF, ASXL1 (N = 7) was the most frequent mutation, followed by TET2 (N = 5).
Fig. 1.
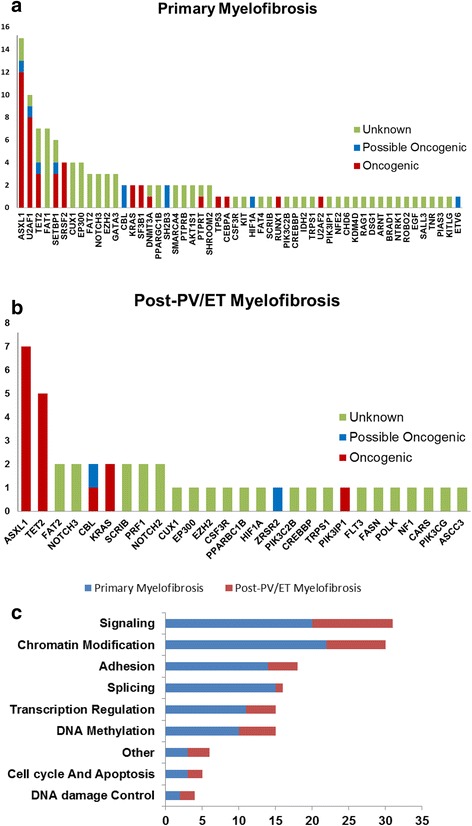
Mutational spectrum in primary myelofibrosis and post-PV/ET myelofibrosis. Frequency of non-driver mutations identified in the sequencing screen in 45 patients with PMF (a) and 17 patients with post-PV/ET MF (b). Frequency of gene mutations involved in common functional pathways (c)
Changes in non-driver genes were grouped into several functional pathways. The most frequent mutation targets were genes associated with signal transduction (N = 31), followed by chromatin modification (N = 30), adhesion (N = 18), RNA splicing (N = 16), transcription (N = 15), DNA methylation (N = 15), cell cycle/apoptosis (N = 5), and DNA damage control (N = 4; Fig. 1c). The mutation frequencies of the mRNA splicing genes were significantly different between the subjects with PMF and those with post-PV/ET (33 vs. 6%; P = 0.015). U2AF1 mutations were significantly more common in the subjects with PMF than those with post-PV/ET MF (22 vs. 0; P = 0.008).
We used copy number-adjusted variant allelic frequencies (VAF) in each subject with a non-driver mutation to determine whether a mutation was in the ancestral clone or whether it arose in a sub-clone. Using statistically significant differences in VAFs, the subjects were classified as representing two different clonal architectures: (1) ancestral clonal only; or (2) ancestral clone and sub-clone(s). Figure 2a–c shows data from three subjects. Frequencies of ancestral clonal only vs. ancestral clone and sub-clone(s) architecture were significantly different between the subjects with PMF and those with post-PV/ET MF (ancestral clonal only: 49 vs. 13%; P = 0.013; Fig. 2d). In the subjects with PMF, the driver and non-driver mutations were ancestral in 14 subjects (e.g., subject 2015-R02413 in Fig. 2a) and sub-clonal in 9 subjects (e.g., subject 2015-R02406 in Fig. 2b). Eleven subjects with post-PV/ET MFs had JAK2 V617F as the only ancestral mutation (e.g., subject G121517R00701 in Fig. 2c).
Fig. 2.
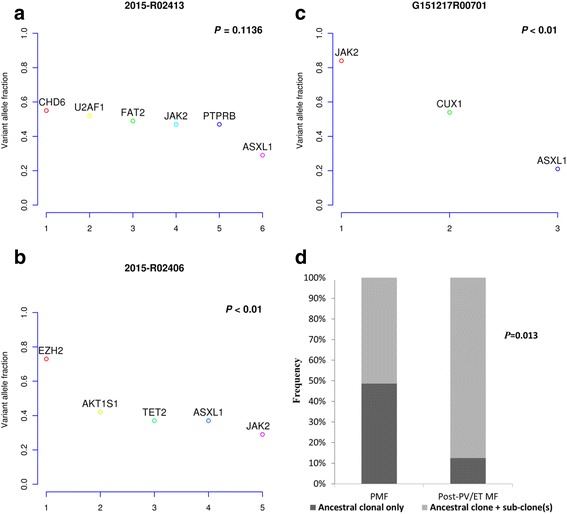
Clonal architecture in PMF and post-PV/ET MF. a In a representative patient with PMF, mutant JAK2 co-occurs with non-driver mutations as ancestral mutations. b In a representative patient with PMF, mutant JAK2 was a sub-clonal mutation and other non-driver mutations precede mutant JAK2. c In a representative patient with post-PV MF, mutant JAK2 was the only ancestral mutation preceding other non-driver mutations. d More PMFs were classified as having an ancestral clonal only architecture than post-PV/ET MF
In conclusion, we found differences in the non-driver mutation profile and the clonal architecture among the subjects with PMF compared with those with post-PV/ET MF. There was a significant difference in the frequency of the mRNA splicing genes among cohorts, similar to data reported by others [2, 10]. In individuals with PMF, approximately 30% of driver mutations co-occur with non-driver mutations as ancestral events, whereas approximately 20% of driver mutations were in sub-clones. In contrast, driver mutations occurred only in the ancestral clone in most subjects with post-PV/ET MF.
Additional files
Clinical and laboratory features of 45 patients with PMF and 17 patients with post-PV/ET MF. Table S2. Gene list of the 190-gene NGS panel. Table S3. Two hundred and twenty-nine high-confidence variants sequenced by the 190-gene NGS panel. (DOCX 43 kb)
Methods (DOCX 26 kb)
Acknowledgements
We would like to thank Dr. Jian Wu (MyGenostic Inc., Beijing, China) and Dr. Na Liu (MyGenostic Inc., Beijing, China) for the bioinformatics analysis.
Funding
This study was supported in part by the National Natural Science Funds (No. 81530008, No. 81370611, No. 81600098, No. 81270585, No. 81470295), the Program for Peking Union Scholars and Innovative Research Team, PUMC Youth Fund and Fundamental Research Funds for the Central Universities (No. 3332016089), and the Science and Technology Project of Tianjin (No. 15ZXLCSY00010). RPG acknowledges the support from the National Institutes of Health Research (NIHR) Biomedical Research Centre funding scheme.
Availability of data and materials
Not applicable. All data are in the manuscript and supplemental materials.
Authors’ contributions
ZJX designed the research, was the principal investigator, prepared the manuscript, and takes primary responsibility for the study; BL acquired, analyzed, and interpreted the data and drafted the manuscript; ZS performed the statistical analyses; PHZ, JQL, and GH acquired the data; ZFX, TJQ, JB, LZ, YZ, and ZJX recruited the subjects; and RPG helped with the data analyses and with the manuscript. All authors read and approved the final manuscript.
Competing interests
RPG is a part-time employee of Celgene Corp. The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study design was approved by the Ethics Committee of the Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, reference KT2015010-EC-1.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ET
Essential thrombocythemia
- MPN
Myeloproliferative neoplasm
- PMF
Primary myelofibrosis
- PV
Polycythemia vera
- VAF
Variant allelic frequency
Contributor Information
Bing Li, Email: libing1106@aliyun.com.
Robert Peter Gale, Email: robertpetergale@alumni.ucla.edu.
Zefeng Xu, Email: xuzefeng@ihcams.ac.cn.
Tiejun Qin, Email: qintiejun@ihcams.ac.cn.
Zhen Song, Email: songzhen@ihcams.ac.cn.
Peihong Zhang, Email: zhangpeihong@ihcams.ac.cn.
Jie Bai, Email: baijie@ihcams.ac.cn.
Lei Zhang, Email: zhanglei1@ihcams.ac.cn.
Yue Zhang, Email: zhangyue@ihcams.ac.cn.
Jinqin Liu, Email: liujinqin@ihcams.ac.cn.
Gang Huang, Email: Gang.Huang@cchmc.org.
Zhijian Xiao, Phone: +86-22-23909184, Email: zjxiao@hotmail.com, Email: zjxiao@medmail.com.cn.
References
- 1.Mesa RA, Green A, Barosi G, Verstovsek S, Vardiman J, Gale RP. MPN-associated myelofibrosis (MPN-MF) Leuk Res. 2011;35(1):12–3. doi: 10.1016/j.leukres.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 2.Rotunno G, Pacilli A, Artusi V, Rumi E, Maffioli M, Delaini F, Brogi G, Fanelli T, Pancrazzi A, Pietra D, Bernardis I, Belotti C, Pieri L, Sant’Antonio E, Salmoiraghi S, Cilloni D, Rambaldi A, Passamonti F, Barbui T, Manfredini R, Cazzola M, Tagliafico E, Vannucchi AM, Guglielmelli P. Epidemiology and clinical relevance of mutations in postpolycythemia vera and postessential thrombocythemia myelofibrosis: a study on 359 patients of the AGIMM group. Am J Hematol. 2016;91(7):681–686. doi: 10.1002/ajh.24377. [DOI] [PubMed] [Google Scholar]
- 3.Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2016; e-pub ahead of print 27 December 2016; doi: 10.1182/blood-2016-10-695940. [DOI] [PubMed]
- 4.Guglielmelli P, Lasho TL, Rotunno G, Score J, Mannarelli C, Pancrazzi A, Biamonte F, Pardanani A, Zoi K, Reiter A, Duncombe A, Fanelli T, Pietra D, Rumi E, Finke C, Gangat N, Ketterling RP, Knudson RA, Hanson CA, Bosi A, Pereira A, Manfredini R, Cervantes F, Barosi G, Cazzola M, Cross NC, Vannucchi AM, Tefferi A, et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28(9):1804–10. doi: 10.1038/leu.2014.76. [DOI] [PubMed] [Google Scholar]
- 5.Li B, Xu J, Wang J, Gale RP, Xu Z, Cui Y, Yang L, Xing R, Ai X, Qin T, Zhang Y, Zhang P, Xiao Z. Calreticulin mutations in Chinese with primary myelofibrosis. Haematologica. 2014;99(11):1697–700. doi: 10.3324/haematol.2014.109249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lussana F, Carobbio A, Salmoraghi S, Guglielmelli P, Vannucchi AM, Bottazzi B, Leone R, Mantovani A, Barbui T, Rambaldi A. Driver mutations (JAK2V617F, MPLW515L/K or CALR), pentraxin-3 and C-reactive protein in essential thrombocythemia and polycythemia vera. J Hematol Oncol. 2017;10(1):54. doi: 10.1186/s13045-017-0425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 8.Passamonti F, Cervantes F, Vannucchi AM, Morra E, Rumi E, Pereira A, Guglielmelli P, Pungolino E, Caramella M, Maffioli M, Pascutto C, Lazzarino M, Cazzola M, Tefferi A. A dynamic prognostic model to predict survival in primary myelofibrosis: a study by the IWG-MRT (International Working Group for Myeloproliferative Neoplasms Research and Treatment) Blood. 2010;115(9):1703–8. doi: 10.1182/blood-2009-09-245837. [DOI] [PubMed] [Google Scholar]
- 9.Gangat N, Caramazza D, Vaidya R, George G, Begna K, Schwager S, Van Dyke D, Hanson C, Wu W, Pardanani A, Cervantes F, Passamonti F, Tefferi A. DIPSS-Plus: a refined Dynamic International PrognosticScoring System (DIPSS) for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count and transfusion status. J Clin Oncol. 2011;29(4):392–7. doi: 10.1200/JCO.2010.32.2446. [DOI] [PubMed] [Google Scholar]
- 10.Brecqueville M, Rey J, Devillier R, Guille A, Gillet R, Adelaide J, Gelsi-Boyer V, Arnoulet C, Chaffanet M, Mozziconacci MJ, Vey N, Birnbaum D, Murati A. Array comparative genomic hybridization and sequencing of 23 genes in 80 patients with myelofibrosis at chronic or acute phase. Haematologica. 2014;99(1):37–45. doi: 10.3324/haematol.2013.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and laboratory features of 45 patients with PMF and 17 patients with post-PV/ET MF. Table S2. Gene list of the 190-gene NGS panel. Table S3. Two hundred and twenty-nine high-confidence variants sequenced by the 190-gene NGS panel. (DOCX 43 kb)
Methods (DOCX 26 kb)
Data Availability Statement
Not applicable. All data are in the manuscript and supplemental materials.


