Abstract
The specification of cell fate is critical for proper cell differentiation and organogenesis. In endocrine tissues, this process leads to the differentiation, often a multistep process, of hormone-producing cells. This process is driven by a combination of transcription factors (TFs) that includes general factor, tissue-restricted, and/or cell-restricted factors. The last 2 decades have seen the discovery of many TFs of restricted expression and function in endocrine tissues. These factors are typically critical for expression of hormone-coding genes as well as for differentiation and proper function of hormone-producing cells. Further, genes encoding these tissue-restricted TFs are themselves subject to mutations that cause hormone deficiencies. Although the model that emerged from these 2 decades is one in which a specific combination of TFs drives a unique cell specification and genetic program, recent findings have led to the discovery of TFs that have the unique property of being able to remodel chromatin and thus modify the epigenome. Most importantly, such factors, known as pioneer TFs, appear to play critical roles in programming the epigenome during the successive steps involved in cell specification. This review summarizes our current understanding of the mechanisms for pioneer TF remodeling of chromatin. Currently, very few TFs that have proven pioneer activity are known, but it will be critical to identify these factors and understand their mechanisms of action if we are to harness the potential of regenerative therapies in endocrinology.
The determination of cell fate is a stepwise process that involves early events of tissue specification followed by the subsequent action of a combination of transcription factors (TFs). In fully differentiated cells, the result of this developmental process is observed at regulatory elements, promoters, and enhancers, where multiple TFs are bound. A unique combination of TFs defines the identity of a cell lineage and its genetic program. Enhancers thus appear like an assemblage of DNA-binding sites for TFs that are either ubiquitous or more restricted in their expression pattern. Endocrine cell-specific genes provide good examples of enhancers driven by a TF combination that includes one or a few tissue- or cell-restricted factors. It is noteworthy that even the most cell-restricted TF is not sufficient on its own to drive cell-specific transcription and that its vital role in transcriptional activation may only be exerted in combination with other factors. The last decades have taught us a lot about the contribution of TFs for cell specificity, signal dependence, and quantitative activation of transcription. However, genome-wide comparisons of active enhancers in different cells established that a cell's identity and gene expression program are a reflection of the repertoire of enhancers active in a given cell. Hence, the simplistic model of a TF combination to explain cell identity is not sufficient, and some epigenetic mechanism must account for the choice of active enhancers (and genes) in a given cell (reviewed in Ref. 1).
It was suspected for a long time that tissues of different developmental origins are specified early in development by global regulators of transcription and/or chromatin organization. One early concept that supported this model is that of selector genes, which were defined in the mid-1970s based on work in Drosophila (2). Selector gene expression in early developmental domains specifies the subsequent fates that can be derived from a given developmental domain. It was assumed and later shown that the action of selector genes is accompanied by global changes in chromatin organization, including rearrangement of the accessible domains of the chromatin-embedded genome. The notion that some regulatory factors could change genome accessibility in chromatin was a necessary correlate of these models. In this context, the notion of pioneer TFs evolved as factors that have the unique ability to alter the chromatin environment, either increase/open or decrease/reduce the accessibility of a network of regulatory sequences, hence making them accessible (or not) for the action of other TFs. The concept of pioneer TFs thus represents a mechanism that selector genes may use to affect the ultimate fate of cells deriving from a developmental field marked by their expression.
Although these concepts have been around for some time, only in the last few years were specific TFs proven to have pioneer activity. A renewed interest in pioneer TFs was triggered by the realization that cell fate could be reprogrammed toward an embryonic stem cell fate such as that in induced pluripotent stem (iPS) cells. Further, the possibility of reprogramming iPS cells toward specific fates, such as pancreatic β cells or other endocrine cells for regenerative therapies opens tremendous opportunities. Hence, there is considerable interest in identifying pioneer TFs and understanding their unique properties. The fact that a unique cascade of pioneer factors may dictate a cell fate would thus show how to reprogram cells for therapeutic purposes.
In this review, I will first briefly outline the molecular basis for transcriptional control of cell fate, in particular through the combinatorial use of groups of TFs to set the stage in which pioneer factors exert their action. These aspects have been abundantly reviewed, and they define the result of the cell fate specification process and hence the result achieved after pioneer TF action. I will then discuss 2 notions of pioneer action that represent different mechanisms and that have been labeled as either passive or active pioneer action in recent reviews (3).
Both mechanisms of the pioneering action are relevant for endocrine function. An example of a passive pioneer action is the role of forkhead box A (FoxA) TFs in estrogen receptor (ER) action, whereas an active pioneer role is exerted by Pax7 to specify the intermediate (relative to anterior) pituitary tissue and its constituent lineage.
Transcriptional Control of Cell Fate
Although most genes rely on the concerted action of multiple enhancers for their expression, individual enhancers often epitomize one feature of its target gene's expression, be it tissue specificity, hormone activation, and/or temporal window of expression. Individual enhancers thus represent functional units for control of gene expression. The properties of enhancers are usually shown in transgenic mouse assays that reveal their restricted spectrum of activity (4, 5), for example, in terminally differentiated lineages of the pituitary, pancreas, adrenal, or thyroid (6–9). Typically, cell-specific enhancers use the same set of TFs as those that direct cell fate: the study of such enhancers has thus led to identification of many tissue-specific TFs and provided insight on their mechanism of action, such as Pit1 in the pituitary (10), Pdx1 in pancreas (7), and Nkx2.1 and Pax8 in thyroid (9). In addition, cell-specific TFs further determine cell identity and gene expression programs, such as Tpit in pituitary proopiomelanocortin (POMC) cells (11–13), SF1 in steroid-producing tissues (8), or Nkx6.1 in β cells (7). The detailed analyses of enhancers targeted by these tissue and cell-restricted factors have highlighted the combinatorial use of factors with related developmental history for setting up lineage-specific transcription. Numerous protein-protein interactions between DNA-binding TFs and with their positive and negative coregulators are critical for assembly of protein complexes required for activity. Typical enhancers represent 1 to 2 kb of DNA that is marked by unique coregulators and chromatin marks (Figure 1). For example, the general coactivator p300 marks enhancers poised for activity (4).
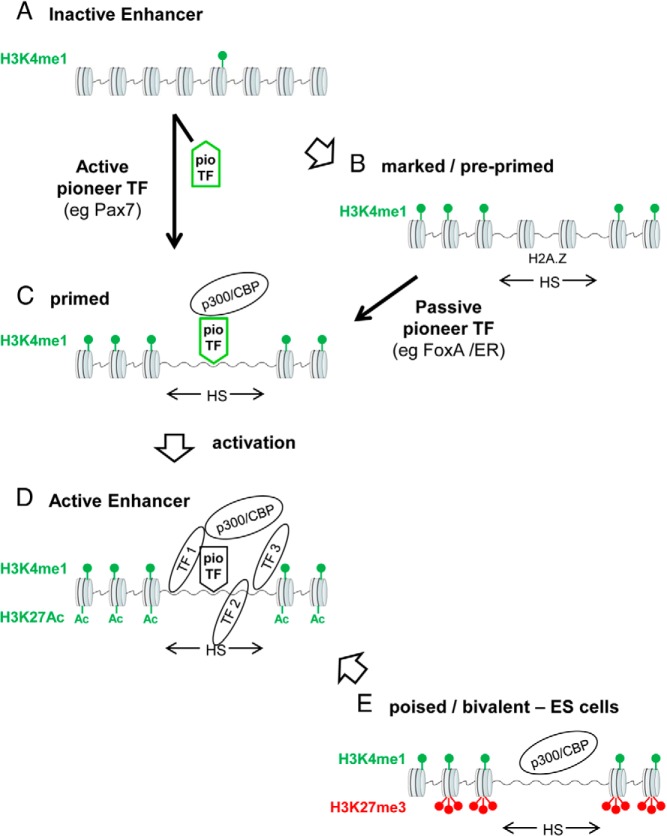
Figure 1.
Chromatin remodeling, enhancer activation, and pioneer transcription factors. A, In differentiated cells, inactive enhancers have no distinctive mark that allows their identification. They have a low level of H3K4me1. B, A subset of enhancers is marked or “preprimed” by the presence of H3K4me1 at the borders of the enhancer. These enhancers also appear to have more dynamic nucleosomes marked by the presence of the variant histone H2A.Z, and they may be revealed by their sensitivity (HS) to DNase1 or by FAIRE. The process that is responsible for this enhancer marking is not clear and may involve the action of an active pioneer transcription factor (pioTF) that has yet to be identified. Such preprimed enhancers may be targets of passive pioneer TFs that facilitate the recruitment of other TFs. An example of this situation is the activation of marked enhancers by the pioneer factor FoxA that facilitates recruitment of the ER. C, Primed enhancers have an accessible chromatin structure revealed by DNase hypersensitivity or FAIRE (HS) together with flanking H3K4me1, and they are usually occupied by the general transcriptional coactivator p300/cAMP response element-binding protein-binding protein (CBP). Inactive enhancers can be primed through the action of active pioneer TFs that directly access their target DNA sequence on nucleosomal chromatin and lead to chromatin opening. The active pioneer TF Pax7 fulfills such a role in intermediate pituitary cells. D, The activation of primed enhancers results from recruitment of various TFs and coactivators, and this is associated with H3K27Ac. E, A particular class of enhancers are observed in ES cells, in which these so-called “poised” or “bivalent” enhancers exhibit marks of both activated and repressed chromatin, namely H3K4me1 flanking the enhancer together with the H3K27me3 mark that blocks enhancer activity. They also harbor p300/CBP, but it appears to be inactive in this context. These bivalent enhancers are often associated with key developmental regulatory genes. Their activation requires H3K27me3 demethylation (by proteins such as UTX) and subsequent K27 acetylation by p300/CBP.
Active enhancers include a region of accessible DNA revealed either through use of DNase I sensitivity or by the more recent technique of formaldehyde-assisted isolation of regulatory elements (FAIRE) (14). When applied genome-wide together with high-throughput sequencing (seq), these techniques of DNase-seq and FAIREseq build a snapshot of the genome's region of greater physical accessibility, including enhancers and promoters (15). For active enhancers, these regions of accessible DNA are flanked by 2 histone marks, namely, monomethyl lysine 4 histone H3 (H3K4me1) and acetylated lysine 27 histone H3 (H3K27Ac) (1, 16, 17). Thus, a bimodal pattern of these 2 marks flanking a region of accessible DNA delineates active enhancers (Figure 1D). To some extent, the height of these features in chromatin immunoprecipitation sequencing (ChIPseq) profiles is correlated with enhancer activity, as revealed by the introduction (or activation) of a new TF binding the enhancer (18). In addition, active enhancers make physical links with the promoters of their target genes. These links involve structural proteins such as the cohesins but also a regulator of signal-activated transcription, the Mediator complex (19). Active promoters are also marked by H3K4 methylation, in particular by H3K4me3 as opposed to the H3K4me1 mark at enhancers; both active promoters and enhancers show enrichment of H3K4me2, but this mark is not as discriminatory as the monomethyl and trimethyl marks (17).
Thus, enhancers are the targets of TF combinations that direct lineage-specific transcription, and it was argued that such combinatorial action may suffice to account for most cell fate determination as proposed for macrophage-related lineages (20). In such cases, it is presumed that a subset of active enhancers are targeted, implying that their chromatin organization and accessibility were preset. This also implies that at some point in the developmental history of these lineages, the relevant set of active enhancers had to be activated, and at this critical developmental decision point pioneer TFs are likely to be involved.
Active and Passive Pioneers
Historically, the notion of pioneer TFs has referred to factors that access and “open” condensed chromatin to remodel its structure and allow access to other TFs. This generic definition does not discriminate between the initial and final states of chromatin, which may differ substantially. Hence, factors that trigger qualitative, rather than just quantitative, changes in chromatin structure best fit the definition of true pioneer TFs. Those were labeled active pioneers by Zaret and Carroll (3) to differentiate them from another usage of the term pioneer. For example, TFs that lead to the appearance of bimodal histone H3 marks flanking enhancers fit this definition.
The term pioneer was also used to describe the action of the FoxA1 TF that promotes recruitment of ER to its genomic targets (reviewed in Ref. 21). This usage came from the observation that a large proportion of ER recruitment sites determined by ChIPseq also harbor FoxA1 (22, 23) and that FoxA1 promotes ER recruitment and action, not the reverse (24). This action was labeled as passive pioneer. Because not all ER- or FoxA1-binding sites overlap, it is likely that other factors are involved in promoting ER recruitment (25). Notwithstanding the true active pioneer activity of FoxA1 (discussed below), it appears that FoxA1 recruitment occurs mostly at genomic loci that already exhibit H3K4 methylation and FAIRE signals (Figure 1, B and C), although the intensity of these marks is increased by FoxA1 and ER recruitment (24, 26–28). Also, these loci already contain the histone H2A.Z (Figure 1B) that marks active regulatory sequences (29). Further, FoxA1 appears to have a maintenance role for these marks (24, 26). Thus, FoxA1 binding appears to occur at already accessible putative enhancer regions, but this binding leads to further accessibility and enhanced histone methylation marks. FoxA1 was also shown to stabilize recruitment of the androgen and glucocorticoid receptors, and hence it may facilitate the recruitment of a variety of nuclear receptors (25, 30–32). Other TFs that facilitate ER or androgen receptor recruitment are the GATA factors (30, 33) and the homeobox factor Pbx1, which also interacts directly with ER (3, 34). Nuclear receptors themselves may act as pioneers as they were shown to enhance chromatin accessibility upon enhancer binding; for example, this was shown for glucocorticoid receptor (35), ER (36), peroxisome proliferator-activated receptor γ (37), and the ecdysone receptor (38). However, it is not clear whether these actions are associated with qualitative changes in chromatin organization in addition to increased accessibility revealed by DNase-seq or FAIREseq.
The term “passive pioneer” was coined to reflect this interaction that occurs rapidly and does not require, but may enhance, chromatin remodeling (3). Thus, passive pioneers act on previously “marked” chromatin (Figure 1B) and further enhance and maintain the marks of active enhancers, such as the H3K4me1 marks that flank FAIRE peaks (Figure 1C). FoxA1 exerts these activities for ER and glucocorticoid receptor, but it also has active pioneer activity, ie, opening of condensed chromatin, as discussed below.
Of pioneers and Cell Fate (Re)-Programming
Beyond the selector gene concept, the first molecular evidence for TF-dependent chromatin remodeling involved in cell fate specification was provided by the work of Cirillo et al (39) on the role of FoxA1 in specification of endoderm fates. Since that time, much has been learned about the mechanisms and protein complexes involved in chromatin remodeling, and many reviews have dealt with this, either from the perspective of chromatin marks (1, 40, 41) or from that of the protein complexes responsible for chromatin remodeling (42–44). The salient features of our current understanding of chromatin marks and remodeling are summarized below to provide the backdrop within which the action of TFs operates.
The features of active enhancers as described above include regions of accessible DNA flanked by peaks of H3K4me1 and H3K27Ac (16, 17). They also exhibit a peak of recruitment for the general coactivator p300 (4). In contrast, the same enhancer sequences condensed in their inaccessible or closed chromatin state may typically harbor the repressive marks H3K27me3 and/or H3K9me3 (16). This later mark is primarily associated with heterochromatin. Similarly, active promoters are marked by the presence of H3K4me3, among other marks (17). How are these different chromatin marks introduced or erased from chromatin? Genes encoding proteins of large complexes responsible for implementation of these activating or repressive marks were first identified through genetic studies in Drosophila. Thus, the trithorax complex is responsible for introduction of activating marks into chromatin and in mammals, there are 4 similar complexes containing the mixed-lineage leukemia (MLL) 1 to 4 proteins at their core (for reviews, see Refs. 44, 45). The Drosophila polycomb (Pc) complex is responsible for introduction of repressive marks in chromatin and the mammalian equivalent complexes are the PRC1 and PRC2 complexes (reviewed in Refs. 42, 43, 46). Although the presence of activating and repressive marks in chromatin together with the protein complexes responsible for introducing these marks suggests a binary chromatin world, it was also realized that some enhancers, in particular enhancers for key developmental genes, appear to be poised for activation (Figure 1E) and contain both the activating H3K4me1 and repressive H3K27me3 marks (41, 47, 48). Upon activation by the appropriate developmental signals, it appears that the repressive H3K7me3 mark is lost and the poised enhancers become active. Clearly, the simplistic binary view of chromatin marks reflects our still limited understanding of the numerous marks and their interplay; it is likely that a continuum of marks and readers of these marks is active to continuously interpret and modify the epigenome.
The trithorax and MLL complexes create and maintain an active chromatin environment
The first mammalian trithorax homolog to be identified was MLL1, and it was identified at translocations associated with different leukemias (44). There are four MLL genes, MLL1 to MLL4, and they are fulfilling nonredundant functions because they target different genomic targets. The MLL proteins are found in complexes containing some common proteins such as Ash2L, RBBP5, and WDR5 and also complex-specific proteins, and current evidence suggests that there are multiple forms of these complexes of yet unknown specificity and function. All MLL complexes appear to direct H3K4 trimethylation. The MLL1/2 complexes associated with the tumor suppressor menin (49) were shown to be critical for H3K4 trimethylation of Hox genes (50), whereas the MLL3/4 complexes are dispensable for this function. In contrast, MLL3/4 complexes are involved in H3K4 trimethylation of retinoic acid receptor target genes (51), and they may be more present at intergenic regions containing enhancers (52) and thus be particularly important for H3K4 monomethylation of enhancers. In addition, the latter complexes include the protein UTX (KDM10A) that removes the H3K27me3 repressive mark; this activity may serve to counteract the repressive action of Pc-related complexes. In addition to the MLL complexes, mammalian cells also have Set1a/b complexes that mediate H3K4 trimethylation. Collectively, these enzyme complexes have been primarily associated with H3K4 trimethylation, but the mechanism driving their recruitment to specific genomic targets, either promoters and/or enhancers, and their enzymatic activation remain poorly understood.
Although the MLL complexes appear to fulfill the function of histone writers, another set of protein complexes constitute the readers of activating histone marks to facilitate transcription. These include the SWI/SNIF complexes containing the related ATPases Brg1 and Brm that bind to acetylated histones via their bromodomain and mediate chromatin remodeling. There are also numerous complexes containing CHD proteins (CHD1–CHD6) that bind H3K4me3 via their chromodomains and regulate a variety of processes favoring gene expression, such as deposition of histone H3.3 and counteraction of Pc repression.
The repressive Pc complexes PRC1 and PRC2
The 2 PRC complexes fulfill complementary functions with PRC 2 as the writer and PRC 1 as the reader (42, 43). The PRC 2 complex is composed of 3 core proteins, Ezh2 (enhancer of zest 2) or its homolog Ezh1, Eed (embryonic ectoderm development), and Suz12 (suppressor of zest 12). This complex has methylase activity that results in H3K27 monomethylation, dimethylation, and trimethylation (53, 54). The enzyme activity is contained in the Ezh protein, but the other proteins are required for catalytic activity; in addition, Eed binds H3K27me3, and this function may serve the propagation role of the PRC1 and PRC2 complexes, ie, spreading of the repressive H3K27me3 mark to cover an entire locus (55, 56).
The PRC1 complex is the reader and effector. It contains a ring finger protein (either Ring1a/RNF1 or Ring1b/RNF2); the RNF proteins are E3 ubiquitin ligases, and they ubiquitinate lysine 119 of H2A. H2AK119Ub has been suggested to prevent methylation of H3K4 (57, 58). It was also suggested that it may impair the eviction of the H2A-H2B dimers from nucleosomes that occur during transcription elongation (59). The PRC1 complex also includes a chromodomain protein of the Cbx family; the homolog of Drosophila Pc. These proteins bind H3K27me3 and H3K9me3. Finally, the PRC complex also contains one member of the Pcgf family of ring finger proteins. These proteins are related to the Drosophila protein Psc, and they act as cofactors for Ring1A and Ring1B activity.
The PRC complexes are present at repressed genes and, in particular, over the CpG islands that are often present upstream of the gene (60, 61). In ES cells, it was found that thousands of genes playing very different functions are repressed by PRC complexes. These genes have different organizations and do not share a common sequence motif that may act to recruit PRC complexes. The exact mechanisms that recruit PRC complexes to specific loci remain under active investigation. Although the PRC2 complex appears to have affinity for CpG islands, possibly through the interaction with associated proteins, recruitment of PRC1 to chromatin depends in part on the affinity of Cbx proteins for H3K27me3. For both complexes, some reports have suggested recruitment to specific target sequences through interaction with DNA-binding TFs (42).
Condensed chromatin and accessibility of DNA
The highly condensed heterochromatin is associated with methyl cytosines and the histone mark H3K9me3. In general, repressed genes and enhancers are marked by the H3K27me3 modification, and bivalent or poised enhancers, in particular, are marked by both this repressive mark and the activating H3K4me1 mark (47, 62); the poised enhancers also miss the activating mark H3K27Ac (Figure 1E). These bivalent enhancers were mostly observed in ES cells, and they also appear to recruit both PRC and MLL complexes. Whether pioneer TFs can access DNA in these different forms of repressed chromatin remains unknown. It is possible that different classes of pioneer TFs may access different forms of repressed chromatin. However, the small number of TFs currently known as chromatin-opening pioneers target a large number of loci and trigger extensive chromatin remodeling.
Currently Documented Pioneer Factors
There are still relatively few TFs that have well documented pioneer activity, namely factors shown to “open” chromatin and permit accessibility to other TFs. These factors may also have a negative impact on chromatin, leading to establishment of repressive chromatin environments, but this aspect is still even less understood than the activating function described below for the known well-characterized pioneer TFs.
Forkhead FoxA factors and endoderm determination
The FoxA1 and FoxA2 genes are expressed in foregut endoderm, and they were shown to be essential for liver development (39). These factors, together with Gata4, were shown to first occupy the liver-specific albumin enhancer and hence were suggested to act as pioneers for this enhancer. Direct assessment of the ability of these factors to bind compacted chromatin and create regions of DNase hypersensitivity indicated that FoxA1 is the most active in this respect (39). In addition to FoxA1, FoxE2 and FoxO were also shown to open compacted chromatin. Further, the Caenorhabditis elegans homolog of FoxA, PHA-4, can trigger decompaction of chromosomal domains (63) and recruitment of the histone variant of H2A.Z to promoters (64). Analysis of PHA-4 action in C elegans suggested that the sites with highest affinity for this factor are activated earlier in development than those with lesser affinity. DNA-binding affinity may thus be a determinant of the pioneering ability of TFs. This idea was not born out by analyses of the FoxA1 target sites identified in mouse liver, but these analyses may not have discriminated sites of FoxA action where FoxA plays either pioneering or regular transcription activator functions. Although FoxA1 and FoxA2 were shown to be critical for embryonic liver development presumably through their pioneering action, they are not required to maintain local chromatin organization and a third member of this family, FoxA3, compensates in part in later liver development (65). The pioneering activity of FoxA factors has been associated with the DNA-binding domain of these factors but also with a C-terminal domain of the proteins that interact with core histones: this dual interaction with DNA and chromatin proteins appeared critical for in vitro chromatin opening (39). Pioneer TFs may thus have a unique chromatin interaction property that allows them to access DNA in compacted nucleosomal chromatin.
Pax7 as selector of pituitary intermediate lobe melanotrope identity
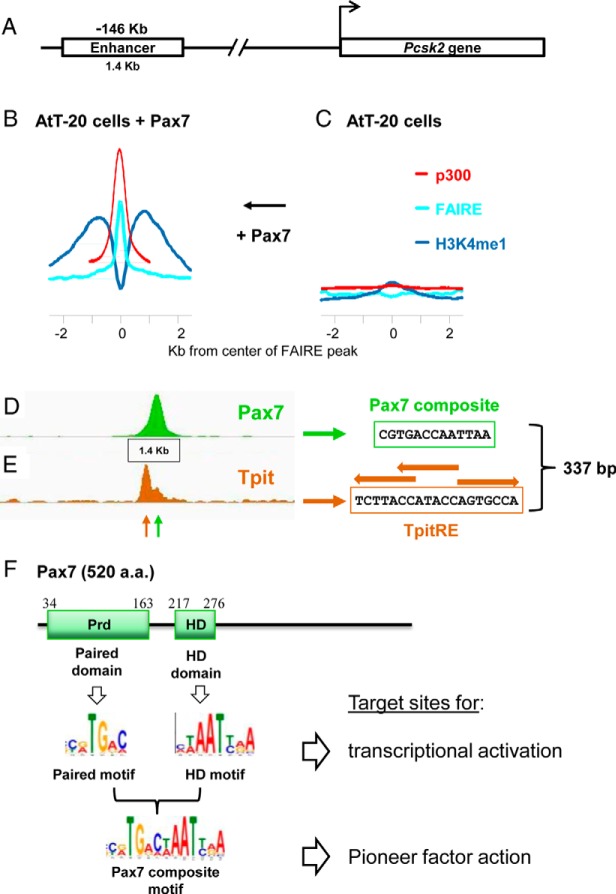
The paired-homeodomain TF Pax7 was well investigated for its role in the transition of muscle progenitor cells into the differentiation pathway (66). Its restricted expression in the pituitary intermediate lobe suggested that it may play a role in differentiation of the POMC-expressing melanotrope cells that constitute the endocrine cells of this tissue. The other pituitary compartment, the anterior lobe, also contains a lineage expressing the POMC gene, the corticotropes, and prior work had shown that terminal differentiation of both POMC-expressing lineages is accomplished by the Tbox factor, Tpit (11). Pax7 was shown to not only act as a positive regulator of the melanotrope fate but also to repress the corticotrope fate, such that the Pax7 mutant pituitary intermediate lobe cells switch fate from melanotrope to corticotrope (67). Pax7 is expressed before Tpit in melanotropes and, in fact, its expression overlaps transiently with the pituitary stem cell progenitor marker Sox2. Gain-of-function experiments in the corticotrope model cell line AtT-20 showed that Pax7 can reprogram these cells to express melanotrope markers and that this reprogramming entails significant chromatin remodeling (Figure 2, A–C). In fact, Pax7 action leads to either appearance or disappearance of >6000 Tpit-binding sites genome-wide. Hence, Pax7 action on chromatin has a major effect on genome accessibility for Tpit (Figure 2, D and E). In particular, novel Tpit binding subsequent to Pax7 action is associated with increased accessibility of the chromatin (Figure 2B compared with 2C) as assessed by FAIREseq, and this chromatin opening is associated with appearance of the bimodal marks of H3K4me1 that typically flank active enhancers (67). The >3000 sites that undergo chromatin opening after Pax7 action include enhancers that direct melanotrope-specific gene expression, such as the protein convertase 2 (PCSK2) gene that is activated after Pax7 action on an enhancer located 146 kb upstream of its initiation site (67). This enhancer is a target of Tpit action (Figure 2, A, B, D, and E) but before Pax7-dependent chromatin remodeling (Figure 2C), Tpit does not have access to its canonical palindromic binding site within the PCSK2 −146-kb enhancer, clearly showing that Tpit does not have the pioneer activity that is the hallmark of Pax7.
Figure 2.
The active pioneer transcription factor Pax7 opens the chromatin structure at intermediate pituitary melanotrope-specific enhancers. A, The protein convertase 2 (PCSK2) gene is specifically expressed in pituitary intermediate lobe melanotropes and not in the anterior lobe corticotropes, the other POMC-expressing lineage. Expression of this gene is critically dependent on an enhancer sequence that is located at −146 kb and that includes 1.4 kb of evolutionarily conserved sequences. B and C, Chromatin changes at enhancers pioneered by Pax7. The average distribution of chromatin marks at enhancers similar to the PCSK2 −146-kb enhancer in corticotrope AtT-20 cells (C) where the enhancers are inactive and after their activation through the pioneering action of Pax7 (B). The action of Pax7 on these enhancers results in chromatin opening as revealed by FAIRE and by the appearance of a bimodal distribution of H3K4me1 flanking the enhancer. Activation is also accompanied by recruitment of p300. D and E, Recruitment of Pax7 (D) and Tpit (E) is revealed by ChIPseq in AtT-20 cells expressing Pax7; neither protein is present at these positions in normal cells. Pax7 binds a composite sequence that is the hallmark of the pioneering targets of Pax7, whereas Tpit binds a complex palindrome (half-site motifs indicated by brown arrows over the sequence) similar to that of other Tpit-binding sites. The Pax7 and Tpit binding sites are separated by 337 bp and a small peak of Tpit recruitment coincident with the Pax7 peak reveals direct interactions between these proteins. F, Structure of Pax7 showing the presence of two DNA-binding domains, the Prd and HD. Each of these DNA-binding domains binds a unique sequence motif and either or both of these motifs are found at most Pax7 binding sites observed by ChIPseq in AtT-20 cells (67). Some Pax7 ChIPseq peaks harbor a unique composite Pax7 target sequence that includes the Prd and HD motifs side by side. This composite target site is uniquely associated with sites of pioneering by Pax7, such as the PCSK2 −146-kb enhancer where Pax7 binding leads to chromatin remodeling and enhancer activation. a.a., amino acids.
Interestingly, sites of Pax7 pioneering were found to contain a novel composite DNA target sequence that includes, next to each other, the consensus binding sites for its paired (Prd) domain abutting the consensus motif recognized by the homeodomain (HD) (Figure 2F). Prior work had indicated that either of these DNA sequence motifs was associated with Pax7-dependent transcription, and, hence, either of them appeared to be sufficient for the standard transcriptional action of Pax7. It thus appears that the pioneering activity of Pax7 involves a particular DNA interaction with this longer composite target sequence (67). It will be interesting to see how Pax7 interaction with the pioneer composite target sequence differs from the interaction of Pax7 with its other target sequences, either the Prd or HD DNA sites, and more specifically whether higher affinity and/or different Pax7 protein conformations account for the unique pioneering properties.
The pluripotency factors have pioneer activity
The 3 so-called pluripotency TFs that are sufficient to reprogram fibroblasts into stem-like iPS cells were recently shown by direct genome-wide analyses to have pioneer activity (68). Within the first 48 hours after transfection, the pluripotency factors Oct4, Sox2, and Klf4 target genome regions that are in condensed (closed) chromatin that does not harbor a particular pattern of histone modification. One striking chromatin feature was the identification of large megabase domains that are not initially targeted by pluripotency factors (within 48 hours) but that become accessible later and include some key genes for the pluripotent ES cell phenotype. These large fibroblast-specific regions exhibit high levels of H3K9me3 that are lost when they become accessible in ES cells. The 3 pluripotency factors thus exhibit pioneer activity, but this is a slow process, particularly within the megabase domains of condensed chromatin marked by H3K9me3. Because all 3 factors are required for reprogramming, no data on the pioneering effect of individual (or their pairwise combination for that matter) factors at specific loci are available. Deciphering the steps required for the pioneering action of these factors represents the next milestone for understanding the underlying mechanism for pioneer factors to change the chromatin environment.
A fourth factor, c-Myc, increases the efficiency and speed of reprogramming by the other 3 factors. Indeed, Myc stabilizes the recruitment of the other 3 factors at the critical loci for reprogramming. Rather than supporting a previous hypothesis suggesting that by promoting cell survival, c-Myc might allow more time for rare events required for reprogramming, it now appears that c-Myc achieves its effect on reprogramming through direct interaction with the other pluripotency factors and stabilization of their genomic recruitment. Much remains to be determined about the mechanism of pioneering by the 3 pluripotency factors, but Soufi et al (68) highlighted the fact that FoxA, Oct4, Sox2, and Klf4 all bind DNA on one surface, leaving the opposing surface free to interact with other proteins, possibly within nucleosomes. These DNA-binding properties may be important for the ability of the factors to recognize specific DNA sequences in the condensed nucleosomal chromatin.
Perspectives and Conclusion
At this time, many questions are arising about pioneer factors and their mode of action, as so little is known about so few factors! The importance of understanding how these factors achieve their task of reshaping the epigenome is critically important if we are to control the process of reprogramming and use it for therapeutically useful objectives. More than ever, it is evident that an appropriately programmed chromatin environment is the key to programming differentiated cells that may be useful in regenerative medicine as at the same time we discover the numerous alterations of the epigenome that are associated with disease, in particular with cancer. The unique properties that allow pioneer TFs to access their DNA targets in condensed chromatin represents at the same time a liability unless there are limitations on the ability of different pioneer factors to access different types of “closed” chromatin. Although the sequential action of pioneer factor cascades during normal development may be an important feature to achieve the result observed in differentiated cells, the brute force expression of these factors in tissue culture cells may yield cells with not only characteristics similar to those of the normal differentiated cells but also some aberrant epigenetic features that may represent liabilities in a therapeutic setting. We now have tools to reprogram cells, but much needs to be learned about the epigenome and its reprogramming before the power of pioneer TFs can be used knowingly and appropriately.
A comparison of the unique structural aspects of the interaction of pioneer factors with their target DNA in chromatin to the structural aspects of the interaction of the same factors with free DNA represents a key question to address for an understanding of the specificity of the pioneering action. Indeed, the factors identified so far for their pioneering activity also play roles as regular TFs involved in maintenance of gene expression for entire gene networks. The work on Pax7 presented a paradigm to distinguish these 2 types of activities because a unique target sequence, the composite Pax7 motif, is associated with sites of pioneering in contrast to sites of Pax7 action as a regular TF where either or both of the 2 alternate Pax7 target sequences, the Prd and HD motifs, are found (Figure 2F). Is DNA affinity the sole determinant of this difference: because the Pax7 composite target sequence is longer than either of its Prd or HD halves, it is likely that the interaction with the Pax7 composite motif is stronger. The possibility that affinity for sequences that are closer to consensus may be important for PHA-4, the C elegans homolog of FoxA, has been proposed based on the correlation between timing of expression and target site affinity (69), but reiterated binding sites may also contribute to pioneering (63). However, this correlation did not seem to hold in mouse cells (70). Alternatively, the interaction of a pioneer factor with a specific DNA sequence (such as the Pax7 composite target) or chromatin environment may lead to the recruitment of chromatin remodeling proteins that are specifically required to initiate the opening of condensed chromatin. It is indeed a question of whether the initial events in pioneering require a unique chromatin remodeling machinery by comparison with the numerous complexes and enzymes that have been involved in maintenance or enhancement of active vs repressed chromatin states. In view of the very slow process of chromatin remodeling after the action of pioneer factors (in the range of days), it is possible that the usual chromatin maintenance complexes may not suffice to initiate major remodeling either locally at the site of pioneer factor binding or more broadly when changes in large chromatin domains are involved such as those described after the action of the pluripotency factors (68). Is time required for cells to go through cell division and offer the possibility of reprogramming after DNA replication (71) or is time required for some stochastic rare permissive event to occur and enable the pioneer factor to shift the balance from repressive to active chromatin conformation?
It will also be of prime importance to identify the primary targets of pioneering: is this first occurring at promoters and genes or at their distant enhancers? Are chromatin domains targeted rather than specific promoters or enhancers? What proportion of the pioneering events involves specific genes as opposed to large domains? One could imagine that during development, all these scenarios play a role at different key developmental steps and possibly that different pioneer factors fulfill different functions in relation to large domains (like the pluripotency factors) or specific genes (like Pax7).
Finally, what is (are) the chromatin substrate(s) on which pioneer factors are acting? Is there a unique substrate or many different ones accessible to different subsets of pioneer factors? The large domains marked by H3K9me3 identified by Soufi et al (68) and remodeled after the action of the 3 pluripotency factors may represent a unique aspect of reprogramming fibroblast into stem cells. In contrast, during development, the opening of such regions may occur in a stepwise fashion where cascades of TFs lead to progressive opening of subdomains or specific genes. The iPS system has been intensely investigated in recent years, and it is yielding important information on this process, but it will be equally important to understand the normal developmental process to assess to what extent the reprogramming of iPS cells is similar or not to the normal generation of pluripotent stem cells.
Acknowledgments
I thank the many colleagues who have contributed to my work over the years, both laboratory members and collaborators from all around the world. I am grateful to Tabasum Abdul-Rasul for her expert assistance. Work in my laboratory was supported by grants from the Canadian Institutes of Health Research.
This work was supported by grants from the Canadian Institutes for Health Research to J.D.
Disclosure Summary: The author has nothing to disclose.
Funding Statement
This work was supported by grants from the Canadian Institutes for Health Research to J.D.
Footnotes
- ChIPseq
- chromatin immunoprecipitation sequencing
- ER
- estrogen receptor
- FAIRE
- formaldehyde-assisted isolation of regulatory elements
- H3K4me1
- monomethyl-lysine 4 histone H3
- H3K27Ac
- acetylated-lysine 27 histone H3
- HD
- homeodomain
- iPS
- induced pluripotent stem
- MLL
- mixed-lineage leukemia
- Pc
- polycomb
- POMC
- proopiomelanocortin
- Prd
- paired
- seq
- sequencing
- TF
- transcription factor.
References
- 1. Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. García-Bellido A. Genetic control of wing disc development in Drosophila. Ciba Found Symp. 1975;29:161–182. [DOI] [PubMed] [Google Scholar]
- 3. Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cotney J, Leng J, Oh S, et al. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drouin J. Pituitary development. In: Melmed S, ed. The Pituitary. New York, NY: Elsevier-Academic Press; 2010:3–19. [Google Scholar]
- 7. Mastracci TL, Sussel L. The endocrine pancreas: insights into development, differentiation, and diabetes. Wiley Interdiscip Rev Dev Biol. 2012;1:609–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parker KL, Rice DA, Lala DS, et al. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res. 2002;57:19–36. [DOI] [PubMed] [Google Scholar]
- 9. De Felice M, Di Lauro R. Minireview: Intrinsic and extrinsic factors in thyroid gland development: an update. Endocrinology. 2011;152:2948–2956. [DOI] [PubMed] [Google Scholar]
- 10. Scully KM, Rosenfeld MG. Pituitary development: regulatory codes in mammalian organogenesis. Science. 2002;295:2231–2235. [DOI] [PubMed] [Google Scholar]
- 11. Lamolet B, Pulichino AM, Lamonerie T, et al. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell. 2001;104:849–859. [DOI] [PubMed] [Google Scholar]
- 12. Pulichino AM, Vallette-Kasic S, Tsai JP, Couture C, Gauthier Y, Drouin J. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 2003;17:738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pulichino AM, Vallette-Kasic S, Couture C, et al. Human and mouse Tpit gene mutations cause early onset pituitary ACTH deficiency. Genes Dev. 2003;17:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods. 2009;48:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song L, Zhang Z, Grasfeder LL, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. [DOI] [PubMed] [Google Scholar]
- 18. Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47:38–49. [DOI] [PubMed] [Google Scholar]
- 19. Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010;467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer. 2012;12:381–385. [DOI] [PubMed] [Google Scholar]
- 22. Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. [DOI] [PubMed] [Google Scholar]
- 23. Laganière J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguère V. From the cover: Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA. 2005;102:11651–11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eeckhoute J, Lupien M, Meyer CA, et al. Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res. 2009;19:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. John S, Sabo PJ, Thurman RE, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sérandour AA, Avner S, Percevault F, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gévry N, Hardy S, Jacques PE, et al. Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 2009;23:1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belikov S, Astrand C, Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol. 2009;29:5413–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M. Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 2007;67:6477–6483. [DOI] [PubMed] [Google Scholar]
- 34. Magnani L, Ballantyne EB, Zhang X, Lupien M. PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 2011;7:e1002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voss TC, Schiltz RL, Sung MH, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madsen MS, Siersbaek R, Boergesen M, Nielsen R, Mandrup S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol. 2014;34:939–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shlyueva D, Stelzer C, Gerlach D, et al. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell. 2014;54:180–192. [DOI] [PubMed] [Google Scholar]
- 39. Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. [DOI] [PubMed] [Google Scholar]
- 40. Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. [DOI] [PubMed] [Google Scholar]
- 43. Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. [DOI] [PubMed] [Google Scholar]
- 45. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwartz YB, Pirrotta V. A new world of polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–864. [DOI] [PubMed] [Google Scholar]
- 47. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. [DOI] [PubMed] [Google Scholar]
- 48. Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hendy GN, Kaji H, Canaff L. Cellular functions of menin. Adv Exp Med Biol. 2009;668:37–50. [DOI] [PubMed] [Google Scholar]
- 50. Wu M, Wang PF, Lee JS, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goo YH, Sohn YC, Kim DH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herz HM, Mohan M, Garruss AS, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shen X, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hansen KH, Bracken AP, Pasini D, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. [DOI] [PubMed] [Google Scholar]
- 56. Margueron R, Justin N, Ohno K, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2004;101:15603–15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. [DOI] [PubMed] [Google Scholar]
- 59. Zhou W, Zhu P, Wang J, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ku M, Koche RP, Rheinbay E, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie polycomb-binding sites. Proc Natl Acad Sci USA. 2007;104:5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fakhouri TH, Stevenson J, Chisholm AD, Mango SE. Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 2010;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Updike DL, Mango SE. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet. 2006;2:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li Z, Schug J, Tuteja G, White P, Kaestner KH. The nucleosome map of the mammalian liver. Nat Struct Mol Biol. 2011;18:742–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28:225–238. [DOI] [PubMed] [Google Scholar]
- 67. Budry L, Balsalobre A, Gauthier Y, et al. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 2012;26:2299–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. [DOI] [PubMed] [Google Scholar]
- 70. Tuteja G, Jensen ST, White P, Kaestner KH. Cis-regulatory modules in the mammalian liver: composition depends on strength of Foxa2 consensus site. Nucleic Acids Res. 2008;36:4149–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]