Abstract
The concept that the gut microbiota serves as a virtual endocrine organ arises from a number of important observations. Evidence for a direct role arises from its metabolic capacity to produce and regulate multiple compounds that reach the circulation and act to influence the function of distal organs and systems. For example, metabolism of carbohydrates results in the production of short-chain fatty acids, such as butyrate and propionate, which provide an important source of nutrients as well as regulatory control of the host digestive system. This influence over host metabolism is also seen in the ability of the prebiotic inulin to influence production of relevant hormones such as glucagon-like peptide-1, peptide YY, ghrelin, and leptin. Moreover, the probiotic Lactobacillus rhamnosus PL60, which produces conjugated linoleic acid, has been shown to reduce body-weight gain and white adipose tissue without effects on food intake. Manipulating the microbial composition of the gastrointestinal tract modulates plasma concentrations of tryptophan, an essential amino acid and precursor to serotonin, a key neurotransmitter within both the enteric and central nervous systems. Indirectly and through as yet unknown mechanisms, the gut microbiota exerts control over the hypothalamic-pituitary-adrenal axis. This is clear from studies on animals raised in a germ-free environment, who show exaggerated responses to psychological stress, which normalizes after monocolonization by certain bacterial species including Bifidobacterium infantis. It is tempting to speculate that therapeutic targeting of the gut microbiota may be useful in treating stress-related disorders and metabolic diseases.
Microbiome science has prospered in recent years, and the exciting discoveries arising from this surge in interest have forced a reevaluation of our perception of the trillions of microorganisms which reside in the human gastrointestinal (GI) tract in particular. The gut microbiota performs a number of essential protective, structural, and metabolic functions for host health, including food processing, digestion of complex host-indigestible polysaccharides, pathogen displacement, and synthesis of vitamins (1–3). As well as a direct action on the gut mucosa and the enteric nervous system (ENS), the metabolic output of the gut microbiota gives it a reach well beyond the local GI compartment. Thus, considering the ability to influence the function of distal organs and systems, in many respects, the gut microbiota resembles an endocrine organ (4, 5).
Through this lens, the microbiota produces numerous chemicals of a hormonal nature that are released into the bloodstream and act at distal sites (see Table 1). The targets for these substances are not just the local ENS but many other organs including the brain. It releases its hormonal products into interstitial tissue to be picked up by blood and lymph capillaries, and these secretions are usually effective in low concentrations on target organs or tissues remote from the enteric microbiota. As well as providing an important direct source of humoral agents, both active compounds and precursors, which act at distal sites (see Figure 1), the microbiota also plays an indirect role in regulating complex endocrine networks. Moreover, specific members of the overall microbial community can respond to hormones secreted by the host (6, 7). Taken together with the specialized functions performed by the gut microbiota and despite the obvious physical dissimilarity, its ability to function as a collective to influence other organs within the host and, in turn, to be responsive to the secretions of other host organs satisfies the most important conditions of any conceptual definition of an organ (8). It is these qualities that have seen the term “virtual organ” assigned to the gut microbiota (5).
Table 1.
Candidate Hormones of the Gut Microbiota
| Class | Examples | Functions | Comment | Reference |
|---|---|---|---|---|
| SCFAs | Acetate | Energy source | Directly produced by bacteria; epigenetic and receptor-mediated effects; CNS effects linked to autism-like behaviors | 63, 84, 94 |
| Butyrate | Host metabolism | |||
| Propionate | Signaling molecules | |||
| Neurotransmitters | Serotonin Dopamine Noradrenaline GABA |
Mood, emotion, cognition, reward (CNS) Motility/secretion (ENS) |
Can be directly produced by bacteria (see Table 2) or indirectly regulated | 61, 188, 200 |
| Precursors to neuroactive compounds | Tryptophan | Precursor to: 5-HT | Kynurenine is itself a metabolite of tryptophan, production subject to regulation by microbiota | 135, 137, 138, 194 |
| Kynurenine l-Dopa |
Kynurenic acid, quinolinic acid, Dopamine | |||
| Bile acids | Secondary bile acids | Antimicrobial | Some effects mediated by bile acid receptors | 5, 101 |
| Host metabolism | ||||
| Choline metabolites | Trimethylamine | Lipid metabolism (choline) | Metabolized in the liver to trimethylamine-N-oxide, linked to cardiovascular disease | 37, 102, 103 |
| HPA hormones | Cortisol | Stress response | Indirect regulation; HPA endocrine abnormalities prominent in stress-related psychiatric disorders | 177 |
| Host metabolism | ||||
| Anti-inflammatory | ||||
| Wound healing | ||||
| GI hormones | Ghrelin | Host metabolism | Indirect regulation; possibly mediated by SCFAs via enteroendocrine cells | 70, 128 |
| Leptin | Appetite regulation | |||
| Glucagon-like peptide-1 PYY |
GI motility/secretion |
Figure 1.
Hormone-like metabolites produced or regulated by the gut microbiota. Microbial metabolites such as SCFAs with signaling functions are secreted into the gut lumen, transported across the epithelial barrier, and transported to the effector organs, including the brain, via the bloodstream. The gut microbiota is also capable of producing or releasing neurotransmitters such as serotonin or regulating the availability of precursors such as tryptophan. The microbiota also regulates the bioavailability of choline and its metabolites.
In this review, we consider the main features of the gut microbiota that enable this endocrine capacity and render it susceptible to modification, focusing on development, structure, and function as well as the key hormonal mediators produced. Using specific examples, we explore the regulation of glucose metabolism and obesity together with the impact on the stress system, especially on the hypothalamic-pituitary-adrenal (HPA) axis, and the influence on brain and behavior. Having examined the key endocrine functions this collection of microorganisms regulates in the host, we also consider the implications for health and disease.
The Diverse Gut Microbiome: A Complex Endocrine Organ
Unlike other endocrine systems or organs (see Figure 2), which secrete a single or at most a small number of humoral agents, the gut microbiota has the potential to produce hundreds of products. From a morphological and biochemical perspective, it is far larger and more biochemically heterogeneous than any other endocrine organ in man (9). In fact, the biochemical complexity of the gut microbiota even exceeds that of the brain, and many of the hormones produced by the microbiota are also neurotransmitters within the central nervous system (CNS). For example, γ-aminobutyric acid (GABA), the most important inhibitory transmitter in the brain is produced by several lactobacilli (10), whereas monoamines such as noradrenaline, dopamine, and serotonin are also produced by certain strains of bacteria (11, 12) (See Table 2).
Figure 2.
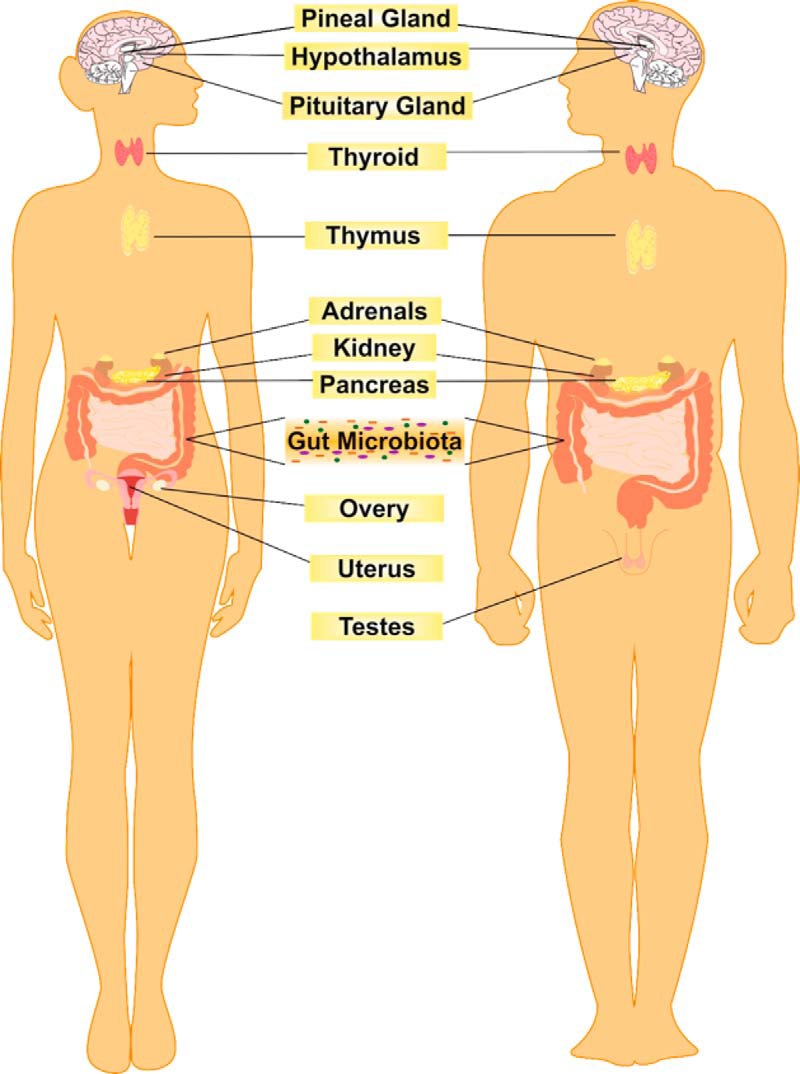
The gut microbiota as an endocrine organ. The endocrine system is primarily composed of glands that each produce a single or at most a small number of humoral agents. However, the microbiota produces numerous chemicals of a hormonal nature that are released into the bloodstream and act at distal sites. This biochemical capacity arises from the vast and diverse array of microbial cells that constitute the gut microbiota.
Table 2.
Examples of Neurotransmitter-Producing or -Releasing Bacterial Strains
| Neurotransmitter | Bacterial Strain | Reference |
|---|---|---|
| Serotonin | Lactococcus lactis subsp. cremoris (MG 1363) | 201 |
| L. lactis subsp. lactis (IL1403) | 201 | |
| Lactobacillus plantarum (FI8595) | 201 | |
| Streptococcus thermophilus (NCFB2392) | 201 | |
| Escherichia coli K-12 | 156 | |
| Morganella morganii (NCIMB, 10466) | 202 | |
| Klebsiella pneumoniae (NCIMB, 673) | 202 | |
| Hafnia alvei (NCIMB, 11999) | 202 | |
| Dopamine | Bacillus cereus | 33 |
| B. mycoides | 33 | |
| B. subtilis | 33 | |
| Proteus vulgaris | 33 | |
| Serratia marcescens | 33 | |
| S. aureus | 33 | |
| E.coli | 33 | |
| E. coli K-12 | 156 | |
| M. morganii (NCIMB, 10466) | 202 | |
| K. pneumoniae (NCIMB, 673) | 202 | |
| H. alvei (NCIMB, 11999) | 202 | |
| Noradrenaline | B. mycoides | 33 |
| B. subtilis, | 33 | |
| P. vulgaris | 33 | |
| S. marcescens | 33 | |
| E. coli K-12 | 156 | |
| GABA | L. brevis DPC6108 | 10 |
| B. adolescentis DPC6044 | 10 | |
| B. dentium DPC6333 | 10 | |
| B. dentium NFBC2243 | 10 | |
| B. infantis UCC35624 | 10 | |
| L. rhamnosus YS9 | 71 | |
| Acetylcholine | L. plantarum | 122 |
| Histamine | L. lactis subsp. cremoris (MG 1363) | 201 |
| L. lactis subsp. lactis (IL1403) | 201 | |
| L. plantarum (FI8595) | 201 | |
| S. thermophiles (NCFB2392) | 201 | |
| M. morganii (NCIMB, 10466) | 202 | |
| K. pneumoniae (NCIMB, 673) | 202 | |
| H. alvei (NCIMB, 11999) | 202 |
This biochemical capacity arises from the vast and diverse array of microbial cells, with an approximate weight of 1 to 2 kg in an average adult (13). The number of cells is far greater than the number of host cells such that an estimated 90% of cells found in the human body are actually of prokaryotic origin, derived from at least 40 000 bacterial species in 1800 genera (14, 15). The genomic complexity of this virtual endocrine organ is enormous, and in the adult, approximately 8 million genes are represented in a small number of phyla (16). However, most of our gut bacteria are not readily culturable in the laboratory, and there is potential for selective growth bias toward specific organisms (17). Thus, in many ways, our appreciation of the functional repertoire of the gut microbiota has been constrained and distorted by the culture-based techniques that have dominated microbiology until relatively recently (18). An increased understanding of the key microbiota genes involved in its various functions is essential to parse and beneficially exploit the endocrine role of the gut microbiota.
Structure and Development of the Gut Microbiome
Our knowledge of the gut microbiome is rapidly expanding, and this is due to the emergence and rapid technological advances in culture-independent techniques, particularly genomic approaches (19, 20). This is probably best exemplified by large-scale metagenomic-based studies such as National Institutes of Health-funded Human Microbiome Project (http://commonfund.nih.gov/hmp), the European-funded Metagenomics of the Human Intestinal Tract (http://www.metahit.eu) consortium, and the International Human Microbiome Consortium (http://www.human-microbiome.org). Together, these studies have helped determine the breadth of microbial variation and function across large populations, revealed a high interindividual diversity and stability over time, and established associations between microbiota alterations (both in terms of composition and stability) and disease states (21–23).
In adults, there is a stable and diverse gut microbiome, dominated by Bacteriodetes and Firmicutes phyla, and there seems to be a high metagenomic abundance of a housekeeping core with specific functional capabilities (21). These features of compositional stability and diversity are considered essential for health but are also readily amenable to potentially deleterious alterations. Moreover, the developmental trajectory from birth, which may prime for health later in life, is also fragile (24). Interestingly, and in contrast to the elaboration of other endocrine organs, large-scale colonization of the infant gut commences only at birth when the infant is first exposed to a complex microbiota and its initial microbiome is seeded from maternal sources. Existing data indicate that babies born by cesarean section develop a different microbiota from their vaginally delivered counterparts with aberrant short-term immune responses and a greater long-term risk of developing immune diseases (25–27). Whether or not cesarean section delivery affects the subsequent endocrine capacity of the gut microbiota is uncertain but likely, and higher rates of diabetes in such children have been established (26). The microbiome of unweaned infants is simple with alterations dictated by, for example, weaning or infection (28). After 1 to 3 years of age, a complex adult-like microbiome is evident, and this adult-type microbiome is functionally specialized for energy harvest from particular diets (29). Homeostatic mechanisms within the microbiota become less effective in the elderly, and it is clear that the microbiota diverges between those who age healthily and those whose health deteriorates with age (30, 31).
The concept of bacterial enterotypes within the gut ecosystem has recently emerged (32), and these alternative stable states of the gut microbiota have differential functional capabilities and, by extension, potentially differential endocrine capabilities. Driven by species composition, these enterotypes are marked out by a relatively high abundance of either Bacteroides, Prevotella, or Ruminococcus species independently of body mass index, age, or gender (14). The Bacteroides and Prevotella enterotype have subsequently been associated with diets enriched for protein/animal fat and carbohydrate, respectively, and appear to be somewhat resilient to short-term dietary interventions (34). However, caution is advisable and a debate persists regarding the clarity of these enterotypes and whether they might more accurately be considered gradients (35). This view is supported by a study based on Human Microbiome Project data in which a proportion of intermediate subjects were identified without such discrete boundaries (36). Nevertheless, the concept of an endocrine profile dependent on specific microbiome states is appealing and warrants further investigation.
Microbiota and Host Metabolism
The endocrine range of the gut microbiota may at least partially explain the links between this microbial community and the development of obesity, cardiovascular disease, and metabolic syndromes (37). The functional interactions allowing such an important contribution to host metabolism and energy homeostasis are manifold and allow for a key role in regulating insulin sensitivity, fat storage, adiposity, and body weight (see Figure 3) (38). Bäckhed and co-workers (39) showed that mice raised in absence of microorganisms, termed germ-free (GF), had less total body fat than mice raised conventionally, despite the fact that GF animals had higher caloric intake. When GF mice are colonized with the microbiota of a conventional mouse, they show a robust increase in body fat and become glucose-intolerant even with a reduction in food intake. Bäckhed and co-workers (40) also demonstrated that GF mice fed a high-fat, high-carbohydrate Western diet for 8 weeks gained significantly less weight than mice raised conventionally and were protected against diet-induced glucose intolerance and insulin resistance.
Figure 3.
The gut microbiota and host metabolism. A, GF animals are protected from high-fat diet-induced obesity. A number of important studies have demonstrated that GF animals gain less weight than their conventionally colonized counterparts when fed a high-fat/high-sugar Western-style diet. GF animals also have less total body fat than mice raised conventionally, and when colonized with the microbiota of a conventional mouse, they show a robust increase in body fat. Moreover, obesity is associated with alterations in the composition of the microbiota. B, GF animals adopt the phenotype of the microbiota donor. Previously GF mice display an obese phenotype on receipt of a microbiota transplant from obese mice or when humanized with a microbiota from an obese individual. Similarly, GF animals lose weight upon receipt of a microbiota transfer from animals who have exhibited rapid weight loss after gastric bypass surgery.
Interestingly, previously GF mice display an obese phenotype on receipt of a microbiota transplant from obese mice (41, 42) and lose weight upon receipt of a microbiota transfer from animals who had exhibited rapid weight loss after gastric bypass surgery (43). In line with this, a recent study showed transmission of the donor's metabolic phenotype using GF mice that received a microbiota transfer from monozygotic twins discordant for obesity. Notably, when cohousing mice who had received an obese twin's microbiota with mice containing the lean co-twin's microbiota, the development of increased body mass and obesity-associated metabolic profile was prevented, possibly due to the invasion of specific members of Bacteroidetes (44). Taken together with the evidence that GF animals appear to be protected against metabolic diseases, these studies suggest a possible causal relationship between the gut microbiota and the emergence of obesity (45).
Whether changes in microbiota composition are the cause or the consequence of obesity and what particular community configurations or members are important to the development of obesity have been studied intensively. In genetically obese leptin-deficient ob/ob mice, this characteristic has been associated with a reduction in the abundance of Bacteroidetes and a proportional increase in Firmicutes, 2 of the major bacterial phyla in the gut microbiota (46). Similar changes in microbiota composition have been observed in wild-type mice fed a high-fat/high-sugar Western diet (41). The functional consequences of these compositional shifts might well be reflected in differential biochemical profiles and functional metabolic outcomes, with the shift toward Firmicutes, for example, promoting more efficient absorption of calories and subsequent weight gain (47).
In line with these principles, Turnbaugh and co-workers (203) identified a core microbiome of microbial genes shared among individuals and found that variations from that core are associated with obesity (203). However, comparisons of the abundance of the 2 major bacterial phyla in the obese vs lean microbiome in animals and humans, have often produced conflicting results. Although several studies have reported similar increases in the ratio between Firmicutes and Bacteroidetes in obese individuals (48–50), other studies have not observed a change in the Firmicutes to Bacteroidetes ratio (51–53). For example, one study reported reductions in Bifidobacterium spp. and increases in specific members of Firmicutes (eg, Staphylococcus) and Proteobacteria (eg, Enterobacteriaceae) in overweight pregnant women (49) whereas another study found enrichment in Prevotellaceae, a group within the Bacteroidetes phylum, in obese individuals (51). These disparities highlight the importance of considering the multitude of potential confounding factors such as diet, age, degree of obesity, demographic geography, and population size as well as technique and methodology used to profile the gut microbiota when comparing studies (19). Indeed, high-fat diets can influence the murine microbiome composition independently of obesity (54). Thus, although much of the preclinical data are convincing, it remains unclear to date from clinical studies if the altered microbiota precedes the development of obesity or is a consequence of dietary intake and differences in host physiology (55).
In this context, some important observations can be noted. Olanzapine, an atypical antipsychotic that causes weight gain and type 2 diabetes, has been shown to alter the composition of the gut microbiota in rats as assessed by pyrosequencing technology (56). Administration of vancomycin (a broad-spectrum antibiotic) to mice with diet-induced obesity resulted in changes in Firmicutes, Bacteroidetes, and Proteobacteria with an improvement in metabolic abnormalities (57). Furthermore, a fatty acid-based dietary intervention (using an oleic acid-derived compound) has been shown to offset a high-fat diet-induced dysbiosis of the gut and improve weight gain measures in obese mice (58). Conversely, transfer of the microbiomes from women in the third trimester of pregnancy, characterized by reduced microbial diversity and enrichment in Proteobacteria and Actinobacteria, to GF mice induced symptoms of metabolic syndrome in the mice (59). It seems likely then that at least in some cases of obesity, the microbiota might be a causal factor and a therapeutic target for symptom improvement.
However, it is important to acknowledge that obesity is likely multifactorial, with both central and peripheral mechanisms underlying its pathogenesis, including hypothalamic dysfunction (60). It is currently unclear whether CNS control of food intake is subject to microbial influences, endocrine or otherwise, at relevant brain centers (61). Future studies geared toward identifying important microbial genes instead of focusing solely on characterizing gut microbiota composition are required to more accurately describe the obese microbiome and predict its response after therapeutic intervention. Nevertheless, there are a number of candidate hormones, either produced or regulated by the gut microbiota, which might mediate the impact that the gut microbiota can have on host metabolism.
Candidate Hormones of the Gut Microbiota: Focus on Short-Chain Fatty Acids
Short-chain fatty acids (SCFAs) are the major products of the bacterial fermentation of carbohydrates and proteins in the gut (62, 63). In many ways, they represent the signature hormones of the microbiota and may mediate many of the functions assigned to the microbiota through classical endocrine signaling. Receptors and transporters for SCFAs are expressed in the GI tract, where they appear to be of relevance to GI function (64–69). For example, SCFAs may modulate both enteroendocrine serotonin (5-HT) secretion (5) and peptide YY (PYY) release, an important neuropeptide at multiple levels of the gut-brain axis (70).
Fiber metabolized by the gut microbiota can increase the concentration of circulating SCFAs (2). These circulating SCFAs, such as butyrate and propionate, travel to sites far removed from their site of production and can be carried by monocarboxylate transporters, which are abundantly expressed at the blood-brain barrier (72–75). This provides a plausible mechanism through which they can cross the blood-brain barrier and enter the CNS. Once available in the CNS, they can be taken up via these same monocarboxylate transporters on glia and neurons (76–78), and they are thought to comprise a major energy source in cellular metabolism, particularly during early brain development (79). Interestingly, they can modulate intracellular calcium levels in neutrophils, suggesting a role in cell signaling (80–82) and, via regulation of tyrosine hydroxylase gene expression, can potentially affect neurotransmitter synthesis (83). Of note, intraventricular administration of propionic acid in rats induces a variety of behavioral alterations of relevance to autism, although it is unclear whether this occurs via similar mechanisms to the periphery (84). It is worth noting that G-protein coupled receptor (GPR)41, a receptor activated by propionic acid, is highly expressed in rat brain tissue (85). However, it remains to be definitively established whether alterations in intestinal microbiota-derived SCFAs are actually reflected at physiologically relevant concentrations in the CNS.
Although there is patently a role for these microbial metabolites beyond the local regulation of energy homeostasis (86), this is the context in which they have been most studied. The main SCFAs are acetic, propionic, and n-butyric acids, occurring roughly in molar ratios of 60:20:20 in the colon (87). Through their absorption and metabolism, the host is able to salvage energy from foodstuffs, particularly resistant starch and fibers that are not digested in the upper part of the GI tract. The main site for SCFA production and absorption is the proximal large intestine, where the fermentation of the undigested food by colonic bacteria occurs at high rates. Bacteria that produce SCFAs include, but are not limited to, Bacteroides, Bifidobacterium, Propionibacterium, Eubacterium, Lactobacillus, Clostridium, Roseburia, and Prevotella (63).
SCFAs have a multiplicity of other effects in the body and affect epithelial cell transport and metabolism, epithelial cell growth and differentiation, and hepatic control of lipid and carbohydrates while providing energy sources for muscles and kidneys as well as the heart and brain (88). Epithelial cells in the distal colon derive 60% to 70% of their energy requirements from bacterial fermentation products (89). As indicated above, SCFAs also act as signaling molecules. Propionate, acetate, and to a lesser extent butyrate, are ligands for at least 2 GPRS, GPR41 and GPR 43, which are broadly expressed, including in the distal small intestine, colon, and adipocytes (90, 91). SCFA interaction with GPR43 can profoundly affect inflammatory responses. For example, mice treated with oral acetate showed a substantial decrease in inflammation. This protection was mediated by acetate binding to GPR43, because acetate had no effect in GPR43-deficient mice. Furthermore, it was shown that GPR43 exhibited enhanced expression in neutrophils and eosinophils, suggesting that SCFA-GPR43 signaling is one of the molecular pathways whereby commensal bacteria regulate immune and inflammatory responses (92). GPR43 is also induced during adipocyte differentiation and exhibits increased levels during high-fat feeding in rodents, indicating that GPR43 may also affect adipocyte function (93). Hong et al (94) demonstrated that acetate and propionate act on lipid accumulation and inhibition of lipolysis mainly through GPR43. GPR41 has been shown to be implicated in microbiota-dependent regulation of host adiposity and leptin production (95).
Butyrate is known to exhibit many important physiological functions in eukaryotic cells (63). One of the most recognized cellular mechanisms for the action of butyrate is its effects on histone acetylation (96), where the inhibition of histone deacetylase facilitates hyperacetylation of histone proteins to occur, thus facilitating the access of DNA repair enzymes. Interestingly, sodium butyrate has been demonstrated to elicit an antidepressant effect in the murine brain (97). When injected systemically, sodium butyrate induced a short-lasting transient acetylation of histones in frontal cortex and hippocampus in conjunction with dynamic changes in expression of the brain-derived neurotrophic factor, thereby resulting in an antidepressant-like behavioral response (97). Antidepressants can suppress cortisol in patients with melancholic depression, but whether butyrate has an impact on the HPA axis is not known.
Enhanced carbohydrate utilization from nondigestible plant polysaccharides is known to result in higher production of SCFAs that contribute to total energy in the host. In GF mice monoassociated with the probiotic strains Lactobacillus plantarum WCFS1 (98) and Lactobacillus johnsonii NCC533 (99), an upregulation of genes involved in carbohydrate transport and metabolism was observed. Although increased efficiency to hydrolyze different complex sugars in L. plantarum WCFS1 resulted in higher production of fumarate and alcohol (98), it led to a longer colonization period of L. johnsonii NCC533 in the mouse gut (99). Consumption of fermented milk containing multiple probiotic strains led to enrichment of enzymes catalyzing carbohydrates into propionate in both monozygotic twins and gnotobiotic mice containing a model human microbiota community (100).
Other Candidate Hormones of the Gut Microbiota: Relevance for Host Metabolism and Disease
The gut microbiota is involved in the transformation of primary bile acids and, by doing so, may control the host profile of agents involved in lipid and glucose metabolism (5, 101). Interestingly, bile acids have an antimicrobial function, and GF animals have more bile acids than their conventionally raised counterparts (37). Secondary bile acids produced microbially may also enter the circulation to interact with similar receptors that mediate the effects of the parent compounds (eg, farnesoid X receptor-α) on glucose homeostasis (5). Microbial metabolic activities also include the metabolism of choline, important for lipid metabolism, to trimethylamine (37). Once synthesized by the intestinal microbiota, trimethylamine can be further metabolized in the liver to trimethylamine-N-oxide, which, when present in the circulation at sufficient concentrations, can contribute to the development of cardiovascular disease (102, 103). Moreover, reducing the bioavailability of choline can contribute to nonalcoholic fatty liver disease and altered glucose metabolism, both in mice and humans (104–106). These and other mechanisms implicate the hormone-like secretions of the intestinal microbiota in liver disease, through what has been termed the gut-liver axis (107).
Manipulation of the Gut Microbiota: Probiotics, Prebiotics, and Host Metabolism
If the composition of the gut microbiota changes, then theoretically, the endocrine output should also be modified. This does seem to be the case when membership of the microbiota is directly modified, albeit transiently, by probiotic strains or indirectly via prebiotics, which then promote the growth of specific beneficial strains over others. Strain-specific biochemicals may then boost the endocrine capacity of the endogenous gut microbiota and act like hormones to influence host metabolism. Alternatively, the transient alteration of the microbiota composition may nonspecifically engender an alteration in the overall endocrine output from this virtual organ or produce beneficial effects by alternative, possibly unknown, mechanisms.
Conjugated linoleic acid (CLA) is a naturally occurring derivative of linoleic acid found in foods and dairy products and has been shown to increase metabolic rates in mice (108, 109). The CLA-producing probiotic strain, L. rhamnosus PL60, has been reported to reduce body weight gain and white adipose tissue mass with no effect on food intake in high-fat diet-fed mice. The effect was coupled to higher expression of uncoupling protein-2, whereas expression of fatty acid synthase (fas) and serum leptin and glucose levels were reduced (110). Another probiotic strain that produces CLA, Lactobacillus plantarum PL62, also resulted in reduced body weight gain and glucose levels in diet-induced obese mice (111). Other CLA-producing strains such as L. paracasei NFBC 338 (112) and Bifidobacterium breve NCIMB 702258 (113) can also modulate the fatty acid composition of host adipose tissue.
Probiotics have been shown to reduce adipocyte size in different adipose depots (114, 115), which is considered an important parameter in assessing their antiobesity potential. The putative mechanisms put forth are increased fecal excretion of neutral sterols and bile acids, decreased lymphatic absorption of triglycerides, phospholipids, and cholesterol (116), or increased lipolysis (117). In the 3T3-L1 cell line, incubation with L. plantarum KY1032 cell-free extract resulted in reduced adipogenesis (118), and incubation with the insoluble fraction from fermented kefir resulted in reduced adipocyte differentiation (119). Administration of L. paracasei NCC2461 to rats increased sympathetic nerve activity in white and brown adipose tissue, resulting in higher thermogenesis in brown adipose tissue and increased lipolysis in white adipose tissue (120). Supplementation with L. paracasei F19 resulted in reduced total body fat and decreased triglyceride levels in different lipoprotein fractions in mice fed a high-fat diet (121). In addition, both GF and conventionally raised mice supplemented with L. paracasei F19 had increased serum levels of angiopoietin-like 4, a microbially regulated lipoprotein lipase inhibitor that regulates lipid deposition into adipocytes (40, 121). Administration of L. paracasei F19 and Lactobacillus acidophilus NCFB1748 to GF mice resulted in enrichment of probiotic strains in the ileum compared with the colon and upregulation of insulin-sensitizing hormones, adipsin and adiponectin. Decreased expression of resistin-like β, known to induce insulin resistance, was also reported (123). Apoe−/− mice that were supplemented with Lactobacillus reuteri ATCC exhibited reduced body weight gain, reduced adipose and liver weight, and increased expression of carnitine palmitoyl transferase1a (cpt1a), suggesting higher hepatic β-oxidation (124).
Inulin or inulin-type prebiotics, when taken orally, reach the colon intact where they undergo bacterial fermentation and stimulate the growth of Bifidobacteria species (125). These types of prebiotic can influence production of relevant GI hormones, possibly via SCFAs, such as glucagon-like peptide-1, peptide YY (PYY), ghrelin, and leptin (126–128). Ghrelin in particular has attracted much attention as a hunger hormone (60). This allows the microbiota to exert an influence over not just host metabolism but also food intake and appetite regulation (129).
Candidate Hormones of the Gut Microbiota: Focus on Tryptophan Supply
The ability of the gut microbiota to influence brain and behavior is considered a new frontier of research (61, 130). Although this cross-talk can occur via a number of mechanisms that take advantage of gut-brain axis scaffolding, including neural routes (see Figure 4), humoral options also exist (3). In this context, circulating concentrations of tryptophan, the amino acid precursor for the signaling molecule 5-HT, appears to be under the influence of the gut microbiota. 5-HT is a key neurotransmitter in the gut-brain axis, both at the level of the ENS (131) and in the CNS (132). Moreover, its physiological repertoire is extensive with a widespread receptor distribution and extends to cardiovascular function including blood pressure regulation (133), bladder control, and platelet aggregation (134).
Figure 4.

Neuronal pathways of the gut-brain axis. In addition to hormonal cross-talk, the gut microbiota can influence brain and behavior by recruiting the neuronal pathways of the gut-brain axis. This term describes the bidirectional communication network between the gut and the brain. The CNS and ENS communicate along vagal and autonomic pathways to modulate many GI functions. Mood and various cognitive processes can mediate top-down/bottom-up signaling. Vagal afferent nerves transmit to central brain regions in response to numerous signaling molecules in the gut that can be regulated or secreted by the gut microbiota. This vagal innervation is essential for homeostasis and provides both motor and sensory innervation for several key functions including satiety, nausea, sensations of visceral pain, sphincter operation, and peristalsis. Spinal afferent nerves from the GI tract also project to the dorsal horn of the spinal cord. Not shown in this diagram are the HPA axis and the immune systems, which are also important components of the gut-brain axis.
Studies in GF animals indicate that the peripheral availability of tryptophan in the circulation, critical for CNS 5-HT synthesis, is regulated by the gut microbiota, and these elevated plasma tryptophan concentrations can be normalized after colonization of the GF animals (135). Augmentation is possible after administration of the probiotic Bifidobacterium infantis (136). The mechanism underpinning the regulation of circulating tryptophan concentrations by the bacteria in our gut is unclear but may involve control over degradation of tryptophan along an alternative and physiologically dominant metabolic route, the kynurenine pathway (137, 138). Indoleamine-2,3-dioxgenase (IDO) and tryptophan-2,3-diogenase (TDO), the enzymes that catalyze the initial metabolic step in this pathway, are subject to immune and glucocorticoid control, respectively. A decreased ratio of kynurenine to tryptophan, indicating decreased IDO/TDO activity, in GF animals connects this pathway to the reported alterations (135). Moreover, increased IDO activity is observed after the associated chronic GI inflammation that arises after infection with Trichuris muris (139). Importantly, metabolites produced downstream of kynurenine are neuroactive, such as kynurenic acid and quinolinic acid (137). The ability of the microbiota to regulate influential precursors such as tryptophan and kynurenine likely contributes significantly to its ability to sway CNS and ENS neurotransmission.
The relevance of these preclinical findings to the clinical situation is probably best gauged from studies in irritable bowel syndrome (IBS), a functional GI disorder that has been linked to alterations in the gut microbiota (140). Accordingly, increased IDO activity has been reported in both male and female IBS populations (141–143). Mechanistically, degradation of tryptophan can ensue after activation of Toll-like receptors (TLRs) (144, 145), which have altered expression and activity in both clinical IBS populations (146, 147) and animal models of the disorder (148). A differential TLR-specific pattern of kynurenine production in IBS has also been reported (142).
Other explanations are also possible, and in addition to the utilization of tryptophan to satisfy the growth requirements for bacteria (149), tryptophan is also recruited by a bacteria-specific tryptophanase enzyme for indole production (150, 151). Bacteroides fragilis harbors this enzyme and has recently been linked to GI abnormalities in autism spectrum disorders (152). Certain bacteria are capable of tryptophan biosynthesis via enzymes such as tryptophan synthase (153, 154), whereas some bacterial strains are also proficient in serotonin production from tryptophan, at least in vitro (11, 155, 156).
Microbiota, Stress, and the HPA Axis
The use of GF animals has provided one of the most significant insights into the role of the microbiota in regulating the development of the HPA axis. Subsequent comparison with their conventionally colonized counterparts allows inferences to be drawn regarding the morphological and physiological parameters that may be under the influence of the developing microbiota. However, in the absence of the resident enteric microbiota, key members of the TLR family have low or absent expression profiles in the gut, thus compromising appropriate neuroendocrine responses to pathogens (157). For example, intact TLR4 signaling is required for plasma corticosterone release (158) and the TLR4 knockout mouse does not respond appropriately to lipopolysaccharide from gram-negative bacteria with an activation of the HPA axis (159).
Pivotal studies by Sudo and colleagues (160) provide insight into the role of the intestinal microbiota in the development of the HPA axis. In GF mice, a mild restraint stress induces an exaggerated release of corticosterone and ACTH compared with the specific pathogen-free controls. This has since been replicated independently, again after a mild stressor, in both mice (135) and rats (161). This aberrant stress response in GF mice is partially reversed by colonization with fecal matter from specific pathogen-free animals and fully reversed by monoassociation with B. infantis in a time-dependent manner (160). This study clearly demonstrated that the microbial content of the gut is critical to the development of an appropriate stress response later in life and also that there is a narrow window in early life where colonization must occur to ensure normal development of the HPA axis.
Other aspects of this relationship need to be teased apart and additional indices of the HPA axis assessed for a more detailed characterization of the microbial influence on the stress response. This includes evaluating corticosterone output after different grades of stressor as well as recording not just peak cortisol concentrations but also recovery from the stressful insult. In this regard, the sustained HPA response to an acute stress, the Trier Social Stress Test (TSST), in subjects with IBS may be informative (162, 163). Although it is unclear whether this is specifically related to the proposed alteration in the gut microbiota in this disorder, it will nevertheless be interesting to interrogate in GF animals whether the microbiota affects not just the ability to mount an appropriate response but also the ability to turn that response off and indeed whether these animals habituate to repeated stressors.
Endocrine abnormalities have long been associated with psychiatric illness, with hyperactivity of the HPA axis a prominent feature of at least some subtypes of major depression (164–166). In IBS, a significant comorbidity, such abnormalities are more inconsistent but still regularly reported (140, 163, 167). It is currently unknown whether alterations in the microbiota are associated with psychiatric illness such as depression, although that prospect is increasingly coming into focus. However, it is long known that stress and the HPA axis can influence the composition of the gut microbiome. The functional consequences of such changes are only now being understood.
Maternal separation, an early-life stressor that can result in long-term HPA changes also has long-term effects on the microbiome (168, 169). Analysis of the 16S rRNA diversity in adult rats exposed to maternal separation for 3 hours per day from postnatal days 2 to 12 revealed a significantly altered fecal microbiome when compared with the nonseparated control animals (169). A study using bacterial tag encoded FLX amplicon pyrosequencing demonstrated that the community structure of microbiota from mice exposed to a prolonged restraint stressor was significantly different from the community structure found in nonstressed control mice (170). More recently, using the same approach, repeated social stress has been shown to decrease the relative abundance in bacteria of the genus Bacteroides, while increasing the relative abundance of bacteria in the genus Clostridium in the cecum (171). The stressor also increased circulating levels of IL-6 and monocyte chemoattractant protein-1 (MCP-1), which were significantly correlated with stressor-induced changes to 3 bacterial genera (ie, Coprococcus, Pseudobutyrivibrio, and Dorea).
Demonstrating changes in baseline glucocorticoid levels in either rodents or man based on single time point serum samples, which can be highly variable (172), is notoriously unreliable. This may contribute to the lack of consensus across different studies, although the stress-induced corticosterone response in rodents is considered more robust and reliable. In a maternal separation model, Desbonnet et al (173) found behavioral changes with B. infantis treatment but no reduction in corticosterone. Using a similar model, Gareau et al (174) found that feeding a Lactobacillus strain reduced corticosterone levels and McKernan et al (175) found that B. infantis administration reduced corticosterone levels, although the reduction did not reach statistical significance.
In a recent clinical study, healthy volunteers were given Lactobacillus helveticus R0052 and Bifidobacterium longum R0175 in combination or placebo in a double-blind, randomized parallel-group study for 30 days (176). The 24-hour urinary free cortisol output was reduced with probiotic treatment. No other HPA axis measures were taken. However, no in-depth analysis in man of HPA activity in response to a course of any putative probiotic has been undertaken either in healthy subjects or patient populations. Nonetheless, based on the currently available data, Dinan and Cryan (177) concluded that developmental studies and those involving stress-related disorders should include the gut microbiota as an important regulator of the HPA axis, and failure to do so can result in the introduction of a significant confounding variable.
Microbial Endocrinology
Not only do bacteria in the gut produce hormone-like substances and regulate hormonal output, they can also potentially respond to the hormonal secretions of the host. Lyte and colleagues (6, 7, 12) have termed this area of study Microbial Endocrinology, and although it has mostly focused on hormonally driven bacterial growth in the context of pathogens and infectious processes, it is also of relevance to the composition and distribution of the intestinal microbiota (7). Elevations in noradrenaline concentrations after acute stress can, for example, stimulate the growth of nonpathogenic commensal Escherichia coli (178) as well as other gram-negative bacteria (179). This has implications for stress-related disorders, and as we have emphasized, could have knock-on consequences for the hormonal output of the microbiota that in turn might modify host behavior and perpetuate microbiota-driven pathological states. Clearly, the host-microbiota cross-talk is complex, and we should not consider the contribution of either partner in isolation.
Implications and Perspectives
Long neglected, the gut microbiota can no longer be considered a bystander in health and disease. We have described the clear links between the composition of our intestinal microbiota, its functional endocrine interactions, and the development of obesity, cardiovascular disorders, and metabolic syndromes as well as stress-related psychiatric disorders (see Figure 5). Integrating these important observations is a demanding proposition but an extremely worthwhile venture with many potential benefits to society.
Figure 5.
Implications of endocrine output of the gut microbiota in health and disease. The hormonal output of the gut microbiota is vital for host health and well-being. In addition to the local impact in the GI tract and ENS, it is also critical for control of host metabolism and the normal development of the immune system. Through as yet unknown mechanisms, the gut microbiota also seems to be able to regulate glucocorticoid production in the HPA axis. Similarly, through controlling the availability of the serotonin precursor tryptophan, the gut microbiota can potentially influence brain and behavior. Metabolites produced by the gut microbiota from choline can also affect the cardiovascular system.
There is much current interest in the diagnostic potential of assessing alterations in the microbial ecology of the gut. One of the most frequently reported findings reported in the literature in both clinical and preclinical studies are alterations in the Firmicutes to Bacteroidetes ratio in disease states such as obesity and IBS (41, 48, 180). Although a consensus has still to become apparent on the reliability of these alterations as a biosignature, there is broad agreement that we need to move toward establishing a causal role for any alterations in composition that are present (181). In addition, preclinical studies suggest the presence of melancholic microbes with the ability to influence psychiatric disorders such as depression and anxiety (182, 183). Tracking a compositional dysbiosis is a complex process and the repercussions in terms of a functional endocrine output remains to be defined. Given the broad span of impact exerted by the candidate hormones of the gut microbiota, urgent attention should be devoted to this topic, which could also spawn novel therapeutic targets.
An important consideration to bear in mind when evaluating microbiota data from clinical studies is the caveat that much of what we understand is based on the assessment of fecal specimens. It is likely that fecal, luminal, and mucosal bacterial populations represent distinct ecological niches in addition to the considerable variation along the known gradient of bacteria in the GI tract (18). The microbial communities in the small intestine, for example, have consequently received much less attention yet are likely to be proportionally much more affected after probiotic consumption (184). Moreover, the relationship between an altered microbiota composition in the fecal compartment and microbe-mucosa interactions remains to be fully defined. Clearly, there are many practical logistical reasons that favor fecal sampling protocols, but reliance on this source may need to be reconsidered if we are to capture the true complexity of the gut microbiota and the clear distinction between the mucosa-associated and lumen-residing microbiota (19). This has implications for our understanding of the gut microbiota as an endocrine organ and by extension its role in health and disease, which are as yet not fully understood.
Deliberately manipulating the gut microbiota is an appealing therapeutic strategy and, in the light of our discussion, could be considered as a viable option to treat stress-related psychiatric illnesses as well as metabolic dysfunction and eating disorders. Certainly, it seems that the gut microbiota is very plastic, and the composition can be rapidly altered when the diet is changed. For example, Turnbaugh and colleagues (185) showed that switching from a low-fat, polysaccharide-rich diet to a high-fat, high-sugar diet shifted the structure of the microbiota within a single day and changed the representation of metabolic pathways in the microbiome. Probiotics have also shown utility in the treatment of some features of IBS, for example (186) and boast a favorable safety profile (187). An extension of this concept is the promise of psychobiotics to exert beneficial psychotropic effects (188). As indicated above, prebiotics can modulate hormonal output and have also been considered in the treatment of GI disorders (189, 190). Fecal microbiota transplants are currently attracting much attention for the treatment of recurrent Clostridium difficile infections, and this approach may ultimately filter into the treatment of other disorders as well (191–193). One could argue, based on the likely vast metabolic capacity in such a collection of bacteria, that this is akin to a large-scale hormone replacement therapy.
This strategy is not without its challenges. We should bear in mind that individual bacteria or community structures can exert pleiotropic effects while the concept of functional redundancy also applies. It will also be interesting to see whether we can artificially produce specific gut microbiota compositions that accurately deliver, for example, precise circulating or regional tryptophan concentrations. Similarly, it may be possible to use this approach to control the bioavailability of l-dopa with implications for the treatment of Parkinson's disease (194). We do not yet know whether such a strategy might prove a double-edged sword such that beneficial effects in one domain might engender deleterious consequences in another, altered choline availability for example.
Outside of antibiotics, probiotics, and prebiotics, less consideration has been given to the potential impact of currently available therapeutics on the intestinal microbiota. As indicated above, recent preclinical studies suggest that olanzapine may also alter the microbiota, reducing the levels of Proteobacteria and Actinobacteria by day 21 of treatment with a parallel trend toward an increase in the Firmicutes levels (56). Importantly, many of these effects can be attenuated by concurrent antibiotic administration in rats (195). Some members of the selective serotonin reuptake inhibitors may also possess antimicrobial activity (196).
It is interesting to note some apparent contradictions, arising from studies in GF animals in particular, pertaining to the view of the gut microbiota as an essential endocrine organ. Chief among these is the fact that however abnormal these animals are, not only do they survive in the absence of this virtual organ, they also live longer than their conventionally colonized counterparts (197). This contrasts with the potentially life-threatening nature of endocrine organ malfunction. For example, adrenocortical insufficiency in Addison's disease requires lifelong glucocorticoid and mineralocorticoid replacement therapy for survival and well-being (198). We should, however, point out that the diets of GF animals are customized and supplemented with various factors usually provided by the microbiota and essential to the survival of the host such as vitamin K (199). That the entire hormonal repertoire of the microbiota does not need to be replaced in GF animals is likely due to the fact that many of the intestinal secretions of the microbiota are not bacteria-specific, whereas other hormones at distal sites are under the influence of the microbiota but are host-derived. The exaggerated stress response of GF mice in terms of corticosterone output is also somewhat at odds with their fearless nature in various behavioral tests such as the light/dark box, which are interpreted as indicating reduced anxiety (135). Adding to this complexity is that GF rats do display increases in anxiety-like behaviors that seem to better correspond with their exaggerated stress response (161). However, such contradictions probably emphasize some of the difficulties inherent in translating from the preclinical arena to the clinical domain rather than contradicting the endocrine nature of the microbiota.
Conclusions
The microbiota has the capacity to produce a diverse range of compounds that play a major role in regulating the activity of distal organs including the brain. The influence of the microbiota in regulating metabolic activity is now recognized with increasing evidence suggesting involvement in glucose and weight regulation. Microbial signatures that increase the risk of diabetes mellitus and obesity have been put forward. Likewise, a role for the microbiota in the regulation of the HPA axis has been established as well as the serotonergic system via modulation of tryptophan availability. Unlike other endocrine organs, the microbiota has intense plasticity and can alter dramatically and rapidly in response to diet. With increased understanding of the key microbiota genes involved in endocrine regulation, it may be possible to use probiotics or other modulatory means to treat or prevent the metabolic syndrome and stress-related disorders.
Acknowledgments
We acknowledge the contribution of Dr Romain Gosselin in the production of Figure 4.
A literature review was performed for English language publications using PubMed, Science Direct, Google Scholar, and other online sources using combinations of words such as: gut bacteria, endocrinology, microbiome, host metabolism, behavior, gastrointestinal microbiota, hormone, HPA, short-chain fatty acid, neurotransmitter, tryptophan, and serotonin. The bibliographies of relevant studies and systematic reviews were also considered. Studies were excluded if the abstract and title contained sufficient information to render it irrelevant. All potentially eligible studies were discussed in detail by all authors before selection for inclusion.
The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan. The authors and their work were supported by SFI (Grants SFI/12/RC/2273, 02/CE/B124, and 07/CE/B1368) and by the Health Research Board (HRB) through Health Research Awards (Grant HRA_POR/2011/23 to T.G.D., J.F.C., and G.C.). The Centre has conducted studies in collaboration with several companies including GSK, Pfizer, Wyeth, and Mead Johnson. J.F.C. is also funded by the European Community's Seventh Framework Programme (Grant FP7/2007–2013, Grant Agreement 201 714). G.C. is supported by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (Grant 20771).
This article is not an exhaustive review, and space constraints have prevented the inclusion of all relevant studies.
Disclosure Summary: T.G.D. has until recently been on the Board of Alimentary Health. Each of the authors has spoken at meetings sponsored by food and pharmaceutical companies.
Funding Statement
The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan. The authors and their work were supported by SFI (Grants SFI/12/RC/2273, 02/CE/B124, and 07/CE/B1368) and by the Health Research Board (HRB) through Health Research Awards (Grant HRA_POR/2011/23 to T.G.D., J.F.C., and G.C.). The Centre has conducted studies in collaboration with several companies including GSK, Pfizer, Wyeth, and Mead Johnson. J.F.C. is also funded by the European Community's Seventh Framework Programme (Grant FP7/2007–2013, Grant Agreement 201 714). G.C. is supported by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (Grant 20771).
Footnotes
- CLA
- conjugated linoleic acid
- CNS
- central nervous system
- GABA
- γ-aminobutyric acid
- GF
- germ-free
- GI
- gastrointestinal
- GPR
- G-protein coupled receptor
- HPA
- hypothalamic-pituitary-adrenal
- 5-HT
- serotonin
- IBS
- irritable bowel syndrome
- IDO
- indoleamine-2,3-dioxgenase
- PYY
- peptide YY
- SCFA
- short-chain fatty acid
- TLR
- Toll-like receptor.
References
- 1. Neish AS. Mucosal immunity and the microbiome. Ann Am Thorac Soc. 2014;11(Suppl 1):S28–S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. [DOI] [PubMed] [Google Scholar]
- 3. Grenham S, Clarke G, Cryan J, Dinan TG. Brain-gut-Microbe communication in health and disease. Front Physiol. 2011;2:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forsythe P, Sudo N, Dinan T, Taylor VH, Bienenstock J. Mood and gut feelings. Brain Behav Immun. 2010;24:9–16. [DOI] [PubMed] [Google Scholar]
- 5. Evans JM, Morris LS, Marchesi JR. The gut microbiome: the role of a virtual organ in the endocrinology of the host. J Endocrinol. 2013;218:R37–R47. [DOI] [PubMed] [Google Scholar]
- 6. Freestone PP, Sandrini SM, Haigh RD, Lyte M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. 2008;16:55–64. [DOI] [PubMed] [Google Scholar]
- 7. Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 2004;12:14–20. [DOI] [PubMed] [Google Scholar]
- 8. Lyte M. The microbial organ in the gut as a driver of homeostasis and disease. Med Hypotheses. 2010;74:634–638. [DOI] [PubMed] [Google Scholar]
- 9. Russell WR, Duncan SH, Flint HJ. The gut microbial metabolome: modulation of cancer risk in obese individuals. Proc Nutr Soc. 2013;72:178–188. [DOI] [PubMed] [Google Scholar]
- 10. Barrett E, Ross RP, O'Toole PW, Fitzgerald GF, Stanton C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113:411–417. [DOI] [PubMed] [Google Scholar]
- 11. Lyte M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: Microbial endocrinology in the design and use of probiotics. BioEssays. 2011;33:574–581. [DOI] [PubMed] [Google Scholar]
- 12. Lyte M. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 2013;9:e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forsythe P, Kunze WA. Voices from within: gut microbes and the CNS. Cell Mol Life Sci. 2013;70:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Avershina E, Storrø O, Øien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol. 2014;87:280–290. [DOI] [PubMed] [Google Scholar]
- 17. Fraher MH, O'Toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9:312–322. [DOI] [PubMed] [Google Scholar]
- 18. O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO reports. 2006;7:688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clarke G, O'Toole PW, Dinan TG, Cryan JF. Characterizing the gut microbiome: role in brain-gut function. In: Coppola G, ed. The OMICS: Applications in Neuroscience. Oxford, UK: Oxford University Press; 2014:265–287. [Google Scholar]
- 20. Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet. 2013;29:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clarke G, O'Mahony S, Dinan T, Cryan J. Priming for health: gut microbiota acquired in early life regulates physiology, brain and behaviour [published online May 17, 2014]. Acta Paediatr. doi:10.1111/apa.12674. [DOI] [PubMed] [Google Scholar]
- 25. Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. [DOI] [PubMed] [Google Scholar]
- 26. Cho CE, Norman M. Cesarean section and development of the immune system in the offspring. Am J Obstet Gynecol. 2013;208:249–254. [DOI] [PubMed] [Google Scholar]
- 27. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. [DOI] [PubMed] [Google Scholar]
- 29. Lepage P, Leclerc MC, Joossens M, et al. A metagenomic insight into our gut's microbiome. Gut. 2013;62:146–158 [DOI] [PubMed] [Google Scholar]
- 30. Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. [DOI] [PubMed] [Google Scholar]
- 32. Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. [DOI] [PubMed] [Google Scholar]
- 33. Tsavkelova EA, Botvinko IV, Kudrin VS, Oleskin AV. Detection of neurotransmitter amines in microorganisms with the use of high-performance liquid chromatography. Dokl Biochem. 2000;372:115–117. [PubMed] [Google Scholar]
- 34. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeffery IB, Claesson MJ, O'Toole PW, Shanahan F. Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol. 2012;10:591–592. [DOI] [PubMed] [Google Scholar]
- 36. Huse SM, Ye Y, Zhou Y, Fodor AA. A core human microbiome as viewed through 16S rRNA sequence clusters. PloS One. 2012;7:e34242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. [DOI] [PubMed] [Google Scholar]
- 38. Clarke SF, Murphy EF, Nilaweera K, et al. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes. 2012;3:186–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 43. Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5:178ra141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Greiner T, Bäckhed F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab. 2011;22:117–123. [DOI] [PubMed] [Google Scholar]
- 46. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kallus SJ, Brandt LJ. The intestinal microbiota and obesity. J Clin Gastroenterol. 2012;46:16–24. [DOI] [PubMed] [Google Scholar]
- 48. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. [DOI] [PubMed] [Google Scholar]
- 49. Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and methanogens in anorexic patients. PloS One. 2009;4:e7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Santacruz A, Collado MC, García-Valdés L, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104:83–92. [DOI] [PubMed] [Google Scholar]
- 51. Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duncan SH, Lobley GE, Holtrop G, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond). 2008;32:1720–1724. [DOI] [PubMed] [Google Scholar]
- 53. Schwiertz A, Taras D, Schäfer K, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. [DOI] [PubMed] [Google Scholar]
- 54. Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. [DOI] [PubMed] [Google Scholar]
- 56. Davey KJ, O'Mahony SM, Schellekens H, et al. Gender-dependent consequences of chronic olanzapine in the rat: effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacology (Berl). 2012;221:155–169. [DOI] [PubMed] [Google Scholar]
- 57. Murphy EF, Cotter PD, Hogan A, et al. 2012 Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut [DOI] [PubMed] [Google Scholar]
- 58. Wald A. Irritable bowel syndrome–diarrhoea. Best Pract Res Clin Gastroenterol. 2012;26:573–580. [DOI] [PubMed] [Google Scholar]
- 59. Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schellekens H, Finger BC, Dinan TG, Cryan JF. Ghrelin signalling and obesity: at the interface of stress, mood and food reward. Pharmacol Ther. 2012;135:316–326. [DOI] [PubMed] [Google Scholar]
- 61. Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. [DOI] [PubMed] [Google Scholar]
- 62. Kovatcheva-Datchary P, Arora T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract Res Clin Gastroenterol. 2013;27:59–72. [DOI] [PubMed] [Google Scholar]
- 63. Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int. 2012;95:50–60. [DOI] [PubMed] [Google Scholar]
- 64. Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki SI, Kuwahara A. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol. 2008;59(Suppl 2):251–262. [PubMed] [Google Scholar]
- 65. Karaki S, Tazoe H, Hayashi H, et al. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. J Mol Histol. 2008;39:135–142. [DOI] [PubMed] [Google Scholar]
- 66. Tazoe H, Otomo Y, Karaki S, et al. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed Res. 2009;30:149–156. [DOI] [PubMed] [Google Scholar]
- 67. Milligan G, Stoddart LA, Smith NJ. Agonism and allosterism: the pharmacology of the free fatty acid receptors FFA2 and FFA3. Br J Pharmacol. 2009;158:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. 2013;13:869–874. [DOI] [PubMed] [Google Scholar]
- 69. Dass NB, John AK, Bassil AK, et al. The relationship between the effects of short-chain fatty acids on intestinal motility in vitro and GPR43 receptor activation. Neurogastroenterol Motil. 2007;19:66–74. [DOI] [PubMed] [Google Scholar]
- 70. Holzer P, Reichmann F, Farzi A. Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides. 2012;46:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin Q. Submerged fermentation of Lactobacillus rhamnosus YS9 for gamma-aminobutyric acid (GABA) production. Braz J Microbiol. 2013;44:183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Steele RD. Blood-brain barrier transport of the alpha-keto acid analogs of amino acids. Fed Proc. 1986;45:2060–2064. [PubMed] [Google Scholar]
- 73. Vijay N, Morris ME. Role of monocarboxylate transporters in drug delivery to the brain. Curr Pharm Des. 2014;20:1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kekuda R, Manoharan P, Baseler W, Sundaram U. Monocarboxylate 4 mediated butyrate transport in a rat intestinal epithelial cell line. Dig Dis Sci. 2013;58:660–667. [DOI] [PubMed] [Google Scholar]
- 75. Maurer MH, Canis M, Kuschinsky W, Duelli R. Correlation between local monocarboxylate transporter 1 (MCT1) and glucose transporter 1 (GLUT1) densities in the adult rat brain. Neurosci Lett. 2004;355:105–108. [DOI] [PubMed] [Google Scholar]
- 76. Pellerin L, Bergersen LH, Halestrap AP, Pierre K. Cellular and subcellular distribution of monocarboxylate transporters in cultured brain cells and in the adult brain. J Neurosci Res. 2005;79:55–64. [DOI] [PubMed] [Google Scholar]
- 77. Pierre K, Pellerin L. Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J Neurochem. 2005;94:1–14. [DOI] [PubMed] [Google Scholar]
- 78. Pellerin L. How astrocytes feed hungry neurons. Mol Neurobiol. 2005;32:59–72. [DOI] [PubMed] [Google Scholar]
- 79. Rafiki A, Boulland JL, Halestrap AP, Ottersen OP, Bergersen L. Highly differential expression of the monocarboxylate transporters MCT2 and MCT4 in the developing rat brain. Neuroscience. 2003;122:677–688. [DOI] [PubMed] [Google Scholar]
- 80. Naccache PH, Faucher N, Caon AC, McColl SR. Propionic acid-induced calcium mobilization in human neutrophils. J Cell Physiol. 1988;136:118–124. [DOI] [PubMed] [Google Scholar]
- 81. Nakao S, Fujii A, Niederman R. Alteration of cytoplasmic Ca2+ in resting and stimulated human neutrophils by short-chain carboxylic acids at neutral pH. Infect Immun. 1992;60:5307–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakao S, Moriya Y, Furuyama S, Niederman R, Sugiya H. Propionic acid stimulates superoxide generation in human neutrophils. Cell Biol Int. 1998;22:331–337. [DOI] [PubMed] [Google Scholar]
- 83. DeCastro M, Nankova BB, Shah P, et al. Short chain fatty acids regulate tyrosine hydroxylase gene expression through a cAMP-dependent signaling pathway. Brain Res Mol Brain Res. 2005;142:28–38. [DOI] [PubMed] [Google Scholar]
- 84. Macfabe DF. Short-chain fatty acid fermentation products of the gut microbiome: implications in autism spectrum disorders. Microb Ecol Health Dis. 2012;23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bonini JA, Anderson SM, Steiner DF. Molecular cloning and tissue expression of a novel orphan G protein-coupled receptor from rat lung. Biochem Biophys Res Commun. 1997;234:190–193. [DOI] [PubMed] [Google Scholar]
- 86. Ichimura A, Hirasawa A, Hara T, Tsujimoto G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostagland Other Lipid Mediat. 2009;89:82–88. [DOI] [PubMed] [Google Scholar]
- 87. Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22:763–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cummings JH, Englyst HN. Gastrointestinal effects of food carbohydrate. Am J Clin Nutr. 1995;61:938S–945S. [DOI] [PubMed] [Google Scholar]
- 89. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. [DOI] [PubMed] [Google Scholar]
- 90. Brown AJ, Goldsworthy SM, Barnes AA, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. [DOI] [PubMed] [Google Scholar]
- 91. Xiong Y, Miyamoto N, Shibata K, et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci USA. 2004;101:1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ge H, Li X, Weiszmann J, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149:4519–4526. [DOI] [PubMed] [Google Scholar]
- 94. Hong YH, Nishimura Y, Hishikawa D, et al. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005;146:5092–5099. [DOI] [PubMed] [Google Scholar]
- 95. Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kruh J. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem. 1982;42:65–82. [DOI] [PubMed] [Google Scholar]
- 97. Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55–64. [DOI] [PubMed] [Google Scholar]
- 98. Marco ML, Peters TH, Bongers RS, et al. Lifestyle of Lactobacillus plantarum in the mouse caecum. Environ Microbiol. 2009;11:2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Denou E, Pridmore RD, Berger B, Panoff JM, Arigoni F, Brüssow H. Identification of genes associated with the long-gut-persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J Bacteriol. 2008;190:3161–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. McNulty NP, Yatsunenko T, Hsiao A, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol. 2014;30:120–127. [DOI] [PubMed] [Google Scholar]
- 102. Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Visschers RG, Luyer MD, Schaap FG, Olde Damink SW, Soeters PB. The gut-liver axis. Curr Opin Clin Nutr Metab Care. 2013;16:576–581. [DOI] [PubMed] [Google Scholar]
- 108. West DB, Delany JP, Camet PM, Blohm F, Truett AA, Scimeca J. Effects of conjugated linoleic acid on body fat and energy metabolism in the mouse. Am J Physiol. 1998;275:R667–672. [DOI] [PubMed] [Google Scholar]
- 109. O'Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol. 2012;152:189–205. [DOI] [PubMed] [Google Scholar]
- 110. Lee HY, Park JH, Seok SH, Baek MW, Kim DJ, Lee KE, Paek KS, Lee Y, Park JH. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta. 2006;1761:736–744. [DOI] [PubMed] [Google Scholar]
- 111. Lee K, Paek K, Lee HY, Park JH, Lee Y. Antiobesity effect of trans-10,cis-12-conjugated linoleic acid-producing Lactobacillus plantarum PL62 on diet-induced obese mice. J Appl Microbiol. 2007;103:1140–1146. [DOI] [PubMed] [Google Scholar]
- 112. Rosberg-Cody E, Stanton C, O'Mahony L, et al. Recombinant lactobacilli expressing linoleic acid isomerase can modulate the fatty acid composition of host adipose tissue in mice. Microbiology. 2011;157:609–615. [DOI] [PubMed] [Google Scholar]
- 113. Wall R, Ross RP, Shanahan F, et al. Impact of administered bifidobacterium on murine host fatty acid composition. Lipids. 2010;45:429–436. [DOI] [PubMed] [Google Scholar]
- 114. Sato M, Uzu K, Yoshida T, et al. Effects of milk fermented by Lactobacillus gasseri SBT2055 on adipocyte size in rats. Br J Nutr. 2008;99:1013–1017. [DOI] [PubMed] [Google Scholar]
- 115. Takemura N, Okubo T, Sonoyama K. Lactobacillus plantarum strain no. 14 reduces adipocyte size in mice fed high-fat diet. Exp Biol Med (Maywood). 2010;235:849–856. [DOI] [PubMed] [Google Scholar]
- 116. Hamad EM, Sato M, Uzu K, et al. Milk fermented by Lactobacillus gasseri SBT2055 influences adipocyte size via inhibition of dietary fat absorption in Zucker rats. Br J Nutr. 2009;101:716–724. [DOI] [PubMed] [Google Scholar]
- 117. Kadooka Y, Ogawa A, Ikuyama K, Sato M. The probiotic Lactobacillus gasseri SBT2055 inhibits enlargement of visceral adipocytes and upregulation of serum soluble adhesion molecule (sICAM-1) in rats. Int Dairy J. 2011;21:623–627. [Google Scholar]
- 118. Park DY, Ahn YT, Huh CS, Jeon SM, Choi MS. The inhibitory effect of Lactobacillus plantarum KY1032 cell Extract on the adipogenesis of 3T3-L1 cells. J Med Food. 2011;14:670–675. [DOI] [PubMed] [Google Scholar]
- 119. Ho JN, Choi JW, Lim WC, Kim MK, Lee IY, Cho HY. Kefir inhibits 3T3-L1 adipocyte differentiation through down-regulation of adipogenic transcription factor expression. J Sci Food Agr. 2013;93:485–490. [DOI] [PubMed] [Google Scholar]
- 120. Tanida M, Shen J, Maeda K, et al. High-fat diet-induced obesity is attenuated by probiotic strain Lactobacillus paracasei ST11 (NCC2461) in rats. Obes Res Clin Pract. 2008;2:159–169. [DOI] [PubMed] [Google Scholar]
- 121. Aronsson L, Huang Y, Parini P, et al. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4). PloS One. 2010;5:e13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Stanaszek PM, Snell JF, O'Neill JJ. Isolation, extraction, and measurement of acetylcholine from Lactobacillus plantarum. Appl Environ Microbiol. 1977;34:237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Nerstedt A, Nilsson EC, Ohlson K, et al. Administration of Lactobacillus evokes coordinated changes in the intestinal expression profile of genes regulating energy homeostasis and immune phenotype in mice. Br J Nutr. 2007;97:1117–1127. [DOI] [PubMed] [Google Scholar]
- 124. Fak F, Bäckhed F. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe−/− mice. PloS One. 2012;7:e46837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kelly G. Inulin-type prebiotics–a review: part 1. Altern Med Rev. 2008;13:315–329. [PubMed] [Google Scholar]
- 126. Delzenne NM, Cani PD, Neyrinck AM. Modulation of glucagon-like peptide 1 and energy metabolism by inulin and oligofructose: experimental data. J Nutr. 2007;137:2547S–2551S. [DOI] [PubMed] [Google Scholar]
- 127. Tarini J, Wolever TM. The fermentable fibre inulin increases postprandial serum short-chain fatty acids and reduces free-fatty acids and ghrelin in healthy subjects. Appl Physiol Nutr Metab. 2010;35:9–16. [DOI] [PubMed] [Google Scholar]
- 128. Dockray GJ. Gastrointestinal hormones and the dialogue between gut and brain [published online March 17, 2014]. J Physiol. doi:10.1113/jphysiol.2014.270850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Darzi J, Frost GS, Robertson MD. Do SCFA have a role in appetite regulation? Proc Nutr Soc. 2011;70:119–128. [DOI] [PubMed] [Google Scholar]
- 130. Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. [DOI] [PubMed] [Google Scholar]
- 131. Mawe GM, Hoffman JM. Serotonin signalling in the gut–functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2013;10:473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ruddick JP, Evans AK, Nutt DJ, Lightman SL, Rook GA, Lowry CA. Tryptophan metabolism in the central nervous system: medical implications. Expert Rev Mol Med. 2006;8:1–27. [DOI] [PubMed] [Google Scholar]
- 133. Watts SW, Morrison SF, Davis RP, Barman SM. Serotonin and blood pressure regulation. Pharmacol Rev. 2012;64:359–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Annu Rev Med. 2009;60:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Clarke G, Grenham S, Scully P, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666–673. [DOI] [PubMed] [Google Scholar]
- 136. Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J Psychiatr Res. 2008;43:164–174. [DOI] [PubMed] [Google Scholar]
- 137. Schwarcz R, Bruno JP, Muchowski PJ, Wu HQ. Kynurenines in the mammalian brain: when physiology meets pathology. Nat Rev Neurosci. 2012;13:465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Stone TW, Stoy N, Darlington LG. An expanding range of targets for kynurenine metabolites of tryptophan. Trends Pharmacol Sci. 2013;34:136–143. [DOI] [PubMed] [Google Scholar]
- 139. Bercik P, Verdu EF, Foster JA, et al. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology. 2010;139:2102–2112.e1. [DOI] [PubMed] [Google Scholar]
- 140. Clarke G, Quigley EM, Cryan JF, Dinan TG. Irritable bowel syndrome: towards biomarker identification. Trends Mol Med. 2009;15:478–489. [DOI] [PubMed] [Google Scholar]
- 141. Clarke G, Fitzgerald P, Cryan JF, Cassidy EM, Quigley EM, Dinan TG. Tryptophan degradation in irritable bowel syndrome: evidence of indoleamine 2,3-dioxygenase activation in a male cohort. BMC Gastroenterol. 2009;9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Clarke G, McKernan DP, Gaszner G, Quigley EM, Cryan JF, Dinan TG. A distinct profile of tryptophan metabolism along the kynurenine pathway downstream of toll-like receptor activation in irritable bowel syndrome. Front Pharmacol. 2012;3:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Fitzgerald P, Cassidy Eugene M, et al. Tryptophan catabolism in females with irritable bowel syndrome: relationship to interferon-gamma, severity of symptoms and psychiatric co-morbidity. Neurogastroenterol Motil. 2008;20:1291–1297. [DOI] [PubMed] [Google Scholar]
- 144. Schroecksnadel K, Winkler C, Wirleitner B, Schennach H, Fuchs D. Aspirin down-regulates tryptophan degradation in stimulated human peripheral blood mononuclear cells in vitro. Clin Exp Immunol. 2005;140:41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Schroecksnadel S, Sucher R, Kurz K, Fuchs D, Brandacher G. Influence of immunosuppressive agents on tryptophan degradation and neopterin production in human peripheral blood mononuclear cells. Transpl Immunol. 2011;25:119–123. [DOI] [PubMed] [Google Scholar]
- 146. Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011;106:329–336. [DOI] [PubMed] [Google Scholar]
- 147. McKernan DP, Gaszner G, Quigley EM, Cryan JF, Dinan TG. Altered peripheral toll-like receptor responses in the irritable bowel syndrome. Aliment Pharmacol Ther. 2011;33:1045–1052. [DOI] [PubMed] [Google Scholar]
- 148. McKernan DP, Nolan A, Brint EK, et al. Toll-like receptor mRNA expression is selectively increased in the colonic mucosa of two animal models relevant to irritable bowel syndrome. PloS One. 2009;4:e8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Milligan TW, Doran TI, Straus DC, Mattingly SJ. Growth and amino acid requirements of various strains of group B streptococci. J Clin Microbiol. 1978;7:28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology. 2013;159:402–410. [DOI] [PubMed] [Google Scholar]
- 151. Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. [DOI] [PubMed] [Google Scholar]
- 152. Hsiao EY. Immune dysregulation in autism spectrum disorder. Int Rev Neurobiol. 2013;113:269–302. [DOI] [PubMed] [Google Scholar]
- 153. Raboni S, Bettati S, Mozzarelli A. Tryptophan synthase: a mine for enzymologists. Cell Mol Life Sci. 2009;66:2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Yanofsky C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA. 2007;13:1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Jimenez E, Ladero V, Chico I, et al. Antibiotic resistance, virulence determinants and production of biogenic amines among enterococci from ovine, feline, canine, porcine and human milk. BMC Microbiol. 2013;13:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Shishov VA, Kirovskaia TA, Kudrin VS, Oleskin AV. Amine neuromediators, their precursors, and oxidation products in the culture of Escherichia coli K-12 [in Russian]. Prikl Biokhim Mikrobiol. 2009;45:550–554. [PubMed] [Google Scholar]
- 157. Wang YW, Devkota S, Musch MW, et al. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PloS One. 2010;5:e13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Gosselin D, Rivest S. MyD88 signaling in brain endothelial cells is essential for the neuronal activity and glucocorticoid release during systemic inflammation. Mol Psychiatry. 2008;13:480–497. [DOI] [PubMed] [Google Scholar]
- 159. Zacharowski K, Zacharowski PA, Koch A, et al. Toll-like receptor 4 plays a crucial role in the immune-adrenal response to systemic inflammatory response syndrome. Proc Natl Acad Sci USA. 2006;103:6392–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Sudo N, Chida Y, Aiba Y, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Crumeyrolle-Arias M, Jaglin M, Bruneau A, et al. Absence of the gut microbiota enhances anxiety-like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology. 2014;42:207–217. [DOI] [PubMed] [Google Scholar]
- 162. Allen AP, Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Biological and psychological markers of stress in humans: focus on the Trier Social Stress Test. Neurosci Biobehav Rev. 2014;38:94–124. [DOI] [PubMed] [Google Scholar]
- 163. Kennedy PJ, Quigley E, Cryan JF, Dinan TG, Clarke G. A sustained hypothalamic-pituitary-adrenal axis response to acute psychosocial stress in irritable bowel syndrome [published online March 14, 2014]. Psychol Med. doi:10.1017/S003329171400052X. [DOI] [PubMed] [Google Scholar]
- 164. Dinan TG, Scott LV. Anatomy of melancholia: focus on hypothalamic-pituitary-adrenal axis overactivity and the role of vasopressin. J Anat. 2005;207:259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Gold PW, Chrousos GP. Melancholic and atypical subtypes of depression represent distinct pathophysiological entities: CRH, neural circuits, and the diathesis for anxiety and depression. Mol Psychiatry. 2013;18:632–634. [DOI] [PubMed] [Google Scholar]
- 166. Gold PW, Gabry KE, Yasuda MR, Chrousos GP. Divergent endocrine abnormalities in melancholic and atypical depression: clinical and pathophysiologic implications. Endocrinol Metab Clin North Am. 2002;31:37–62, vi [DOI] [PubMed] [Google Scholar]
- 167. Kennedy PJ, Clarke G, Quigley EM, Groeger JA, Dinan TG, Cryan JF. Gut memories: towards a cognitive neurobiology of irritable bowel syndrome. Neurosci Biobehav Rev. 2012;36:310–340. [DOI] [PubMed] [Google Scholar]
- 168. Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol. 1999;35:146–155. [PubMed] [Google Scholar]
- 169. O'Mahony SM, Marchesi JR, Scully P, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263–267. [DOI] [PubMed] [Google Scholar]
- 170. Bailey MT, Dowd SE, Parry NM, Galley JD, Schauer DB, Lyte M. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun. 2010;78:1509–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun. 2011;25:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Young EA, Abelson J, Lightman SL. Cortisol pulsatility and its role in stress regulation and health. Front Neuroendocrinol. 2004;25:69–76. [DOI] [PubMed] [Google Scholar]
- 173. Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–1188. [DOI] [PubMed] [Google Scholar]
- 174. Gareau MG, Jury J, MacQueen G, Sherman PM, Perdue MH. Probiotic treatment of rat pups normalises corticosterone release and ameliorates colonic dysfunction induced by maternal separation. Gut. 2007;56:1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. McKernan DP, Fitzgerald P, Dinan TG, Cryan JF. The probiotic Bifidobacterium infantis 35624 displays visceral antinociceptive effects in the rat. Neurogastroenterol Motil. 2010;22:1029–1035.e1268. [DOI] [PubMed] [Google Scholar]
- 176. Messaoudi M, Lalonde R, Violle N, et al. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011;105:755–764. [DOI] [PubMed] [Google Scholar]
- 177. Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37:1369–1378. [DOI] [PubMed] [Google Scholar]
- 178. Freestone PP, Williams PH, Haigh RD, Maggs AF, Neal CP, Lyte M. Growth stimulation of intestinal commensal Escherichia coli by catecholamines: a possible contributory factor in trauma-induced sepsis. Shock. 2002;18:465–470. [DOI] [PubMed] [Google Scholar]
- 179. Lyte M, Ernst S. Catecholamine induced growth of gram negative bacteria. Life Sci. 1992;50:203–212. [DOI] [PubMed] [Google Scholar]
- 180. Jeffery IB, O'Toole PW, Öhman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. [DOI] [PubMed] [Google Scholar]
- 181. de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45–S56. [DOI] [PubMed] [Google Scholar]
- 182. Dinan TG, Cryan JF. Melancholic microbes: a link between gut microbiota and depression? Neurogastroenterol Motil. 2013;25:713–719. [DOI] [PubMed] [Google Scholar]
- 183. Park AJ, Collins J, Blennerhassett PA, et al. Altered colonic function and microbiota profile in a mouse model of chronic depression. Neurogastroenterol Motil. 2013;25:733–e575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Booijink CC, Zoetendal EG, Kleerebezem M, de Vos WM. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2007;2:285–295. [DOI] [PubMed] [Google Scholar]
- 185. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Clarke G, Cryan JF, Dinan TG, Quigley EM. Review article: probiotics for the treatment of irritable bowel syndrome–focus on lactic acid bacteria. Aliment Pharmacol Ther. 2012;35:403–413. [DOI] [PubMed] [Google Scholar]
- 187. Shanahan F. A commentary on the safety of probiotics. Gastroenterol Clin North Am. 2012;41:869–876. [DOI] [PubMed] [Google Scholar]
- 188. Dinan TG, Stanton C, Cryan JF. Psychobiotics: a novel class of psychotropic. Biol Psychiatry. 2013;74:720–726. [DOI] [PubMed] [Google Scholar]
- 189. Quigley EM. Prebiotics and probiotics: their role in the management of gastrointestinal disorders in adults. Nutr Clin Pract. 2012;27:195–200. [DOI] [PubMed] [Google Scholar]
- 190. Guarner F, Khan AG, Garisch J, et al. ; World Gastroenterology Organisation. World Gastroenterology Organisation Global Guidelines: probiotics and prebiotics October 2011. J Clin Gastroenterol. 2012;46:468–481. [DOI] [PubMed] [Google Scholar]
- 191. Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9:88–96. [DOI] [PubMed] [Google Scholar]
- 192. Kelly CP. Fecal microbiota transplantation–an old therapy comes of age. N Engl J Med. 2013;368:474–475. [DOI] [PubMed] [Google Scholar]
- 193. Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology. 2013;145:946–953. [DOI] [PubMed] [Google Scholar]
- 194. Lyte M. Microbial endocrinology as a basis for improved l-DOPA bioavailability in Parkinson's patients treated for Helicobacter pylori. Med Hypotheses. 2010;74:895–897. [DOI] [PubMed] [Google Scholar]
- 195. Davey KJ, Cotter PD, O'Sullivan O, et al. Antipsychotics and the gut microbiome: olanzapine-induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Transl Psychiatry. 2013;3:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Munoz-Bellido JL, Munoz-Criado S, Garcìa-Rodrìguez JA. Antimicrobial activity of psychotropic drugs: selective serotonin reuptake inhibitors. Int J Antimicrob Agents. 2000;14:177–180. [DOI] [PubMed] [Google Scholar]
- 197. Coates ME. Gnotobiotic animals in research: their uses and limitations. Lab Animals. 1975;9:275–282. [DOI] [PubMed] [Google Scholar]
- 198. Napier C, Pearce SH. Autoimmune Addison's disease. Presse Med. 2012;41:e626–e635. [DOI] [PubMed] [Google Scholar]
- 199. Hirayama K, Uetsuka K, Kuwabara Y, Tamura M, Itoh K. Vitamin K deficiency of germfree mice caused by feeding standard purified diet sterilized by gamma-irradiation. Exp Anim. 2007;56:273–278. [DOI] [PubMed] [Google Scholar]
- 200. Hyland NP, Cryan JF. A gut feeling about GABA: focus on GABA(B) receptors. Front Pharmacol. 2010;1:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Ozogul F, Kuley E, Ozogul Y, Ozogul I. The function of lactic acid bacteria on biogenic amines production by food-borne pathogens in arginine decarboxylase broth. Food Sci Technol Res. 2012;18:795–804. [Google Scholar]
- 202. Ozogul F. Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. Eur Food Res Technol. 2004;219:465–469. [Google Scholar]
- 203. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]





