Proline, glutamic acid, and leucine-rich protein 1 (PELP1) has been ascribed a large number of cellular functions, including roles in regulation of steroid receptor (SR) signaling (1), SR cross talk, cell cycle regulation (2), ribosome biogenesis (3), apoptosis, and differentiation (4). PELP1 is critical during development, as evidenced by embryonic lethality of PELP1 knockdown. PELP1 may act as a protooncogene, with its dysregulation linked to therapy resistance in hormonal cancers (1, 5–9).
How does PELP1 perform these various cellular roles? PELP1 neither binds specific sequences on the DNA nor is involved in signal transduction at the plasma membrane not has any known enzymatic activity. The explanation for the ascribed roles of PELP1 may lie in its extensive social network with other protein partners. PELP1 has multiple structural motifs that enable its direct interaction with a number of key proteins (Figure 1) (10); by functioning as a scaffold, PELP1 “enables” cross talk between its interacting proteins (11–14). The interactome of PELP1 is extensive and includes SRs, chromatin-modifying enzymes, cellular kinases, splicing factors, and transcriptional regulators (Figure 2). PELP1 likely indirectly but critically influences cell signaling and cellular processes. This review critically evaluates the molecular basis of the role of PELP1 in hormonal signaling and cancer.
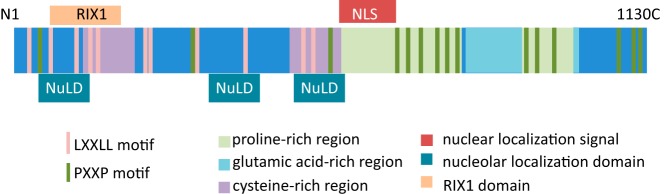
Figure 1. Primary domain structure of PELP1.
PELP1 has 11 LXXLL motifs for interactions with SRs, 2 proline-rich regions and PXXP domains for interactions with signaling proteins, a glutamic acid-rich carboxy region for interactions with histones. PELP1 also contains 2 cysteine-rich regions, which could possibly form zinc fingers for protein interactions, a nuclear localization signal (NLS), 3 nucleolar localization domains (NuLD), and a RIX1 domain.
Figure 2. PELP1 as a scaffolding protein.
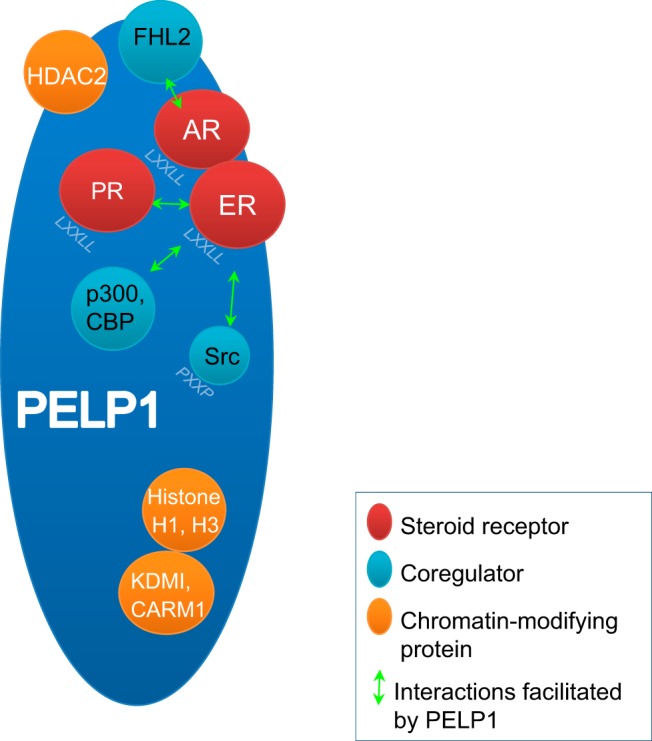
PELP1 interacts with SRs such as AR, ER, and PR through its LXXLL motif, SR coregulator proteins such as FHL2, p300, CBP, and Src, and chromatin-modifying proteins such as KDM1, coactivator-associated arginine methyltransferase 1 (CARM1), and histones through its glutamic acid-rich region. The interactome of PELP1 enables cross talk between these proteins and with the DNA.
Identification of PELP1
PELP1 was originally identified as a Src homology 2 (Sh2)-binding protein in a glutathione-S-transferase-pull down screening study (15). It was later purified, peptide sequenced (16), and a cDNA clone isolated from a HeLa cDNA library (1). The resulting 160-kDa protein was found to be rich in proline, glutamic acid and leucine, which constitute about 40% of the total protein and contribute to the eponymous name. Because overexpression of PELP1 enhanced estradiol-dependent transcriptional activation from the estrogen receptor (ER) response element, PELP1 was designated as a coactivator of ERα (17).
Primary Structure and the Interactome of PELP1
PELP1 is a 160-kDa scaffolding protein with several motifs commonly found in coregulators (uniprot.org/uniprot/Q8IZL8). The primary structure offers significant clues to its putative function. A domain scan using an online tool (http://scansite.mit.edu) revealed several protein-interacting domains on PELP1 (Figure 1). Among them is a strong nuclear localization sequence (centered at amino acid 640) (1) and 3 nucleolar localization domains (amino acids 74–152, 423–489, and 569–639). Unsurprisingly, PELP1 is largely nuclear in most studies (1).
PELP1 contains ten distinct LXXLL motifs (L is leucine, x is any residue, also known as nuclear receptor box), which enable its interaction with SRs (18). Interaction of PELP1 has been documented with ER, androgen receptor (AR) (11, 19), estrogen-related receptor-α (20), and retinoid X receptor (RXR) (4) and may be enhanced with ligand binding to the SR (1, 14). Each LXXLL motif within PELP1 has unique flanking sequences, which may dictate a differential affinity for each SR (21). For example, ERα preferentially interacts with the fourth and fifth LXXLL motifs on PELP1 (21). In addition, PELP1 may interface with SRs through motifs other than the LXXLL motif, for example, PELP1 interacts with glucocorticoid receptor (GR) through its proline- and glutamic acid-rich regions (12).
PELP1 also has several potential Src-binding sites, which account for its initial isolation in a Src-pull down. Mechanistic studies have shown that a proline-rich region in the N terminus of PELP1 (PxxP, where P is a proline, x is any residue) interacts with the SH3 motif of c-Src. In addition, the SH2 motifs of PELP1 represent high-affinity interaction sites for c-Src. The close proximity of nuclear receptor boxes and c-Src-binding sites on PELP1 may enable cross talk between c-Src and SRs by forming complexes (19, 21). Such complexes of AR, Src, and PELP1 were noted in androgen-dependent LNCaP cells upon androgenic stimulation and constitutively present in androgen-independent LNCaP cells (19).
PELP1 does not have an affinity for distinct DNA sequences but interacts with the chromatin. The highly acidic glutamic acid-rich domain within the C terminus of PELP1 enables its interaction with basic chromatin and recruitment of histone-modifying enzymes (22, 23). Thus, PELP1 may have a functional role on chromatin remodeling by displacing histone H1 (22).
Finally, dispersed throughout PELP1 are many cognate phosphorylation sites for potential regulation of its activity by kinases. PELP1 serves as a substrate for many kinases, including protein kinase A (24) and cyclin-dependent kinase (CDK) (25). The importance and the role of PELP1 phosphorylation in cellular processes are not known.
Altered Expression of PELP1 in Cancer
The expression of PELP1 is deregulated in a variety of hormonal cancers, including breast (1, 26, 27), ovarian (28), endometrial (29, 30), and prostate (6) cancers. Copy number alterations of PELP1 gene have been noted in a small fraction of these cancers (over 2% of prostate cancers, 1.5% of ovarian cancers, and 1% of breast cancers, current analysis of The Cancer Genome Atlas databases, May 2015). In breast tumors, PELP1 expression appears to be positively correlated with tumor grade and invasiveness, and inversely correlated with disease-free survival (27, 31, 32). Of note, high PELP1 expression and low ER, progesterone receptor (PR), and AR expression were observed in invasive breast cancers (27). Altered subcellular localization of PELP1 has been observed in a subset of breast and endometrial cancer (26, 30). PELP1 expression was also linked to high tumor grade in astrocytic brain tumors (33). Although PELP1 expression was higher in colon and salivary gland tumors compared with normal tissues, no correlation could be established to tumor grade or aggression (34–36). Mutations in PELP1 have been detected in various cancers but not adequately studied.
Role in Genomic SR Signaling
Initial characterization revealed an abundance of PELP1 in the mammary epithelium, where ERα is predominant (1). PELP1 was then demonstrated to be a coactivator of estradiol-induced ER-driven gene expression. PELP1 can enhance or repress SR transactivation depending on cellular context and ligand concentration (12). For example, in A549 cells that express significant endogenous GR, PELP1 inhibited GR transactivation. In contrast, in HEK293 cells that express very low levels of endogenous GR, PELP1 enhanced ligand-dependent GR transactivation. Biphasic behavior of exogenously expressed signaling molecules (ie, often when overexpressed) is indicative of scaffold activity, where stoichiometric concentrations of the interacting components that participate within a protein complex may facilitate maximal signal output, whereas too much of a particular player (ie, in this case, the scaffold itself) may interfere with signal output via sequestration of 1 of more of the interacting components (37). Suspected scaffold activity may be examined by physiologic manipulation of the levels of 1 protein (ie, altering total PELP1 levels using small interfering RNA (siRNA) or short hairpin RNA knockdown or exogenous PELP1 overexpression) while maintaining the same levels of its interacting partners.
PELP1 interaction with SRs is mediated through its multiple LXXLL domains, and the sequences flanking each LXXLL domain mediate its specificity for individual SRs. PELP1 may also interact with SRs through non-LXXLL domains For example, exogenous overexpression of fragments of GR in HEK293 cells revealed that PELP1 interacts with both the amino and the C termini of GR. PELP1 was able to regulate both activation function 1 and activation function 2 transactivation functions of GR through these differential interactions (12).
The role of PELP1 in genomic signaling mediated by multiple SRs has been well characterized. For example, PELP1 cooperates with the orphan SR, estrogen-related receptor-α to increase aromatase gene expression (20). Knockdown of PELP1 affects the genomic signaling mediated by several SRs, with multiple mechanisms proposed. PELP1 may enable SR genomic signaling via its interactions with traditional SR-transcriptional coactivators, such as and the closely related CREB binding protein (CBP) (Figure 3A) (38). PELP1 may recruit both SRs and their specific coactivators, thereby decreasing the entropy of such interactions within a cell via “forced” proximity. PELP1 may also stabilize SR-coregulator complexes that otherwise interact weakly in the absence of PELP1 (Figure 3A). PELP1 may also induce chromatin remodeling at ER target gene promoters by displacing histone H1 (22). PELP1 mediates methyl modifications of histone H3 at gene promoters via interactions with histone demethylase KDM1 (lysine-specific histone demethylase 1A) (23) or coactivator-associated arginine methyltransferase 1 (CARM1) (Figure 3A) (39). PELP1 may enable SRs to interact with some coregulators, even the SRs and coregulators cannot directly interact. For example, PELP1 aids the AR-coregulator four-and-a-half lens intrinsic membrane protein 2 (FHL2) that lacks motifs for direct interactions with AR by binding both AR and FHL2 at the N-terminal region (1–600 amino acids); the close proximity of these proteins on PELP1 enables the coregulator and AR to “functionally interact” without a direct physical interaction (11). PELP1 further aids in FHL2 transactivation function by binding other proteins through its SH3, PDZ, and WW motifs at its C-terminal end (Figure 3B). Thus, PELP1 may serve as a platform or “node” for the recruitment and organization of transcription complexes.
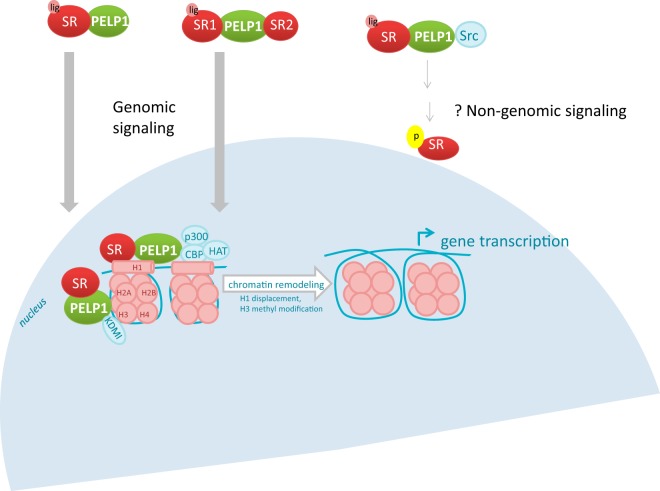
Figure 3. Role of PELP1 in SR signaling.
SR bound to PELP1 is translocated into the nucleus, where it binds chromatin. PELP1 facilitates transcription of SR-regulated genes by recruiting coregulators p300, CBP, and HAT to the chromatin that displace histone H1, and by interacting with KDM1 that causes methyl modifications on histone H3. In the absence of ligand, PELP1 may couple an unliganded SR to a liganded SR and activate genomic signaling. In the cytoplasm, PELP1 interacts with both Src and SRs. However, the role of PELP1 in nongenomic signaling needs to be robustly validated.
Role in Nongenomic SR Signaling
Although PELP1 is predominantly localized in the nucleus, its localization to the cytoplasm has been observed in a subset of breast tumors (26, 40). In the cytoplasm, PELP1 may couple SRs with kinase signaling cascades, as shown for AR, which enables interaction with signaling proteins such as G proteins (14) and Src (19). Mutations of the PxxP domains on PELP1 resulted in the loss of ER-Src interaction and estrogen-induced MAPK activation (13). Overexpression of cytoplasmic PELP1 resulted in the rapid induction of MAPK and AKT signaling pathways in a hormone-independent manner, which in turn phosphorylated ER (Figure 3) (26, 41). Complexes of AR, PELP1, and Src have been observed in LNCaP cells; however, their role in Src signaling has not been validated (19). The role for PELP1 in nongenomic SR signaling in mammalian cells has been proposed but needs robust validation and remains unproven.
Role in SR Cross Talk
PELP1 functions as a bridge for SRs, enabling the activation of 1 SR to activate another SR, even in the absence of the ligand for the second SR. For example, in prostate cancer cells, in the absence of androgens, PELP1 facilitates estradiol binding to ER to the unliganded AR and activates AR signaling in these cells (Figure 3) (6). Knockdown of PELP1 abrogated the coupling of liganded ER signal to unliganded AR (6). Similarly, in breast cancer cells, PELP1 facilitated cross talk between ER and PR, in the absence of the PR ligand progestin (8). Gene expression analyses indicated that the interaction between PR, PELP1, and ER enabled the activation of a subset of ER-target genes (8). Together, these data indicated that PELP1 might enable a liganded SR to influence the activity of another unliganded SR or vice versa. This finding has significant implications for resistance pathways in endocrine cancers, where the ligands of specific SRs are targeted.
Role in Chromatin Modification
PELP1 participates as a transcriptional coactivator/corepressor of nonnuclear receptors activator protein 1, nuclear factor κB, ternary complex factor/serum response factor (2). The glutamic acid-rich domain of PELP1 enables its interaction with the chromatin and interaction with basic histones (Figure 2). Although PELP1 does not directly modify the histones, its recruitment of histone modifiers enables histone modification, for example, the leucine-rich N-terminal region of PELP1 recruits histone deacetylase 2 to the chromatin, which, in turn, deacetylateslysine residues on core histones (2). PELP1 then binds to the hypoacetylated histones and protects them from acetylation, thereby hindering chromatin remodeling and influencing gene transcription.
Biological Roles of PELP1
PELP1 may play critical roles in a variety of biological processes, including proliferation, cell cycle regulation, and apoptosis. Most of these proposed roles have been inferred from overexpression or knockdown experiments. However, because PELP1 appears to be critical for cellular survival, siRNA-mediated knockdowns of PELP1 expression rarely exceeds 80% of protein level and significant PELP1 expression level can still be detected (6, 14, 42). To our knowledge, there is no single cell line that does not express PELP1. At this time, it is unclear whether cells genetically engineered to not express PELP1 using clustered regularly interspaced short palindromic repeats or transcription activator-like effector nuclease techniques will survive. The true extent and role of PELP1 in these cellular processes is thus not evaluable.
Proliferation
PELP1 has been shown to be important for both estrogen-induced and estrogen-independent proliferation in breast cancer cells (7, 20, 28, 29, 43, 44). In ER-positive breast cancers, PELP1 expression correlated with tumor size and mitotic count (27). Knockdown of PELP1 decreased growth of both ER-positive and ER-negative breast and ovarian cancer cells (7, 28, 43). Overexpression of PELP1 in MCF7 cells increased its estradiol-dependent and estradiol-independent proliferation and induced cellular transformation (44). PELP1 may enhance proliferation of breast cancer cells by increasing local estrogen synthesis (20, 45). In MCF7 cells, through the Src and phosphatidylinositide 3-kinases pathways, PELP1 increased aromatase expression, which in turn may regulate estrogen levels (20). Additionally, PELP1 mediates cross talk between PR and ER, which sensitizes cells to estradiol and IGF-1 (8). Together, these data support the oncogenic potential of PELP1 when dysregulated by overexpression and/or mislocalization.
Cell Cycle Regulation
PELP1 has been shown to bind the critical cell cycle switch, the retinoblastoma protein (pRb) (46). PELP1 actuates the phosphorylation of pRb at Ser807 and Ser811 (46). PELP1 itself is a target for CDKs and cyclin D1, which is involved in pRb phosphorylation and activation (46, 47). Further evidence for its role in the cell cycle comes from Xenopus oocytes through its interaction with membrane G proteins, specifically Gβγ (14). Knockdown of PELP1 in these oocytes decreased Gβγ-signaling, overcame the meiotic arrest, increased maturation of oocytes and activated the MAPK pathway.
Apoptosis
In response to stressors, such as chemotherapeutic drugs and ionizing radiation, DNA damage response kinases phosphorylate PELP1 at Ser1033 (48). Phosphorylated PELP1 binds tumor suppressor p53 and modulates p53 phosphorylation and acetylation states, which in turn affects its role in DNA damage response. PELP1 knockdown results in defective p53 signaling in response to stressors and affects stress-induced apoptosis. Overexpression of PELP1 induces apoptosis through caspase-mediated cleavage of the DNA repair enzyme, poly ADP ribose polymerase, in response to TNF-α (40). Additionally, PELP1 interacts with both RXR homodimers and RXR-peroxisome proliferator-activated receptor γ heterodimers and induces apoptotic activity (4).
Ribosome Biogenesis
PELP1 contains an RIX1 domain and sequences for nucleolar localization and is present in the nucleolus during S and G2 phases of the cell cycle, when ribosomal transcription occurs (3). Nucleolar localization of PELP1 is dependent on cell cycle progression and may be regulated by phosphorylation by CDK. PELP1 also interacts with 28S rRNA precursors and the 28S rRNA maturation factor, LAS1-like protein, and may regulate larger subunit maturation (49, 50).
Tumor Migration and Metastasis
PELP1 is highly expressed in all breast cancer cells. However, quantitative expression is higher in metastatic breast cancer than in node-negative tumors. Knockdown of PELP1 altered several epithelial to mesenchymal transition markers, including those that influence cell adhesion, migration, motility, and proliferation, and is associated with a loss of migration potential of breast cancer cells, both in vitro and in mice models (7). PELP1 also modulates microRNAs (miR-200 and miR-141) (51) involved in metastasis, possibly by recruiting histone-modifying enzymes at their promoters.
Role in Therapy Resistance
The role of PELP1 in a number of cellular processes in both ER-positive and ER-negative breast cancer cells in both hormone-dependent and hormone-independent pathways may enable a central role in therapy resistance. For example, PELP1 increased ER driven gene expression in MCF7 cells in response to either epidermal growth factor or tamoxifen, even in the absence of estrogen (24). Mutational analyses revealed that EGF and tamoxifen induced phosphorylation of PELP1 at 3 serine residues (S350, S415, and S613) by protein kinase A, which in turn activated ER-mediated genomic signaling. PELP1 localization in the cytoplasm resulted in poor response to tamoxifen in transgenic mouse models (31). The ability of PELP1 to bridge SRs with nongenomic signaling mediated through players like Src and PI3 kinases may induce therapy resistance (9). Additionally, through its role in SR cross talk, PELP1 may activate ER signaling, even in the absence of estrogen or progesterone ligand (9, 26). PELP1-mediated cross talk of PR-B with ER was found to activate a subset of ER target genes associated with luminal-B and survival phenotype and may be an underlying mechanism of resistance to hormone therapy and tamoxifen in breast cancer (8). Similar processes may be important for other hormone-dependent or hormonally influenced cancers such as ovarian and breast cancers.
Targeting PELP1
The involvement of PELP1 in several cellular processes and its deregulation in hormonal cancers make it attractive as a therapeutic target. However, any agent targeting PELP1 must be evaluated for toxicity, as multiple SR-dependent and SR-independent pathways may be involved. An ideal agent would selectively target PELP1 in tumor cells.
To date, nanoliposomal formulations of siRNA against PELP1 have successfully been used to decrease tumor growth and metastatic tumor nodules in ovarian xenograft tumor models (42). Alternatively, targeting PELP1-coupled nongenomic signaling pathways, including via the Src inhibitor, dasatinib, may be effective against PELP1-mediated hormone resistant tumors (ie, due to its ability to block ER-PELP1-Src axis) (9).
Recently, we demonstrated the use of peptidomimetics to inhibit prostate cancer cell proliferation by specifically blocking PELP1 interactions with AR (52). The peptidomimetics were designed to mimic the interface between AR and PELP1 and blocked the interaction between AR and PELP1. Importantly, these peptidomimetics do not affect PELP1 protein expression or its interaction with other protein partners. Because PELP1 appears to be central to AR function, administration of the peptidomimetic blocked AR-driven signaling and AR-driven proliferation in these cancer cells both in vitro and as xenografts in vivo. Importantly, these peptidomimetics did not show any evidence of toxicity in mice at the therapeutic doses. These results evince the possibility of using peptidomimetics as a strategy to target PELP1 without accompanying toxicity. Further, specific PELP1-protein interactions may be targeted with peptidomimetics designed to block either individual interactions or subsets of interactions.
Conclusions and Future Directions
There is mounting evidence that PELP1 plays key roles in several biological processes and its deregulation leads to the development of cancers. PELP1 appears to mediate these functions by serving a scaffold that brings critical protein partners together for heightened signaling and transcriptional activity. PELP1 is largely nuclear and appears to interact with chromatin and modulate histones through its recruitment of histone-modifying enzymes. In hormonally dependent cancers, PELP1 appears to be critical for both hormone-dependent and hormone-independent activities of multiple SRs. The function of PELP1 appears to be largely dependent on its “social” ability to interact with a large multitude of proteins. PELP1 functions may be both context-dependent and cell-line specific, depending upon its distinct interactome. Although the role of PELP1 in SR-mediated genomic signaling has been widely validated, its role in SR-driven nongenomic signaling remains controversial and unproven. The central scaffolding role of PELP1 makes it an attractive target and selective targeting of specific interactions within the “social network” of PELP1 may be possible with peptidomimetics.
Acknowledgments
Funding was provided by the Congressionally Directed Medical Research Programs (W81XWH-12-0288 and W81XH-13-2-0093), the Cancer Prevention Research Institute of Texas (DP150096), and the Mimi and John Cole Foundation.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
Funding was provided by the Congressionally Directed Medical Research Programs (W81XWH-12-0288 and W81XH-13-2-0093), the Cancer Prevention Research Institute of Texas (DP150096), and the Mimi and John Cole Foundation.
Footnotes
- AR
- androgen receptor
- CBP
- CREB binding protein
- CDK
- cyclin-dependent kinase
- ER
- estrogen receptor
- FHL2
- four-and-a-half lens intrinsic membrane protein 2
- GR
- glucocorticoid receptor
- KDM1A
- lysine-specific histone demethylase 1A
- PELP1
- proline, glutamic acid, and leucine-rich protein 1
- PR
- progesterone receptor
- pRb
- retinoblastoma protein
- RXR
- retinoid X receptor
- Sh2
- Src homology 2
- siRNA
- small interfering RNA
- SR
- steroid receptor.
References
- 1. Vadlamudi RK, Wang RA, Mazumdar A, et al. . Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem. 2001;276(41):38272–38279. [DOI] [PubMed] [Google Scholar]
- 2. Choi YB, Ko JK, Shin J. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. J Biol Chem. 2004;279(49):50930–50941. [DOI] [PubMed] [Google Scholar]
- 3. Gonugunta VK, Nair BC, Rajhans R, Sareddy GR, Nair SS, Vadlamudi RK. Regulation of rDNA transcription by proto-oncogene PELP1. PLoS One. 2011;6(6):e21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh RR, Gururaj AE, Vadlamudi RK, Kumar R. 9-cis-retinoic acid up-regulates expression of transcriptional coregulator PELP1, a novel coactivator of the retinoid X receptor α pathway. J Biol Chem. 2006;281(22):15394–15404. [DOI] [PubMed] [Google Scholar]
- 5. Bagheri-Yarmand R, Mandal M, Taludker AH, et al. . Etk/Bmx tyrosine kinase activates Pak1 and regulates tumorigenicity of breast cancer cells. J Biol Chem. 2001;276(31):29403–29409. [DOI] [PubMed] [Google Scholar]
- 6. Yang L, Ravindranathan P, Ramanan M, et al. . Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol Endocrinol. 2012;26(4):550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roy S, Chakravarty D, Cortez V, et al. . Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res. 2012;10(1):25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daniel AR, Gaviglio AL, Knutson TP, et al. . Progesterone receptor-B enhances estrogen responsiveness of breast cancer cells via scaffolding PELP1- and estrogen receptor-containing transcription complexes. Oncogene. 2015;34(4):506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vallabhaneni S, Nair BC, Cortez V, et al. . Significance of ER-Src axis in hormonal therapy resistance. Breast Cancer Res Treat. 2011;130(2):377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gonugunta VK, Miao L, Sareddy GR, Ravindranathan P, Vadlamudi R, Raj GV. The social network of PELP1 and its implications in breast and prostate cancers. Endocr Relat Cancer. 2014;21(4):T79–T86. [DOI] [PubMed] [Google Scholar]
- 11. Nair SS, Guo Z, Mueller JM, et al. . Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol. 2007;21(3):613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kayahara M, Ohanian J, Ohanian V, Berry A, Vadlamudi R, Ray DW. MNAR functionally interacts with both NH2- and COOH-terminal GR domains to modulate transactivation. Am J Physiol Endocrinol Metab. 2008;295(5):E1047–E1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boonyaratanakornkit V. Scaffolding proteins mediating membrane-initiated extra-nuclear actions of estrogen receptor. Steroids. 2011;76(9):877–884. [DOI] [PubMed] [Google Scholar]
- 14. Haas D, White SN, Lutz LB, Rasar M, Hammes SR. The modulator of nongenomic actions of the estrogen receptor (MNAR) regulates transcription-independent androgen receptor-mediated signaling: evidence that MNAR participates in G protein-regulated meiosis in Xenopus laevis oocytes. Mol Endocrinol. 2005;19(8):2035–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park I, Chung J, Walsh CT, Yun Y, Strominger JL, Shin J. Phosphotyrosine-independent binding of a 62-kDa protein to the src homology 2 (SH2) domain of p56lck and its regulation by phosphorylation of Ser-59 in the lck unique N-terminal region. Proc Natl Acad Sci USA. 1995;92(26):12338–12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joung I, Strominger JL, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci USA. 1996;93(12):5991–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wong CW, McNally C, Nickbarg E, et al. . Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc Natl Acad Sci USA. 2009;106(33):14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plevin MJ, Mills MM, Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci. 2005;30(2):66–69. [DOI] [PubMed] [Google Scholar]
- 19. Unni E, Sun S, Nan B, et al. . Changes in androgen receptor nongenotropic signaling correlate with transition of LNCaP cells to androgen independence. Cancer Res. 2004;64(19):7156–7168. [DOI] [PubMed] [Google Scholar]
- 20. Rajhans R, Nair HB, Nair SS, et al. . Modulation of in situ estrogen synthesis by proline-, glutamic acid-, and leucine-rich protein-1: potential estrogen receptor autocrine signaling loop in breast cancer cells. Mol Endocrinol. 2008;22(3):649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barletta F, Wong CW, McNally C, Komm BS, Katzenellenbogen B, Cheskis BJ. Characterization of the interactions of estrogen receptor and MNAR in the activation of cSrc. Mol Endocrinol. 2004;18(5):1096–1108. [DOI] [PubMed] [Google Scholar]
- 22. Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64(18):6416–6423. [DOI] [PubMed] [Google Scholar]
- 23. Nair SS, Nair BC, Cortez V, et al. . PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-α target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11(6):438–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagpal JK, Nair S, Chakravarty D, et al. . Growth factor regulation of estrogen receptor coregulator PELP1 functions via protein kinase A pathway. Mol Cancer Res. 2008;6(5):851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nair BC, Nair SS, Chakravarty D, et al. . Cyclin-dependent kinase-mediated phosphorylation plays a critical role in the oncogenic functions of PELP1. Cancer Res. 2010;70(18):7166–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vadlamudi RK, Manavathi B, Balasenthil S, et al. . Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005;65(17):7724–7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Habashy HO, Powe DG, Rakha EA, et al. . The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2010;120(3):603–612. [DOI] [PubMed] [Google Scholar]
- 28. Dimple C, Nair SS, Rajhans R, et al. . Role of PELP1/MNAR signaling in ovarian tumorigenesis. Cancer Res. 2008;68(12):4902–4909. [DOI] [PubMed] [Google Scholar]
- 29. Wan J, Li X. PELP1/MNAR suppression inhibits proliferation and metastasis of endometrial carcinoma cells. Oncol Rep. 2012;28(6):2035–2042. [DOI] [PubMed] [Google Scholar]
- 30. Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89(12):6130–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar R, Zhang H, Holm C, Vadlamudi RK, Landberg G, Rayala SK. Extranuclear coactivator signaling confers insensitivity to tamoxifen. Clin Cancer Res. 2009;15(12):4123–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar R, Gururaj AE, Vadlamudi RK, Rayala SK. The clinical relevance of steroid hormone receptor corepressors. Clin Cancer Res. 2005;11(8):2822–2831. [DOI] [PubMed] [Google Scholar]
- 33. Kefalopoulou Z, Aviles-Olmos I, Foltynie T. Critical aspects of clinical trial design for novel cell and gene therapies. Parkinsons Dis. 2011;2011:804041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grivas PD, Tzelepi V, Sotiropoulou-Bonikou G, Kefalopoulou Z, Papavassiliou AG, Kalofonos H. Expression of ERα, ERβ and co-regulator PELP1/MNAR in colorectal cancer: prognostic significance and clinicopathologic correlations. Cell Oncol. 2009;31(3):235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vadlamudi RK, Balasenthil S, Sahin AA, et al. . Novel estrogen receptor coactivator PELP1/MNAR gene and ERβ expression in salivary duct adenocarcinoma: potential therapeutic targets. Hum Pathol. 2005;36(6):670–675. [DOI] [PubMed] [Google Scholar]
- 36. Williams MD, Roberts D, Blumenschein GR Jr, et al. . Differential expression of hormonal and growth factor receptors in salivary duct carcinomas: biologic significance and potential role in therapeutic stratification of patients. Am J Surg Pathol. 2007;31(11):1645–1652. [DOI] [PubMed] [Google Scholar]
- 37. Ferrell JE., Jr What do scaffold proteins really do? Sci STKE. 2000;2000(52):pe1. [DOI] [PubMed] [Google Scholar]
- 38. Gururaj AE, Peng S, Vadlamudi RK, Kumar R. Estrogen induces expression of BCAS3, a novel estrogen receptor-α coactivator, through proline-, glutamic acid-, and leucine-rich protein-1 (PELP1). Mol Endocrinol. 2007;21(8):1847–1860. [DOI] [PubMed] [Google Scholar]
- 39. Mann M, Cortez V, Vadlamudi R. PELP1 oncogenic functions involve CARM1 regulation. Carcinogenesis. 2013;34(7):1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rayala SK, Mascarenhas J, Vadlamudi RK, Kumar R. Altered localization of a coactivator sensitizes breast cancer cells to tumor necrosis factor-induced apoptosis. Mol Cancer Ther. 2006;5(2):230–237. [DOI] [PubMed] [Google Scholar]
- 41. Rayala SK, Hollander Pd, Balasenthil S, et al. . Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) interacts with PELP1 and activates MAPK. J Biol Chem. 2006;281(7):4395–4403. [DOI] [PubMed] [Google Scholar]
- 42. Chakravarty D, Roy SS, Babu CR, et al. . Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis. Clin Cancer Res. 2011;17(8):2250–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cortez V, Mann M, Tekmal S, et al. . Targeting the PELP1-KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res. 2012;14(4):R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res. 2007;67(11):5505–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vadlamudi RK, Rajhans R, Chakravarty D, et al. . Regulation of aromatase induction by nuclear receptor coregulator PELP1. J Steroid Biochem Mol Biol. 2010;118(4–5):211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Balasenthil S, Vadlamudi RK. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem. 2003;278(24):22119–22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Müller H, Lukas J, Schneider A, et al. . Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1994;91(8):2945–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nair BC, Krishnan SR, Sareddy GR, et al. . Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell Death Differ. 2014;21(9):1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30(6):1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castle CD, Cassimere EK, Denicourt C. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol Biol Cell. 2012;23(4):716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roy SS, Gonugunta VK, Bandyopadhyay A, et al. . Significance of PELP1/HDAC2/miR-200 regulatory network in EMT and metastasis of breast cancer. Oncogene. 2014;33(28):3707–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bartlett DL, Liu Z, Sathaiah M, et al. . Oncolytic viruses as therapeutic cancer vaccines. Mol Cancer. 2013;12(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]