SUMMARY
The eukaryotic translation factor eIF5A, originally identified as an initiation factor, was later shown to promote translation elongation of iterated proline sequences. Using a combination of ribosome profiling and in vitro biochemistry, we report a much broader role for eIF5A in elongation and uncover a critical function for eIF5A in termination. Ribosome profiling of an eIF5A-depleted strain reveals a global elongation defect, with abundant ribosomes stalling at many sequences, not limited to proline stretches. Our data also show ribosome accumulation at stop codons and in the 3′ UTR, suggesting a global defect in termination in the absence of eIF5A. Using an in vitro reconstituted translation system, we find that eIF5A strongly promotes the translation of the stalling sequences identified by profiling and increases the rate of peptidyl-tRNA hydrolysis more than 17-fold. We conclude that eIF5A functions broadly in elongation and termination, rationalizing its high cellular abundance and essential nature.
In Brief
In this manuscript, Schuller et al. characterize the activity of the translation factor eIF5A. They show that eIF5A accelerates peptidyl transfer at most, if not all, sequences and promotes peptide release. These expanded roles for eIF5A help to explain its essential nature and high abundance in eukaryotes.
INTRODUCTION
In addition to the core set of factors required for protein synthesis, many auxiliary proteins stimulate specific processes in the translation cycle. One such protein, eIF5A, was originally identified nearly 40 years ago as a factor that stimulates formation of the first peptide bond between Met-tRNA and puromycin (Benne and Hershey, 1978; Kemper et al., 1976; Schreier et al., 1977). More recent work from several groups identified eIF5A as having a more general role in elongation by conditionally depleting eIF5A from yeast cells in vivo and performing polysome analyses and ribosome transit measurements (Gregio et al., 2009; Henderson and Hershey, 2011; Saini et al., 2009). eIF5A is an essential gene in eukaryotes, and the nature of its critical cellular function has been a subject of ongoing exploration.
eIF5A is a small, highly expressed protein containing only 157 amino acids and is post-translationally modified with hypusine at a conserved lysine residue (Dever et al., 2014; Park et al., 1981). This hypusine modification is critical for eIF5A function in vivo and in assays of Met-puromycin formation (Park et al., 1991, 2011; Park, 1989; Saini et al., 2009). Recent biochemical work with the bacterial homolog of eIF5A, called EFP, revealed that EFP functions to promote the translation of polyproline containing peptides that stall the ribosome (in addition to Met-puromycin formation) (Doerfel et al., 2013; Glick et al., 1979; Glick and Ganoza, 1975; Ude et al., 2013). Proline residues were shown to be poor substrates for peptide bond formation, likely due to the unique geometry that they assume, thereby leading to slower ribosome elongation (Pavlov et al., 2009; Wohlgemuth et al., 2008). Subsequent work showed that eIF5A similarly stimulates translation of polyproline peptides (with as few as two prolines) in eukaryotes and that this function is highly dependent on its hypusine modification (Gutierrez et al., 2013).
While EFP and eIF5A appear to have similar biochemical functions in translation elongation, the essential nature of eIF5A in eukaryotic cells remains poorly understood. Given the documented role in polyproline synthesis and that ~10% of yeast genes contain stretches of three or more consecutive proline residues (and that 95 proteins in yeast contain four or more consecutive Pro residues as compared with only 9 in E. coli [Doerfel et al., 2013]), it was rationalized that the global defect in elongation observed upon eIF5A depletion (Saini et al., 2009) resulted from defects in translation mediated by polyproline motifs (Gutierrez et al., 2013). These bioinformatic data led to the suggestion that eIF5A is essential in eukaryotes because of the relative abundance of polyproline motifs in these organisms.
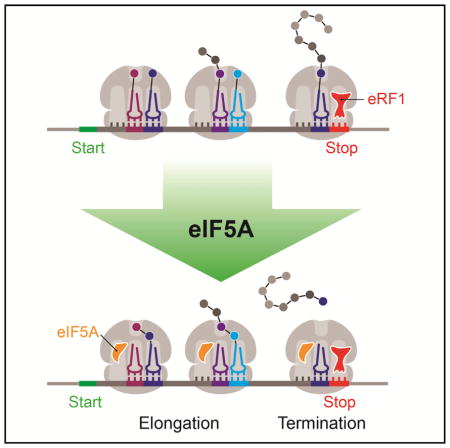
Here we explore the in vivo and in vitro function of eIF5A in both translation elongation and termination. Through a combination of ribosome profiling and biochemistry in a reconstituted translation system, we find that eIF5A has a broader role in elongation than previously understood, and we uncover a critical function for eIF5A in translation termination. Importantly, we show that eIF5A functions to stimulate translation elongation in many peptide contexts, certainly not limited to proline stretches, and to accelerate the rate of peptidyl-tRNA hydrolysis by eRF1 during termination. Our findings suggest that eIF5A is an obligate translation factor acting on many (if not all) translating ribosomes, thereby rationalizing its essential nature and high abundance in eukaryotic cells.
RESULTS
Conditional Depletion of eIF5A by an Auxin-Inducible Degron
Given that eIF5A is encoded by an essential gene, we had to develop a method for conditional depletion in order to observe its in vivo function. Initial efforts to perform ribosome profiling with a temperature-sensitive allele were largely unsuccessful due to poor reproducibility and global changes in gene expression introduced by the temperature shift. Not only does the shift to the non-permissive temperature alter the transcriptional landscape, but we suspected that it also might affect translation elongation in a way that could confound analyses of protein synthesis (Grousl et al., 2013).
We constructed a yeast strain in which both the transcription of eIF5A and the degradation of the protein can be manipulated by inducers added to the growth media. In this strain, eIF5A is expressed from the GAL1 promoter; transcription levels are high in the presence of galactose and low in the presence of glucose. To promote rapid turnover of the protein, we fused eIF5A with a mini auxin-inducible degron (mAID) tag; in the presence of auxin, this domain recruits the E3-ubiq-uitin ligase TIR1, which ubiquitinates the mAID-eIF5A fusion protein for proteasome degradation (Nishimura and Kanemaki, 2014).
By inhibiting transcription and promoting turnover of the protein, this strategy effectively depletes eIF5A and broadly recapitulates observations using other methods (Gutierrez et al., 2013; Saini et al., 2009). Although the eIF5A-degron fusion protein (eIF5Ad) is expressed in our system at somewhat lower levels than endogenous eIF5A, it supports cell viability (Figures S1A and S1B). Following a switch to glucose media to shut off transcription and the addition of auxin to deplete eIF5A, cell growth arrests after ~10 hr (Figure S1C). The timing of this arrest is consistent with the expectation that eIF5A is essential for cell growth: after ~8 hr, we were unable to detect the eIF5A protein on immunoblots (Figure S1A). For additional validation, we analyzed polysome profiles and observed that after 10 hr of induction, eIF5Ad cells showed an increase in the polysome/monosome ratio compared to WT cells (Figure S1D), as previously reported (Saini et al., 2009).
eIF5A Depletion Causes Redistribution of Ribosomes toward the 5′ End of Genes
We generated libraries of ribosome footprints from the WT and eIF5Ad strains after 10 hr of conditional growth. We did not pre-treat the culture with antibiotics to arrest translation (Ingolia et al., 2009), because adding cycloheximide to the media has been shown to cause sequence-specific ribosome pausing in vivo (Hussmann et al., 2015). Instead, lysates were flash frozen in liquid nitrogen, lysed, and thawed in lysis buffer containing cycloheximide to prevent lingering elongation during lysis (Guydosh and Green, 2014; Weinberg et al., 2016). We generated two biological replicates of the WT and eIF5Ad libraries and obtained ~15–25 million mapped reads for each. The number of ribosome footprints per gene is highly reproducible between biological replicates (Pearson’s r = 0.96 for eIF5Ad cells, 0.99 for WT cells) (Figure S1E).
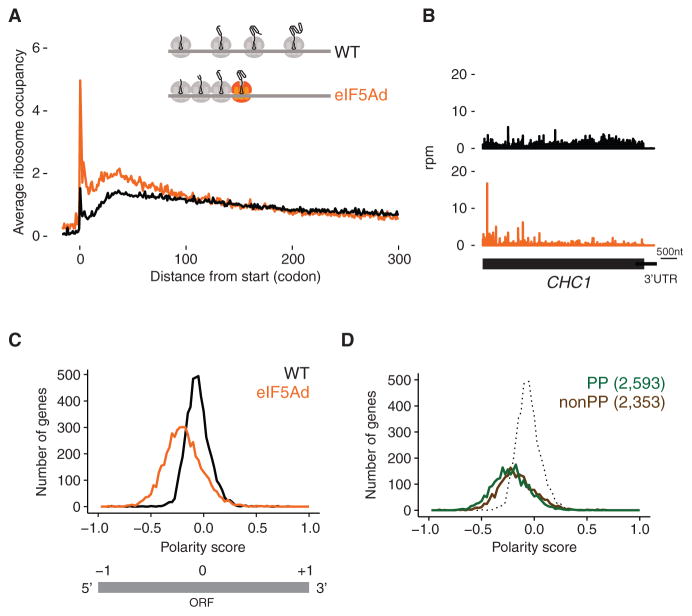
Our previous observation that depletion of eIF5A increases the fraction of ribosomes in polysomes led to the argument that eIF5A is a general translation factor (Saini et al., 2009). To get a genome-wide view of this phenomenon in our ribosome profiling data, we plotted the average ribosome occupancy of all genes aligned at start codons. In eIF5Ad cells, we observed an increase in ribosome occupancy in the first ~50 codons relative to the wild-type strain and a decrease in downstream occupancy (Figure 1A). These trends are readily understood by examining specific genes where we observe higher ribosome occupancy in the 5′ end relative to the 3′ end (Figure 1B). This phenomenon is consistent with a general elongation defect: an enrichment of ribosomes at the 5′ end of genes was previously observed during amino acid starvation or treatment with antibiotics that inhibit elongation, both of which result in global ribosome pausing (Gerashchenko and Gladyshev, 2014; Ingolia et al., 2009; Subramaniam et al., 2014).
Figure 1. Polar Distribution of Ribosomes toward 5′ End of Genes in eIF5Ad Cells.
(A) Average ribosome occupancy from all genes aligned at start codons for WT (black line) and eIF5Ad cells (orange line) with schematic depicting queuing of ribosomes upstream of a paused ribosome (colored orange). Ribosome occupancy was normalized to show a mean value of 1 for each codon.
(B) Example of ribosome occupancy along CHC1 gene in WT.1 (black) and eIF5Ad.1 (orange) cells. Rpm, reads per million.
(C) Distributions of polarity scores for 4,946 genes from WT and eIF5Ad cells are plotted, for genes with at least 64 reads per dataset in ORFs (top panel). Schematic representation of polarity score (bottom panel).
(D) Distributions of polarity scores for genes containing Pro-Pro dipeptide motifs (green) and genes lacking these motifs (brown) in eIF5Ad cells. WT distribution (includes both PP and non-PP) is included for reference (dotted). Numbers in parentheses denote the gene numbers included in the analysis. See also Figures S1 and S2.
To quantify the differences in the position of ribosomes along transcripts, we computed a polarity score for every gene (see STAR Methods). This metric assigns a value between −1 and +1 to each gene based on the distribution of ribosome footprints along it. We excluded ribosome densities at both ends of genes (15 nucleotides) from our analysis to avoid known artifacts introduced by start and stop codons (Young et al., 2015). Enrichment of ribosome occupancy at the 5′ end of a gene gives a polarity score between −1 and 0, while enrichment toward the 3′ end of a gene results in a score between 0 and +1. This metric yields information about the balance of ribosome occupancy across the gene (5′ to 3′), but not about the fine details of the distribution. For example, two very different distributions of ribosome density could both yield a polarity score of 0: enrichment at the center of a gene or an equal enrichment at both ends. Nevertheless, we find that this metric reveals broad trends in our data that provide insight into eIF5A function. Strikingly, the distribution of ribosome occupancy in eIF5Ad cells is shifted to the left relative to the distribution in WT cells (Figures 1C and S2A). This quantitative metric nicely summarizes the skewed ribosome distribution observed in specific genes and in plots of average ribosome occupancy (Figures 1A and 1B).
The accumulation of ribosomes at the 5′ ends of genes in eIF5Ad cells is consistent with pauses in elongation followed by queuing of upstream ribosomes (as depicted in the model in Figure 1A). Given eIF5A’s ability to resolve pausing at polyproline stretches, we hypothesized that the loss of eIF5A could explain these pauses in elongation. To test this idea, we divided yeast genes into two (roughly equal) subsets, one containing Pro-Pro dipeptide motifs and the other lacking these motifs. We chose to separate our data based on possession of Pro-Pro motifs because it was the minimal motif on which eIF5A was known to function (Gutierrez et al., 2013). We were surprised to find that both subsets showed a significant shift to the left, which is indicative of marked enrichment in ribosome occupancy at the 5′ end (Figures 1D and S2B), independent of the presence or absence of Pro-Pro motifs. This striking genome-wide observation of ribosome pausing suggests that eIF5A may function broadly in eukaryotic cells and may not simply relieve pausing at polyproline motifs.
eIF5A Alleviates Ribosome Pausing at Poly-Pro Motifs
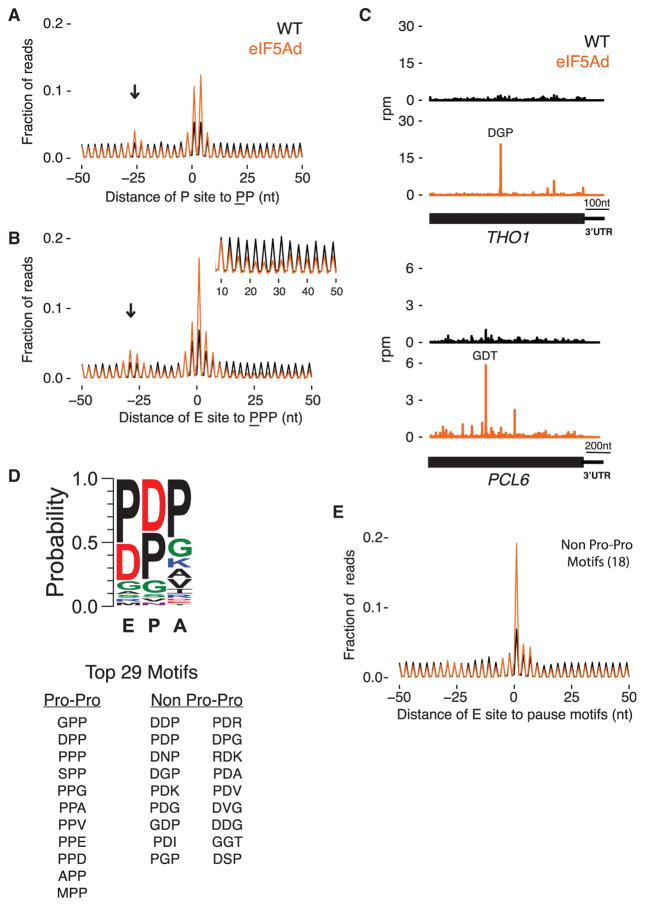
As a first step in analyzing translation pauses, we started with the known role of eIF5A in resolving pausing at polyproline stretches. We computed the average ribosome occupancy at two or three consecutive Pro codons in well-expressed genes and found the peak was ~2-fold higher in eIF5Ad cells than in WT cells (Figures 2A and 2B). We observed two major peaks in the diproline plot at the center of the average gene plot (Figure 2A); one peak corresponds to pauses with the Pro codons positioned in the P and A sites, while the second corresponds to pauses with the Pro codons positioned in the E and P sites. These observations suggest that forming the first Pro-Pro peptide bond is challenging without eIF5A and that forming downstream peptide bonds is challenging when Pro-Pro dipeptide is engaged in the exit channel. These findings are consistent with previous biochemical and profiling studies of EFP in the E. coli system (Doerfel et al., 2013; Ude et al., 2013; Woolstenhulme et al., 2015).
Figure 2. eIF5A Alleviates Ribosome Pausing at More Than Poly-Pro Motifs.
(A) Average ribosome occupancy centered at diproline motifs with the underlined Pro in the P site of the ribosome. Excluding motifs in genes with less than 64 reads per dataset in ORFs, 5,920 and 5,939 positions are averaged in WT and eIF5Ad, respectively. Arrow indicates stacked ribosomes.
(B) Similar to (A), average plot centered at triproline motifs with underlined Pro in the E site of the ribosome. Inset: close-up view of the ribosome occupancy 10–50 nt downstream of the triproline motif.
(C) Ribosome footprints on THO1 and PCL6 genes in WT.1 (black) and eIF5Ad.1 (orange) cells. Motifs of highest ribosome pausing are denoted. Rpm, reads per million.
(D) Peptide motif associated with ribosome pausing in eIF5Ad cells, using motifs with pause score greater than 10 and weighted by pause score ratio of eIF5Ad/WT. Top 29 motifs are listed.
(E) Average ribosome occupancy centered at 5,478 pause sites that match the 18 non-Pro-Pro pausing motifs identified in eIF5Ad cells. See also Figure S3.
Pauses at polyproline stretches affect the ribosome distribution surrounding the pausing motifs as well. We observed increased ribosome occupancy ~30 nt upstream of the polypro-line motifs (Figures 2A and 2B, arrows) due to trailing ribosomes forming a queue behind those paused at the polyproline motif. In addition, ribosome occupancy downstream of polyproline motifs is strongly depleted, presumably because downstream ribosomes continue elongating, leaving empty mRNA behind them (Figure 2B, inset). This depletion is more pronounced for triproline motifs than for diproline motifs, consistent with prior observations that a diproline motif does not reduce protein levels upon loss of eIF5A activity; an effect on protein expression is only seen with three or four consecutive Pro codons (Gutierrez et al., 2013).
Our observations of strong pauses, stacked ribosomes upstream of the paused ribosome, and depletion of downstream density after PPP motifs are very similar to the effects seen in our earlier work upon deletion of the EFP gene in bacteria (Woolstenhulme et al., 2015).
We note that pausing at polyproline motifs is evident in WT cells, suggesting that these motifs are problematic even in the presence of factors that evolved to mitigate their impact on elongation (Pavlov et al., 2009; Wohlgemuth et al., 2008). Proline residues have been associated with ribosome pausing in several eukaryotic-based ribosome profiling studies (Artieri and Fraser, 2014; Ingolia et al., 2011), suggesting that the endogenous activity of eIF5A may not be sufficient to wholly relieve ribosome pausing at polyproline motifs in these cells.
Widespread Ribosome Pausing in eIF5Ad Cells
In our analyses of ribosome occupancy on individual genes, we also noticed many strong pauses in eIF5Ad cells at positions that do not encode polyproline motifs (e.g., THO1 and PCL6 in Figure 2C). These additional pauses may explain the polarity effects of eIF5A depletion on genes lacking polyproline motifs. We explored this phenomenon by computing pause scores for all 8,000 tripeptide motifs. In eIF5Ad cells, 445 tripeptide motifs showed at least a 2-fold increase in pause scores compared to WT cells, while 29 motifs have a pause score 10-fold or greater than the average density of the gene (Figures 2D and S3A; Table S1).
The top 29 tripeptide motifs show a consensus peptide sequence with enrichment of Pro or Asp in the E and the P sites and Pro in the A site; Gly is modestly enriched in all three sites (Figure 2D). Despite the importance of Pro in the consensus sequence, 18 of these 29 motifs surprisingly do not contain a Pro-Pro sequence. To demonstrate the impact of eIF5A on these identified sequences, we show an average gene analysis of ~5,500 sites corresponding to the 18 non-Pro-Pro motifs that reveals a level of pausing upon eIF5A depletion (Figure 2E) comparable to that observed at PPP (Figure 2B). Moreover, strong pauses are observed at the four tripeptide motifs completely lacking Pro (DVG, DDG, GGT, and RDK; Figure S3B). Taken together, our findings demonstrate that eIF5A has a much broader role in alleviating ribosome pausing than previously thought.
eIF5A Promotes Translation of Stalling Sequences In Vitro
Ribosome profiling is not a time-resolved experiment, and so there is no way of knowing whether all positions are impacted by the depletion of eIF5A. Profiling only reveals which motifs are more strongly impacted than others. To address this, we performed experiments in vitro using a reconstituted translation system to ask whether eIF5A is directly involved in promoting slow steps in translation elongation. We first tested the ability of our purified eIF5A to accelerate the reaction between Met-tRNA and the antibiotic puromycin, since eIF5A was originally identified using this assay (Benne and Hershey, 1978; Kemper et al., 1976; Schreier et al., 1977). Pelleted 80S initiation complexes programmed with a simple mRNA (MFN-Stop) with [35S]-Met-tRNAiMet loaded in the ribosomal P site were mixed with puromycin (Puro) in the presence and absence of eIF5A, and formation of the Met-Puro product was observed over time. As anticipated, we found that post-translationally hypusinated eIF5A greatly enhances the rate of Met-Puro formation, while un-modified eIF5A has a more subtle effect (Figure S4C). This experiment documents for the first time a >100-fold rate enhancement for this reaction resulting from the addition of modified eIF5A.
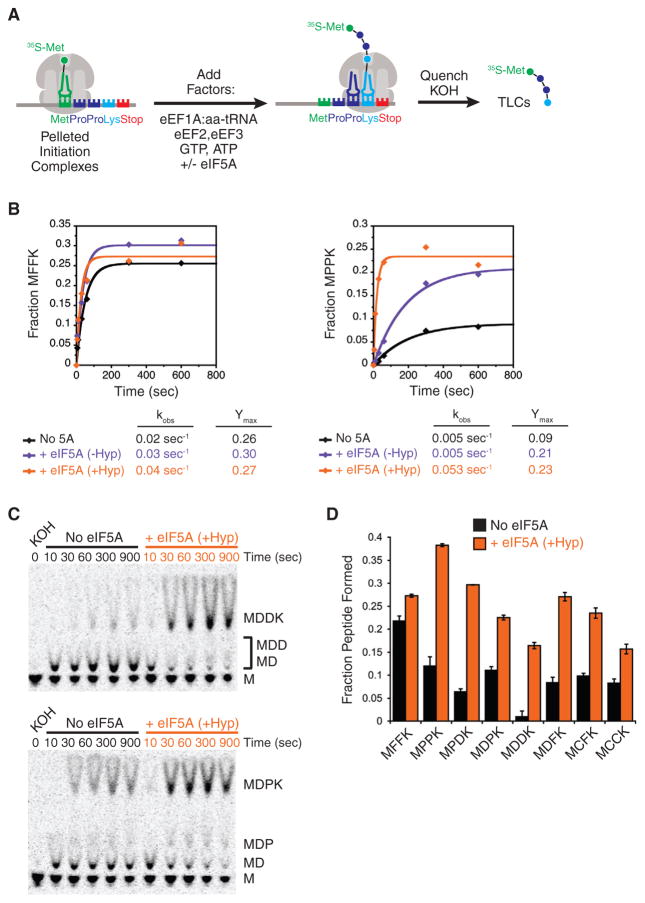
With this confirmation of robust in vitro activity, we tested the role of eIF5A in the translation of peptides containing several stalling motifs that were identified in our profiling experiments. While our profiling analysis described above focused on identifying tripeptide stalling motifs, we were also able to identify pausing motifs at the dipeptide level in our profiling data (Table S2), and because these motifs are simpler to characterize biochemically, we chose to focus efforts on these for the in vitro assays. In these experiments, pelleted 80S initiation complexes programmed with a simple mRNA (MXXK-Stop where X is any amino acid) containing [35S]-Met-tRNAiMet were incubated with elongation factors and the appropriate tRNAs (eEF1A:aa-tRNA, eEF2, eEF3), and peptide formation was measured over time (Figure 3A). Each peptide contains the problematic dipeptide being analyzed (e.g., Pro-Pro) followed by a C-terminal lysine that helps us to resolve the tetrapeptide product using an electrophoretic TLC system.
Figure 3. eIF5A Stimulates Translation of Stalling Motifs In Vitro.
(A) Schematic representation of in vitro elongation reactions using reconstituted translation system. Pelleted 80S initiation complexes are incubated with elongation factors, aminoacyl-tRNAs, GTP, ATP, and eIF5A. Reactions are quenched with KOH and peptide products resolved by electrophoretic TLCs.
(B) Elongation kinetics for Phe-Phe- and Pro-Pro-containing peptides (MFFK and MPPK) in the presence and absence of eIF5A and hypusination modification.
(C) Representative TLCs for elongation kinetics of dipeptide stalling motifs Asp-Asp (MDDK) and Asp-Pro (MDPK).
(D) Comparison of elongation endpoints for all peptides analyzed in presence and absence of eIF5A. Error bars represent the standard deviation from three replicate experiments. See also Figure S4.
As we previously reported (Gutierrez et al., 2013), eIF5A is essential for robust synthesis of a Pro-Pro-containing peptide (MPPK), while it has a negligible effect on a Phe-Phe-containing peptide (MFFK). For the MPPK reaction, we see that the stimulatory effect of eIF5A is highly dependent on the hypusine modification (Figure 3B): unmodified eIF5A increases the reaction endpoint (amount of peptide formed, Ymax) but hypusinated eIF5A increases both the endpoint and the rate of the reaction (kobs, compared to unmodified eIF5A). The endpoint defects are due to substantial peptidyl-tRNA drop-off in these slow peptidyl-transfer reactions (Figure S4D); similar trends for short peptidyl-tRNAs have been reported by others (Doerfel et al., 2013; Katoh et al., 2016). These data argue that the presence of unmodified eIF5A stabilizes the complex against peptidyl-tRNA drop-off but that the hypusine modification is critical for maximal rate enhancement.
We next evaluated the role of eIF5A in promoting elongation through several dipeptide stalling motifs identified from our ribosome profiling data (Table S2), focusing on hyspuinated eIF5A as this provided maximal stimulation of the Pro-Pro-containing peptide. In what follows, we limit our analysis to the endpoint of the reaction, given the difficulty of separating the rate for the reaction of interest from the competing drop-off reaction in observed rate constants. Since several of the strongest dipeptide stalling motifs contained aspartic acid, we measured the synthesis of the MDDK and MDPK peptides with and without eIF5A (Figure 3C); eIF5A strongly promoted synthesis of both peptides. We then analyzed the effects of eIF5A for several other peptides containing combinations of proline, aspartic acid, cysteine, and phenylalanine. We anticipated that several of these motifs would induce strong pauses (DD, DP, and PD), while others would induce little or no stalling (CC and CF) based on their relative pause scores (Table S2). Somewhat surprisingly, we found that eIF5A has a stimulatory effect on elongation for all the peptides tested (Figure 3D). This result points to the fact that our profiling data only measure relative pausing levels between peptide motifs and not absolute levels, as were measured in our in vitro experiments.
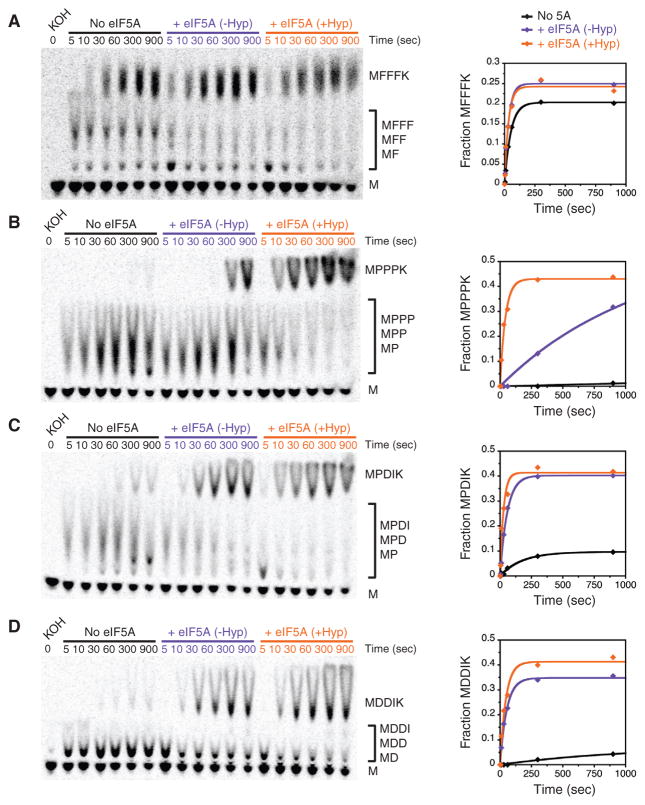
While these experiments gave us insight into the breadth of stalling motifs that are rescued by eIF5A, we wanted to further investigate the role of hypusine in resolving these translation pauses. We chose to probe the requirement of eIF5A hypusine modification using several tripeptide motifs: one with polyphenylalanine as a control (MFFFK), one with polyproline (MPPPK), one with a single proline (MPDIK), and one with no prolines (MDDIK). We find that eIF5A does not stimulate polyphenylalanine synthesis (MFFFK), while it is critical for polyproline synthesis (MPPPK) (Figures 4A and 4B). As observed for diproline (Figure 3B), non-hypusinated eIF5A increases the MPPPK reaction endpoint, while hypusinated eIF5A is critical for maximal endpoint and rate enhancement. Interestingly, we find that hypusine is less important for translation of other non-polyproline stalling motifs, such as PDI and DDI (Figures 4C and 4D). Taken together, our in vitro and ribosome profiling data suggest that eIF5A plays a substantial role in the rate of translation elongation at most (if not all) sites in the transcriptome, although hypusine modification may be required only for resolving polyproline stalls.
Figure 4. Hypusine Is Required for Polypro-line Elongation In Vitro.
(A–D) Translation elongation kinetics for the following tripeptide motifs in the presence and absence of eIF5A and hypusination modification: (A) Phe-Phe-Phe (MFFFK), (B) Pro-Pro-Pro (MPPPK), (C) Pro-Asp-Ile (MPDIK), (D) Asp-Asp-Ile (MDDIK). Time points were quantified and reaction progression fitted to a single exponential. See also Figure S4.
eIF5A Promotes Translation Termination
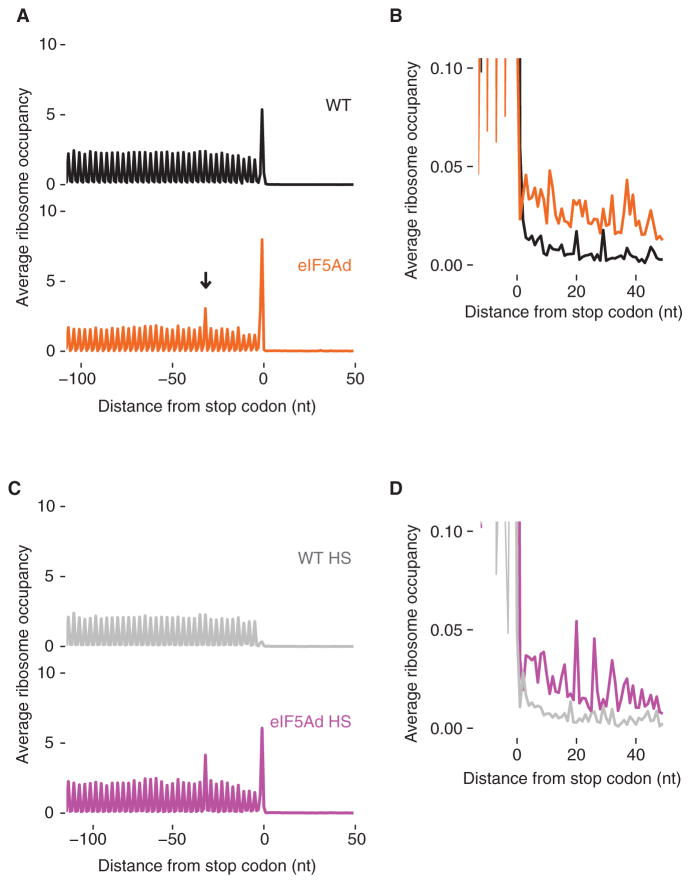
Recent biochemical and structural studies have shown that eIF5A binds to the E site of the ribosome where it stabilizes the peptidyl-tRNA for nucleophilic attack by the incoming amino acid on the A-site tRNA (Gutierrez et al., 2013; Melnikov et al., 2016; Schmidt et al., 2016). We wondered if eIF5A also promotes peptidyl-tRNA hydrolysis by eRF1, given that both of these reactions would benefit from a well-ordered pep-tidyl-tRNA and active site geometry. Turning again to our profiling data, we computed the average ribosome occupancy of all genes aligned at stop codons (Figure 5A) and observed several major differences between the WT and eIF5Ad strains. First, the stop codon peak in the eIF5Ad strain is ~2-fold higher than that seen in the WT strain; indeed, it is accompanied by a secondary peak ~30 nt upstream, presumably due to ribosomes stacked behind the terminating ribosome. Second, ribosome occupancy in the 3′ UTR is modestly increased (Figure 5B). Both of these observations are consistent with defects in translation termination in the eIF5Ad strain.
Figure 5. Ribosomes Accumulate at Stop Codons and in 3′ UTRs in eIF5Ad Cells.
(A) Metagene analysis of translation termination. Average ribosome occupancy from all genes aligned at their stop codons for WT and eIF5Ad cells. Arrow denotes stacked ribosomes ~30 nt upstream the stop codon peak.
(B) Overlay and close-up view of (A), showing accumulated ribosomes in 3′ UTRs in eIF5Ad cells.
(C) Similar to (A), metagene plot of stop codons for WT and eIF5Ad lysates treated with high-salt buffer.
(D) Similar to (B), close-up view of 3′ UTR regions for WT and eIF5Ad lysates after high-salt wash. See also Figure S5.
To ask whether ribosome occupancy at stop codons and in the 3′ UTR reflects elongating or scanning (post-termination) ribosomes, we treated lysates with a high-salt buffer that releases ribosomes lacking a nascent polypeptide chain (Blobel and Sabatini, 1971; Mills et al., 2016). As expected, elongating ribosomes in coding regions and the stacked ribosomes located ~30 nt upstream of stop codons (in eIF5Ad cells) are not sensitive to the high-salt wash. In contrast, the high-salt wash released ~90% of ribosomes at stop codons in WT cells (Figure 5C, top panel), suggesting that most ribosomes at stop codons have released their polypeptide chains and are waiting for the subunits to be split and recycled. On the other hand, only ~25% of ribosomes at stop codons were washed away in eIF5Ad cells (Figure 5C, bottom panel), suggesting that a majority of these ribosomes have yet to properly terminate and release the nascent peptide. After the high-salt wash, the stop codon peak in eIF5Ad cells is dramatically higher (~12-fold) than that seen in WT cells (Figure 5C). Importantly, eIF5Ad cells still show increased levels of 3′ UTR ribosomes after clearing by salt washing (Figure 5D), suggesting that the 3′ UTR-localized ribosomes in eIF5Ad cells are actually translating. These observations are consistent with the fact that defective termination can result in ribosome read-through or frameshifting at stop codons (Dever and Green, 2012; Pande et al., 1995; Stansfield et al., 1995).
We also asked whether eIF5A stimulates peptide release generally or if it has a stronger effect in specific contexts. First, we analyzed the effect of the final amino acid, as this has been shown to affect rates of peptide release (Björnsson et al., 1996; Woolstenhulme et al., 2013). We find that while eIF5A stimulates peptide release in all contexts, termination at certain C-terminal amino acids (e.g., Thr and Val; Figure S5A) is somewhat more affected by eIF5A. We also investigated the role of the nucleotide after the stop codon, a position that has been argued to be important for eRF1 binding (Brown et al., 1990, 2015; Matheisl et al., 2015), but we find no robust trends (Figure S5B). This finding is in line with our previous observation in vitro that the fourth nucleotide in the stop codon has only a minor effect on eRF1-mediated release (Eyler et al., 2013). Taken together, the profiling results point to globally inefficient termination in eIF5Ad cells, resulting in the accumulation of terminating ribosomes at stop codons and translating ribosomes in the 3′ UTR regions.
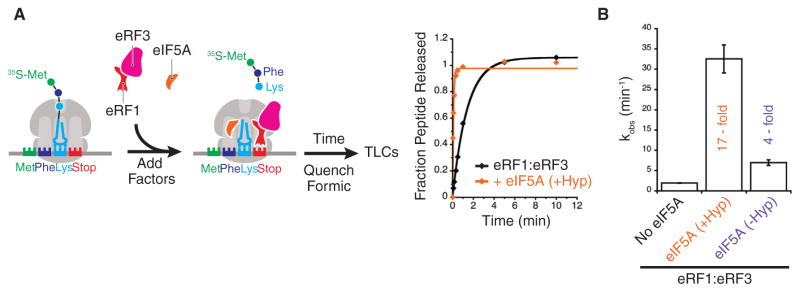
eIF5A Stimulates Peptidyl-tRNA Hydrolysis by eRF1 In Vitro
Given that termination is inhibited in cells when eIF5A is depleted, we wanted to ask whether eIF5A has a direct effect on the rate of peptidyl-tRNA hydrolysis using our in vitro system. We purified elongation complexes with [35S]-Met-Phe-Lys-tRNALys in the P site and a stop codon (UAA) in the A site, incubated them with eRF1:eRF3, and followed the rate of hydrolysis of the [35S]-Met-Phe-Lys (MFK) peptide from tRNALys over time (Figure 6A). When hypusinated eIF5A was added to the termination reaction with eRF1:eRF3, the rate of peptidyl-tRNA hydrolysis was stimulated 17-fold from a rate of 1.9 min−1 to 32.5 min−1 (Figure 6B). The hypusine modification of eIF5A is critical for this activity, as unmodified eIF5A has only a 4-fold effect on this same reaction. While our earlier work showed a minor effect (1.7-fold) of eIF5A in a peptide release assay (Saini et al., 2009), the dynamic range of our biochemical system has increased over time. Because these experiments were performed with saturating amounts of eIF5A (Figure S6), we conclude that the stimulatory effects that we document are at the level of kcat.
Figure 6. eIF5A Stimulates eRF1-Mediated Peptidyl Hydrolysis.
(A) Schematic for in vitro termination assays. Met-Phe-Lys elongation complexes containing an A-site stop codon (UAA) are reacted with eRF1:eRF3:GTP in the presence and absence of eIF5A. Time points are quenched with formic acid, peptide products resolved by electrophoretic TLCs, and rates quantified.
(B) Rates of peptidyl hydrolysis by eRF1:eRF3 in presence and absence of eIF5A and hypusination modification. Error bars represent the standard deviation from at least three replicate experiments. See also Figures S6 and S7.
Given that our elongation complexes were purified prior to the peptide hydrolysis assay by pelleting them over a sucrose cushion, we were concerned that the E-site tRNA might dissociate in the cushion, creating a situation where eIF5A binding to the E site might be more important than it is during elongation within cells. To rule out this possibility, we performed the same termination assay in a “one-pot” manner, omitting the pelleting step, and found that eIF5A still robustly stimulated the termination reaction (Figures S7A and S7B). These data suggest that the rate of E-site tRNA departure is fast enough in a typical elongation reaction to provide access to the E site for eIF5A. Finally, we performed the termination assay with a catalytically inactive GGQ→AGQ eRF1 to confirm that eIF5A was stimulating hydrolysis through the canonical eRF1-mediated reaction. Indeed, this eRF1 mutant completely abolishes peptidyl-tRNA hydrolysis even in presence of eIF5A (Figure S7C). Taken together, our in vitro biochemistry and ribosome profiling provide independent support that eIF5A plays a crucial role in translation termination by stimulating the catalytic activity of eRF1.
eIF5A Acts Globally while EFP Is Proline Specific
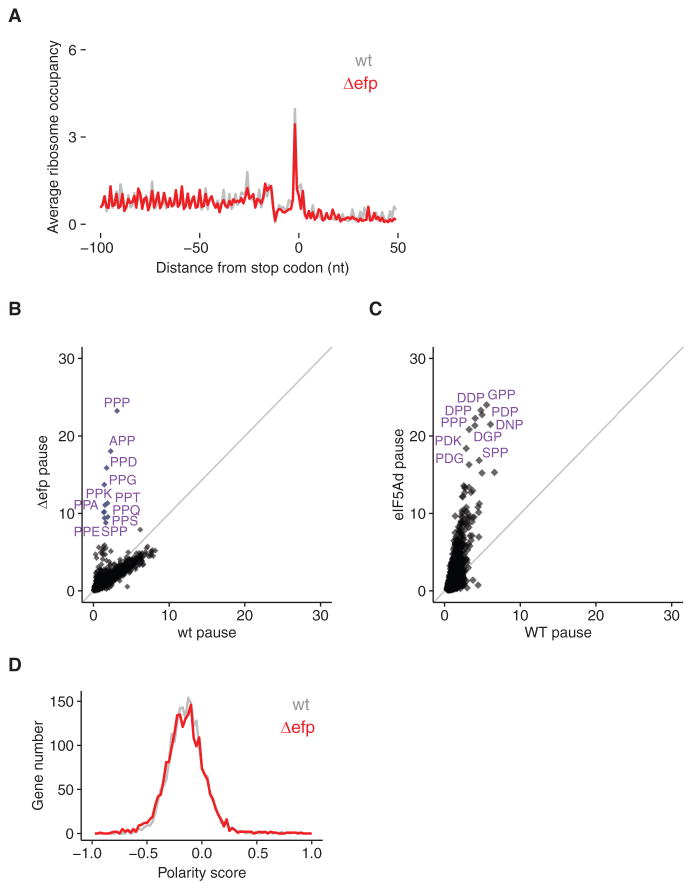
In light of this critical function for eIF5A in translation termination, we wondered if EFP might play a similar role in termination in bacterial translation. We performed the same analysis of stop codon-aligned ribosome footprints with data from Δefp cells (Woolstenhulme et al., 2015) and found no evidence that the loss of EFP enhances pausing at stop codons (Figure 7A). Additionally, recent biochemical work using an E. coli in vitro reconstituted translation system has shown that EFP does not stimulate the rate of release of fMet-Pro or fMet-Gly from tRNA by RF1 or RF2 (Pierson et al., 2016). These data argue that the function of eIF5A in translation termination is unique to eIF5A.
Figure 7. eIF5A Has a More General Role Than EFP.
(A) Metagene analysis of translation termination for WT and Δefp E. coli cells shows neither stacked ribosomes (~30 nt upstream of stop codons) nor 3′ UTR ribosomes.
(B) Comparison of tripeptide pausing in WT and Δefp E. coli cells. Pause scores of tripeptide motifs are plotted using E. coli WT and Δefp datasets (Woolstenhulme et al., 2015). Motifs with fewer than 30 occurrences in E. coli transcriptome are excluded from the analysis. Each dot represents one tripeptide motif; 6,018 motifs are included. Motifs with pause score higher than 8 are labeled. The diagonal line indicates the distribution expected for no enrichment.
(C) Comparison of tripeptide pausing in WT and eIF5Ad cells. Pause scores of 6,022 tripeptide motifs (Table S1) are plotted for WT and eIF5Ad cells, for motifs with more than 100 occurrences in yeast transcriptome. Ten tripeptide motifs with highest pause scores upon eIF5A depletion are labeled. The diagonal line indicates the distribution expected for no enrichment.
(D) Distributions of polarity scores for 2,186 genes from WT and Δefp E. coli cells are plotted, showing no significant difference. See also Figures S2 and S3.
We also revisited the activities of eIF5A and EFP in promoting peptidyl transfer during elongation by directly comparing their pausing spectra. It is well established that both proteins resolve pauses at polyproline stretches (Doerfel et al., 2013; Elgamal et al., 2014; Gutierrez et al., 2013; Hersch et al., 2013; Peil et al., 2013; Starosta et al., 2014; Ude et al., 2013; Woolstenhulme et al., 2015). In our eIF5A depletion strain, we identified many tripeptide motifs that pause the ribosome (Table S1), greatly expanding the scope of substrates that eIF5A acts on. Many of these sequences contain no proline at all, but rather combinations of aspartic acid, glycine, and other amino acids. Strong pausing at such motifs was not observed in previous studies of Δefp E. coli cells (Elgamal et al., 2014; Woolstenhulme et al., 2015). While the absolute intensity of pauses in these studies cannot be compared directly, differences in the relative enrichment of the pause scores in mutant strains lead to a striking observation. In Δefp cells, only the motifs with Pro-Pro lie off the diagonal and are enriched, compared to those motifs in wild-type E. coli cells (Figure 7B). In contrast, upon eIF5A depletion, the entire distribution of pausing motifs is reoriented more vertically, which is indicative of increased levels of pausing across a majority of tripeptide motifs in the absence of eIF5A (Figure 7C).
This observation explains the defect in global elongation rate upon eIF5A depletion that was previously reported (Saini et al., 2009) and that we observe by ribosome profiling (Figure 1C). In Δefp cells we find no evidence of a global redistribution of ribosomes (Figure 7D), but we identify genes that contain the strongest Pro-Pro dipeptide pausing motifs early in the gene to have an effect on the overall distribution of ribosome occupancy (Figure S2C). These comparisons argue that the global effects of eIF5A on translation elongation are unique to eIF5A, while the role of EFP is limited to translation of genes containing proline stretches.
DISCUSSION
eIF5A and EFP were originally identified because they promote formation of (f)Met-puromycin in a simplified system for studying translation (Benne and Hershey, 1978; Glick et al., 1979; Glick and Ganoza, 1975; Kemper et al., 1976; Schreier et al., 1977), and only later were they shown to promote translation elongation during Pro-Pro bond formation (Doerfel et al., 2013; Gutierrez et al., 2013; Ude et al., 2013). In this study, we report a much broader role for eIF5A in promoting elongation at many (perhaps all) sites in the transcriptome. From ribosome profiling of an eIF5A depletion strain, we identified many tripeptide motifs that substantially pause the ribosome (pause score >10) (Figure 2; Table S1) and showed that these widespread pauses lead to a global defect in elongation as observed by ribosome redistribution toward the 5′ end of genes (Figure 1). Our data help to explain prior observations of global defects in elongation (Saini et al., 2009) and support a model where eIF5A is a very general elongation factor. In contrast, EFP is much more specialized, limited to a role in enhancing the rate of translation of Pro-Pro motifs; the loss of EFP does not lead to global defects in elongation (Figure 7).
In this study we also uncovered a role for eIF5A in termination, increasing the rate of eRF1-mediated peptidyl-tRNA hydrolysis. In vivo, our ribosome profiling reveals a global defect in termination in the absence of eIF5A (Figure 5), while our previous study of E. coli cells lacking EFP shows no such effect (Figure 7). We also showed in vitro that eIF5A stimulates eRF1-mediated peptide release by 17-fold (Figure 6) and that this enhanced rate functions through canonical GGQ-hydrolysis (Figure S7C). In contrast to eIF5A, EFP was shown to have no function in RF1- or RF2-mediated peptidyl hydrolysis (Pierson et al., 2016). These observations support a model where eIF5A functions globally to stimulate slow reactions at the ribosomal peptidyl transferase center (PTC) while EFP is more specialized for several specific slow peptide motifs.
The differences in substrate specificity of these two proteins may be explained at a structural level. In the structure bound to the 70S ribosome, EFP makes extensive contacts with the P-site peptidyl-tRNA (Blaha et al., 2009). These contacts allow EFP to recognize specific substrate tRNAs containing a unique structural element in the D-arm of both tRNAPro and tRNAfMet (Katoh et al., 2016). In the recent structures of eIF5A bound in the ribosome (Melnikov et al., 2016; Schmidt et al., 2016), direct contact with the D-arm of the P-site tRNA is not observed. Based on the broader role for eIF5A we present here, including biochemical data to support a role for eIF5A function at many peptide sequences (Figures 3 and 4), we do not believe that eIF5A recognizes specific tRNAs as EFP does.
Previous biochemical probing and structural studies have shown that eIF5A binding in the ribosomal E site likely stabilizes the CCA-end of the peptidyl-tRNA (Gutierrez et al., 2013; Melnikov et al., 2016; Schmidt et al., 2016). These observations suggest that when a ribosome encounters problematic motifs (slow peptidyl transfer or eRF1-mediated hydrolysis), the ribosomal E site becomes vacant; prolonged E-site vacancy may allow eIF5A to bind and resolve ribosome pausing. From our data we find eIF5A to alleviate ribosome stalls that occur because of particular amino acid sequences, at the level of promoting slow peptide bond formation (Figures 2 and 3). Interestingly, these sequences are not selected against in S. cerevisiae genome (data not shown). While other mechanisms of ribosome stalling have been described involving codon optimality or tRNA identity/abundance, we find no evidence for these types of stalling in our data. For example, we analyzed our profiling data after eIF5A depletion for any stalling effects correlated with codon optimality (Radhakrishnan et al., 2016; Sabi and Tuller, 2014) and saw no correlation (Figure S3C). Similarly, we found no evidence for stalling at recently identified codon pairs that inhibit translation (Gamble et al., 2016) (Figure S3D; data not shown). Taken together, our data suggest that eIF5A is specific for resolving stalls due to slow peptidyl transfer at the ribosomal PTC in the 60S subunit, rather than any events related to the efficiency of tRNA binding or decoding.
Our reported function for eIF5A in termination, in addition to its role in alleviating widespread elongation pausing, may account for both its great abundance and its essential nature in eukaryotic cells. In S. cerevisiae, eIF5A is one of the most well-expressed proteins with >273,000 copies per cell (Kulak et al., 2014). In fact, eIF5A is in the top 50 highest expressing genes in S. cerevisiae, S. pombe, and HeLa cells (Kulak et al., 2014), with levels equivalent to that of ribosomes (von der Haar, 2008). In addition to its abundance, eIF5A interacts with 80S ribosomes very strongly, with an approximate dissociation constant of 9 nM (Rossi et al., 2016), suggesting that it could interact with all ribosomes that have an empty E site (eIF5A cellular concentration is approximately 8–15 μM). Given its critical and diverse roles in both translation elongation and termination, these observations lead us to suspect that most, if not all, ribosomes interact with eIF5A throughout the elongation and termination phases of translation.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mAID | MBL International Corporation | M214-3 |
| Anti-Pgk1 monoclonal | Life Technologies-Novex | 459250 |
| Anti-eIF5A | Dr. Thomas Dever | Saini et al., 2009 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cycloheximide | Sigma-Aldrich | C1988 |
| SuperaseIn | Ambion | AM2694 |
| RNase I | Ambion | AM2294 |
| Superscript III | Invitrogen | 56575 |
| GlycoBlue | ThermoFisher | AM9515 |
| Protease Inhibitor Cocktail | Sigma-Aldrich | P8215 |
| IAA (Indole-3-acetic acid sodium salt) | Sigma-Aldrich | I5148 |
| T4 Polynucleotide Kinase | New England Biosciences | M0201L |
| T4 RNA Ligase 2, Truncated | New England Biosciences | M0242L |
| CircLigase ssDNA Ligase | Epicenter | CL4115K |
| Zeba spin desalting column (2 mL) | ThermoFisher | 89889 |
| TLC cellulose plate | EMD Millipore | 1055770001 |
| Ni-NTA agarose | QIAGEN | 30210 |
| HiPrep 26/60 Sephacryl S200 | GE Healthcare Life Sciences | 17119501 |
| Superose Increase 6 10/300GL | GE Healthcare Life Sciences | 29091596 |
| tRNAPhe (yeast) | Sigma | R4018 |
| Initiator tRNAiMet (yeast) | tRNA Probes (College Station, TX) | MI-03 |
| Lys-tRNALys (yeast) | tRNA Probes (College Station, TX) | L-60 |
| Cys-tRNACys (yeast) | tRNA Probes (College Station, TX) | C-60 |
| EasyTag L-35S-Methionine | Perkin-Elmer | NEG709A005MC |
| Critical Commercial Assays | ||
| Ribo-Zero Gold rRNA Removal Kit (Yeast) | Illumina | MRZY1306 |
| IMPACT Kit | New England Biosciences | E6901S |
| BL21-CodonPlus (DE3)-RIPL | Agilent | NC9122855 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE89704 |
| R64-1-1 S288C sacCer3 Genome Assembly | Saccharomyces Genome Database Project | http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/ |
| Yeast ncRNA Gene Database | Saccharomyces Genome Database Project | http://downloads.yeastgenome.org/sequence/S288C_reference/rna/archive/rna_coding_R64-1-1_20110203.fasta.gz |
| Mendeley Database | This paper | http://dx.doi.org/10.17632/nmkd3jbhx7.1 |
| Experimental Models: Organisms/Strains | ||
| BY4741 | Dharmacon | YSC1048 |
| Δanb1 | Dharmacon | Clone ID: 6845 |
| BY4741; AHD1p-OsTIR1-URA3 | This paper | yCW30 – WT |
| Δanb1; Δhyp2; mAID-HYP2-pAG413GAL; AHD1p-OsTIR1-URA3 | This paper | yCW33 – eIF5Ad |
| Oligonucleotides | ||
| Please refer to Table S3 for oligonucleotides used in this study | N/A | |
| Software and Algorithms | ||
| Bowtie 1.0.0 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net |
| CutAdapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| ImageQuant TL | GE Healthcare Life Sciences | Version 8.1 |
| Kaleidagraph | Synergy Software | Version 4.1.3 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Please direct any requests for further information or reagents to the lead contact, Rachel Green (ragreen@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains and growth conditions
As shown in Figure S1, WT and eIF5Ad strains were grown in YPGR (0.2% galactose+ 0.2% raffinose) overnight at 30°C, harvested by centrifugation, washed and resuspended in YPD (0.2% glucose) medium containing 0.5 mM auxin (Sigma) to an OD600 of 0.003 for WT and 0.15 for eIF5Ad. Both WT and eIF5Ad cells were grown in the presence of auxin for 10 hr (to deplete eIF5A completely), harvested by fast filtration, and then flash frozen in liquid nitrogen. It took ~1 min to collect ~800 mL culture. Cell pellets were ground with droplets of footprint lysis buffer [20 mM Tris-Cl (pH8.0), 140 mM KCl, 1.5 mM MgCl2, 1% Triton X-100, 0.1 mg/mL cycloheximide] in a Spex 6870 freezer mill.
METHOD DETAILS
Yeast strain construction
ADH1p-OsTIR1 cassette was amplified from pMK200 (Nishimura and Kanemaki, 2014) and inserted in HO locus in BY4741 and Δanb1 strains (GE Dharmacon), resulting in yCW30 (WT strain: MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 HO::ADH1p-OsTIR1-URA3) and yCW31 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 anb1::KanMX HO::ADH1p-OsTIR1-URA3)), respectively. After transforming mAID-HYP2-pAG413GAL into yCW31, the genomic copy of hyp2 was deleted by a NatMX4 cassette, resulting in yCW33 (eIF5Ad strain: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 HO::adh1p-OsTIR1-URA3 anb1::KanMX hyp2::NatMX4 mAID-HYP2-pAG413GAL).
Preparation of ribosome footprint libraries
After clarification, 25 OD260 units of lysates were treated with 375 units of RNaseI (Ambion) for 1 hr at RT. For high-salt wash experiments, same amount of lysates were treated with 1 M KCl (final conc.) on ice for 15 min, and desalted by passing through desalting a column (Zeba spin, Thermo Fisher Scientific). The resulting cell lysates were then treated with RNaseI as mentioned above. Monosomes were isolated by sucrose gradients. The extracted RNA was size-selected from 15% denaturing PAGE gels, cutting between 15–34 nt. An oligonucleotide adaptor was ligated to the 3′ end of footprints. After rRNA depletion using RiboZero (Illumina), reverse transcription, circularization and PCR amplification. Footprint libraries were sequenced on a HiSeq2500 machine at facilities at Johns Hopkins Institute of Genetic Medicine.
Purification of translation factors
Translation initiation factors eIF2, eIF1, eIF1A, eIF5, and eIF5b were purified from both S. cerevisiae (eIF2) and E. coli using reported procedures (Acker et al., 2007). Purification of eEF1A from S. cerevisiae also followed a previously reported protocol (Eyler and Green, 2011).
During purification of eEF2 and eEF3 from S. cerevisiae we identified the presence of low-level (though highly active) contaminating eIF5A. Endogenous eIF5A is very abundant, and can be enriched from yeast lysate by both Ni-NTA and amylose affinity chromatography (see Figure S6 for yeast-purified His-MBP activity). In order to isolate eEF2 and eEF3 in the absence of eIF5A, we took great care to sacrifice any contaminated fractions during purification to yield a highly pure protein. After purification, individual fractions were tested for MPPK synthesis in the absence of eIF5A as a test for contamination. Those fractions that did not stimulate MPPK formation were used in subsequent assays.
eEF2 was purified using a C-terminal His6 tag from strain TKY675 (Jørgensen et al., 2002). eEF2 was purified by Ni-NTA chromatography in buffer containing 20 mM HEPES pH 7.4, 10% glycerol, 300 mM KCl, and 5 mM 2-mercaptoethanol. Then the protein was diluted to 30 mM KCl and further purified by cation exchange chromatography using a gradient from 30–150 mM KCl. Fractions containing eEF2 (and no eIF5A) were pooled, concentrated, and further purified by gel filtration using a HiPrep 26/60 Sephacryl S-200 column (GE Healthcare Life Sciences) for best separation of eEF2 and contaminant eIF5A. Fractions containing eEF2 were individually concentrated, and stored in gel filtration buffer containing 20 mM HEPES pH 7.4, 100 mM KOAc pH 7.6, 10% glycerol, and 2 mM DTT.
eEF3 was purified using a N-terminal His6 tag from strain TKY702 (Andersen et al., 2004). eEF3 was purified by Ni-NTA chromatography in buffer containing 25 mM Tris pH 7.5, 10 mM Imidazole, 1 M KCl, 10% glycerol, and 5 mM 2-mercaptoethanol. After elution, protein was further purified by gel filtration using a Superose 6 10/300GL column (GE Healthcare Life Sciences). Fractions containing eEF3 were individually concentrated, and stored in gel filtration buffer containing 20 mM HEPES pH 7.4, 100 mM KOAc pH 7.6, 10% glycerol, and 2 mM DTT.
Eukaryotic release factor eRF1 (and AGQ mutant) was purified from E. coli with a His6 tag as previously described (Shoemaker et al., 2010). Release factor eRF3 was purified from E. coli using the IMPACT Protein Purification System (New England Biolabs) as previously described (Eyler et al., 2013).
Expression and purification of eIF5A
Expression and purification of recombinant eIF5A from E. coli was performed as previously reported (Gutierrez et al., 2013). Briefly, a plasmid expressing His6-eIF5A, or a plasmid co-expressing His6-eIF5A with modification enzymes Dys1 and Lia1 was transformed into BL21-CodonPlus (DE3)-RIPL cells (Agilent). Proteins were expressed and purified by Ni-NTA affinity chromatography and anion exchange chromatography.
To confirm eIF5A hypusination, proteins were analyzed by MALDI-TOF mass spectrometry (Voyager DE-STR MALDI-TOF). As shown in Figure S4B, unmodified eIF5A purified from E. coli showed a major peak with a calculated weight of 17804, consistent with the predicted mass of unmodified His6-eIF5A. Analysis of His6-eIF5A purified from E. coli expressing Dys1 and Lia1 modification enzymes revealed a shift in molecular weight to 17901, consistent with modification.
Purification of tRNAs and mRNAs
Phenylalanine-specific tRNA from S. cerevisiae was purchased from Sigma. Initiator methionine, lysine and cysteine-specific tRNAs were purchased from tRNA Probes (College Station, TX). Proline, aspartic acid, and isoleucine tRNAs were isolated from S. cerevisiae bulk tRNA using 3′ biotinylated oligos (IDT) as previously described (Yokogawa et al., 2010).
Oligo for tRNAPro(UGG): 5′ – CCAAAGCGAG AATCATACCA CTAGAC – 3′ Biotin-TEG. Oligo for tRNAAsp(GUC): 5′ – GACAAG CGCCCATTCTGACCATTAAAC – 3′ Biotin-TEG.
Oligo for tRNAIle(AAU): 5′ – ATTAGCACGGTGCCTTAACCAACTGGGC – 3′ Biotin-TEG.
Isolated tRNAPro, tRNAAsp, and tRNAIle were subject to CCA-adding reaction (Gutierrez et al., 2013). All tRNAs were charged using a S100 extract (Eyler and Green, 2011).
Model mRNAs for in vitro translation were transcribed using T7 RNA polymerase and contain the following sequence: 5′ – GAAU CUCUCUCUCUCUCU AUG XXX XXX XXX UAA CUCUCUCUCUCUCUC – 3′ (underlined G is T7 start, bold is open reading frame, XXX = generic codon). In elongation experiments, codons used are as follows: Phe (UUC), Pro (CCA), Asp (GAC), Ile (AUU), Cys (UGC), and Lys (AAA). All mRNAs were gel purified using 10% TBE-Urea gels.
In vitro 80S initiation and elongation complex formation
80S initiation complexes were formed as previously described (Eyler and Green, 2011) with minor differences. Briefly, 3 pmol of 35S-Met-tRNAiMet was mixed with 25 pmol of eIF2 and 1 mM GTP in 1X Buffer E (20 mM Tris pH 7.5, 100 mM KOAc pH 7.6, 2.5 mM Mg(OAc)2, 0.25 mM Spermidine, and 2 mM DTT) for 10 min at 26°C. Next a mixture containing 25 pmol 40S subunits, 200 pmol T7-synthesized mRNA template, 150 pmol eIF1, and 150 pmol eIF1A in 1X Buffer E was added for 5 min. To form the 80S complex, a mixture containing 25 pmol 60S subunits, 125 pmol eIF5, 125 pmol eIF5b, and 1 mM GTP in 1X Buffer E was added for 1 min. Complexes were then mixed 1:1 with buffer E containing 17.5 mM Mg(OAc)2 to yield a final magnesium concentration of 10 mM. Ribosomes were then pelleted through a 600 μL sucrose cushion containing 1.1 M sucrose in buffer E with 10 mM Mg(OAc)2 using a MLA-130 rotor (Beckmann) at 75,000 rpm for 1 hr at 4°C. After pelleting, ribosomes were resuspended in 25 μL of 1X Buffer E containing 10 mM Mg(OAc)2 and stored at −80°C.
To form elongation complexes for peptide release experiments, initiation complexes were formed as described above and elongated before pelleting. To create the eEF1A ternary complex, a mixture containing 50 pmol eEF1A, 30 pmol aa-tRNA, and 1mM GTP was incubated at 26°C for 15 min. Ternary complex for each required tRNA was mixed with 80S initiation complexes for 5 min to allow for peptidyl transfer. Subsequently a mixture containing 50 pmol eEF2, 75 pmol eEF3, 2.5 mM GTP, and 5.0 mM ATP was added to promote elongation and subsequent rounds of peptidyl transfer for 7 min. Elongated complexes were then mixed 1:1 with buffer E containing 17.5 mM Mg(OAc)2 and pelleted as described above.
In vitro reconstituted translation elongation
Translation elongation reactions were performed in 1X Buffer E (20 mM Tris pH 7.5, 100 mM KOAc pH 7.6, 2.5 mM Mg(OAc)2, 0.25 mM Spermidine, and 2 mM DTT). Limited amounts of 80S initiation complexes (3 nM) were mixed with purified eEF1A (1 μM), aa-tRNA (500 nM), eEF2 (1 μM), eEF3 (1 μM), ATP (3 mM) and GTP (2 mM) in the presence or absence of eIF5A (1 μM). Reactions were incubated at 26°C and time points quenched with 250 mM KOH. Peptide formation was monitored by electrophoretic TLC (Millipore). TLC plates were equilibrated with pyridine acetate buffer (5 mL pyridine, 200 mL acetic acid in 1 l, pH 2.8) before electrophoresis at 1200 V for 28 min. Plates were developed using a Typhoon FLA 9500 Phosphorimager system (GE Healthcare Life Sciences) and quantified using ImageQuantTL (GE Healthcare Life Sciences). Time courses were fit to single exponential kinetics using Kaleidagraph (Synergy Software).
In vitro reconstituted eRF1:eRF3 peptidyl hydrolysis
Peptide release assays were performed in 1X Buffer E (same as elongation reactions). Limited amounts of 80S elongation complexes (3 nM) were mixed with purified eRF1 (4 μM), eRF3 (6 μM) and GTP (1 mM) in the presence or absence of eIF5A (1 μM). Reactions were incubated at 26°C and time points quenched with 5% formic acid. TLC plates were equilibrated with pyridine acetate buffer (5 mL pyridine, 200 mL acetic acid in 1 l, pH 2.8) before electrophoresis at 1200 V for 18 min. Plates were developed, quantified, and fit to single exponential kinetics as described for elongation reactions.
In vitro Met-Puromycin assay
Reactions containing 2 nM initiation complexes and 1 μM eIF5A in 1X Buffer E (20 mM Tris pH 7.5, 100 mM KOAc pH 7.6, 2.5 mM Mg(OAc)2, 0.25 mM Spermidine, and 2 mM DTT) were incubated at 26°C in the presence of 5 mM puromycin. Time points over the course of 90 min were quenched with 250 mM KOH and analyzed by electrophoretic TLC (Millipore). TLC plates were equilibrated with pyridine acetate buffer (5 mL pyridine, 200 mL acetic acid in 1 l, pH 2.8) before electrophoresis at 1200 V for 15min. Plates were developed using a Typhoon FLA 9500 Phosphorimager system (GE Healthcare Life Sciences) and quantified using ImageQuantTL (GE Healthcare Life Sciences). Time courses were fit to single exponential kinetics using Kaleidagraph (Synergy Software).
In vitro analysis of peptidyl-tRNA drop-off using PTH
Translation elongation reactions were performed as described in the primary methods in the presence of 27 μM peptidyl-tRNA hydrolase (PTH) to monitor drop-off of peptidyl-tRNAs from translating ribosomes. PTH can only cleave peptidyl-tRNAs that have been fully released from the ribosome (Figure S4D, note zero time point). Time points for drop-off products were quenched with 10% formic acid, and peptide formation was monitored by KOH quench at the start and end of the reaction. Reaction time points were analyzed by electrophoretic TLC in pyridine acetate buffer (see above) at 1200 V for 30 min.
QUANTIFICATION AND STATISTICAL ANALYSIS
Read preparation and alignment
The R64-1-1 S288C reference genome assembly (SacCer3) from the Saccharomyces Genome Database Project was used for all analyses. Adaptor sequence (CTGTAGGCACCATCAAT) was first removed from demultiplexed reads using Cutadapt (Martin, 2011), and low quality reads (any position with Phred score less than 20) were discarded. Then alignment to the RNA gene database FASTA file (http://downloads.yeastgenome.org/sequence/S288C_reference/rna/archive/rna_coding_R64-1-1_20110203.fasta.gz) was performed to remove noncoding RNAs. The resulting reads were then aligned to the genome using 3′ end mapping, and the reads that failed to be mapped were aligned to the annotated splice junctions Finally, reads left were trimmed of consecutive As from their 3′ ends and realigned to the genome and the splice junction. Read length between 25–34 nt was used for the following analyses. The mapped reads were normalized to reads per million (rpm) using total number of mapped reads. Alignment was performed using Bowtie 1.0.0 (Langmead et al., 2009) using the following parameters: ‘-v 2 –y –a –m 1 –best –strata –S –p 4’. All other analyses were performed using software custom written in Python 2.7 and R 3.3.1.
Ribosome profiling datasets for Δefp (Woolstenhulme et al., 2015) were downloaded from the GEO (GSE64488) and the Escherichia coli MG1655 reference genome NC_000913.2 was used for all E. coli ribosome profiling analyses.
Analysis of aligned reads
Calibrated by using start and stop codons of coding sequences, we defined that the first 5′ end nucleotide of ribosome A site to the 3′ end of the footprint is separated by 12 nt. In general, −14 shift (the center of P site) is used for start codons (Figure 1A), pause motifs and quantitation of coding sequences (Figures 1C, 1D, 2, and 7), and −11 shift (center of ribosomal A site) is used for stop codons (Figure 5).
The polarity at position i in a gene of length l is defined as follows:
where
The terms di and wi are the ribosome density and the normalized distance from the center of a gene at position i, respectively. Polarity score for a gene is the total sum of pi at each position. To avoid known artifacts around start and stop codon peaks, we excluded the first and the last 15 nt of coding sequences from our analysis. Genes with more than 64 reads/dataset in coding sequences were plotted.
Pause scores for peptide motifs (Figures 2D, 7B, and 7C) were calculated by taking the ratio of the ribosome density in a 3-nt window at the motif over the overall density in the coding sequence (excluding the first and the last 100 nt). Genes with less than 64 reads/dataset in coding sequences (excluding the first and the last 100 nt) are excluded from the analysis. The same threshold was used for metagene plots (Figures 1A and 5) and metacodon plots (Figures 2A, 2B, and 2E). Logo was weighted by pause score ratio of eIF5Ad/WT (Table S1). For metagene plots, ribosome density at each position is normalized by the overall ribosome density of coding sequence. Genes with features that are smaller than the window size (50 nt upstream of start codons and 900 nt of coding sequences for Figure 1A, and 100 nt upstream of stop codons and 50 nt of 3′ UTRs for Figure 5) were excluded from the analysis.
DATA AND SOFTWARE AVAILABILITY
Accession numbers
Sequencing data were deposited in the GEO database under the accession number GSE89704.
TLC and gel images are deposited in the Mendeley database at the following link: http://dx.doi.org/10.17632/nmkd3jbhx7.1.
Supplementary Material
Highlights.
Depletion of eIF5A leads to global translation elongation and termination defects
eIF5A alleviates stalling on many motifs besides polyproline tracts
eIF5A stimulates eRF1-mediated peptidyl-tRNA hydrolysis in translation termination
Acknowledgments
We would like to thank members of the Green lab for their helpful discussions. This work was supported by the NIH (GM110113 to A.R.B. and GM059425 to R.G.) and HHMI (R.G.).
Footnotes
Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.03.003.
AUTHOR CONTRIBUTIONS
A.P.S. performed the in vitro biochemical experiments. C.C.-C.W. performed the ribosome profiling experiments. A.R.B. and T.E.D. initiated early ribosome profiling experiments. A.P.S., C.C.-C.W., A.R.B., and R.G. wrote the manuscript with input from T.E.D.
References
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- Andersen CF, Anand M, Boesen T, Van LB, Kinzy TG, Andersen GR. Purification and crystallization of the yeast translation elongation factor eEF3. Acta Crystallogr D Biol Crystallogr. 2004;60:1304–1307. doi: 10.1107/S0907444904010716. [DOI] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014;24:2011–2021. doi: 10.1101/gr.175893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Björnsson A, Mottagui-Tabar S, Isaksson LA. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci USA. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524:493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Gutierrez E, Shin BS. The hypusine-containing translation factor eIF5A. Crit Rev Biochem Mol Biol. 2014;49:413–425. doi: 10.3109/10409238.2014.939608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Elgamal S, Katz A, Hersch SJ, Newsom D, White P, Navarre WW, Ibba M. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 2014;10:e1004553. doi: 10.1371/journal.pgen.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Green R. Distinct response of yeast ribosomes to a miscoding event during translation. RNA. 2011;17:925–932. doi: 10.1261/rna.2623711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Wehner KA, Green R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem. 2013;288:29530–29538. doi: 10.1074/jbc.M113.487090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016;166:679–690. doi: 10.1016/j.cell.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Chládek S, Ganoza MC. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur J Biochem. 1979;97:23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Malcova I, Pompach P, Frydlova I, Slaba R, Senohrabkova L, Novakova L, Hasek J. Heat shock-induced accumulation of translation elongation and termination factors precedes assembly of stress granules in S. cerevisiae. PLoS ONE. 2013;8:e57083. doi: 10.1371/journal.pone.0057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A, Hershey JW. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108:6415–6419. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SJ, Wang M, Zou SB, Moon KM, Foster LJ, Ibba M, Navarre WW. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio. 2013;4:e00180–e13. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen R, Carr-Schmid A, Ortiz PA, Kinzy TG, Andersen GR. Purification and crystallization of the yeast elongation factor eEF2. Acta Crystallogr D Biol Crystallogr. 2002;58:712–715. doi: 10.1107/s0907444902003001. [DOI] [PubMed] [Google Scholar]
- Katoh T, Wohlgemuth I, Nagano M, Rodnina MV, Suga H. Essential structural elements in tRNA(Pro) for EF-P-mediated alleviation of translation stalling. Nat Commun. 2016;7:11657. doi: 10.1038/ncomms11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–12. [Google Scholar]
- Matheisl S, Berninghausen O, Becker T, Beckmann R. Structure of a human translation termination complex. Nucleic Acids Res. 2015;43:8615–8626. doi: 10.1093/nar/gkv909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Shin BS, Rigger L, Yusupova G, Micura R, Dever TE, Yusupov M. Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J Mol Biol. 2016;428:3570–3576. doi: 10.1016/j.jmb.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Wangen J, Green R, Ingolia NT. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 2016;17:1–10. doi: 10.1016/j.celrep.2016.08.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kanemaki MT. Rapid depletion of budding yeast proteins via the fusion of an auxin-inducible degron (AID) Curr Protoc Cell Biol. 2014;64:20.9.1–20.9.16. doi: 10.1002/0471143030.cb2009s64. [DOI] [PubMed] [Google Scholar]
- Pande S, Vimaladithan A, Zhao H, Farabaugh PJ. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol Cell Biol. 1995;15:298–304. doi: 10.1128/mcb.15.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH. The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J Biol Chem. 1989;264:18531–18535. [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Wolff EC, Smit-McBride Z, Hershey JW, Folk JE. Comparison of the activities of variant forms of eIF-4D. The requirement for hypusine or deoxyhypusine. J Biol Chem. 1991;266:7988–7994. [PubMed] [Google Scholar]
- Park JH, Dias CA, Lee SB, Valentini SR, Sokabe M, Fraser CS, Park MH. Production of active recombinant eIF5A: reconstitution in E.coli of eukaryotic hypusine modification of eIF5A by its coexpression with modifying enzymes. Protein Eng Des Sel. 2011;24:301–309. doi: 10.1093/protein/gzq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Lassak J, Atkinson GC, Virumäe K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci USA. 2013;110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson WE, Hoffer ED, Keedy HE, Simms CL, Dunham CM, Zaher HS. Uniformity of peptide release is maintained by methylation of release factors. Cell Rep. 2016;17:11–18. doi: 10.1016/j.celrep.2016.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen YH, Martin S, Alhusaini N, Green R, Coller J. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell. 2016;167:122–132. e9. doi: 10.1016/j.cell.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Barbosa NM, Galvão FC, Boldrin PE, Hershey JW, Zanelli CF, Fraser CS, Valentini SR. Evidence for a negative cooperativity between eIF5A and eEF2 on binding to the ribosome. PLoS ONE. 2016;11:e0154205. doi: 10.1371/journal.pone.0154205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabi R, Tuller T. Modelling the efficiency of codon-tRNA interactions based on codon usage bias. DNA Res. 2014;21:511–526. doi: 10.1093/dnares/dsu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Becker T, Heuer A, Braunger K, Shanmuganathan V, Pech M, Berninghausen O, Wilson DN, Beckmann R. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2016;44:1944–1951. doi: 10.1093/nar/gkv1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J Mol Biol. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I, Akhmaloka, Tuite MF. A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr Genet. 1995;27:417–426. doi: 10.1007/BF00311210. [DOI] [PubMed] [Google Scholar]
- Starosta AL, Lassak J, Peil L, Atkinson GC, Woolstenhulme CJ, Virumäe K, Buskirk A, Tenson T, Remme J, Jung K, Wilson DN. A conserved proline triplet in Val-tRNA synthetase and the origin of elongation factor P. Cell Rep. 2014;9:476–483. doi: 10.1016/j.celrep.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, O’Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- von der Haar T. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol. 2008;2:87. doi: 10.1186/1752-0509-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 2016;14:1787–1799. doi: 10.1016/j.celrep.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 2015;11:13–21. doi: 10.1016/j.celrep.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T, Kitamura Y, Nakamura D, Ohno S, Nishikawa K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010;38:e89. doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3’UTRs in vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.