Abstract
The purpose of this study was to find disease-associated genes and potential mechanisms in head and neck squamous cell carcinoma (HNSCC) with deoxyribonucleic acid microarrays. The gene expression profiles of GSE6791 were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were obtained with packages in R language and STRING constructed protein–protein interaction (PPI) network of the DEGs with combined score >0.8. Subsequently, module analysis of the PPI network was performed by Molecular Complex Detection plugin and functions and pathways of the hub gene in subnetwork were studied. Finally, overall survival analysis of hub genes was verified in TCGA HNSCC cohort. A total of 811 DEGs were obtained, which were mainly enriched in the terms related to extracellular matrix (ECM)–receptor interaction, ECM structural constituent, and ECM organization. A PPI network was constructed, consisting of 401 nodes and 1,254 edges and 15 hub genes with high degrees in the network. High expression of 4 genes of the 15 genes was associated with poor OS of patients in HNSCC, including PSMA7, ITGA6, ITGB4, and APP. Two significant modules were detected from the PPI network, and the enriched functions and pathways included proteasome, ECM organization, and ECM–receptor interaction. In conclusion, we propose that PSMA7, ITGA6, ITGB4, and APP may be further explored as potential biomarkers to aid HNSCC diagnosis and treatment.
Keywords: head and neck squamous cell carcinoma, interaction network, prognostic biomarkers, function and pathway analysis
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide, with ~650,000 new cases and nearly 350,000 patient deaths from HNSCC annually.1 Prognosis remains poor, and the 5-year survival rates for HNSCC patients continue to be <50%. Local tumor recurrence, distant metastasis, and therapeutic resistance appear to be the major contributing factors for this low survival rate.2
Previously identified biomarkers can help in predicting the prognosis of HNSCC. However, their clinical application is limited. Currently, there is no evidence-based recommendation for altering the treatment of patients with HNSCC by the expression of individual biomarkers.3 Therefore, it is crucial to investigate the molecular mechanisms involved in proliferation, apoptosis, and invasion of HNSCC and discover more effective biomarkers of HNSCC to improve diagnosis and prevention of the disease.
Currently, genetic and genomics research is developing rapidly, which helps us to understand the potential mechanisms of some diseases.4,5 For example, microarray analysis is widely used in the field of cancer genetics research, which may measure gene expression on a genome-wide scale simultaneously.6
In the present study, the biological informatics approach was used to analyze the gene expression profiles in HNSCC, and functional analysis was performed to identify differentially expressed genes (DEGs) between HNSCC and normal control. Subsequently, network analysis was applied for the DEGs and a protein–protein interaction (PPI) network was constructed; then, we investigated whether the hub gene of the subnetwork could reduce the overall survival (OS) in TCGA database. Through analyzing their biological functions, pathways, and OS, we may bring to light the underlying mechanisms of HNSCC development and identify the potential candidate biomarkers for diagnosis, prognosis, and drug targets.
Materials and methods
Microarray data
Microarray expression profiles of GSE67917 were downloaded from Gene Expression Omnibus database for identifying DEGs of HNSCC. GSE6791, which was already deposited in GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array, Santa Clara, CA, USA), consisted of 42 HNSCC samples and 14 normal epithelial samples.
Data preprocessing and identification of DEGs
The raw array data were subjected to background correction and quartile data normalization. Then, the DEGs between HNSCC samples and normal controls were identified using the empirical Bayes approach in linear models for the microarray data (limma) package.8 |log FC| >1 and P<0.05 were selected as the cutoff criterion.
Functional and pathway enrichment analysis of DEGs
The Database for Annotation, Visualization, and Integrated Discovery (DAVID),9 which is a comprehensive set of functional annotation tools, has been used for systematic and integrative analysis of large gene lists. In this work, the significant gene ontology (GO) biological process terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the identified DEGs were performed using DAVID database with the thresholds of P<0.05.
Modules from the PPI network
To evaluate the interactive relationships among DEGs, the DEGs were mapped to the Search Tool for the Retrieval of Interacting Genes(STRING) database.10 Then, the interaction relationships of DEGs were selected to construct the PPI network (combined score >0.8) and visualized using Cytoscape.11 The Molecular Complex Detection (MCODE) plugin12 in Cytoscape was used to screen the modules of PPI network, using cutoff values as follows: MCODE scores >15 and number of nodes >15. Moreover, the function and pathway enrichment analysis of DEGs in each module was performed using DAVID.
Survival analysis of the hub gene
OS analysis was performed using HNSCC samples from the TCGA dataset and mRNA Z-score data files were downloaded from the cBioPortal.13 Patients were classified into high or low expression based on whether Z-score expression was > median (high) or < median (low). Based on these categories, log-rank analysis and Kaplan–Meier plots were produced using Prism Software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Identification of DEGs
The total number of samples analyzed was 42 HNSCC samples, along with 14 normal epithelial samples. After data preprocessing, DEG analysis was performed using the limma software package. A total of 811 genes were identified after the analyses of GSE6791, including 550 upregulated and 261 downregulated genes.
GO and KEGG pathway enrichment analyses
We uploaded all 811 DEGs to the online software DAVID to identify overrepresented GO categories and KEGG pathways. GO analysis results showed that the most overrepresented GO terms in biological processes were enriched in extracellular matrix (ECM) organization, antigen processing and presentation of exogenous peptide antigen via major histocompatibility class I, transporter associated with antigen processing-dependent, and collagen catabolic process. In addition, the most enriched GO terms in molecular function and cellular component were threonine-type endopeptidase activity and extracellular exosome, respectively. On the other hand, the most enriched KEGG pathway terms were as follows: ECM–receptor interaction, amebiasis, proteasome, focal adhesion, and small cell lung cancer (Table 1).
Table 1.
Functional and pathway enrichment analysis of upregulated and downregulated DEGs in HNSCC
| ID | Go term | Count | P-value |
|---|---|---|---|
| GO_function | |||
| GO_BP:0030198 | ECM organization | 43 | 2.07E–18 |
| GO_BP:0002479 | Antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent | 23 | 5.77E–15 |
| GO_BP:0030574 | Collagen catabolic process | 21 | 1.10E–12 |
| GO_BP:0060337 | Type I interferon signaling pathway | 20 | 1.13E–11 |
| GO_BP:0031145 | Anaphase-promoting complex-dependent catabolic process | 21 | 8.10E–11 |
| GO_CC:0070062 | Extracellular exosome | 213 | 2.62E–20 |
| GO_CC:0005576 | Extracellular region | 129 | 1.88E–13 |
| GO_CC:0005615 | Extracellular space | 113 | 4.42E–13 |
| GO_CC:0005578 | Proteinaceous ECM | 41 | 1.51E–12 |
| GO_CC:0031012 | ECM | 43 | 2.31E–12 |
| GO_MF:0004298 | Threonine-type endopeptidase activity | 11 | 4.09E–09 |
| GO_MF:0005201 | ECM structural constituent | 17 | 1.38E–08 |
| GO_MF:0005518 | Collagen binding | 14 | 1.05E–06 |
| GO_MF:0004252 | Serine-type endopeptidase activity | 28 | 1.18E–05 |
| GO_MF:0005515 | Protein binding | 429 | 1.27E–05 |
| KEGG_PATHWAY | |||
| Hsa:04512 | ECM–receptor interaction | 23 | 1.23E–10 |
| Hsa:05146 | Amebiasis | 25 | 2.11E–10 |
| Hsa:03050 | Proteasome | 15 | 1.25E–08 |
| Hsa:04510 | Focal adhesion | 30 | 2.76E–07 |
| Hsa:05222 | Small cell lung cancer | 16 | 1.52E–05 |
Note: Top five terms were selected according to P-value.
Abbreviations: DEGs, differentially expressed genes; HNSCC, head and neck squamous cell carcinoma; GO, gene ontology; BP, biological process; ECM, extracellular matrix; MHC, major histocompatibility; TAP, transporter associated with antigen processing; CC, cellular component; MF, molecular function.
Coexpression network analysis of DEGs
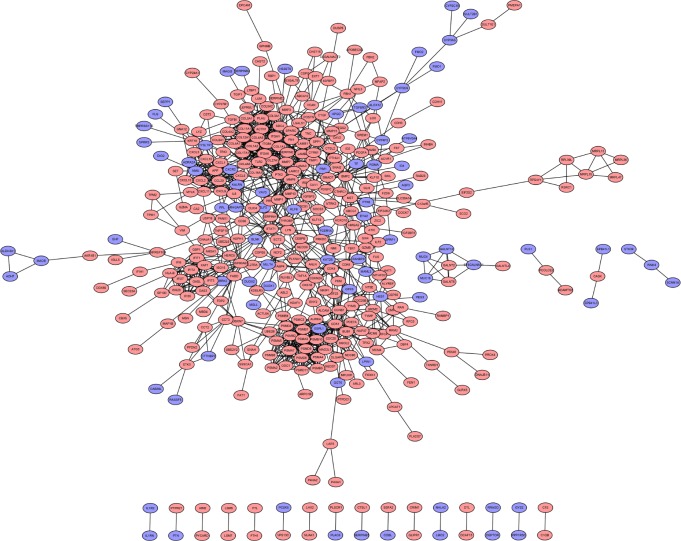
To interpret the biological meaning of the identified DEGs, we constructed a coexpression network for the DEGs with a combined score >0.8 and with significant interaction relation composed of 401 nodes and 1,254 edges by STRING database analysis (Figure 1). From the coexpression network of the selected DEGs, the top 15 hub genes were determined according to the number of the interacting edges: CDK1, PTK2, ITGAV, APP, COL1A1, MMP9, AURKA, BMP2, ITGB4, CDC20, SDC4, COL1A2, ITGA6, PSMA7, and STAT1 (Table 2). The distinct modules of 401 DEGs and their interacting genes were further identified by the MCODE using Cytoscape software. Among the modules, two subnetworks with >15 nodes were selected (Figure 2), and enrichment analysis showed that the genes in the subnetworks were mainly associated with proteasome, ECM–receptor interaction, protein digestion and absorption, and focal adhesion (Table 3).
Figure 1.
PPI network of differentially expressed genes.
Notes: Blue represents downregulated DEGs; red represents upregulated DEGs.
Abbreviations: PPI, protein–protein interaction; DEGs, differentially expressed genes.
Table 2.
The hub genes that had a degree >22 in PPI network
| Gene | Regulation | Degree |
|---|---|---|
| CDK1 | Up | 36 |
| PTK2 | Up | 34 |
| ITGAV | Up | 29 |
| APP | Up | 28 |
| COL1A1 | Up | 27 |
| MMP9 | Up | 27 |
| AURKA | Up | 26 |
| BMP2 | Up | 26 |
| ITGB4 | Up | 26 |
| CDC20 | Up | 25 |
| SDC4 | Up | 25 |
| COL1A2 | Up | 23 |
| ITGA6 | Up | 23 |
| PSMA7 | Up | 23 |
| STAT1 | Up | 23 |
Abbreviation: PPI, protein–protein interaction.
Figure 2.
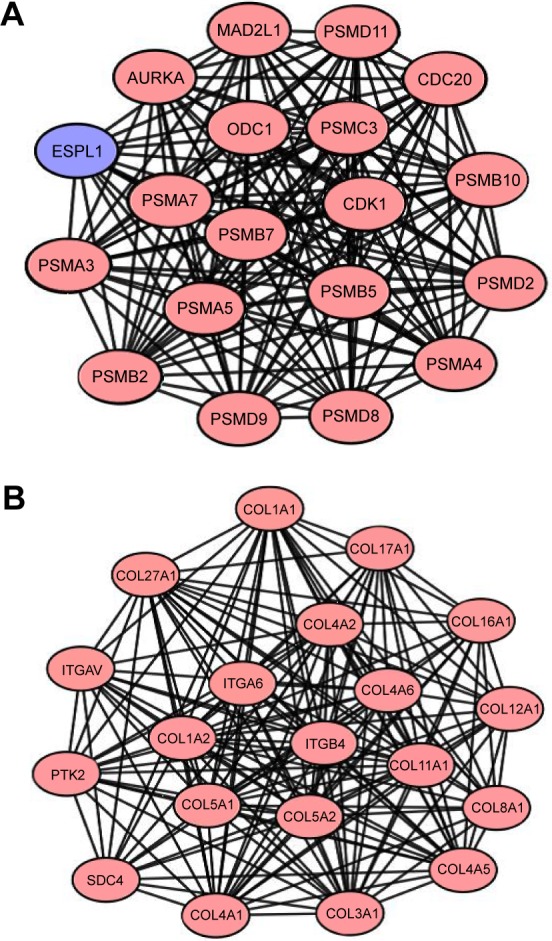
Functional modules in the PPI network.
Notes: From PPI networks of DEGs with combined score >0.8, we clustered two functional modules, using MCODE: module 1 (A) and module 2 (B). Blue represents downregulated DEGs; red represents upregulated DEGs.
Abbreviations: PPI, protein–protein interaction; DEGs, differentially expressed genes; MCODE, Molecular Complex Detection.
Table 3.
Functional and pathway enrichment analysis of the DEGs in modules
| ID | Description | Count | P-value |
|---|---|---|---|
| Module 1 | |||
| GO_BP:0031145 | Anaphase-promoting complex-dependent catabolic process | 17 | 1.73E–36 |
| GO_BP:0051436 | Negative regulation of ubiquitin protein ligase activity involved in mitotic cell cycle | 16 | 4.09E–34 |
| GO_BP:0051437 | Positive regulation of ubiquitin protein ligase activity involved in regulation of mitotic cell cycle transition | 16 | 1.27E–33 |
| GO_BP:0006521 | Regulation of cellular amino acid metabolic process | 14 | 2.99E–30 |
| GO_BP:0043161 | Proteasome-mediated ubiquitin-dependent protein catabolic process | 16 | 8.04E–27 |
| GO_CC:0000502 | Proteasome complex | 12 | 5.82E–24 |
| GO_CC:0005839 | Proteasome core complex | 8 | 2.77E–17 |
| GO_CC:0005829 | Cytosol | 19 | 4.58E–14 |
| GO_CC:0005654 | Nucleoplasm | 16 | 2.87E–10 |
| GO_CC:0005634 | Nucleus | 18 | 1.40E–08 |
| GO_MF:0004298 | Threonine-type endopeptidase activity | 8 | 2.90E–17 |
| GO_MF:0005515 | Protein binding | 18 | 1.49E–05 |
| Hsa:03050 | Proteasome | 12 | 2.16E–21 |
| Module 2 | |||
| GO_BP:0030198 | ECM organization | 17 | 5.98E–29 |
| GO_BP:0030574 | Collagen catabolic process | 12 | 7.33E–23 |
| GO_BP:0030199 | Collagen fibril organization | 7 | 2.78E–12 |
| GO_BP:0071230 | Cellular response to amino acid stimulus | 7 | 9.11E–12 |
| GO_BP:0007155 | Cell adhesion | 9 | 1.70E–08 |
| GO_CC:0005788 | Endoplasmic reticulum lumen | 15 | 1.42E–24 |
| GO_CC:0005581 | Collagen trimer | 11 | 5.76E–19 |
| GO_CC:0005576 | Extracellular region | 15 | 1.27E–11 |
| GO_CC:0031012 | ECM | 9 | 2.85E–10 |
| GO_CC:0005578 | Proteinaceous ECM | 8 | 5.96E–09 |
| GO_MF:0005201 | ECM structural constituent | 11 | 4.30E–20 |
| GO_MF:0048407 | Platelet-derived growth factor binding | 5 | 3.76E–10 |
| GO_MF:0038132 | Neuregulin binding | 3 | 1.20E–05 |
| Hsa:04512 | ECM–receptor interaction | 15 | 5.56E–25 |
| Hsa:04974 | Protein digestion and absorption | 13 | 4.91E–20 |
| Hsa:04510 | Focal adhesion | 15 | 1.76E–19 |
| Hsa:05146 | Amebiasis | 12 | 7.43E–17 |
| Hsa:04151 | PI3K-Akt signaling pathway | 15 | 2.72E–16 |
| Hsa:05222 | Small cell lung cancer | 7 | 3.22E–08 |
Abbreviations: DEGs, differentially expressed genes; GO, gene ontology; BP, biological process; MHC, major histocompatibility; TAP, transporter associated with antigen processing; CC, cellular component; ECM, extracellular matrix; MF, molecular function.
Hub genes were validated as an independent predictor for OS in the TCGA cohort
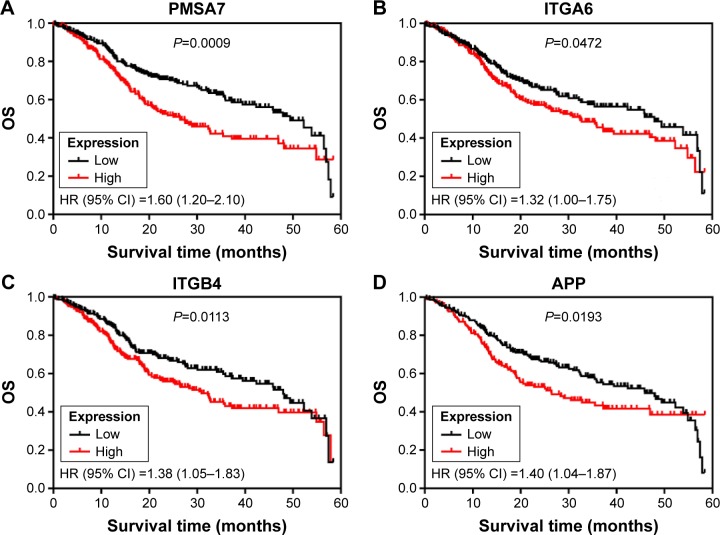
We subsequently sought to assess the significance of expression of 15 hub genes in HNSCC. Therefore, the relation between expression of 15 hub genes and OS in the TCGA HNSCC cohort (461 patients) was verified, and the patients were divided into low or high expression groups according to the median expression. Our results showed that poor OS was associated only in those patients with high expression of PSMA7 (HR: 1.60 [1.20–2.10], P=0.0009) in the TCGA HNSCC cohort, as well as ITGA6 (HR: 1.32 [1.00–1.75], P=0.0472), ITGB4 (HR: 1.38 [1.05–1.83], P=0.0113), and APP (HR: 1.40 [1.04–1.87], P=0.0113; Figure 3).
Figure 3.
Kaplan–Meier curves depicting OS in the TCGA HNSCC cohort with high and low expression of PMSA7 (A), ITGA6 (B), ITGB4 (C) and APP (D), respectively. Abbreviations: OS, overall survival; HNSCC, head and neck squamous cell carcinoma; HR, hazard ratio; CI, confidence interval.
Conclusion
Despite advances in surgical, chemotherapy, and medical therapy, the overall mortality of HNSCC has remained virtually unchanged over the past decades. The lethality of HNSCC is mainly due to difficulties in detecting it at an early stage and the lack of effective treatments for patients in advanced stages. Interestingly, bioinformatics plays a major role in the analysis and interpretation of genomic and proteomic data.14 For example, some researchers focus on bioinformatics, nanogenomics, and nanoproteomics aspects of contemporary nanodentistry and summarize some proteomics and proteogenomics approaches for oral diseases.15,16 Therefore, in the present study, we attempted to utilize comprehensive bioinformatics methods to explore the potential molecular mechanism of HNSCC to improve survival rate and prevention.
In this study, a total of 811 DEGs were screened, consisting of 550 upregulated genes and 261 downregulated genes. Moreover, we selected two significant modules with several key DEGs (like PSMA7, ITGA6, and ITGB4) in HNSCC regulatory network, and functional enrichment analyses showed that these key DEGs were mainly enriched in ECM–receptor interaction, which is closely related to cancer. Finally, survival analysis of these hub genes revealed that four overexpressed genes were significantly correlated with poor OS of patients in the TCGA HNSCC cohort, and these included PSMA7, ITGA6, ITGB4, and APP.
The data showed that PSMA7 is involved in “module 1” of the gene coexpression network, which is enriched in the proteasome pathway. Many studies have suggested that proteasome promotes the degradation of oxidatively damaged proteins that play a role in the cell cycle and transcription, which are essential for cancer improvement. Previously, it was reported that PSMA7 inhibits the proliferation, tumorigenicity, and invasion of human lung adenocarcinoma cells.17 Similar results also showed that high expression of PSMA7 is associated with liver metastasis in colorectal cancer.18 Besides, Hu et al also found depletion of PSMA7 inhibited cell growth, invasion, and migration in RKO cells and strongly suppressed the tumorigenic ability of RKO cells in vivo.19 Taken together, we speculate that the overexpression of PSMA7 may contribute to HNSCC progression and correlate with a poor prognosis.
On the other hand, ITGA6 and ITGB4, which are found in “module 2” in PPI network, were associated with the ECM–receptor interaction pathway, and belong to the integrin family, which participates in cell adhesion as well as cell surface-mediated signaling. Interactions between cells and the ECM could lead to the direct or indirect control of cellular processes of adhesion, migration, differentiation, proliferation, and apoptosis.20 As previously reported, silencing of ITGA6 genes significantly inhibited cell migration and invasion in head and neck cancer cells and hepatocellular carcinoma cells.21,22 Similarly high ITGA6 expression was shown to enhance invasion in models of metastatic breast cancer.23 Moreover, Kwon et al24 found ITGA6 is a possible target for antibody-related diagnostic and therapeutic modalities in esophageal squamous cell carcinoma. Meanwhile, ITGB4 regulates migration and invasion in models of metastatic prostate cancer.25 Moreover, Masugi et al26 found that knockdown of ITGB4 reduced the migration and invasion and that upregulation of ITGB4 promoted cell scattering and motility in pancreatic ductal adenocarcinoma cells. Besides, our study shows that ITGB4 was associated with poor prognosis in HNSCC; similar results have also been shown in pancreatic ductal adenocarcinoma patients.27 Together, we speculate that ITGA6 and ITGB4 in ECM–receptor interaction signaling pathway may play a significant role in HNSCC.
Amyloid-β precursor protein (APP) is the highly conservative single transmembrane protein with a receptor-like structure that has been shown to be involved in Alzheimer disease,28 but its function in normal physiological is unclear. Interestingly, APP is increased in many different cancers, such as colon cancer, pancreatic cancer, and thyroid cancer.29–31 Lim et al32 found that overexpression of APP is found both in malignant breast cancer cell lines and in human breast cancer tissues, and APP could regulate cell growth, apoptosis, and motility of breast cancer, possibly via engagement of AKT-mediated signaling pathways. Similarly, APP could promote cell growth in pancreatic cancer cells.31 In addition, Ko et al33 found a significant increase of APP in an oral squamous cell carcinoma (OSCC) tissue and also that OSCC patients with high mRNA levels of APP had poor prognoses. The abovementioned studies show that APP may be involved in the pathogenesis of malignant tumors by affecting cell growth or apoptosis, thereby supporting our findings.
In summary, the current study was intended to identify DEGs with comprehensive bioinformatics analysis to find the potential biomarkers and predict progression of diseases. We found that hub genes of complex networks, such as PSMA7, ITGA6, ITGB4, and APP, may be exploited as a prognostic tool for HNSCC. Finally, our results suggested that proteasome and ECM–receptor interaction may be important in the development of HNSCC. However, further experimental studies are still required to prove our findings and determine the potential clinical value of these as biomarkers.
Acknowledgments
The project was supported by the Guangdong Natural Science Foundation of China (2015A030313309).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Sturgis EM. Human Papillomavirus as a marker of the natural history and response to therapy of head and neck squamous cell carcinoma. Semin Radiat Oncol. 2012;22(2):128–242. doi: 10.1016/j.semradonc.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Orlando B, Bragazzi N, Nicolini C. Bioinformatics and systems biology analysis of genes network involved in OLP (Oral Lichen Planus) pathogenesis. Arch Oral Biol. 2013;58(6):664–673. doi: 10.1016/j.archoralbio.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Lakhani SR, Ashworth A. Microarray and histopathological analysis of tumours: the future and the past? Nat Rev Cancer. 2001;1(2):151–157. doi: 10.1038/35101087. [DOI] [PubMed] [Google Scholar]
- 6.Rays M, Chen Y, Su YA. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14(4):457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 7.Pyeon D, Newton MA, Lambert PF, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67(10):4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Seattle, WA: Springer; 2005. pp. 397–420. [Google Scholar]
- 9.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Suppl 1):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright JT, Hart TC. The genome projects: implications for dental practice and education. J Dent Educ. 2002;66(5):659–671. [PubMed] [Google Scholar]
- 15.Bragazzi NL, Pechkova E, Nicolini C. Proteomics and proteogenomics approaches for oral diseases. Adv Protein Chem Struct Biol. 2014;95:125–162. doi: 10.1016/B978-0-12-800453-1.00004-X. [DOI] [PubMed] [Google Scholar]
- 16.Nicolini C, Bragazzi N. Nanogenomics and nanoproteomics for personalized nanotheranostics for oral and colorectal cancer. Per Med. 2015;13(1):9–11. doi: 10.2217/pme.15.46. [DOI] [PubMed] [Google Scholar]
- 17.Tan JY, Huang X, Luo YL. PSMA7 inhibits the tumorigenicity of A549 human lung adenocarcinoma cells. Mol Cell Biochem. 2012;366(1–2):131–137. doi: 10.1007/s11010-012-1290-2. [DOI] [PubMed] [Google Scholar]
- 18.Hu XT, Chen W, Wang D, et al. The proteasome subunit PSMA7 located on the 20q13 amplicon is overexpressed and associated with liver metastasis in colorectal cancer. Oncol Rep. 2008;19(2):441–446. [PubMed] [Google Scholar]
- 19.Hu XT, Chen W, Zhang FB, et al. Depletion of the proteasome subunit PSMA7 inhibits colorectal cancer cell tumorigenicity and migration. Oncol Rep. 2009;22(5):1247–1252. doi: 10.3892/or_00000561. [DOI] [PubMed] [Google Scholar]
- 20.Hansen NU, Genovese F, Leeming DJ, Karsdal MA. The importance of extracellular matrix for cell function and in vivo likeness. Exp Mol Pathol. 2015;98(2):286–294. doi: 10.1016/j.yexmp.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, Nohata N, Hanazawa T, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109(10):2636–2645. doi: 10.1038/bjc.2013.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lv G, Lv T, Qiao S, et al. RNA interference targeting human integrin α6 suppresses the metastasis potential of hepatocellular carcinoma cells. Eur J Med Res. 2013;18:52. doi: 10.1186/2047-783X-18-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks DLP, Schwab LP, Krutilina R, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26. doi: 10.1186/s12943-016-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon J, Lee TS, Lee HW, et al. Integrin alpha 6: a novel therapeutic target in esophageal squamous cell carcinoma. Int J Oncol. 2013;43(5):1523–1530. doi: 10.3892/ijo.2013.2097. [DOI] [PubMed] [Google Scholar]
- 25.Banyard J, Chung I, Migliozzi M, et al. Identification of genes regulating migration and invasion using a new model of metastatic prostate cancer. BMC Cancer. 2014;14:387. doi: 10.1186/1471-2407-14-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masugi Y, Yamazaki K, Emoto K, et al. Upregulation of integrin β4 promotes epithelial-mesenchymal transition and is a novel prognostic marker in pancreatic ductal adenocarcinoma. Lab Invest. 2015;95(3):308–319. doi: 10.1038/labinvest.2014.166. [DOI] [PubMed] [Google Scholar]
- 27.Damhofer H, Medema JP, Veenstra VL, et al. Assessment of the stromal contribution to Sonic Hedgehog-dependent pancreatic adenocarcinoma. Mol Oncol. 2013;7(6):1031–1042. doi: 10.1016/j.molonc.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng JY, Kataoka H, Itoh H, Koono M. Amyloid β protein precursor is involved in the growth of human colon carcinoma cell in vitro and in vivo. Int J Cancer. 2001;92(1):31–39. [PubMed] [Google Scholar]
- 30.Krause K, Karger S, Sheu SY, et al. Evidence for a role of the amyloid precursor protein in thyroid carcinogenesis. J Endocrinol. 2008;198(2):291–299. doi: 10.1677/JOE-08-0005. [DOI] [PubMed] [Google Scholar]
- 31.Hansel DE, Rahman A, Wehner S, Herzog V, Yeo CJ, Maitra A. Increased expression and processing of the Alzheimer amyloid precursor protein in pancreatic cancer may influence cellular proliferation. Cancer Res. 2003;63(21):7032–7037. [PubMed] [Google Scholar]
- 32.Lim S, Yoo BK, Kim HS, et al. Amyloid-beta precursor protein promotes cell proliferation and motility of advanced breast cancer. BMC Cancer. 2014;14:928. doi: 10.1186/1471-2407-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko SY, Lin SC, Chang KW, et al. Increased expression of amyloid precursor protein in oral squamous cell carcinoma. Int J Cancer. 2004;111(5):727–732. doi: 10.1002/ijc.20328. [DOI] [PubMed] [Google Scholar]