Abstract
In the dairy industry, cow health and farmer profits depend on the balance between diet (ie, nutrient composition, daily intake) and metabolism. This is especially true during the transition period, where dramatic physiological changes foster vulnerability to immunosuppression, negative energy balance, and clinical and subclinical disorders. Using an Agilent microarray platform, this study examined changes in the transcriptome of bovine polymorphonuclear leukocytes (PMNLs) due to prepartal dietary intake. Holstein cows were fed a high-straw, control-energy diet (CON; NEL = 1.34 Mcal/kg) or overfed a moderate-energy diet (OVE; NEL = 1.62 Mcal/kg) during the dry period. Blood for PMNL isolation and metabolite analysis was collected at −14 and +7 days relative to parturition. At an analysis of variance false discovery rate <0.05, energy intake (OVE vs CON) influenced 1806 genes. Dynamic Impact Approach bioinformatics analysis classified treatment effects on Kyoto Encyclopedia of Genes and Genomes pathways, including activated oxidative phosphorylation and biosynthesis of unsaturated fatty acids and inhibited RNA polymerase, proteasome, and toll-like receptor signaling pathway. This analysis indicates that processes critical for energy metabolism and cellular and immune function were affected with mixed results. However, overall interpretation of the transcriptome data agreed in part with literature documenting a potentially detrimental, chronic activation of PMNL in response to overfeeding. The widespread, transcriptome-level changes captured here confirm the importance of dietary energy adjustments around calving on the immune system.
Keywords: Transition cow, gene expression, neutrophil, nutrigenomics
Introduction
The transition into lactation is a volatile period in dairy cow health, largely due to the shifts in energy demands and nutrient partitioning that occur after calving.1,2 Traditionally, a depression in dry matter (DM) intake is observed postpartum, although cow energy requirements increase dramatically to support milk production. The result is a negative energy balance (NEB) that can precede a host of health complications.3 Appropriate nutritional management throughout the lactation cycle, especially preceding calving (ie, the dry period), is therefore essential to ensuring short-term and long-term herd health and productivity.
In the past, nutritional strategies to alleviate postpartum NEB included overfeeding by increasing prepartum dietary energy.3,4 Although prepartum energy intake of dairy cows does influence postpartum health, recent studies have revealed that it tends to have a negative rather than positive impact. Overfeeding energy prepartum can deepen NEB and increase fat mobilization and deposition in liver, ketone production, and incidence of metabolic disorders, whereas controlling energy intake improves these outcomes.5–7 Some studies also suggest that milk yield8 and reproductive measures4,9 following calving are enhanced when prepartum energy is restricted. Prepartal dietary intake may even yield more substantial metabolic effects than dietary composition.10
In addition, recent studies demonstrate that transition period adaptations can compromise the immune system, further exposing cows to postpartum disorders and increased susceptibility to disease.11 Diversion of maintenance energy in favor of lactation, as well as increasing cortisol and estrogen levels, may drive immunosuppression near calving, leaving cows susceptible to pathogens.12,13 Therefore, the importance of researching the immune system in peripartal cows cannot be understated. As first responders of the innate immune system and coordinators of the adaptive immune response,14 polymorphonuclear leukocytes (PMNLs) are representative immune cells. Understanding their behavior during the transition period may provide insights into cow health status.
Although some transcriptome-level work evaluating bovine PMNL has already been conducted,12,15,16 these have largely focused on the effect of parturition or parturition-related conditions, eg, glucocorticoid levels. As proposals for new transition cow management strategies arise, it is necessary to evaluate the PMNL transcriptome in context of nutrition and management trials, such that immunologic health is maintained or improved with shifting practices. The present data were therefore generated from PMNL in cows fed higher or control-energy diets to illustrate the impacts of current dry period diets (ie, greater energy intake) versus emerging recommendations (lower energy intake) on immunity during transition. We hypothesized that adaptation of PMNL in overfed cows would consist of significant transcription-level changes.
Materials and Methods
Animals and treatments
Cows used for this study were from the overfeeding study of Khan et al, and details of animal care and treatment have been outlined in previous publications.17,18 Briefly, 8 cows were assigned to 1 of the 2 groups which differed in dietary energy levels fed throughout the 45-day dry period. One group received at least 100% calculated NEL (CON; 1.34 Mcal/kg DM) from a diet high in wheat straw, whereas the other received over 140% calculated NEL (OVE; 1.62 Mcal/kg DM) from a corn silage–based diet. Both diets were fed once daily (0600 hours) using an individual gate feeding system (American Calan, Northwood, NH, USA). All cows were housed in a ventilated, enclosed barn for the entire dry period. After calving, cows were fed a common lactation diet (NEL = 1.69 Mcal/kg DM) as total mixed ration once daily (0600 hours) and housed in a tie-stall barn. Milking occurred twice daily (0400 and 1600 hours).
PMNL isolation and RNA isolation
The specific details of these procedures are included in the Supplemental File.
Microarrays
Complementary RNA synthesis and labeling, fragmentation, hybridization, and slide scanning
The specific details of these procedures are included in the Supplemental File. Array data are publicly available in the Gene Expression Omnibus database (Series ID: GSE95677).
Statistical analysis
Gene analysis
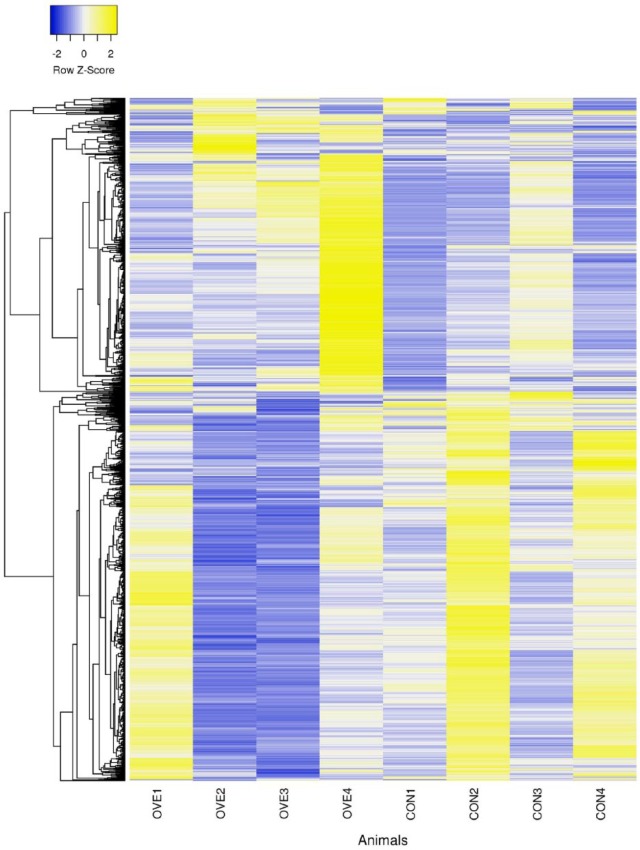
Microarray statistics were analyzed using SAS (SAS Institute Inc., Cary, NC, USA). Data from the 8 microarrays were adjusted using Lowess normalization and array centering to account for dye and array effects. A mixed model with repeated measures was fitted to the normalized log2-transformed adjusted ratios using PROC MIXED. The model included fixed effects of time (−14 and +7 days), diet (CON and OVE), and interaction of time × diet. Cow was a random effect. Raw P values were adjusted to account for multiple comparisons using Benjamini and Hochberg’s false discovery rate (FDR).19 Significant differences in gene expression were considered at FDR < 0.05. Only genes with degree of freedom ⩾7 were considered. The signal intensity (average between days −14 and +7) of the differentially expressed genes (DEGs) for each animal was represented as a heat map (Figure 1). Genes were clustered using the average linkage method, with Pearson coefficient as distance measurement. Results below will focus on transcriptome differences due to the main effect of diet.
Figure 1.
Heat-map representation of signal intensity (average days −14 and +7) for differentially expressed genes (DEGs) in polymorphonuclear leukocytes of individual cows overfed energy (OVE) versus control fed (CON). The DEGs were clustered using the average linkage method, with distances calculated as Pearson coefficients.
Pathway analysis
The Dynamic Impact Approach (DIA) was used to analyze Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on DEGs between dietary groups (OVE vs CON). Complete DIA methodology has already been reported,20 but overall, the model illustrates impact (relevance) and flux (direction of impact) for KEGG pathways. Full data including Entrez Gene ID, Oligo Gene ID, FDR (<0.05), P value (<.05), and fold change (FC) were entered into the DIA. Information for pathways was only provided when ⩾30% of annotated genes were covered by the data set.
Results of the final pathway analysis are compiled in Tables 1 and 2 and Figures 2 and 3.
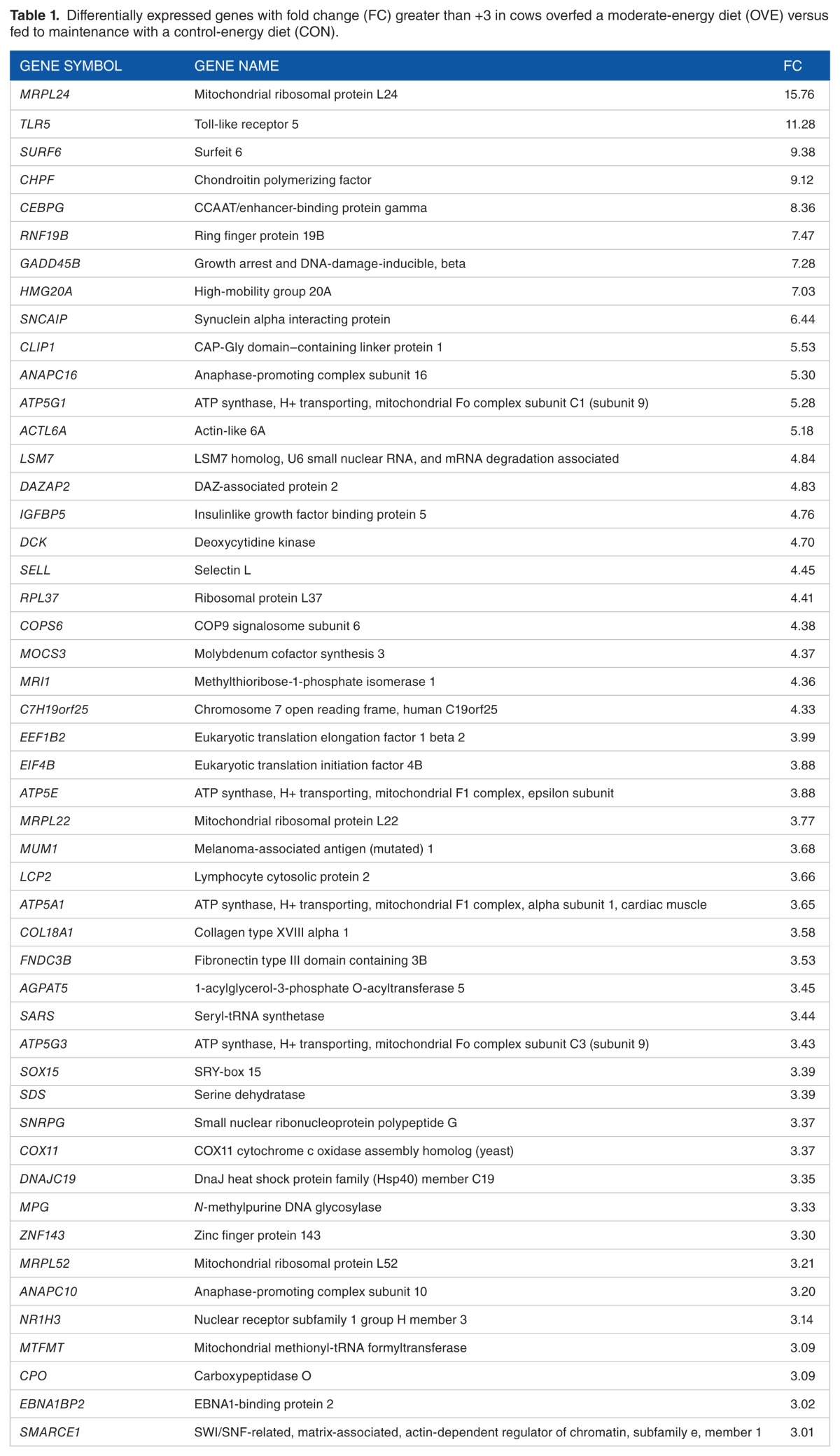
Table 1.
Differentially expressed genes with fold change (FC) greater than +3 in cows overfed a moderate-energy diet (OVE) versus fed to maintenance with a control-energy diet (CON).
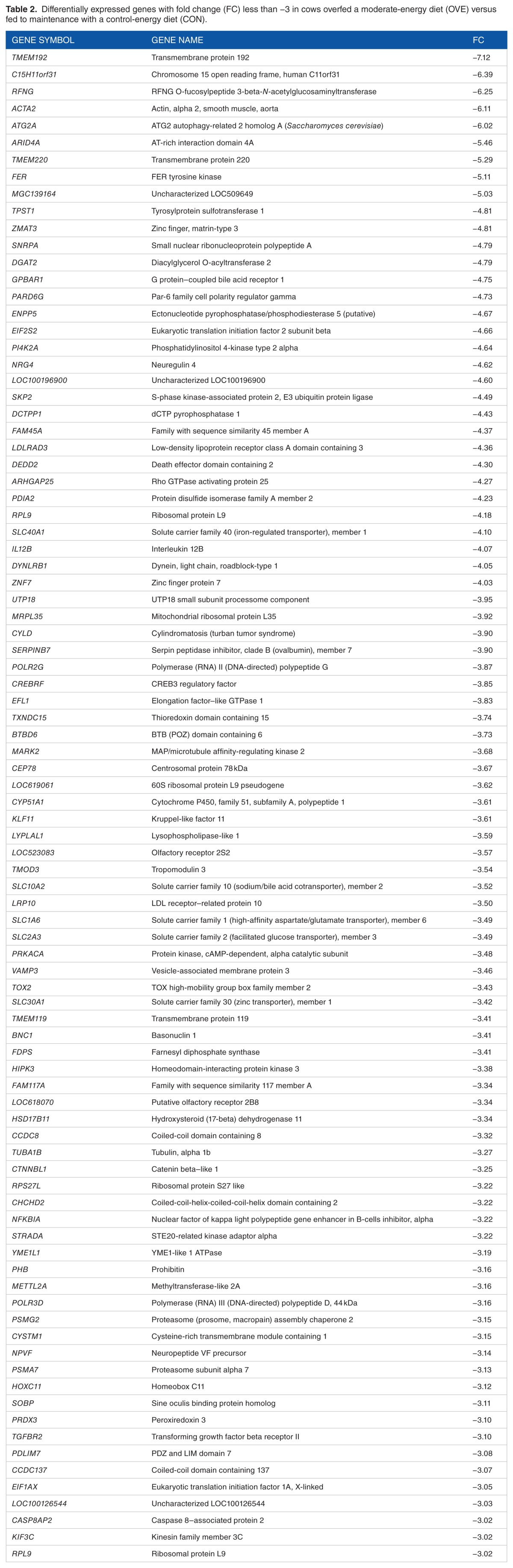
Table 2.
Differentially expressed genes with fold change (FC) less than −3 in cows overfed a moderate-energy diet (OVE) versus fed to maintenance with a control-energy diet (CON).
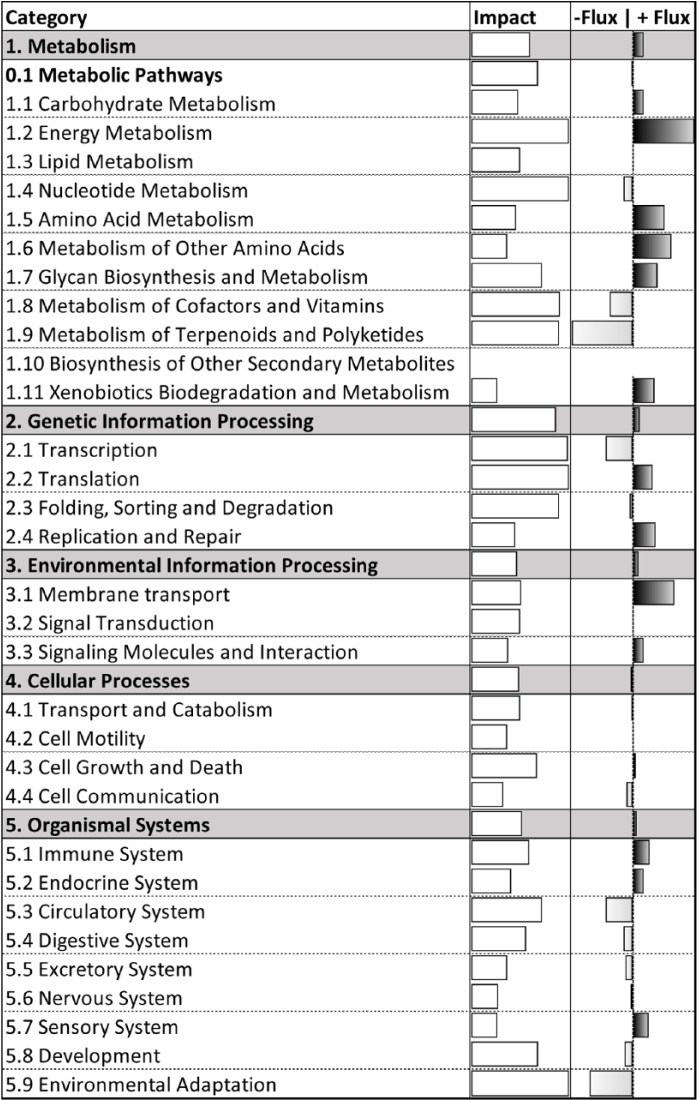
Figure 2.
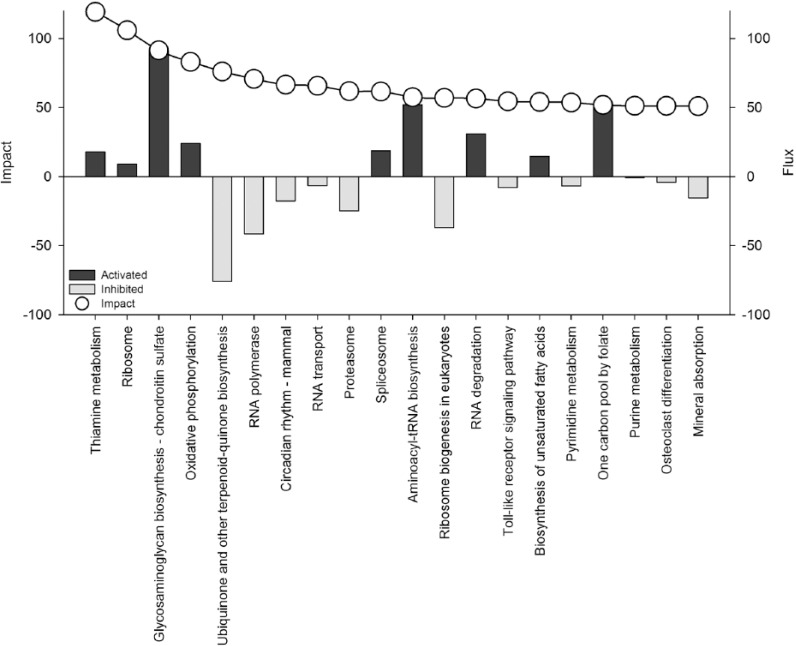
Summary of treatment effects on all Kyoto Encyclopedia of Genes and Genomes pathways in polymorphonuclear leukocytes of overfed (OVE) versus control-fed (CON) cows, divided by category and subcategory. The second column of white bars indicates impact on a relative scale of 0 to 50. The third column of gray and black bars indicates flux, or direction of impact, on a scale of −25 to +25, where gray bars represent negative flux (−25 to 0) and black bars represent positive flux (0 to +25).
Figure 3.
Top 20 affected Kyoto Encyclopedia of Genes and Genomes pathways in polymorphonuclear leukocytes of overfed (OVE) versus control-fed (CON) cows, ranked as such.
Results and Discussion
Due to the abundance of literature on transition period (ie, time effect) adaptations, discussion here will focus on the effect of treatment, ie, overfeeding versus feeding to meet requirements in the close-up dry period on PMNL gene expression. We chose to analyze the samples collected at −14 and 7 days relative to parturition to avoid interference of parturition by sampling too close to it prepartum and to observe transcriptomic changes in the first week of lactation when cows are most susceptible to diseases.21 Data on 43 target genes measured via reverse transcription-polymerase chain reaction (RT-PCR) at −14, 7, and 14 days relative to parturition are reported in Zhou et al.18
Overall, 1806 DEGs were entered into the DIA for pathway analysis. There were 1000 upregulated and 806 down-regulated genes. Of these, 49 upregulated genes had an FC > +3 (see Table 1) and 90 downregulated genes had an FC < −3 (see Table 2).
KEGG analysis
Data for pathways within human diseases categories are omitted from analysis results. Full summary of impact and flux for KEGG categories and subcategories can be reviewed in Figure 2. The top 20 affected pathways, ranked by impact in Figure 3, are discussed by category below.
Metabolism
Energy metabolism
It is known that PMNLs primarily depend on glycolysis to produce energy.22 Therefore, it is surprising that the oxidative phosphorylation pathway was upregulated in OVE cows. However, regardless of glucose availability, as examined in guinea pig PMNLs, an increase in oxygen uptake and respiration occurs in phagocytic cells compared with those in the resting state.23,24 Increases in respiration during phagocytosis were also detected in chicken PMNLs.25 This indicates that, across species, oxygen utilization helps promote PMNL phagocytosis. In addition, in anaerobic environments, glucose consumption or glycogen breakdown (ie, retrieval of glucose for glycolysis in either substrate-rich or substrate-poor environments) increases for phagocytic cells compared with an aerobic environment.24 In other words, alternate sources of energy from oxidative phosphorylation may be available under aerobic conditions. In anaerobic environments, however, glycolysis is primarily responsible for meeting the increased energy demand.
Taken together, the above results demonstrate a potential preference for oxidative phosphorylation during phagocytosis. This preference may correspond to the so-called oxidative burst that allows PMNLs to eliminate phagocytosed particles via reactive oxygen species (ROS). This is supported by findings of Fossati et al26 that complex V of the mitochondrial respiratory chain, or adenosine triphosphatase, is involved in functions related to cell death regulation and respiratory burst. Inhibition of this unit by oligomycin inhibited respiratory burst and chemotaxis. Therefore, upregulation of the oxidative phosphorylation pathway here might indicate higher phagocytic activity of PMNLs, ie, a more activated immune system. This also agrees with earlier quantitative PCR findings that OVE cows had higher expression of genes associated with adhesion, migration, and phagocytosis.18
Another possibility remains open; that is, that adenosine triphosphate (ATP) produced in the mitochondria via oxidative phosphorylation is not formed for cellular consumption but rather for signaling purposes. Purinergic signaling (ie, through ligands such as ATP, adenosine, and other purine metabolites) via P2Y2 receptors has been implicated in neutrophil chemotaxis.27,28 In fact, it has recently been suggested that the extracellular ATP which fuels chemotaxis in human neutrophils is produced specifically by PMNL mitochondria.29 Chemical uncoupling of mitochondria prevented ATP release from stimulated neutrophils, although it did not change the intracellular ATP levels. In the same study, inhibiting mitochondrial ATP also reduced ROS production, demonstrating that oxidative phosphorylation also participates in oxidative burst as speculated above.29 If this mechanism holds true for bovine, it may help reconcile the high levels of oxidative phosphorylation found here in OVE cows, with previous (and current) evidence of glycolysis as the primary energy production pathway in PMNLs. Because purine metabolism was also altered in this study, purinergic signaling is further explored in the “Nucleotide Metabolism” section below.
Lipid metabolism
Biosynthesis of unsaturated fatty acids was induced in OVE cows compared with CON. This can be partly explained by significant upregulation of stearoyl-CoA desaturase (SCD), the enzyme responsible for transforming stearic acid into oleic acid.30 Once in the circulation, eg, from adipose tissue lipolysis, oleic acid may stimulate degranulation31 and superoxide production in PMNLs.32 This indicates that oleic acid signaling could provoke a greater level of PMNL activation in OVE cows, despite abundant evidence in other species that it can be both pro- and anti-inflammatory.33 More research is needed on signaling action of oleic acid in bovine PMNLs. Further research would also be beneficial in understanding the effects of SCD in bovine PMNLs as in rodent liver SCD plays a role in lipid oxidation and accumulation.34
The upregulation of this pathway could also help describe the increased expression of ALOX5AP and PLA2G4A postcalving in OVE cows.18 These enzymes are responsible for freeing arachidonic acid from membrane phospholipids and, further, deriving leukotriene B4 (LTB4) from the polyunsaturated fatty acid intermediate.35 Leukotriene B4 enhances inflammatory processes, including PMNL migration36 and adhesion,37 eosinophil recruitment,38 and generation of ROS.39 It is noteworthy, however, that LTB4 can also exert prosurvival effects in PMNLs through activation of nuclear factor κB (NF-κB).40 In this group of cows, NF-κB followed a similar gene expression pattern to ALOX5AP and PLA2G4A through the peripartal period.18 Therefore, it is possible that unsaturated fatty acids produce more eicosanoids and drive persistent PMNL activation and survival (ie, chronic inflammation) in OVE cows postcalving, even when no clinical disease is present.
Nucleotide metabolism
Both purine metabolism and pyrimidine metabolism were downregulated in OVE versus CON cows. As mentioned above, changes to purine metabolism may be related to the processing of ATP as a chemotactic signal.27 Because release of ATP is a signal of cellular distress or death, it can induce ROS production, signaling, and migration of immune cells.41 Both ATP42 and adenosine43 have been implicated in delaying neutrophil apoptosis. In this study, suppression of purine metabolism could be an adaptation to conserve ATP and other purinergic signals, feeding the inflammatory state. The affected genes in this pathway support this hypothesis.
Among the downregulated genes are ectonucleotide pyrophosphate (ENPP1) and nucleotide triphosphatase (NTPCR), both enzymes that hydrolyze purine metabolites (ie, adenosine monophosphate [AMP] to adenosine and ATP to adenosine diphosphate, respectively) and regulate their signaling capacity.44 Upregulation of deoxycytidine kinase (DCK) and adenosine kinase (ADK) is consistent with this immune profile. Deoxycytidine kinase is a salvage enzyme for deoxynucleosides which allows for DNA repair in nonproliferating immune cells. This enzyme is normally less active in PMNLs than in monocytes or lymphocytes,45 such that its more than 4-fold upregulation in OVE versus CON cows indicates that PMNLs may have had greater DNA repair and survival capacity. In the future, this could be verified by measuring protein expression and/or enzyme activity alongside gene expression.
Adenosine kinase can also affect PMNL function through purine metabolism; it is responsible for creating AMP by phosphorylating adenosine.46 Therefore, higher expression of ADK may indicate reduced adenosine concentrations. Because adenosine has historically been reported as an anti-inflammatory molecule,47 lower adenosine concentrations would again point to aggravated inflammation. Specifically, the interaction of ADK and adenosine has implications in PMNL adhesion and accumulation.46,48 Although this may seem incongruous with its antiapoptotic effect in PMNLs, Walker et al43 suggested that apoptosis can be regarded as a neutrophil function.
Pyrimidines may also participate in immune function and signaling, and in this study, pyrimidine metabolism followed a similar pattern to that of purines. The release of uridine triphosphate (UTP) from PMNL and other cell types can stimulate PMNL through the same P2Y2 receptor to a lesser extent than ATP. When added in combination with chemoattractants to human neutrophils, UTP has been shown to augment intracellular calcium and ROS levels, as well as cell aggregation.49,50 Therefore, it could be expected that, as with ATP, genes that help maintain UTP levels would increase. This appeared to be the case in OVE cows by upregulation of uridine-cytidine kinase (UCK2). If OVE PMNLs are, in fact, hyperactivated, production of ROS would damage DNA. Maintaining a pool of both purines and pyrimidines would aid DNA repair and cell survival. This effect is illustrated by upregulated DCK, as in purine metabolism, and downregulated deoxycytidine triphosphate pyrophosphatase (DCTPP) and ectonucleoside triphosphate diphosphohydrolase (ENTPD8).
The above changes indicate a potential accumulation of repair nucleotides and precursors by conserving phosphorylated forms of the bases.51 Further evidence of potentially increased ROS and DNA damage is indicated by the downregulation of thymidine kinase (TK2), a mitochondrial DNA salvage enzyme whose activity is known to be downregulated during oxidative stress.52 Based on these results, more research on purine and pyrimidine metabolism could be beneficial to clarify the relationship between PMNL activation and survival.
Glycan biosynthesis and metabolism
Chondroitin sulfate (CS) is one of the several glycosaminoglycan chains which modify proteins through glycosylation. After alteration, proteoglycan products are either secreted by the cell or retained in the plasma membrane.53 Recent evidence revealed that CS in both the extracellular matrix and the membrane may contribute to neutrophil activation.53
Recently, the role of free CS on neutrophils has been tested. Incubation of PMNL with CS amplified ROS production in response to interleukin 8 (IL-8).54 Thus, there is potential that OVE PMNL with increased CS biosynthesis secretes the proteoglycan as a pro-inflammatory autocrine signal. This molecule is also involved in neutrophil-activating pathways from the membrane. The carbohydrate portion of CS forms binding sites on the receptor for platelet factor 4 (PF4), an α-chemokine that promotes exocytosis, even independently of IL-8 or calcium mobilization.55 In contrast, the sulfate portion of CS has been identified as a binding site for human leukocyte elastase and cathepsin G.56 Both of these proteases activate cytokines and coordinate neutrophil responses once released from PMNL granules.57 This study underscores a relationship between CS and the immune status of the cow, such that increased CS bio-synthesis corresponded to more activated neutrophils. However, the specific mechanisms of action for CS-moderated responses in bovine neutrophils need further investigation.
Metabolism of cofactors and vitamins
Thiamine metabolism was the most affected pathway in this study. Although thiamine has functions in energy metabolism, eg, as a cofactor in carbohydrate catabolism, it also has antioxidant capacity. The active metabolite of thiamine, thiamine pyrophosphate, is known to protect against oxidative stress in liver,58,59 cardiac muscle,60 and ovaries61 in rats. Thiamine also exerts antioxidant activity directly on neutrophils by inhibiting the peroxidase/H2O2/halide system. These data, rather than denoting an inhibition of neutrophil function, are indicative that thiamine increases neutrophil motility by promoting chemoattraction over antimicrobial signals.62 Thiamine also prevents oxidation of components of PMNL membranes, eg, sulfhydryl groups, by inhibiting the peroxidase system.63 We speculate that the reduction in cell activation and damage in favor of cell migration through thiamine could be a regulatory mechanism that contributes to PMNL longevity and more chronic inflammation in OVE cows.
One carbon pool by folate was also upregulated in OVE cows. The upregulated genes driving this change encode mitochondrial proteins, methylene tetrahydrofolate dehydrogenase 2 (MTHFD2), and mitochondrial methionyl-tRNA formyltransferase (MTFMT), indicating that the diet impact on folate metabolism was localized to the mitochondria. MTHFD2 is instrumental in converting methylene tetrahydrofolate (THF) to formate in the mitochondria. Mitochondrial formate can be transported to the cytosol and used in purine synthesis, thus sparing cytosolic pools of THF for use in transferring 1-carbon groups, the other main function accomplished by cytosolic counterpart MTHFD1.64,65 As discussed above, purine synthesis (and metabolism) is important in modulating the immune response of PMNLs. However, the present data reveal that OVE PMNLs may experience more 1-carbon transfer through the methionine cycle.66 This could have broad effects, although it has been speculated that methionine availability in PMNL enhances immune function.67
The enzyme encoded by MTFMT catalyzes the formylation of methionyl-tRNA, which corresponds to the start codon in mitochondrial protein synthesis.68 Because formylated methionyl-tRNA is used to initiate synthesis of nearly all mitochondrial proteins, it is hard to correlate the increment in MTFMT with a particular outcome. However, it may hint at a more active mitochondria, and therefore, a more active cell. This argument could be strengthened by a deeper analysis of mitochondrial protein expression and activity.
The last cofactor pathway, ubiquinone and other terpenoidquinone biosynthesis, was downregulated in response to OVE. After some disagreement as to whether ubiquinone is involved in PMNL ROS production,69,70 it has been demonstrated that ubiquinone, or coenzyme Q, is associated with neutrophil granules71 and can, in fact, have an inhibitory effect on super-oxide production via membrane signaling.72 At the same time, ubiquinone is required in the electron transport chain of mitochondria, which produces ATP and can produce ROS as byproducts.73 Therefore, the inhibition of ubiquinone biosynthesis could have dual effects on superoxide formation. This is further complicated by the fact that oxidative phosphorylation was increased in OVE cows, leading to an expectation that, overall, elements of the respiratory chain would be upregulated, not downregulated. Nevertheless, the affected genes of this pathway are primarily related to production of ubiquinone for oxidative phosphorylation. These contrasting results indicate that more research is needed to evaluate ubiquinone synthesis and use during periods of elevated mitochondrial activity in PMNLs.
Genetic information processing
Transcription and translation
The instinctive relationship between transcription and translation, formed by the central dogma, DNA to RNA to protein, allows for a better view of cellular function when the 2 processes are evaluated together. Therefore, the results of relevant pathways will be interpreted together in the following section.
First, the RNA polymerase pathway was downregulated in OVE cows. Although RNA polymerases are responsible for the transcription of DNA to RNA,74 this result does not necessarily mean that there was less overall transcription. In eukaryotes, there are 3 distinct RNA polymerases (each with a particular function regarding the different types of RNA, and each of these was affected in this study). Simplistically, polymerase I transcribes ribosomal RNA (rRNA), polymerase II transcribes messenger RNA (mRNA), and polymerase III transcribes transfer RNA (tRNA).75 Furthermore, these transcription complexes are composed of heterogeneous subunits, which can individually affect the transcription complex; decreases in the availability of any one subunit can prevent complex assembly.76 Some subunits are conserved between polymerases and/or across species, whereas others contribute to their differential functions.77 Because the downregulation of the RNA polymerase pathway in this study was driven by changes to the expression of subunit-encoding genes, the specific roles of those genes, and consequences for downstream pathways, will be expanded when possible. Highly affected pathways upstream of RNA polymerase will also be considered when relevant.
Thus far, it seems that the DEG in OVE cows support greater ROS production and immune activation. Under such conditions, these cells may experience a degree of oxidative stress,78 and previous inquiries into PMNL gene expression revealed transcription-level differences to this effect.18 Polymerase I activity (ie, synthesis of rRNA) is known to be downregulated under oxidative stress.79 The only polymerase I gene altered in this pathway was POLR1D, which encodes the smaller of 2 α subunits in polymerase I cores.80 This subunit is essential to polymerase assembly and function.81 In fact, mutation of this gene in humans causes a disturbance to rRNA transcription and ribosomal biogenesis, characterized by craniofacial defects.82 Therefore, downregulation of the gene in OVE cows could certainly inhibit function of polymerase I. This is supported by a corresponding downregulation of the ribosome biogenesis in eukaryotes pathway.
Ribosome biogenesis is particularly important for growing and dividing cells to perform efficient protein synthesis.76 Because neutrophils are terminally differentiated with normally short life spans,83 this process may simply be less biologically important than in other cell types. However, this does not account for differential expression of relevant genes between OVE and CON cows. Another conflict regarding this idea is that in OVE cows, ribosome biogenesis was downregulated, but the ribosome pathway itself was upregulated. This discordance could reflect a distinction between the activity of cytoplasmic and mitochondrial ribosomes, which are believed to have evolved different functions.84 Recall that genes from the folate pathway pointed to an increase in formylated methionyl-tRNA, which initiates most mitochondrial protein synthesis.68 Potentially, mitochondrial proteins or functions (eg, oxidative phosphorylation) become more important in the adaptation to overfeeding, whereas more basic cellular proteins or functions are suppressed.
An analysis of each affected mitochondrial and cytoplasmic gene in the ribosome pathway is beyond the scope of this article. However, one result that stands out is a 15-fold upregulation in mitochondrial ribosomal protein L24 (MRPL24). There is currently little research dedicated to the gene or protein it encodes; however, due to the dramatic change in expression brought on by overfeeding, significance of this gene to PMNLs and/or bovine cells should be further investigated. In addition, another noteworthy mitochondrial ribosomal protein, MRPL18, was upregulated. It has recently been uncovered that MRPL18 induces heat shock protein translation under stressful conditions.84 Because this could potentially support the hypothesis that OVE PMNLs have increased cell survival, more in-depth research appears warranted.
As argued above, assuming that OVE PMNLs produce more ROS and are exposed to greater oxidative stress, cellular protein synthesis could be downregulated to an extent.85 In this scenario, subunits of polymerase II (responsible for mRNA transcription) would likely experience some level of transcriptional control themselves. If less mRNAs are being translated into proteins, it would be a cellular waste to continue producing transcription machinery. Upstream mechanisms of this regulation could be multifactorial, but one possibility is that polymerase I repression (represented here by downregulation of POLR1D) contributed to feedback signals on polymerase II activity, as Laferte et al86 discovered.
In this study, 2 polymerase II genes, POLR2G and POLR2L, were downregulated in OVE cows. The subunit from POLR2G participates in the complex which induces a conformational change in polymerase structure, underscoring its importance to transcription initiation.87 Similarly, the protein encoded by POLR2L has been identified as a key protein to yeast cell viability for its role in transcription.88 If downregulation of this gene can also negatively affect the viability of bovine PMNLs, it may be of interest to consider how and to what extent it can affect the life span of PMNLs, which otherwise seemed to be increased in OVE cows.
As a logical response to lower polymerase II gene expression, OVE cows also experienced suppression of the RNA transport pathway. RNA transport allows mRNA transcribed in the nucleus to be processed and translated later in the cytoplasm.89 With lower mRNA transcription, subsequent processing steps obviously become less important. Multiple genes downregulated in RNA transport are also eukaryotic translation initiation factors, without which proper assembly of the translation initiation complex is not possible.90 Along with lower ribosome biogenesis, the flux of this pathway introduces the idea that both transcription and translation are inhibited in OVE cows.
Polymerase III was also critically affected in OVE cows by changes to 2 subunits used in initiation of tRNA transcription: POLR3C and POLR3G. They comprise 2 parts of a trimer that confers stability to the preinitiation complex and is essential to recruitment of the polymerase to segments of DNA.91,92 Unlike other changes in RNA polymerase gene expression, these subunits were upregulated in OVE cows. Not surprisingly, there was a corresponding increase in aminoacyl-tRNA biosynthesis. Surprisingly, although these findings are consistent with each other, they are inconsistent with other transcription and translation data.
Cherkasov et al93 have recently published findings regarding yeast cell cytoplasmic stress granules. Interestingly, some of the findings from stress granules display similarities to the neutrophils in OVE cows, including increases in aminoacyl-tRNA synthetases, as well as proteins involved in ribosome biogenesis and translation. However, it should be kept in mind that sequestering these proteins in heat-stressed yeast makes them less available for cellular functions. Therefore, OVE PMNLs resemble those stressed eukaryotic cells, in that ribosome biogenesis and translation initiation were inhibited (as mentioned above), yet, differ regarding tRNA. Where yeast accumulated more tRNA synthetases in stress granules, and therefore could create fewer tRNA for protein synthesis, OVE PMNLs had increased tRNA synthetase expression, which should increase protein synthesis. Another level of complexity is added when one considers that other transcription and translation-related molecules, eg, polymerase II subunits and initiation factors, were downregulated in OVE cows. It is not clear why regulation of polymerase III and tRNA biosynthesis were not coordinated with other genetic information processing pathways. Whether this could be attributable to differences in mitochondrial and cytoplasmic translation requirements, as postulated for the ribosome pathway, requires further research.
By the same token, upregulation of the spliceosome pathway in OVE cows indicates greater pre-mRNA processing that should facilitate translation. Many small nuclear ribonucleoproteins, Sm proteins, LSm proteins, and 1 heat shock protein, HSPA1A, were among the genes upregulated in OVE cows. These components are recruited to form the spliceosome on pre-mRNA molecules, assist in removing introns and ligating exons, then disassemble for recycling.94 The spliceosome is important for cell viability, an idea which has been demonstrated by studies in human cancer research,95,96 and fits with the idea that OVE PMNLs have enhanced survival. Nonetheless, it is a surprising result given that mRNA transcription, transport, and even translation seemed suppressed. It is striking that these changes mirror those surrounding ribosome; the reason(s) why major ribonuclear units such as the ribosome and spliceosome are induced in OVE cows, yet closely related pathways do not support their functions, is not known.
Folding, sorting, and degradation
Alternatively, 1 pathway that did concur with spliceosome was RNA degradation. Although increased RNA degradation may normally cause concern for the validity of microarray results derived from RNA, it should be noted that more than 1 gene upregulated in this pathway is an LSm gene, which has already been identified as a spliceosome component.94 Other upregulated genes such as exoribonuclease DIS3 act mainly in the nucleus, and, thus, may also play a role in processing premature mRNA.97 Then, it is reasonable to suggest that RNA degradation may simply account for the cleavage and breakdown of introns or unstable nuclear mRNA, rather than untranslated exons. The high impact on this pathway serves as a reminder that the transcriptome should not be strictly interpreted as actual biological activity without the supporting proteomic work because many changes occur between the expression of genes and proteins.98
Posttranslationally, the cellular profile can be partly controlled by the ubiquitin-proteasome system. Highly specific ubiquitination marks target proteins for degradation by the proteasome to invoke quality control and normal turnover.99 Because the proteasome can be localized to many cellular locations and is responsible for degrading proteins involved in all aspects of cell growth, signaling, and death,100 changes in its activity do not speak to one end result. Despite this, the proteasome pathway (as well as the lower affected ubiquitin-mediated proteolysis pathway) was downregulated in OVE cows, which at least suggests that these cells required greater protein conservation. This would be especially important if OVE PMNLs were experiencing decreased rates of protein synthesis. The process of ubiquitination and proteolysis via this system is also ATP-requiring99,101; thus, reduced expression of proteasome subunits may occur partly to conserve ATP, an effort that we also detected in nucleotide metabolism pathways in OVE PMNLs.
The impact on proteasome may also relate with previous observations and hypotheses of increased mitochondrial activity and protein synthesis in the PMNL. It was previously thought that individual proteases were more relevant in the mitochondria than the proteasome.102 Instead, recent experiments by Lehmann et al103 reveal a high level of ubiquitination on mitochondria-specific proteins and presence of multiple proteasomal components within the mitochondria. If the proteasome does govern more protein turnover in the mitochondria, its downregulation may corroborate the activating effect of OVE on PMNL that leads to prioritization of mitochondrial function.
Organismal systems
Immune system and development
Although it is counterintuitive that osteoclast differentiation should be highly affected in terminally differentiated PMNL, this can be explained by examining the group of genes implicated. The KEGG pathways are species specific but not tissue specific. Many of the osteoclast differentiation genes are involved in cell cycle or growth and differentiation signaling but are also shared by the toll-like receptor (TLR) signaling pathway, where their functions are primarily tied to immunity. Due to the high proportion of similar genes, both pathways had the same overall direction of impact. For this reason, these 2 pathways and their KEGG categories have been combined and will mostly be discussed within context of TLR signaling.
This article has so far emphasized the activation of OVE PMNL, which makes it surprising that the TLR signaling pathway, typically associated with immune activation, was downregulated. Toll-like receptors are transmembrane proteins that recognize pathogen-associated molecular patterns and initiate cascades of pro-inflammatory cytokine signals.104 In OVE cows, 2 specific TLR genes were downregulated—TLR4, which recognizes lipopolysaccharide of gram-negative bacteria105 and TLR5, which recognizes bacterial flagellin.106 Given the evidence above that OVE PMNLs are more activated, the downregulation of TLR4/5 implies that this response is pathogen independent and therefore inappropriate. Lower expression of TLR and related signaling molecules may also cause these PMNLs to later be less responsive toward real pathogens, such as the gram-negative bacteria that can cause acute, environmental mastitis, among other diseases.107 Other pro-inflammatory signaling molecules, downstream of TLR or related networks, were downregulated in both the TLR signaling and osteoclast differentiation pathways. Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3KCG), signal transducer and activator of transcription (STAT1), and mitogen-activated protein kinase kinase kinase 7 (MAP3K7 or TAK1) were all dually reported.
PI3KCG encodes a subunit of PI3K that is critical for PIP3 signaling, chemokinesis, neutrophil accumulation, and ROS production.108 It is interesting that this key gene is downregulated, impairing inflammation, although the PMNL generally seemed activated. Other pathways that have similar activation but run parallel to PI3K, such as Jak/STAT and MAPK,109 should be examined as alternate means to achieve PMNL activation. In fact, the PI3K/Akt and Jak/STAT pathways have similar primary functions: inflammation and neutrophil survival,110 and both alternate pathways are well-represented within the DEG.
Overall, the Jak/STAT cascade seems activated. The osteoclast differentiation pathway reveals a downregulation of suppressor of cytokine signaling 3 (SOCS3)—a Jak inhibitor111—with concurrent upregulation of JAK1. In addition, interferon regulatory factor 9 (IRFN9) was upregulated. After activation by Jak1,112 STAT1 and STAT2 complex with IRFN9 to promote pro-inflammatory transcription due to interferon signaling.113 Transcription of a different set of immune-related genes can even occur by combination of STAT2 and IRFN9, but in the absence of STAT1,114 which could potentially explain down-regulation of STAT1 in an activated PMNL profile.
MAP3K7 is central to another complex cascade, mediating the effects of cytokine (eg, IL-1β) and toll-like signals on pro-inflammatory gene transcription by MAPK, JNK, and NF-κB.115 This particular gene may have a negative regulator function in mouse neutrophils, although this contrasts with data from other immune cells and human PMNL115,116 and needs to be confirmed in bovine. However, in an inhibitory role, downregulation of MAP3K7 would tend to fit with patterns of pro-inflammatory activation in OVE cows, including upregulated gene expression of myeloid differentiation primary response 88 (MYD88) upstream (in TLR signaling pathway) and partners RELA (in both pathways) and NF-κB18 downstream.117
Although upregulation of MYD88 suggests activation of the MyD88-dependent pathway, downregulation of preceding cytokines (eg, IL1B and IL12B) and receptors (eg, TLR4, TLR5, interferon receptor IFNAR2, and transforming growth factor β receptor TGFBR2) may indicate alternative ways of stimulation for this pathway. Saturated fatty acids, eg, are known to stimulate pro-inflammatory signaling through TLRs,118,119 and this mechanism is exacerbated by ROS production.120 In OVE cows with higher serum nonesterified fatty acids and ROS,17 saturated fatty acids could very well be a part of pathogen-independent activation.
At some point, negative feedback may act to limit inflammation and prevent stress-induced PMNL apoptosis in OVE cows.121 Upregulation of NF-κB inhibitor (NFKBIA; in both pathways) could be one example of how and where this feedback influences NF-κB–induced inflammation. With down-regulation of most protein-management systems such as proteasome, this feedback likely results in more important transcription-level changes, emphasizing the relevance of the transcriptome seen here.
Digestion
Mineral absorption was downregulated in OVE cows. Due to its importance in neutrophilic responses, this section will focus on calcium (Ca2+)-related effects. Free Ca2+ signaling is central to PMNL functions such as chemotaxis, degranulation, and ROS production.122 It is also closely related to mitochondrial ATP production and purinergic signaling,29 pathways already recognized for their importance to OVE cows.
Ca2+ release from intracellular stores is stimulated by chemokine and G protein–coupled receptor signaling.123 When Ca2+ is released, a new influx is required to replenish stores.124 In neutrophils, this influx occurs through the transient receptor potential family of transporters,125 where TRPM7 is the primary transporter in mammals.122 Downregulation of TRPM7 and TRPV6 indicates a lower capability to take up Ca2+, and therefore, lower potential for Ca2+-mediated immune responses. This contrasts with previous results, especially those of metabolic pathways, which strongly support the idea that OVE cows have more active PMNL. A couple of possibilities exist to resolve this disagreement. First, that the microarray provides a snapshot of the entire transcriptome at just 2 time points. As with any other signaling pathway, changes captured could represent activated functions of the pathway as well as negative feedback. Downregulation of Ca2+ channel expression could be another example of feedback meant to limit inflammation in a chronically activated system. Another possibility is that there is, in fact, greater PMNL activation before calving (as suggested by Zhou et al18), but that the downregulation of mineral transporters here represents a stronger postpartum effect of Ca2+ partitioning to the mammary gland. The demand for Ca2+ during early lactation is such that all intact cows developed postpartum hypocalcemia in one study comparing mastectomized versus intact cows.126 The same study also concluded that metabolic demands of the mammary gland contributed to general loss of immune cell functions. The specific inhibitory effect of parturition (ie, induction of lactation) on Ca2+ signaling in monocytes has also been documented.127 Therefore, genes relevant to Ca2+ signaling in PMNL may be better considered from a time effect or interaction perspective as well.
Environmental adaptation
Circadian rhythm was downregulated in OVE cows. Although this may seem peripheral to immediate immune function, it is suggested that circadian rhythm actually plays a role in the function of bovine neutrophils.128 Thus, this pathway could help explain the changes in OVE PMNL behavior. Two key regulating factors to circadian rhythm are the cryptochrome (CRY) and period (PER) proteins. These 2 proteins dimerize and provide negative feedback for their own cyclic gene transcription.129 The level of activity from these proteins is tightly controlled by phosphorylation, dephosphorylation, and ultimately proteolysis.129,130 In OVE cows, cryptochrome gene CRY1 was downregulated, whereas E3 ubiquitin ligase component RBX1 was upregulated. This combination suggests that there was an inhibition of the circadian rhythm in the PMNL. Regulatory gene casein kinase 1ε (CSNK1E) was also downregulated, suggesting that there was less phosphorylation (ie, inhibition) to PER131 and some balance to control of the system even as it was altered. However, as PER does not act without CRY, and as there are additional levels of control by phosphorylation, it is likely that the system was still shifted. Dysregulation of circadian rhythm could, thus, be one contributor to abnormal PMNL function, and it would be useful to elucidate the mechanism by which OVE interacts with this system. This potentially has great importance to the transition cow because the dry period is already associated with photoperiod manipulation, which can also influence the immune system and circadian rhythm.129,132
Conclusions
This study investigated whether dietary energy intake in the dry period affected immune function, as represented by the neutrophil transcriptome. Both metabolic and nonmetabolic PMNL functions were highly affected by the difference in energy level. Together, the above results portray that feeding higher energy diets in the close-up period may prolong PMNL life span and heighten nonpathogenic inflammation. With more activated and longer circulating immune cells, it is likely that OVE cows experience some degree of chronic inflammation. This confirms results from a previous target gene expression study (using RT-PCR) of the same cows, and other experiments which document poorer health outcomes in overfed transition cows.
Although no clinical disease or production differences were detected in these cows, chronic inflammation may result in subclinical or long-term health conditions that should be considered. In addition, this study has highlighted a few topics which would benefit from further examination. Of specific interest to the immune function of transition cows might be the expanding role of the mitochondria in active PMNL, purinergic signaling, and the contrasting results for translation-related and TLR-related genes and pathways, including how they contribute to cell survival. Generating appropriate data will require more bovine-specific and neutrophil-specific work because many differences may exist between bovine PMNL and yeast or human line cells, which mostly contribute to current literature relevant to the KEGG pathways. Future research on these specific topics as well as more cow-level outcomes would confirm whether overfeeding creates any immune or metabolic disadvantage to the transition cow. If true, controlling or restricting dry period, dietary energy may be better justified, from a holistic standpoint, and nutrition trials will be useful to pinpoint more ideal energy levels.
Supplementary Materials
Footnotes
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 669 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Portions of this work was financially supported by National Research Initiative competitive grant 2007-35204-17758 and National Institute of Food and Agriculture (Washington, DC; ILLU-538-914). Alea Agrawal received a Jonathan Baldwin Turner MS fellowship from the University of Illinois (Urbana-Champaign).
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
JJL designed the cow study. MJK, DEG and MVR performed the experiments, gene function and pathway analysis. AA performed gene function and pathway analysis, interpreted the data, and wrote the manuscript. SLZ performed statistical analysis of the transcriptome data. JSO and JJL participated in data interpretation and revision of the paper. All authors reviewed and approved the final version of the manuscript.
REFERENCES
- 1.Bell AW. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J Anim Sci. 1995;73:2804–2819. doi: 10.2527/1995.7392804x. [DOI] [PubMed] [Google Scholar]
- 2.Drackley JK, ADSA Foundation Scholar Award Biology of dairy cows during the transition period: the final frontier? J Dairy Sci. 1999;82:2259–2273. doi: 10.3168/jds.s0022-0302(99)75474-3. [DOI] [PubMed] [Google Scholar]
- 3.Grummer RR, Mashek DG, Hayirli A. Dry matter intake and energy balance in the transition period. Vet Clin North Am Food Anim Pract. 2004;20:447–470. doi: 10.1016/j.cvfa.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Beever DE. The impact of controlled nutrition during the dry period on dairy cow health, fertility and performance. Anim Reprod Sci. 2006;96:212–226. doi: 10.1016/j.anireprosci.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Dann HM, Litherland NB, Underwood JP, et al. Diets during far-off and close-up dry periods affect periparturient metabolism and lactation in multiparous cows. J Dairy Sci. 2006;89:3563–3577. doi: 10.3168/jds.S0022-0302(06)72396-7. [DOI] [PubMed] [Google Scholar]
- 6.Janovick NA, Boisclair YR, Drackley JK. Prepartum dietary energy intake affects metabolism and health during the periparturient period in primiparous and multiparous Holstein cows. J Dairy Sci. 2011;94:1385–1400. doi: 10.3168/jds.2010-3303. [DOI] [PubMed] [Google Scholar]
- 7.Urdl M, Gruber L, Obritzhauser W, Schauer A. Metabolic parameters and their relationship to energy balance in multiparous Simmental, Brown Swiss and Holstein cows in the periparturient period as influenced by energy supply pre- and post-calving. J Anim Physiol Anim Nutr (Berl) 2015;99:174–189. doi: 10.1111/jpn.12178. [DOI] [PubMed] [Google Scholar]
- 8.Huang W, Tian Y, Wang Y, et al. Effect of reduced energy density of close-up diets on dry matter intake, lactation performance and energy balance in multiparous Holstein cows. J Anim Sci Biotechnol. 2014;5:30. doi: 10.1186/2049-1891-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso FC, LeBlanc SJ, Murphy MR, Drackley JK. Prepartum nutritional strategy affects reproductive performance in dairy cows. J Dairy Sci. 2013;96:5859–5871. doi: 10.3168/jds.2013-6759. [DOI] [PubMed] [Google Scholar]
- 10.Douglas GN, Overton TR, Bateman HG, 2nd, Dann HM, Drackley JK. Prepartal plane of nutrition, regardless of dietary energy source, affects peri-parturient metabolism and dry matter intake in Holstein cows. J Dairy Sci. 2006;89:2141–2157. doi: 10.3168/jds.S0022-0302(06)72285-8. [DOI] [PubMed] [Google Scholar]
- 11.Sordillo LM, Raphael W. Significance of metabolic stress, lipid mobilization, and inflammation on transition cow disorders. Vet Clin North Am Food Anim Pract. 2013;29:267–278. doi: 10.1016/j.cvfa.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Burton JL, Madsen SA, Chang LC, et al. Gene expression signatures in neutrophils exposed to glucocorticoids: a new paradigm to help explain “neutrophil dysfunction” in parturient dairy cows. Vet Immunol Immunopathol. 2005;105:197–219. doi: 10.1016/j.vetimm.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Goff JP, Horst RL. Physiological changes at parturition and their relationship to metabolic disorders. J Dairy Sci. 1997;80:1260–1268. doi: 10.3168/jds.S0022-0302(97)76055-7. [DOI] [PubMed] [Google Scholar]
- 14.Kumar V, Sharma A. Neutrophils: Cinderella of innate immune system. Int Immunopharmacol. 2010;10:1325–1334. doi: 10.1016/j.intimp.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Burton JL, Madsen SA, Yao J, Sipkovsky SS, Coussens PM. An immunogenomics approach to understanding periparturient immunosuppression and mastitis susceptibility in dairy cows. Acta Vet Scand. 2001;42:407–424. [PubMed] [Google Scholar]
- 16.Madsen SA, Chang LC, Hickey MC, Rosa GJ, Coussens PM, Burton JL. Microarray analysis of gene expression in blood neutrophils of parturient cows. Physiol Genomics. 2004;16:212–221. doi: 10.1152/physiolgenomics.00121.2003. [DOI] [PubMed] [Google Scholar]
- 17.Khan MJ, Jacometo CB, Graugnard DE, et al. Overfeeding dairy cattle during late-pregnancy alters hepatic PPARα-regulated pathways including hepatokines: impact on metabolism and peripheral insulin sensitivity. Gene Regul Syst Bio. 2014;8:97–111. doi: 10.4137/GRSB.S14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z, Bu DP, Vailati Riboni M, et al. Prepartal dietary energy level affects peripartal bovine blood neutrophil metabolic, antioxidant, and inflammatory gene expression. J Dairy Sci. 2015;98:5492–5505. doi: 10.3168/jds.2014-8811. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 20.Bionaz M, Periasamy K, Rodriguez-Zas SL, Hurley WL, Loor JJ. A novel dynamic impact approach (DIA) for functional analysis of time-course omics studies: validation using the bovine mammary transcriptome. PLoS ONE. 2012;7:e32455. doi: 10.1371/journal.pone.0032455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingvartsen KL, Moyes K. Nutrition, immune function and health of dairy cattle. Animal. 2013;7:112–122. doi: 10.1017/S175173111200170X. [DOI] [PubMed] [Google Scholar]
- 22.Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol. 2010;184:4062–4068. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oren R, Farnham AE, Saito K, Milofsky E, Karnovsky ML. Metabolic patterns in three types of phagocytizing cells. J Cell Biol. 1963;17:487–501. doi: 10.1083/jcb.17.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sbarra AJ, Karnovsky ML. The biochemical basis of phagocytosis. I. Metabolic changes during the ingestion of particles by polymorphonuclear leukocytes. J Biol Chem. 1959;234:1355–1362. [PubMed] [Google Scholar]
- 25.Penniall R, Spitznagel JK. Chicken neutrophils: oxidative metabolism in phagocytic cells devoid of myeloperoxidase. Proc Natl Acad Sci U S A. 1975;72:5012–5015. doi: 10.1073/pnas.72.12.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fossati G, Moulding DA, Spiller DG, Moots RJ, White MR, Edwards SW. The mitochondrial network of human neutrophils: role in chemotaxis, phagocytosis, respiratory burst activation, and commitment to apoptosis. J Immunol. 2003;170:1964–1972. doi: 10.4049/jimmunol.170.4.1964. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Corriden R, Inoue Y, et al. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science. 2006;314:1792–1795. doi: 10.1126/science.1132559. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Yao Y, Sumi Y, et al. Purinergic signaling: a fundamental mechanism in neutrophil activation. Sci Signal. 2010;3:ra45. doi: 10.1126/scisignal.2000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao Y, Ledderose C, Seier T, et al. Mitochondria regulate neutrophil activation by generating ATP for autocrine purinergic signaling. J Biol Chem. 2014;289:26794–26803. doi: 10.1074/jbc.M114.572495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes-Quiroz ME, Alba G, Saenz J, et al. Oleic acid modulates mRNA expression of liver X receptor (LXR) and its target genes ABCA1 and SREBP1c in human neutrophils. Eur J Nutr. 2014;53:1707–1717. doi: 10.1007/s00394-014-0677-0. [DOI] [PubMed] [Google Scholar]
- 31.Hidalgo MA, Nahuelpan C, Manosalva C, et al. Oleic acid induces intracellular calcium mobilization, MAPK phosphorylation, superoxide production and granule release in bovine neutrophils. Biochem Biophys Res Commun. 2011;409:280–286. doi: 10.1016/j.bbrc.2011.04.144. [DOI] [PubMed] [Google Scholar]
- 32.Badwey JA, Curnutte JT, Karnovsky ML. cis-Polyunsaturated fatty acids induce high levels of superoxide production by human neutrophils. J Biol Chem. 1981;256:12640–12643. [PubMed] [Google Scholar]
- 33.Carrillo C, Cavia Mdel M, Alonso-Torre S. Role of oleic acid in immune system; mechanism of action; a review. Nutr Hosp. 2012;27:978–990. doi: 10.3305/nh.2012.27.4.5783. [DOI] [PubMed] [Google Scholar]
- 34.Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase as a new drug target for obesity treatment. Obes Rev. 2005;6:169–174. doi: 10.1111/j.1467-789X.2005.00177.x. [DOI] [PubMed] [Google Scholar]
- 35.Brock TG, Peters-Golden M. Activation and regulation of cellular eicosanoid biosynthesis. ScientificWorldJournal. 2007;7:1273–1284. doi: 10.1100/tsw.2007.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith MJ, Ford-Hutchinson AW, Bray MA. Leukotriene B: a potential mediator of inflammation. J Pharm Pharmacol. 1980;32:517–518. doi: 10.1111/j.2042-7158.1980.tb12985.x. [DOI] [PubMed] [Google Scholar]
- 37.Coxon A, Rieu P, Barkalow FJ, et al. A novel role for the beta 2 integrin CD11b/CD18 in neutrophil apoptosis: a homeostatic mechanism in inflammation. Immunity. 1996;5:653–666. doi: 10.1016/s1074-7613(00)80278-2. [DOI] [PubMed] [Google Scholar]
- 38.Tager AM, Dufour JH, Goodarzi K, Bercury SD, von Andrian UH, Luster AD. BLTR mediates leukotriene B(4)-induced chemotaxis and adhesion and plays a dominant role in eosinophil accumulation in a murine model of peritonitis. J Exp Med. 2000;192:439–446. doi: 10.1084/jem.192.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo CH, You HJ, Cho SH, et al. Leukotriene B(4) stimulates Rac-ERK cascade to generate reactive oxygen species that mediates chemotaxis. J Biol Chem. 2002;277:8572–8578. doi: 10.1074/jbc.M104766200. [DOI] [PubMed] [Google Scholar]
- 40.Barcellos-de-Souza P, Canetti C, Barja-Fidalgo C, Arruda MA. Leukotriene B(4) inhibits neutrophil apoptosis via NADPH oxidase activity: redox control of NF-κB pathway and mitochondrial stability. Biochim Biophys Acta. 2012;1823:1990–1997. doi: 10.1016/j.bbamcr.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Kawamura H, Kawamura T, Kanda Y, Kobayashi T, Abo T. Extracellular ATP-stimulated macrophages produce macrophage inflammatory protein-2 which is important for neutrophil migration. Immunology. 2012;136:448–458. doi: 10.1111/j.1365-2567.2012.03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughan KR, Stokes L, Prince LR, et al. Inhibition of neutrophil apoptosis by ATP is mediated by the P2Y11 receptor. J Immunol. 2007;179:8544–8553. doi: 10.4049/jimmunol.179.12.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker BA, Rocchini C, Boone RH, Ip S, Jacobson MA. Adenosine A2a receptor activation delays apoptosis in human neutrophils. J Immunol. 1997;158:2926–2931. [PubMed] [Google Scholar]
- 44.Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Terai C, Wasson DB, Carrera CJ, Carson DA. Dependence of cell survival on DNA repair in human mononuclear phagocytes. J Immunol. 1991;147:4302–4306. [PubMed] [Google Scholar]
- 46.Rosengren S, Bong GW, Firestein GS. Anti-inflammatory effects of an adenosine kinase inhibitor. Decreased neutrophil accumulation and vascular leakage. J Immunol. 1995;154:5444–5451. [PubMed] [Google Scholar]
- 47.Barletta KE, Ley K, Mehrad B. Regulation of neutrophil function by adenosine. Arterioscler Thromb Vasc Biol. 2012;32:856–864. doi: 10.1161/ATVBAHA.111.226845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firestein GS, Bullough DA, Erion MD, et al. Inhibition of neutrophil adhesion by adenosine and an adenosine kinase inhibitor. The role of selectins. J Immunol. 1995;154:326–334. [PubMed] [Google Scholar]
- 49.Seifert R, Wenzel K, Eckstein F, Schultz G. Purine and pyrimidine nucleotides potentiate activation of NADPH oxidase and degranulation by chemotactic peptides and induce aggregation of human neutrophils via G proteins. Eur J Biochem. 1989;181:277–285. doi: 10.1111/j.1432-1033.1989.tb14722.x. [DOI] [PubMed] [Google Scholar]
- 50.Tuluc F, Bredetean O, Brailoiu E, et al. The priming effect of extracellular UTP on human neutrophils: role of calcium released from thapsigargin-sensitive intracellular stores. Purinergic Signal. 2005;1:359–368. doi: 10.1007/s11302-005-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Requena CE, Perez-Moreno G, Ruiz-Perez LM, Vidal AE, Gonzalez-Pacanowska D. The NTP pyrophosphatase DCTPP1 contributes to the homoeostasis and cleansing of the dNTP pool in human cells. Biochem J. 2014;459:171–180. doi: 10.1042/BJ20130894. [DOI] [PubMed] [Google Scholar]
- 52.Sun R, Eriksson S, Wang L. Zidovudine induces downregulation of mitochondrial deoxynucleoside kinases: implications for mitochondrial toxicity of antiviral nucleoside analogs. Antimicrob Agents Chemother. 2014;58:6758–6766. doi: 10.1128/AAC.03613-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pantazaka E, Papadimitriou E. Chondroitin sulfate-cell membrane effectors as regulators of growth factor-mediated vascular and cancer cell migration. Biochim Biophys Acta. 2014;1840:2643–2650. doi: 10.1016/j.bbagen.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Schlorke D, Thomas L, Samsonov SA, Huster D, Arnhold J, Pichert A. The influence of glycosaminoglycans on IL-8-mediated functions of neutrophils. Carbohydr Res. 2012;356:196–203. doi: 10.1016/j.carres.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 55.Petersen F, Bock L, Flad HD, Brandt E. A chondroitin sulfate proteoglycan on human neutrophils specifically binds platelet factor 4 and is involved in cell activation. J Immunol. 1998;161:4347–4355. [PubMed] [Google Scholar]
- 56.Campbell EJ, Owen CA. The sulfate groups of chondroitin sulfate- and heparan sulfate-containing proteoglycans in neutrophil plasma membranes are novel binding sites for human leukocyte elastase and cathepsin G. J Biol Chem. 2007;282:14645–14654. doi: 10.1074/jbc.M608346200. [DOI] [PubMed] [Google Scholar]
- 57.Adkison AM, Raptis SZ, Kelley DG, Pham CT. Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest. 2002;109:363–371. doi: 10.1172/JCI13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turan MI, Siltelioglu Turan I, Mammadov R, Altinkaynak K, Kisaoglu A. The effect of thiamine and thiamine pyrophosphate on oxidative liver damage induced in rats with cisplatin. Biomed Res Int. 2013;2013:783809. doi: 10.1155/2013/783809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yilmaz I, Demiryilmaz I, Turan MI, Cetin N, Gul MA, Suleyman H. The effects of thiamine and thiamine pyrophosphate on alcohol-induced hepatic damage biomarkers in rats. Eur Rev Med Pharmacol Sci. 2015;19:664–670. [PubMed] [Google Scholar]
- 60.Polat B, Suleyman H, Sener E, Akcay F. Examination of the effects of thiamine and thiamine pyrophosphate on Doxorubicin-induced experimental cardiotoxicity. J Cardiovasc Pharmacol Ther. 2015;20:221–229. doi: 10.1177/1074248414552901. [DOI] [PubMed] [Google Scholar]
- 61.Demiryilmaz I, Sener E, Cetin N, Altuner D, Akcay F, Suleyman H. A comparative investigation of biochemical and histopathological effects of thiamine and thiamine pyrophosphate on ischemia-reperfusion induced oxidative damage in rat ovarian tissue. Arch Pharm Res. 2013;36:1133–1139. doi: 10.1007/s12272-013-0173-8. [DOI] [PubMed] [Google Scholar]
- 62.Theron A, Anderson R, Grabow G, Meiring JL. In vitro and in vivo stimulation of neutrophil migration and lymphocyte transformation by thiamine related to inhibition of the peroxidase/H2O2/halide system. Clin Exp Immunol. 1981;44:295–303. [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson R, Jones PT. Increased leucoattractant binding and reversible inhibition of neutrophil motility mediated by the peroxidase/H2O2/halide system: effects of ascorbate, cysteine, dithiothreitol, levamisole and thiamine. Clin Exp Immunol. 1982;47:487–496. [PMC free article] [PubMed] [Google Scholar]
- 64.Christensen KE, Mackenzie RE. Mitochondrial methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate cyclohydrolase, and formyltetrahydrofolate synthetases. Vitam Horm. 2008;79:393–410. doi: 10.1016/S0083-6729(08)00414-7. [DOI] [PubMed] [Google Scholar]
- 65.Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 66.Field MS, Kamynina E, Stover PJ. MTHFD1 regulates nuclear de novo thy-midylate biosynthesis and genome stability. Biochimie. 2016;126:27–30. doi: 10.1016/j.biochi.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li C, Batistel F, Osorio JS, Drackley JK, Luchini D, Loor JJ. Peripartal ru-men-protected methionine supplementation to higher energy diets elicits positive effects on blood neutrophil gene networks, performance and liver lipid content in dairy cows. J Anim Sci Biotechnol. 2016;7:18. doi: 10.1186/s40104-016-0077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vial L, Gomez P, Panvert M, Schmitt E, Blanquet S, Mechulam Y. Mitochondrial methionyl-tRNAfMet formyltransferase from Saccharomyces cerevisiae: gene disruption and tRNA substrate specificity. Biochemistry. 2003;42:932–939. doi: 10.1021/bi026901x. [DOI] [PubMed] [Google Scholar]
- 69.Crawford DR, Schneider DL. Identification of ubiquinone-50 in human neutrophils and its role in microbicidal events. J Biol Chem. 1982;257:6662–6668. [PubMed] [Google Scholar]
- 70.Cross AR, Jones OT, Garcia R, Segal AW. The subcellular localization of ubi-quinone in human neutrophils. Biochem J. 1983;216:765–768. doi: 10.1042/bj2160765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee JA. Histochemical detection of ubiquinone in neutrophil polymorphonuclear leucocyte granules. Exp Cell Biol. 1986;54:89–93. doi: 10.1159/000163348. [DOI] [PubMed] [Google Scholar]
- 72.Kanno T, Utsumi T, Takehara Y, et al. Inhibition of neutrophil-superoxide generation by alpha-tocopherol and coenzyme Q. Free Radic Res. 1996;24:281–289. doi: 10.3109/10715769609088025. [DOI] [PubMed] [Google Scholar]
- 73.Sarewicz M, Osyczka A. Electronic connection between the quinone and cytochrome C redox pools and its role in regulation of mitochondrial electron transport and redox signaling. Physiol Rev. 2015;95:219–243. doi: 10.1152/physrev.00006.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burton ZF. The Old and New Testaments of gene regulation. Evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD. Transcription. 2014;5:e28674. doi: 10.4161/trns.28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viktorovskaya OV, Schneider DA. Functional divergence of eukaryotic RNA polymerases: unique properties of RNA polymerase I suit its cellular role. Gene. 2015;556:19–26. doi: 10.1016/j.gene.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- 77.Lalo D, Carles C, Sentenac A, Thuriaux P. Interactions between three common subunits of yeast RNA polymerases I and III. Proc Natl Acad Sci U S A. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernabucci U, Ronchi B, Lacetera N, Nardone A. Influence of body condition score on relationships between metabolic status and oxidative stress in periparturient dairy cows. J Dairy Sci. 2005;88:2017–2026. doi: 10.3168/jds.S0022-0302(05)72878-2. [DOI] [PubMed] [Google Scholar]
- 79.Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- 80.Yao Y, Yamamoto K, Nishi Y, Nogi Y, Muramatsu M. Mouse RNA polymerase I 16-kDa subunit able to associate with 40-kDa subunit is a homolog of yeast AC19 subunit of RNA polymerases I and III. J Biol Chem. 1996;271:32881–32885. doi: 10.1074/jbc.271.51.32881. [DOI] [PubMed] [Google Scholar]
- 81.Larkin RM, Guilfoyle TJ. A 14-kDa Arabidopsis thaliana RNA polymerase III subunit contains two alpha-motifs flanked by a highly charged C terminus. Gene. 1996;172:211–215. doi: 10.1016/0378-1119(96)00030-3. [DOI] [PubMed] [Google Scholar]
- 82.Dauwerse JG, Dixon J, Seland S, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 83.Tak T, Tesselaar K, Pillay J, Borghans JA, Koenderman L. What’s your age again? determination of human neutrophil half-lives revisited. J Leukoc Biol. 2013;94:595–601. doi: 10.1189/jlb.1112571. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Gao X, Coots RA, Conn CS, Liu B, Qian SB. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat Struct Mol Biol. 2015;22:404–410. doi: 10.1038/nsmb.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 86.Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kimura M, Ishihama A. Involvement of multiple subunit-subunit contacts in the assembly of RNA polymerase II. Nucleic Acids Res. 2000;28:952–959. doi: 10.1093/nar/28.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woychik NA, Young RA. RNA polymerase II subunit RPB10 is essential for yeast cell viability. J Biol Chem. 1990;265:17816–17819. [PubMed] [Google Scholar]
- 89.Kireeva ML, Kashlev M, Burton ZF. RNA polymerase structure, function, regulation, dynamics, fidelity, and roles in gene expression. Chem Rev. 2013;113:8325–8330. doi: 10.1021/cr400436m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 91.Kenneth NS, Marshall L, White RJ. Recruitment of RNA polymerase III in vivo. Nucleic Acids Res. 2008;36:3757–3764. doi: 10.1093/nar/gkn272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Renaud M, Praz V, Vieu E, et al. Gene duplication and neofunctionalization: POLR3G and POLR3GL. Genome Res. 2014;24:37–51. doi: 10.1101/gr.161570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cherkasov V, Grousl T, Theer P, et al. Systemic control of protein synthesis through sequestration of translation and ribosome biogenesis factors during severe heat stress. FEBS Lett. 2015;589:3654–3664. doi: 10.1016/j.febslet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 94.Staley JP, Woolford JL., Jr Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr Opin Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsu TY, Simon LM, Neill NJ, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Quidville V, Alsafadi S, Goubar A, et al. Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res. 2013;73:2247–2258. doi: 10.1158/0008-5472.CAN-12-2501. [DOI] [PubMed] [Google Scholar]
- 97.Tomecki R, Dziembowski A. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA. 2010;16:1692–1724. doi: 10.1261/rna.2237610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 100.Wojcik C, Di Napoli M. Ubiquitin-proteasome system and proteasome inhibition: new strategies in stroke therapy. Stroke. 2004;35:1506–1518. doi: 10.1161/01.STR.0000126891.93919.4e. [DOI] [PubMed] [Google Scholar]
- 101.Groll M, Clausen T. Molecular shredders: how proteasomes fulfill their role. Curr Opin Struct Biol. 2003;13:665–673. doi: 10.1016/j.sbi.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 102.Kaser M, Langer T. Protein degradation in mitochondria. Semin Cell Dev Biol. 2000;11:181–190. doi: 10.1006/scdb.2000.0166. [DOI] [PubMed] [Google Scholar]
- 103.Lehmann G, Udasin RG, Ciechanover A. On the linkage between the ubiquitin-proteasome system and the mitochondria. Biochem Biophys Res Commun. 2016;473:80–86. doi: 10.1016/j.bbrc.2016.03.055. [DOI] [PubMed] [Google Scholar]
- 104.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 105.Paramo T, Piggot TJ, Bryant CE, Bond PJ. The structural basis for endotoxin-induced allosteric regulation of the Toll-like receptor 4 (TLR4) innate immune receptor. J Biol Chem. 2013;288:36215–36225. doi: 10.1074/jbc.M113.501957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shibata T, Takemura N, Motoi Y, et al. PRAT4A-dependent expression of cell surface TLR5 on neutrophils, classical monocytes and dendritic cells. Int Immunol. 2012;24:613–623. doi: 10.1093/intimm/dxs068. [DOI] [PubMed] [Google Scholar]
- 107.Hogan J, Larry Smith K. Coliform mastitis. Vet Res. 2003;34:507–519. doi: 10.1051/vetres:2003022. [DOI] [PubMed] [Google Scholar]
- 108.Deladeriere A, Gambardella L, Pan D, Anderson KE, Hawkins PT, Stephens LR. The regulatory subunits of PI3Kγ control distinct neutrophil responses. Sci Signal. 2015;8:ra8. doi: 10.1126/scisignal.2005564. [DOI] [PubMed] [Google Scholar]
- 109.Futosi K, Fodor S, Mocsai A. Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int Immunopharmacol. 2013;17:638–650. doi: 10.1016/j.intimp.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vier J, Groth M, Sochalska M, Kirschnek S. The anti-apoptotic Bcl-2 family protein A1/Bfl-1 regulates neutrophil survival and homeostasis and is controlled via PI3K and JAK/STAT signaling. Cell Death Dis. 2016;7:e2103. doi: 10.1038/cddis.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 112.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 113.Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fink K, Grandvaux N. STAT2 and IRF9: beyond ISGF3. JAKSTAT. 2013;2:e27521. doi: 10.4161/jkst.27521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ajibade AA, Wang HY, Wang RF. Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 2013;34:307–316. doi: 10.1016/j.it.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 116.Ajibade AA, Wang Q, Cui J, et al. TAK1 negatively regulates NF-κB and p38 MAP kinase activation in Gr-1+CD11b+ neutrophils. Immunity. 2012;36:43–54. doi: 10.1016/j.immuni.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stevens MG, Van Poucke M, Peelman LJ, et al. Anaphylatoxin C5a- induced toll-like receptor 4 signaling in bovine neutrophils. J Dairy Sci. 2011;94:152–164. doi: 10.3168/jds.2010-3358. [DOI] [PubMed] [Google Scholar]
- 118.Rocha DM, Caldas AP, Oliveira LL, Bressan J, Hermsdorff HH. Saturated fatty acids trigger TLR4-mediated inflammatory response. Atherosclerosis. 2016;244:211–215. doi: 10.1016/j.atherosclerosis.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 119.Wong SW, Kwon MJ, Choi AM, Kim HP, Nakahira K, Hwang DH. Fatty acids modulate Toll-like receptor 4 activation through regulation of receptor dimerization and recruitment into lipid rafts in a reactive oxygen species- dependent manner. J Biol Chem. 2009;284:27384–27392. doi: 10.1074/jbc.M109.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang S, Rutkowsky JM, Snodgrass RG, et al. Saturated fatty acids activate TLR-mediated proinflammatory signaling pathways. J Lipid Res. 2012;53:2002–2013. doi: 10.1194/jlr.D029546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matsuzawa A, Ichijo H. Redox control of cell fate by MAP kinase: physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochim Biophys Acta. 2008;1780:1325–1336. doi: 10.1016/j.bbagen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 122.Wang CH, Rong MY, Wang L, et al. CD147 up-regulates calcium-induced chemotaxis, adhesion ability and invasiveness of human neutrophils via a TRPM-7-mediated mechanism. Rheumatology (Oxford) 2014;53:2288–2296. doi: 10.1093/rheumatology/keu260. [DOI] [PubMed] [Google Scholar]
- 123.Heiner I, Eisfeld J, Luckhoff A. Role and regulation of TRP channels in neu-trophil granulocytes. Cell Calcium. 2003;33:533–540. doi: 10.1016/s0143-4160(03)00058-7. [DOI] [PubMed] [Google Scholar]
- 124.Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 125.Itagaki K, Kannan KB, Livingston DH, Deitch EA, Fekete Z, Hauser CJ. Store-operated calcium entry in human neutrophils reflects multiple contributions from independently regulated pathways. J Immunol. 2002;168:4063–4069. doi: 10.4049/jimmunol.168.8.4063. [DOI] [PubMed] [Google Scholar]
- 126.Nonnecke BJ, Kimura K, Goff JP, Kehrli ME., Jr Effects of the mammary gland on functional capacities of blood mononuclear leukocyte populations from periparturient cows. J Dairy Sci. 2003;86:2359–2368. doi: 10.3168/jds.S0022-0302(03)73829-6. [DOI] [PubMed] [Google Scholar]
- 127.Kimura K, Reinhardt TA, Goff JP. Parturition and hypocalcemia blunts calcium signals in immune cells of dairy cattle. J Dairy Sci. 2006;89:2588–2595. doi: 10.3168/jds.S0022-0302(06)72335-9. [DOI] [PubMed] [Google Scholar]
- 128.Nebzydoski SJ, Pozzo S, Nemec L, Rankin MK, Gressley TF. The effect of dexamethasone on clock gene mRNA levels in bovine neutrophils and lymphocytes. Vet Immunol Immunopathol. 2010;138:183–192. doi: 10.1016/j.vetimm.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 129.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ella K, Csepanyi-Komi R, Kaldi K. Circadian regulation of human peripheral neutrophils. Brain Behav Immun. 2016;57:209–221. doi: 10.1016/j.bbi.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 131.Zheng X, Sowcik M, Chen D, Sehgal A. Casein kinase 1 promotes synchrony of the circadian clock network. Mol Cell Biol. 2014;34:2682–2694. doi: 10.1128/MCB.01571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Auchtung TL, Salak-Johnson JL, Morin DE, Mallard CC, Dahl GE. Effects of photoperiod during the dry period on cellular immune function of dairy cows. J Dairy Sci. 2004;87:3683–3689. doi: 10.3168/jds.S0022-0302(04)73507-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.