Abstract
Human GH binds to its receptor (GHR) on target cells and activates multiple intracellular pathways, leading to changes in gene expression, differentiation, and metabolism. GHR deficiency is associated with growth and metabolic disorders whereas increased GHR expression has been reported in certain cancers, suggesting that the GHR gene requires tight controls. Several regulatory mechanisms have been found within its 5′-untranslated region (UTR) promoter and coding regions. However, the 3′-UTR has not been previously examined. MicroRNAs (miRNAs) are small (19–22 nucleotides) noncoding RNAs that downregulate gene expression mainly through targeting the 3′-UTR of mRNAs and enhancing their degradation or inhibiting translation. In the present study, we investigated whether miRNAs regulate GHR expression. To define putative miRNA binding sites in the GHR 3′-UTR, we used multiple in silico prediction tools, analyzed conservation across species and the presence of parallel sites in GH/IGF axis-related genes, and searched for reports linking miRNAs to GHR-related physiological or pathophysiological activities. To test prioritized sites, we cotransfected a wild-type GHR 3′-UTR luciferase reporter vector as well as miRNA binding site mutants into HEK293 cells with miRNA mimics. Furthermore, we tested whether the miRNAs altered endogenous GHR mRNA and protein levels in HEK293 cells and in 2 cancer cell lines (MCF7 and LNCaP). Our experiments have identified miRNA (miR)-129–5p, miR-142–3p, miR-202, and miR-16 as potent inhibitors of human GHR expression in normal (HEK293) and cancer (MCF7 and LNCaP) cells. This study paves the way for the development of miRNA inhibitors as therapeutic agents in GH/GHR-related pathophysiologies, including cancer.
Human GH is essential for normal musculoskeletal development in children; in addition, it has important regulatory effects on protein, carbohydrate, and lipid metabolism at all stages of life (1, 2). It functions by binding to a dimer of its high-affinity receptor (GH receptor [GHR]) on target cells, leading to phosphorylation of associated JAK2 tyrosine kinases as well as the receptor itself. The subsequent activation of multiple intracellular signaling pathways culminates in the biological actions of GH: changes in gene expression, enhanced proliferation, blocking of apoptosis, differentiation, and metabolic activity (3).
The ability of GH to exert its biological effects is intimately linked to the number and functional status of GHRs in target tissues. Individuals with low GHR levels or a dysfunctional GHR do not respond normally to GH; not only are they short, but they also have decreased bone mineral density and increased adiposity, with a greater risk of osteoporosis, lipid disorders, and cardiovascular disease (4). Persons with enhanced GH response, due to increased GH secretion or elevated functional GHR levels in target tissues, exhibit excessive growth and very abnormal metabolic activities, leading to an increased incidence of cardiomyopathies, hypertension, diabetes, and several types of cancers: leukemia, breast, prostate, colorectal, and gastric cancers (5, 6). Thus, to prevent these major medical morbidities, GHR expression must be tightly regulated at every stage of life.
The human GHR gene is located at chromosome 5p13.1-p12, where it spans more than 300 kb (7–9). It contains several noncoding 5′-untranslated region (UTR) exons with multiple splice variants that give rise to at least 14 different mRNAs, each with a unique 5′-UTR but all of which code for the same protein due to splicing into the same site upstream of the translation start site in the first coding exon, exon 2 (9–12). Transcription of the GHR gene results in an ∼4.5-kb mRNA (13). This GHR transcript is more than twice the minimum 1.9 kb necessary to encode the 638-amino-acid signal/receptor peptide molecule; most of the excess size is due to the presence of an ∼2.5-kb 3′-UTR within the GHR mRNA (14).
There have been extensive studies of how GHR gene expression is regulated at its multiple 5′-UTR promoters by our lab (15–18) as well as others (19). However, potential regulation at the 3′-UTR has not been examined. 3′-UTRs of mRNAs are well-known to be critical for the targeting of transcripts to specific subcellular compartments and for translational control (20). More recently, microRNAs (miRNAs) have been shown to be posttranscriptional regulators of gene expression, acting mainly via the 3′-UTRs of mRNAs (21, 22). The miRNAs are naturally occurring, 19- to 22-nucleotide-long, noncoding RNAs; nucleotides 2 to 8 at the 5′-end are known as the seed sequence, whereas the remaining nucleotides are the flanking region (23). The miRNAs function in the form of ribonucleoprotein complexes known as miRNA-induced silencing complexes (24). The miRNAs direct the miRNA-induced silencing complexes to sites primarily in the 3′-UTR of target mRNAs, the specificity of which is defined by both the miRNA seed sequence and the flanking region. The complex subsequently inhibits protein synthesis by mRNA degradation and/or the arrest of mRNA translation (21, 25, 26). Computational analysis indicates that more than 60% of protein-coding genes may be directly modulated by miRNAs (27), and accumulating evidence indicates that miRNAs play a central role in controlling a broad range of biological activities including embryonic development, cell proliferation, metabolic homeostasis, and apoptosis (28–33).
To understand whether miRNAs play a role in regulating human GHR expression, we have undertaken an analysis of miRNA effects on the GHR mRNA 3′-UTR. Our primary model was the human embryonic HEK293 cell line; however, we extended our studies to include 2 well-known cancer cell lines, MCF7 (breast) and LNCaP (prostate), and their corresponding normal cell lines, MCF10a and PNT1a, to determine whether the miRNA effects might be applicable to cancers linked to GHR overexpression. There is an extensive literature showing that GHR mRNA and protein are increased 2- to 5-fold in breast carcinomas (34–36), prostatic carcinomas (37–41), colorectal adenomas and adenocarcinomas (34, 42, 43), gastric adenocarcinomas (44), and hepatic carcinomas (34, 45–47) compared with their corresponding control tissues, suggesting that the GH/GHR axis is involved in progression of these cancers.
Materials and Methods
PCR amplification and cloning of the wild-type GHR 3′-UTR into the pmiR-luciferase vector
The following primers were used to amplify a 2293-bp sequence of the human GHR 3′-UTR using 10ng of genomic DNA as a template: forward primer (SacI) 5′-cgagctcaattgactggggcaataacg-3′ and reverse primer (MluI) 5′-cgacgcgtaaactgccagacacaactagtca-3′. A 2-step PCR assay was run using the Phusion High Fidelity DNA polymerase kit (Thermo Scientific) under the following conditions: an initial denaturation at 98°C for 2 minutes followed by 5 cycles of 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 150 seconds and then 30 cycles of 98°C for 10 seconds, 60°C for 4 minutes, and 72°C for 150 seconds. The PCR product was inserted into the SacI and MluI sites of the pmiR-REPORT luciferase vector (Applied Biosystems/Invitrogen Life Sciences) downstream of the luciferase gene under the CMV promoter. Before its use, the vector insert was sequenced at the McGill Genome Centre to ensure no mutations or deletions had occurred.
Mutagenesis
The QuikChange Lightning site-directed mutagenesis kit (Stratagene) was used to mutate the seed sequence at each of the miRNA binding sites under study. The pmiR-Luc-GHR 3′-UTR vector was the template, and overlapping pairs of primers were designed using the Stratagene Primer Design Program (www.genomics.agilent.com/primerDesignProgram.jsp) (Supplemental Table 1). To ensure that the mutated sequences did not form a new miRNA binding site, they were checked using the TargetScan Custom version 4.2 in silico program (www.targetscan.org/vert_42/seedmatch.html) (48).
Cell culture conditions and transfections
HEK293 cells were cultured in DMEM with 10% heat-inactivated fetal bovine serum (FBS), whereas MCF7, LNCaP, and PNT1a cells were grown in RPMI medium with 10% non–heat-inactivated FBS. All media were supplemented with 25mM HEPES, 50U/mL penicillin and 1.6 mg/mL gentamycin. MCF10a cells were grown in Mammary Epithelial Cell Basal Medium (MEBM) media and supplemented with the following growth factors: 0.4% bovine pituitary extract, 0.1% hydrocortisone, 0.1% recombinant human epidermal growth factor, 0.1% insulin, and 10μM forskolin (Lonza). Cells were grown at 37°C with 5% CO2 in air.
For GHR RNA and protein studies, cells were plated at 2.5–3 × 105 cells per well in 6-well plates in duplicate. The following day, the media were replaced with antibiotic-free media supplemented with 10% FBS and 50nM miRNA mimics (synthetic double-stranded miRNA-like RNA fragments from Dharmacon, division of Thermo Scientific) were transfected using the Dharmafect-1 transfection reagent for HEK293 and MCF7 cells and Dharmafect-2 for LNCaP cells, according to the manufacturer's protocol. Total RNA was isolated for quantitative RT-PCR analysis 24 hours after transfection, whereas proteins were extracted from a parallel set of cells after 48 hours.
For luciferase reporter gene assays, HEK293 cells were plated at 50 × 103 cells per well in 24-well plates. The pmiR-Luc vector (50 ng) containing the wild-type GHR 3′-UTR or a mutant version in addition to 1nM to 100nM miRNA mimics or 50nM to 100nM miRCURY LNA miRNA Power inhibitors (Exiqon) were cotransfected into cells using the Dual-Transfecting reagent (Dharmacon). The pmiR-REPORT β-galactosidase vector (50 ng) (Applied Biosystems) was cotransfected into all wells to normalize for transfection efficiency; this vector was designed to work in parallel with the pmiR-REPORT luciferase vector. Experiments were performed a minimum of 3 times with each mimic or the inhibitors.
Luciferase and β-galactosidase measurements
For luciferase experiments, 48 hours after transfection, cells were washed once with cold PBS and harvested in 200 μL lysis buffer (0.05% Nonidet P-40, 0.01 M dithiothreitol in 0.1M Tris [pH 8]) for 15 minutes at room temperature. Lysate (10 μL) was dispensed into 96-well microtiter plates, and luciferase activity was assayed after injecting 100 μL/well of 1× luciferin solution (0.1mM coenzyme A, 2.5mM ATP, 1× luciferin in 5mM Tris-HCl [pH 7.9]). For the β-galactosidase assay, 10 μL of the lysate was dispensed into 96-well microtiter plates, mixed with 100 μL Tropix Galacton-Star substrate diluted to 1× in Galacto-Star reaction buffer diluent (Applied Biosystems) and incubated for 1 hour at room temperature in the dark. Luciferase and β-galactosidase activities were measured in a bioluminometer (GloMax). Assays were performed in triplicate. Data were initially normalized to β-galactosidase activity and then expressed as a ratio of luciferase activity over empty pmiR-Luc vector values.
RNA and miRNA extractions and cDNA synthesis
Total RNA, including small RNA (miRNA), was extracted from the cells using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. RNA quantity was assessed using the Nanodrop Spectrophotometer ND-1000 (Thermo Scientific), and RNA integrity was verified using the Bio-Rad bioanalyzer. For cDNA amplification assays, 1 μg RNA from each sample was used for cDNA synthesis using the QuantiTect reverse transcription kit (QIAGEN) according to the manufacturer's instructions.
Quantitative PCR
GHR mRNA levels were measured in triplicate using the SYBR Green PCR assay (QIAGEN), proprietary human GHR primers (QIAGEN), and the LC480 quantitative PCR instrument (Roche). No template and no RT controls were included in each assay. Human B2 microglobulin (B2M) mRNA was used as an internal control to normalize between samples. The fold expression of the GHR mRNA in cells treated with specific miRNA mimics relative to negative control-treated samples was determined using the 2−ΔΔCt (where Ct is cycle threshold) method. The final data represent a minimum of 3 experiments.
Endogenous miRNA expression in HEK293, MCF10a, MCF7, PNT1a, and LNCaP cells
Total RNA (1 μg/20 μL reaction) from 4 independent experiments using different cell passages of the 5 cell lines were converted to cDNA using the miScript RT-PCR assay (QIAGEN) following the manufacturer's instructions. The miRNA miScript Primer assays (QIAGEN) were then used to quantify specific mature miRNAs present in each cell line tested. Specific mature miRNA primers were purchased from QIAGEN (Supplemental Table 2). The PCRs were performed in triplicate on the Roche LightCycler 480 under the following conditions: 15 minutes at 95°C, followed by 35 cycles of 15 seconds at 94°C, 30 seconds at 58°C, and 30 seconds at 70°C. U6B (small nuclear) RNA was used as an endogenous control to normalize between samples. No RT and water were used as negative controls for the PCR, whereas RNA from cells transfected with miRNA mimics was used as a positive control for undetected miRNAs. Data for the miRNA profile in HEK293 cells are presented as 2−ΔCt, where ΔCt = (Ct of target miRNA) minus (Ct U6B), whereas mature miRNA profiles in MCF7 and LNCaP cells were presented as 2−ΔΔCt, where ΔCt = (Ct of target miRNA) − (Ct U6B), and ΔΔCt = (ΔCt of target miRNA) in cancer cells − (ΔCt of target miRNA) in normal cells. MCF10a normal mammary gland/breast epithelial cells were used to normalize MCF7 cells, and PNT1a normal prostate epithelial cells were the control for LNCaP cells.
Immunoblotting
Forty-eight hours after transfection with miRNA mimics, the cells were washed with cold PBS and lysed in cold RIPA buffer containing 50mM Tris-HCl (pH 8), 150mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS as well as protease and phosphatase inhibitors (Roche). Protein concentration was measured by DC protein assays (Bio-Rad). Total proteins (25 μg) were separated on 8% SDS-PAGE gels at 100 V for 2.5 hours. The proteins were then electroblotted onto polyvinylidene difluoride membranes (Thermo Scientific) at 300 mA for 1 hour. Membranes were blocked in 1% Tween/Tris-buffered saline with 5% skim milk for 2 hours at room temperature or overnight at 4°C. For GHR detection, blocked membranes were immunoblotted with the primary antibody (B12 mouse anti-GHR; Santa Cruz Biotechnology) diluted 1:1000 in blocking buffer overnight at 4°C. After extensive washing with Tween/Tris-buffered saline, the membranes were incubated with horseradish peroxidase-labeled antimouse (1:5000) secondary antibodies (Cell Signaling Technology) diluted in blocking buffer for 1 hour at room temperature. Signals were detected using the ECL Prime Western blotting detection kit (GE Healthcare Amersham, division of Thermo Scientific) and exposure on HyBlot CL autoradiography films (Denville Scientific). The membranes were subsequently stripped using 0.1% SDS in Tris with 0.7% β-mercaptoethanol at 50°C and reprobed using a primary antibody against calnexin (1:5000; BD Biosciences) and the horseradish peroxidase-labeled antimouse secondary antibody (1:5000; Cell Signaling), as a control for protein loading. Two other control proteins were tested, β-tubulin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and gave similar results. Developed films were scanned, and a densitometric analysis of bands was carried out using the ImageJ program (National Institutes of Health) (49).
Statistical analysis
The results are expressed as mean ± SE from at least 3 independent experiments. All statistical analyses were performed initially with ANOVA; for subsequent group comparisons, the Tukey-Kramer multiple-comparisons test was used. Data were considered statistically significant when P < .05. All statistical tests were performed using GraphPad InSTAT software.
Results
Mapping the human GHR 3′-UTR sequence for potential miRNA binding sites
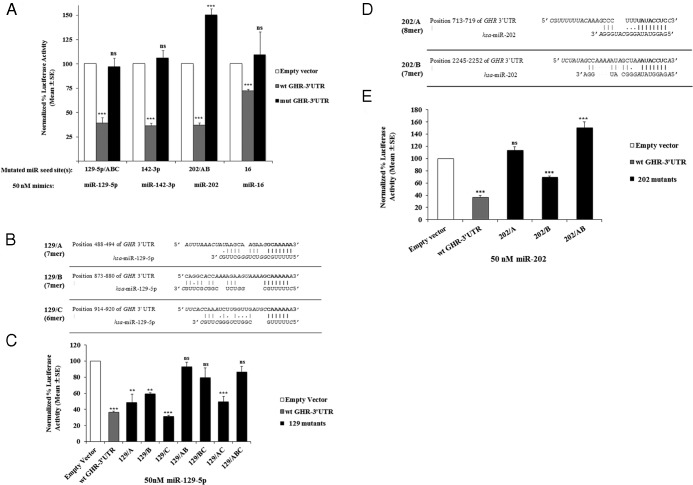
To identify putative miRNA target sites within the 3′-UTR of the human GHR gene, multiple miRNA target prediction programs were used: TargetScan version 5.2 and later version 6.2 (Lewis et al, www.targetscan.org) (48), PicTar (Krek et al, http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi) (50), miRDB (Wang et al, http://www.mirdb.org/miRDB/) (51, 52), Microcosm Targets (Rehmsmeier et al, http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) (53), and MiRanda (John et al, http://www.microrna.org/microrna/home.do) (54). The miRNA sites were prioritized based on the fact that they were identified by 2 or more in silico programs, the sites are conserved across several species, there are homologous sites in GH/IGF-1 axis-related genes, and there are reports linking specific miRNAs to GHR-related physiological or pathophysiological activities (Figure 1 and Supplemental Table 3). The final set included sites for miR-129–5p, which has 3 potential binding sites (denoted by A, B, and C at 488–494, 873–880, and 914–920, respectively, counting from the first nucleotide after the stop codon in exon 10); miR-142–3p (located at 583–590); miR-202, a member of the Let7 family that is present only in mammals and has 2 sites (A and B at 713–719 and 2245–2252, respectively); the miR-15/16 family (at 2092–2099); and miR-135 (at 2294–2300, the most 3′ site).
Figure 1.

Putative miRNA binding sites in the human GHR 3′-UTR. Schematic representation of the 3′-UTR sequence from the human GHR mRNA indicating the potential miRNA binding sites within the sequence tested during the present study. A, B, and C denote multiple binding sites for the same miR.
Endogenous levels of miRNAs in HEK293 cells
The endogenous levels of these miRNAs were determined in HEK293 cells by quantitative PCR; the data are presented as fold of ΔCt values normalized to U6B RNA (Supplemental Figure 1). All of the miR-15/16 family members studied were expressed in HEK293 cells with miR-15b and -16 the highest; miR-129–5p was at the limit of detection, whereas miR-142–3p, -202 and -135 were not detectable. Cells transfected with specific mimics were used as positive controls to validate the assays for the low and undetectable miRNAs.
In vitro testing for miRNA regulation of the human GHR 3′-UTR
To determine whether any of the prioritized miRNA sites were able to regulate the GHR 3′-UTR, we created a pmiR-Luc-GHR 3′-UTR reporter vector by cloning 2293 nucleotides of the possible 2454 nucleotides, spanning all of the putative miRNA sites (Figure 1). Reporter assays were performed using transient cotransfections of the reporter vector along with miRNA mimics in HEK293 cells. MiR-129–5p, -142–3p, -202 and, -16 showed significant inhibitory effects on the pmiR-Luc-GHR 3′-UTR vector, whereas other members of the miR-16 family (miR-15a, -103, -107, and -503) and Let7-b and -e as well as miR-135 had very low to no effects (Figure 2 and data not shown). Because it was possible that the lower effect of miR-16 was due to the high endogenous levels of miR-15/16 family members present in HEK293 cells (Supplemental Figure 1), we carried out reporter assays with 50nM miRNA inhibitors against 3 of the most highly expressed members in combination: miR-15a, -15b and –16. Indeed, results from 4 independent experiments inhibiting these 3 miRNAs showed a significant (59%, P < .05) upregulation of luciferase activity when normalized to the negative inhibitor control (data not shown).
Figure 2.

Effect of miRNAs on the GHR 3′-UTR luciferase reporter vector. Effects of 1nM to 100nM miR-129–5p, miR-142–3p, and miR-202 and 10nM to 100nM miR-16 on the pmiR-Luc-GHR 3′-UTR reporter vector were examined in HEK293 cells. Data are presented as mean ± SE (n = 5). All results were normalized to the empty Luc reporter vector and to negative mimic control treatment at each mimic dose followed by the calculation of luciferase to β-galactosidase ratios. Results for the first 3 miRNAs were highly significant by both ANOVA and Tukey's group comparison tests, whereas the miR-16 data were significant for ANOVA (P < .05) but not Tukey's group comparisons. This is likely due to the relatively high level of endogenous miR-16 in the HEK293 cells. *, P ≤ .05; ***, P ≤ .001.
Determining the specificity of the miRNA sites in the human GHR 3′-UTR
To confirm whether the miRNAs that had inhibitory effects in the reporter assays were acting through their predicted sites, reporter vectors with mutations of the binding sites were tested in parallel with the wild-type vector. Details of each mutation and the mutagenesis primers are in Supplemental Table 1. In the case of multiple binding sites for a miRNA (miR-129–5p and -202), simple and compound mutations were created.
MiR-142–3p and -16
Both of these miRNAs have a single predicted perfect seed site (8-mer) in the GHR 3′-UTR (Diana miRNA prediction tools [DIANA-microT-CDS web server version 5.0]). Mutating the corresponding potential binding sites resulted in full recovery of the luciferase activity, confirming that the observed repression in luciferase activity with the wild-type vector was due to effects of these miRNAs at their predicted sites (Figure 3A).
Figure 3.
Reporter assays of wild-type (wt) vs mutant (mut) human GHR 3′-UTR vectors. Panel A, Effects of 50nM miRNA mimics on wild-type vs mutant GHR 3′-UTR reporter vectors. Results are presented as mean ± SE (n = 4 for all but miR-202/AB n = 5). Panel B, Schematic representation of hybridization between miR-129–5p and its 3 putative GHR 3′-UTR binding sites (A, B and C); the seed sites are bold, noncomplementary nucleotides are italicized, lines represent Watson-Crick complementarity, and dots represent U to G wobble. Panel C, Reporter assays of wild-type GHR 3′-UTR and the predicted miR-129–5p single, double, and triple mutants. Data are presented as mean ± SE (n = 4). Panel D, Schematic representation of hybridization between miR-202 and its 2 putative binding sites in the GHR 3′-UTR (A and B); the seed sites are bold, noncomplementary nucleotides are italicized, lines represent Watson-Crick complementarity, and dots represent U to G wobble. Panel E, Normalized data from luciferase assays of single and double mutants of miR-202 binding sites compared with the wild-type vector. Results are presented as mean ± SE (n = 5). **, P ≤ .01; ***, P ≤ .001; ns, not significant.
MiR-129–5p
This miRNA has 3 putative binding sites in the 3′-UTR of the GHR gene (Figure 1). To test the contribution of each site to the total effect observed by the reporter assay, single mutants (miR-129–5p/A, -129–5p/B, and -129–5p/C), double mutants (miR-129–5p/AB, -129–5p/AC, and -129–5p/BC). and the triple mutant (miR-129–5p/ABC) were created and assayed with miR-129–5p mimics along with the wild-type vector. These assays showed that both sites A and B are the major contributors to the repression observed in the wild type, although site C has a minor contribution. However, it took the triple mutant to fully relieve the effects of miR-129–5p repression (Figure 3, A–C). Interestingly, the strength of the individual sites correlates with predicted seed weighting; sites A and B are 7-mers, whereas site C is a 6-mer (Figure 3B).
MiR-202
MiR-202 belongs to the Let-7 family of miRNAs. It has two predicted binding sites in the GHR 3′-UTR denoted by A and B (Figure 1). Single miR-202/A and miR-202/B mutants in addition to the double mutant (miR-202/AB) were created and cotransfected into HEK293 cells along with miR-202 mimics. Mutating site A abolished the repression completely, whereas mutating site B relieved the repression by only ∼37% (Figure 3D). Interestingly, mutating the 2 sites simultaneously (miR-202/AB) resulted in an ∼50% increase in expression above controls (Figures 3, A, D, and E). These results suggest that site A is sufficient and necessary for the inhibition of GHR expression by miR-202, whereas site B acts cooperatively with site A to enhance its inhibitory effects on the 3′-UTR. Again the predicted seed weighting for these 2 sites (A is an 8-mer and B is a 7-mer) correlates with the effectiveness of each site (Figure 3D).
The effect of individual miRNAs on endogenous GHR mRNA and protein levels in HEK293 cells
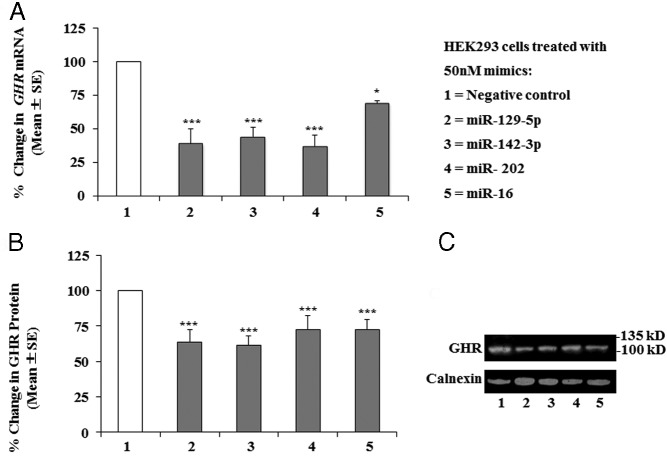
To test whether miR-129–5p, -142–3p, -202, and -16 affect the endogenous levels of GHR mRNA in HEK293 cells, we carried out quantitative RT-PCR assays after transfections with miRNA mimics vs negative control mimics. Similar to the results from the reporter assays, the aforementioned miRNAs significantly downregulated the GHR mRNA levels compared with controls: by ∼60% with miR-129–5p, -142–3p, and -202 and ∼30% with miR-16 (Figure 4A).
Figure 4.
Effect of miRNAs on endogenous GHR mRNA and protein in HEK293 cells. A, Expression of human total GHR mRNA in HEK293 cells transfected with 50nM miR-129–5p, miR-142–3p, miR-202, and miR-16 mimics. The data are presented as mean ± SE of 3 individual experiments for all but miR-142–3p (n = 5); duplicates were assayed within each experiment. Statistically significant differences between the negative control and miR-treated cells were found for miR-129–5p, miR-142–3p, miR-202 (***, P ≤ .001) and miR-16 (*, P ≤ .05). B, Composite results for n = 4 individual experiments (mean ± SE) showing statistically significant differences between negative control and miR-treated cells: ***, P ≤ .001 for all treatments. C, Representative Western blot of GHR and calnexin protein levels in HEK293 cells after transfections with 50nM miR-129–5p, miR-142–3p, miR-202, and miR-16 mimics.
Because the ultimate functional effect of miRNAs on gene expression is the alteration of protein levels, we measured the effect of the same miRNAs on the endogenous GHR protein levels in HEK293 cells by Western blotting. An antibody against the intracellular domain of GHR was used that recognizes a band of ∼100 kDa, corresponding to the mature GHR. Densitometry results from the Western blots were normalized to calnexin and to negative control-treated cells (representative blot in Figure 4B). Composite results of 4 independent experiments showed statistically significant decreases in GHR protein levels of ∼ 40% when treated with miR-129–5p or miR-142–3p and ∼30% when treated with either miR-202 or miR-16 (Figure 4C).
The effect of individual miRNAs on endogenous GHR mRNA and protein levels in MCF7 and LNCaP cells
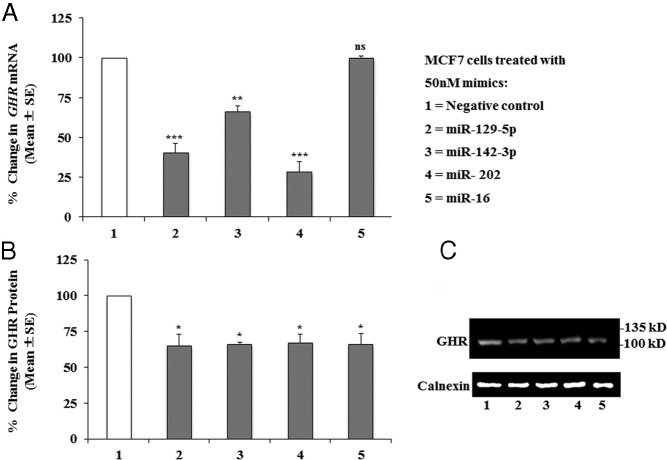
To determine whether the effect of miR-129–5p, -142–3p, -202, and -16 on GHR expression observed in HEK293 cells extends to other cell lines, we tested the effect of these 4 miRNAs on 2 other cell lines: MCF7, a mammary gland breast cancer cell line, and LNCaP, a human prostatic carcinoma cell line. Transfecting MCF7 cells with 50nM mimics of each miRNA resulted in decreases of 30% to 70% in GHR mRNA and ∼35% decreases in GHR protein in response to miR-129–5p, miR-142–3p, and miR-202 (Figure 5, A–C). Surprisingly, miR-16 significantly decreased the levels of GHR protein but not mRNA. It is possible that a degradation at the mRNA level occurred at an earlier time point, before the 24-hour collection, or that, in MCF7 cells, miR-16 acts primarily to inhibit GHR mRNA translation.
Figure 5.
Effect of miRNAs on endogenous GHR mRNA and protein expression in MCF7 cells. A, Expression of human total GHR mRNA in MCF7 cells transfected with 50nM of each mimic. The data are presented as mean ± SE of 4 individual experiments for all but miR-202 (n = 5); duplicates were assayed within each experiment. Statistically significant differences between the control and miR-treated cells were found for miR-129–5p, miR-202 (***, P ≤ .001), and miR-142–3p (**, P ≤ .01), whereas miR-16 had no effect (not significant [ns]). B, GHR protein levels were determined by Western blot and normalized to calnexin. Normalized values are shown relative to the negative mimics control treatments (100%). Composite results for n = 4 individual experiments (mean ± SE) showing statistically significant differences between negative control and miR-treated cells: *, P ≤ .05 for all treatments. C, Representative Western blot of GHR and calnexin protein levels in MCF7 cells after transfections with 50nM miR-129–5p, miR-142–3p, miR-202, and miR-16 mimics.
Transfecting LNCaP cells with 50nM of each miRNA led to significant 43% to 60% decreases in GHR mRNA levels in response to miR129–5p, miR-202, and miR-16, whereas miR-142–3p had no effect (Figure 6A); the GHR protein levels were significantly decreased by 35%–74% with all 4 miRNAs (Figures 6B and 6C). Thus, in LNCaP cells, miR-142–3p may have an earlier effect on GHR mRNA degradation or primarily targets GHR mRNA translation. Interestingly, in the LNCaP cells, the GHR antibody used for studies of all of the cell lines detected not only the major isoform (∼100 kDa) band but also a slightly higher ∼120-kDa molecular mass band. This second band may correspond to a more fully glycosylated form of GHR (Figure 6C and Supplemental Figure 2C).
Figure 6.
Effect of miRNAs on endogenous GHR mRNA and protein expression in LNCaP cells. A, Expression of human total GHR mRNA in LNCaP cells transfected with 50nM of each mimic. The data are presented as mean ± SE of 3 individual experiments for all but miR-129–5p (n = 5) and miR-202 (n = 4); duplicates were assayed within each experiment. Statistically significant differences between the control and miR-treated cells were found for miR-129–5p, miR-202, and miR-16 (***, P ≤ .001) but not for miR-142–3p (not significant [ns]). B, GHR protein levels were determined by Western blot and normalized to calnexin. Normalized values are shown relative to the negative mimics control treatments (100%). Composite results for n = 4 individual experiments (mean ± SE) showing statistically significant differences between negative control and miR-treated cells for miR-129–5p, miR-202 (***, P ≤ .001), miR-142–3p, and miR-16 (**, P ≤ .01) for all treatments. C, Representative Western blot of GHR and calnexin protein levels in LNCaP cells after transfections with 50nM miR-129–5p, miR-142–3p, miR-202, and miR-16 mimics.
Differential expression of endogenous levels of miR-129–5p, miR-142–3p, miR-202, and miR-16 and GHR protein in MCF10a, MCF7, PNT1a, and LNCaP cells
To further investigate whether the 4 miRNAs might be involved in regulating GHR expression in MCF7 and LNCaP cancer cells, we first compared the endogenous levels of these miRNAs in the cancer cells vs their corresponding normal control cells, MCF10a (normal breast epithelium) and PNT1a (normal prostate epithelium). Indeed, the miRNA profile showed significantly decreased levels of miR-129–5p, -202, and -16 in both MCF7 and LNCaP cells compared with their controls, whereas miR-142–3p was not detected in any of the cell lines tested (Supplemental Figure 2A). These results are in accordance with the miRNA expression database (www.microRNA.org) (55) for MCF7 and MCF10a; there were no comparable miRNA profiles for either the LNCaP or PNT1a cells.
We next measured GHR protein by Western blots: GHR was increased 4.7-fold in MCF7 cells when normalized to MCF10a and 3.4-fold in LNCaP cells compared with PNT1a (Supplemental Figure 2, B and C). Thus, our results demonstrate an inverse correlation between endogenous miR-129–5p, -202, and -16 and GHR expression in the 2 cancer cell lines relative to their normal counterparts.
Discussion
In this study, we identified a new mechanism by which human GHR expression is regulated in both normal and cancer cells: miRNAs. Our results show miR-129–5p, miR-142–3p, miR-202, and miR-16 to be significant inhibitors, acting through specific GHR 3′-UTR sites. In general, there was a parallel decrease in GHR mRNA and protein by these 4 miRNAs in all 3 cell lines examined, suggesting that the primary mechanism by which they are acting is by degrading the GHR mRNA.
Initially, to prioritize a list of miRNAs that might regulate human GHR expression, we searched the literature for prior evidence of strong direct or inverse correlations between the expression of GHR and that of individual miRNAs in both physiological and pathophysiological contexts (Supplemental Table 3). Although we were able to find a few links in relation to cell proliferation, apoptosis, angiogenesis, and diurnal rhythms, most miRNA papers have used high-throughput microarrays to identify cancer-specific miRNA fingerprints in a spectrum of different cancers, including breast (56), prostate (57, 58), hepatocellular (59, 60), lung (61–63), colon (64, 65), and gastric carcinomas (66). Results from these studies and others have shown that the miRNA expression profile in malignant cells is significantly different from that in healthy control cells, suggesting that alterations in miRNA genes play a critical role in the pathophysiology of many human cancers (67). Thus, much of the discussion will focus on the potential relevance of our findings to GHR in cancer.
MiR-129–5p
Our results demonstrated a potent repressive effect of miR-129–5p on GHR mRNA and protein expression in HEK293, MCF7, and LNCaP cells. Even more significant was our finding of an inverse correlation between mature miR-129–5p and GHR levels in both MCF7 and LNCaP cells compared with their respective normal control cells (MCF10a and LNCaP). Further experiments are needed to study the functional relevance of this miRNA in primary breast and prostate tumors and whether a decrease in GHR by enhanced expression of this miRNA might be of therapeutic benefit.
Bandrés et al (68) has identified miR-129–5p (a known inducer of G1 phase arrest) (69) as one of the miRNAs significantly downregulated in 15 colorectal cancer cell lines compared with a human normal colon cell line. In a more recent study, Karaayvaz et al (70) found that the level of miR-129–5p expression was significantly decreased in samples from 22 human colorectal tumor tissues compared with their paired normal controls. Moreover, the same group studied a set of 61 colorectal specimens from different stages of the disease and found that the expression of miR-129–5p was significantly reduced in patients with stage 3 and 4 of the disease compared with normal or adenoma tissues (70). Although Karaayvaz et al (70) did not examine GHR levels in these tumors, multiple studies have reported the overexpression of GHR in human colorectal cancer tissues and cell lines, suggesting a role for GHR in the progression of colorectal carcinomas (43, 71, 72). Our data suggest that the inverse relationship between miR-129–5p and GHR in colorectal cancers should be studied further to determine whether, in fact, overexpression of this miRNA in colorectal cells would downregulate GHR expression and halt or reverse progression of these tumors. Interestingly, in addition to GHR, miR-129–5p has putative binding sites on the STAT5b and IGF-1 mRNA 3′-UTRs, suggesting that it could affect multiple steps in the GHR signaling pathway in these tumors as well as normal cells (Supplemental Table 3).
MiR-142–3p
In the present study, we observed significant inhibitory effects of miR-142–3p on GHR expression in normal as well as cancer cell lines. Although miR-142–3p expression was not detected in any of these cell lines under our experimental conditions, we did observe that it was induced in HEK293 cells in response to serum starvation (data not shown), suggesting a role for miR-142–3p in regulation of metabolic systems and/or the cell cycle.
In a recent study by Shende et al (73), miR-142–3p was shown to be downregulated in colon cancer tissues; in addition, its overexpression acted as a tumor suppressor, inhibiting the growth of colorectal cancer cell lines. As mentioned earlier, GHR has been strongly implicated in the progression of colorectal cancers. Our finding of a robust inhibitory effect of not only miR-129–5p but also miR-142–3p on GHR expression indicates that the interactions between both of these miRNAs and GHR in colorectal tumors now need to be verified and explored experimentally.
MiR-142–3p has also been shown to be involved in regulation of the circadian clock by targeting mRNAs of the clock gene BMAL1. The expression level of miR-142–3p oscillates in a circadian rhythm in mouse NIH3T3 cells (74) and the suprachiasmatic nuclei (73). In addition, expression of the MIR-142 gene is controlled by CLOCK/BMAL1 heterodimers, suggesting a negative feedback loop (73). Studies by Itoh et al (75) and Zvonic et al (76) have reported that GHR mRNA levels in murine liver, skeletal muscle, and calvarial bone are expressed in a diurnal manner. Although we have data demonstrating that the circadian regulators D-site binding protein (DBP) and E4 binding protein 4 (E4BP4) can regulate GHR expression through sites in at least two of the human GHR 5′-UTR promoters (Kenth and Goodyer, unpublished), our present data suggest that it would be interesting to investigate whether miR-142–3p also plays a role in controlling the normal physiological circadian expression of GHR.
MiR-202
Similar to our findings for miR-129–5p, we observed a strong inhibitory effect of miR-202 on GHR expression in HEK293, MCF7, and LNCaP cells. In addition, there was an inverse correlation between miR-202 and GHR levels in both MCF7 and LNCaP cells compared with their respective controls. These results provide support for further investigations of the role that this potential tumor suppressor might play in in breast and prostate cancers.
Expression of miR-202 (a known repressor of MYCN) (77) has also been reported to be downregulated in gastric cancer, both in cell lines and tumor tissues (78). Moreover, overexpression of miR-202 in 2 gastric cancer cell lines, MNK-28 and BGC-823, markedly suppressed cell proliferation and induced cell apoptosis both in vitro and in vivo (78). As with colorectal tumors, multiple studies have reported GHR overexpression in gastric carcinomas; experimental results indicate that GHR expression is significantly higher in primary gastric adenocarcinoma than in normal gastric mucosa and is significantly correlated with tumor differentiation and tumor grade (44, 79–81). These data as well as our present findings strongly suggest that further investigations of miR-202/GHR gene interactions in primary gastric cancers are in order. As observed for miR-129–5p, miR-202 may also target several members of the GH/GHR axis (GHRHR, GHR, IGF-1, IGF-IR, and CIS) and, thus, could affect the axis at multiple levels (Supplemental Table 3).
MiR-16
MiR-16 belongs to the miR-15/107 group of miRNA genes. MiR-15a and -16 were the first miRNAs linked to a cancer; deletion of their genes at 13q14.3 has been reported in more than 50% of chronic lymphocytic leukemia cases, suggesting that they function as tumor suppressors (82). GHR is widely expressed in human lymphocytes and many leukemic cell lines (83–85), and when Calin et al (86) carried out a microarray analysis of the genes modulated by overexpressing miR-15a/16–1 in a leukemic cell line, they found a significant downregulation of the GHR gene, providing the first suggestion that miRNAs might regulate GHR expression. Decreased expression of miR-15a/16 was subsequently reported in multiple adenomas as well as breast and prostate cancers (87–93).
Data from our present study are in line with these reports: we demonstrated a decrease in mature miR-16 expression in both the MCF7 breast cancer and the LNCaP prostate cancer cells compared with their corresponding controls. In addition, we observed a significant inhibitory effect of miR-16 on GHR expression in HEK293, MCF7, and LNCaP cells. Together these data suggest that a parallel aberrant low expression of miR-16 and high expression of GHR are likely involved in progression of breast and prostate cancers and certain leukemias.
In addition to miR-129–5p and miR-202, miR-16 may also target several genes in the GH/GHR axis: GHR, IGF-1, IGF-IR, and IGF-2R (Supplemental Table 3). Interestingly, the promoter for the MIR-15b/16–2 genes has been shown to be negatively regulated by Signal Transducer and Activator 5 (STAT5), a downstream effector of GH-activated GHR, resulting in decreased mature miR-15 and miR-16 expression levels (94). These findings suggest that there may be a coordinated regulation allowing for communication between the level of GH/GHR axis activity and miR15/16 expression via STAT5.
Although our results suggest that these 4 miRNAs may be important regulators of GHR expression, this does not exclude a role for other miRNAs. The limitation in miRNA prediction of true targets is due to the complex nature of the miRNA-mRNA interactions; as a result, the available prediction programs have many false-positive results. Indeed, we found that other predicted miRNAs, such as miR-135, members of the Let7 family, and other members of the miR-16 family (103, 107, and 503), did not have any effect on the GHR 3′-UTR luciferase activity. Thus, other determinants (eg, flanking regions) likely contribute to the effectiveness of the predicted miRNAs. Developmental stages constitute another layer of miRNA gene regulation complexity whereby these 4 or other miRNAs could play a temporal role in the regulation of GHR expression. Finally, according to a new miRNA prediction tool (DIANA-microT-CDS web server version 5.0; http:www.microrna.gr/microT-CDS) (95, 96), there are additional miRNA binding sites for the miR-16 and Let7 families, including miR-202, in the coding region sequence of exon 10 of the human GHR mRNA, which might play a role. Although miRNAs have been shown to have effects at sites outside the 3′-UTR region of mRNAs (96), the exon 10 sites may be more important in regulating expression of the truncated forms of GHR: these represent up to 10% of total GHR and occur due to alternative splicing in exon 9, converting the entire exon 10 to 3′-UTR status (13).
In summary, our experiments have demonstrated, for the first time, significant inhibitory effects of miRNAs on human GHR mRNA and protein levels in both normal and cancer cell lines. In addition, the expression of these 4 miRNAs is decreased in cancer cell lines compared with normal cells and is inversely correlated with GHR. Thus, this study paves the way for the development of miRNA inhibitors as therapeutic agents in GH/GHR-related pathophysiologies, including cancers.
Acknowledgments
We thank Dr Aimee Ryan for her critical review of the manuscript.
This study was supported by operating funds from the Canadian Institutes of Health Research (to C.G.G.) and by a studentship from the Division of Endocrinology and Metabolism at McGill University (to S.E.). C.G.G. is a member of the Research Institute of McGill University Health Centre, which is supported in part by Le Fonds de Recherche du Québec–Santé (FRQS).
Disclosure Summary: The authors have nothing to disclose.
Funding Statement
This study was supported by operating funds from the Canadian Institutes of Health Research (to C.G.G.) and by a studentship from the Division of Endocrinology and Metabolism at McGill University (to S.E.). C.G.G. is a member of the Research Institute of McGill University Health Centre, which is supported in part by Le Fonds de Recherche du Québec–Santé (FRQS).
Footnotes
- Ct
- cycle threshold
- FBS
- fetal bovine serum
- GHR
- GH receptor
- miRNA
- microRNA
- UTR
- untranslated region.
References
- 1. Veldhuis JD, Roemmich JN, Richmond EJ, et al. . Endocrine control of body composition in infancy, childhood, and puberty. Endocr Rev. 2005;26(1):114–146. [DOI] [PubMed] [Google Scholar]
- 2. Lichanska AM, Waters MJ. New insights into growth hormone receptor function and clinical implications. Horm Res. 2008;69(3):138–145. [DOI] [PubMed] [Google Scholar]
- 3. Lichanska AM, Waters MJ. How growth hormone controls growth, obesity and sexual dimorphism. Trends Genet. 2008;24(1):41–47. [DOI] [PubMed] [Google Scholar]
- 4. Brooks AJ, Waters MJ. The growth hormone receptor: mechanism of activation and clinical implications. Nat Rev Endocrinol. 2010;6(9):515–525. [DOI] [PubMed] [Google Scholar]
- 5. Savage MO, Attie KM, David A, Metherell LA, Clark AJ, Camacho-Hubner C. Endocrine assessment, molecular characterization and treatment of growth hormone insensitivity disorders. Nat Clin Pract Endocrinol Metab. 2006;2(7):395–407. [DOI] [PubMed] [Google Scholar]
- 6. Perry JK, Liu DX, Wu ZS, Zhu T, Lobie PE. Growth hormone and cancer: an update on progress. Curr Opin Endocrinol Diabetes Obes. 2013;20(4):307–313. [DOI] [PubMed] [Google Scholar]
- 7. Barton DE, Foellmer BE, Wood WI, Francke U. Chromosome mapping of the growth hormone receptor gene in man and mouse. Cytogenet Cell Genet. 1989;50(2–3):137–141. [DOI] [PubMed] [Google Scholar]
- 8. Godowski PJ, Leung DW, Meacham LR, et al. . Characterization of the human growth hormone receptor gene and demonstration of a partial gene deletion in two patients with Laron-type dwarfism. Proc Natl Acad Sci U S A. 1989;86(20):8083–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodyer CG, Zogopoulos G, Schwartzbauer G, Zheng H, Hendy GN, Menon RK. Organization and evolution of the human growth hormone receptor gene 5′-flanking region. Endocrinology. 2001;142(5):1923–1934. [DOI] [PubMed] [Google Scholar]
- 10. Orlovskii IV, Sverdlova PS, Rubtsov PM. Fine structure, expression and polymorphism of the human growth hormone receptor gene [in Russian]. Mol Biol (Mosk). 2004;38(1):29–39. [PubMed] [Google Scholar]
- 11. Pekhletsky RI, Chernov BK, Rubtsov PM. Variants of the 5′-untranslated sequence of human growth hormone receptor mRNA. Mol Cell Endocrinol. 1992;90(1):103–109. [DOI] [PubMed] [Google Scholar]
- 12. Wei Y, Rhani Z, Goodyer CG. Characterization of growth hormone receptor messenger ribonucleic acid variants in human adipocytes. J Clin Endocrinol Metab. 2006;91(5):1901–1908. [DOI] [PubMed] [Google Scholar]
- 13. Ross RJ, Esposito N, Shen XY, et al. . A short isoform of the human growth hormone receptor functions as a dominant negative inhibitor of the full-length receptor and generates large amounts of binding protein. Mol Endocrinol. 1997;11(3):265–273. [DOI] [PubMed] [Google Scholar]
- 14. Edens A, Talamantes F. Alternative processing of growth hormone receptor transcripts. Endocr Rev. 1998;19(5):559–582. [DOI] [PubMed] [Google Scholar]
- 15. Goodyer CG, Rhani Z, Zheng H. Expression of the hepatic specific V1 messenger ribonucleic acid of the human growth hormone receptor gene is regulated by hepatic nuclear factor (HNF)-4alpha2 and HNF-4alpha8. Mol Endocrinol. 2008;22(2):485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kenth G, Puzhko S, Goodyer CG. Human growth hormone receptor gene expression is regulated by Gfi-1/1b and GAGA cis-elements. Mol Cell Endocrinol. 2011;335(2):135–147. [DOI] [PubMed] [Google Scholar]
- 17. Erman A, Wabitsch M, Goodyer CG. Human growth hormone receptor (GHR) expression in obesity: II. Regulation of the human GHR gene by obesity-related factors. Int J Obes (Lond). 2011;35(12):1520–1529. [DOI] [PubMed] [Google Scholar]
- 18. Wei Y, Puzhko S, Wabitsch M, Goodyer CG. Transcriptional regulation of the human growth hormone receptor (hGHR) gene V2 promoter by transcriptional activators and repressor. Mol Endocrinol. 2009;23(3):373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu JH, Schwartzbauer G, Kazlman A, Menon RK. Role of the Sp family of transcription factors in the ontogeny of growth hormone receptor gene expression. J Biol Chem. 1999;274(48):34327–34336. [DOI] [PubMed] [Google Scholar]
- 20. Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19(9):465–474. [DOI] [PubMed] [Google Scholar]
- 21. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 22. Xie X, Lu J, Kulbokas EJ, et al. . Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. [DOI] [PubMed] [Google Scholar]
- 24. Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21(3):452–460. [DOI] [PubMed] [Google Scholar]
- 25. Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. [DOI] [PubMed] [Google Scholar]
- 26. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- 27. Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38(3):323–332. [DOI] [PubMed] [Google Scholar]
- 28. Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6(9):e24248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karp X, Ambros V. Developmental biology. Encountering microRNAs in cell fate signaling. Science. 2005;310(5752):1288–1289. [DOI] [PubMed] [Google Scholar]
- 31. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. [DOI] [PubMed] [Google Scholar]
- 32. Poy MN, Eliasson L, Krutzfeldt J, et al. . A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. [DOI] [PubMed] [Google Scholar]
- 33. Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. 2013;51(4):759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagano M, Chastre E, Choquet A, Bara J, Gespach C, Kelly PA. Expression of prolactin and growth hormone receptor genes and their isoforms in the gastrointestinal tract. Am J Physiol. 1995;268(3 Pt 1):G431–G442. [DOI] [PubMed] [Google Scholar]
- 35. Mertani HC, Garcia-Caballero T, Lambert A, et al. . Cellular expression of growth hormone and prolactin receptors in human breast disorders. Int J Cancer. 1998;79(2):202–211. [DOI] [PubMed] [Google Scholar]
- 36. Gebre-Medhin M, Kindblom LG, Wennbo H, Törnell J, Meis-Kindblom JM. Growth hormone receptor is expressed in human breast cancer. Am J Pathol. 2001;158(4):1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chopin LK, Veveris-Lowe TL, Philipps AF, Herington AC. Co-expression of GH and GHR isoforms in prostate cancer cell lines. Growth Horm IGF Res. 2002;12(2):126–136. [DOI] [PubMed] [Google Scholar]
- 38. Kölle S, Sinowatz F, Boie G, Temmim-Baker L, Lincoln D. Expression of growth hormone receptor in human prostatic carcinoma and hyperplasia. Int J Oncol. 1999;14(5):911–916. [DOI] [PubMed] [Google Scholar]
- 39. Weiss-Messer E, Merom O, Adi A, et al. . Growth hormone (GH) receptors in prostate cancer: gene expression in human tissues and cell lines and characterization, GH signaling and androgen receptor regulation in LNCaP cells. Mol Cell Endocrinol. 2004;220(1–2):109–123. [DOI] [PubMed] [Google Scholar]
- 40. Sobrier ML, Duquesnoy P, Duriez B, Amselem S, Goossens M. Expression and binding properties of two isoforms of the human growth hormone receptor. FEBS Lett. 1993;319(1–2):16–20. [DOI] [PubMed] [Google Scholar]
- 41. Ballesteros M, Leung KC, Ross RJ, Iismaa TP, Ho KK. Distribution and abundance of messenger ribonucleic acid for growth hormone receptor isoforms in human tissues. J Clin Endocrinol Metab. 2000;85(8):2865–2871. [DOI] [PubMed] [Google Scholar]
- 42. Wu X, Liu F, Yao X, Li W, Chen C. Growth hormone receptor expression is up-regulated during tumorigenesis of human colorectal cancer. J Surg Res. 2007;143(2):294–299. [DOI] [PubMed] [Google Scholar]
- 43. Yang X, Liu F, Xu Z, et al. . Growth hormone receptor expression in human colorectal cancer. Dig Dis Sci. 2004;49(9):1493–1498. [DOI] [PubMed] [Google Scholar]
- 44. Yang X, Huang P, Wang F, Xu Z, Wang X. Growth hormone receptor expression in human primary gastric adenocarcinoma. J Biomed Res. 2012;26(5):307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. García-Caballero T, Mertani HM, Lambert A, et al. . Increased expression of growth hormone and prolactin receptors in hepatocellular carcinomas. Endocrine. 2000;12(3):265–271. [DOI] [PubMed] [Google Scholar]
- 46. Liu JP, Wang HT, Ou QJ, et al. . Expression of growth hormone receptor in hepatocellular carcinoma and its significance [in Chinese]. Ai Zheng. 2003;22(3):298–301. [PubMed] [Google Scholar]
- 47. Zogopoulos G, Albrecht S, Pietsch T, et al. . Fetal- and tumor-specific regulation of growth hormone receptor messenger RNA expression in human liver. Cancer Res. 1996;56(13):2949–2953. [PubMed] [Google Scholar]
- 48. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. [DOI] [PubMed] [Google Scholar]
- 49. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krek A, Grün D, Poy MN, et al. . Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. [DOI] [PubMed] [Google Scholar]
- 51. Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24(3):325–332. [DOI] [PubMed] [Google Scholar]
- 53. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Iorio MV, Ferracin M, Liu CG, et al. . MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. [DOI] [PubMed] [Google Scholar]
- 57. Song H, Liu Y, Pan J, Zhao ST. Expression profile analysis reveals putative prostate cancer-related microRNAs. Genet Mol Res. 2013;12(4):4934–4943. [DOI] [PubMed] [Google Scholar]
- 58. Alshalalfa M, Bader GD, Bismar TA, Alhajj R. Coordinate microRNA-mediated regulation of protein complexes in prostate cancer. PLoS One. 2013;8(12):e84261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Y, Lee AT, Ma JZ, et al. . Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283(19):13205–13215. [DOI] [PubMed] [Google Scholar]
- 60. Murakami Y, Yasuda T, Saigo K, et al. . Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. [DOI] [PubMed] [Google Scholar]
- 61. Hayashita Y, Osada H, Tatematsu Y, et al. . A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. [DOI] [PubMed] [Google Scholar]
- 62. Yanaihara N, Caplen N, Bowman E, et al. . Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. [DOI] [PubMed] [Google Scholar]
- 63. Takamizawa J, Konishi H, Yanagisawa K, et al. . Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. [DOI] [PubMed] [Google Scholar]
- 64. Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 65. Schetter AJ, Leung SY, Sohn JJ, et al. . MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Petrocca F, Visone R, Onelli MR, et al. . E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–286. [DOI] [PubMed] [Google Scholar]
- 67. Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16–1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–220. [DOI] [PubMed] [Google Scholar]
- 68. Bandrés E, Cubedo E, Agirre X, et al. . Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu J, Qian J, Li C, et al. . miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9(9):1809–1818. [DOI] [PubMed] [Google Scholar]
- 70. Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4:e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang XD, Liu FK, Xu Z, Li JS. [Growth hormone receptor expression in human colorectal cancer and its implication]. Zhonghua Wei Chang Wai Ke Za Zhi. 2005;8(3):252–254. [PubMed] [Google Scholar]
- 72. Dagnaes-Hansen F, Duan H, Rasmussen LM, Friend KE, Flyvbjerg A. Growth hormone receptor antagonist administration inhibits growth of human colorectal carcinoma in nude mice. Anticancer Res. 2004;24(6):3735–3742. [PubMed] [Google Scholar]
- 73. Shende VR, Neuendorff N, Earnest DJ. Role of miR-142–3p in the post-transcriptional regulation of the clock gene Bmal1 in the mouse SCN. PLoS One. 2013;8(6):e65300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tan X, Zhang P, Zhou L, Yin B, Pan H, Peng X. Clock-controlled mir-142–3p can target its activator, Bmal1. BMC Mol Biol. 2012;13:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Itoh E, Iida K, del Rincon JP, Kim DS, Thorner MO. Diurnal variation in growth hormone receptor messenger ribonucleic acid in liver and skeletal muscle of lit/+ and lit/lit mice. Endocr J. 2004;51(6):529–535. [DOI] [PubMed] [Google Scholar]
- 76. Zvonic S, Ptitsyn AA, Kilroy G, et al. . Circadian oscillation of gene expression in murine calvarial bone. J Bone Miner Res. 2007;22(3):357–365. [DOI] [PubMed] [Google Scholar]
- 77. Buechner J, Tømte E, Haug BH, et al. . Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105(2):296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao Y, Li C, Wang M, et al. . Decrease of miR-202–3p expression, a novel tumor suppressor, in gastric cancer. PLoS One. 2013;8(7):e69756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lincoln DT, Singal PK, Al-Banaw A. Growth hormone in vascular pathology: neovascularization and expression of receptors is associated with cellular proliferation. Anticancer Res. 2007;27(6B):4201–4218. [PubMed] [Google Scholar]
- 80. Lin Y, Li S, Cao P, Cheng L, Quan M, Jiang S. The effects of recombinant human GH on promoting tumor growth depend on the expression of GH receptor in vivo. J Endocrinol. 2011;211(3):249–256. [DOI] [PubMed] [Google Scholar]
- 81. Ran G, Lin Y, Cao P, Cai XT, Li SY. Effect of rhGH on JAK2-STAT3 signal pathway after GHR was down-regulated by siRNA in gastric cancer cell [in Chinese]. Yao Xue Xue Bao. 2013;48(3):435–440. [PubMed] [Google Scholar]
- 82. Calin GA, Sevignani C, Dumitru CD, et al. . Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dinerstein H, Lago F, Goujon L, et al. . The proline-rich region of the GH receptor is essential for JAK2 phosphorylation, activation of cell proliferation, and gene transcription. Mol Endocrinol. 1995;9(12):1701–1707. [DOI] [PubMed] [Google Scholar]
- 84. Baixeras E, Jeay S, Kelly PA, Postel-Vinay MC. The proliferative and antiapoptotic actions of growth hormone and insulin-like growth factor-1 are mediated through distinct signaling pathways in the Pro-B Ba/F3 cell line. Endocrinology. 2001;142(7):2968–2977. [DOI] [PubMed] [Google Scholar]
- 85. Hattori N, Saito T, Yagyu T, Jiang BH, Kitagawa K, Inagaki C. GH, GH receptor, GH secretagogue receptor, and ghrelin expression in human T cells, B cells, and neutrophils. J Clin Endocrinol Metab. 2001;86(9):4284–4291. [DOI] [PubMed] [Google Scholar]
- 86. Calin GA, Cimmino A, Fabbri M, et al. . MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105(13):5166–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rivas MA, Venturutti L, Huang YW, Schillaci R, Huang TH, Elizalde PV. Downregulation of the tumor-suppressor miR-16 via progestin-mediated oncogenic signaling contributes to breast cancer development. Breast Cancer Res. 2012;14(3):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dong JT, Boyd JC, Frierson HF Jr. Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate. 2001;49(3):166–171. [DOI] [PubMed] [Google Scholar]
- 89. Bhattacharya R, Nicoloso M, Arvizo R, et al. . MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69(23):9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Klein U, Lia M, Crespo M, et al. . The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. [DOI] [PubMed] [Google Scholar]
- 91. Lerner M, Harada M, Lovén J, et al. . DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16–1. Exp Cell Res. 2009;315(17):2941–2952. [DOI] [PubMed] [Google Scholar]
- 92. Bonci D, Coppola V, Musumeci M, et al. . The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. [DOI] [PubMed] [Google Scholar]
- 93. Brookman-Amissah N, Nariculam J, Freeman A, et al. . Allelic imbalance at 13q14.2 approximately q14.3 in localized prostate cancer is associated with early biochemical relapse. Cancer Genet Cytogenet. 2007;179(2):118–126. [DOI] [PubMed] [Google Scholar]
- 94. Li G, Miskimen KL, Wang Z, et al. . STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood. 2010;115(7):1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. . DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41(Web Server issue):W169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28(6):771–776. [DOI] [PubMed] [Google Scholar]