Abstract
Although many tissues express estrogen receptor (ER)α, most studies focus on breast cancer where ERα occupies just a small fraction of its total repertoire of potential DNA-binding sites, based on sequence. This raises the question: Can ERα occupy these other potential binding sites in a different context? Ligands, splice variants, posttranslational modifications, and acquired mutations of ERα affect its conformation, which may alter chromatin interactions. To date, literature describes the DNA-binding sites of ERα (the ERα cistrome) in breast, endometrium, liver, and bone, in which the receptor mainly binds to enhancers. Chromosomal boundaries provide distinct areas for dynamic gene regulation between tissues, where the usage of enhancers deviates. Interactions of ERα with enhancers and its transcriptional complex depend on the proteome, which differs per cell type. This review discusses the biological variables that influence ERα cistromics, using reports from human specimens, cell lines, and mouse tissues, to assess whether ERα genomics in breast cancer can be translated to other tissue types.
Historically, estrogen receptor (ER)α biology is a major focus of attention due to its crucial role in breast cancer development, progression and treatment. More recently, ERα biology in several other tissues gained interest, including reproductive tissues such as prostate and endometrium (inner epithelial lining of the uterus), but also nonreproductive tissues like the liver, bone, and brain (Figure 1) (1–4). Current methods that target ERα in breast cancer treatment, affect these tissues differently.
Figure 1. Tissues that are reported to provide genomic interplay between ERα and (putative) pioneer factors.
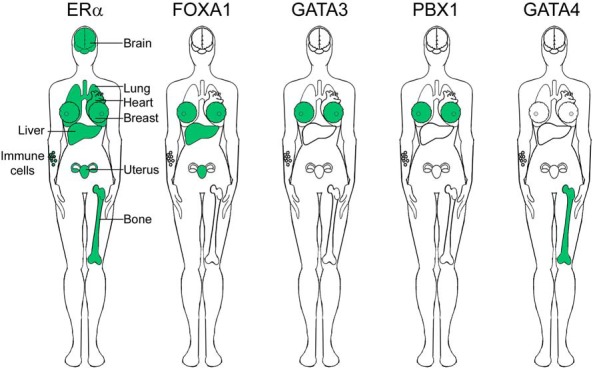
For references, see text.
Breast cancer is the most diagnosed cancer in women worldwide, with 1.67 million new cases and over half a million deaths, each year (5). Clinical studies report that 75% of breast tumors express ERα, a hormone-dependent transcription factor that is essential for tumor growth (6, 7). To block ERα-dependent tumor growth, breast cancer patients often receive tamoxifen. This small molecule inhibitor competes with estrogen to bind ERα. Although tamoxifen blocks tumor growth in breast cancer, it acts as an agonist for ERα in endometrium and osteoblasts, leading to increased risk for endometrial cancer (8–10) and increased bone density (11, 12), respectively. Thus, by targeting ERα in breast cancer, many other tissues are affected: sometimes this is beneficial, sometimes this is harmful (Table 1).
Table 1.
Examples of Estrogen's Effects Throughout the Body and the Effects of Tamoxifen
| Tissue Target | Effects of Estrogen | Effects of Tamoxifen |
|---|---|---|
| Breast | Stimulates growth (7, 130) | Blocks tumor growth (131–134) |
| Endometrium | Stimulates growth (135) | Increases risk for endometrial cancer (8–10) |
| Prostate | Controls sperm concentration (136) | increases sperm density (137) |
| Bone | Protects against osteoporosis (138): Maintains balance between bone-forming osteoblasts and bone-resorbing osteoclasts (139, 140) | Protects against osteoporosis (11, 12, 43, 141, 142) |
| Liver | Protects against diabetes: Improves glucose tolerance and insulin sensitivity (143, 144) | Increases risk for fatty liver (145, 146) |
| Brain | Protects against cognitive decline (147–149) | Decreases cognitive function (40–42) |
| Heart and vascular system | Protects against heart attacks (150–152): Decreases atherosclerosis (153–158), and widens blood vessels (159–161) | Protects against cardiovascular disease in postmenopausal women (162) |
| Lung | Promotes lung function by regulating alveolar size (163); possibly increases risk of lung cancer (164–166) | Decreases risk of death in lung cancer patients (167) |
| Immune system | Protects against allergic reactions (168, 169) | Antiallergic and immunosuppressive (169–172) |
Despite many reports on the molecular mechanism of ERα in breast cancer, we lack knowledge on the genomic action of ERα in many other tissues. Although over a century of clinical studies illustrate that ERα biology is essential throughout the body, molecular studies are comparatively new with genome-wide ERα-binding studies that are only technically feasible since the last decade (reviewed by Flach et al [13]). Genomic studies are crucial to determine the interplay between ERα and chromatin, which at specific locations, regulates genes in a tissue-specific manner.
Here, we review genomic data of ERα in multiple tissue types to compare their cistromic repertoires, and to highlight the effects that endocrine treatment of breast cancer has on this. New data provide opportunities to compare genomic activity of ERα in different physiological contexts, as multiple studies report on the genomic behavior of ERα in breast, endometrium, bone, and liver. We choose to discuss ERα's cistrome, and exclude that of ERβ due to limited cistromic data on the latter. We focus on five topics: 1) the effects of ligands on ERα; 2) tissue-specific isoforms and ligand-independent conformational changes of ERα; 3) the genomic distribution of ERα in various tissues; 4) the dynamic chromosomal architecture that influences ERα; and finally 5) the tissue-specific differences in proteome required for ERα's interaction with the chromatin. A better understanding of how drugs that target ERα in breast cancer affect other tissues provides a rationale for improving tailored endocrine treatment.
How Do Different Ligands Affect ERα Throughout the Body?
Estrogens affect many different tissues that involve both healthy physiological and pathological processes. A link between ovarian function (the main source of estrogens in premenopausal women) and breast cancer was first reported in 1882 when the breast tumor of a woman regressed as she went into menopause (14). This observation eventually led to the concept of ovariectomy as a treatment for breast cancer. And although a third of breast cancer patients benefited from this (15), it associated with a high mortality rate (16).
Currently, endocrine therapies represent the mainstay for hormonal intervention of breast cancer treatment. Small molecule ligands, such as fulvestrant and tamoxifen, compete with estrogens to bind ERα's ligand-binding domain. Fulvestrant targets the ERα for proteosomal degradation (17), whereas tamoxifen alters coregulator recruitment (18). Alternatively, aromatase inhibitors are prescribed to block estrogen synthesis.
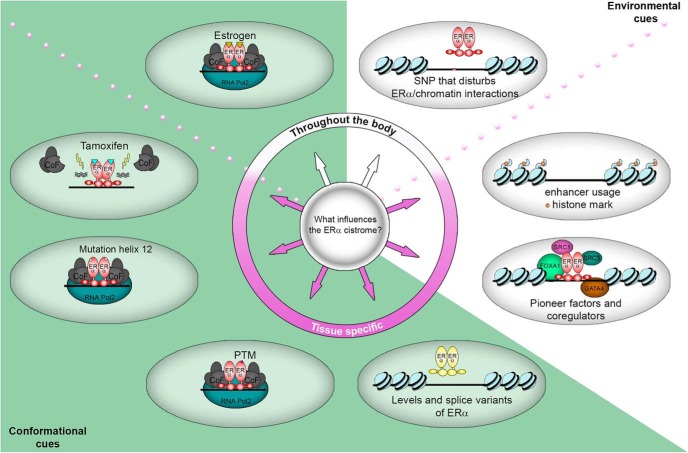
Aromatases, members of the cytochrome P450 superfamily, convert androgens into estrogens (19). Mainly the ovaries in premenopausal women, but also fat cells (20–22) and skin cells (23), express aromatases. Likewise, P450 enzymes convert tamoxifen into its active metabolites (24). Single nucleotide polymorphisms (SNPs) in genes that encode P450 enzymes may increase or decrease enzymatic activity for the conversion of androgens into estrogens (or small competitive molecules into their active metabolites), and thus alter their concentration (25).
The bloodstream carries estrogens, bound mainly to sex hormone-binding globulin (26) or serum albumin (27–29), to various organs. When unbound, estrogens diffuse through cell membranes and activate ERα (29). This causes a string of events as ERα dissociates from chaperones, binds the chromatin, and recruits coregulators (30) to regulate gene expression. In this way, estrogens drive development of female secondary sexual characteristics such as breast maturation (31), ovulation (32) and endometrial thickening (33), but also sometimes oncogenesis. Although initially linked to reproductive organs, estrogens also play many roles in nonreproductive organs, including bone density, liver metabolism and cognitive function (Table 1). Estrogens affect distinct genes depending on these tissues (34, 35).
ERα contains multiple domains including a DNA-binding domain, a hinge region and a ligand-binding domain. Within the ligand-binding domain lies helix12, which is crucial for the interaction with coregulators. Helix12 adapts its conformation when ligands bind ERα. How this structure is altered depends on the ligand: ERα in complex with agonists mediates interaction with coregulators, whereas ERα in complex with antagonists inhibits these interactions (Figure 2) (36, 37) and instead recruits other interacting partners to the complex (18). Although this alternative composition of helix12 explains tamoxifen's antagonistic effects in breast cancer, tamoxifen's agonistic features remain obscure.
Figure 2. An overview of reported factors that influence the ERα cistrome.
Conformational cues (green zone) alter the conformation of ERα, thereby influencing its potential to interact with the chromatin and interaction partner(s), whereas environmental cues by the chromatin (white zone) affect the capacity of genomic regions to bind ERα. Some cues provide opportunities for ERα to bind throughout the body (white part of the circle), whereas other cues occur in a tissue-specific manner (purple part of the circle). PTM: Posttranslational Modification.
After the success of tamoxifen in the treatment of breast cancer, novel small molecule inhibitors followed, such as raloxifene. Like tamoxifen, these new drugs compete for the ligand-binding domain of ERα. Both tamoxifen and raloxifene require interaction with amino acid D351 of the ligand-binding domain of ERα for their estrogenic/antiestrogenic properties (38). Raloxifene has a side chain that shields D351 of the ERα, which renders the complex antiestrogenic (38, 39). This occurs due to a raloxifene-induced relocation of helix12 so that coactivators required for agonistic effects no longer bind. Tamoxifen lacks this specific side chain, causing D351 to allosterically influence activation of the receptor (37). Currently, third generation antiestrogens, including lasofoxifene are being investigated for their clinical effects.
The influence of ERα exceeds breast cancer as illustrated by both physiological and pathological effects of hormones throughout the body. Many studies report that endocrine therapies disrupt beneficial effects of estrogen in nonreproductive organs (Table 1). Tamoxifen for example, increases risk for endometrial cancer (8–10) and associates with cognitive decline in a subset of patients (40–42), whereas aromatases decrease bone density (43). To prevent harmful effects of estrogens, while maintaining its benefits, requires knowledge on the genomic action of ERα for each different physiological context. But although multiple clinical and molecular studies report on estrogens and endocrine therapies to affect several tissues, many lack genomic data to describe the impact of endocrine intervention on the cistrome of ERα.
How Do Ligand-Independent Conformational Changes of ERα Affect Its Cistrome?
Different tissues express different levels of ERα. Estrogens (44–46) and other hormones (45) regulate ERα levels but little is known about the transcription factors involved. Epigenetic mechanisms, such as DNA methylation and histone acetylation, regulate ERα expression (47). ERα expression levels may not only influence its cistrome but also affect the detection of ERα binding that can be measured by current techniques.
Most studies generate data with antibodies that are unable to distinguish variants of the receptor, such as splice variants, posttranslational modifications, or mutations. ERα variants influence both the activation of ERα and its downstream effects on gene regulation. These variants might differ in levels in a tissue-specific fashion and thus add a layer of regulation to the cistromic repertoire of ERα.
Isoforms may be differentially expressed per tissue due to alternative splicing and promoter usage (Figure 2). The prevalent splice variants of ERα are 66, 46, and 36 kDa. ERα-66 contains six domains, including a ligand-binding domain and an activation domain 1. ERα-46 lacks the activation domain 1 and ERα-36 lacks both the activation domain 1 as well as most of the ligand-binding domain (48, 49).
Studies in mice on RNA levels of ERα variants (50) show that the female reproductive organs mainly produce ERα-66, whereas nonreproductive tissues also express it, but at lower levels. The heart, both of female and male mice, mostly expresses ERα-46, whereas ERα-36 is prevalent in kidney and liver of female mice only. Many of these splice variants named above however, have yet to be validated on the protein level in these tissues, both in mice and in humans.
The extracellular environment influences signaling pathways within the cell, which differs per tissue and may modify ERα posttranslationally. Hence, it can alter ERα's cistrome and transcriptional capacity (Figure 2). Examples of such posttranslational modifications include phosphorylation, acetylation and S-nitrosylation. Phosphorylation of ERα at serines 104, 106, and 118 influences its ligand-independent activation (51, 52), whereas phosphorylation at S305 redirects ERα to new transcriptional start sites (53) and allows cofactors to bind in the presence of tamoxifen, leading to agonistic effects (54, 55). Acetylation at lysine 266 and lysine 268 increases transcriptional activity of ERα (56, 57), whereas S-nitrosylation of cysteines in the DNA-binding domain inhibits it (58). However, it remains undetermined whether the latter posttranslational modifications on ERα also give rise to an altered cistrome.
Acquired mutation of the gene that encodes ERα (ESR1), which occurs in approximately 20% of metastasized breast cancers, may also influence the ERα cistrome. This acquired mutation generally occurs at Y537, D538 or both, in helix12 of the ligand-binding domain. Due to these mutations, helix12 adapts a more estrogen-like conformation that creates a constitutively active ERα (Figure 2) (59–61). Whether these mutations alter the ERα cistrome as compared with wildtype receptor, and whether other tissue-specific cancers also produce mutations in ESR1 on this type of scale, remains unexplored.
How Is ERα Distributed Across the Genome in Various Tissues?
The number of ERα-binding sites in MCF-7 cells increases upon estrogen or tamoxifen treatment, in comparison with hormone depletion. In case of a short treatment, ERα binds the same chromatin sites irrespective of the ligand, although signal intensity is typically highest for estrogen treatment (62). Upon prolonged tamoxifen treatment (in the order of months) (63), the ERα cistrome shifts and the MCF-7 cells acquire tamoxifen resistance as they regain proliferative potential despite treatment (62). These data illustrate the dynamic nature of ERα-binding sites.
ERα sites vary between primary breast tumors, and also between breast cancer cell lines (64–67). When it comes to breast cancer patients, these differences in ERα cistrome enable patient stratification on outcome, highlighting the clinical significance of ERα cistromics.
To date, public genome-wide data to describe ERα patterns in healthy mammary tissue exist only for mammary glands from healthy-6-week-old mice. Similar to human breast cancer (62, 64, 66–69), ERα occupies mostly enhancers in healthy mouse mammary glands, at DNA motifs for ERα (ESR1) but also other transcription factors, such as Transcription Factor AP-2 (activating enhancer binding protein 2, TFAP2) and Jun (70) (both proteins that were previously found to facilitate ERα action in breast cancer cell line MCF-7 [71–73]). Although genomic studies on human breast cancers identified thousands of ERα-binding sites (at DNA regions with strong enrichment for forkhead motifs) (64, 67), genomic data on healthy mice mammary glands show only hundreds of ERα-binding sites (lacking strong enrichment for forkhead motifs) (70).
It remains unclear how the difference in ERα sites of breast cancer compared with healthy tissue affects tumor biology. The higher amount of ERα sites in breast cancer potentially relates to TNFα signaling, which regulates interactions of forkhead box protein A1 (FOXA1) with the chromatin (74) and expands the number of ERα-binding sites in breast cancer cells (67, 74). However, the contrasts between mammary glands derived from healthy mice and breast cancer patients have yet to be confirmed by other studies because technical factors such as antibody specificity between different species, available tissue material, and bioinformatic thresholds, potentially influence the data.
Genomic studies in cell lines reported little resemblance in ERα cistromics between breast cancer cell line T47D and endometrial cancer cell line Ishikawa. ERα shares only 19% of binding sites between these cell lines (75–77), with deviating estrogen-responsive gene expression profiles. Shared ERα-binding sites contain high-affinity estrogen receptor elements (EREs), lack DNA methylation, and gain accessibility upon estrogen treatment (75). In contrast, cell type-specific ERα-binding sites lack high-affinity EREs and display specific DNA methylation at accessible chromatin. Cell type-specific ERα sites also show distinct DNA motifs, such as forkhead and GATA-binding protein (GATA)3 motifs at T47D-unique regions, and E26 transformation specific (ETS) protein motifs in Ishikawa-unique regions. But because ETS factors interact with ERα in MCF-7 cells (78), differences in motifs between these two cell lines might have little physiological implications, and therefore require biological validation in multiple models.
A translational study identified ERα sites in several endometrial tumors from breast cancer patients who received tamoxifen, and compared these with breast tumors (79). The data show both unique and shared binding sites between endometrial tumors and breast cancer. The ERα cistrome in these tamoxifen-associated endometrial tumors locate mainly at distal intergenic regions and introns, containing acetylation of histone H3 at lysine 27 (a marker for activity) and RNA polymerase II, suggesting occupancy at active enhancers. The ERα cistromes between these 2 reproductive tissues show much resemblance, reinforcing the question how tamoxifen blocks cell proliferation in one tissue while stimulating proliferation in the other.
Healthy mouse uteri are estrogen-responsive, and their ERα cistromes contain not only motifs previously found in breast cancer, but also unique motifs. ERα-binding sites in the uterus triple in numbers after estrogen injection of ovariectomized mice, locating mainly at introns and distal intergenic regions that contain RNA polymerase II (80). ERα sites with an ERE contain motifs of other nuclear receptor family members, whereas ERα sites lacking ERE motifs show motifs for HOX homeodomain-protein transcription factors and their cofactor pre-B-cell leukemia transcription factor 1 (PBX1) (previously identified as a putative pioneer factor in breast cancer) (81). Although the increase of binding sites resemble the genomic behavior of ERα in breast cancer cell lines, the motifs are very different, suggesting tissue-specificity of ERα interactions with the chromatin.
Genomic ERα data in liver (82) show differences and similarities with the tissues described above. Similarly, ERα-binding sites locate at distal intergenic and intronic regions that contain EREs as well as motifs for forkhead, activator protein 1, and ETS factors. In addition, the expression of ERα-target genes increases upon estrogen treatment. In contrast to what was found in other tissues, liver tissue contains ERα-binding sites proximal to genes involved in energy metabolism.
Thus far, the ERα cistrome has been reported in breast, endometrium, bone, and liver (Table 2). More tissues can be tested but some will have obstacles such as the brain, where biopsies are either taken postmortem (cut off from normal blood supply), or from diseased tissue (thus enriching for abnormalities). Another obstacle is that some tissues have very low levels of ERα as described above, which makes it more difficult to measure its cistrome. Cell lines allow for manipulation to identify proteins that mediate ERα's function, but the number of ERα-positive cell lines in various tissues is limited. Consequently, many parts of the ERα cistrome are uninvestigated and require innovative approaches to overcome these obstacles.
Table 2.
Overview of Public Genomic ERα-Binding Sites in Different Cell Types
| Tissue | Model | Method | Main Binding Regions |
|---|---|---|---|
| Breast | MCF-7 (69, 71, 81, 173) | ChIP-seq | Enhancer + intron |
| T-47D (173) | |||
| Patient tumors (64, 67) | |||
| Mouse (70) | |||
| Endometrium | Ishikawa (79, 174) | ChIP-seq | Enhancer + intron |
| Patient tumors (79) | |||
| Mouse | |||
| Bone | ERα-U2OS (119)a | ChIP-on-chip | Enhancer + intron |
| Liver | Mouse (82) | ChIP-on-chip | Enhancer + intron |
This U2OS cell line expresses ERα exogenously.
Can Chromosomal Architecture Influence ERα Distribution?
The increased number of ERα-binding sites upon estrogen induction in MCF-7 cells and mouse uteri illustrates the dynamic nature of ERα cistromics (80, 83). This dynamic nature of ERα is in part facilitated by the surrounding chromatin, which needs to be accessible for ERα to bind. Chromatin organization is essential for proper gene regulation as shown in acute myeloid leukemia (84) as well as malformation of limbs (85), in which disruptions of chromosomal boundaries at topologically associated domains cause inappropriate gene expression. These chromosomal boundaries confine regions that require coordinated regulation, thereby shielding other regions that require a different mode of regulation (86). Chromosomal boundaries are stable across cell types (86) but can be disrupted during oncogenesis, which may potentially affect the ERα cistrome and change estrogen-mediated gene expression.
In healthy tissues, chromosome boundaries are stable across cell types (86), but the regions within each domain are dynamic so that they can regulate genes according to their cell type. Within chromosomal boundaries, each region can contain multiple genes and regulatory elements such as enhancers and promoters. Enhancers control cell type specificity of gene expression, and although many enhancers are inactive in certain cells, they do function in other cells (87, 88) or respond to stimulation (89).
Active enhancers are essential for ERα action. A CRISPR-Cas9 dropout screen in the breast cancer cell lines MCF-7 and T47D identified ERα bound enhancers required for proliferation. These data suggest individual ERα sites to have substantial downstream effects on cell proliferation (90).
When regulatory elements of the genome differ per tissue, enhancer-binding transcription factors, such as ERα, will follow this divergent enhancer-activity (Figure 2). This is exemplified by data that show ERα binds near genes involved in osteoblast differentiation in bone (91), luminal breast cancer-defining genes in breast cancer (62, 92), and energy metabolism in liver (82). Thus, the chromosomal architecture defines the tissue-specific cistrome of ERα through tissue-specific enhancer-usage (88). Still, because many tissue types are relatively understudied, ERα could be more promoter-centered in yet unexplored tissues or during specific stages of tissue development.
As described above, ERα mainly occupies distal enhancers (in the reported tissues breast, endometrium, bone, and liver) and requires chromatin looping to interact with proximal promoters of genes to regulate expression (93–97). Chromosomal looping involves CCCTC-binding factor (CTCF), a ubiquitously expressed transcription factor that confines genes that require coregulation (98, 99), and defines ERα action (100). Irrespective of hormonal treatment, CTCF binds genomic regions that ERα also occupies and that associate with estrogen-regulated genes. CTCF occupies cell line-specific ERα sites more often than ERα sites that are shared between multiple breast cancer cell lines (101), suggesting that through looping, CTCF modulates the ERα cistrome in a cell line-specific fashion.
Genomic architectural studies provide valuable details about the “infrastructure” of the chromatin and its dynamic properties within the boundaries of topologically associated domains. How differences in chromatin state between cell types originate, such as differential enhancer usage, remains unknown. Tissue-specific proteomes may play a role in this process and thus affect the ERα cistrome.
How Can Other Transcription Factors Facilitate the ERα Cistrome?
Transcription factors facilitate the architectural make-up of the chromatin, with deviating expression levels among tissues. However, genomic data that indicates their direct involvement in ERα complexes, and cistrome, is lacking in many tissues (Figure 2).
As mentioned above, when ERα binds the chromatin, it recruits cofactors. These cofactors include family members of the p160 family such as stereoid receptor coactivator 1 (SRC1) (102), SRC2 (103), and SRC3 (104–107). One study investigated the varying responses of tissues to tamoxifen and found that levels of SRC1 differ per tissue (108). Yet, because coregulators follow ERα to the DNA, they are unlikely to define the genomic regions of chromatin interactions.
In luminal epithelial breast cells (67, 109), ERα requires FOXA1 to facilitate estrogen-mediated gene regulation (110) and to drive cell proliferation (62, 69). FOXA1, which depends on enhancers that are marked with dimethylation of histone H3 at lysine 4 (111), was the first pioneer factor for ERα to be identified (69). Pioneer factors bind inaccessible chromatin and make it more accessible, so that other transcription factors may bind. Clinical studies report that FOXA1 associates with a good prognosis in breast cancer patients (112). SNPs at sites of genomic interplay between ERα and FOXA1 associate with breast cancer risk (Figure 1) (113). These reports imply that FOXA1 facilitates ERα-mediated gene expression in breast cancer.
Like breast cancer, endometrial tumors express FOXA1, which associates with a favorable outcome in endometrial cancer patients (114, 115). Comparative cistromics of ERα between breast cancer and tamoxifen-associated endometrial cancer suggests ERα and FOXA1 facilitate tamoxifen-stimulatory effects in endometrial cancer development (79). These data show that FOXA1 and ERα expression in endometrial tumors, from women with a history of breast cancer, associates with the interval time between breast cancer and endometrial cancer in tamoxifen-treated breast cancer patients only. In addition, tumors of breast and tamoxifen-associated endometrial cancer patients share binding events between ERα and FOXA1. These sites are mainly at enhancers and cluster with other enhancer-bound transcription factors in the endometrial cancer cell line Ishikawa.
The liver expresses FOXA1 and FOXA2, which facilitate the activity of both ERα and the androgen receptor. The liver is greatly influenced by the hormonal environment as illustrated by sexual dimorphic features of hepatocellular carcinoma, which predominates in men (5, 116, 117). These sexual dimorphic features revers in mice that lack FOXA1 and FOXA2, as hepatocellular carcinoma predominate in females instead (3). Correspondingly, the Serpina6-rs1998056-SNP, which locates at a site of genomic interplay between ERα and FOXA1, increases the risk for hepatocellular carcinoma in women (118). These data suggest FOXA1 and FOXA2 are crucial for hormonal regulation in the liver.
In contrast to breast, endometrium, and liver, the human osteoblasts cell line U2OS lacks FOXA1 and requires GATA4 instead to facilitate genomic ERα function (119). This study used U2OS cells that expressed ERα exogenously (ERα-U2OS). Upon estrogen treatment, GATA4 binds chromatin before ERα, and its knockdown reduces ERα binding, suggesting a pioneer-like function for GATA4. Unlike FOXA1, GATA4 creates active enhancers by recruiting histone methyltransferases at enhancers, leading to H3K4me2 (91). Thus, although GATA4 and FOXA1 both bind the DNA before ERα, they operate in different fashions.
Although ERα binds mainly to enhancers with EREs in MCF-7 and ERα-U2OS, only 15% of ERα-binding sites overlap between them (119). Less than 10% of genes that are estrogen-responsive in MCF-7 cells respond to estrogen in ERα-U2OS. Instead, ERα-U2OS expresses many other genes upon estrogen stimulation.
Different tissues express different pioneer factors, which may alter ERα cistromics as exemplified by the osteoblast cell line ERα–U2OS and the breast cancer cell line MCF-7. However, these findings require further validation by other model systems because the ERα-U2OS model is intrinsically artificial. To justifiably generalize observations when comparing different organs, supportive data in multiple cell lines or primary tissues per tissue type are essential.
ERα-binding sites differ between tissues even if they do express the same pioneer factor, suggesting one pioneer factor alone is insufficient to explain deviations in the ERα cistrome (114, 115, 120). Instead, it is likely that multiple proteins, which may vary per tissue, are in fact responsible. Several molecular studies identified other (putative) pioneer factors, including GATA3 (121), activatin protein (AP) 2γ (71), and PBX1 (81), which facilitate ERα to bind the chromatin and drive breast cancer development. These transcription factors potentially function alone or together to create synergy for gene regulation.
PBX1 has been linked to breast cancer (81), ovarian cancer (122) and prostate cancer (123), but was also linked to endometrial development (124). Cistromic studies measured PBX1-binding sites in breast cancer cell lines and associated those with ERα-binding sites. In addition, PBX1 was found cytoplasmatic in endometrial cells during development (124). Hence, expression of transcription factors alone is insufficient to claim a role in ERα cistrome regulation and instead require molecular and cistromic confirmations, as has thus far mostly been done in breast cancer.
Jointly, FOXA1 and GATA3 are sufficient to drive ERα-dependent transcriptional programs. GATA3 defines ERα-positive luminal breast cancer (110), in which it is frequently mutated (125), and correlates with good prognosis (109). When introducing GATA3, ERα, and FOXA1 simultaneously to ERα-negative cell lines (MDA-MB231 and BT-549), cells respond to hormonal stimuli as they proliferate and express hormone-responsive genes (126).
Activation of other steroid hormone receptors affect ERα genomic action through direct interaction. Progesterone Receptor binds ERα upon hormone stimulation, and redistributes ERα over the genome in the breast cancer cell line MCF-7 (127). Thus, in addition to the cell's proteome, the hormonal environment (beside estrogen) controls the location of ERα-binding sites.
Some ERα-positive tissues lack certain (putative) pioneer factors (Figure 1), suggesting they play a role in tissue-specific gene regulation. In addition, ERα binds other hormone receptors that can influence its cistrome. Taken together, cell-specific proteomes allow for a cell-specific ERα cistrome.
Concluding Remarks
Estrogens play a crucial role in sexual development and protect against osteoporosis, diabetes and cognitive decline. When breast cancer patients receive tamoxifen to stop breast tumor growth, they gain bone mineral density (128), but risk endometrial cancer (10) and cognitive decline (40–42). These observations led to structure-based drug design in search of other competitive inhibitors such as raloxifene and lasofoxifene. Aromatase inhibitors can be prescribed as well, but these perturb many beneficial functions of estrogen, such as protecting against heart attack, cognitive decline and osteoporosis (Table 1). Consequently, an ideal endocrine therapeutic approach of blocking ERα would involve a more tissue-tailored mode-of-action.
The structural conformation of ERα determines its ability to interact with the chromatin and with interaction partners. As described above, this conformation of the receptor depends on ligand-binding, splice variants, posttranslational modifications and acquired mutations. Beside these structural conformations, ERα-binding events depend on enhancer activity, SNPs that disturb chromatin interactions, and other transcription factors. Taken together, these biological variables determine the ERα cistrome (Figure 2), which differs per context.
Comparative studies of ERα cistromics may identify similarities and differences between tissues, enabling selective targeting of the receptor by small-molecule design. An example of this lies in the concept of targeting FOXA1 (129), which theoretically abrogates ERα action in breast, endometrium and liver while leaving ERα unaffected in osteoblasts. In this manner, therapy manipulates ERα target tissue only, leaving the receptor unaffected in other tissues. This type of treatment may pave the way for fully tissue-selective endocrine therapeutics.
Acknowledgments
We thank the support of Pink Ribbon, KWF Dutch Cancer Society, the Netherlands Organization of Scientific Research (NWO), and The Netherlands Cancer Institute.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
We thank the support of Pink Ribbon, KWF Dutch Cancer Society, the Netherlands Organization of Scientific Research (NWO), and The Netherlands Cancer Institute.
Footnotes
- CTCF
- CCCTC-binding factor
- ER
- estrogen receptor
- ERE
- estrogen receptor element
- ESR1
- gene that encodes for estrogen receptor α
- ETS
- E26 transformation specific
- FOXA1
- forkhead box protein A1
- GATA
- GATA-binding protein
- PBX1
- pre-B-cell leukemia transcription factor 1
- SNP
- single nucleotide polymorphism
- SRC
- steroid receptor coactivator.
References
- 1. Couse JF, Lindzey J, Grandien K, Gustafsson JA, Korach KS. Tissue distribution and quantitative analysis of estrogen receptor-α (ERα) and estrogen receptor-β (ERβ) messenger ribonucleic acid in the wild-type and ERα-knockout mouse. Endocrinology. 1997;138:4613–4621. [DOI] [PubMed] [Google Scholar]
- 2. Ropero AB, Eghbali M, Minosyan TY, Tang G, Toro L, Stefani E. Heart estrogen receptor α: distinct membrane and nuclear distribution patterns and regulation by estrogen. J Mol Cell Cardiol. 2006;41:496–510. [DOI] [PubMed] [Google Scholar]
- 3. Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zwart W, Terra H, Linn SC, Schagen SB. Cognitive effects of endocrine therapy for breast cancer: keep calm and carry on? Nat Rev Clin Oncol. 2015;12:597–606. [DOI] [PubMed] [Google Scholar]
- 5. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. [DOI] [PubMed] [Google Scholar]
- 6. Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. [DOI] [PubMed] [Google Scholar]
- 7. Allred DC, Brown P, Medina D. The origins of estrogen receptor α-positive and estrogen receptor α-negative human breast cancer. Breast Cancer Res. 2004;6:240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. Endometrial cancer in tamoxifen-treated breast cancer patients: findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86:527–537. [DOI] [PubMed] [Google Scholar]
- 9. Lahti E, Blanco G, Kauppila A, Apaja-Sarkkinen M, Taskinen PJ, Laatikainen T. Endometrial changes in postmenopausal breast cancer patients receiving tamoxifen. Obstet Gynecol. 1993;81:660–664. [PubMed] [Google Scholar]
- 10. van Leeuwen FE, Benraadt J, Coebergh JW, et al. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343:448–452. [DOI] [PubMed] [Google Scholar]
- 11. Galea GL, Meakin LB, Sugiyama T, et al. Estrogen receptor α mediates proliferation of osteoblastic cells stimulated by estrogen and mechanical strain, but their acute down-regulation of the Wnt antagonist Sost is mediated by estrogen receptor β. J Biol Chem. 2013;288:9035–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nuttall ME, Stroup GB, Fisher PW, Nadeau DP, Gowen M, Suva LJ. Distinct mechanisms of action of selective estrogen receptor modulators in breast and osteoblastic cells. Am J Physiol Cell Physiol. 2000;279:C1550–C1557. [DOI] [PubMed] [Google Scholar]
- 13. Flach KD, Zwart W. The first decade of estrogen receptor cistromics in breast cancer. J Endocrinol. 2016;229:R43–R56. [DOI] [PubMed] [Google Scholar]
- 14. Nunn TW. On Cancer of the Breast. London, United Kingdom: J. & A. Churchill; 1882. p71. [Google Scholar]
- 15. Boyd S. On oophorectomy in cancer of the breast. BMJ. 1900;2:1161–1167. [Google Scholar]
- 16. Love RR, Philips J. Oophorectomy for breast cancer: history revisited. J Natl Cancer Inst. 2002;94:1433–1434. [DOI] [PubMed] [Google Scholar]
- 17. Reid G, Hübner MR, Métivier R, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. [DOI] [PubMed] [Google Scholar]
- 18. Mohammed H, Taylor C, Brown GD, Papachristou EK, Carroll JS, D'Santos CS. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11:316–326. [DOI] [PubMed] [Google Scholar]
- 19. Simpson ER, Mahendroo MS, Means GD, et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–355. [DOI] [PubMed] [Google Scholar]
- 20. Hemsell DL, Grodin JM, Brenner PF, Siiteri PK, MacDonald PC. Plasma precursors of estrogen. II. Correlation of the extent of conversion of plasma androstenedione to estrone with age. J Clin Endocrinol Metab. 1974;38:476–479. [DOI] [PubMed] [Google Scholar]
- 21. MacDonald PC, Edman CD, Hemsell DL, Porter JC, Siiteri PK. Effect of obesity on conversion of plasma androstenedione to estrone in postmenopausal women with and without endometrial cancer. Am J Obstet Gynecol. 1978;130:448–455. [DOI] [PubMed] [Google Scholar]
- 22. Edman CD, MacDonald PC. Effect of obesity on conversion of plasma androstenedione to estrone in ovulatory and anovulator young women. Am J Obstet Gynecol. 1978;130:456–461. [DOI] [PubMed] [Google Scholar]
- 23. Harada N. A unique aromatase (P-450AROM) mRNA formed by alternative use of tissue-specific exons 1 in human skin fibroblasts. Biochem Biophys Res Commun. 1992;189:1001–1007. [DOI] [PubMed] [Google Scholar]
- 24. Jordan VC, Collins MM, Rowsby L, Prestwich G. A monohydroxylated metabolite of tamoxifen with potent antioestrogenic activity. J Endocrinol. 1977;75:305–316. [DOI] [PubMed] [Google Scholar]
- 25. Preissner SC, Hoffmann MF, Preissner R, Dunkel M, Gewiess A, Preissner S. Polymorphic cytochrome P450 enzymes (CYPs) and their role in personalized therapy. PLoS One. 2013;8:e82562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan CC, Woolever CA, Bhavnani BR. Transport of equine estrogens: binding of conjugated and unconjugated equine estrogens with human serum proteins. J Clin Endocrinol Metab. 1985;61:499–507. [DOI] [PubMed] [Google Scholar]
- 27. Anderson JN, Peck EJ Jr, Clark JH. Nuclear receptor-estrogen complex: in vivo and in vitro binding of estradiol and estriol as influenced by serum albumin. J Steroid Biochem. 1974;5:103–107. [DOI] [PubMed] [Google Scholar]
- 28. Dunn JF, Nisula BC, Rodbard D. Transport of steroid hormones: binding of 21 endogenous steroids to both testosterone-binding globulin and corticosteroid-binding globulin in human plasma. J Clin Endocrinol Metab. 1981;53:58–68. [DOI] [PubMed] [Google Scholar]
- 29. Södergård R, Bäckström T, Shanbhag V, Carstensen H. Calculation of free and bound fractions of testosterone and estradiol-17 β to human plasma proteins at body temperature. J Steroid Biochem. 1982;16:801–810. [DOI] [PubMed] [Google Scholar]
- 30. Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. [DOI] [PubMed] [Google Scholar]
- 31. Bocchinfuso WP, Korach KS. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia. 1997;2:323–334. [DOI] [PubMed] [Google Scholar]
- 32. Schomberg DW, Couse JF, Mukherjee A, et al. Targeted disruption of the estrogen receptor-α gene in female mice: characterization of ovarian responses and phenotype in the adult. Endocrinology. 1999;140:2733–2744. [DOI] [PubMed] [Google Scholar]
- 33. Tibbetts TA, Mendoza-Meneses M, O'Malley BW, Conneely OM. Mutual and intercompartmental regulation of estrogen receptor and progesterone receptor expression in the mouse uterus. Biol Reprod. 1998;59:1143–1152. [DOI] [PubMed] [Google Scholar]
- 34. Tang S, Han H, Bajic VB. ERGDB: estrogen responsive genes database. Nucleic Acids Res. 2004;32:D533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang S, Zhang Z, Tan SL, et al. KBERG: knowledge base for estrogen responsive genes. Nucleic Acids Res. 2007;35:D732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brzozowski AM, Pike AC, Dauter Z, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. [DOI] [PubMed] [Google Scholar]
- 37. Shiau AK, Barstad D, Loria PM, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. [DOI] [PubMed] [Google Scholar]
- 38. Liu H, Park WC, Bentrem DJ, et al. Structure-function relationships of the raloxifene-estrogen receptor-α complex for regulating transforming growth factor-α expression in breast cancer cells. J Biol Chem. 2002;277:9189–9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levenson AS, Jordan VC. The key to the antiestrogenic mechanism of raloxifene is amino acid 351 (aspartate) in the estrogen receptor. Cancer Res. 1998;58:1872–1875. [PubMed] [Google Scholar]
- 40. Collins B, Mackenzie J, Stewart A, Bielajew C, Verma S. Cognitive effects of hormonal therapy in early stage breast cancer patients: a prospective study. Psychooncology. 2009;18:811–821. [DOI] [PubMed] [Google Scholar]
- 41. Jenkins V, Shilling V, Fallowfield L, Howell A, Hutton S. Does hormone therapy for the treatment of breast cancer have a detrimental effect on memory and cognition? A pilot study. Psychooncology. 2004;13:61–66. [DOI] [PubMed] [Google Scholar]
- 42. Schilder CM, Seynaeve C, Beex LV, et al. Effects of tamoxifen and exemestane on cognitive functioning of postmenopausal patients with breast cancer: results from the neuropsychological side study of the tamoxifen and exemestane adjuvant multinational trial. J Clin Oncol. 2010;28:1294–1300. [DOI] [PubMed] [Google Scholar]
- 43. Eastell R, Adams JE, Coleman RE, et al. Effect of anastrozole on bone mineral density: 5-year results from the anastrozole, tamoxifen, alone or in combination trial 18233230. J Clin Oncol. 2008;26:1051–1057. [DOI] [PubMed] [Google Scholar]
- 44. Ihionkhan CE, Chambliss KL, Gibson LL, Hahner LD, Mendelsohn ME, Shaul PW. Estrogen causes dynamic alterations in endothelial estrogen receptor expression. Circ Res. 2002;91:814–820. [DOI] [PubMed] [Google Scholar]
- 45. Pinzone JJ, Stevenson H, Strobl JS, Berg PE. Molecular and cellular determinants of estrogen receptor α expression. Mol Cell Biol. 2004;24:4605–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaneko KJ, Furlow JD, Gorski J. Involvement of the coding sequence for the estrogen receptor gene in autologous ligand-dependent down-regulation. Mol Endocrinol. 1993;7:879–888. [DOI] [PubMed] [Google Scholar]
- 47. Yang X, Phillips DL, Ferguson AT, Nelson WG, Herman JG, Davidson NE. Synergistic activation of functional estrogen receptor (ER)-α by DNA methyltransferase and histone deacetylase inhibition in human ER-α-negative breast cancer cells. Cancer Res. 2001;61:7025–7029. [PubMed] [Google Scholar]
- 48. Flouriot G, Brand H, Denger S, et al. Identification of a new isoform of the human estrogen receptor-α (hER-α) that is encoded by distinct transcripts and that is able to repress hER-α activation function 1. EMBO J. 2000;19:4688–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. Identification, cloning, and expression of human estrogen receptor-α36, a novel variant of human estrogen receptor-α66. Biochem Biophys Res Commun. 2005;336:1023–1027. [DOI] [PubMed] [Google Scholar]
- 50. Irsik DL, Carmines PK, Lane PH. Classical estrogen receptors and ERα splice variants in the mouse. PLoS One. 2013;8:e70926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thomas RS, Sarwar N, Phoenix F, Coombes RC, Ali S. Phosphorylation at serines 104 and 106 by Erk1/2 MAPK is important for estrogen receptor-α activity. J Mol Endocrinol. 2008;40:173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen D, Washbrook E, Sarwar N, et al. Phosphorylation of human estrogen receptor α at serine 118 by two distinct signal transduction pathways revealed by phosphorylation-specific antisera. Oncogene. 2002;21:4921–4931. [DOI] [PubMed] [Google Scholar]
- 53. de Leeuw R, Flach K, Bentin Toaldo C, et al. PKA phosphorylation redirects ERα to promoters of a unique gene set to induce tamoxifen resistance. Oncogene. 2013;32:3543–3551. [DOI] [PubMed] [Google Scholar]
- 54. Zwart W, Griekspoor A, Berno V, et al. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERα towards co-activator SRC-1. EMBO J. 2007;26:3534–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Michalides R, Griekspoor A, Balkenende A, et al. Tamoxifen resistance by a conformational arrest of the estrogen receptor α after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. [DOI] [PubMed] [Google Scholar]
- 56. Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL. Acetylation of estrogen receptor α by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol. 2006;20:1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguère V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor α. Mol Endocrinol. 2010;24:1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garbán HJ, Márquez-Garbán DC, Pietras RJ, Ignarro LJ. Rapid nitric oxide-mediated S-nitrosylation of estrogen receptor: regulation of estrogen-dependent gene transcription. Proc Natl Acad Sci USA. 2005;102:2632–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-α: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73:6856–6864. [DOI] [PubMed] [Google Scholar]
- 60. Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45:1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Knowlden JM, Hutcheson IR, Jones HE, et al. Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology. 2003;144:1032–1044. [DOI] [PubMed] [Google Scholar]
- 64. Jansen MP, Knijnenburg T, Reijm EA, et al. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res. 2013;73:6632–6641. [DOI] [PubMed] [Google Scholar]
- 65. Ross-Innes CS, Stark R, Holmes KA, et al. Cooperative interaction between retinoic acid receptor-α and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zwart W, Flach KD, Rudraraju B, et al. SRC3 phosphorylation at Serine 543 is a positive independent prognostic factor in ER positive breast cancer. Clin Cancer Res. 2016;22:479–491. [DOI] [PubMed] [Google Scholar]
- 67. Ross-Innes CS, Stark R, Teschendorff AE, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zwart W, Koornstra R, Wesseling J, Rutgers E, Linn S, Carroll JS. A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics. 2013;14:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. [DOI] [PubMed] [Google Scholar]
- 70. Nautiyal J, Steel JH, Mane MR, et al. The transcriptional co-factor RIP140 regulates mammary gland development by promoting the generation of key mitogenic signals. Development. 2013;140:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tan SK, Lin ZH, Chang CW, et al. AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011;30:2569–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Petz LN, Ziegler YS, Loven MA, Nardulli AM. Estrogen receptor α and activating protein-1 mediate estrogen responsiveness of the progesterone receptor gene in MCF-7 breast cancer cells. Endocrinology. 2002;143:4583–4591. [DOI] [PubMed] [Google Scholar]
- 73. Carroll JS, Meyer CA, Song J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 74. Franco HL, Nagari A, Kraus WL. TNFα signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell. 2015;58:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gertz J, Savic D, Varley KE, et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Korch C, Spillman MA, Jackson TA, et al. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gertz J, Reddy TE, Varley KE, Garabedian MJ, Myers RM. Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome Res. 2012;22:2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kalet BT, Anglin SR, Handschy A, et al. Transcription factor Ets1 cooperates with estrogen receptor α to stimulate estradiol-dependent growth in breast cancer cells and tumors. PLoS One. 2013;8:e68815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Droog M, Nevedomskaya E, Kim Y, et al. Comparative cistromics reveals genomic crosstalk between FOXA1 and ER-α in tamoxifen-associated endometrial carcinomas. Cancer Res. 2016;76:3773–3784. [DOI] [PubMed] [Google Scholar]
- 80. Hewitt SC, Li L, Grimm SA, et al. Research resource: whole-genome estrogen receptor α binding in mouse uterine tissue revealed by ChIP-seq. Mol Endocrinol. 2012;26:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gao H, Fält S, Sandelin A, Gustafsson JA, Dahlman-Wright K. Genome-wide identification of estrogen receptor α-binding sites in mouse liver. Mol Endocrinol. 2008;22:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Welboren WJ, van Driel MA, Janssen-Megens EM, et al. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. [DOI] [PubMed] [Google Scholar]
- 85. Lupiáñez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mercer EM, Lin YC, Benner C, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ostuni R, Piccolo V, Barozzi I, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. [DOI] [PubMed] [Google Scholar]
- 90. Korkmaz G, Lopes R, Ugalde AP, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34:192–198. [DOI] [PubMed] [Google Scholar]
- 91. Miranda-Carboni GA, Guemes M, Bailey S, et al. GATA4 regulates estrogen receptor-α-mediated osteoblast transcription. Mol Endocrinol. 2011;25:1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zwart W, Theodorou V, Kok M, Canisius S, Linn S, Carroll JS. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 2011;30:4764–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Barnett DH, Sheng S, Charn TH, et al. Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 2008;68:3505–3515. [DOI] [PubMed] [Google Scholar]
- 94. Deschênes J, Bourdeau V, White JH, Mader S. Regulation of GREB1 transcription by estrogen receptor α through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J Biol Chem. 2007;282:17335–17339. [DOI] [PubMed] [Google Scholar]
- 95. Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hsu PY, Hsu HK, Singer GA, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20:733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pan YF, Wansa KD, Liu MH, et al. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283:32977–32988. [DOI] [PubMed] [Google Scholar]
- 98. Kim TH, Abdullaev ZK, Smith AD, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci USA. 2007;104:7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chan CS, Song JS. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res. 2008;68:9041–9049. [DOI] [PubMed] [Google Scholar]
- 101. Ross-Innes CS, Brown GD, Carroll JS. A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics. 2011;12:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Oñate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. [DOI] [PubMed] [Google Scholar]
- 103. Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. [DOI] [PubMed] [Google Scholar]
- 105. Chen H, Lin RJ, Schiltz RL, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. [DOI] [PubMed] [Google Scholar]
- 106. Suen CS, Berrodin TJ, Mastroeni R, Cheskis BJ, Lyttle CR, Frail DE. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J Biol Chem. 1998;273:27645–27653. [DOI] [PubMed] [Google Scholar]
- 107. Torchia J, Rose DW, Inostroza J, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. [DOI] [PubMed] [Google Scholar]
- 108. Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. [DOI] [PubMed] [Google Scholar]
- 109. Hisamatsu Y, Tokunaga E, Yamashita N, et al. Impact of GATA-3 and FOXA1 expression in patients with hormone receptor-positive/HER2-negative breast cancer. Breast Cancer. 2015;22:520–528. [DOI] [PubMed] [Google Scholar]
- 110. Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. [DOI] [PubMed] [Google Scholar]
- 111. Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mehta RJ, Jain RK, Leung S, et al. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Cancer Res Treat. 2012;131:881–890. [DOI] [PubMed] [Google Scholar]
- 113. Cowper-Sal lari R, Zhang X, Wright JB, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44:1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP. FOXA1: growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer. 2007;120:1013–1022. [DOI] [PubMed] [Google Scholar]
- 115. Tangen IL, Krakstad C, Halle MK, et al. Switch in FOXA1 status associates with endometrial cancer progression. PLoS One. 2014;9:e98069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nakatani T, Roy G, Fujimoto N, Asahara T, Ito A. Sex hormone dependency of diethylnitrosamine-induced liver tumors in mice and chemoprevention by leuprorelin. Jpn J Cancer Res. 2001;92:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Naugler WE, Sakurai T, Kim S, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. [DOI] [PubMed] [Google Scholar]
- 118. Shen N, Gong J, Wang Y, et al. Integrative genomic analysis identifies that SERPINA6-rs1998056 regulated by FOXA/ERα is associated with female hepatocellular carcinoma. PLoS One. 2014;9:e107246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. Unique ERα cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22:2393–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang J, Bao W, Qiu M, et al. Forkhead-box A1 suppresses the progression of endometrial cancer via crosstalk with estrogen receptor α. Oncol Rep. 2014;31:1225–1234. [DOI] [PubMed] [Google Scholar]
- 121. Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013;23:12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Park JT, Shih IeM, Wang TL. Identification of Pbx1, a potential oncogene, as a Notch3 target gene in ovarian cancer. Cancer Res. 2008;68:8852–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kikugawa T, Kinugasa Y, Shiraishi K, et al. PLZF regulates Pbx1 transcription and Pbx1-HoxC8 complex leads to androgen-independent prostate cancer proliferation. Prostate. 2006;66:1092–1099. [DOI] [PubMed] [Google Scholar]
- 124. Dintilhac A, Bihan R, Guerrier D, et al. PBX1 intracellular localization is independent of MEIS1 in epithelial cells of the developing female genital tract. Int J Dev Biol. 2005;49:851–858. [DOI] [PubMed] [Google Scholar]
- 125. Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kong SL, Li G, Loh SL, Sung WK, Liu ET. Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol. 2011;7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mohammed H, Russell IA, Stark R, et al. Progesterone receptor modulates ERα action in breast cancer. Nature. 2015;523:313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Love RR, Mazess RB, Barden HS, et al. Effects of tamoxifen on bone mineral density in postmenopausal women with breast cancer. N Engl J Med. 1992;326:852–856. [DOI] [PubMed] [Google Scholar]
- 129. Nakshatri H, Badve S. FOXA1 as a therapeutic target for breast cancer. Expert Opin Ther Targets. 2007;11:507–514. [DOI] [PubMed] [Google Scholar]
- 130. Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. [DOI] [PubMed] [Google Scholar]
- 131. Fisher B, Costantino J, Redmond C, et al. A randomized clinical trial evaluating tamoxifen in the treatment of patients with node-negative breast cancer who have estrogen-receptor-positive tumors. N Engl J Med. 1989;320:479–484. [DOI] [PubMed] [Google Scholar]
- 132. Powles TJ, Hardy JR, Ashley SE, et al. A pilot trial to evaluate the acute toxicity and feasibility of tamoxifen for prevention of breast cancer. Br J Cancer. 1989;60:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wolmark N, Redmond C, Fisher B. A comparison of two and three years of adjuvant tamoxifen. Horm Res. 1989;32(suppl 1):166–168. [DOI] [PubMed] [Google Scholar]
- 134. Sutherland RL, Hall RE, Taylor IW. Cell proliferation kinetics of MCF-7 human mammary carcinoma cells in culture and effects of tamoxifen on exponentially growing and plateau-phase cells. Cancer Res. 1983;43:3998–4006. [PubMed] [Google Scholar]
- 135. Pollard JW, Pacey J, Cheng SV, Jordan EG. Estrogens and cell death in murine uterine luminal epithelium. Cell Tissue Res. 1987;249:533–540. [DOI] [PubMed] [Google Scholar]
- 136. Hess RA, Bunick D, Lee KH, et al. A role for oestrogens in the male reproductive system. Nature. 1997;390:509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kotoulas IG, Cardamakis E, Michopoulos J, Mitropoulos D, Dounis A. Tamoxifen treatment in male infertility. I. Effect on spermatozoa. Fertil Steril. 1994;61:911–914. [DOI] [PubMed] [Google Scholar]
- 138. Torgerson DJ, Bell-Syer SE. Hormone replacement therapy and prevention of nonvertebral fractures: a meta-analysis of randomized trials. JAMA. 2001;285:2891–2897. [DOI] [PubMed] [Google Scholar]
- 139. Krum SA, Miranda-Carboni GA, Hauschka PV, et al. Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J. 2008;27:535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Imai Y, Youn MY, Kondoh S, et al. Estrogens maintain bone mass by regulating expression of genes controlling function and life span in mature osteoclasts. Ann NY Acad Sci. 2009;1173(suppl 1):E31–E39. [DOI] [PubMed] [Google Scholar]
- 141. Grey AB, Stapleton JP, Evans MC, Tatnell MA, Ames RW, Reid IR. The effect of the antiestrogen tamoxifen on bone mineral density in normal late postmenopausal women. Am J Med. 1995;99:636–641. [DOI] [PubMed] [Google Scholar]
- 142. Marttunen MB, Hietanen P, Tiitinen A, Ylikorkala O. Comparison of effects of tamoxifen and toremifene on bone biochemistry and bone mineral density in postmenopausal breast cancer patients. J Clin Endocrinol Metab. 1998;83:1158–1162. [DOI] [PubMed] [Google Scholar]
- 143. Louet JF, LeMay C, Mauvais-Jarvis F. Antidiabetic actions of estrogen: insight from human and genetic mouse models. Curr Atheroscler Rep. 2004;6:180–185. [DOI] [PubMed] [Google Scholar]
- 144. Bryzgalova G, Gao H, Ahren B, et al. Evidence that oestrogen receptor-α plays an important role in the regulation of glucose homeostasis in mice: insulin sensitivity in the liver. Diabetologia. 2006;49:588–597. [DOI] [PubMed] [Google Scholar]
- 145. Ogawa Y, Murata Y, Nishioka A, Inomata T, Yoshida S. Tamoxifen-induced fatty liver in patients with breast cancer. Lancet. 1998;351:725. [DOI] [PubMed] [Google Scholar]
- 146. Nishino M, Hayakawa K, Nakamura Y, Morimoto T, Mukaihara S. Effects of tamoxifen on hepatic fat content and the development of hepatic steatosis in patients with breast cancer: high frequency of involvement and rapid reversal after completion of tamoxifen therapy. AJR Am J Roentgenol. 2003;180:129–134. [DOI] [PubMed] [Google Scholar]
- 147. Rivera CM, Grossardt BR, Rhodes DJ, Rocca WA. Increased mortality for neurological and mental diseases following early bilateral oophorectomy. Neuroepidemiology. 2009;33:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Luine VN, Khylchevskaya RI, McEwen BS. Effect of gonadal steroids on activities of monoamine oxidase and choline acetylase in rat brain. Brain Res. 1975;86:293–306. [DOI] [PubMed] [Google Scholar]
- 149. Sandstrom NJ, Williams CL. Memory retention is modulated by acute estradiol and progesterone replacement. Behav Neurosci. 2001;115:384–393. [PubMed] [Google Scholar]
- 150. Wang F, He Q, Sun Y, Dai X, Yang XP. Female adult mouse cardiomyocytes are protected against oxidative stress. Hypertension. 2010;55:1172–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Lerner DJ, Kannel WB. Patterns of coronary heart disease morbidity and mortality in the sexes: a 26-year follow-up of the Framingham population. Am Heart J. 1986;111:383–390. [DOI] [PubMed] [Google Scholar]
- 152. Pérez-López FR, Chedraui P, Gilbert JJ, Pérez-Roncero G. Cardiovascular risk in menopausal women and prevalent related co-morbid conditions: facing the post-Women's Health Initiative era. Fertil Steril. 2009;92:1171–1186. [DOI] [PubMed] [Google Scholar]
- 153. Hodgin JB, Krege JH, Reddick RL, Korach KS, Smithies O, Maeda N. Estrogen receptor α is a major mediator of 17β-estradiol's atheroprotective effects on lesion size in Apoe−/− mice. J Clin Invest. 2001;107:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Adams MR, Kaplan JR, Manuck SB, et al. Inhibition of coronary artery atherosclerosis by 17-β estradiol in ovariectomized monkeys. Lack of an effect of added progesterone. Arteriosclerosis. 1990;10:1051–1057. [DOI] [PubMed] [Google Scholar]
- 155. Bourassa PA, Milos PM, Gaynor BJ, Breslow JL, Aiello RJ. Estrogen reduces atherosclerotic lesion development in apolipoprotein E-deficient mice. Proc Natl Acad Sci USA. 1996;93:10022–10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Elhage R, Arnal JF, Pieraggi MT, et al. 17 β-estradiol prevents fatty streak formation in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 1997;17:2679–2684. [DOI] [PubMed] [Google Scholar]
- 157. Haarbo J, Leth-Espensen P, Stender S, Christiansen C. Estrogen monotherapy and combined estrogen-progestogen replacement therapy attenuate aortic accumulation of cholesterol in ovariectomized cholesterol-fed rabbits. J Clin Invest. 1991;87:1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Holm P, Stender S, Andersen HO, Hansen BF, Nordestgaard BG. Antiatherogenic effect of estrogen abolished by balloon catheter injury in cholesterol-clamped rabbits. Arterioscler Thromb Vasc Biol. 1997;17:1504–1511. [DOI] [PubMed] [Google Scholar]
- 159. Caulin-Glaser T, García-Cardeña G, Sarrel P, Sessa WC, Bender JR. 17 β-estradiol regulation of human endothelial cell basal nitric oxide release, independent of cytosolic Ca2+ mobilization. Circ Res. 1997;81:885–892. [DOI] [PubMed] [Google Scholar]
- 160. Mendelsohn ME, Karas RH. The protective effects of estrogen on the cardiovascular system. N Engl J Med. 1999;340:1801–1811. [DOI] [PubMed] [Google Scholar]
- 161. Lantin-Hermoso RL, Rosenfeld CR, Yuhanna IS, German Z, Chen Z, Shaul PW. Estrogen acutely stimulates nitric oxide synthase activity in fetal pulmonary artery endothelium. Am J Physiol. 1997;273:L119–L126. [DOI] [PubMed] [Google Scholar]
- 162. Grey AB, Stapleton JP, Evans MC, Reid IR. The effect of the anti-estrogen tamoxifen on cardiovascular risk factors in normal postmenopausal women. J Clin Endocrinol Metab. 1995;80:3191–3195. [DOI] [PubMed] [Google Scholar]
- 163. Massaro D, Massaro GD. Estrogen regulates pulmonary alveolar formation, loss, and regeneration in mice. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1154–L1159. [DOI] [PubMed] [Google Scholar]
- 164. Mollerup S, Jørgensen K, Berge G, Haugen A. Expression of estrogen receptors α and β in human lung tissue and cell lines. Lung Cancer. 2002;37:153–159. [DOI] [PubMed] [Google Scholar]
- 165. Pietras RJ, Márquez DC, Chen HW, Tsai E, Weinberg O, Fishbein M. Estrogen and growth factor receptor interactions in human breast and non-small cell lung cancer cells. Steroids. 2005;70:372–381. [DOI] [PubMed] [Google Scholar]
- 166. Stabile LP, Davis AL, Gubish CT, et al. Human non-small cell lung tumors and cells derived from normal lung express both estrogen receptor α and β and show biological responses to estrogen. Cancer Res. 2002;62:2141–2150. [PubMed] [Google Scholar]
- 167. Bouchardy C, Benhamou S, Schaffar R, et al. Lung cancer mortality risk among breast cancer patients treated with anti-estrogens. Cancer. 2011;117:1288–1295. [DOI] [PubMed] [Google Scholar]
- 168. Priyanka HP, Krishnan HC, Singh RV, Hima L, Thyagarajan S. Estrogen modulates in vitro T cell responses in a concentration- and receptor-dependent manner: effects on intracellular molecular targets and antioxidant enzymes. Mol Immunol. 2013;56:328–339. [DOI] [PubMed] [Google Scholar]
- 169. Babina M, Kirn F, Hoser D, et al. Tamoxifen counteracts the allergic immune response and improves allergen-induced dermatitis in mice. Clin Exp Allergy. 2010;40:1256–1265. [DOI] [PubMed] [Google Scholar]
- 170. Joffroy CM, Buck MB, Stope MB, Popp SL, Pfizenmaier K, Knabbe C. Antiestrogens induce transforming growth factor β-mediated immunosuppression in breast cancer. Cancer Res. 2010;70:1314–1322. [DOI] [PubMed] [Google Scholar]
- 171. Nalbandian G, Paharkova-Vatchkova V, Mao A, Nale S, Kovats S. The selective estrogen receptor modulators, tamoxifen and raloxifene, impair dendritic cell differentiation and activation. J Immunol. 2005;175:2666–2675. [DOI] [PubMed] [Google Scholar]
- 172. Sthoeger ZM, Bentwich Z, Zinger H, Mozes E. The beneficial effect of the estrogen antagonist, tamoxifen, on experimental systemic lupus erythematosus. J Rheumatol. 1994;21:2231–2238. [PubMed] [Google Scholar]
- 173. Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res. 2007;67:6477–6483. [DOI] [PubMed] [Google Scholar]
- 174. ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. [DOI] [PubMed] [Google Scholar]