Abstract
Muscle fitness is an important determinant of health and disease. However, the molecular mechanisms involved in the coordinate regulation of the metabolic and structural determinants of muscle endurance are still poorly characterized. Herein, we demonstrate that estrogen-related receptor α (ERRα, NR3B1) is essential for skeletal muscle fitness. Notably, we show that ERRα-null animals are hypoactive and that genetic or therapeutic disruption of ERRα in mice results in reduced exercise tolerance. Mice lacking ERRα also exhibited lactatemia at exhaustion. Gene expression profiling demonstrates that ERRα plays a key role in various metabolic processes important for muscle function including energy substrate transport and use (Ldhd, Slc16a1, Hk2, and Glul), the tricarboxylic acid cycle (Cycs, and Idh3g), and oxidative metabolism (Pdha1, and Uqcrq). Metabolomics studies revealed impairment in replenishment of several amino acids (eg, glutamine) during recovery to exercise. Moreover, loss of ERRα was found to alter the expression of genes involved in oxidative stress response (Hmox1), maintenance of muscle fiber integrity (Trim63, and Hspa1b), and muscle plasticity and neovascularization (Vegfa). Taken together, our study shows that ERRα plays a key role in directing transcriptional programs required for optimal mitochondrial oxidative potential and muscle fitness, suggesting that modulation of ERRα activity could be used to manage metabolic myopathies and/or promote the adaptive response to physical exercise.
Skeletal muscle is the largest organ system of the human body, accounting for more than 40% of the mass of a given nonobese individual. It is important not only for locomotion and physical strength but also for whole-body energy metabolism and substrate turnover. It is therefore not surprising that muscle fitness is an important determinant of health and disease.
Muscle endurance, strength, and fatigability depend on a variety of factors including substrate uptake and handling for energy production, mitochondrial function, and composition of the contractile machinery (1–3). Skeletal muscle fibers can be broadly classified based on their contractile properties as either slow-twitch (type I) or fast-twitch (type II) and are tightly associated with morphological, metabolic, and functional properties. More specifically, slow-twitch muscles, also called red muscles, are rich in mitochondria and capillary supply, rely largely on mitochondrial fatty acid oxidation for ATP production, and are specialized for endurance activity (4). In contrast, fast-twitch muscles, also referred to as white muscles, are more glycolytic and are more important for phasic activity (4). Skeletal muscles exhibit remarkable plasticity in that they can undergo changes in fiber-type composition, mitochondrial biogenesis, and energy metabolic pathways to adapt to physiological factors such as physical activity and exercise. However, the molecular mechanisms involved in the coordinate regulation of the metabolic and structural determinants of muscle fitness and endurance are still poorly characterized.
Numerous studies have implicated estrogen-related receptor α (ERRα, NR3B1), an orphan nuclear receptor belonging to the subfamily of classic steroid hormone receptors (5), in the regulation of a wide array of metabolic programs (6–8). Anatomical profiling of ERRα expression showed that the receptor segregated with tissues with high energy demands such as the heart, kidneys, intestinal tract, skeletal muscle, and brown adipose tissue, suggesting an involvement of ERRα in transducing metabolic signals to regulate bioenergetics processes (9–11). Indeed, ERRα was subsequently shown to be essential for the high levels of mitochondrial biogenesis and oxidative capacity of the brown adipose tissue to provide the energy necessary for thermogenesis (12). In addition, ERRα was shown to play a critical role in cardiac function. Notably, hearts of ERRα-deficient animals are smaller and have decreased expression of genes involved in energy substrate oxidation, ATP synthesis, and phosphate transfer and in other aspects of heart function including contractile and structural properties, resulting in reduced energetic reserve capacity and accelerated failure in response to hemodynamic stressors (13, 14). Collectively, these and subsequent studies highlighted an important role for the receptor in several aspects of energy metabolism in both normal and cancer cells (6, 15, 16).
Despite the observed link between ERRα, the control of metabolic gene programs and mitochondrial biogenesis in highly oxidative tissues, the roles of ERRα in skeletal muscle function are still poorly characterized. In vitro, during myogenesis, endogenous expression of ERRα increases to regulate gene programs involved in specialized myocyte function (eg, sarcomeric protein expression) and mitochondrial biogenesis as well as glucose and fatty acid oxidation (17–19). In addition, at a later stage of differentiation, ERRα was shown to be important for the oxidative capacity of the myotubes (17). It was also recently suggested that ERRα has a role in skeletal muscle regeneration in response to cardiotoxin injury (17–19). Taken together, these findings support a role for the receptor in muscle growth and regeneration. However, the global impact of ERRα deficiency on skeletal muscle function has not yet been explored. Herein, we used a physiological genomics/metabolomics approach to investigate the impact of the absence of ERRα on muscle fitness. Our results demonstrate that ERRα controls transcriptional programs that are essential for exercise tolerance and muscle fitness.
Materials and Methods
Animals
Mice were housed and maintained in a pathogen-free housing facility at McGill University, and all mouse manipulations were performed in accordance with the McGill Facility and Canadian Council on Animal Care. The generation of ERRα-knockout (KO) animals was previously described (20). The KO animals were derived from a pure FVB genetic background. For all animal studies, 2- to 3-month-old mice were used. For compound 29 (C29) treatments, wild-type (WT) mice were randomly assigned to receive daily ip injections of C29 (10 mg/kg) (OmegaChem Inc) diluted in Ringer's solution (containing 5.2% polyethylene glycol and 5.2% Tween 80) or vehicle alone (n = 12 for each group). Ten days after the first injection, muscle endurance was assessed.
Treadmill exercise
One day before the treadmill experiments, animals (n = 12 for each group, male and female) were acclimatized to treadmill running (Columbus Instruments) for 5 minutes on a 0% degree grade and at a speed of 5 m/min. The day of the exercise stress test, mice ran on a treadmill with a 5° inclination uphill at a speed of 5 m/min for the first 5 minutes. The speed was increased by 5 m/min every subsequent 5 minutes until a maximal speed of 25 m/min or until exhaustion was reached. The treadmill experiment was stopped when mice stayed for 5 seconds continuously on the electrical grid. A maximal score of 2000 m was given to mice that did not reach exhaustion before this distance was reached. Time to exhaustion and total running distance were determined using the apparatus. Work (joules) and power (watts) were calculated, with work being the product of body weight (kilograms), gravity (9.81 m/s2), vertical speed (meters per second times angle), and time (seconds) and power calculated as the product of body weight (kilograms), gravity (9.81 m/s2) and vertical speed (meters per second times angle).
Metabolic cage measurements
Spontaneous activity and energy metabolism studies were performed on mice (n = 8 for each group, male and female) under a consistent temperature (25°C) using an indirect calorimeter (Oxymax; Columbus Instruments). The day before the experiment, mice were weighed and placed in individual chambers at 25°C. After acclimation, metabolic parameters (oxygen consumption [VO2] and carbon dioxide production [VCO2]) were recorded for 48 hours. Exhaust air from each chamber was sampled at 10-minute intervals for a period of 30 seconds, and sample air was sequentially passed through O2 and CO2 sensors for determination of O2 and CO2 content. VO2 and VCO2 values were normalized with respect to body weight and VO2, VCO2, respiratory exchange ratio (RER), and heat were calculated using Oxymax software included with the calorimeter. The voluntary locomotive activity was quantified using the infrared beam interruptions in horizontal (X) directions (XAMB). To control for circadian fluctuations in activity and metabolism, experiments were always started at Zeitgeber time 6 to 8.
Blood and tissue metabolite measurements
Blood glucose and lactate were measured before and immediately after exercise from tail lateral vein blood using a OneTouch Ultra2 glucose meter (LifeScan) and Lactate Scout (Lactate.com), respectively. Liver and skeletal muscle (gastrocnemius and soleus) glycogen levels were assessed using an available quantification kit (Abcam; ab169558). Serum and skeletal muscle (gastrocnemius and soleus) triglyceride and free fatty acid levels were determined using available quantification kits (Abcam; ab65341 and ab65336, respectively). Skeletal muscle (gastrocnemius and soleus) α-ketoglutarate dehydrogenase enzyme activity was determined using an available quantification kit (BioVision; K678–100).
Metabolomics analyses
Metabolite measurements were performed on liver and skeletal muscle (gastrocnemius and soleus) by liquid chromatography tandem mass spectrometry in collaboration with the Goodman Cancer Research Centre Metabolomics Core Facility (http://gcrcmetabolomics.lab.mcgill.ca). Specific metabolite transitions for quantifier/qualifier ions and electrospray ionization source modes are listed in Supplemental Table 1. Detailed methodology is provided in Supplemental Materials and Methods.
Histology
Tissues were fixed with 10% formalin followed by paraffin embedding. Serial 4-μm sections were immunostained with CD31. Slides were scanned with an Aperio ScanScope instrument (Aperio Technologies Inc) and viewed with Aperio's ImageScope software, and CD31 immunostaining was analyzed using an optimized Aperio algorithm (n = 8 per condition). Histology experiments were performed in collaboration with the Goodman Cancer Research Centre Histology Core Facility.
Expression analyses
Total RNA from mouse tissues was extracted using the RNeasy tissue mini kit and the RNeasy fibrous tissue mini kit (QIAGEN) for liver and gastrocnemius samples, respectively. mRNA was reverse-transcribed into cDNA using Superscript (Invitrogen) and quantified by quantitative RT-PCR (qRT-PCR) on a LightCycler 480 instrument (Roche) using LightCycler 480 SYBR Green I master reagents (Roche). The relative expression was normalized to Rplp0. Gene-specific primers used for qRT-PCR analysis are listed in Supplemental Table 2.
Microarray preparation and analyses
Microarray analyses were performed on gastrocnemius muscle isolated from mice before exercise (sedentary) and 2 hours after exercise (1 run on a treadmill to exhaustion) at the McGill University Génome Québec Innovation Centre. Samples were run on Affymetrix Genechip Mouse gene version 2.0 ST arrays following Affymetrix's standard procedures (n = 3 per condition). The data were analyzed using Expression Console and Transcriptome Analysis Console software (Affymetrix, Inc). We considered linear 1.2-fold changes and a P value threshold of .05 as the cutoff to identify differentially expressed genes (Supplemental Table 3). Ingenuity Pathway Analysis (IPA) software was used to identify enriched canonical pathways.
Statistical analyses and accession numbers
Bars in the graphical data represent means ± SEM. Data were compared by the Student's unpaired two-tailed t test or by log-rank Mantel-Cox test, and P values ≤ .05 were considered statistically significant. Microarray data are available in the NCBI's Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE61712.
Results
ERRα-deficient mice are hypoactive and exhibit decreased exercise capacity
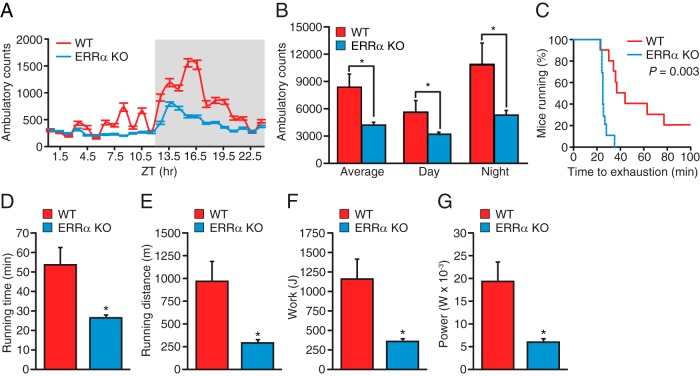
Phenotypic characterization of the ERRα-null mice revealed significantly reduced heart, gastrocnemius, soleus, and quadricep muscle mass relative to WT mice (Figure 1, A–D). Because reduction of muscle mass may impact global locomotion and muscle endurance, we thus investigated the consequence of ablation of ERRα expression in mice on their physical activity and exercise capacity. To this end, we first monitored spontaneous locomotor activity of WT and ERRα-null mice over a 24-hour period in comprehensive laboratory animal monitoring systems (CLAMS) metabolic cages. ERRα-deficient animals were found to exhibit significant hypoactivity during both their light and dark cycles, with a similar decrease in physical activity in both cycles compared with control mice (Figure 2, A and B). We next determined whether the decreased global locomotion observed in the ERRα-null mice translated into reduced treadmill performance during a forced exercise challenge. The mice were allowed to run until exhaustion or until a cutoff distance of 2000 m was achieved. Mice lacking ERRα ran for a significantly shorter time relative to WT mice (26 vs 54 minutes), demonstrating a reduced exercise tolerance in these mice (Figure 2, C and D). Accordingly, the ERRα-KO mice ran for a significantly shorter distance and performed less work compared with the WT mice (Figure 2, E and F). Moreover, loss of ERRα resulted in a significantly lower short-term high-intensity performance, referred to herein as power (Figure 2G). Given that studies regarding skeletal muscle have reported a gender effect (21), physical activity and exercise capacity were also assessed in female mice. Female ERRα-null mice were found to have reduced voluntary physical activity and a diminished endurance capacity and short-term high-intensity performance in a manner similar to that observed using male mice (Supplemental Figure 1, A–G). Taken together, the data clearly identifiy ERRα as a key player in exercise tolerance.
Figure 1. Decreased muscle mass in ERRα-KO mice.
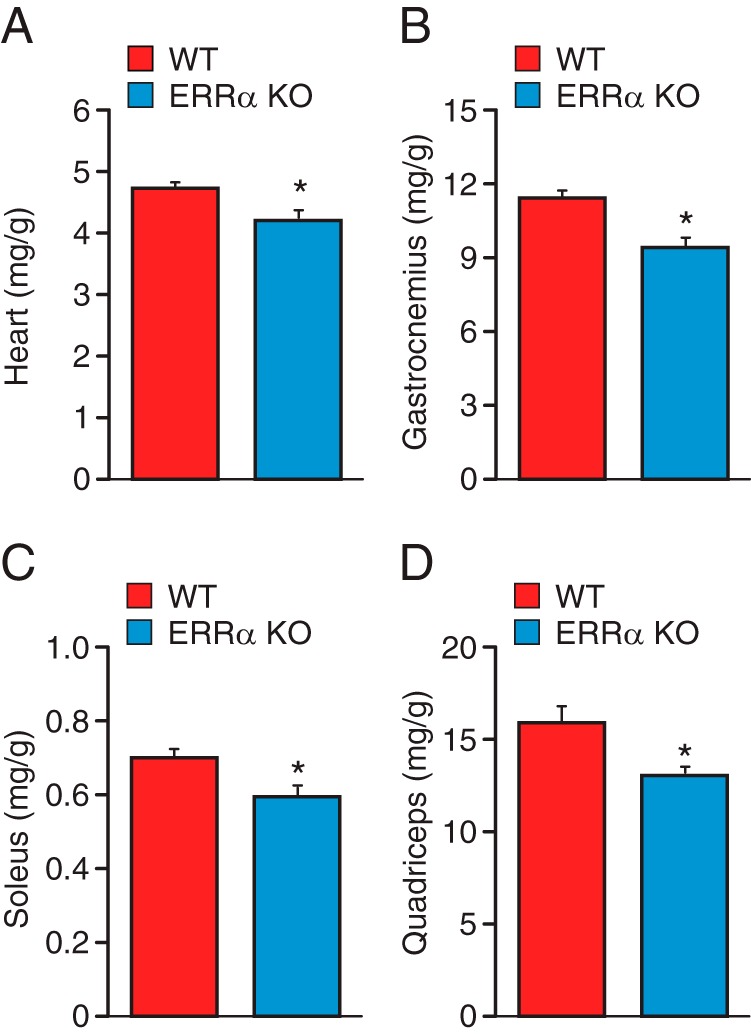
A–D, Indexed mass to body weight ratios of heart (A), gastrocnemius (B), soleus (C), and quadriceps (D) in 3-month old WT and ERRα-null mice. Data are expressed as means ± SEM; n = 12 mice per group. *, P < .05 by unpaired t test.
Figure 2. Mice lacking ERRα exhibit an abnormal skeletal muscle functional phenotype.
A, Circadian patterns of voluntary locomotive activity were recorded in a comprehensive cage monitoring system. B, Average spontaneous physical activity of male WT and KO mice (n = 8 per group). C–G, Measure of muscle endurance with dynamic fiber contractions. Two-month-old male WT and KO mice were run on a treadmill with a 5° inclination and increasing speed to exhaustion (n = 12 per group). Endurance capacity (C), time (D), distance (E), work (F), and power (G) were calculated from the individual performances. A maximal score of 2000 m was given to mice that did not get exhausted. Values are expressed as means ± SEM (B and D–G). *, P < .05 by log-rank Mantel-Cox test (C) or by unpaired t test (B and D–G).
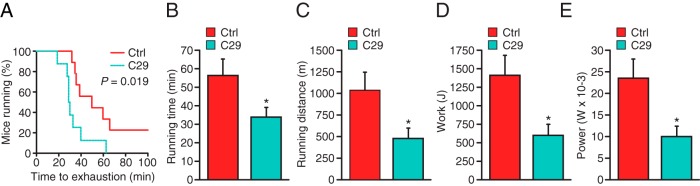
Next, the highly selective ERRα inverse agonist (C29) (22) was used to validate our findings that the genetic loss of ERRα lowers the exercise capacity of mice. To this end, mice administered C29 daily over a 10-day period were allowed to run to exhaustion on a treadmill. Consistent with the results observed with the ERRα-null animals, C29-treated WT male mice were able to run for only 34 minutes on average until exhaustion compared with 56 minutes for control-treated mice (Figure 3, A and B). The distance ran, work, and power generated by the C29-treated male animals were also significantly lower than those of the control mice (Figure 3, C–E). Similar results were obtained in C29-treated female mice (Supplemental Figure 1, H–L). Overall, the results demonstrate that genetic or pharmacological inhibition of ERRα impairs exercise capacity in both male and female mice.
Figure 3. Pharmacological inhibition of ERRα in mice limits exercise tolerance.
Two-month old male C29-treated or control mice were run on a treadmill with a 5° inclination and increasing speed to exhaustion. A–E, Endurance capacity (A), time (B), distance (C), work (D), and power (E) were calculated from the individual performances. A maximal score of 2000 m was given to mice that did not get exhausted (n = 12 per group). Values are expressed as means ± SEM (B–E). *, P < .05 by log-rank Mantel-Cox test (A) or by unpaired t test (B–E).
ERRα-deficient mice display alterations in energy production
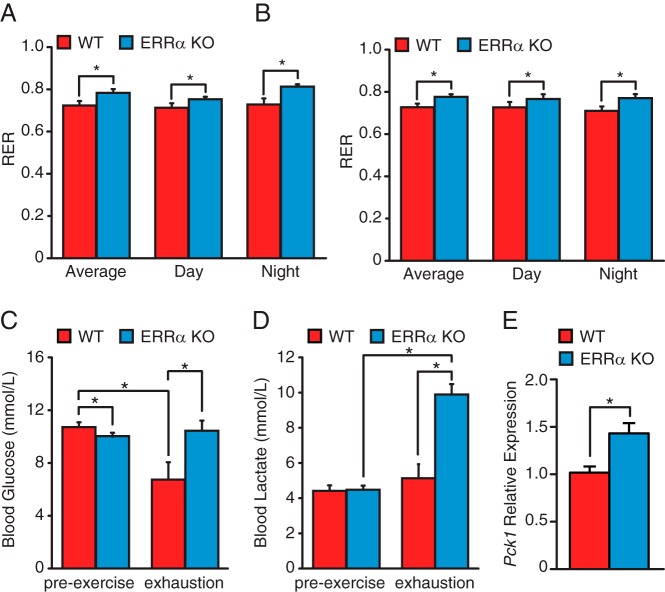
Differences in endurance are frequently associated with alterations in energy production. Thus, we sought to investigate the basal global metabolism of the ERRα-null animals using indirect calorimetry. Although no significant differences in VO2, VCO2, and energy expenditure were observed between ERRα-null mice and WT littermates of both genders (Supplemental Figure 3), a significant difference in the RER could be observed between the 2 genotypes (Figure 4, A and B). The RER was higher for both genders in the KO animals, suggesting that mice lacking ERRα rely more on carbohydrates than lipids as substrates for energy production. We next measured blood lactate and glucose levels in the mice before and immediately after exhaustive exercise as further indicators of glycolytic and oxidative metabolism. ERRα-KO mice were found to have a small pre-exercise decrease in blood glucose levels compared with WT animals, supporting a greater global reliance on carbohydrates in these mice (Figure 4C). In WT mice, exhaustive exercise was found to lower blood glucose levels without a concomitant increase in lactate levels (Figure 4, C and D). In sharp contrast, exhausted ERRα-null mice were found to have a significant accumulation in blood lactate levels, although no change in glucose levels was observed (Figure 4, C and D). These results suggest a deficiency in lactate handling due to a defect in oxidative metabolism and/or gluconeogenesis in the mutant mice. Previous reports have shown a repressive role of ERRα on gluconeogenic gene expression (23, 24). In agreement with these observations, livers of ERRα-null mice were found to have increased transcript levels of Pck1 (Figure 4E), suggesting that the accumulation of lactate in the blood of ERRα-null mice is likely due to impaired mitochondrial oxidative metabolism, not gluconeogenesis.
Figure 4. Increased use of carbohydrates for energy production in ERRα-KO mice.
A and B, The RER in male (A) and female (B) mice was measured during a 24-hour period by indirect calorimetry in WT and ERRα-KO mice. C and D, Blood glucose (C) and lactate (D) levels in WT and ERRα-KO male mice before and after exercise. Data are expressed as means ± SEM; n = 8 mice per group. *, P < .05 by unpaired t test. E, qRT-PCR analysis of hepatic Pck1 expression in WT and ERRα-null mice under basal conditions. Data are normalized to Rplp0 levels and expressed as means ± SEM. *, P < .05 by unpaired t test.
ERRα regulates energetic gene programs in muscle
To further understand the molecular basis for the metabolic defects and diminished exercise capacity observed in ERRα-KO mice in addition to understanding the role of ERRα in the molecular adaptation to exercise, gene expression profiling studies were performed on gastrocnemius muscle isolated before and 2 hours after acute endurance exercise in KO and WT animals. Experiments were performed on gastrocnemius muscle because this is a major calf muscle, containing a combination of fiber types (slow- and fast-twitch), which is used for both powerful bursts of activity and sustained but slower movements (25). Because acute exercise alters the transcriptome with maximal changes in gene expression generally occurring between 3 and 12 hours after exercise (26), we selected a 2-hour time point after exercise to uncover ERRα-dependent early response genes.
First, we compared the genes that were differentially regulated in the ERRα-null mice (vs WT mice) before and after exercise. Gene expression analyses identified 772 modulated genes in the pre-exercised ERRα-KO gastrocnemius and 857 genes after exercise (Figure 5A). Functional analyses of the differentially regulated genes at before and after exercise using IPA software revealed a large set of ERRα-dependent genes involved in mitochondrial dysfunction and energy metabolism (eg, oxidative phosphorylation and the tricarboxylic acid [TCA] cycle). Many genes were deregulated in the KO animals after exercise, indicating that ERRα has a role in transducing metabolic signals to regulate energetic gene programs in response to exercise in skeletal muscle (Figure 5B). Importantly, under both basal conditions and after acute exercise, a significant subset of ERRα target genes important for mitochondrial energy metabolism was found downregulated including Cycs, Idh3g, and Pdha1 (Figure 5C). Cycs encodes a central component of the mitochondrial electron transport chain and Idh3g encodes an enzyme regulating the rate-limiting step of the TCA cycle. Pdha1 encodes a subunit of the pyruvate dehydrogenase complex, which provides the primary link between glycolysis and the TCA cycle. In further support of a diminished mitochondrial oxidative capacity in the ERRα-deficient animals, an accumulation of the TCA cycle intermediates, citrate, cis-aconitate, and α-ketoglutarate, was observed in the skeletal muscle of these mice after exercise (Figure 5D). In addition, decreased levels of the TCA cycle intermediates, succinate and malate, were found in the ERRα-null mice after exercise (Figure 5D), suggesting a block in the enzymatic activity of the α-ketoglutarate dehydrogenase (α-KGDH) complex in these mice responsible for the conversion of α-ketoglutarate to succinyl coenzyme A. As shown in Figure 5E, mice lacking ERRα displayed a significant decrease in α-KGDH activity after exercise. A summary of the altered skeletal muscle TCA cycle metabolism found in the ERRα-null skeletal muscle after acute exhaustive exercise is illustrated in Figure 5F.
Figure 5. Loss of ERRα impairs metabolic and exercise-dependent transcriptional programs in skeletal muscle.
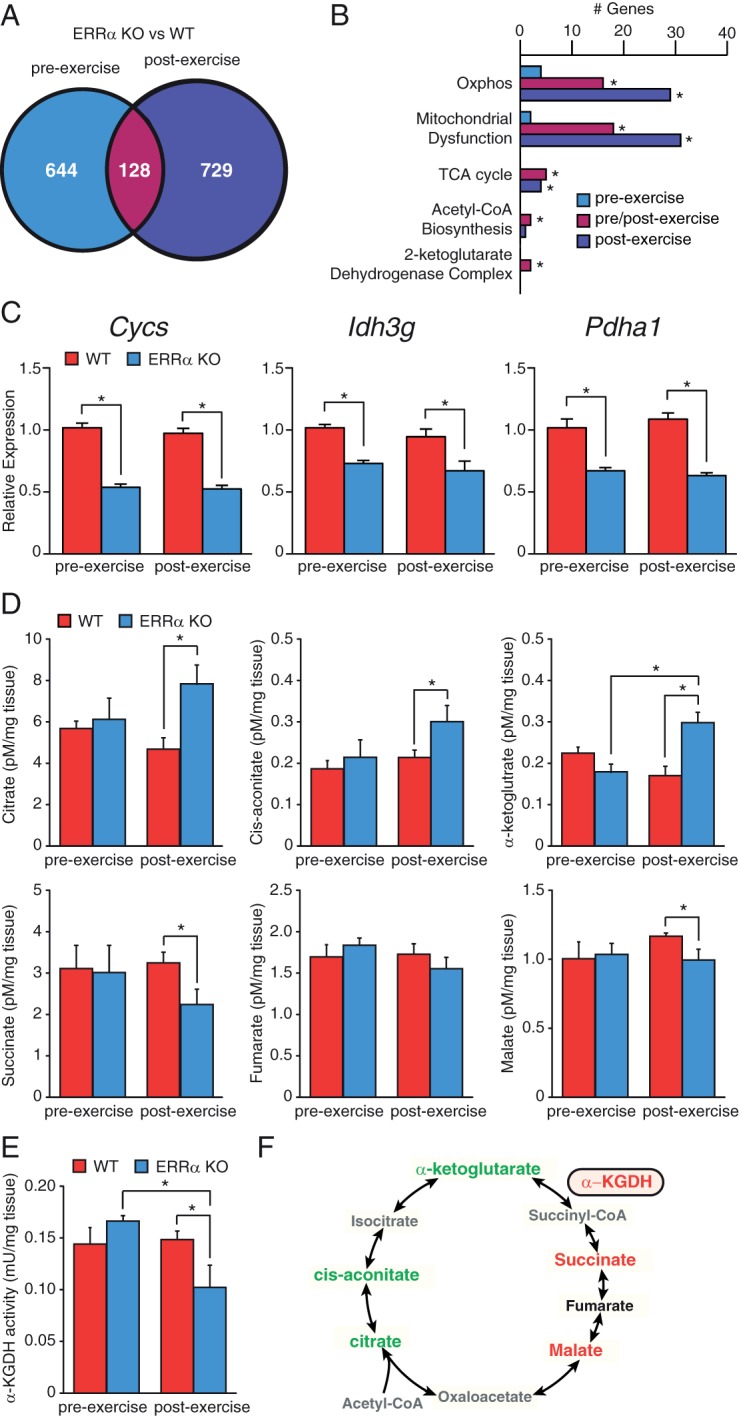
A, Venn diagrams illustrating the overlap in the number of altered genes in ERRα-null gastrocnemius before and after exercise. B, The number of altered genes in ERRα-KO mice within metabolic pathways identified by IPA analysis are shown. *P < .05. C, qRT-PCR analysis of metabolic genes that are ERRα-dependent in gastrocnemius isolated from WT and ERRα-null before and after exercise. Data are normalized to Rplp0 levels and expressed as means ± SEM. *, P < .05 by unpaired t test. D, Quantification of TCA cycle intermediates by liquid chromatography tandem mass spectrometry in skeletal muscle collected from WT and ERRα-KO mice before and after exercise*, P < .05 by unpaired t test. E, Quantification of α-ketoglutarate dehydrogenase (α-KGDH) enzyme activity in skeletal muscle collected from WT and ERRα-KO mice before and after exercise*, P < .05 by unpaired t test. F, Schematic summarizing the TCA cycle alterations identified in ERRα-null skeletal muscle after exercise. Green, increased metabolite levels; red, decreased metabolite levels and α-KGDH enzyme activity (boxed); black, no change; gray, not measured.
Given that peroxisome proliferator-activated receptor γ-coactivator-1 (PGC-1)α and PGC-1β are master regulators of mitochondrial energy metabolism and are coactivators of ERRα, we next examined the gastrocnemius transcript levels of Ppargc1a and Ppargc1b (18, 27, 28). A small increase in Ppargc1a levels was found in the ERRα-null muscle under basal conditions possibly to compensate for the absence of ERRα (Supplemental Figure 4). Although it is well established that exercise induces PGC-1α expression (29), loss of ERRα strongly impaired the induction of PGC-1α transcript levels after exercise (Supplemental Figure 4). In contrast, no significant differences in the expression of PGC-1β mRNA levels were observed in mice lacking ERRα before or after exercise (Supplemental Figure 4). Taken together, the data demonstrate a reduction in the ability of gastrocnemius muscle in ERRα-null mice to sustain a genetic program required to increase mitochondrial oxidative capacity underlying the decreased ability of these mice to endure aerobic exercise.
Loss of ERRα alters energy substrate availability and metabolism
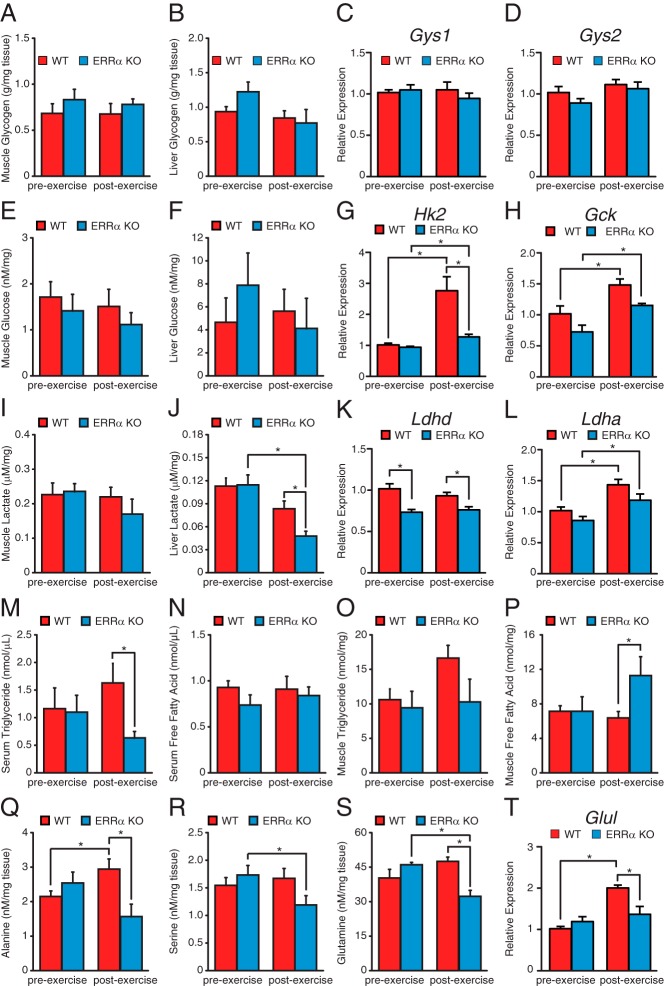
We next investigated the metabolism of the primary energy substrates used in exercised muscle. To this end, the levels of glycogen, glucose, lactate, triglyceride, and amino acids before and 2 hours after acute endurance exercise in ERRα-KO and WT animals were determined (Figure 6). Glycogen is a readily mobilized storage for glucose primarily in the muscle and liver and is of central importance as the first source of energy for muscle contractions, especially during high-intensity exercise. Loss of ERRα had no effect on skeletal muscle or hepatic glycogen content before or after exercise (Figure 6, A and B). Accordingly, we observed no difference in the mRNA levels of the rate-limiting muscle and hepatic glycogen storage enzymes, Gys1 and Gys2, respectively (Figure 6, C and D). As observed for glycogen, WT and ERRα-null mice were found to have similar amounts of muscle and hepatic glucose content (Figure 6, E and F). However, in response to exercise, loss of ERRα significantly impaired the induction of Hk2, the predominant muscle enzyme responsible for committing glucose to the glycolytic pathway. A similar trend was observed for Gck in the liver (Figure 6H). In addition, an increase in both muscle and liver Pdk4 levels were found in WT but not ERRα-null mice after exercise (Supplemental Figure 5, A and B). The data suggest that although glucose levels were restored 2 hours after exercise in both WT and ERRα-null mice, WT mice have an increased potential for anaerobic glycolysis. Although no differences in muscle lactate levels were found in ERRα-KO animals, livers lacking ERRα had significantly lower lactate levels after exercise (Figure 6, I and J). Decreased expression of Ldhd, the predominant muscle lactate dehydrogenase isoform responsible for the conversion of lactate to pyruvate in the final step of anaerobic glycolysis, was found in ERRα-null mice (Figure 6K). A similar trend was observed for the gene encoding the predominant liver Ldh isoform, Ldha (Figure 6L). Interestingly, the induction of the lactate transporter gene Slc16a1 was lost in the muscle in the absence of ERRα (Supplemental Figure 5C). A similar trend was observed in the liver (Supplemental Figure 5D). The reduction in the expression of the lactate transporter and decreased mitochondrial oxidative capacity implies a reduced uptake and oxidation of lactate as a source of energy in the muscle of ERRα-null mice likely contributing to the observed accumulation of circulating lactate.
Figure 6. Measurement of fuel sources in WT and ERRα-null mice before and after exercise.
A and B, Quantification of muscle (A) and liver (B) glycogen levels in WT and ERRα-KO mice before and after exercise. C and D, qRT-PCR analysis of the predominant gastrocnemius (C) and liver (D) glycogen synthase gene in WT and ERRα-null mice before and after exercise. Data are normalized to Rplp0 levels and expressed as means ± SEM. E and F, Quantification of muscle (E) and liver (F) glucose levels in WT and ERRα-KO mice before and after exercise. G and H, qRT-PCR analysis of the predominant gastrocnemius (G) and liver (H) hexokinase gene in WT and ERRα-null mice before and after exercise. Data are normalized to Rplp0 levels and expressed as means ± SEM. *, P < .05 by unpaired t test. I and J, Quantification of muscle (I) and liver (J) lactate levels in WT and ERRα-KO mice before and after exercise. *, P < .05 by unpaired t test. K and L, qRT-PCR analysis of lactate dehydrogenase (LDH) genes in gastrocnemius (K) and liver (L) WT and ERRα-null mice before and after exercise. Data are normalized to Rplp0 levels and expressed as means ± SEM. *, P < .05 by unpaired t test. M–P, Quantification of circulating and skeletal muscle triglyceride and free fatty acids levels in WT and ERRα-KO mice before and after exercise. *, P < .05 by unpaired t test. Q–S, Quantification of skeletal muscle amino acid levels in WT and ERRα-KO mice before and after exercise. *, P < .05 by unpaired t test. T, qRT-PCR analysis of Glul expression in the gastrocnemius of WT and ERRα-null mice before and after exercise. Data are normalized to Rplp0 levels and expressed as means ± SEM. *, P < .05 by unpaired t test.
Furthermore, although no differences in circulating free fatty acids were observed, ERRα-null mice were found to have significantly lower serum triglyceride levels after exercise (Figure 6, M and N). In addition, although there was a tendency for higher muscle triglyceride levels in WT mice after exercise, there was significantly higher free fatty acid levels in ERRα-KO mice (Figure 6, O and P). No difference in mRNA levels of the major skeletal muscle lipid transporter Fabp3 was observed between WT and ERRα-null mice (Supplemental Figure 5E). However, ERRα-null muscles had decreased expression of Acadvl encoding an enzyme involved in fatty acid oxidation, possibly reflecting the accumulation of free fatty acids observed in the animals after exercise (Supplemental Figure 5F) (30).
Finally, we observed that the levels of several amino acids including alanine, serine, and glutamine were not replenished after acute exhaustive exercise in the ERRα-KO skeletal muscle compared with WT (Figure 6, Q–S, and Supplemental Table 4). Of interest, the induction of the mRNA levels of the gene responsible for glutamine synthesis, Glul, after exercise in WT mice was lost in the ERRα-null muscle, reflecting the impaired restoration of glutamine levels in these mice (Figure 6T). Collectively, our findings suggest that the availability, transformation, or replenishment of several substrates important for energy production is altered in the ERRα-KO animals.
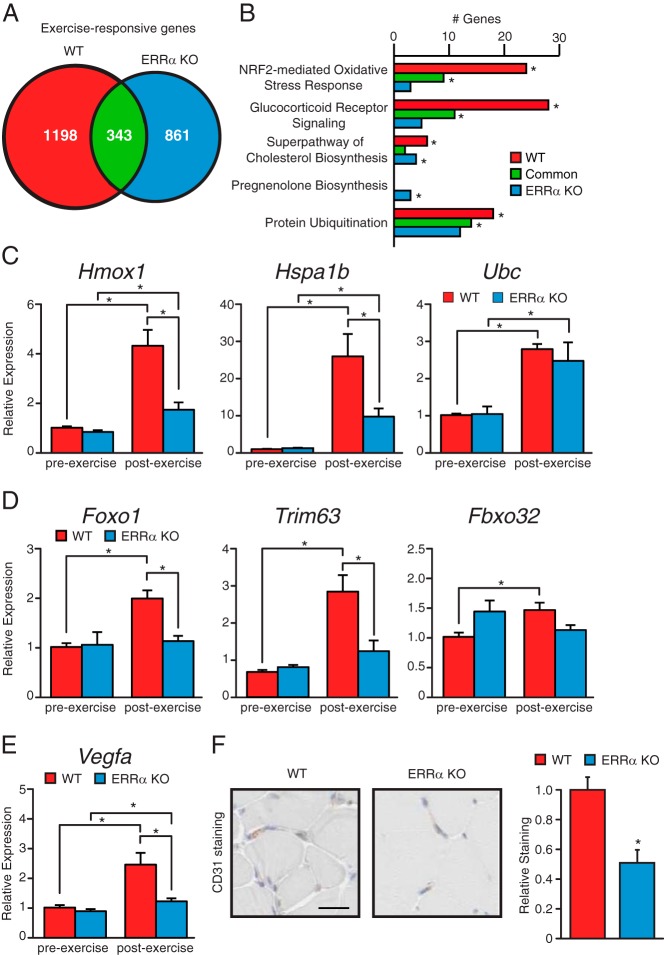
ERRα is implicated in several aspects of muscle fitness
We next explored whether other factors that could also contribute to the diminished voluntary physical activity and dynamic muscle endurance of the KO mice. To this end, we compared the genes that were differentially regulated in gastrocnemius muscle upon exercise between WT and ERRα-KO mice. First, a lower number of exercise-responsive genes were found in the ERRα-KO animals compared with WT (1204 vs 1541) (Figure 7A). Functional analysis of the differentially regulated genes using IPA notably revealed a higher enrichment of genes implicated in the Nuclear respiratory factor 2 (NRF2)-mediated oxidative stress response, glucocorticoid (GC) receptor signaling, cholesterol biosynthesis and protein ubiquitination in the exercised WT animals (Figure 7B). In response to exercise, ERRα-null mice were found to have several genes significantly modulated in the pregnenolone biosynthesis pathway. Pregnenolone is synthesized from cholesterol and is a precursor for several steroid hormones including GCs. The NRF2 pathway plays an important role in limiting muscle damage induced by excessive oxidative stress after exercise (31, 32). GC receptor signaling aids in the maintenance of muscle fiber integrity and the disposition of altered organelles through the regulation of the autophagic machinery (33, 34). Although Hmox1 and Hspa1b, involved, respectively, in oxidative stress response and GC receptor signaling, were induced in both WT and ERRα-null mice, the degree of induction was significantly blunted in the absence of ERRα (Figure 7C). The plasticity of skeletal muscle is highlighted by the remodeling and substantial changes in mass via the ubiquitin proteasome system in response to altered physical activity levels (35). Although the levels of Ubc, encoding ubiquitin C, were induced in a similar manner in both WT and ERRα-null mice after exercise (Figure 7C), the induction of the genes involved in muscle atrophy and in the ubiquitin-proteasome system, Foxo1, Trim63, and Fbxo32, in response to exercise was lost in ERRα-null mice (Figure 7D).
Figure 7. Loss of ERRα impairs exercise-dependent transcriptional programs in skeletal muscle.
A, Venn diagrams illustrating the overlap in number of exercise-responsive genes in gastrocnemius muscle between WT and ERRα-KO mice. B, The number of exercise-responsive genes associated with various biological processes identified by IPA are shown for WT and ERRα-KO gastrocnemius. *P < .05. C–E, qRT-PCR analysis of genes identified in B. Data are normalized to Rplp0 levels and expressed as means ± SEM. *, P < .05 by unpaired t test. F, CD31 staining and quantification of WT and ERRα-null gastrocnemius cross-sections. *, P < .05 by unpaired t test.
Furthermore, we identified a potential role for ERRα in exercise-induced angiogenesis, a common adaptive response to exercise training in skeletal muscle (36). Angiogenesis is necessary to carry the increasing oxygen demand needed for oxidative metabolism during exercise. After exercise, we observed a significantly greater induction in the mRNA level of the angiogenic factor Vegfa in WT animals compared with the ERRα-null mice (Figure 7E). In addition, evaluation of vascularization of gastrocnemius muscles showed reduced density of vessels and lower positive staining with the CD31 endothelial marker (Figure 7F). Taken together, the data implicate ERRα in the regulation of additional genetic programs important for muscle fitness.
Discussion
Recent studies have established vital roles for nuclear receptors in the regulation of muscle energy metabolism and exercise-dependent muscle remodeling and thus the potential for the development of exercise mimetics or pharmacological compounds that could enhance the adaptive response in exercisers (26, 37). In the current work, we demonstrated that the nuclear receptor ERRα, although seemingly dispensable for fiber type specification, is a critical transcription factor regulating physiological processes in muscle that are key determinants of performance, global motor activity, and adaptation to exercise-induced stress.
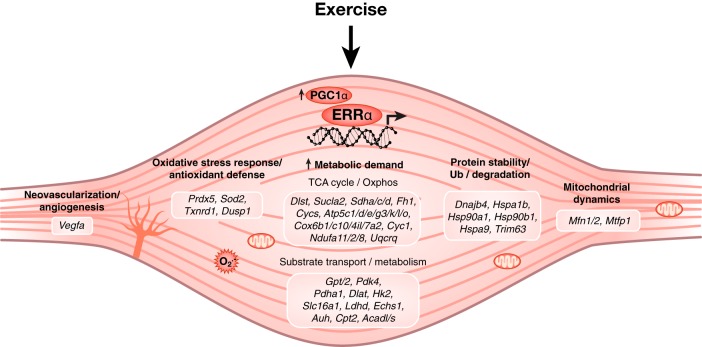
Our work first shows that ablation of ERRα expression in mice affects their muscle mass, as shown by reduced size of several muscles in the transgenic animals. Genetic or therapeutic inhibition of ERRα in mice resulted in a striking decrease in both physical activity and exercise capacity. Gene expression profiling, metabolic, and metabolomics studies demonstrate an essential role for ERRα in muscle metabolic function. Mice lacking ERRα have reduced expression of many genes involved in mitochondrial oxidative metabolism and the TCA cycle, resulting in the accumulation of several TCA cycle intermediates including citrate and α-ketoglutarate in response to the high energetic demands of exercise. In addition, ERRα-null mice exhibited decreased α-KGDH enzyme activity after acute exhaustive exercise, further demonstrating loss of mitochondrial metabolic homeostasis in these mice. Moreover, we show that the absence of ERRα affects the expression of genes associated with substrate transport/uptake and metabolism such as Slc16a1, Ldhd, Glul, and Hk2. Accordingly, exhausted ERRα-null mice exhibited lactatemia, supporting the reduced exercise tolerance observed in these mice. Furthermore, circulating triglycerides and several amino acids in the muscle including alanine and glutamine were not replenished during recovery to exercise. Mechanistically, it should be noted that our previous functional genomics studies identified ERRα as a direct regulator of numerous genes involved in the metabolic pathways described herein (38–42) (Figure 8 and Supplemental Table 5).
Figure 8. ERRα is essential for skeletal muscle fitness.
Schematic representation of biological processes coordinated by ERRα in exercise-induced muscle remodeling. Our model includes the well-characterized exercise-induced expression of the ERRα coactivator PGC1-α, also observed in this study (Supplemental Figure 4). All genes described in this figure are known direct transcriptional targets of ERRα (Supplemental Table 5).
Like ERRα, the ERR coactivator PGC-1α has been shown to play a key role in exercise performance. Studies with muscle-specific PGC-1α transgenic or null mice demonstrate that PGC-1α expression is associated with improved exercise tolerance associated with increased mitochondrial gene expression and activity and resistance to exercise-induced blood lactate accumulation via downregulation of Ldha and upregulation of Ldhb and Slc16a1 expression (43–45). Although we observed no differences in Ldha or Ldhb transcript levels in ERRα-null gastrocnemius, decreased mRNA levels of Ldhd were observed in these mice. Because the loss of ERRα resulted in decreased Slc16a1 expression and blocked the induction of this gene after exercise, our data suggest that ERRα promotes muscle lactate uptake and oxidation for the generation of energy as found for PGC-1α (43). Although the authors did not find an ERR binding consensus motif in the promoter of Slc16a1, ERRα is indeed recruited to this gene at a consensus ERR response element (ERRE), TGACCTTGG, located just 444 bp downstream of the transcription start site of the gene (39).
Aside from the involvement of ERRα in metabolic adaptations to exercise, we found that the receptor is important for the modulation of a set of genes that are needed to reduce exercise-induced muscle damage. In particular, these ERRα-dependent pathways work to counteract localized oxidative stress and inflammation, 2 processes associated with impaired muscular performance. Specifically, functional analysis of the exercise-responsive genes in the muscle of ERRα-null mice identified an impaired NRF2-mediated oxidative stress response (Figure 7B) (31, 32). This result is consistent with previous observations showing that ERRα facilitates the repair and functional recovery of skeletal muscle in response to cardiotoxin-induced injury (17–19). In addition, we found a number of ERRα-dependent genes involved in skeletal muscle plasticity including the ubiquitin proteasome pathway (Trim63, Hspa1b, and Hspa9) (Figure 8 and Supplemental Table 5). Thus, our findings implicate ERRα in tissue remodeling and changes in muscle mass to adapt to physiological factors such as exercise (35). Finally, we show that ERRα is involved in skeletal muscle vascularization, as indicated by decreased staining for the endothelial marker in the gastrocnemius of the ERRα-null mice and in skeletal muscle neovascularization, as shown by the reduced induction of the potent angiogenic factor Vegfa after exercise. Of note, transgenic overexpression of ERRγ was shown to be sufficient to enable anaerobic muscles to acquire enhanced oxidative capacity and dense vasculature (46). In that study, the morphological remodeling was linked to the induction of genes involved in oxidative phosphorylation, fatty acid oxidation, and proangiogenic genes, more specifically Vegfa. Thus, as previously demonstrated in the heart (14, 47), ERRα and ERRγ orchestrate common transcriptional programs to enhance mitochondrial oxidative potential required for exercise performance and fitness. However, in contrast to muscle overexpressing ERRγ (46), global loss of ERRα had no significant effect on myofibrillar gene expression after an acute exercise regimen. Our results thus suggest that ERRα is dispensable for fiber type specification in skeletal muscle as previously observed for the ERR coactivators PGC-1α and PGC-1β (40).
Collectively, our study demonstrates that the nuclear receptor ERRα, although not required for fiber type composition, is essential for the coordinate regulation of metabolic and stress response programs necessary for muscle fitness and endurance. Altered muscle function and metabolism are an underlying factor in many pathological conditions and chronic diseases. The observation that the ERRα inverse agonist C29 diminishes running endurance in a manner similar to the genetic ablation of ERRα suggests that, in contrast, a full ERRα agonist could possibly serve to potentiate the adaptive response to exercise stimuli. In particular, our results suggest that modulation of ERRα activity could provide a new therapeutic avenue in the prevention and management of muscle wasting and weakness in many disease states and conditions including aging, cancer cachexia, sepsis, denervation, chronic kidney or heart failure, and muscular dystrophies.
Acknowledgments
We thank C. Ouellet and M. Ghahremani for their technical assistance, Drs D. Avizonis and G. Bridon for metabolomics analyses, and S. Labrecque for figure design.
M.-C.P. was supported in part by the Natural Sciences and Engineering Research Council of Canada and the Fonds de Recherche Santé Québec. The study was supported by a grant from the Canadian Institutes for Health Research (MOP-64275) to V.G. The metabolomics Core Facility is supported by the Canada Foundation for Innovation (project number 21875), the Dr John R. and Clara M. Fraser Memorial Trust and the Terry Fox Foundation (TFF Oncometabolism Team Grant 116128) and McGill University.
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
M.-C.P. was supported in part by the Natural Sciences and Engineering Research Council of Canada and the Fonds de Recherche Santé Québec. The study was supported by a grant from the Canadian Institutes for Health Research (MOP-64275) to V.G. The metabolomics Core Facility is supported by the Canada Foundation for Innovation (project number 21875), the Dr John R. and Clara M. Fraser Memorial Trust and the Terry Fox Foundation (TFF Oncometabolism Team Grant 116128) and McGill University.
Footnotes
- C29
- compound 29
- ERRα
- estrogen-related receptor α
- GC
- glucocorticoid
- IPA
- Ingenuity Pathway Analysis
- α-KGDH
- α-ketoglutarate dehydrogenase
- KO
- knockout
- NRF2
- nuclear respiratory factor 2
- PGC-1
- peroxisome proliferator-activated receptor γ-coactivator-1
- qRT-PCR
- quantitative RT-PCR
- RER
- respiratory exchange ratio
- TCA
- tricarboxylic acid
- VCO2
- carbon dioxide production
- VO2
- oxygen consumption
- WT
- wild-type.
References
- 1. Booth FW, Thomason DB. Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol Rev. 1991;71:541–585. [DOI] [PubMed] [Google Scholar]
- 2. Hawley JA, Holloszy JO. Exercise: it's the real thing! Nutr Rev. 2009;67:172–178. [DOI] [PubMed] [Google Scholar]
- 3. Yan Z, Okutsu M, Akhtar YN, Lira VA. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J Appl Physiol. 2011;110:264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–1531. [DOI] [PubMed] [Google Scholar]
- 5. Giguère V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. [DOI] [PubMed] [Google Scholar]
- 6. Deblois G, Giguère V. Functional and physiological genomics of estrogen-related receptors (ERRs) in health and disease. Biochim Biophys Acta. 2011;1812:1032–1040. [DOI] [PubMed] [Google Scholar]
- 7. Eichner LJ, Giguère V. Estrogen related receptors (ERRs): a new dawn in the control of mitochondrial gene networks. Mitochondrion. 2011;11:544–552. [DOI] [PubMed] [Google Scholar]
- 8. Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sladek R, Bader JA, Giguère V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17:5400–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ichida M, Nemoto S, Finkel T. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor γ coactivator-1 alpha (PGC-1alpha). J Biol Chem. 2002;277:50991–50995. [DOI] [PubMed] [Google Scholar]
- 12. Villena JA, Hock MB, Chang WY, Barcas JE, Giguère V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A. 2007;104:1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huss JM, Imahashi K, Dufour CR, et al. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. [DOI] [PubMed] [Google Scholar]
- 14. Dufour CR, Wilson BJ, Huss JM, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. [DOI] [PubMed] [Google Scholar]
- 15. Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. [DOI] [PubMed] [Google Scholar]
- 16. Deblois G, Giguère V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer. 2013;13:27–36. [DOI] [PubMed] [Google Scholar]
- 17. Murray J, Huss JM. Estrogen-related receptor α regulates skeletal myocyte differentiation via modulation of the ERK MAP kinase pathway. Am J Physiol Cell Physiol. 2011;301:C630–C645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shao D, Liu Y, Liu X, et al. PGC-1 beta-regulated mitochondrial biogenesis and function in myotubes is mediated by NRF-1 and ERR alpha. Mitochondrion. 2010;10:516–527. [DOI] [PubMed] [Google Scholar]
- 19. Wang SC, Myers S, Dooms C, Capon R, Muscat GE. An ERRβ/γ agonist modulates GRalpha expression, and glucocorticoid responsive gene expression in skeletal muscle cells. Mol Cell Endocrinol. 2010;315:146–152. [DOI] [PubMed] [Google Scholar]
- 20. Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguère V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol Cell Biol. 2003;23:7947–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshioka M, Boivin A, Bolduc C, St-Amand J. Gender difference of androgen actions on skeletal muscle transcriptome. J Mol Endocrinol. 2007;39:119–133. [DOI] [PubMed] [Google Scholar]
- 22. Patch RJ, Searle LL, Kim AJ, et al. Identification of diaryl ether-based ligands for estrogen-related receptor α as potential antidiabetic agents. J Med Chem. 2011;54:788–808. [DOI] [PubMed] [Google Scholar]
- 23. Dufour CR, Levasseur MP, Pham NH, et al. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herzog B, Cardenas J, Hall RK, et al. Estrogen-related receptor alpha is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem. 2006;281:99–106. [DOI] [PubMed] [Google Scholar]
- 25. Gleeson TT, Putnam RW, Bennett AF. Histochemical, enzymatic, and contractile properties of skeletal muscle fibers in the lizard Dipsosaurus dorsalis. J Exp Zool. 1980;214:293–302. [DOI] [PubMed] [Google Scholar]
- 26. Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. [DOI] [PubMed] [Google Scholar]
- 27. Austin S, St-Pierre J. PGC1α and mitochondrial metabolism–emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. [DOI] [PubMed] [Google Scholar]
- 28. Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. [DOI] [PubMed] [Google Scholar]
- 29. Baar K, Wende AR, Jones TE, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. [DOI] [PubMed] [Google Scholar]
- 30. Yoshioka M, Doucet E, St-Pierre S, et al. Impact of high-intensity exercise on energy expenditure, lipid oxidation and body fatness. Int J Obes Relat Metab Disord. 2001;25:332–339. [DOI] [PubMed] [Google Scholar]
- 31. Miller CJ, Gounder SS, Kannan S, et al. Disruption of Nrf2/ARE signaling impairs antioxidant mechanisms and promotes cell degradation pathways in aged skeletal muscle. Biochim Biophys Acta. 2012;1822:1038–1050. [DOI] [PubMed] [Google Scholar]
- 32. Muthusamy VR, Kannan S, Sadhaasivam K, et al. Acute exercise stress activates Nrf2/ARE signaling and promotes antioxidant mechanisms in the myocardium. Free Radic Biol Med. 2012;52:366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waddell DS, Baehr LM, van den Brandt J, et al. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–E797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Masiero E, Agatea L, Mammucari C, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. [DOI] [PubMed] [Google Scholar]
- 35. Murton AJ, Constantin D, Greenhaff PL. The involvement of the ubiquitin proteasome system in human skeletal muscle remodelling and atrophy. Biochim Biophys Acta. 2008;1782:730–743. [DOI] [PubMed] [Google Scholar]
- 36. Gustafsson T, Kraus WE. Exercise-induced angiogenesis-related growth and transcription factors in skeletal muscle, and their modification in muscle pathology. Front Biosci. 2001;6:D75–D89. [DOI] [PubMed] [Google Scholar]
- 37. Fan W, Atkins AR, Yu RT, Downes M, Evans RM. Road to exercise mimetics: targeting nuclear receptors in skeletal muscle. J Mol Endocrinol. 2013;51:T87–T100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Charest-Marcotte A, Dufour CR, Wilson BJ, et al. The homeobox protein Prox1 is a negative modulator of ERRα/PGC-1α bioenergetic functions. Genes Dev. 2010;24:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chaveroux C, Eichner LJ, Dufour CR, et al. Molecular and genetic crosstalks between mTOR and ERRα are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metab. 2013;17:586–598. [DOI] [PubMed] [Google Scholar]
- 40. Zechner C, Lai L, Zechner JF, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sonoda J, Laganière J, Mehl IR, et al. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev. 2007;21:1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tremblay AM, Dufour CR, Ghahremani M, Reudelhuber TL, Giguère V. Physiological genomics identifies estrogen-related receptor alpha as a regulator of renal sodium and potassium homeostasis and the renin-angiotensin pathway. Mol Endocrinol. 2010;24:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Summermatter S, Santos G, Pérez-Schindler J, Handschin C. Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A. Proc Natl Acad Sci U S A. 2013;110:8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Handschin C, Chin S, Li P, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. [DOI] [PubMed] [Google Scholar]
- 45. Arany Z, Foo SY, Ma Y, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. [DOI] [PubMed] [Google Scholar]
- 46. Narkar VA, Fan W, Downes M, et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alaynick WA, Kondo RP, Xie W, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. [DOI] [PubMed] [Google Scholar]