Abstract
The expression of canonical histones is normally coupled to DNA synthesis during the S phase of the cell cycle. Replication-dependent histone mRNAs do not contain a poly(A) tail at their 3′ terminus, but instead possess a stem-loop motif, the binding site for the stem-loop binding protein (SLBP), which regulates mRNA processing, stability, and relocation to polysomes. Here we show that the thyroid hormone can increase the levels of canonical histones independent of DNA replication. Incubation of mouse embryonic fibroblasts with T3 increases the total levels of histones, and expression of the thyroid hormone receptor β induces a further increase. This is not restricted to mouse embryonic fibroblasts, because T3 also raises histone expression in other cell lines. T3 does not increase histone mRNA or SLBP levels, suggesting that T3 regulates histone expression by a posttranscriptional mechanism. Indeed, T3 enhanced translational efficiency, inducing relocation of histone mRNA to heavy polysomes. Increased translation was associated with augmented transcription of the eukaryotic translation initiation factor 4 γ2 (EIF4G2). T3 induced EIF4G2 protein and mRNA levels and the thyroid hormone receptor bound to the promoter region of the Eif4g2 gene. Induction of EIF4G2 was essential for T3-dependent histone induction, because depletion of this factor abolished histone increase. These results point out the importance of the thyroid hormones on the posttranscriptional regulation of histone biosynthesis in a cell cycle–independent manner and also suggest the potential regulation of eukaryotic translation by the modulation of the initiation factor EIF4G2, which also operates in the translation of canonical mRNAs.
Nucleosomes, which fold chromosomal DNA, contain 2 molecules each of the core histones H2A, H2B, H3, and H4 (1). Histones are subjected to a great variety of posttranslational modifications, which normally occur on the amino-terminal and carboxy-terminal histone “tail” domains, which play an essential role in controlling gene expression, DNA repair, or chromosome condensation (2). Maintenance of chromosomal integrity requires accurate coordination of DNA replication with histone synthesis (3). Both histone excess and deficiency can have deleterious effects, and the cells have developed multiple regulatory systems to control histone levels. Thus, mammalian cells sequester the excess of histones accumulating during replication stress, and the histone chaperone Asf1 (antisilencing function 1) plays a key role in this process (4).
Canonical histones, ie, the four core histones as well as the linker H1 histone found between nucleosomes, are encoded by a family of replication-dependent histone genes. In most cells, the expression of cell cycle–regulated histone mRNAs is tightly coupled to DNA synthesis occurring during the S phase. This is achieved by the precise regulation of mRNA synthesis, processing, and stability (5, 6). Replication-dependent histone mRNAs are the only known cellular mRNAs that do not contain a poly(A) tail at their 3′ terminus, but instead exhibit a stem-loop structure that is highly conserved. This stem-loop motif is the binding site for the stem-loop binding protein (SLBP), which regulates pre-mRNA processing, mRNA stability, and localization of the mRNA to polyribosomes (polysomes) (7, 8). Although histone mRNAs have a different 3′ end than the polyadenylated mRNAs, they can be efficiently translated by a similar mechanism and with a similar efficiency. At the 5′ mRNA, the cap binding subunit eukaryotic initiation factor 4E interacts with the scaffold protein eukaryotic translation initiation factor 4 γ (EIF4G). The 3′ histone mRNA end is the binding site of SLBP and SLBP-binding protein 1 (SLIP1). SLIP1 interacts with EIF4G, acting as a bridge between the 2 mRNA ends mediating the mRNA circularization and thus enhancing translation efficiency (9). In addition, histone mRNAs are bound to polysomes through SLBP, also allowing high translational efficiencies (10).
The thyroid hormones (THs) T3 and its precursor T4, are essential for growth and development. Among other functions, they increase the basal metabolic rate and affect almost every aspect of cellular physiology as a result of their actions on fat, carbohydrate, and protein metabolism (11). THs largely exert their actions through the binding to nuclear thyroid hormone receptors (THRs), which normally regulate gene expression by binding to thyroid hormone response elements (TREs) located in regulatory regions of target genes (12, 13). There are 2 genes encoding THRs (THRA and THRB) and 4 THR isoforms designated THRA1, THRB1, THRB2, and THRB3 that are able to bind hormone. THRA1 is widely expressed, showing higher levels of expression in brain, cardiac, and skeletal tissue. THRB1 is predominantly expressed in brain, liver, and kidney. THRB2 expression is circumscribed to hypothalamus, pituitary, retina, and inner ear, and THRB3 is predominantly expressed in kidney, liver, and lungs (12–14).
Although the regulation of DNA synthesis and cell growth by THs has been known for decades, little is known about their actions on histone expression during or outside the S phase of the cell cycle. Here we describe the effects of TH on bulk histone levels in various cell models. TH treatment and the overexpression of THRB1 induced an increase in histone levels not coupled to DNA synthesis. Regulation occurred by a posttranscriptional mechanism. TH treatment did not change the amount of histone mRNAs but increased the amount of histone mRNAs bound to polysomes. TH induced the expression of EIF4G2 (also known as aging-associated protein 1), 1 of the 2 isoforms of EIF4G with essential roles in histone biosynthesis. The analysis of the Eif4g2 promoter by chromatin immunoprecipitation (ChIP) showed the specific binding of THRB to a region upstream to the transcriptional initiation site of the Eif4g2 gene. Moreover, the depletion of Eif4g2 by means of small interfering RNA (siRNA) prevented the TH-mediated induction of histone expression.
Materials and Methods
Cell culture, transfections, and chemical reagents
Immortal mouse embryonic fibroblasts (MEFs) obtained from TP53-knockout mice were a gift from M. Serrano (CNIO). MEFs expressing either the thyroid hormone receptor β1 (THRB) or α1 (THRA) isoforms in an stable manner were obtained as described previously by retroviral transduction and posterior selection with puromycin (15). Spontaneously immortalized MEFs obtained from knockout mice lacking both Thr genes were a gift from J. Samarut (Lyon, France). These cells were also transduced with THRB or THRA (15). Parental NIH-3T3 cells and cells expressing THRB in a stable manner were described previously (16). The rat pituitary cell line GH4C1 and the human colocarcinoma cell line HCT-116 were also used. Cells were grown under standard conditions in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, and antibiotics. Before hormone treatment, cells were grown in medium containing 10% TH–depleted fetal bovine serum by treatment with resin AG-1-X8 (Bio-Rad). Cells were plated at a density of 16 000/cm2 and incubated for the times indicated in the presence or absence of T3 or GC-1 (5 nM). siRNA transfections were performed by using the TransIT-X2 reagent (Myrus), following the manufacturer's instructions. siRNA duplexes SR30004 (control) and SR421103A, B, and C (mouse Eif4g2) were purchased from Origene.
Protein extraction, Western blotting, and acidic extraction of histones
Cells were harvested and lysed in triple-detergent lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.02% sodium azide, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonylfluoride, 2 μg/mL pepstatin, 2 μg/mL aprotinin, 2 μg/mL leupeptin, and phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich). Next, 10 μg of protein lysates were mixed with Laemmli sample buffer, boiled, and loaded onto 8% or 12% SDS-PAGE. Western blotting and protein detection were performed as described previously (15). Antibodies used were tubulin (clone DM1A; Sigma-Aldrich T6199), H3 and H4, (Abcam ab1791 and ab10158), EIF4G2 (donated by C. de Haro (CBM, Madrid, Spain), and anti-SLBP (donated by W.F. Marzluff, University of North Carolina, Chapel Hill, North Carolina). Acid extraction of histones and electrophoresis were performed as described previously (17).
Polysome separation
Separation of free and ribosome-bound mRNA was performed by centrifugation in linear sucrose gradients. Cells were harvested, washed with 1× PBS, and lysed in a mixture containing 1 mL of ice-cold NP-40 buffer (10 mM Tris-HCl [pH 8], 140 mM NaCl, 1.5 mM MgCl2, and 0.5% NP-40), 20 μL of 1 M dithiothreitol, 10 μL of 40 U/μL RNasin (Invitrogen), and 100 μL of 5% sodium deoxycholate. Lysates were centrifuged during 10 seconds at 13 000 rpm, and supernatants were transferred to a new tube containing 13.3 μL of heparin (50 mg/mL), 15 μL of 10 mg/mL cycloheximide, and 10 μl of 0.1 M phenylmethylsulfonylfluoride. This mixture was centrifuged for 5 minutes at 13 000 rpm, and the supernatant was layered onto a sucrose gradient (15%–40% [w/v]) supplemented with 10 mM Tris-HCl (pH 7.5), 140 mM NaCl, 1.5 mM Mg2Cl, 10 mM dithiothreitol, 100 μg/mL cycloheximide, and 0.5 mg/mL heparin. Gradients were centrifuged for 90 minutes at 4°C in an SW41 rotor at 39 000 rpm and fractionated into 20 fractions in tubes containing 10 μL of 10 mg/mL Proteinase K, 25 μL of 20% SDS, and 12 μL of 0.5 M EDTA (pH 8). Fractions were extracted once with phenol-chloroform, and the RNA was precipitated overnight at −80°C with ethanol, glycogen, and 0.3 M sodium acetate. Precipitated RNAs were dissolved in H2O, quantified, and analyzed by quantitative real-time PCR (qRT-PCR).
qRT-PCR
RNA extraction, reverse transcription, and qRT-PCR were performed as described previously (15). The sequences of the oligonucleotides used in this study were the following: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-ACAGTCCATGCCATCACTGCC-3′ (forward) and 5′-GCCTGCTTCACCACCTTCTTG-3′ (reverse); 18S, 5′-GTAACCCGTTGAACCCCATT-3′ (forward) and 5′-CCATCCAATCGGTAGTAGCG-3′ (reverse); H3, 5′-AGTGCTCACCAGCTTGCTTT-3′ (forward) and 5′-GCTCGGTCGACTTCTGGTAG-3′ (reverse); and H4, 5′-AGGTTCTCCGCGATAACATC-3′ (forward) and 5′-GTGCTCCGTGTAGGTGACG-3′ (reverse).
ChIP assay
Cells were plated in 150-mm dishes and on the next day were treated with 2.5 μM α-amanitin (A2263; Sigma-Aldrich) in serum-free medium for 2.5 hours. Cells were then washed and treated with 5 nM T3 for 2 hours, fixed and lysed following the specifications of the Upstate kit (catalog no. 17–295), and sonicated in a Bioruptor UCD-200TM (Diagenode). In each immunoprecipitation, 2 to 3 × 106 cells and 2 μg of the following antibodies were used: normal rabbit IgGs (sc-2027; Santa Cruz Biotechnology, Inc), H3 (ab1791; Abcam), and anti-THRB serum (described in Ref. 15). DNA was purified and precipitated. Immunoprecipitated DNA was used for qRT-PCR amplification of the regions encompassing the mouse Eif4g2 promoter from −647 to −352 and from −199 to −41 with respect to the transcription initiation site, The primers used were 5′-AAACACTTCTCAAGCCAGCC-3′ (forward) and 5′-AAGAACCTTTTCCTCGCCTC-3′ (reverse) for the distal region and 5′-GCCTTCGCGAATATGGCTTT-3′ (forward) and 5′-CGACCCACTAGAGCCTCC-3′(reverse) for the proximal region. Results were normalized and are presented as a fraction of the input.
Cell cycle analysis
Cells were fixed and stained with propidium iodide under standard conditions. Acquisition was carried out with a FACScan system and software. Cell cycle analysis was performed by using ModFit 3.1 software, and the corresponding plots were prepared with FlowJo software.
Statistical analysis
The statistical significance of the data was determined by applying a two-tailed Student t test. P values of <.05 are considered significant. Statistics were calculated with Prism 5 software (GraphPad Software). All results are presented in the figures as means ± SD.
Results
T3 increases bulk histone levels
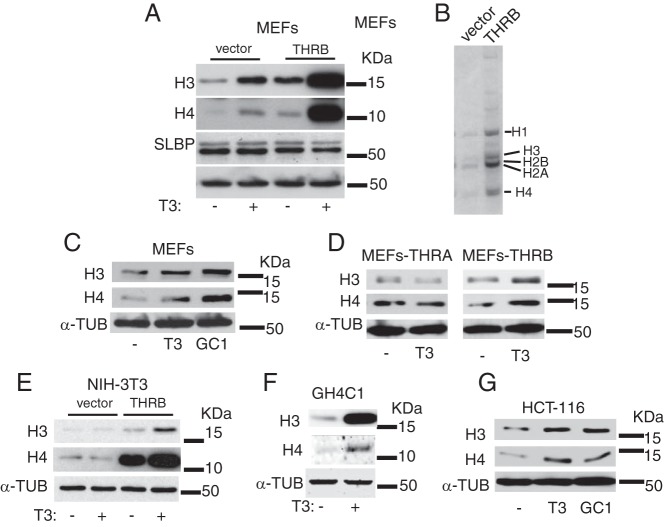
MEFs express THRs (both THRA and THRB) and respond to TH (15). We have previously reported the characterization of MEFs in terms of occurrence of DNA damage and cellular senescence upon exposure to T3 (15). We have also observed that the overexpression of THRB in MEFs drastically exacerbated those effects. In the course of the molecular characterization of those phenomena, we observed an unexpected and rapid increase in the bulk of histones upon T3 treatment. As shown in Figure 1A, a 48-hour treatment with T3 induced the expression of histones H3 and H4 in immortalized MEFs lacking TP53, and the overexpression of THRB significantly magnified this effect. Surprisingly, the T3-dependent increase in histone levels was not associated with an elevation in the expression of SLBP, a key regulator of histone biosynthesis (6). The enrichment in histone bulk mediated by the receptor suggested biosynthesis alterations rather than technical artifacts. This prompted us to purify total histones by acidic extraction and to analyze them by gel electrophoresis. As shown in Figure 1B, when histones from a similar number of cells were applied to the gel, the content of all core histones (H2A, H2B, H3, and H4) and that of the linker histone H1 were significantly higher in THRB-expressing cells than in control cells. Therefore, all canonical histones are induced in MEFs by the receptor. To analyze whether histone induction could also be mediated by THRA, we incubated MEFs with the THRB-specific ligand GC-1. As illustrated in Figure 1C, GC-1 was as effective as T3 in inducing histone levels in MEFs. Furthermore, T3 increased histone H3 and histone H4 in THR-knockout MEFs transduced with THRB, but not with THRA, indicating that only THRB mediates the ability of T3 to induce histone expression (Figure 1D).
Figure 1. Effects of TH on the expression of histones.
A, Western blot analysis of the levels of histones H3 and H4 and the SLBP in MEFs. Cells were treated in the presence or absence of 5 nM T3 for 48 hours before protein extraction. Vector, cells stably transfected with an empty vector; THRB, cells stably transfected with a THRB vector. α-Tubulin was used as a loading control. B, Total histones from 7 × 105 control and THRB-expressing MEFs were purified by acidic extraction. Samples were loaded onto an SDS-PAGE gel and stained subsequently with Coomassie Blue. C, Western blots of histones H3 and H4 in MEFs treated with equal amounts of T3 and GC-1 (5 nM). D, Histone levels in control and T3-treated THR-knockout MEFs stably transfected with THRB or THRA vectors. E, Levels of histone H3 and H4 in control and T3-treated NIH-3T3 cells. F, Histone levels of untreated and T3-treated GH4C1 cells. G, histone levels in HCT-116 cells treated with T3 or GC-1. α-Tubulin (α-TUB) was used as loading control.
Incubation of NIH-3T3 cells, another murine cell line, with T3 for 48 hours did not produce histone induction. This is not surprising because these cells express low THR levels and are unresponsive to T3 unless the receptor is expressed (16). However, the stable expression of THRB also increased H3 and H4 levels very strongly in NIH-3T3 cells, even in the absence of exogenously added T3, and incubation with the hormone induced a further increase (Figure 1E). We then extended the analysis to other nonmurine cells lines such as the rat pituitary cell line GH4C1, which expresses high endogenous levels of both THR isoforms and is highly responsive to T3, and the human colon carcinoma cell line HCT-116. Again, the treatment with T3 gave rise to a significant increase in histone levels (Figure 1F), suggesting a common and conserved phenomenon. Furthermore, GC-1 also increased histone levels in HTC-116 cells, reinforcing the idea that THRB is the receptor isoform responsible (Figure 1G).
Effect of T3 on histone bulk is due to posttranscriptional mechanisms
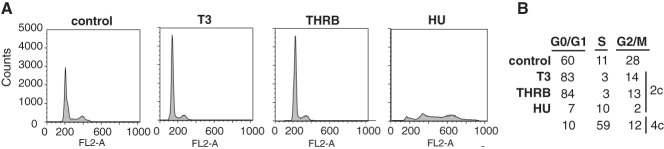
Histone protein synthesis increases during the S phase by transcriptional and posttranscriptional mechanisms to attain fast packing of the newly replicated DNA into chromatin (18). We then analyzed the possibility that the effect of T3 on histone abundance could be a consequence of accumulation of cells in the S phase. However, the DNA profile of the MEFs showed that, in contrast with the results obtained after incubation with hydroxyurea, which promotes fork stalling and induces DNA replication stress arresting cells in the S phase, T3-treated and THRB-expressing cells exhibited significant G0/G1 arrest, dismissing the possibility that the differences in the histone bulk were due to S phase arrest (Figure 2). This result was consistent with the observed expression of SLBP, which remained unchanged under the different conditions (Figure 1A), ruling out the possibility that the alterations in histone content were due to biosynthesis events normally occurring during DNA replication.
Figure 2. T3 does not induce S phase arrest.
A, Cell cycle analysis by flow cytometry of MEFs treated for 24 hours with 5 nM T3 or with 0.1 mM hydroxyurea (HU). DNA contents were also measured in cells transfected with THRB. B, Distribution of cells (percentage) throughout the cell cycle phases. c, haploid number of chromosomes.
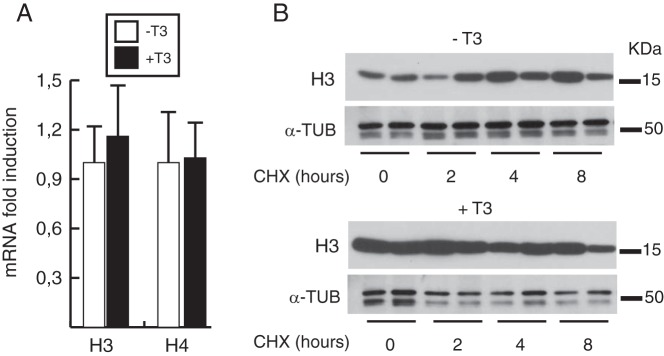
To determine the molecular mechanisms behind this effect, we quantified the mRNA levels of representative histones in T3-treated and untreated MEFs (Figure 3A). We found no significant differences between the 2 conditions, suggesting a posttranscriptional mechanism for T3 induction of histone protein levels. The augmented histone protein content without changes in histone mRNA levels could reflect an increased histone half-life in T3-treated cells. However, it is known that histones can have extremely long half-lives (19, 20), making it very unlikely that a short treatment with the hormone could lead to a detectable increase in bulk histone content. Furthermore, after cycloheximide treatment, we corroborated the observation that histones did not decay for an 8-hour period in the absence of new protein synthesis and we did not find reduced histone turnover in T3-treated cells (Figure 3B).
Figure 3. T3 does not alter histone mRNA levels or histone turnover.
A, Histone mRNA levels were determined by qRT-PCR in control MEFs and in cells treated with T3 for 48 hours. B, Cells were treated with or without T3 for 24 hours before addition of 50 μg/mL cycloheximide (CHX). H3 levels were analyzed by Western blotting at the times indicated for CHX treatment. α-Tubulin (α-TUB) was used as a loading control.
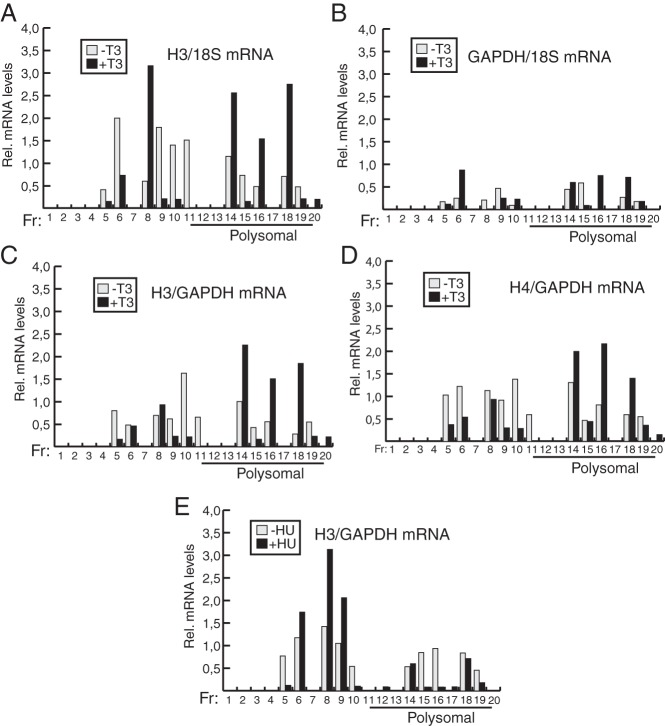
After dismissing the possibility that T3 could increase histone mRNA levels or the histone protein half-life, the most likely explanation for the effect of the hormone was increased mRNA translational efficiency. To demonstrate that translation of histone mRNA is regulated by T3, we next analyzed the distribution of histone H3 and H4 mRNAs on polyribosomes by sucrose gradient ultracentrifugation. As shown in Figure 4A, the hormone altered the distribution of H3 mRNA throughout the gradient. T3 increased mRNA levels in 3 high-density fractions, corresponding with heavy polyribosomes, both expressed relative to 18S RNA (Figure 4A) and to GAPDH mRNA, used as a negative control (Figure 4C). GAPDH mRNA showed a rather homogeneous distribution throughout the gradient that was not significantly altered in the presence of T3 (Figure 4B). Similar results were obtained with H4 mRNA translation, because T3 also increased the amount of mRNA associated with heavy polysomes (Figure 4D). In contrast with the effect of T3, which caused a shift to the high-density fractions, after incubation with hydroxyurea, H3 mRNA sedimented predominantly in low-density fractions, where no polysomal mRNA is present (Figure 4E). These results show conclusively that histone mRNA translation is facilitated by T3.
Figure 4. T3 increases translational efficiency of histone H3 and H4 mRNA.
A, Distribution of histone H3 mRNA relative to 18S RNA throughout a sucrose gradient was analyzed in control MEFs and in MEFs treated with T3 for 48 hours. After the cell lysis, the cytoplasmic fraction was loaded onto a sucrose gradient and fractionated by centrifugation, the RNA of each fraction was purified and analyzed for H3 mRNA and 18S RNA levels by qRT-PCR. The figure shows 20 fractions (Fr) collected from the top of the gradient. The bottom fractions correspond to heavy polysomes. B, GAPDH mRNA relative to 18S RNA values in the same fractions, C, Relative histone H3 mRNA values with respect to the GAPDH mRNA levels shown in B. D, Histone H4 mRNA relative to GAPDH mRNA values. E, Distribution of histone H3 mRNA in cells treated with 0.1 mM hydroxyurea (HU) for 48 hours.
EIF4G2 is up-regulated by T3
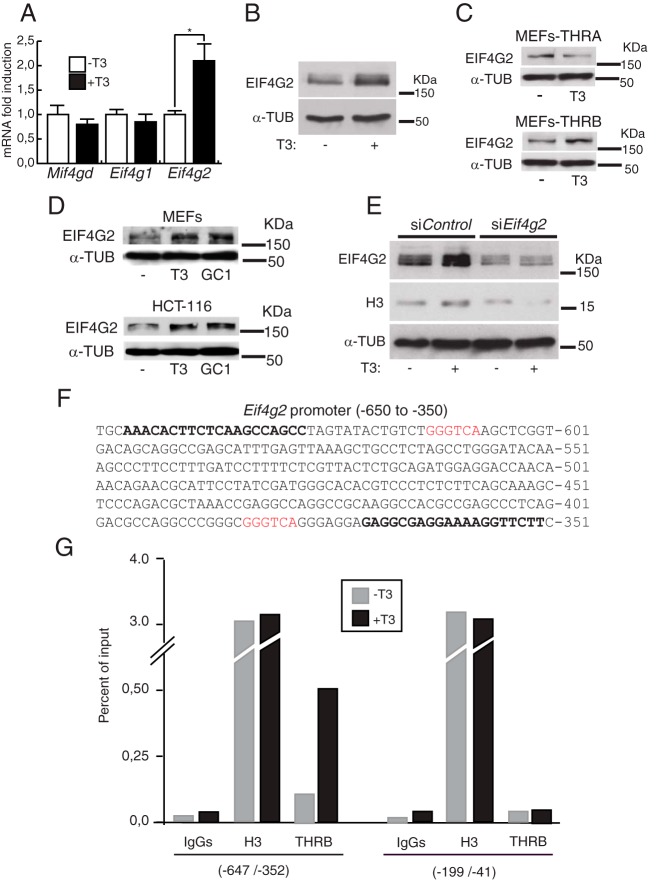
Efficient translation of cell-cycle regulated histones with 3′ stem-loop mRNAs requires specific factors such as SLBP and SLIP1 and the scaffold initiation factor EIF4G, also used for translation of polyadenylated mRNAs. Two isoforms of EIF4G, EIF4G1 and EIF4G2, are found in mammalian cells. We analyzed by qRT-PCR the effect of T3 on transcript levels of the key eukaryotic translation initiation factors Eif4g1 and Eif4g2, and of Mif4gd (Slip1). The results shown in Figure 5A indicate that T3 induced specifically the levels of Eif4g2 mRNA after 24 hours of treatment. The expression of EIF4G2 protein was also analyzed by Western blotting, finding a correlation with its mRNA levels (Figure 5B). T3 increased the Eif4g2 levels in THR-knockout MEFs transduced with THRB, but not with THRA (Figure 5C). Moreover, this induction was also found in MEFs and in HTC-116 cells treated with GC-1 (Figure 5D). This finding indicates that EIF4G2 induction by the hormone is not restricted to fibroblasts and that THRB is the receptor isoform that mediates the increased expression of the initiating factor.
Figure 5. EIF4G2 induction is involved in TH-induced expression of histones.
A, Relative mRNA levels of Slip1, Eif4g1, and Eif4g2 between T3-treated (24 hours) and untreated MEFs. B, Protein levels of EIF4G2 in treated (24 hours) and untreated cells. C, EIF4G2 levels in control and T3-treated THR-knockout MEFs expressing either THRA or THRB. D, EIF4G2 levels in MEFs and HTC-116 cells treated with T3 or GC-1, as indicated. E, Effect of EIF4G2 depletion by means of siRNA on histone H3 expression. MEFs were transfected with the siRNA and treated with T3 for 48 hours. F, Eif4g2 promoter sequence (−650 to −350) showing the 2 putative TRE hemisites found (in red) and in bold the sequence of the primers used in the ChIP experiments. Cells were treated for 2 hours with T3 and analyzed by ChIP with normal IgGs, histone H3, and THRB antibodies. The amplified DNA corresponds to the −647/−352 promoter region and to a proximal promoter region (−199/−41) used as a negative control.
To confirm the relevance of EIF4G2 induction in the effects exerted by the hormone on histone levels, we used siRNA-mediated silencing. MEFs transfected with control siRNAs and treated with T3 showed the expected increase in the levels of EIF4G2 and of histone H3 after 48 hours. However, cells transfected with Eif4g2-specific siRNA exhibited a reduction in the levels of EIF4G2 with a concomitant reduction in histone H3 levels (Figure 4E), indicating that EIF4G2 induction is required for the T3-mediated increase in bulk histones. In the search for a transcriptional mechanism responsible for this action of T3, we explored the Eif4g2 promoter and performed ChIP assays. Sequence analysis revealed the presence of 2 putative half-sites of the TRE upstream from the transcription initiation site (Figure 5F). ChIP assays showed constitutive recruitment of THRB, which was further induced by T3, to the region −647/−352 encompassing those elements, whereas H3 abundance was not altered by the hormone. The promoter region −199/−41 that does not contain putative elements was used as a negative control, and, as illustrated in Figure 5G, THRB did not bind to this proximal region. This finding suggests transcriptional control of EIF4G2 expression by the THR by binding of the receptor to the identified elements.
Discussion
Our results provide a mechanism of regulation of histone mRNA translation by THs. Previous results from our laboratory described the effect of THs on DNA damage and cellular senescence in MEFs (15). The characterization of the effect of T3 in these cells showed an unexpected increase in the total amount of canonical histones. This phenomenon seems to be well conserved, as it was observed in cell lines from different species under the same experimental conditions. The effect is mediated by THRB and not by THRA.
Chronic DNA damage due to telomere shortening during replicative senescence decreases histone H3 and H4 biosynthesis in cultured human fibroblasts (21). This effect in late passage cells is opposite to the increase observed by us in T3-treated fibroblasts that undergo senescence as a consequence of oxidative DNA damage after a short time of hormone exposure (15). The mechanism is also different, because telomeric shortening coincides with a reduction in the levels of the SLBP and histone chaperones, whereas no changes in SLBP levels are found in T3-treated fibroblasts.
The biological significance of the cell cycle–independent enhanced synthesis of histones is still unclear. Changes in histone-DNA equilibrium can induce DNA damage and impair DNA replication (22, 23). Therefore, it could participate in the DNA damage and reduced proliferation observed in T3-treated MEFs (15). However, a T3-dependent histone increase also occurs in pituitary GH4C1 cells, in which the hormone promotes proliferation (24). Therefore, cells must possess a mechanism to buffer the pool of soluble histones not incorporated into chromatin to avoid the deleterious consequences of histone excess when TH signaling is stimulated. Histone chaperones such as Asf1, which provide a buffering system for the histone excess generated in response replication stress (4), might also operate in this process.
The expression of canonical histones is normally associated with DNA synthesis occurring during the S phase of the cell cycle (6) and relies on the binding of SLBP to the stem-loop structure of the 3′ end of nonpolyadenylated histones mRNAs. SLBP and histone mRNA levels are rapidly increased as cells approach the S phase and are reduced at its conclusion (18, 25). Our results show that no significant changes in SLBP levels occur upon T3 treatment and that the hormone induces cell cycle arrest in G0/G1, excluding the possibility that the alterations on histone expression were associated with S phase arrest. In addition, we observed no differences in histone degradation. These data and the fact that the half-lives of histones can be extremely long (3, 26), led us to exclude differences in protein degradation as the origin of the increased histone levels after hormone treatment.
The amounts of representative H3 and H4 mRNAs also remained unaltered in the presence of T3, suggesting a posttranscriptional mechanism behind the observed effects. Efficient translation of histone mRNAs is achieved by mRNA circularization mediated by protein-protein interactions between SLIP1, bound to the 3′ end through SLBP, and EIF4G, which interacts with the cap-binding protein (27). In addition, SLBP directs histone mRNAs to polysomes (8), thus further increasing translation efficiency. Our results support the observation that T3 induces the relocation or distribution of histone mRNAs to heavy polysomes, thus increasing their translation efficiency in a cell cycle–independent manner. The mechanism responsible might imply T3-induced repositioning of SLBP to polysomes.
In addition to this, the hormone induced the expression of EIF4G2 by a transcriptional mechanism involving the recruitment of THRB to the Eif4g2 promoter. We found 2 potential half-sites in a region upstream of the transcription initiation site that could be used by the receptor to activate transcription. The relocation of the mRNAs to polysomes together with the increase in EIF4G2 might enhance and sustain T3-dependent stimulation of the translational capacity of histone mRNAs. Because of the global role of EIF4G in the initiation of translation, T3 might have a more general role in the translational control. However, T3 does not induce a generalized increase in protein synthesis in MEFs, because the translational efficiency of GAPDH was not significantly altered by the hormone, SLBP was not altered, and the levels of proteins used as a loading control were not affected. How specificity is obtained despite the increased levels of the general translation factor EIF4G requires further investigation.
To maintain normal cell growth, the control of protein synthesis and degradation by TH could be crucial. At physiological concentrations, THs can stimulate the synthesis as well as the degradation of proteins. Early studies demonstrated the effect of THs on protein synthesis both in vivo and in vitro cell-free systems (28–31). From these studies it appears that the increase in protein synthesis upon hormone stimulation was due to a primary effect on polypeptide assembly at the level of translation followed by a stimulation of transcription of the protein-synthesizing machinery. Taken together, our results fit into the picture of translational control outlined in these initial studies. A number of later studies have described the effects on polypeptide assembly mediated by nongenomic actions of THs at the level of the phosphoinositide 3 kinase−Akt (protein kinase B)−mammalian target of rapamycin pathway (32–36). Among other functions, this pathway promotes cell growth and protein synthesis through regulation of mammalian target of rapamycin. These actions induce G1 cell cycle progression through signaling via p70 S6 kinase and inhibition of eukaryotic translation initiation factor 4E–binding protein 1. However, it is very unlikely that these nongenomic actions operate in the effects presented here because of the kinetics and the cell cycle context in which they take place.
Acknowledgments
This work was supported by the Ministerio de Economía y Competitividad (Grant BFU2011–28058), the Instituto de Salud Carlos III (Grant RD12/0036/0030), and the Comunidad de Madrid (Grant S2011/BMD-2328 to A.A), and by the Instituto de Salud Carlos III (Grant MPY-1038/14 to A.Z.).
Author contributions: A.Z. and A.A. conceived the project and designed experiments; A.Z., V.G.-C. and R.V. performed the experiments; A.Z., V.G.-C., and A.A. analyzed the data; and A.Z. and A.A. wrote the article.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This work was supported by the Ministerio de Economía y Competitividad (Grant BFU2011–28058), the Instituto de Salud Carlos III (Grant RD12/0036/0030), and the Comunidad de Madrid (Grant S2011/BMD-2328 to A.A), and by the Instituto de Salud Carlos III (Grant MPY-1038/14 to A.Z.).
Footnotes
- ChIP
- chromatin immunoprecipitation
- EIF4G
- eukaryotic translation initiation factor 4 γ
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- MEFs
- mouse embryo fibroblasts
- qRT-PCR
- quantitative real-time PCR
- siRNA
- small-interfering RNA
- SLBP
- stem-loop binding protein
- SLIP1 (MIF4GD)
- SLBP-binding protein 1
- THR
- thyroid hormone receptor
- TRE
- thyroid hormone response element.
References
- 1. Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. [DOI] [PubMed] [Google Scholar]
- 2. Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 3. Bonner WM, Wu RS, Panusz HT, Muneses C. Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry. 1988;27:6542–6550. [DOI] [PubMed] [Google Scholar]
- 4. Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. [DOI] [PubMed] [Google Scholar]
- 5. Jaeger S, Barends S, Giegé R, Eriani G, Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. [DOI] [PubMed] [Google Scholar]
- 6. Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. [DOI] [PubMed] [Google Scholar]
- 8. Whitfield ML, Kaygun H, Erkmann JA, Townley-Tilson WH, Dominski Z, Marzluff WF. SLBP is associated with histone mRNA on polyribosomes as a component of the histone mRNP. Nucleic Acids Res. 2004;32:4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol Cell Biol. 2008;28:1182–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gorgoni B, Andrews S, Schaller A, Schümperli D, Gray NK, Müller B. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lazar MA. Thyroid hormone action: a binding contract. J Clin Invest. 2003;112:497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. [DOI] [PubMed] [Google Scholar]
- 13. Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aranda A, Alonso-Merino E, Zambrano A. Receptors of thyroid hormones. Pediatr Endocrinol Rev. 2013;11:2–13. [PubMed] [Google Scholar]
- 15. Zambrano A, García-Carpizo V, Gallardo ME, et al. The thyroid hormone receptor β induces DNA damage and premature senescence. J Cell Biol. 2014;204:129–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García-Silva S, Aranda A. The thyroid hormone receptor is a suppressor of ras-mediated transcription, proliferation, and transformation. Mol Cell Biol. 2004;24:7514–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. [DOI] [PubMed] [Google Scholar]
- 18. Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. [DOI] [PubMed] [Google Scholar]
- 19. Toyama BH, Hetzer MW. Protein homeostasis: live long, won't prosper. Nat Rev Mol Cell Biol. 2013;14:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Commerford SL, Carsten AL, Cronkite EP. Histone turnover within nonproliferating cells. Proc Natl Acad Sci USA. 1982;79:1163–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. [DOI] [PubMed] [Google Scholar]
- 23. Groth A, Corpet A, Cook AJ, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. [DOI] [PubMed] [Google Scholar]
- 24. Hinkle PM, Kinsella PA. Thyroid hormone induction of an autocrine growth factor secreted by pituitary tumor cells. Science. 1986;234:1549–1552. [DOI] [PubMed] [Google Scholar]
- 25. Mullen TE, Kaygun H, Marzluff WF. Chapter 2. Cell-cycle regulation of histone mRNA degradation in Mammalian cells: role of translation and oligouridylation. Methods Enzymol. 2008;449:23–45. [DOI] [PubMed] [Google Scholar]
- 26. Tsvetkov S, Ivanova E, Djondjurov L. Metabolic behaviors of the core histones in proliferating Friend cells. Exp Cell Res. 1989;180:94–105. [DOI] [PubMed] [Google Scholar]
- 27. Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. [DOI] [PubMed] [Google Scholar]
- 28. Sokoloff L, Kaufman S. Effects of thyroxin on amino acid incorporation into protein. Science. 1959;129:569–570. [DOI] [PubMed] [Google Scholar]
- 29. Sokoloff L, Kaufman S. Thyroxine stimulation of amino acid incorporation into protein. J Biol Chem. 1961;236:795–803. [PubMed] [Google Scholar]
- 30. Sokoloff L, Kaufman S, Gelboin HV. Thyroxine stimulation of soluble ribonucleic acid bound amino acid transfer to microsomal protein. Biochim Biophys Acta. 1961;52:410–412. [DOI] [PubMed] [Google Scholar]
- 31. Sokoloff L, Roberts PA, Januska MM, Kline JE. Mechanisms of stimulation of protein synthesis by thyroid hormones in vivo. Proc Natl Acad Sci USA. 1968;60:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cao X, Kambe F, Moeller LC, Refetoff S, Seo H. Thyroid hormone induces rapid activation of Akt/protein kinase B-mammalian target of rapamycin-p70S6K cascade through phosphatidylinositol 3-kinase in human fibroblasts. Mol Endocrinol. 2005;19:102–112. [DOI] [PubMed] [Google Scholar]
- 33. Furuya F, Hanover JA, Cheng SY. Activation of phosphatidylinositol 3-kinase signaling by a mutant thyroid hormone beta receptor. Proc Natl Acad Sci USA. 2006;103:1780–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kenessey A, Ojamaa K. Thyroid hormone stimulates protein synthesis in the cardiomyocyte by activating the Akt-mTOR and p70S6K pathways. J Biol Chem. 2006;281:20666–20672. [DOI] [PubMed] [Google Scholar]
- 35. Moeller LC, Cao X, Dumitrescu AM, Seo H, Refetoff S. Thyroid hormone mediated changes in gene expression can be initiated by cytosolic action of the thyroid hormone receptor beta through the phosphatidylinositol 3-kinase pathway. Nucl Recept Signal. 2006;4:e020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hiroi Y, Kim HH, Ying H, et al. Rapid nongenomic actions of thyroid hormone. Proc Natl Acad Sci USA. 2006;103:14104–14109. [DOI] [PMC free article] [PubMed] [Google Scholar]